Abstract
The transcriptional coactivator PGC-1α is a master regulator of energy metabolism and adaptive thermogenesis in the brown fat cell. PGC-1α is a short-lived protein, and the molecular components that control PGC-1α turnover and their functional importance in energy metabolism are largely unknown. Here we performed a luciferase-based overexpression screen and identified a Ring-finger-containing protein, RNF34, as a specific E3 ubiquitin ligase for PGC-1α. RNF34 is a nuclear protein that interacts with and ubiquitinates PGC-1α to promote its turnover. Interestingly, RNF34 binds to the C-terminal half of PGC-1α and targets it for degradation independently of the previously identified N-terminal phosphodegron motif. In brown fat cells, knockdown of RNF34 increases the endogenous PGC-1α protein level, uncoupling protein 1 (UCP1) expression, and oxygen consumption, while the opposite effects are observed in brown fat cells ectopically expressing wild-type RNF34 but not in cells expressing the ligase activity-defective mutant. Moreover, cold exposure and β3-adrenergic receptor signaling, conditions that induce PGC-1α expression, suppress RNF34 expression in the brown fat cell, indicating a physiological relevance of this E3 ligase in thermogenesis. Our results reveal that RNF34 is a bona fide E3 ubiquitin ligase for PGC-1α and negatively regulates brown fat cell metabolism.
INTRODUCTION
Metabolic programs are to a large extent controlled at the transcriptional level, in which the transcriptional coactivator PGC-1α is at the central node. Through its interaction with and coactivation of functionally distinct transcription factors, PGC-1α regulates tissue-specific metabolic pathways, including mitochondrial oxidative metabolism and adaptive thermogenesis in the brown fat cell, gluconeogenesis in the liver, and mitochondrial oxidative metabolism and muscle fiber specification in the skeletal muscle (13, 20, 23). The functions of PGC-1α in these tissues are highly coordinated with environmental cues and/or internal nutrient levels and are modulated by several mechanisms (6, 23). For example, in the brown fat cell, in response to cold exposure, transcription of PGC-1α mRNA is robustly induced, which leads to increased uncoupling protein 1 (UCP1) expression and heat production (14, 20). More recently, we identified the transcriptional regulator twist-1 as a negative-feedback component that interacts with and directly antagonizes PGC-1α activity in the brown fat cell (18).
Posttranslational modification of PGC-1α is another key mechanism that fine-tunes PGC-1α's function (6, 23). Phosphorylation and acetylation are the two best-characterized modifications to PGC-1α, and their functional impacts have been studied in skeletal muscle cells and/or liver cells. Phosphorylation of PGC-1α by AMPK and p38 mitogen-activated protein kinase (MAPK) increases PGC-1α activity and enhances mitochondrial oxidative metabolism in skeletal muscle cells (9, 19). On the other hand, phosphorylation by Akt and Clk2 suppresses PGC-1α activity and reduces hepatic gluconeogenesis (12, 22). PGC-1α is also known to be acetylated by GCN5 and deacetylated by NAD-dependent Sirt1. Deacetylation of PGC-1α by Sirt1 activates PGC-1α and promotes both gluconeogenesis in the liver and mitochondrial oxidative metabolism in the skeletal muscle (1, 2, 7, 24), whereas acetylation of PGC-1α by GCN5 has the opposite effects (7, 11). Overall, these modifications to PGC-1α allow precise metabolic control that coordinates with cellular energy needs.
PGC-1α is a short-lived protein and is presumably targeted for degradation by the ubiquitin-proteasome pathway. In contrast to phosphorylation and acetylation, ubiquitination of PGC-1α has not been well studied. In the ubiquitination system, E3 ligases determine substrate specificity by interacting with target proteins and catalyzing the transfer of ubiquitin to them. E3 ligases are grouped into three classes: the single-subunit Ring-finger type, the multisubunit Ring-finger type, and the HECT-domain type (3). Recently, the multisubunit E3 ligase SCFFbw7 was shown to promote PGC-1α ubiquitination and degradation. SCFFbw7 recognizes a phosphodegron in PGC-1α, which consists of four residues located in the N terminus of PGC-1α that are phosphorylated by Gsk3β and p38 MAPK (16). However, the functional relevance of this Fbw7-mediated ubiquitination in PGC-1α target gene expression and energy metabolism is unknown. In this regard, insulin signaling inactivates Gsk3β and is thus expected to stabilize PGC-1α protein. This is at odds with the fact that PGC-1α function is augmented during fasting and is suppressed by insulin signaling (12, 22, 28, 33) and suggests that additional mechanisms exist to regulate PGC-1α turnover.
Given the noted presence of brown fat depots in adult humans, its inverse correlation with human obesity (5, 15, 26, 30, 31), and the central role of PGC-1α in brown fat metabolism, it would be of great importance to identify enzymes that modify PGC-1α and regulate its function in brown fat cells. In this report, we demonstrate such a role for RNF34, which acts as a single-subunit Ring-finger type E3 ubiquitin ligase for PGC-1α.
MATERIALS AND METHODS
E3 ligase screen and luciferase reporter assays.
A total of 264 cDNA expression clones involved in protein ubiquitination and degradation were obtained from Origene (Rockville, MD). A 10-ng volume of each clone, along with 25 ng of Gal4- upstream activation sequence (UAS)-luciferase plasmid, 5 ng of Gal4 DNA-binding domain (DBD)-PGC-1α plasmids, and 5 ng of β-galactosidase plasmid, was used to transfect Cos-7 cells plated in 96-well plates. Luciferase activity was measured 48 h after transfection and normalized with β-galactosidase activity. For other luciferase assays, Cos-7 cells plated in 48-well plates were transfected with the indicated plasmids. Vector plasmids were used to adjust the total amount of plasmids per well.
Ubiquitination assay and coimmunoprecipitation.
For an in vivo ubiquitination assay, HEK293 cells were transfected with hemagglutinin (HA)–PGC-1α and Flag-ubiquitin plasmids. Cells were treated with MG132 proteasome inhibitor (10 μM) for 6 h prior to lysis. Cells were lysed in buffer A (250 mM NaCl, 50 mM Tris [pH 7.5], 0.5% Triton X-100, 5% glycerol) containing10 μM MG132. Cell extracts were incubated with anti-Flag antibody beads (catalog no. A2220; Sigma) for 2 h. The beads were washed four times with buffer B (250 mM NaCl, 50 mM Tris [pH 7.5], 0.1% Triton X-100, 5% glycerol) containing 10 μM MG132. Immunoprecipitates were probed with an anti-HA antibody (catalog no. 12013819001; Roche). To detect the interaction between RNF34 and PGC-1α, HEK293 cells were transfected with the indicated plasmids and coimmunoprecipitation was performed as described previously (18). Anti-PGC-1α antibody was obtained from Santa Cruz (catalog no. SC13067).
Immunofluorescence.
HA-PGC-1α and Flag-RNF34 plasmids were transfected into HEK293 cells cultured in a 24-well plate. Two days after transfection, cells were rinsed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 20 min. The fixed cells were incubated with 0.1% Triton X-100–PBS for 10 min and 5% goat serum–PBS for 30 min. The cells were incubated with anti-HA (catalog no. SC7392; Santa Cruz) (1:200) and anti-Flag (catalog no. F7425; Sigma) (1:200) antibodies at 4°C overnight, followed by an incubation with fluorophore-conjugated secondary antibodies for 1 h at room temperature. All images were recorded and processed under equivalent conditions.
Adenoviruses.
Adenoviruses for overexpression and knockdown were generated by using the AdEasy-1 system (8). All adenoviruses were purified with cesium chloride ultracentrifugation, and virus titers were determined in HEK 293 cells by scoring green fluorescent protein (GFP)-positive cells. RNF34 short hairpin RNA (shRNA) targeting sequences are available upon request.
Brown fat adipocytes and gene expression.
Brown fat primary cells were isolated and cultured essentially as described previously (10). Immobilized brown fat preadipocytes were generated previously (18). Brown fat cell differentiation was performed as described previously (18). Adenovirus infection was performed using differentiated brown fat cells unless otherwise indicated. GFP control adenovirus was added, when needed, to ensure similar degrees of infection in different samples. The infection efficiency was monitored by GFP fluorescence. Two days after infection, gene expression was measured by quantitative reverse transcription-PCR (qRT-PCR) and normalized to the level of U36B4. Primer sequences are available upon request.
Assays of endogenous PGC-1α protein level and oxygen consumption.
These experiments were performed in differentiated brown fat cells 2 days after adenovirus infection. Nuclear extracts were isolated as described previously (25), and the PGC-1α protein level was determined by Western blotting using an anti-PGC-1α antibody (catalog no. SC13067; Santa Cruz). To measure oxygen consumption, cells were treated with CL-316243 β3-adrenergic agonist (10 μM) for 6 h. Equal number of cells were trypsinized, pelleted, and resuspended in PBS supplemented with 25 mM glucose, 1 mM pyruvate, and 2% bovine serum albumin (BSA). The respiration rate was measured with a Clark-type electrode (YSI model 5300) at 37°C and normalized to cell numbers.
RNF34 expression in brown fat.
Wild-type mice were placed at 4°C or room temperature for 5 h, and brown fat and white fat were isolated. CL-316243 β3-adrenergic receptor agonist (catalog no. 100507-726; VWR) dissolved in PBS was intraperitoneally (i.p.) injected into wild-type mice (0.5 mg/kg body weight) daily for 6 days. Brown fat and white fat were isolated.
Statistical analysis.
The Student's (two-tailed) t test was used for statistical analysis. A P value <0.05 was considered to indicate statistical significance. Data are presented as means ± standard errors of the means.
RESULTS
Identification of RNF34 as an E3 ubiquitin ligase for PGC-1α.
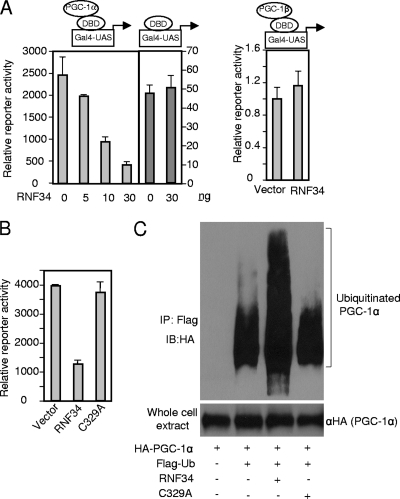
Previously we have shown that twist-1 is a crucial physiological negative regulator of brown fat metabolism. Twist-1 interacts with the C-terminal half of PGC-1α and powerfully inhibits its transcriptional activity. Interestingly, as a secondary consequence of this interaction, PGC-1α protein is stabilized (18). These results raise the possibility of the existence of an E3 ubiquitin ligase that binds to the C-terminal half of PGC-1α and targets it for degradation. To identify this potential E3 ligase, we performed a luciferase activity-based screening. Briefly, we obtained a collection of mammalian expression cDNA clones that are involved in protein ubiquitination and degradation pathways. These clones were arrayed in 96-well plates. Among them, approximately 100 clones have been shown or are predicted to be E3 ubiquitin ligases. Cos-7 cells were cultured in the arrayed plates and cotransfected with plasmids containing a full-length PGC-1α fused to the Gal4 DNA-binding domain (DBD-PGC-1α), along with luciferase plasmids driven by the Gal4-UAS promoter. Two clones were found to robustly suppress the transcriptional activity of PGC-1α: one was Fbw7, as would be expected (16); the second was RNF34. RNF34 specifically suppressed PGC-1α activity in a dose-dependent manner but had no effect on the basal activity of Gal4-DBD alone or on the activity of PGC-1β (Fig. 1A). RNF34 is a Ring-finger-containing protein predicted to be an E3 ligase, but its substrate(s) has not been identified. Ring-finger E3 ligases contain a conserved cysteine residue, located in the Ring-finger domain, that is essential for their ligase activity. Mutation of this cysteine residue (C329A) in RNF34 abolished its suppressive effect on PGC-1α activity (Fig. 1B), indicating that RNF34 might be an E3 ligase for PGC-1α. To test this idea, HEK293 cells were transfected with HA-PGC-1α, RNF34, or the C329A mutant and Flag-ubiquitin in the presence of the proteasome inhibitor MG132. Ubiquitinated proteins were immunoprecipitated with anti-Flag beads. Immunoblotting with anti-HA revealed that wild-type RNF34, but not the C329A mutant, strongly increased the level of ubiquitinated PGC-1α (Fig. 1C). These results suggest that RNF34 is an E3 ubiquitin ligase for PGC-1α.
Fig 1.
RNF34 is an E3 ubiquitin ligase for PGC-1α. (A) Left panel, dose-dependent inhibition of PGC-1α transcriptional activity by RNF34. Right panel, RNF34 does not decrease the transcriptional activity of PGC-1β. Cos-7 cells were transfected with RNF34, β-galactosidase (β-Gal) (10 ng), Gal4-UAS-luciferase reporter (50 ng), and Gal4 DBD-PGC-1α, Gal4 DBD-PGC-1β, or DBD alone (10 ng). Luciferase activity was measured and normalized to β-galactosidase activity. Error bars represent standard errors of the means; n = 3. (B) E3 ligase-dependent inhibition of PGC-1α transcriptional activity by RNF34. Experiments were performed as described for panel A. Gal4-PGC-1α, 10 ng; RNF34 or C329A mutant, 30 ng. Error bars represent standard errors of the means; n = 3. (C) RNF34 promotes ubiquitination of PGC-1α in vivo. HEK293 cells were transfected with the indicated plasmids. Ubiquitinated PGC-1α species were precipitated with Flag antibody beads and probed with an anti-HA antibody. IP, immunoprecipitation; IB, immunoblot.
RNF34 promotes PGC-1α degradation.
We examined whether RNF34 controls the steady-state level of PGC-1α protein (Fig. 2A). HEK293 cells were transfected with PGC-1α and RNF34 or the C329A mutant. Expression of RNF34 rendered the PGC-1α protein almost undetectable, whereas the C329A mutant had little effect. Moreover, the reduction in the level of PGC-1α protein caused by RNF34 was completely recovered upon addition of the proteasome inhibitor MG132, suggesting that RNF34 directs PGC-1α for proteasome-mediated degradation. On the other hand, RNF34 did not affect the protein level of PGC-1β or peroxisome proliferator-activated receptor alpha (PPARα) (Fig. 2B). To ensure that the observed effect of RNF34 on PGC-1α protein degradation occurs in intact cells, not after cell lysis, we performed immunofluorescence staining experiments (Fig. 2C). Exogenous HA-PGC-1α was uniformly distributed in the nucleus and was readily detectable when cells were cotransfected with an empty vector or the C329A mutant. However, upon coexpression of wild-type RNF34, which is mainly localized in the nucleus, HA-PGC-1α was barely detectable in the vast majority of cells. Thus, RNF34 regulates the steady-state level of PGC-1α protein in cells. To further test the hypothesis that the decreased protein level of PGC-1α caused by RNF34 is due to increased proteolysis, we treated cells with cycloheximide to inhibit protein translation and monitored the turnover of preexisting PGC-1α protein. As shown in Fig. 2D, expression of RNF34 accelerated the turnover of PGC-1α protein and considerably shortened its half-life. Taken together, the data presented in Fig. 1 and 2 suggest that RNF34 is a bona fide E3 ligase for PGC-1α and directs PGC-1α for proteasomal degradation.
Fig 2.
RNF34 promotes PGC-1α degradation. (A) RNF34 decreases the steady-state level of PGC-1α protein. HEK293 cells were transfected with the indicated plasmids, and total cell extracts were analyzed by Western blotting. Proteasome inhibitor MG132 (10 μM) was added, as indicated, to the culture medium 6 h prior to lysis. (B) RNF34 does not affect the steady-state protein level of PGC-1β or PPARα. HEK293 cells were transfected with the indicated plasmids, and cell extracts were probed with the indicated antibodies. (C) Immunofluorescence staining of ectopically expressed PGC-1α and RNF34 in HEK293 cells. DAPI, 4′,6-diamidino-2-phenylindole. (D) RNF34 accelerates the turnover of PGC-1α protein. HEK 293 cells were transfected with PGC-1α along with vector or Flag-RNF34. At 24 h later, the cells were added to 6-well plates. The next day, cycloheximide (500 μM) was added to the medium, the cells were harvested at the indicated time points, and the PGC-1α protein level was determined. The two images had different exposure times. The relative level of PGC-1α protein at each time point is plotted as a percentage of the amount at the 0 time point.
Regulation of PGC-1α ubiquitination by endogenous RNF34.
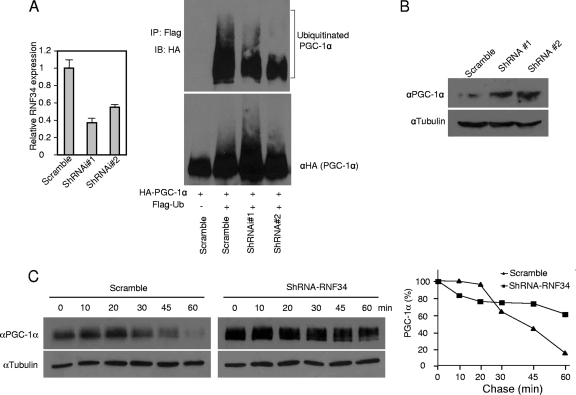
Addition of the proteasomal inhibitor MG132 to HEK293 cells led to a high basal level of PGC-1α ubiquitination (Fig. 1C and Fig. 3A), indicating the presence of E3 ligase(s) for PGC-1α. To examine whether endogenous RNF34 plays a part, we generated two RNF34 shRNA knockdown adenoviral constructs. Infection of HEK293 cells with the adenoviruses resulted in a 50% to 60% knockdown of RNF34 expression (Fig. 3A). We measured ubiquitination of exogenous PGC-1α in these cells. Knockdown of RNF34 reduced the level of ubiquitinated PGC-1α (Fig. 3A) and, as a consequence, increased the steady-state level of PGC-1α and extended its half-life compared to the scramble control (Fig. 3B and C). These results suggest that endogenous RNF34 participates in ubiquitination and degradation of PGC-1α protein.
Fig 3.
Regulation of PGC-1α ubiquitination by endogenous RNF34. (A) HEK293 cells were infected with adenoviruses expressing RNF34 shRNA knockdown constructs. Left panel, endogenous RNF34 mRNA levels were measured. Right panel, 24 h after infection, cells were transfected with HA-PGC-1α and Flag-ubiquitin. Proteasome inhibitor MG132 (10 μM) was added to the culture medium 6 h prior to lysis. Ubiquitinated PGC-1α species were precipitated with Flag antibody beads and probed with an anti-HA antibody. (B) HEK293 cells were infected with RNF34 shRNA adenoviruses. The next day, cells were transfected with PGC-1α. Steady-state PGC-1α protein was measured by Western blotting with an anti-PGC-1α antibody. (C) HEK293 cells were infected with RNF34 shRNA adenoviruses, followed by transfection with PGC-1α. At 24 h later, the cells were added to 6-well plates. The next day, cycloheximide (500 μM) was added to the medium, and the cells were harvested at the indicated time points and the PGC-1α protein level was determined. The relative level of PGC-1α protein at each time point is plotted as a percentage of the amount at the 0 time point. We noted that the turnover rates of PGC-1a differed among independent experiments, possibly due to cultured-cell conditions.
RNF34 binds to the C-terminal half of PGC-1α and targets it for degradation.
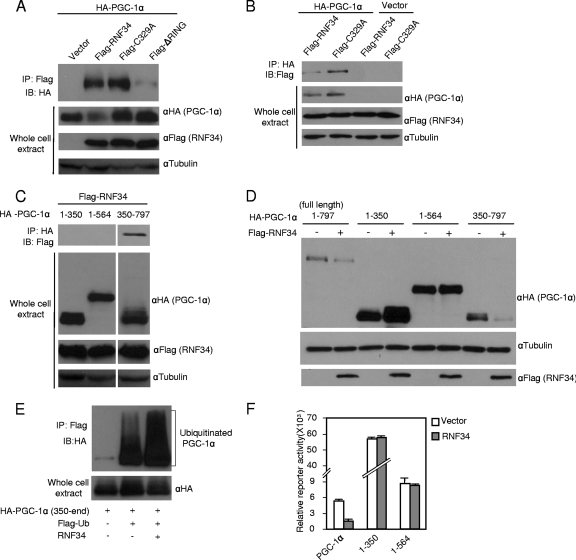
We next carried out a series of experiments to investigate how PGC-1α is targeted by RNF34 for ubiquitination and degradation. We expressed PGC-1α and RNF34 in HEK293 cells. Coimmunoprecipitation experiments revealed that PGC-1α was present in the RNF34 immunocomplex and vice versa (Fig. 4A and B), indicating that these two proteins interact in cells and that RNF34 acts as a single-subunit Ring-finger E3 ligase for PGC-1α. While the C329A mutant of RNF34 was fully capable of interacting with PGC-1α, removing the Ring-finger domain abolished their interaction (Fig. 4A). Thus, both recognition and ubiquitination of its substrate, PGC-1α, by RNF34 are mediated by its Ring-finger domain.
Fig 4.
RNF34 interacts with PGC-1α and targets the C-terminal half of PGC-1α for degradation. (A, B) RNF34 interacts with PGC-1α, and the Ring-finger domain is required for the interaction. HEK 293 cell was transfected with plasmids as indicated. (C) RNF34 interacts with the C-terminal half of PGC-1α. Note that different amounts (aa 1 to 350, 2.5 μg; 1 to 564, 4 μg; 350 to 797, 7.5 μg) of the PGC-1α deletion constructs were used to transfect HEK293 cells to ensure that they expressed similar levels of PGC-1α protein for immunoprecipitation. The two images were from the same exposed film and were processed identically. (D) RNF34 decreases the steady-state level of the C-terminal half of PGC-1α protein. HEK293 cells were cotransfected with PGC-1α and RNF34 or vector. (E) RNF34 promotes ubiquitination of the C-terminal half of PGC-1α. (F) RNF34 does not decrease the transcriptional activity of N-terminal PGC-1α constructs. Full-length PGC-1α or N-terminal fragments fused with Gal4-DBD were transfected along with RNF34 into Cos-7 cells as described for Fig. 1B. Luciferase activity was measured and normalized to β-galactosidase activity. Error bars represent standard errors of the means; n = 3.
To identify the region in PGC-1α that is important for its interaction with RNF34, we expressed PGC-1α truncation constructs along with RNF34 and performed coimmunoprecipitation experiments. Figure 4C shows that the C-terminal half (amino acids [aa] 350 to 797) of PGC-1α interacted with RNF34. Expression of RNF34 decreased the steady-state protein level of the C-terminal half PGC-1α construct, but not those of the N-terminal constructs, and promoted ubiquitination on the C-terminal region of PGC-1α (Fig. 4D and E). These results suggest that the C-terminal half of PGC-1α is responsible for the interaction with RNF34 and subsequent RNF34-mediated degradation and that the N-terminal half of PGC-1α is dispensable. This is in agreement with the lack of suppressive effect of RNF34 on the N-terminal half of PGC-1α constructs, which harbor PGC-1α's transcriptional activity (Fig. 4F).
Ubiquitination and degradation of PGC-1α protein by RNF34 are independent of phosphodegron and Fbw7.
The previously identified phosphodegron contains four sites phosphorylated by Gsk3β and p38 MAPK in the N-terminal half of PGC-1α (16). Our data presented above suggest that RNF34-mediated degradation of PGC-1α might not require the phosphorylation of these four sites. To test this more rigorously, we coexpressed RNF34 and a PGC-1α mutant in which the four phosphorylation sites were changed to alanine (PGC-1α-4A). RNF34 was still able to efficiently promote the degradation of this PGC-1α mutant (Fig. 5A). In agreement with previous studies (16), expression of the Fbw7 E3 ligase had no effect on this mutant (Fig. 5A). These results clearly show that RNF34-mediated PGC-1α degradation is independent of the N-terminal phosphodegron in a manner that is distinct from that of Fbw7.
Fig 5.
Ubiquitination and degradation of PGC-1α protein by RNF34 are independent of phosphodegron and Fbw7. (A) HEK293 cells were transfected with PGC-1α or a PGC-1α mutant in which the four phosphorylation sites targeted by p38 MAPK and Gsk3b were mutated (PGC-1α-4A), along with RNF34 or Fbw7. The steady-state PGC-1α protein level was determined by Western blotting with an antibody against PGC-1α. (B) HEK293 cells were infected with lentivirus expressing Fbw7 shRNA. Ubiquitination of PGC-1α protein by RNF34 was assayed in the stable knockdown cells as described for Fig. 1C. (C) Stable Fbw7 knockdown cells were transfected with PGC-1α and RNF34 plasmids. Steady-state PGC-1α protein was determined by Western blotting with an antibody against PGC-1α.
However, it still remained a possibility that ubiquitination and degradation of PGC-1α by RNF34 might require a triggering event somehow initiated by Fbw7. To rule out this possibility, we examined RNF34-mediated PGC-1α ubiquitination and degradation in an Fbw7 knockdown background. RNF34 promoted a similar degree of increase of PGC-1α ubiquitination (Fig. 5B) and a similar level of degradation (Fig. 5C) in Fbw7 knockdown cells relative to scramble cells, indicating that RNF34 is fully functional in the absence of Fbw7. As expected, knockdown of Fbw7 itself led to a decrease of PGC-1α ubiquitination (compare second and fifth lanes of Fig. 5B) and an increase in the PGC-1α protein level (compare first and third lanes of Fig. 5C).
RNF34 regulates PGC-1α target gene expression and brown fat cell metabolism.
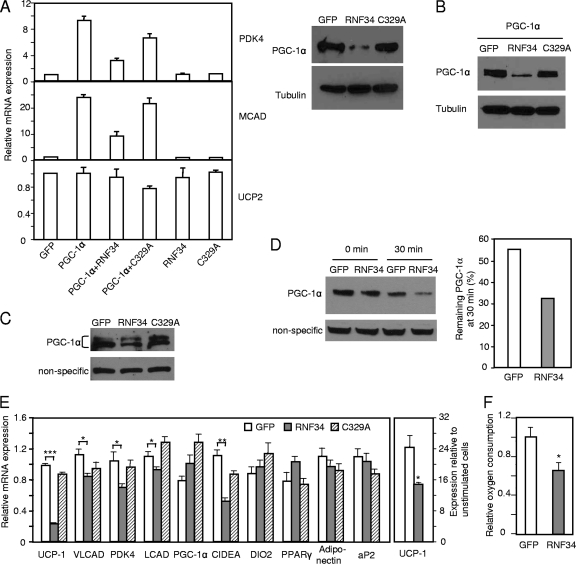
We determined whether RNF34-regulated PGC-1α degradation has any impact on PGC-1α target gene expression. The rat FAO cell line is highly responsive to exogenous PGC-1α expression, resulting in a robust increase in PGC-1α target gene expression. We thus took advantage of this system to test the functional antagonism of RNF34 with respect to PGC-1α. Adenoviral expression of PGC-1α in FAO cells induced the expression of PGC-1α target genes, including PDK4 and MCAD. This induction was strongly negated by coexpression of wild-type RNF34 but not by coexpression of the C329A mutant (Fig. 6A), in agreement with their different effects on the PGC-1α protein level (Fig. 6A). The expression of UCP2, which is not a PGC-1α target gene and was included as a control, was not affected by RNF34. The results suggest a specific modulation of PGC-1α target gene expression by RNF34.
Fig 6.
RNF34 regulates PGC-1α target gene expression and brown fat cell metabolism. (A) RNF34 inhibits PGC-1α-mediated target gene expression in FAO cells. Left panel, FAO cells were infected with the indicated adenoviruses and gene expression was determined by qRT-PCR. Right panel, FAO cells were infected with the indicated adenoviruses and the PGC-1α protein level was determined. (B) Exogenous RNF34 decreases the level of exogenous PGC-1α protein in brown fat cells. Brown fat preadipocytes were differentiated and then infected with PGC-1α and RNF34 or C329A mutant adenoviruses. The PGC-1α protein level was determined. (C) Differentiated brown fat adipocytes were infected with RNF34 or C329A mutant adenoviruses. Nuclear protein was isolated, and the endogenous level of PGC-1α protein was determined. (D) Differentiated brown fat adipocytes were infected with RNF34 adenoviruses. Cells were treated with cycloheximide (500 μM) for 0 or 30 min. Nuclear protein was then isolated, and the endogenous level of PGC-1α protein was determined. (E) Differentiated brown fat adipocytes were infected with RNF34 or C329A mutant adenoviruses. Gene expression was determined. Left panel, unstimulated cells. Right panel, cells were treated with β3-adrenergic receptor agonist CL-316243 (10 μM) for 6 h. Error bars represent standard errors of the means of the results determined in 3 samples. ∗, P < 0.05; ∗∗, P < 0.005; ∗∗∗, P < 0.0001. (F) Differentiated brown fat adipocytes were infected with RNF34 adenoviruses. Oxygen consumption was measured with a Clark-type electrode. Error bars represent standard errors of the means of the results determined in 2 experiments. ∗, P < 0.05.
We next determined the role of RNF34 in a more physiological setting. One of the predominant functions of PGC-1α is its control of brown fat metabolism. We previously generated immobilized brown fat cells that expressed a high level of PGC-1α and UCP1 after differentiation (18). We first validated the idea that the brown fat cells operate this RNF34 E3 ligase system by showing that ectopically expressed RNF34 promoted the degradation of ectopically expressed PGC-1α (Fig. 6B). We then examined whether overexpression of RNF34 has any effect on the endogenous PGC-1α. Endogenous PGC-1α protein from brown fat adipocytes sometimes, but not always, migrates as a doublet band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Figure 6C shows that overexpression of RNF34 in differentiated brown fat cells decreased the endogenous PGC-1α protein level and that the C329A mutant had no effects. When cells were treated with cycloheximide for 30 min, the turnover of preexisting PGC-1α protein was enhanced by RNF34 expression (Fig. 6D). Importantly, expression of RNF34 did not decrease the level of endogenous PGC-1α mRNA (Fig. 6E). A fraction of PGC-1α protein appeared to be resistant to RNF34-mediated degradation in these cells. We speculate that this pool of PGC-1α might be in a particular protein complex that renders PGC-1α inaccessible to RNF34; one such complex candidate is PGC-1α and twist-1 interaction. Nevertheless, the decrease of PGC-1α protein levels mediated by RNF34 led to downregulation of PGC-1α target genes and reduction of oxygen consumption (Fig. 6E and F). The gene most affected was UCP1. Our data are consistent with the observation that acute knockdown of PGC-1α decreases UCP1 expression in immobilized brown fat cells (18) (see Fig. S1 in the supplemental material). We note that Uldry et al. reported that UCP1 expression remains unchanged in immobilized brown fat cells generated from PGC-1α knockout mice (29); this may reflect a possible compensatory response that occurs in the preadipocytes in vivo or during the tedious in vitro cell purification process. Importantly, brown fat cell differentiation was not affected by RNF34 expression, as judged by Oil-Red O staining (see Fig. S2 in the supplemental material) and expression of general adipocyte marker PPARγ, adiponectin, aP2, and brown fat-specific markers PGC-1α and DIO2 (Fig. 6E), further confirming a direct effect of RNF34 on PGC-1α target gene expression. As expected, overexpression of RNF34 also suppressed the induction of UCP1 expression elicited by a β3-adrenergic receptor agonist (Fig. 6E). Taken together, these results show that ectopic expression of RNF34 in brown fat cells (in an E3 ligase activity-dependent manner) enhances endogenous PGC-1α protein turnover and hence negatively regulates its target gene expression.
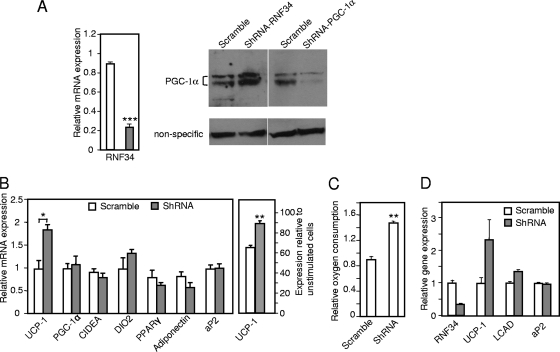
An important question is whether endogenous RNF34 is required for the regulation of brown fat cell function. We differentiated the immobilized brown fat cells and infected them with RNF34 shRNA adenoviruses. As in our overexpression experiments, administration of RNF34 knockdown viruses at a multiplicity of infection (MOI) of 2,000 at differentiation day 3 or 4 was sufficient for 100% infection efficiency, as indicated by GFP expression results (see Fig. S3 in the supplemental material). Knockdown of RNF34 increased the endogenous PGC-1α protein level without affecting its mRNA level (Fig. 7A and B). Accordingly, RNF34 knockdown cells showed higher level of both basal and β3-adrenergic agonist-induced UCP1 expression (Fig. 7B) and had a higher oxygen consumption rate (Fig. 7C) without affecting adipocyte differentiation (Fig. 7B; see also Fig. S2 in the supplemental material). We also knocked down RNF34 in primary brown fat cells and found that these cells showed similarly increased UCP1 expression (Fig. 7D). These results suggest that endogenous RNF34 plays an indispensable role in regulation of brown fat cell metabolism.
Fig 7.
Endogenous RNF34 is required for the regulation of brown fat cell function. (A) Differentiated brown fat adipocytes were infected with RNF34 shRNA adenoviruses. Left panel, the endogenous level of RNF34 mRNA was determined. ∗∗∗, P < 0.0001. The right panel shows endogenous PGC-1α protein levels in RNF34 knockdown cells and in PGC-1α knockdown cells from the same SDS-PAGE gel. Open bar, scramble; closed bar, shRNA. (B) Differentiated brown fat adipocytes were infected with RNF34 shRNA adenoviruses. Gene expression was determined. Left panel, unstimulated cells. Right panel, cells were treated with β3-adrenergic receptor agonist CL-316243 (10 μM) for 6 h. Error bars represent standard errors of the means of the results determined in 3 samples. ∗, P < 0.05, ∗∗, P < 0.005. (C) Differentiated brown fat adipocytes were infected with RNF34 shRNA adenoviruses. Oxygen consumption was measured with a Clark-type electrode. Error bars represent standard errors of the means of the results determined in 3 experiments. ∗∗, P < 0.005. (D) Primary brown fat cells were differentiated and then infected with RNF34 shRNA adenoviruses. Gene expression was determined. Error bars represent standard errors of the means of the results determined in 2 samples.
Regulation of RNF34 expression in brown fat cells.
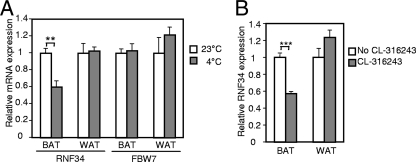
Given the modulation of PGC-1α expression by temperature (20) and our observations regarding the role of RNF34 in brown fat cell metabolism, we examined whether cold exposure affects RNF34 expression. We found that, after exposure for 5 h at 4°C, RNF34 expression in the brown fat cells was significantly decreased (Fig. 8), whereas PGC-1α expression was strongly induced (20). Interestingly, Fbw7 expression remained unchanged (Fig. 8). Moreover, suppression of RNF34 expression by cold exposure did not occur in the white fat cells. Cold sympathetically activates β3-adrenergic receptor signaling in the brown fat cells. We therefore treated wild-type mice with the β3-adrenergic receptor agonist for 6 days at room temperature. This treatment similarly reduced RNF34 expression in the brown fat cells but not in the white fat cells (Fig. 8), suggesting that suppression of RNF34 expression by cold exposure is mediated by β3-adrenergic receptor pathway. The data further establish the physiological relevance of RNF34 in regulation of PGC-1α function and indicate a negative role of RNF34 in adaptive thermogenesis.
Fig 8.
Regulation of RNF34 expression in the brown fat cells. (A) Wild-type mice were maintained at 4°C for 5 h, and RNF34 expression and Fbw7 expression were compared with the expression results for wild-type mice maintained at room temperature (n = 5 per group). Error bars represent standard errors of the means. ∗∗, P < 0.005. (B) Wild-type mice were i.p. injected with β3-adrenergic receptor agonist CL-316243 (0.5 mg/kg body weight) daily for 6 days, and RNF34 expression was compared with that seen with untreated wild-type mice (n = 8 or 9 mice per group). Error bars represent standard errors of the means. ∗∗∗, P < 0.0001.
DISCUSSION
As a central regulator in energy metabolism, PGC-1α is modulated by a plethora of mechanisms. Among them is posttranslational modification. Phosphorylation and acetylation/deacetylation of PGC-1α and their functional impacts on systematic metabolism have been well documented (6, 23). Here, using an unbiased screening technique, we identified RNF34 as an E3 ubiquitin ligase that ubiquitinates PGC-1α protein and controls its turnover. Importantly, through both ectopic expression and knockdown experiments, we showed that RNF34 regulates the endogenous level of PGC-1α protein, PGC-1α target gene expression, and cellular respiration rate in brown fat cells. These effects are not associated with any change of PGC-1α mRNA expression and are not observed when a ligase-defective point mutant of RNF34 is expressed. Thus, our results demonstrate a critical role of this RNF34-mediated modification of PGC-1α in energy expenditure.
Besides RNF34, the SCFFbw7 complex is another E3 ubiquitin ligase for PGC-1α (16). The distinctions between the two ligases are evident. First, the interaction with and degradation of PGC-1α by Fbw7 require phosphorylation of four residues located in the N terminus of PGC-1α, first by p38 MAPK-mediated priming phosphorylation and second by Gsk3β-mediated phosphorylation (16). Interestingly, p38 MAPK, by phosphorylating three of the four residues, was shown instead to stabilize PGC-1α (19). Moreover, this phosphodegron recognition motif is conserved among PGC-1 members, indicating that Fbw7 might be an E3 ligase for PGC-1β as well (16). On the other hand, RNF34 targets the C-terminal half of PGC-1α for degradation, independently of the phosphorylation of these four residues, and RNF34 does not regulate PGC-1β turnover. Second, Fbw7 is a haploinsufficient tumor suppressor that targets multiple substrates for degradation, including the proto-oncoproteins Myc, Jun, Notch, and cyclin E and the lipogenic regulator SREBP (32). Third, in contrast to RNF34, Fbw7 expression in the brown fat cells is not regulated by environmental temperature or by β3-adrenergic receptor signaling. Fourth, the functional relevance of Fbw7-mediated ubiquitination of PGC-1α in PGC-1α target gene expression and energy metabolism is unknown. Liver-specific Fbw7 knockout mice display massive hepatic deposition of triglycerides and increased levels of SREBP protein but no increase of PGC-1α protein levels (17). Thus, the metabolic axis for Fbw7 in the liver appears to be the regulation of lipogenesis through SREBP. We found that overexpression of Fbw7 in brown fat cells slightly decreases the endogenous PGC-1α protein level (see Fig. S4 in the supplemental material). Whether Fbw7 is involved in brown fat metabolism remains to be determined.
An intriguing observation is that cold exposure suppresses RNF34 expression in the brown fat cells. This suppression is mediated by β3-adrenergic receptor signaling. Among numerous regulators or modifiers of PGC-1α, RNF34 is the only one reported to date to be regulated by environmental temperature. The results indicate that ubiquitination of PGC-1α by RNF34 is likely to be coordinated with environmental cues and strongly suggest an in vivo physiological function of RNF34 in thermogenesis. This expression pattern presents a striking contrast with that of PGC-1α, whose transcription is induced by the presence of either cold exposure or β3-adrenergic signaling. Thus, in the cold, PGC-1α activity is maximized not only by induction of its mRNA but also by possible stabilization of its protein, ensuring sufficient heat production to maintain body temperature. Currently, the detailed mechanism underlying the temperature modulation of RNF34 expression is unclear. The modulation might not be cell-autonomous, as treatment of brown fat cells in vitro with the β3-adrenergic receptor agonist has no effect on RNF34 expression (our unpublished data). Interestingly, Cidea, a lipid droplet protein that also negatively regulates brown fat metabolism (4, 21, 34), displays a similar pattern of regulation. Cidea expression in the brown fat cells is suppressed by cold exposure through β3-adrenergic signaling (27) but not in brown fat cells in vitro (our unpublished data).
Phosphorylation and acetylation/deacetylation of PGC-1α are critical for PGC-1α-regulated energy metabolism in liver and skeletal muscle. Here we identify ubiquitination of PGC-1α by RNF34 as a regulatory module operating in brown fat cells. It should be interesting to investigate whether this RNF34-mediated ubiquitination coordinates with and/or affects the dynamics of other modifications of PGC-1α. Although our studies were performed using isolated brown fat cells, the regulation of RNF34 expression by environmental temperature strongly indicates a potential role of this E3 ligase in brown fat metabolism in vivo, which is planned to be fully investigated using tissue-specific RNF34 knockout mice in the future. Given the inverse correlation between brown fat activity and human obesity, inhibition of RNF34 ligase activity might be useful to promote energy expenditure and counteract obesity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Keiji Tanaka for the ubiquitin constructs and Jenny Bennanti for comments on the manuscript.
These studies were funded by grants from American Diabetes Association and NIH/NIDDK (R01DK076118).
Footnotes
Published ahead of print 7 November 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Cantó C, et al. 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458:1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cantó C, et al. 2010. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 11:213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cardozo T, Pagano M. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5:739–751 [DOI] [PubMed] [Google Scholar]
- 4. Christianson JL, Boutet E, Puri V, Chawla A, Czech MP. 2010. Identification of the lipid droplet targeting domain of the Cidea protein. J. Lipid Res. 51:3455–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cypess AM, et al. 2009. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fernandez-Marcos PJ, Auwerx J. 2011. Regulation of PGC-1{alpha}, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 93:884S–90S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerhart-Hines Z, et al. 2007. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 26:1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He TC, et al. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 95:2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jäger S, Handschin C, St.-Pierre J, Spiegelman BM. 2007. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. U. S. A. 104:12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klein J, et al. 1999. Beta(3)-adrenergic stimulation differentially inhibits insulin signaling and decreases insulin-induced glucose uptake in brown adipocytes. J. Biol. Chem. 274:34795–34802 [DOI] [PubMed] [Google Scholar]
- 11. Lerin C, et al. 2006. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 3:429–438 [DOI] [PubMed] [Google Scholar]
- 12. Li X, Monks B, Ge Q, Birnbaum MJ. 2007. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature 447:1012–1016 [DOI] [PubMed] [Google Scholar]
- 13. Lin J, Handschin C, Spiegelman BM. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 1:361–370 [DOI] [PubMed] [Google Scholar]
- 14. Lin J, et al. 2004. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell 119:121–135 [DOI] [PubMed] [Google Scholar]
- 15. Nedergaard J, Bengtsson T, Cannon B. 2007. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 293:E444–452 [DOI] [PubMed] [Google Scholar]
- 16. Olson BL, et al. 2008. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 22:252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Onoyama I, et al. 2011. Fbxw7 regulates lipid metabolism and cell fate decisions in the mouse liver. J. Clin. Invest. 121:342–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pan D, Fujimoto M, Lopes A, Wang YX. 2009. Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell 137:73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puigserver P, et al. 2001. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol. Cell 8:971–982 [DOI] [PubMed] [Google Scholar]
- 20. Puigserver P, et al. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829–839 [DOI] [PubMed] [Google Scholar]
- 21. Puri V, et al. 2008. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc. Natl. Acad. Sci. U. S. A. 105:7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodgers JT, Haas W, Gygi SP, Puigserver P. 2010. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab. 11:23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. 2008. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 582:46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodgers JT, et al. 2005. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434:113–118 [DOI] [PubMed] [Google Scholar]
- 25. Schreiber E, Matthias P, Muller MM, Schaffner W. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seale P, Lazar MA. 2009. Brown fat in humans: turning up the heat on obesity. Diabetes 58:1482–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimizu T, Yokotani K. 2009. Acute cold exposure-induced down-regulation of CIDEA, cell death-inducing DNA fragmentation factor-alpha-like effector A, in rat interscapular brown adipose tissue by sympathetically activated beta3-adrenoreceptors. Biochem. Biophys. Res. Commun. 387:294–299 [DOI] [PubMed] [Google Scholar]
- 28. Southgate RJ, et al. 2005. PGC-1alpha gene expression is down-regulated by Akt-mediated phosphorylation and nuclear exclusion of FoxO1 in insulin-stimulated skeletal muscle. FASEB J. 19:2072–2074 [DOI] [PubMed] [Google Scholar]
- 29. Uldry M, et al. 2006. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 3:333–341 [DOI] [PubMed] [Google Scholar]
- 30. van Marken Lichtenbelt WD, et al. 2009. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360:1500–1508 [DOI] [PubMed] [Google Scholar]
- 31. Virtanen KA, et al. 2009. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360:1518–1525 [DOI] [PubMed] [Google Scholar]
- 32. Welcker M, Clurman BE. 2008. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8:83–93 [DOI] [PubMed] [Google Scholar]
- 33. Yoon JC, et al. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131–138 [DOI] [PubMed] [Google Scholar]
- 34. Zhou Z, et al. 2003. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat. Genet. 35:49–56 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.