Abstract
Telomeres are coated by shelterin, a six-subunit complex that is required for protection and replication of chromosome ends. The central subunit TIN2, with binding sites to three subunits (TRF1, TRF2, and TPP1), is essential for stability and function of the complex. Here we show that TIN2 stability is regulated by the E3 ligase Siah2. We demonstrate that TIN2 binds to Siah2 and is ubiquitylated in vivo. We show using purified proteins that Siah2 acts as an E3 ligase to directly ubiquitylate TIN2 in vitro. Depletion of Siah2 led to stabilization of TIN2 protein, indicating that Siah2 regulates TIN2 protein levels in vivo. Overexpression of Siah2 in human cells led to loss of TIN2 at telomeres that was dependent on the presence of the catalytic RING domain of Siah2. In contrast to RNAi-mediated depletion of TIN2 that led to loss of TRF1 and TRF2 at telomeres, Siah2-mediated depletion of TIN2 allowed TRF1 and TRF2 to remain on telomeres, indicating a different fate for shelterin subunits when TIN2 is depleted posttranslationally. TPP1 was lost from telomeres, although its protein level was not reduced. We speculate that Siah2-mediated removal of TIN2 may allow dynamic remodeling of the shelterin complex and its associated factors during the cell cycle.
INTRODUCTION
Telomere integrity is essential for chromosome stability. Telomeres are composed of duplex TTAGGG repeats that end as a 3′ single-stranded overhang and shelterin, a six-subunit complex that is required for the protection and replication of chromosome ends (41). Shelterin localizes to telomeres via the duplex repeat binding proteins TRF1 and TRF2, related proteins containing a C-terminal myb DNA-binding domain and an internal TRFH (TRF homology) homodimerization domain (7–9, 15, 19). TRF1 and TRF2 do not interact directly but are linked by a common binding partner, TIN2 (24, 31, 32, 37, 53). TIN2 binds to a third shelterin subunit, TPP1 (24, 38, 54), that in turn recruits the 3′ single-stranded overhang binding POT1 protein (5, 39). The sixth subunit, RAP1, binds to TRF2 (34). Although the shelterin complex can exist as a unit, individual subunits have distinct functions and (in some cases) unique mechanisms to regulate their stability on telomeres.
The six shelterin subunits are abundant at telomeres and localize there throughout the cell cycle. In contrast, there are a number of shelterin accessory proteins that are much less abundant at telomeres and may associate only transiently to modulate telomere function (41). Most of the accessory factors that have been identified thus far bind to TRF1 or TRF2. The TRFH domain serves as a docking site for target proteins containing FXLXP motifs (for binding TRF1) and YXLXP motifs (for binding TRF2) (14). TIN2 binds to the TRFH domain in TRF1 with its C-terminal FXLXP motif, whereas it binds to the hinge domain in TRF2 with its N-terminal domain. The shelterin accessory factor, the telomerase inhibitor PinX1 (56), uses an FXLXP motif to bind the TRFH domain in TRF1 in the same fashion as TIN2 (14). TRF1 also has partners such as the poly(ADP-ribose) polymerase tankyrase 1 that binds to its unique aminoterminal domain (45).
TRF1 serves as a negative regulator of telomere length by inhibiting access of telomerase to chromosome ends (2, 48). Overexpression of TRF1 leads to telomere shortening, whereas loss of TRF1 from telomeres leads to telomere elongation (48). Posttranslational modification of TRF1 can modulate telomere length by regulating TRF1 stability and localization to telomeres. For example, PARsylation of TRF1 by overexpression of its tankyrase 1 accessory factor in the nucleus leads to TRF1 eviction from telomeres (44) and telomere elongation by telomerase (16). Non-telomere-bound TRF1 is ubiquitylated and degraded by the proteasome (13). TRF1's shelterin binding partner TIN2 appears to protect it from degradation; depletion of TIN2 by small interfering RNA (siRNA) leads to loss of TRF1 from telomeres (52) and degradation of TRF1 by the proteasome (11). Other posttranslational modifications to TRF1 such as phosphorylation (30) and deubiquitylation (3) have been shown to influence TRF1 stability via degradation by the proteasome.
The proteasome degrades proteins in a manner dependent on its recognition of a polyubiquitin signal. The ubiquitin-proteasome degradation pathway consists of a series of enzymatic reactions involving an ubiquitin-activating protein (E1), an ubiquitin-conjugating protein (E2); and an ubiquitin ligase (E3) (22). The E3 ligase confers target specificity; it associates with the E2 and the substrate to catalyze transfer of ubiquitin. The E3s, the largest and most complex of the enzymes in the pathway, determine the specificity of ubiquitylation. The members of a major class of structurally distinct E3 ligases possess an E2-binding catalytic RING domain (18). RING domain E3s come in the form of single polypeptides such as Siah (a designation derived from “seven in absentia homolog”) (28), where the RING and substrate binding domains reside in the same protein, or in the form of multiprotein complexes such as SCF (Skp1-Cul1-F box protein-Rbx1) (12), where the RING and substrate-binding domains reside in different proteins, the Rbx1 and F box proteins, respectively.
Two E3 ligases have been previously described that mediate ubiquitylation and degradation of TRF1: RLIM, a RING H2 zinc finger E3 ligase that binds at a position adjacent to the myb domain in TRF1 (21) and localizes to the nucleus (21, 29), and the SCF ubiquitin E3 ligase Fbx4, which binds to the TRFH domain in TRF1 (33) and localizes to the cytoplasm (36). Fbx4 binds to TRF1 by the use of an atypical small GTPase domain (55). Comparison of the crystal structures of the TRF1-Fbx4 complex with the TRF1-TIN2 complex indicated that TIN2 binding to TRF1 could interfere with Fbx4 binding to TRF1 (55). In support of this idea, RNA interference (RNAi)-mediated depletion of TIN2 led to a reduction in TRF1 protein levels in a manner dependent on Fbx4 (55). Taken together, the results of those studies indicate that TIN2 binding to TRF1 prevents its ubiquitylation and degradation by Fbx4.
But the question remains: how is TIN2 itself regulated? Is association of TIN2 with TRF1 and telomeres subject to posttranslational regulation? Here we report the identification of the E3 ligase Siah as a TIN2 binding partner. Siah has been implicated in a range of cellular processes, including DNA damage, cell proliferation, hypoxia, and tumorigenesis (20, 40, 51). In human cells, Siah exists in two closely related isoforms (Siah 1 and Siah 2) that interact with a diverse group of substrates to promote their degradation. We show here that Siah2 directly ubiquitylates TIN2 in vitro and regulates TIN2 protein levels in vivo. Posttranslational degradation of TIN2 by Siah2 leaves TRF1 (and TRF2) protein levels and localization to telomeres intact. We speculate that Siah2-induced removal of TIN2 could be a mechanism to expose binding sites at telomeres to shelterin accessory factors, such as PinX1, that bind to the same site on TRF1 as TIN2.
MATERIALS AND METHODS
Yeast two-hybrid screening and assays.
A two-hybrid screening was performed using the Saccharomyces cerevisiae L40 reporter strain (23), a HeLa cell or human fetal liver cDNA library (Clontech), and LexA-TIN2-C bait plasmid (pBTTIN2-C; TIN2 amino acids [aa] 180 to 354) or LexA-TIN2N (pBTTIN2-N; TIN2 aa 1 to 195) cloned into pBTM116 vector (4) according to Clontech Matchmaker protocol. TIN2-C.K280X and TIN2-C.Q298Rfs were generated as described previously (10). Siah1 (aa 148 to 282), Siah2 (aa 126 to 324), and full-length TRF1 and TPP1 were cloned into pACT2 (Clontech). Two-hybrid interactions were scored according to the number of minutes required for the color change as follows: +++, 15 min; ++, 30 min; +, 45 min. Interactions were confirmed by two or more independent experiments.
Plasmids.
FlagTIN2 contained full-length TIN2 cloned into 3× Flag–cytomegalovirus 10 (CMV-10) (Sigma). The Flag TIN2-K280X mutation was generated as described previously (10). MycSiah2 contained full-length Siah2 from pcDNA3-Siah2 (kindly provided by Eric Fearon) (28) cloned into the EcoRI and XhoI sites in p3xMyc-2A (Sigma). MycΔNSiah2 contained aa 126 to 324 of Siah2 cloned into the EcoRI site of p3xMyc-2A. Glutathione S-transferase–TIN2 (GST-TIN2) contained full-length TIN2 cloned into the EcoRI and SalI sites of pGEX-5x-1. Sumo-Siah2 contained full-length Siah2 cloned into the BamHI and XhoI sites of pET28b-Sumo-6×His vector (kindly provided by Ming Lei) (14).
Cell lines.
HeLaI.2.11 cells are a HeLa-derived clonal cell line (49). Supertelomerase HeLa cells were generated by transduction with retroviral vectors encoding hTR and hTERT (kindly provided by Joachim Lingner) (17).
Ubiquitylation reaction in vitro.
In vitro ubiquitylation reactions were carried out in ubiquitination buffer (50 mM Tris [pH 7.4], 5 mM MgCl2, 2 mM dithiothreitol [DTT]) with 100 ng of human recombinant E1 (Boston Biochem), 200 ng of human recombinant E2 UbcH5b (Boston Biochem), and 10 μg of hemagglutinin-ubiquitin (HA-ubiquitin) (Boston Biochem). Sumo-Siah2 and GST-TIN2 were expressed and purified from Escherichia coli BL21 cells according to standard protocols. Sumo-6×His-Siah2 (250 ng) and GST-TIN2 (200 ng) were used in 25-μl ubiquitylation reaction mixtures incubated at 30°C for 2 h. Reactions were terminated by the addition of 4× sodium dodecyl sulfate (SDS) loading buffer and processed for immunoblot analysis as described below.
RNA isolation and RT-PCR.
RNA was isolated from HelaI.2.11 cells transfected for 48 h with the siSIAH2 or siGFP siRNAs by the use of an RNeasy kit (Qiagen) with DNase I for column digestion (Qiagen) according to the manufacturer's specifications. RNA (500 ng) was converted to cDNA by the use of an oligo(dT) primer and a SuperScript III first-strand synthesis system for reverse transcriptase PCR (RT-PCR) (Invitrogen) according to the manufacturer's specifications. cDNA (3 μl) from the RT reaction mixture was amplified by PCR using primer pair 5′-TTTCCCTGTAAGTATGCCACCAC (forward) and 5′-GTTCCCATTCAACTCCAGTCTG (reverse) for SIAH2 and primer pair 5′-ACAACAGCCTCAAGATCATCAG (forward) and 5′-GGTCCACCACTGACACGTTG (reverse) for GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
siRNA transfection.
siRNA transfections were performed for 48 h with Oligofectamine (Invitrogen) according to the manufacturer's protocol. The final concentration of siRNA was 100 nM. The following siRNAs (synthesized by Dharmacon Research Inc.) were used: Siah2 (5′-CCAAUGCCGCCAGAAGUUG-3′) and GFP Duplex I.
Plasmid transfection.
Cells were transfected for 16 h with FlagTIN2, MycSiah2, HA-ubiquitin (pMT123; kindly provided by Dirk Bowmann) (47), FlagTIN2.K280X, MycSiah2ΔN, or 3× Myc-2A vector (Sigma), and Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. For coimmunoprecipitation experiments, MG132 (10 μm) was added 4 h prior to harvest.
Immunoprecipitation.
Cells were lysed in 1 ml (per 15-cm-diameter dish) of buffer C (20 mM HEPES [pH 7.9], 0.2% Nonidet P-40, 0.42 M KCl, 0.1 mM EDTA, 1 mM DTT, 5 mM MgCl2, 25% glycerol, protease inhibitor cocktail [Sigma]) on ice for 1 h and then pelleted at 8,000 × g for 10 min. Supernatants were precleared with protein G-Sepharose (GE Healthcare) rotating at 4°C for 30 min. Nonspecific antibody complexes and protein aggregates were removed by centrifugation, and the supernatant was used for immunoprecipitation analysis or fractionated directly using SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (indicated as input [approximately 0.5% of the amount used in the immunoprecipitation]). Supernatants were incubated with 25 μl of mouse anti-Flag-agarose (Sigma) or 25 μl of anti-Myc-agarose (Sigma) at 4°C with rocking for 3 h. Immunocomplexes were then washed three times with 1.0 ml of buffer D (20 mM HEPES [pH 7.9], 0.1 M KCl, 0.2 mM EDTA, 1 mM DTT, 5 mM MgCl2, 25% glycerol), suspended in sample buffer, fractionated by SDS-PAGE, and processed for immunoblotting as described below.
Whole-cell extracts.
Cells were resuspended in four volumes of buffer C (20 mM HEPES-KOH [pH 7.9], 420 mM KCl, 25% glycerol, 0.1 mm EDTA, 5 mM MgCl2, 0.2% NP-40, 1 mM dithiothreitol, and 2.5% protease inhibitor cocktail [Sigma]) and incubated for 1 h on ice. Suspensions were pelleted at 8,000 × g for 10 min. Supernatant proteins (25 μg, as determined by Bio-Rad protein assay) were fractionated by SDS-PAGE and analyzed by immunoblotting.
Immunoblot analysis.
Immunoblots were incubated with the following primary antibodies: rabbit anti-TIN2 701 (24) (0.5 μg/ml); mouse anti-TRF2 (Imgenex) (2.0 μg/ml); rabbit anti-TPP1 911 (24) (0.2 μg/ml); rabbit anti-Flag (Sigma) (1.0 μg/ml); rabbit anti-HA (Abcam) (0.5 μg/ml); rabbit anti-Myc (Santa Cruz Biotechnologies) (0.8 μg/ml); and mouse anti-α-tubulin ascitic fluid (Sigma) (1:500,000) followed by horseradish peroxidase-conjugated donkey anti-rabbit or anti-mouse IgG (Amersham) (1:2,500). Bound antibody was detected with Super Signal West Pico (Thermo Scientific).
Indirect immunofluorescence.
Cells were processed for indirect immunofluorescence as described previously (16). Briefly, cells were fixed in 2% paraformaldehyde–phosphate-buffered saline (PBS), permeabilized in 0.5% NP-40–PBS, and blocked in 1% bovine serum albumin (BSA)–PBS. Cells were incubated with goat anti-myc (Bethyl) (4.0 μg/ml), rabbit anti-TIN2 701 (0.5 μg/ml), mouse anti-TRF1 serum (1:2,000), mouse anti-TPP1 1146 serum (raised against a peptide containing TPP1 aa 466 to 496) (1:200), or mouse anti-TRF2 (Imgenex) (0.1 μg/ml). Primary antibodies were detected with fluorescein isothiocyanate-, tetramethyl rhodamine isothiocyanate-, or aminomethylcoumarin acetate-conjugated donkey anti-goat, anti-rabbit, or anti-mouse antibodies (Jackson Laboratories) (1:100). DNA was stained with 4′,6-diamino-2-phenylindole (DAPI) (0.2 μg/ml).
Image acquisition.
Images were acquired using a microscope (Axioplan 2; Carl Zeiss, Inc.) with a Plan Apochrome 63× numerical aperture (NA) 1.4 oil immersion lens (Carl Zeiss, Inc.) and a digital camera (catalog no. C4742-95; Hamamatsu Photonics). Images were acquired and processed using Openlab software (Perkin Elmer).
RESULTS
Siah binds TIN2 and directly ubiquitylates TIN2 in vitro.
We identified human Siah1 and Siah2 in three independent yeast two-hybrid screenings using human TIN2 as the bait. A fetal liver cDNA library screening using the C-terminal half of TIN2 (TIN2-C) yielded a Siah2 isolate comprising its C-terminal substrate-binding domain (aa 126 to 324) (Fig. 1A). A HeLa cDNA library screening with TIN2-C identified a Siah1 isolate comprising its C-terminal substrate-binding domain (aa 148 to 282) (Fig. 1A). Finally, the same Siah1 isolate was identified in a HeLa cDNA library screening using the N-terminal half of TIN2 (TIN2-N) as bait (Fig. 1A). Together, the data indicated that TIN2 binds to Siah1 and to Siah2 by the use of its N- or C-terminal domain. Siah1 and Siah2 are highly conserved; they share 90% identity in the substrate binding domain (which binds TIN2) and function equivalently to induce degradation of a number of substrates. We focused on one isoform (Siah2) for the remaining experiments.
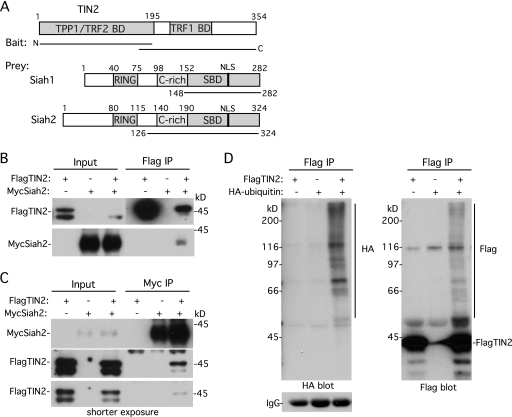
Fig 1.
TIN2 binds Siah and is ubiquitylated in vivo. (A) Schematic representation of TIN2, with the TPP1, TRF2, and TRF1 binding domains (BD) indicated and Siah1 and Siah2 and the RING domain, cysteine-rich (C-rich) domain, substrate-binding domain (SBD), and nuclear localization signal (NLS) indicated. The lines indicate the TIN2 baits and Siah prey. (B and C) TIN2 binds Siah2 in human cells. HeLaI.2.11 cells were transfected with FlagTIN2 and/or MycSiah2. Cell lysates were immunoprecipitated (IP) with anti-Flag (B) or anti-Myc (C) beads and analyzed by immunoblotting with anti-Myc or anti-Flag antibody. (D) TIN2 is ubiquitylated in vivo. HeLaI.2.11 cells were transfected for 16 h with FlagTIN2 and/or HA-ubiquitin. Cell lysates were immunoprecipitated with anti-Flag beads and analyzed by immunoblotting with anti-HA antibody (left panel) or anti-Flag antibody (right panel).
To determine whether full-length TIN2 interacted with full-length Siah2 in human cells, HeLa cells were cotransfected with Flag epitope-tagged TIN2 and Myc epitope-tagged Siah2. As shown in Fig. 1B, MycSiah2 coimmunoprecipitated with FlagTIN2. Conversely, FlagTIN2 coimmunoprecipitated with MycSiah2 (Fig. 1C). Siah2 is an E3 ligase. Its N terminus (containing the catalytic RING domain) binds to the E2 protein, whereas its C terminus binds to the substrate. Interaction (of a number of proteins) with the C-terminal domain of Siah leads to their ubiquitylation and degradation. Indeed, we note that cotransfection of MycSiah2 with FlagTIN2 led to a reduction in FlagTIN2 (see the Input lanes in Fig. 1B and C [shorter exposure]).
We next asked whether TIN2 was ubiquitylated in vivo. HeLa cells were cotransfected with FlagTIN2 and HA epitope-tagged ubiquitin. FlagTIN2 was immunoprecipitated and analyzed by immunoblotting with anti-HA antibody. As shown in Fig. 1D (left panel), a ladder of HA-ubiquitylated TIN2 was detected. Probing the blot with anti-Flag antibody yielded the same ladder of proteins (Fig. 1D, right panel), confirming that they correspond to ubiquitylated FlagTIN2.
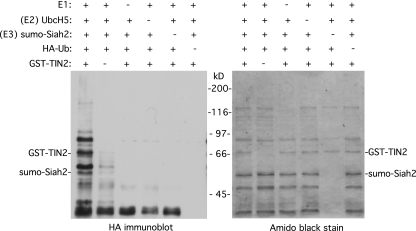
Together, the data described above indicate that TIN2 is ubiquitylated in vivo and suggest Siah2 as a potential E3 ligase. We thus performed the ubiquitylation reaction in vitro with purified components to ask whether Siah2 could directly ubiquitylate TIN2. Recombinant GST-tagged TIN2 and Sumo-tagged Siah2 proteins were purified from E. coli. The in vitro ubiquitylation reaction mixture contained the five following reagents: the substrate GST-TIN2, the E3 ligase sumo-Siah2, purified E1, purified E2 (UbcH5), and HA-ubiquitin. Following incubation, the reaction products were visualized by immunoblotting with anti-HA antibody. As shown in Fig. 2, left panel, a ladder of HA-ubiquitylated TIN2 was detected. Importantly, ubiquitylation depended on each of the five reagents; omission of any one of the five led to loss of the HA-ubiquitylated TIN2 products. These data indicate that Siah2 is an E3 ligase that directly ubiquitylates TIN2.
Fig 2.
Siah2 directly ubiquitylates TIN2 in vitro. Reaction mixtures containing the indicated combinations of E1, E2 (UbcH5), E3 (Sumo-Siah2), GST-TIN2, and HA-ubiquitin were incubated, fractionated on SDS-PAGE, and transferred to nitrocellulose. Proteins were visualized by staining with Amido black (right panel), and ubiquitylated proteins were detected by probing with anti-HA antibody (left panel).
TIN2 has at least two binding sites for Siah.
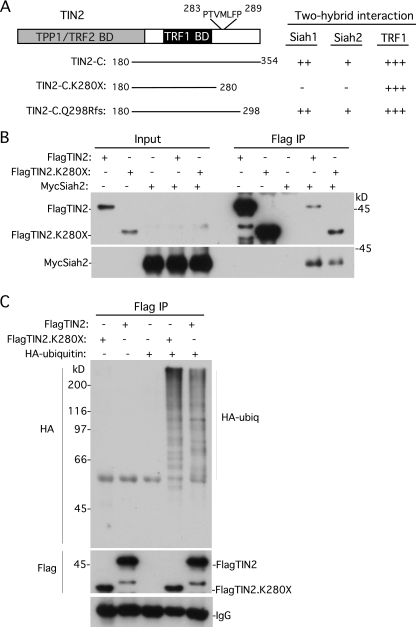
A consensus motif composed of PXAXVXP has been identified in many (but not all) Siah-binding proteins (25, 26). Inspection of the TIN2 amino acid sequence revealed no exact matches (and not even 3 out of 4 matches) to the consensus sequence. However, we did detect many sites throughout the protein that contained 2 out of 4 matches to the consensus sequence. Of particular interest was the most C-terminal (potential) site at amino acid position 283 in TIN2 comprising PTVMLFP, since it falls within the cluster of amino acids that are mutated in TIN2-associated dyskeratosis congenita (42, 50), an inherited bone marrow failure syndrome that is caused by defects in telomere maintenance (6). We showed recently that the PTVML sequence at position 283 constituted a binding site for heterochromatin protein 1 (10). To determine whether this C-terminal domain was required for TIN2 binding to Siah, we generated a C-terminal deletion allele of TIN2-C that was truncated at position 280 (TIN2-C.K280X). As shown in Fig. 3A, truncation at amino acid 280 abrogated binding to Siah1 and to Siah2. In contrast, a TIN2-C allele truncated at amino acid 298 (TIN2-C.Q298Rfs; generated by a frameshift at position Q298) retained Siah binding. These data suggest the PTVMLFP motif as a potential Siah-binding site in the C-terminal domain of TIN2.
Fig 3.
Two-hybrid analysis identified a C-terminal Siah-binding site in TIN2, but it is not essential for Siah2 binding and TIN2 ubiquitylation in vivo. (A) Schematic representation of TIN2 and the indicated TIN2-C alleles. The potential Siah-binding motif PTVMLFP is indicated. Two-hybrid interactions were scored according to the number of minutes required for the color change: +++, 15 min; ++, 30 min; +, 45 min; −, no color change. (B) Full-length TIN2 lacking the C-terminal Siah2-binding site binds to Siah2 by coimmunoprecipitation. HeLaI.2.11 cells were transfected with FlagTIN2, FlagTIN2.K280X, and/or MycSiah2. Cell lysates were immunoprecipitated with anti-Flag beads and analyzed by immunoblotting with anti-Flag or anti-Myc antibody. (C) TIN2 lacking the C-terminal Siah-binding site is ubiquitylated in vivo. HeLaI.2.11 cells were transfected with FlagTIN2, FlagTIN2.K280X, and/or HA-ubiquitin. Cell lysates were immunoprecipitated with anti-Flag beads and analyzed by immunoblotting with anti-HA or anti-Flag antibody.
To determine the importance of this Siah binding site in the context of full-length TIN2, we generated the K280X C-terminal deletion in the full-length FlagTIN2 allele to create FlagTIN2.K280X. FlagTIN2 or FlagTIN2.K280X was cotransfected with MycSiah2 into HeLa cells. As shown in Fig. 3B, FlagTIN2.K280X (like FlagTIN2) coimmunoprecipitated MycSiah2, although in a slightly reduced amount compared to full-length FlagTIN2. Hence, TIN2 binds Siah2 despite deletion of its C-terminal Siah-binding site. These data are consistent with the two-hybrid screening results showing that TIN2 has multiple binding sites for Siah.
To determine whether deletion of the C-terminal Siah binding site in TIN2 affected ubiquitylation in vivo, FlagTIN2 or FlagTIN2.K280X was cotransfected with HA-ubiquitin into HeLa cells. FlagTIN2 or FlagTIN2.K280X was immunoprecipitated and analyzed by immunoblotting with anti-HA antibody. As shown in Fig. 3C, a ladder of HA-ubiquitylated FlagTIN2.K280X (similar in intensity to that of full-length FlagTIN2) was detected, indicating that the N-terminal Siah binding site is sufficient for ubiquitylation of TIN2 in vivo. It is possible that the N- and C-terminal Siah-binding sites of TIN2 play redundant roles in TIN2 binding and ubiquitylation.
Together, our data show that TIN2 contains at least two binding sites for Siah: a C-terminal binding site between amino acids 280 and 298 and one (or more) uncharacterized site in the N-terminal domain that is sufficient for ubiquitylation in vivo. Inspection of aa 1 to 280 in TIN2 revealed 13 weak sites (containing only 2 out of 4 matches to the PXAXVXP consensus sequence) that could potentially bind Siah. Alternatively, TIN2 may contain a noncanonical Siah binding site in its N-terminal domain.
Depletion of Siah2 leads to stabilization of TIN2 in vivo.
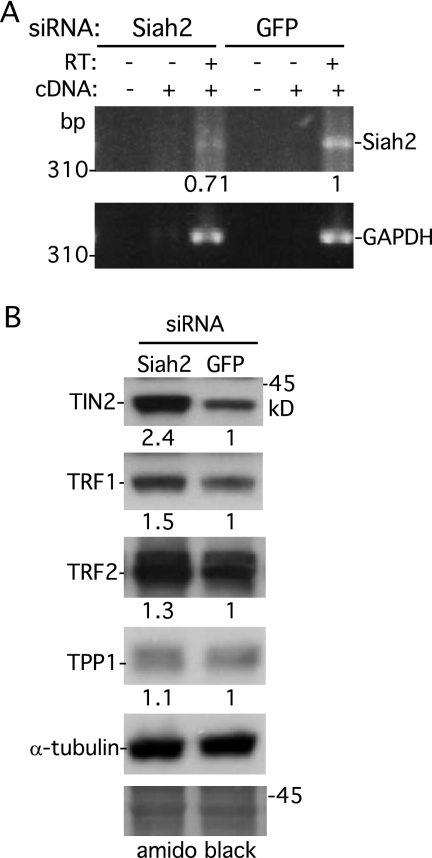
We next asked whether depletion of Siah2 influenced TIN2 stability in vivo. Siah2 was depleted using siRNA and analyzed by RT-PCR. RNA was isolated from HeLa cells following transfection for 48 h with GFP or Siah2 siRNA and used to generate a cDNA fragment by RT-PCR (in the presence or absence of RT) with primers specific to Siah2 or GAPDH (as a control). As shown in Fig. 4A, transfection with Siah2 siRNA led to a reduction in the level of Siah2 (but not GAPDH) cDNA. To determine the effect of Siah2 depletion on TIN2 protein levels, cell extracts were subjected to immunoblot analysis. As shown in Fig. 4B, depletion of Siah2 led to stabilization of TIN2, which is consistent with the notion that Siah2 is an E3 ligase that promotes ubiquitylation and degradation of TIN2 in vivo. We noted a slight stabilization of TRF1 and TRF2 but no effect on TPP1 protein levels. Our observation that depletion of Siah2 leads to stabilization of TIN2 suggests that Siah1 was not compensatory with respect to Siah2 function in these experiments.
Fig 4.
Depletion of Siah2 leads to stabilization of TIN2. (A) RT-PCR analysis of RNA isolated from HeLaI.2.11 cells transfected with GFP or Siah2 siRNA. PCR was performed with primers specific for Siah2 or GAPDH in reactions without cDNA (−) or with cDNA (+) prepared without (−) or with (+) RT. Siah2 DNA levels relative to GAPDH and normalized to the GFP control are indicated below the upper gel. (B) Immunoblot analysis of extracts from HeLaI.2.11 cells transfected with Siah2 or GFP siRNA. The blot was stained with Amido black (bottom panel), and proteins were detected by probing with anti-TIN2 701, anti-TRF1 415, anti-TRF2, anti-TPP1 911, or anti-α-tubulin. Protein levels relative to α-tubulin and normalized to the vector control are indicated between the blots.
Overexpression of Siah2 leads to degradation of TIN2 by the proteasome.
We next sought to determine the effect of overexpression of Siah2 on endogenous TIN2. HeLa cells were transfected with a vector control, MycSiah2, or MycΔNSiah2 (a construct lacking the N-terminal catalytic RING domain), and protein levels were analyzed by immunoblotting. As shown in Fig. 5A, overexpression of MycSiah2 led to a 3.5-fold reduction in the level of TIN2. Overexpression of MycΔNSiah2 led to a reduction in the level of TIN2, but the reduction was smaller (2-fold). We note that MycΔNSiah2 is expressed at extremely high levels compared to MycSiah2, which is consistent with studies showing that Siah regulates its own turnover via its RING domain (27). Although we observed some reduction in the level of TIN2 induced by MycΔNSiah2 expression in this experiment, it may have been nonspecific, as we show below with additional experiments that loss of TIN2 depends on the presence of the RING domain of Siah2.
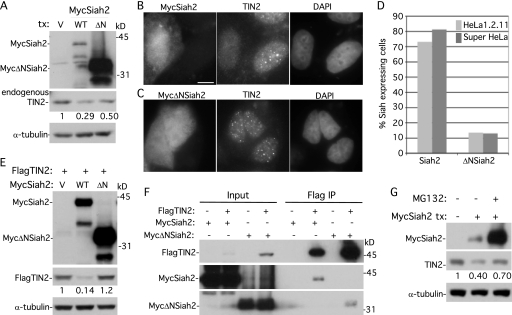
Fig 5.
Overexpression of Siah2 leads to loss of TIN2 at telomeres dependent upon the Siah2 RING domain and on the proteasome. (A) Immunoblot analysis of extracts from supertelomerase HeLa cells transfected with vector (V), MycSiah2 (WT), or MycΔNSiah2 (ΔN) and probed with anti-Myc, anti-TIN2 701, or anti-α-tubulin. (B and C) Immunofluorescence analysis of supertelomerase HeLa cells transfected with MycSiah2 (B) or MycΔNSiah2 (C). Cells were formaldehyde fixed and dually stained with anti-Myc and anti-TIN2 701 antibodies and with DAPI. Bar, 5 μm. (D) Graphical representation of the frequency of Siah2-expressing cells lacking TIN2 at telomeres (n = 126 cells or more each). (E) Immunoblot analysis of extracts from supertelomerase HeLa cells transfected with FlagTIN2 (+) and vector (V), MycSiah2 (WT), or MycΔNSiah2 (ΔN) and probed with anti-Myc, anti-Flag, or anti-α-tubulin. (F) Although MycΔNSiah2 does not affect TIN2 protein levels or localization to telomeres, it nonetheless binds to TIN2 by coimmunoprecipitation. 293T cells were transfected with FlagTIN2 and with MycSiah2 or MycΔNSiah2. Cell lysates were immunoprecipitated with anti-Flag beads and analyzed by immunoblotting with anti-Flag or anti-Myc antibody. (G) Inhibition of the proteasome by MG132 rescues Siah2-induced loss of TIN2. Supertelomerase HeLa cells were transfected for 16 h with vector (−) or MycSiah2 (+). MG132 (10 μm) was added 4 h prior to harvest. Cell extracts were analyzed by immunoblotting with anti-Myc, anti-TIN2 701, and anti-α-tubulin. (A, E, and G) TIN2 protein levels relative to α-tubulin and normalized to the vector control are indicated below the blots.
The effect of Siah2 on TIN2 was more striking in immunofluorescence analysis, where individual MycSiah2-expressing cells can be analyzed. As shown in Fig. 5B and D, overexpression of MycSiah2 led to loss of TIN2 at telomeres, whereas overexpression of MycΔNSiah2 had no effect (Fig. 5C and D). To further confirm that loss of TIN2 was due to the RING domain of Siah2, we analyzed the fate of the FlagTIN2 that was cotransfected with MycSiah2 versus MycΔNSiah2. As shown in Fig. 5E, overexpression of MycSiah2 (but not MycΔNSiah2) led to a reduction in FlagTIN2 protein levels. The inability of MycΔNSiah2 to reduce TIN2 levels was not a result of its inability to bind to TIN2; we showed by coimmunoprecipitation analysis that MycΔNSiah2 retained its capacity to bind to TIN2 (Fig. 5F). Together, these data indicate that Siah2 induces loss of TIN2 at telomeres and that the loss is dependent on the presence of its RING domain.
Finally, to determine whether Siah2-induced loss of TIN2 was due to degradation by the proteasome, we asked whether inhibition of the proteasome would rescue the loss of TIN2. As shown in Fig. 5G, treatment with the proteasome inhibitor MG132 partly restored TIN2 protein levels, indicating that Siah2-induced TIN2 depletion was due to degradation by the proteasome. We note that Siah2 was itself dramatically stabilized by MG132 treatment, which is consistent with studies showing that Siah regulates its own turnover (27). The increased levels of Siah2 seen upon MG132 treatment may explain the inability of MG132 to fully rescue TIN2 levels.
Overexpression of Siah2 leads to loss of TIN2 (but not TRF1 or TRF2) from telomeres.
We showed previously that depletion of TIN2 by siRNA led to TRF1 degradation by the proteasome (11). We thus wondered whether the posttranslational depletion of TIN2 by Siah2 (as observed here) would have a similar effect on TRF1. HeLa cells were transfected with MycSiah2 or MycΔNSiah2, and the protein levels of TIN2's shelterin binding partners were subjected to immunoblot analysis (Fig. 6A). As shown above, transfection with MycSiah2 (but not MycΔNSiah2) led to loss of TIN2. Despite the Siah2-induced reduction in the levels of TIN2, TRF1 protein levels were not affected, suggesting that posttranslational removal of TIN2 by Siah2 (compared to RNAi depletion of TIN2) differentially affected TRF1. To determine whether TRF1 remained on telomeres, we performed triple immunofluorescence analyses. As shown in Fig. 6B and E, under conditions of Siah2-induced loss of TIN2 from telomeres, TRF1 remained on telomeres.
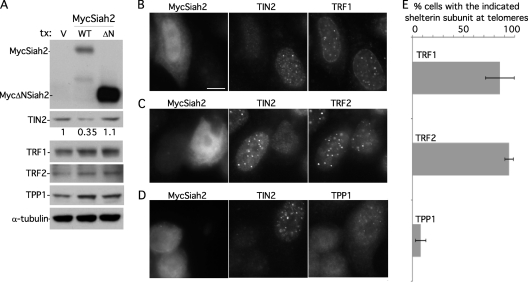
Fig 6.
TRF1 and TRF2, but not TPP1, remain at telomeres upon Siah2-mediated loss of TIN2. (A) Immunoblot analysis shows that MycSiah2 (but not MycΔNSiah2) induces loss of TIN2 protein but not TRF1, TRF2, or TPP1. Results of immunoblot analysis of extracts from supertelomerase HeLa cells transfected with vector (V), MycSiah2 (WT), or MycΔNSiah2 (ΔN) and probed with anti-TIN2 701, anti-TRF1 415, anti-TRF2, anti-TPP1 911, or anti-α-tubulin antibodies are shown. TIN2 protein levels relative to α-tubulin and normalized to the vector control are indicated below the upper blot. (B to D) Immunofluorescence analysis of supertelomerase HeLa cells transfected with MycSiah2. Cells were formaldehyde fixed and triply stained with anti-myc, anti-TIN2 701, and anti-TRF1 (B), anti-TRF2 (C), or anti-TPP1 1146 (D) and DAPI. Bar, 5 μm. (E) Graphical representation of the frequency of Siah2-expressing TIN2-lacking cells with the indicated shelterin component at telomeres. Data represent the means ± standard deviations of the results of three independent experiments (n = 69 cells or more each).
Regarding the fate of TRF2, previous studies showed that the presence of TIN2 siRNA led to loss of TRF2 from telomeres (53). We show here that Siah2-induced loss of TIN2 has no effect on TRF2 protein levels (Fig. 6A) or on TRF2 localization to telomeres (Fig. 6C and E). A recent study demonstrated p53-dependent ubiquitylation and proteasomal degradation of TRF2 by Siah1 (20). This activity may be unique to Siah1 and/or require p53 activation, since we saw no effect of Siah2 overexpression on TRF2.
Finally, we found that TPP1 protein levels were not reduced by Siah2-mediated loss of TIN2 (Fig. 6A). However, in contrast to TRF1 and TRF2, TPP1 was lost from telomeres, which is consistent with previous studies showing that TPP1 localization to telomeres depends on its ability to bind TIN2 (24).
DISCUSSION
Here we show that TIN2, the central component in the shelterin complex, is regulated posttranslationally; it is ubiquitylated by the E3 ligase Siah2 and degraded by the proteasome. We further demonstrate the fate of TIN2's three shelterin-binding partners. TRF1 and TRF2 remain on telomeres (likely via binding to DNA), and their protein levels are unaffected. In contrast, TPP1 is removed from telomeres (as expected, since the bulk of TPP1 associates with telomeres via TIN2), but its protein levels are not reduced. We speculate that posttranslational eviction of TIN2 could serve to transiently remodel the shelterin complex to accommodate a dynamic telomere function.
Consider, for example, the role of TRF1 as a negative regulator of telomere elongation by telomerase (2, 48). Siah2-mediated removal of TIN2 during the appropriate window of the cell cycle could open up a binding site for the telomerase inhibitor PinX1 to bind to TRF1 and inhibit telomere elongation by telomerase (46, 56). At the same time, the associated loss of TPP1 would inhibit recruitment of telomerase to telomeres (1). In another example, TRF1 has been found to be required for replication through telomeric DNA, likely in order to recruit helicases (BLM and RTEL1) to resolve the secondary structure of the G-rich DNA (43). Siah2-mediated removal of TIN2 in S phase could open a binding site on TRF1 for BLM, a TRF1-binding protein (35) that contains a potential TRF1 TRFH-binding motif, to facilitate passage of the replication fork.
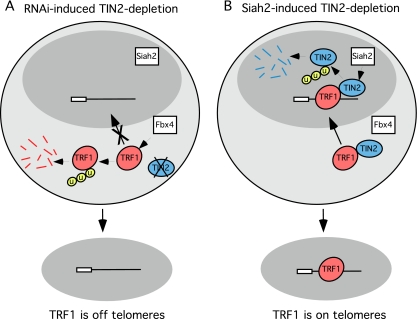
Previous studies have shown that RNAi-mediated depletion of TIN2 led to degradation of TRF1 by the proteasome (11) and, further, that this degradation was mediated by Fbx4 (33, 55). Structural and biochemical analysis suggests that TIN2 protects TRF1 from Fbx4-mediated degradation (55). Surprisingly, however, we found that, when TIN2 is depleted posttranslationally by Siah2 (rather than by RNAi), TRF1 levels and telomeric localization remained intact. Hence, the method of TIN2 depletion influences TRF1 stability. How might this work? We speculate that the protective role of TIN2 operating against Fbx4-mediated degradation is performed with newly synthesized TRF1 in the cytoplasm where Fbx4 is localized (36). When TIN2 is depleted by RNAi, TIN2 protein is not expressed, leaving newly translated TRF1 exposed to Fbx4-mediated ubiquitylation and degradation (Fig. 7A). In contrast, with Siah2-induced degradation of TIN2 protein, newly synthesized TRF1 in the cytoplasm would be protected from degradation by newly synthesized TIN2. Once TRF1 (and TIN2) is transported to telomeres in the nucleus (away from Fbx4), its stability is no longer sensitive to TIN2 being bound. Hence, eviction of TIN2 from telomeres by the nucleus-localized E3 ligase Siah2 would leave TRF1 intact and available to bind shelterin accessory proteins that are crucial for telomere function (Fig. 7B).
Fig 7.
Speculative model of how the fate of TRF1 depends on the method of TIN2 depletion. (A) RNAi-induced TIN2 depletion. TIN2 is not expressed and thus not available to protect TRF1 from degradation by the cytoplasmic E3 ligase Fbx4. (B) Siah2-induced TIN2 depletion. TIN2 is expressed and available in the cytoplasm to protect TRF1 from degradation by the cytoplasmic E3 ligase Fbx4. Upon transport to the nucleus, TIN2 is degraded by the nuclear E3 ligase Siah2. Once in the nucleus, TRF1 does not require TIN2 for protection from Fbx4 and thus remains on telomeres.
ACKNOWLEDGMENTS
We thank Tom Meier and members of the Smith laboratory for comments on the manuscript and helpful discussion. We thank Ben Houghtaling for the initial identification of Siah in two-hybrid screens. We are grateful to Joachim Lingner for supertelomerase HeLa cells and Ming Lei and Eric Fearon for plasmids.
This work was supported by NIH R01 CA116352 and NSF MCB-0543553 grants to S.S.
Footnotes
Published ahead of print 7 November 2011
REFERENCES
- 1. Abreu E, et al. 2010. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 30: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ancelin K, et al. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 22: 3474–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atanassov BS, et al. 2009. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol. Cell 35: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartel PL, Chien C, Sternglanz R, Fields S. 1993. Using the two-hybrid system to detect protein-protein interaction, p 153–179 In Harley D. A. (ed), Cellular interactions in development: a practical approach. IRL Press, Oxford, United Kingdom [Google Scholar]
- 5. Baumann P, Cech TR. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- 6. Bessler M, Wilson DB, Mason PJ. 2010. Dyskeratosis congenita. FEBS Lett. 584: 3831–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bianchi A, Smith S, Chong L, Elias P, de Lange T. 1997. TRF1 is a dimer and bends telomeric DNA. EMBO J. 16: 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bilaud T, et al. 1997. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17: 236–239 [DOI] [PubMed] [Google Scholar]
- 9. Broccoli D, Smogorzewska A, Chong L, de Lange T. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17: 231–235 [DOI] [PubMed] [Google Scholar]
- 10. Canudas S, et al. 2011. A role for heterochromatin protein 1γ at human telomeres. Genes Dev. 25: 1807–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Canudas S, et al. 2007. Protein requirements for sister telomere association in human cells. EMBO J. 26: 4867–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cardozo T, Pagano M. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5: 739–751 [DOI] [PubMed] [Google Scholar]
- 13. Chang W, Dynek JN, Smith S. 2003. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 17: 1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Y, et al. 2008. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 319: 1092–1096 [DOI] [PubMed] [Google Scholar]
- 15. Chong L, et al. 1995. A human telomeric protein. Science 270: 1663–1667 [DOI] [PubMed] [Google Scholar]
- 16. Cook BD, Dynek JN, Chang W, Shostak G, Smith S. 2002. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell. Biol. 22: 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cristofari G, Lingner J. 2006. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 25: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78: 399–434 [DOI] [PubMed] [Google Scholar]
- 19. Fairall L, Chapman L, Moss H, de Lange T, Rhodes D. 2001. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell 8: 351–361 [DOI] [PubMed] [Google Scholar]
- 20. Fujita K, et al. 2010. Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat. Cell Biol. 12: 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Her YR, Chung IK. 2009. Ubiquitin ligase RLIM modulates telomere length homeostasis through a proteolysis of TRF1. J. Biol. Chem. 284: 8557–8566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hershko A, Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67: 425–479 [DOI] [PubMed] [Google Scholar]
- 23. Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15: 3813–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Houghtaling BR, Cuttonaro L, Chang W, Smith S. 2004. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14: 1621–1631 [DOI] [PubMed] [Google Scholar]
- 25. House CM, et al. 2003. A binding motif for Siah ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 100: 3101–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. House CM, et al. 2006. Elucidation of the substrate binding site of Siah ubiquitin ligase. Structure 14: 695–701 [DOI] [PubMed] [Google Scholar]
- 27. Hu G, Fearon ER. 1999. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol. 19: 724–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu G, et al. 1997. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev. 11: 2701–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnsen SA, et al. 2009. Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res. 69: 128–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim MK, et al. 2008. Regulation of telomeric repeat binding factor 1 binding to telomeres by casein kinase 2-mediated phosphorylation. J. Biol. Chem. 283: 14144–14152 [DOI] [PubMed] [Google Scholar]
- 31. Kim SH, et al. 2004. TIN2 mediates functions of TRF2 at human telomeres. J. Biol. Chem. 279: 43799–43804 [DOI] [PubMed] [Google Scholar]
- 32. Kim SH, Kaminker P, Campisi J. 1999. TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ. 2006. The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J. Biol. Chem. 281: 759–768 [DOI] [PubMed] [Google Scholar]
- 34. Li B, Oestreich S, de Lange T. 2000. Identification of human Rap1: implications for telomere evolution. Cell 101: 471–483 [DOI] [PubMed] [Google Scholar]
- 35. Lillard-Wetherell K, et al. 2004. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 13: 1919–1932 [DOI] [PubMed] [Google Scholar]
- 36. Lin DI, et al. 2006. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol. Cell 24: 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu D, O'Connor MS, Qin J, Songyang Z. 2004. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- 38. Liu D, et al. 2004. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6: 673–680 [DOI] [PubMed] [Google Scholar]
- 39. Loayza D, de Lange T. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018 [DOI] [PubMed] [Google Scholar]
- 40. Nakayama K, Qi J, Ronai Z. 2009. The ubiquitin ligase Siah2 and the hypoxia response. Mol. Cancer Res. 7: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Palm W, de Lange T. 2008. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42: 301–334 [DOI] [PubMed] [Google Scholar]
- 42. Savage SA, et al. 2008. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 82: 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sfeir A, et al. 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith S, de Lange T. 2000. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10: 1299–1302 [DOI] [PubMed] [Google Scholar]
- 45. Smith S, Giriat I, Schmitt A, de Lange T. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282: 1484–1487 [DOI] [PubMed] [Google Scholar]
- 46. Soohoo CY, et al. 2011. Telomerase inhibitor PinX1 provides a link between TRF1 and telomerase to prevent telomere elongation. J. Biol. Chem. 286: 3894–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Treier M, Staszewski LM, Bohmann D. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78: 787–798 [DOI] [PubMed] [Google Scholar]
- 48. van Steensel B, de Lange T. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743 [DOI] [PubMed] [Google Scholar]
- 49. van Steensel B, Smogorzewska A, de Lange T. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- 50. Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. 2008. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood 112: 3594–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winter M, et al. 2008. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat. Cell Biol. 10: 812–824 [DOI] [PubMed] [Google Scholar]
- 52. Ye JZ, de Lange T. 2004. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat. Genet. 36: 618–623 [DOI] [PubMed] [Google Scholar]
- 53. Ye JZ, et al. 2004. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 279: 47264–47271 [DOI] [PubMed] [Google Scholar]
- 54. Ye JZ, et al. 2004. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zeng Z, et al. 2010. Structural basis of selective ubiquitination of TRF1 by SCFFbx4. Dev. Cell 18: 214–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou XZ, Lu KP. 2001. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107: 347–359 [DOI] [PubMed] [Google Scholar]