Abstract
Ribosome synthesis entails the formation of mature rRNAs from long precursor molecules, following a complex pre-rRNA processing pathway. Why the generation of mature rRNA ends is so complicated is unclear. Nor is it understood how pre-rRNA processing is coordinated at distant sites on pre-rRNA molecules. Here we characterized, in budding yeast and human cells, the evolutionarily conserved protein Las1. We found that, in both species, Las1 is required to process ITS2, which separates the 5.8S and 25S/28S rRNAs. In yeast, Las1 is required for pre-rRNA processing at both ends of ITS2. It is required for Rrp6-dependent formation of the 5.8S rRNA 3′ end and for Rat1-dependent formation of the 25S rRNA 5′ end. We further show that the Rat1-Rai1 5′-3′ exoribonuclease (exoRNase) complex functionally connects processing at both ends of the 5.8S rRNA. We suggest that pre-rRNA processing is coordinated at both ends of 5.8S rRNA and both ends of ITS2, which are brought together by pre-rRNA folding, by an RNA processing complex. Consistently, we note the conspicuous presence of ∼7- or 8-nucleotide extensions on both ends of 5.8S rRNA precursors and at the 5′ end of pre-25S RNAs suggestive of a protected spacer fragment of similar length.
INTRODUCTION
Ribosomes are essential to all life forms. Ribogenesis is a major metabolic activity requiring the coordinated expression of the core RNA and protein components of the small and large subunits and also of a myriad of trans-acting protein and RNA factors, their maturation, packaging, and transport (reviewed in references 21, 29, and 42). Despite this great complexity, ribogenesis is an extremely robust process. Quality control and fail-safe mechanisms are available at all steps along the assembly pathway to ensure that a sufficient amount of functional ribosomes is provided at all times (reviewed in references 24 and 30).
In eukaryotes, ribogenesis is initiated in the nucleolus. There, at the interface between the cortical side of fibrillar centers (FCs) and the surrounding dense fibrillar components (DFCs), RNA polymerase I is recruited and threaded along the ribosomal DNA (rDNA), producing pre-rRNA transcripts that extend into the DFC, where pre-rRNAs undergo cotranscriptional modification. In fast-growing budding yeast cells, up to 50 to 70% of pre-rRNA molecules are also subjected to cotranscriptional cleavage (3, 23, 28, 36). Cotranscriptional cleavage occurs in internal transcribed spacer 1 (ITS1), separating the precursors destined to mature into the small and large ribosome subunit rRNAs, i.e., the 18S and 5.8S-25S rRNAs, respectively (28, 36, 47). This strategy, involving the cosynthesis of three of the four mature rRNAs from a single long transcript, contributes to coordinating the synthesis of the various components of the translational machinery. This strategy has been extensively conserved throughout evolution and predates the eukaryotes, since the Bacteria and Archaea also synthesize their rRNAs from polycistronic precursors (reviewed in reference 31).
The synthesis of mature rRNAs relies on processing within the external and internal transcribed spacers rather than direct cleavages at the mature endpoints of the molecules. This is also an evolutionarily conserved feature (reviewed in references 11 and 31). Pre-rRNA processing involves numerous ribonucleolytic activities and proceeds through a succession of both endo- and exoribonucleolytic cleavages, leading to the accumulation of a large set of RNA intermediates, with “precursor-product” relationships that are sufficiently stable to be readily detected. Why the formation of the mature 5′ and 3′ rRNA termini is so complicated has remained completely unclear. One possibility is that pre-rRNA processing might integrate other ribosome assembly reactions, such as RNA folding, RNA modification, protein binding, and ribonucleoprotein particle structure remodeling, by providing precise window frames for these reactions to occur. Put simply, cleavages in the spacers according to a strict timetable likely allow a more finely tuned regulation and provide an efficient means of keeping the “assembly line” under surveillance. Whether cleavages at distant sites on pre-rRNA molecules are coordinated and how such functional connections might occur are unclear.
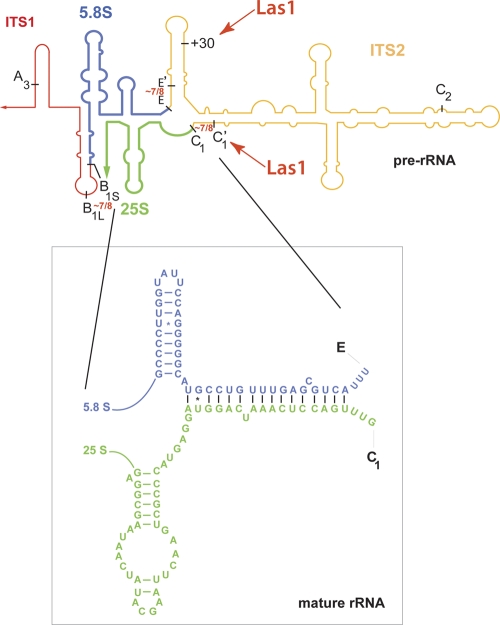
In eukaryotes, within mature 60S subunits, the 3′ end of 5.8S rRNA and 5′ end of 25S/28S rRNA are joined by Watson-Crick base pairing through the formation of a long evolutionarily conserved helix, which requires the precise excision of ITS2 to be formed (Fig. 1). In Bacteria and Archaea, there is no ITS2 and the sequence corresponding to 5.8S rRNA is encoded at the 5′ end of the 23S gene (31). Processing of ITS2 is thus particularly interesting to study from an evolutionary standpoint.
Fig 1.
Is pre-rRNA processing coordinated at distant sites on pre-rRNA molecules? Within mature 60S subunits (inset), the 5.8S and 25S rRNAs are joined by Watson-Crick base pairing, which requires the precise removal of ITS2. Within preribosomes, pre-rRNA processing is functionally connected at distant sites brought together by RNA folding. Evidence is presented in this paper for coordination on both sides of 5.8S rRNA precursors (Rat1-Rai1) and on both sides of ITS2 (Las1). rRNA folding is based on results from references 8 and 49.
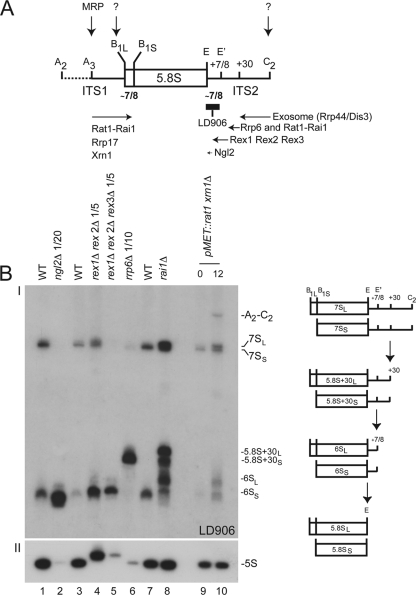
The synthesis of 5.8S rRNA is particularly complex and has been best described in budding yeast (Fig. 2). In a major pathway that generates a short form of 5.8S rRNA, 5.8SS, 5′-end formation starts with endoribonucleolytic cleavage at site A3 in ITS1 (by RNase MRP) and involves progressive trimming of ITS1 extensions by at least three 5′-3′ exoribonucleases (exoRNases) with partially overlapping specificities, namely, the Rat1-Rai1 complex (22), Rrp17 (35), and Xrn1 (22). Rat1 and Rrp17 are both essential and are thought to contribute to the process to various extents via parallel pathways (35). Xrn1 is mostly cytoplasmic but has been shown to be active in the nucleus and nucleolus (22, 25). Via an alternative minor route, longer forms of 5.8S rRNA, 5.8SL, 5′ extended by ∼7 or 8 nucleotides, are generated by direct clipping by an unidentified endoribonuclease (endoRNase) (22). In the steady state in wild-type cells, the ratios of 5.8SS to 5.8SL are about 80:20 in budding yeast and 60:40 in HeLa cells (22; this work). Formation of the 5.8S 3′ end is even more complicated (summarized in Fig. 2). It starts with an endoribonucleolytic cleavage within ITS2, at site C2, by an unknown endo-RNase and requires at least half a dozen 3′-5′ exoRNases, including the core RNA exosome and its catalytic subunit Rrp44/Dis3 (endowed with both endo- and exoRNase activities) (32, 38, 41), the nucleus-specific exosome subunit Rrp6 (6), several members of the RNase D family, the Rex proteins (46), and Ngl2 (15). Trimming by the core exosome stops at position +30 with respect to the mature 3′ end of the 5.8S rRNA. At this point the substrate is “handed over” to Rrp6 and then to the Rex proteins and Ngl2. The final cleavage step occurs in the cytoplasm. There the 6S RNA, a heterogeneous population of precursors extended by ∼6 nucleotides, is converted to mature 5.8S rRNA by Ngl2 (15, 44).
Fig 2.
How is 5.8S rRNA synthesized in budding yeast? (A) Summary cartoon. The cleavage sites (A2 to E) and trans-acting factors involved in 5.8S rRNA maturation are indicated. (B) The successive steps of 5.8S rRNA 3′-end formation are illustrated by a high-resolution Northern blot analysis of different mutants and isogenic control wild-type cells (I). Schematics depicting the structure of each physiological intermediate detected with a probe that overlaps with the 3′ end of 5.8S rRNA (LD906; see panel A) are provided to the right. Total RNA was extracted from ngl2, rai1, rex1 rex2, rex1 rex2 rex3, rrp6, and rat1 xrn1 mutants and analyzed by Northern blotting (see Materials and Methods). Ten micrograms of total RNA was loaded in each lane unless stated otherwise. As a control, panel II was hybridized with a probe specific to the 5S rRNA, as Rex1 and Rex2 have known functions in 5S rRNA 3′-end formation (46).
Lethal in the absence of Ssd1 (Las1) is an evolutionarily conserved protein carrying an ∼150-residue amino-terminal domain of unknown function (see Fig. 3). In human cells, Las1 (Las1L) largely colocalizes with nucleophosmin/B23, a late-acting ribosome synthesis factor. Consistently, it was recently shown to be required for large ribosome subunit synthesis (7). Las1L depletion, like other nucleolar stresses (reviewed in reference 4), induces a tumor surveillance pathway resulting in p53 stabilization, G1 arrest, and ultimately cell death (7). Yeast Las1 has known functions in cell morphogenesis (surface growth) and cell division (13).
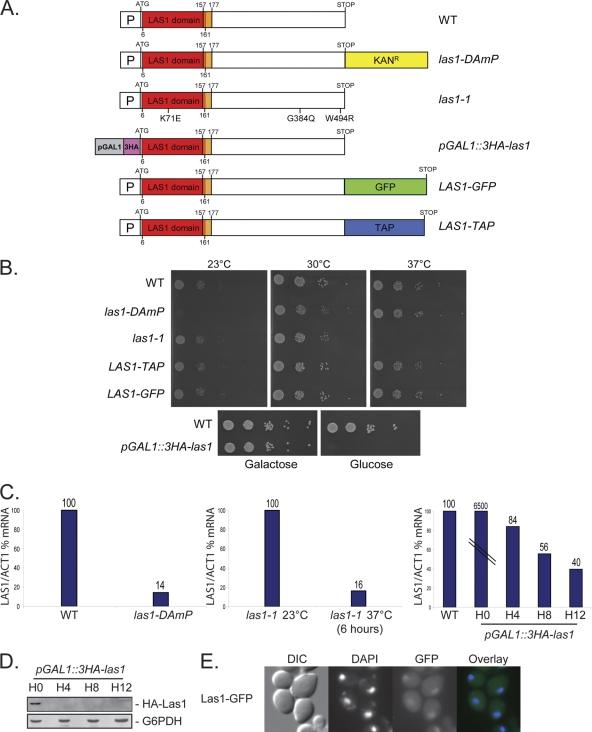
Fig 3.
Description and characterization of the five LAS1 alleles used in this work and subcellular distribution of yeast Las1. (A) Genomic structure of the LAS1 alleles. The LAS1 domain extends from position 6 to 161 and is followed by a negatively charged cluster (positions 157 to 177). P, endogenous LAS1 promoter. In las1-DAmP (decreased abundance by mRNA perturbation) cells (5), a hypomorphic allele was generated by disrupting the 3′ UTR of LAS1 by insertion of an antibiotic resistance cassette. In las1-1 cells, the open reading frame (ORF) of LAS1 was subjected to random PCR mutagenesis and mutations were selected for conferring a severe thermosensitive (TS) growth defect. We established that las1-1 expresses a protein carrying three amino acid substitutions, one in the LAS1 domain (K71E) and two in the carboxy-terminal region (G394Q and W494R). In pGAL::HA-las1, the LAS1 gene was fused at its 5′ end to three copies of the hemagglutinin (HA) epitope and placed under the control of the repressible GAL1 promoter. Genes under the control of pGAL1 are severely downregulated when cells are shifted from galactose- to glucose-based medium. In the LAS1-GFP and LAS1-TAP alleles, a GFP or TAP epitope was directly inserted in the genome of haploid cells at the 3′ end of LAS1 by homologous recombination. WT, wild type. (B) Effects on growth of various conditional las1 alleles. Shown is a drop assay on plates. Serial dilution of yeast cultures were plated at the indicated temperature on complete medium. The las1-DAmP, las1-1, LAS1-TAP, and LAS1-GFP strains were grown on YPD. The pGAL1::3HA-las1 strain was grown at 30°C in YPGSR (galactose) or YPD (glucose). Note that the growth of LAS1-TAP and LAS1-GFP strains is not affected. (C) Steady-state accumulation of LAS1 mRNA in different mutants. The level of LAS1 mRNA was determined by RTqPCR with respect to ACT1. The las1-DAmP strain and its isogenic control were grown to mid-log phase in YPD at 30°C. The las1-1 strain was grown to mid-log phase in YPD at 23°C and transferred to 37°C for 6 h. The pGAL1::3HA-las1 strain was grown to mid-log phase in YPGSR, washed in water, and transferred to YPD; its isogenic wild type was cultured in YPGSR to mid-log phase. The amplicons used in the RTqPCR assay are described in materials and methods in the supplemental material. Note that genes under the control of the pGAL promoter are often overexpressed under permissive conditions; this is the case for LAS1 (see H0, pGAL1::3HA-las1). (D) Steady-state accumulation of the Las1 protein in pGAL1::3HA-las1 cells grown in YPGSR (H0) and transferred to YPD for up to 12 h. Total protein was extracted from equal amounts of cells grown to mid-log phase. For the Western blot analysis, membranes were probed with anti-HA or anti-glucose-6-phosphate dehydrogenase (anti-G6PDH; loading control) antibodies. (E) Subcellular localization of Las1 in cells expressing a functional Las1-GFP fusion (see panel B). Las1 is present mainly in the nucleus but is also detected in the cytoplasm. Cells were inspected by fluorescence microscopy. DIC, differential interference contrast; DAPI, 4′,6-diamidino-2-phenylindole (DNA stain); GFP, green fluorescent protein.
Several discoveries have led us to investigate in budding yeast the involvement of Las1 in ribosome synthesis: (i) the recent finding that human Las1L is involved in large ribosome subunit synthesis (7); (ii) the recent identification of Las1L as a privileged partner of the trimeric complex Pelp1-Tex10-Wdr18, involved in maturation of the large ribosome subunit and its release from the nucleolus and whose nucleolar distribution is actively regulated by SUMO (18); (iii) the recent characterization of Las1 in fission yeast as a physical and functional partner of the polynucleotide kinase Grc3, a protein required for large subunit rRNA synthesis and a partner of the trimeric complex involved in processing of ITS2 (IPI) (27, 37).
We report that Las1 is associated with preribosomes, where it is specifically required for 5.8S rRNA 3′-end formation, precisely at a step requiring Rrp6, and also for the final step of 25S rRNA 5′-end formation. We also report that in human cells as in yeast, 5.8S rRNA 3′-end formation is a multistep process involving ExoSC10 (human Rrp6) and the helicase Skiv2L2 (human Mtr4/Dob1), a known exosome cofactor. Lastly, we show that the Rat1-Rai1 complex is required for processing at both ends of 5.8S rRNA; in particular, we report a novel and quite unexpected indirect requirement for the 5′-3′ exoRNase Rat1/Xrn2 in 5.8S rRNA 3′-end maturation in both yeast and human cells.
MATERIALS AND METHODS
Cell culture and gene inactivation.
Yeast cells were grown according to standard procedures in YPD (2% peptone, 1% yeast extract, 2% glucose). Cultures were kept in mid-log phase at all times by continuous dilution with fresh, prewarmed medium. Unless otherwise stated, cells were grown at 30°C.
To achieve thermoinactivation of Las1, las1-1 cells were grown in YPD at 23°C and transferred to 37°C for up to 6 h. Thermoinactivation of Rat1 in rat1-1 cells required a shift to 37°C for 2 h. To achieve depletion of Las1, pGAL1::3HA-las1 and pGAL1::3HA-las1 rai1Δ cells were grown in YPGSR (yeast extract, peptone, galactose-sucrose-raffinose [2% each]) to mid-log phase, washed in prewarmed water, and transferred to YPD for up to 14 h. To achieve depletion of Rat1, pMET::rat1 xrn1Δ cells were grown in synthetic medium lacking methionine to mid-log phase, methionine was then added at 5 mM final concentration, and the cells were collected after 12 h. The yeast strains used are listed in materials and methods in the supplemental material.
HeLa cells (NIH Aids Research and Reference Reagent Program; catalog no. 153) were grown in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (A&E Scientific), GlutaMAX-I (Gibco), and 1% penicillin-streptomycin (Gibco). Cells were grown at 37°C under 5% CO2.
For each target, 150,000 HeLa cells (in six-well plates) were reverse transfected independently with up to three different specific small interfering RNAs (siRNAs) (Silencer select; Ambion), each at 10 nM concentration. This was done with the Lipofectamine RNAiMAX reagent (Invitrogen) as recommended by the manufacturer. The siRNA sequences used are listed in materials and methods in the supplemental material. Inactivation was carried out for 72 h, after which the cells were harvested in 1 ml Tri Reagent (Ambion) for parallel isolation of total RNA and protein.
RNA extraction, Northern blotting, real-time quantitative PCR (RTqPCR), and primer extension.
RNA extractions from yeast cells and Northern blotting were performed as described previously (7a). The oligonucleotides used for hybridizations are listed in materials and methods in the supplemental material.
Extraction of total RNA from HeLa cells was carried out with Tri Reagent (Ambion) according to the manufacturer's recommendations, except that RNA pellets were first washed with absolute ethanol and then with 75% ethanol.
For Northern blot analysis of low-molecular-weight species, 4 to 5 μg total RNA was resolved on an 8% polyacrylamide gel. For the analysis of higher-molecular-weight species, 5 to 10 μg total RNA was resolved on a 1.2% agarose–6% formaldehyde gel as described previously (7a).
In order to validate the efficiency of mRNA depletion by RTqPCR, 1 μg total yeast RNA or 100 ng total human RNA was treated with 1 μl (1 U) RNase-free DNase I (Fermentas) in a reaction mix containing 1× reaction buffer with MgCl2 for DNase I (Fermentas). For cDNA synthesis, DNase-treated RNA was first incubated in a mix containing 100 μM oligo(dT)18 primer (Fermentas), 100 μM random hexamer primer (Fermentas), and 10 mM deoxynucleoside triphosphates (dNTP) (RNase free; Fermentas). The reaction mixture was incubated for 5 min at 65°C, and the following components were added: 5× first-strand buffer (Life Technologies), 0.1 M dithiothreitol (DTT) (Life Technologies), RiboLock RNase inhibitor (40 U/μl; Fermentas), and SuperScript II reverse transcriptase II (200 U/μl; Life Technologies). The reaction mixture was incubated at 42°C for 2 min, then at 25°C for 10 min, and finally at 42°C for 1 h. cDNAs were then amplified by quantitative PCR in Platinum SYBR green qPCR SuperMix-UDG with ROX (Life Technologies). Amplicons for human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) or yeast actin1 were used as an endogenous control for qPCR analysis. The sequences of the primers used are described in materials and methods in the supplemental material. Data were analyzed with StepOne, version 2.1, software available from Applied Biosystems (Life Technologies), and the comparative threshold cycle (CT) method was used for quantification (40).
Primer extension reactions were performed in the presence of a molar excess of primers with 10 μg of total RNA. The primers are described in materials and methods in the supplemental material. After extension with avian myeloblastosis virus (AMV) reverse transcriptase (Promega), RNA was hydrolyzed and the samples were precipitated and allowed to migrate through 6% sequencing gels.
Northern blot quantification.
Phosphorimager quantitation used a Fuji FLA-7000 and the native Multi Gauge software (version 3.1).
Additional materials and methods are available in the supplemental material.
RESULTS
Description of the LAS1 alleles used in this work.
To characterize functionally the essential LAS1 (YKR063C) gene in yeast, we used five alleles (Fig. 3). Each variant was expressed in haploid cells from the LAS1 locus and was the sole source of the protein. The las1-DAmP (decreased abundance by mRNA perturbation), las1-1 (thermosenstive for growth), and pGAL1::3HA-las1 alleles cause specific alteration of LAS1 expression at the mRNA and/or protein level (Fig. 3C and D). The LAS1-GFP and LAS1-TAP alleles express functional C-terminally epitope-tagged versions of Las1 (Fig. 3B). All alleles but pGAL1::3HA-las1 were expressed from the endogenous LAS1 promoter (Fig. 3). In cells expressing a functional green fluorescent protein (GFP) construct, Las1 was found in the nucleus and the cytoplasm (Fig. 3E).
Las1 is required for large ribosomal subunit accumulation.
To address the involvement of Las1 in ribosome subunit formation, we first characterized polysome profiles by sucrose gradient ultracentrifugation. Exponentially growing cells were treated with cycloheximide to “snap-freeze” the polysomes, which were then resolved on 10 to 50% sucrose gradients. This analysis was conducted in parallel on las1-1 and las1-DAmP cells (Fig. 4 and data not shown).
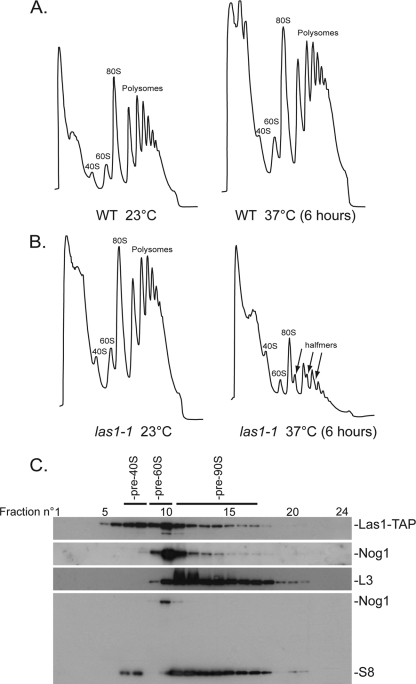
Fig 4.
Yeast Las1 is required for large ribosome subunit maturation, and it comigrates with preribosomes on velocity gradients. (A and B) Polysome profiles of the las1-1 and isogenic wild-type (WT) control strains were produced on 10 to 50% sucrose gradients. Total extracts were prepared from strains grown to mid-log phase in YPD. Cells were treated with cycloheximide (50 μg/ml) for 10 min prior to collection. The profiles are readings of optical density at 254 nm (OD254). The wild type was grown at 23°C and shifted to 37°C for 6 h; the las1-1 strain was grown at 23°C and then transferred to 37°C for 6 h. (C) Yeast Las1 colocalizes with preribosomes on velocity gradients. A total extract of cells expressing a functional Las1-TAP fusion (Fig. 3B) was prepared in 20 mM Tris-HCl (pH 7.4), 50 mM KCl, and 5 mM MgCl2 and fractionated on 10 to 50% sucrose gradients. Total RNA and total protein were extracted from each of the 24 fractions and analyzed by Northern and Western blotting, respectively. Pre-60S ribosomes were detected by Western blotting with an antibody against Nog1. Mature ribosomal subunits were detected by Western blotting with antibodies against the small and large subunits (S8 and L3, respectively). Preribosomes were detected by probing Northern blots for specific precursor RNAs (data not shown).
Isogenic wild-type control strains showed distinct peaks corresponding to 40S, 60S, 80S, and polysomes. Overall, wild-type cells grown at 23°C or shifted to 37°C for 6 h showed similar polysome profiles (Fig. 4A). Transfer of las1-1 cells to 37°C for 6 h led to a striking reduction in 60S (compared to 40S subunits) and to the pronounced accumulation of half-mers (Fig. 4B). Half-mers are kinetically stalled 43S initiation complexes that have threaded onto the mRNA but are unable to initiate translation owing to a lack of cytoplasmic 60S subunits. Many fewer half-mers accumulated in the las1-DAmP strain, but the 60S reduction and the subunit imbalance were visible in these cells also (data not shown). Thus, yeast Las1 is required for large ribosome subunit maturation.
Las1 copurifies with preribosomes that contain the 5′-3′ exo-RNases Rat1-Rai1 and Xrn1.
Next we used velocity gradient centrifugation (Fig. 4C) and affinity purification (Fig. 5) to determine whether Las1 interacts with precursor ribosomes. Total extracts of cells expressing a functional Las1-TAP fusion (Fig. 3B) were fractionated on sucrose gradients, and each fraction was tested for the presence of specific proteins and RNAs by Western and Northern blotting, respectively (Fig. 4C and data not shown). The extract was prepared in nearly physiological ionic strength conditions (20 mM Tris-HCl [pH 7.4], 50 mM KCl, and 5 mM MgCl2). Las1 peaked in fraction 10, where Nog1, a known pre-60S component, was also detected (Fig. 4C). In addition, Las1 colocalized both with lighter fractions, possibly corresponding to subcomplexes of preassembled trans-acting factors, and larger complexes, likely pre-90S ribosomes.
Fig 5.
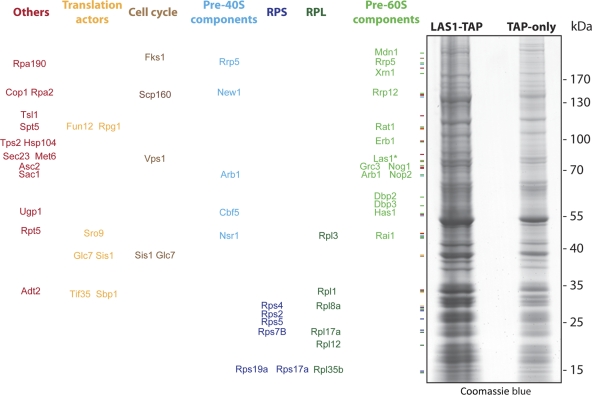
Yeast Las1 copurifies with preribosomes that also contain two 5′-3′ exoRNases (Rat1-Rai1 and Xrn1), three RNA helicases (Dbp2, Dbp3, and Has1), and several preribosome components. Partners of Las1 were identified by affinity purification and mass spectrometry analysis. Yeast cells expressing a functional Las1-TAP fusion were grown exponentially to mid-log phase at 30°C. Total lysates were prepared in solid phase and subjected to affinity purifications (see Materials and Methods). As a control, we used a strain expressing the TAP tag alone (TAP-only). All interactions are listed in Table S1 in the supplemental material. Interactants were identified by LC-MS/MS. Interactions were carefully curated with those identified in the TAP-only control and with an unrelated bait (see Materials and Methods). (Left) Major Las1 interactants sorted and color coded by known or putative function. RPS and RPL, ribosomal proteins from the small and large subunits, respectively. (Right) Eluates from representative Las1-TAP and TAP-only purifications were analyzed by 4 to 12% NuPage Novex Bis-Tris (Invitrogen), and the gel was stained with Coomassie blue. Las1-TAP is marked with an asterisk.
In parallel, total lysates were engaged in a revised one-step affinity purification protocol optimized to capture transient interactions (34, 35, 50). Las1-TAP and its partners were isolated on paramagnetic beads coated with IgGs selectively targeting the protein A moiety of the TAP epitope, and interactants were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Fig. 5; see Table S1 in the supplemental material). Las1-TAP was found to copurify with several ribosome synthesis factors having known or predicted functions in formation of the small subunit, the large one, or both. Additional coprecipitating factors are involved in translation and cell division. Las1-TAP was notably found to copurify with preribosomes that also contained two 5′-3′ exoRNases (the Rat1-Rai1 complex and Xrn1) and several putative RNA helicases (Dpb2, Dbp3, and Has1). Grc3, a known partner of Las1 in fission yeast (27), was also detected. We conclude that Las1 is associated with multiple species of nuclear preribosomes (12, 17, 29). This is consistent with the results of our velocity gradient analysis (Fig. 4C).
Las1 is required for pre-rRNA processing at both ends of ITS2.
Next we wondered whether Las1 is required for pre-rRNA processing. We addressed this question in parallel in las1-1, las1-DAmP, and pGAL::HA-las1 cells (Fig. 6 to 8; see Fig. S2 and S3 in the supplemental material). The pGAL::HA-las1 strain was grown under permissive conditions (galactose-based medium), transferred to nonpermissive conditions (glucose), and analyzed in a time course experiment. Total RNA was extracted, resolved on agarose denaturing gels, and analyzed by Northern blotting with a set of probes specific to precursor or mature rRNAs (Fig. 6; see Fig. S1A in the supplemental material).
Fig 6.
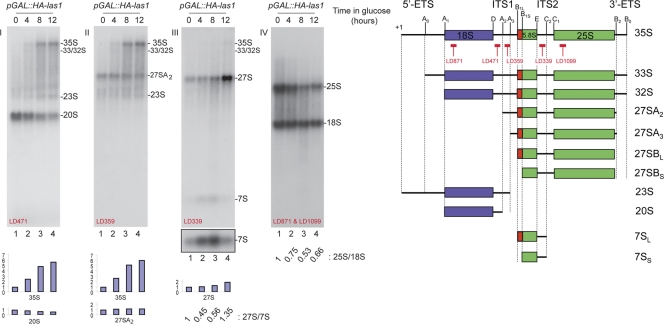
Yeast Las1 is required for large rRNA pre-rRNA processing. Pre-rRNA processing was analyzed by Northern blotting. Total RNA was extracted from pGAL::HA-las1 cells grown to mid-log phase in YPGSR at 30°C, washed in water, and transferred to YPD for up to 12 h. The RNA was separated on a denaturing agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized with a set of oligonucleotide probes specific to mature and precursor rRNAs (see schematics at the right). Probes used are indicated in each panel and on the schematics. At the bottom of panel III is a longer exposure for 7S. Fuji FLA7000 quantitations are summarized in cartoons, and the 27S/7S and 25S/18S ratios are provided (see Table S2 in the supplemental material for details).
Fig 8.
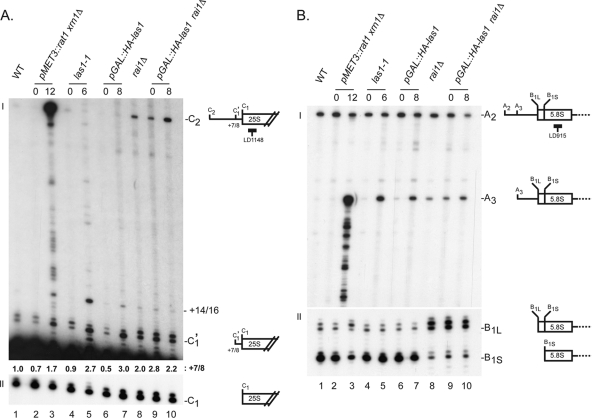
Yeast Las1 is required for the final step of 25S rRNA 5′-end formation. Total RNA extracted from the las1 mutants indicated (see legend to Fig. 7) was analyzed by primer extension performed with oligonucleotide LD1148 (A) or LD915 (B) to analyze pre-rRNA processing in ITS2 or ITS1, respectively. Schematics depict the processing sites and the positions of the oligonucleotides used. The signal observed at position +7 or +8 (C′1) was quantitated in each lane and normalized to the wild type (A, I).
The pGAL::HA-las1 mutant strain showed an altered steady-state accumulation of mature 18S and 25S rRNAs. The 25S rRNA level was much more strongly affected than the 18S rRNA level, suggesting that ITS2 processing is strongly impaired (see 25S/18S ratios in Fig. 6, IV). This was confirmed by the accumulation of the 27S pre-rRNA and concurrent reduction of 7S pre-rRNA (see 27S/7S ratios in Fig. 6, III). This is in agreement with the strong accumulation of half-mers in our polysome analysis of the las1-1 strain (Fig. 4B). The pGAL::HA-las1 mutant strain further showed altered early pre-rRNA processing (cleavage at sites A0, A1, and A2) (Fig. 6, schematics). When these reactions are delayed, 35S pre-rRNA accumulates (a 6-fold increase in 35S was detected in pGAL::HA-las1 cells at the latest time point of our analysis [see quantitation in Table S2 in the supplemental material]). When the A0-A2 cleavages are kinetically delayed, premature cleavage at site A3 in ITS1 generates the 23S RNA (Fig. 6, schematics). This RNA was observed in the pGAL::HA-las1 strain. 23S RNA is not normally further processed but rather polyadenylated mostly by TRAMP5 and degraded by Rrp6 (see reference 47 and references therein). Many mutations affecting large-subunit rRNA synthesis also affect early nucleolar cleavages at sites A0 to A2; this is thought to reflect a feedback regulatory circuit whose molecular basis is unknown.
las1 depletion thus primarily affects the synthesis of large-subunit rRNAs. This conclusion was corroborated with the two other mutant alleles used in this work (las1-1 and las1-DAmP) (see Fig. S2 in the supplemental material). It is not clear at present why, in cells expressing the las1-DAmP allele, the 27S pre-rRNA appeared less stable than in the other mutants (see Fig. S2A, III, in the supplemental material). In the las1-DAmP construct the 3′ untranslated region (UTR) of LAS1 is totally disrupted, and this, in addition to substantially reducing expression at the mRNA level (Fig. 3C), might lead to the production of an aberrant protein (e.g., by translational readthrough), which might selectively target preribosomes for degradation. Consistent with this idea, we found that the 27S pre-rRNA is stabilized in las1-DAmP cells also inactivated for the surveillance component TRAMP5 and that this even led to some growth restoration (see Fig. S2B in the supplemental material). Note that the 7S pre-rRNA was not stabilized in this analysis, indicating that it is not targeted by this surveillance (see Fig. S2B in the supplemental material).
We investigated further pre-rRNA processing in ITS2 by Northern blotting to study 5.8S rRNA 3′-end processing in our different mutants (Fig. 7). Our las1 mutants had a phenotype somewhat reminiscent of that of the rrp6 and rai1 mutants, in that 5.8S+30 accumulated in all cases (Fig. 7A, I, lanes 6 to 15, 1, and 3). The short form 5.8S+30SS remained the major form, indicating that Las1, like Rrp6, does not play a major role in 5.8S rRNA 5′-end maturation (Fig. 7A, I, lanes 6 to 15; see below for a discussion of a minor effect of Las1 at site A3). In addition, forms of 6S rRNAs accumulated also in the normal small form-to-large form (S-to-L) ratio (Fig. 7A, I, lanes 6 to 15). Thus, although the las1 mutations were found to inhibit 5.8S rRNA synthesis, they did not alter strongly the ratio of short to long forms of 5.8S rRNA (Fig. 7B, lanes 6 to 15). In contrast, the rat1 and rai1 mutations, used here as controls, strongly affected this ratio in favor of the long form (Fig. 7B, lanes 2 to 5) (22, 48). This is expected since the Rat1-Rai1 complex is primarily involved in A3-to-B1S processing (Fig. 2).
Fig 7.
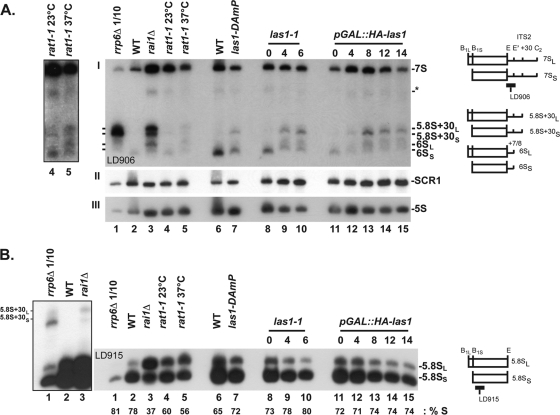
Yeast Las1 is required for 5.8S rRNA 3′-end formation, and the Rat1-Rai1 complex is required for maturation at both ends of 5.8S rRNA precursors. Pre-rRNA processing was analyzed by Northern blotting. Total RNA was extracted from the las1 mutants indicated, separated on denaturing acrylamide gels, transferred to a nylon membrane, and hybridized with oligonucleotide probes specific to precursor and mature rRNAs (see schematics at the right and Materials and Methods). pGAL::HA-las1 cells were grown as described in the legend to Fig. 6; las1-1 cells were grown at 23°C and shifted to 37°C for 6 h; las1-DAmP cells were grown to mid-log phase in YPD at 30°C. (A, I) The blot was hybridized with LD906. Loading controls are provided by SCR1 and 5S hybridizations (A, II and III, respectively). The RNA species annotated with an asterisk was detected only with probes specific to sequences located between sites E and C2, not with probes specific to sequences between A2 and B1, indicating that it corresponds to a long 3′-extended form of 5.8S rRNA. As controls, total RNA was extracted from rai1, rat1, and rrp6 mutants. The rai1Δ and rrp6Δ mutants were grown to mid-log phase in YPD at 30°C, and the rrp6Δ mutant was then shifted to 37°C for 2 h. The rat1-1 mutant was grown to mid-log phase at 23°C and transferred to 37°C for 2 h. The amounts of 5.8S+30 are so high in rrp6 mutants that 10 times less of this RNA was loaded on the gel (lane 1). In each case, a relevant isogenic wild-type strain was used. In lane 2, the WT is YDL793; in lane 6, the WT is YDL1542. The panel to the left shows a darker exposure of lanes 4 and 5. (B) The membrane was probed with oligonucleotide LD915, and the percentage of the short form of 5.8S rRNA (% S) was determined by phosphorimager quantitation.
Importantly, we interpret the accumulation of 5.8S+30 and 6S pre-rRNAs in rai1Δ cells as a strong indication that processing at both ends of 5.8S rRNA is functionally connected (Fig. 7A, lane 3, and B, lane 3; see also Discussion). This conclusion was substantiated with our observation that Rat1 also plays a minor, and most likely indirect, role in 5.8S rRNA 3′-end formation (Fig. 7A, lane 5 [a darker exposure is shown at the left], and 2B, lane 10). Indeed, a low level of RNAs reminiscent of those detected in rai1Δ cells was identified in rat1 mutants upon thermoinactivation or genetic depletion of Rat1. Quite interestingly, we found that human Rat1 (Xrn2) is also involved in 5.8S rRNA 3′-end formation (see below for characterization of ITS2 processing in human cells).
Throughout our experiments, we consistently detected in wild-type cells a 6S RNA whose 3′ end showed some heterogeneity in length, indicating that it is a normal pre-rRNA intermediate (Fig. 2B, lanes 1, 3, and 7, and 7A, lanes 2 and 6). We conclude that 6S is a population of pre-5.8S species extended in 3′ by ∼5 to 8 nucleotides (44). Note that in the analysis presented in Fig. 7, RNA loading was checked by hybridizing the Northern blot with probes specific to the RNA component of SRP (SCR1) and to the 5S rRNA (Fig. 7A, II and III). Also note that some strains with major defects in 5.8S rRNA 5′-end maturation are known to accumulate aberrant pre-rRNA intermediates extending between sites A2 and C2, A3 and C2, and A3 and E, owing to altered pre-rRNA kinetics (discussed in reference 35). None of these species was detected in las1 mutants, again in keeping with the view that Las1 has no major function in 5.8S rRNA 5′-end formation (see Fig. S3 in the supplemental material).
The effects of Las1 mutations on pre-rRNA processing in the internal transcribed spacers were further examined by primer extension (Fig. 8). Extension of a primer hybridizing at the 5′ end of the 25S rRNA made it possible to test the efficiency of trimming between sites C2 and C1 (Fig. 8A). A primer specific to the 5.8S rRNA was used to test cleavages at sites A2 and A3 and the efficiency of cleavage at, and relative use of, B1S and B1L (Fig. 8B).
Depletion or thermoinactivation of Las1 led to the accumulation of discrete pre-rRNA precursors ending at positions +7 or +8 (site C1′) and +14 or +16 with respect to the 5′ end of 25S (Fig. 8A, I, lanes 4 to 7). Products extended by 7 or 8 nucleotides were also detected in the rat1 xrn1 and rai1 mutants (Fig. 8, I, lanes 3 and 8). Importantly, these were also found in wild-type cells and in the rat1 xrn1 mutant grown under permissive conditions (lanes 1 and 2), indicating that they are normal pre-rRNA intermediates (25S′ pre-rRNAs). Las1 thus appeared particularly important for the last maturation step of the 25S rRNA, during which the last ∼7 or 8 nucleotides are removed (cleavage at site C1′, conversion of 25S′ pre-rRNA to 25S rRNA). Products extended by 14 or 16 nucleotides were also detected in the rat1 xrn1 and rai1 mutants (Fig. 8A, I, lanes 3 and 8). As expected, Rat1, assisted by Xrn1, was required for efficient C2-C1 trimming, and inactivation of these exoRNases led to detection of products ending at site C2 and of a striking ladder extending between C2 and C1′ (Fig. 8A, I, lane 3) (22). In the absence of the Rat1 cofactor Rai1, RNAs ending at C2 also accumulated, in keeping with a requirement for this cofactor in C2-C1 processing (Fig. 8A, lane 8). Analysis in ITS1 confirmed that Las1 does not play a major role in 5.8S rRNA 5′-end formation (Fig. 8B), either in the major pathway (no ladder accumulation, in contrast to the rat1 xrn1 control and normal cleavage at B1S) or in the minor pathway (normal cleavage at site B1L). However, RNAs ending at site A3 accumulated in las1 mutants, indicating a minor requirement in ITS1 processing (Fig. 8B, I, lanes 5 and 7). As expected, the B1S/B1L ratio was strikingly in favor of the long form in the rai1 mutant (Fig. 8B, II, lane 8). Finally, all sites tested (A2, A3, B1S, B1L, C1′, C1, and C2) were accurate at the nucleotide level.
In conclusion, yeast Las1 is required for pre-rRNA processing at both ends of ITS2 and the Rat1-Rai1 complex is involved in pre-rRNA processing at both ends of 5.8S rRNA precursors.
Las1 interacts functionally with Rai1.
To test whether Las1 and Rai1 interact functionally, the nonessential RAI1 gene was deleted in a strain expressing the pGAL::HA-las1 allele and the cumulative effects of the two mutations on pre-rRNA processing were tested (Fig. 8; see Fig. S4 in the supplemental material). In permissive conditions, genes under the control of pGAL promoters are known to be overexpressed, and this was the case for LAS1 in the pGAL::HA-las1 rai1Δ strain (a 20-fold increase in expression was estimated by RTqPCR [data not shown]). In these conditions, we found that the accumulation of products ending at site C2 (Fig. 8A, compare lanes 8 and 9) and the accumulation of the 5.8S+30 and 6S pre-rRNAs (compare lanes 2 and 7 in Fig. S4 in the supplemental material) was reduced. Reciprocally, upon genetic depletion of Las1 in the rai1Δ strain, processing products ending at site C2 accumulated more strongly (Fig. 8A, compare lanes 9 and 10). The effect on cleavage at site A3 was also somewhat enhanced in rai1Δ cells depleted of Las1 (Fig. 8B, lanes 9 and 10). Altogether, these observations are suggestive of functional interactions between Las1 and Rai1.
Las1 is required for cell cycle progression.
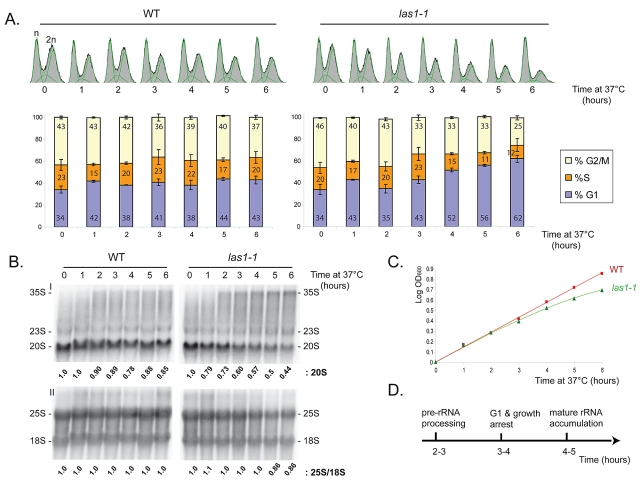
Yeast las1 mutants accumulate unbudded cells (13). This and the recent demonstration that Las1L (human Las1) is involved in p53-dependent G1 arrest (7) led us to investigate at which point in the cell cycle the protein is required in yeast. This was addressed by flow cytometry in las1-1 cells (Fig. 9). The cells were grown at 23°C to mid-log phase and transferred to 37°C for up to 6 h. Samples were collected every hour and analyzed for their contents of cells in the G1, S, and G2/M phases of the cell cycle. As a control, an isogenic wild-type strain was used. Inactivation of las1 led to a substantial and progressive accumulation of cells in G1 (starting between 3 and 4 h) and to a concomitant reduction of cells in S and G2/M (Fig. 9A). The ratio between cells in G1, S, and G2/M was 40:20:40 in the wild type and shifted to 60:10:30 following Las1 inactivation. These trends coincided in time with the appearance of the growth defect (Fig. 9C). A pre-rRNA processing defect was detected about 2 to 3 h after transfer to 37°C (see 20S pre-rRNA quantitation and 35S accumulation in panel B). The steady-state accumulation of mature rRNAs remained unaffected until 5 h after transfer (see 25S/18S ratio). The timing of the establishment of the various inhibitions (pre-rRNA processing, cell division, slowed growth, and accumulation of mature rRNA) is summarized in panel D. We conclude that yeast Las1 is required for the G1/S transition of the cell cycle and that cell cycle arrest is a consequence of the pre-rRNA processing defect rather than of an overall reduction in protein synthesis (see Discussion).
Fig 9.
Yeast Las1 is required for the G1/S transition of the cell cycle. (A) Flow cytometry analysis. The DNA contents (n or 2n) of las1-1 cells and of an isogenic wild-type control, grown to mid-log phase at 23°C and transferred to 37°C for up to 6 h, were determined by flow cytometry. Curve fitting with a dedicated software was used to determine precisely the percentage of cells at each stage of the cell cycle (G1, S, and G2/M). Each point was analyzed in triplicate. (B) Pre-rRNA processing analysis. Total RNA was analyzed by denaturing agarose gel electrophoresis and Northern blotting with probe LD471 (I) or probes LD871 and LD1099 (II) (see Fig. S1A in the supplemental material). The steady-state level of 20S pre-rRNA and the 25S/18S ratio were determined by phosphorimager quantitation. (C) Growth curves. (D) Timing of the different inhibitions.
Characterization of ITS2 processing in human cells.
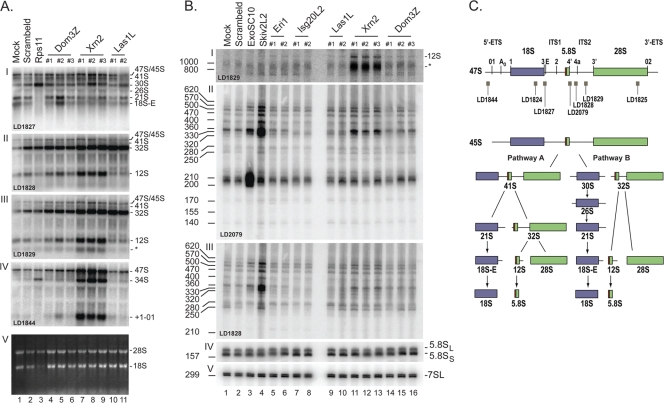
Recent analyses established that human Las1L is required for large subunit rRNA synthesis (7). To precisely characterize the involvement of Las1L in ITS2 processing, we extracted total RNA from HeLa cells treated for 72 h with siRNAs targeting the LAS1L mRNA and analyzed them by Northern blotting (Fig. 10). Analysis of high-molecular-weight RNAs confirmed that Las1L is required for ITS2 processing, as 32S was found to accumulate and 12S to diminish upon Las1L depletion (Fig. 10A, II and III; see quantitation in Table S2 in the supplemental material). The steady-state accumulation of 5.8S rRNA and the ratio of S to L forms (which is 60 to 40 in cultured human cells [see Table S2 in the supplemental material]), however, remained largely unaffected (Fig. 10B, IV). As controls, cells were treated with a nontargeting siRNA (scrambled) and with a silencer specific to the RPS11 mRNA. Rps11 depletion led to the expected striking accumulation of 30S pre-rRNA and to loss of both the 21S and 18S-E pre-rRNAs (Fig. 10A, I, lane 3) (33); this resulted in strong inhibition of 18S rRNA synthesis (Fig. 10A, V, lane 3).
Fig 10.
ITS2 processing in human cells is a multistep process requiring core exosome cofactors and ExoSC10. Total RNA extracted from HeLa cells treated for 72 h with 10 nM siRNA specific to the gene indicated was resolved by denaturing gel electrophoresis and analyzed by Northern blotting. Either 2 or 3 (#1 to #3) individual Silencer Select siRNAs (Ambion) were used. As controls, cells were treated with Lipofectamine alone (mock), a nontargeting silencer (scrambled), or a silencer specific to RPS11. (A) High-molecular-weight RNA analysis on agarose-formaldehyde gels. The probes used were LD1827 (I), LD1828 (II), LD1829 (III), and LD1844 (IV). In panel V, the gel was stained with ethidium bromide. (B) Low-molecular-weight RNA analysis on acrylamide gels. The probes used were LD1829 (I), LD2079 (II), LD1828 (III), and LD2132 (IV). 7SL (LD2133) hybridization provided a loading control. The sizes of the RNA intermediates were precisely estimated by reference to known molecular weight markers. (C) Pre-rRNA processing pathway in human cells. The 18S, 5.8S, and 28S rRNA sequences are embedded within the 5′ and 3′ external transcribed spacers (ETS) and internal transcribed spacer 1 (ITS1) and ITS2. The positions of the oligonucleotide probes used in the hybridizations are indicated. The primary transcript (47S) is cleaved at sites 01 and 02 on both sides of the molecule, generating the 45S pre-rRNA, which is processed by two alternative pathways. In pathway A, site A0, in the 5′-ETS, and site 1, at the 5′ end of 18S rRNA, are cleaved, yielding the 41S pre-rRNA. The 41S pre-rRNA is digested at site 2 within ITS1, separating the RNA precursors destined to become the small and large subunits, the 21S and 32S pre-rRNAs, respectively. The 21S pre-rRNA is cleaved at site E, producing the 18S-E intermediate, which is processed at site 3 into mature 18S rRNA. Processing of the 32S within ITS2 generates the 12S pre-rRNA and the 28S rRNA. The 12S pre-rRNA is digested to 5.8S rRNA by a succession of exoribonucleolytic digestions involving the core exosome and ExoSC10 (B). The detection of stabilized ITS2 fragments ranging in size from 800 and 1,000 nucleotides upon Xrn2 depletion (∗) indicates that additional cleavage occurs within the 5′ portion of ITS2 (site 4a). In alternative pathway B, the 45S pre-rRNA is directly cleaved at site 2 in ITS1, prior to cleavage at sites A0 and 1, generating the 30S and 32S pre-rRNAs. The 30S pre-RNA is cleaved at site A0 to 26S and at site 1 to 21S. In human cells, as in budding yeast, there are two forms, short and long, of 5.8S rRNA (red extension).
In parallel, we targeted the human orthologs of two partners of yeast Las1, namely, the pyrophosphohydrolase Dom3Z (ortholog of Rai1) and its apoenzyme, the 5′-3′ exoRNase Xrn2 (ortholog of Rat1). Xrn2 depletion led to a striking accumulation of the 47S primary transcript and of the 32S and 12S pre-rRNAs (Fig. 10A, III, lanes 7 to 9, and 10B, I, lanes 11 to 13). In addition, Xrn2 depletion led to strong accumulation of 5′ external transcribed spacer (ETS) (+1-01) and ITS2 (asterisk) fragments (Fig. 10A, III, and IV, lanes 7 to 9, and B, I, lanes 11 to 13). Lastly, a species whose size is compatible with direct cleavage of nascent transcripts at site 2, the 34S RNA, was detected (Fig. 10A, IV, lanes 7 to 9). This 34S RNA was also detected, albeit at a lower level, upon Rps11 depletion (Fig. 10A, IV, lane 3). The accumulation of 34S RNA indicates that, as in budding yeast (23S RNA; see above), direct cleavage in ITS1 can occur in human cells in the absence of the early cleavages at sites 01, A0, and 1. Many of the RNA intermediates accumulating upon Xrn2 depletion, including the spacer fragments, were also increased upon Dom3Z depletion (Fig. 10A, lanes 4 to 6), but to a much lesser extent. Note that either two or three different silencers (#1 to #3) were used independently for each target to address possible “off-target” effects and that the efficiency of each depletion was established by RTqPCR and/or Western blotting (see Fig. S5 in the supplemental material).
Processing of ITS2 in human cells is known to generate a 12S pre-rRNA that is trimmed down to the 3′ end of 5.8S rRNA (Fig. 10C). The exact number of steps involved in this processing and the sizes of the pre-rRNA intermediates generated are not known. In addition to Las1L, Dom3Z, and Xrn2, our analysis of low-molecular-weight RNAs (Fig. 10B) included targeting two conserved 3′-5′ exoRNases, Eri1 and Isg20L2, with known or suspected functions in 5.8S rRNA 3′-end formation in mammals (1, 9), the exosome subunit ExoSC10 (human Rrp6), and the ATP-dependent 3′-5′ RNA helicase Skiv2L2 (human Dob1/Mtr4). Skiv2L2 is a core exosome cofactor with known functions in 5.8S rRNA 3′-end formation in yeast and humans (10, 39).
Probing the Northern blots with a radiolabeled antisense oligonucleotide overlapping the 3′ end of 5.8S rRNA (Fig. 10B, II, LD2079; see schematics in panel C) revealed a range of discrete products of up to ∼620 nucleotides, corresponding to 5.8S rRNA extended by up to ∼460 nucleotides. The 3′-extended forms of 5.8S rRNA were all stabilized, to various extents upon inactivation of the exosome cofactor Skiv2L2 (Fig. 10B, II and III, lane 4; see quantitation in Table S2 in the supplemental material); the major product migrated to between 330 and 360. Inactivation of ExoSC10 led to stabilization of some but not all RNA intermediates. The most abundant product was shorter, migrating to between 200 and 210. Consistently, it was detected only with oligonucleotide LD2079, not LD1828. Depletion of all the other candidate proteins tested, including Las1L, caused only minor fluctuations in the accumulation of the various precursors (see quantitation profiling in Table S2 in the supplemental material). Notably, a doublet migrating to between 320 and 330, similar in length to fragments detected upon depletion of exosome components (Fig. 10B, lanes 3 and 4), was detected in cells deprived of Eri1, Isg20L2, Las1L, or Xrn2, but not in those deprived of Dom3Z. Xrn2 depletion led to the striking accumulation of 3′-extended species of 5.8S rRNA migrating at ∼360, indicating that, like its yeast ortholog Rat1 (see above), this protein is also somehow involved in ITS2 processing. Note that this RNA species was not detected with a probe specific to sequences located 5′ to the 5.8S rRNA (data not shown) and that, notably, it was absent from cells depleted of Isg20L2 (Fig. 10B, II, lanes 7 and 8). In the analysis presented in Fig. 10B, hybridization with a probe specific to 7SL, the RNA component of human SRP, provided a loading control (panel V).
We conclude that, in human cells as in budding yeast, ITS2 processing is a multistep process involving both core exosome cofactors and specific exosome subunits. Regarding the tested human homologs of certain yeast trans-acting factors having an essential involvement in ITS2 processing, we conclude that their individual depletion has only moderate effects in human cells. We also conclude that the 5′-3′ exoRNase Xrn2 is involved in spacer fragment degradation (5′-ETS and ITS2) and that it has a likely indirect effect on ITS2 processing, like its yeast ortholog.
DISCUSSION
Yeast Las1 is required for pre-rRNA processing at both ends of ITS2.
The production of mature rRNAs from polycistronic precursors is evolutionarily conserved. The processing of large pre-rRNA molecules into rRNAs has even become more complex in the course of evolution, with a steady increase in both the number of cleavage sites in noncoding spacers and the number of trans-acting factors involved. By its very design, a strategy involving the coproduction of several rRNAs from a single molecule contributes to coordinating the synthesis of ribosome components. Whether the necessary extensive pre-rRNA processing contributes also to regulating ribosome assembly remains to be demonstrated. In any event, nature has retained an extreme solution for synthesizing the mature 5′ and 3′ ends of 5.8S rRNA, since it relies on the use of alternative pathways and involves up to 11 distinct endo- and exoRNases, often with partially overlapping specificities, acting in parallel or sequentially (Fig. 2). By comparison, the sequences corresponding to the 5.8S rRNA are simply directly encoded at the 5′ ends of bacterial and archaeal 23S rRNAs; this appears to be a much simpler solution.
Here we have used a combination of genetic and biochemical approaches to demonstrate that, in budding yeast, the evolutionarily conserved protein Las1 is required for both Rrp6-dependent 5.8S rRNA 3′-end formation and Rat1-dependent 25S rRNA 5′-end formation (Fig. 6 to 8). These maturation events occur on both sides of ITS2, at processing sites 233 nucleotides apart. To our knowledge, no such phenotype has been reported previously. In addition, we found that the Rat1-Rai1 complex is involved in pre-rRNA processing at both ends of 5.8S RNA precursors (Fig. 2 and 7).
The function of Las1 in pre-rRNA processing at the 3′ end of ITS2 suggests that it works in close connection with the 5′-3′ exoRNases Rat1 and Xrn1, known to operate there (14, 19). We show that Las1 copurifies with preribosomes that also contain Rat1 and its cofactor Rai1 (Fig. 5). Xrn1 also copurifies with Las1 (Fig. 5). Las1 mutants, however, show a different phenotype from that of rat1 xrn1 mutants. First, they do not accumulate RNAs ending at cleavage site C2 and they do not generate a ladder of RNA products extending from site C2 to site C1′ (Fig. 8). Rather, Las1 is specifically required for the last trimming steps of ITS2, the conversion of 25S′ pre-rRNAs into 25S rRNA, involving removal of ∼7- to 8-nucleotide extensions (Fig. 8). Second, Las1 does not play a major role in 5.8S rRNA 5′-end formation (Fig. 7 and 8).
The involvement of Las1 in pre-rRNA processing at both ends of ITS2 is consistent with the observed reduced accumulation of 5.8S and 25S rRNAs (Fig. 6 and 7) and 60S subunits (Fig. 4) in las1 mutants and with the association of Las1 with precursor ribosomes (Fig. 4 and 5). The trans-acting factors copurifying with Las1 indicate that it is associated with nucleolar pre-90S ribosomes and with a range of pre-60S ribosomes comprising nuclear and cytoplasmic species (12, 17, 29). In Las1 pulldowns, the presence of trans-acting factors such as Rrp5 and Rrp12, involved in the synthesis of both the small and the large subunits, might explain why early nucleolar cleavages are also delayed in las1 mutants (Fig. 6). Ribosome synthesis factors are available in limited quantity, and their recycling is essential. Las1 might possibly contribute to this recycling, as it copurifies with several of the trans-acting factors involved in this process, such as Mdn1, an AAA-type ATPase distantly related to the motor protein dynein (2, 45).
Yeast Las1 is required for the G1/S transition of the cell cycle.
Pioneering work in the 1970s established that cell division and ribosome synthesis are tightly connected processes. Cells need to achieve a certain size, for which they require active ribosomes, in order to commit to cell division (reviewed in reference 26). This is known as the G1/S transition in yeast and as the restriction point (RP) in mammals. This G1/S (RP) transition is actively monitored by quality control mechanisms (reviewed in references 12 and 20). It was established previously that Las1 is involved in cell division (13), but the point in the cell cycle at which it intervenes was not determined. We find that yeast Las1, like its human ortholog, Las1L, is required for the G1/S transition (Fig. 9). The pre-rRNA processing defects associated with las1 mutations occur faster than inhibition of cell division and the reduction in mature ribosomes. This is compatible with the idea that, at the G1/S transition, quality control mechanisms monitor not only protein synthesis but also pre-rRNA processing.
What is the essential function of Las1? The 5.8S rRNA intermediates that accumulate in las1 mutants accumulate to a greater extent in cells with the nonessential gene rai1 or rrp6 deleted, making loss of Las1 an unlikely cause of lethality. However, las1 mutations additionally lead to a substantial reduction in 25S rRNA accumulation, which is likely sufficient to cause cell death. Further, las1 mutants are defective for early nucleolar cleavages (sites A0 to A2), which are known to be monitored by G1/S transition surveillance mechanisms (for example, mutations in small subunit-processome components), and consistently we showed that cells expressing the las1-1 allele are blocked at this step in cell division.
Human ITS2 processing is a multistep process.
We show that in human cells, as in budding yeast, ITS2 processing is a multistep process. We notably show that 5.8S rRNA 3′-end formation specifically requires the core exosome cofactor Skiv2L2 (human Dob1) and ExoSC10 (Rrp6) (Fig. 10). We further report that Xrn2 (human Rat1) is involved in spacer fragment degradation and contributes, likely indirectly, to 5.8S rRNA 3′-end formation. Having analyzed prime candidates for putative functions in ITS2 processing, we find, quite surprisingly, that depleting cells of any one of these proteins individually has only a limited effect on pre-rRNA processing (Fig. 10). This provides an indication that the situation is going to be far more complex in the human species than in budding yeasts.
How is pre-rRNA processing at distant cleavage sites coordinated? (i) Pre-rRNA processing at the two ends of 5.8S rRNA is coordinated.
As shown here and elsewhere (Fig. 2, 6, and 7) (16, 43, 48), rai1 mutations affect rRNA maturation at both ends of 5.8S rRNA. At the 5′ end, Rai1 inactivation tips the scales in favor of the L form of 5.8S rRNA, in agreement with its function as a cofactor of Rat1. At the 3′ end, it leads to accumulation of 5.8S rRNA species extended by 30 nucleotides, reminiscent of those found in rrp6 mutants. We further report that Rat1 is also involved in 5.8S 3′-end maturation (Fig. 2 and 7). We interpret both these observations as a strong indication that functional interactions exist on both sides of 5.8S rRNA precursors. The folding of the 5.8S rRNA sequence, which brings its 5′ and 3′ ends into close proximity, is compatible with the idea that an RNA processing complex sitting there might affect maturation at both ends (Fig. 1). Such functional interactions might take place in a “functional neighborhood” within pre-60S ribosomes. The existence of such functional neighborhoods is further supported by the observation that Rat1 indirectly affects 5.8S 3′-end formation and its function at this level appears to be conserved in yeast and HeLa cells. In yeast, inactivation of Rat1 leads to low-level accumulation of 3′-extended forms of 5.8S rRNA (Fig. 2 and 7), reminiscent of those found in rai1 mutants. In human cells, depletion of Xrn2 leads to a striking accumulation of 5.8S rRNA extended by 170 nucleotides and migrating around 330 to 360 (Fig. 10).
(ii) Pre-rRNA processing at the two ends of ITS2 is coordinated.
In mature 60S subunits, the 3′ end of the 5.8S rRNA and the 5′ end of the 25S rRNA interact with each other, forming a long, evolutionarily conserved stem that most likely contributes to specifying the mature ends of the rRNAs (Fig. 1, inset). Las1 is required for both 5.8 rRNA 3′-end formation and 25S rRNA 5′-end formation. Again, folding the ITS2 sequence brings these two sequences into close proximity, suggesting that a functional neighborhood might affect these two distant sites (Fig. 1).
Finally, we found it quite striking that ∼7- or 8-nucleotide extensions are detected on pre-rRNA precursors on both sides of the 5.8S rRNA and at the 5′ end of 25S rRNA (Fig. 1). We think that it is suggestive of a mechanism in which this distance is protected by a common RNA processing complex, whose function could be to coordinate spatially and temporally, and possibly to quality prove, pre-rRNA cleavage at distant sites on the primary sequence.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following colleagues for providing reagents: Ambro van Hoof (University of Texas Health Science Center-Houston) for the rex mutants, Giorgio Dieci (Università di Parma) for the anti-Rps8 antibody, Micheline Fromont-Racine (Institut Pasteur, Paris) for the anti-Nog1 antibody, Jonathan Warner (Albert Einstein College, NYC) for the anti-Rpl3 antibody, and Aziz El Hage and David Tollervey (Wellcome Trust Centre for Cell Biology, Edinburgh) for the pMET::rat1 xrn1Δ strain. We are deeply indebted to Christiane Zorbas, Taylor Mullineux, and Lionel Tafforeau for help with the RNA interference depletions.
This work was funded by the FRIA, the FRS-F.N.R.S., the Communauté Française de Belgique (ARC), and the Région Wallone (Cibles).
Footnotes
Published ahead of print 14 November 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Ansel KM, et al. 2008. Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat. Struct. Mol. Biol. 15:523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bassler J, et al. 2010. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell 38:712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8:574–585 [DOI] [PubMed] [Google Scholar]
- 4. Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. 2010. The nucleolus under stress. Mol. Cell 40:216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breslow DK, et al. 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5:711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Briggs MW, Burkard KT, Butler JS. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273:13255–13263 [DOI] [PubMed] [Google Scholar]
- 7. Castle CD, Cassimere EK, Lee J, Denicourt C. 2010. Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis. Mol. Cell. Biol. 30:4404–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a. Colau G, Thiry M, Leduc V, Bordonné R, Lafontaine DLJ. 2004. The small nucle(ol)ar RNA cap trimethyltransferase is required for ribosome synthesis and intact nucleolar morphology. Mol. Cell. Biol. 24:7976–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cote CA, Greer CL, Peculis BA. 2002. Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA 8:786–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coute Y, et al. 2008. ISG20L2, a novel vertebrate nucleolar exoribonuclease involved in ribosome biogenesis. Mol. Cell Proteomics 7:546–559 [DOI] [PubMed] [Google Scholar]
- 10. de la Cruz J, Kressler D, Tollervey D, Linder P. 1998. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 17:1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deutscher MP. 2009. Maturation and degradation of ribosomal RNA in bacteria. Prog. Mol. Biol. Transl. Sci. 85:369–391 [DOI] [PubMed] [Google Scholar]
- 12. Dez C, Tollervey D. 2004. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7:631–637 [DOI] [PubMed] [Google Scholar]
- 13. Doseff AI, Arndt KT. 1995. LAS1 is an essential nuclear protein involved in cell morphogenesis and cell surface growth. Genetics 141:857–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. El Hage A, Koper M, Kufel J, Tollervey D. 2008. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 22:1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Faber AW, Van Dijk M, Raue HA, Vos JC. 2002. Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA 8:1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fang F, Phillips S, Butler JS. 2005. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA 11:1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fatica A, Tollervey D. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313–318 [DOI] [PubMed] [Google Scholar]
- 18. Finkbeiner E, Haindl M, Muller S. 2011. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 30:1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geerlings TH, Vos JC, Raue HA. 2000. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′→3′ exonucleases. RNA 6:1698–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gérus M, Caizergues-Ferrer M, Henry Y, Henras AK. 2011. Crosstalk between ribosome synthesis and cell cycle and its potential implications in human diseases, p. 157–184 In Olson MO. (ed.), Protein reviews, vol. 15. The nucleolus. Springer, New York, NY [Google Scholar]
- 21. Henras AK, et al. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65:2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Henry Y, et al. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13:2452–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DLJ. 2010. The nucleolus: structure/function in RNA metabolism. RNA 1:415–431 [DOI] [PubMed] [Google Scholar]
- 24. Houseley J, LaCava J, Tollervey D. 2006. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7:529–539 [DOI] [PubMed] [Google Scholar]
- 25. Johnson AW. 1997. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 17:6122–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jorgensen P, Tyers M, Warner JR. 2004. Forging the factory: ribosome synthesis and growth control in budding yeast, p 329–370 In Hall M, Raff M, Thomas G. (ed), Cell growth: control of cell size. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Kitano E, Hayashi A, Kanai D, Shinmyozu K, Nakayama JI. 2011Roles of fission yeast Grc3 in ribosomal RNA processing and heterochromatic gene silencing. J. Biol. Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kos M, Tollervey D. 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 37:809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kressler D, Hurt E, Bassler J. 2010. Driving ribosome assembly. Biochim. Biophys. Acta 1803:673–683 [DOI] [PubMed] [Google Scholar]
- 30. Lafontaine DLJ. 2010. A ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes? Trends Biochem. Sci. 35:267–277 [DOI] [PubMed] [Google Scholar]
- 31. Lafontaine DLJ, Tollervey D. 2001. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2:514–520 [DOI] [PubMed] [Google Scholar]
- 32. Lebreton A, Tomecki R, Dziembowski A, Seraphin B. 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456:993–996 [DOI] [PubMed] [Google Scholar]
- 33. O'Donohue MF, Choesmel V, Faubladier M, Fichant G, Gleizes PE. 2010. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 190:853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oeffinger M, et al. 2007. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods 4:951–956 [DOI] [PubMed] [Google Scholar]
- 35. Oeffinger M, et al. 2009. Rrp17p is a eukaryotic exonuclease required for 5′ end processing of pre-60S ribosomal RNA. Mol. Cell 36:768–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Osheim YN, et al. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 16:943–954 [DOI] [PubMed] [Google Scholar]
- 37. Peng WT, et al. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919–933 [DOI] [PubMed] [Google Scholar]
- 38. Schaeffer D, et al. 2009. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 16:56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schilders G, van Dijk E, Pruijn GJ. 2007. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 35:2564–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 41. Schneider C, Leung E, Brown J, Tollervey D. 2009. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 37:1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Strunk BS, Karbstein K. 2009. Powering through ribosome assembly. RNA 15:2083–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sydorskyy Y, et al. 2003. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol. Cell. Biol. 23:2042–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thomson E, Tollervey D. 2010. The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol. Cell. Biol. 30:976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ulbrich C, et al. 2009. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 138:911–922 [DOI] [PubMed] [Google Scholar]
- 46. van Hoof A, Lennertz P, Parker R. 2000. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 19:1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wery M, Ruidant S, Schillewaert S, Lepore N, Lafontaine DL. 2009. The nuclear poly(A) polymerase and exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA 15:406–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xue Y, et al. 2000. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol. 20:4006–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yeh LC, Lee JC. 1990. Structural analysis of the internal transcribed spacer 2 of the precursor ribosomal RNA from Saccharomyces cerevisiae. J. Mol. Biol. 211:699–712 [DOI] [PubMed] [Google Scholar]
- 50. Zhang J, Jr, et al. 2007. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 21:2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.