Abstract
The assembly of prespliceosomes is responsible for selection of intron sites for splicing. U1 and U2 snRNPs recognize 5′ splice sites and branch sites, respectively; although there is information regarding the composition of these complexes, little is known about interaction among the components or between the two snRNPs. Here we describe the protein network of interactions linking U1 and U2 snRNPs with the ATPase Prp5, important for branch site recognition and fidelity during the first steps of the reaction, using fission yeast Schizosaccharomyces pombe. The U1 snRNP core protein U1A binds to a novel SR-like protein, Rsd1, which has homologs implicated in transcription. Rsd1 also contacts S. pombe Prp5 (SpPrp5), mediated by SR-like domains in both proteins. SpPrp5 then contacts U2 snRNP through SF3b, mediated by a conserved DPLD motif in Prp5. We show that mutations in this motif have consequences not only in vitro (defects in prespliceosome formation) but also in vivo, yielding intron retention and exon skipping defects in fission yeast and altered intron recognition in budding yeast Saccharomyces cerevisiae, indicating that the U1-U2 network provides critical, evolutionarily conserved contacts during intron definition.
INTRODUCTION
Intron removal from new transcripts by pre-mRNA splicing is a fundamental feature of all eukaryotes. Such splicing is catalyzed by the spliceosome, a dynamic RNA-protein complex containing >150 proteins and five snRNAs. The assembly of the spliceosome is considered to be a dynamic process with a large number of RNA-RNA and RNA-protein rearrangements (31, 35). In the canonical pathway, U1 snRNP recognizes the pre-mRNA at the 5′ splice site (5′SS); then, U2 snRNP stably binds the branch site (BS) region to form a prespliceosome. U4/5/6 tri-snRNP then joins, after rearrangements, U1 and U4 snRNPs are released, and the remaining U2/5/6 core forms the catalytic spliceosome. Both the early recognition of pre-mRNA and the rearrangements of snRNP structures to form an active conformation are facilitated by DExD/H ATPases, which couple ATP binding/hydrolysis with structural alterations (33, 35).
Two modes for early exon and intron specification have been described: exon definition for short exons flanked by long introns, which mostly appear in vertebrates, and intron definition for short introns, which are often present in lower eukaryotes (5). Interactions between U1 and U2 snRNPs are critical to both exon- and intron-defined phases of spliceosome assembly. In the formation of commitment complexes in budding yeast Saccharomyces cerevisiae, or E complexes in mammals, cross-intron bridging interactions are proposed to connect from Prp40 in U1 snRNP at the 5′SS to SF1/BBP at the branch site or to U2AF at the polypyrimidine tract (PPT), respectively (1, 24). However, in prespliceosome formation, the first ATP-dependent transition, the BS-SF1/BBP interaction (or the PPT-U2AF interaction) is disrupted and replaced by BS-U2 snRNP interactions (28, 33). This exchange of interactions is facilitated by the ATPase Prp5, which has been proposed to unwind the branchpoint-interacting stem-loop (BSL) of U2 snRNA and allow for U2-BS pairing (21). At this stage, there is little information as to how the 5′SS-BS connection is maintained. We previously reported that Schizosaccharomyces pombe Prp5 (SpPrp5) is part of a physical bridge between U1 and U2 snRNPs (42), which are multicomponent complexes, each with one snRNA and >10 proteins (14); we have now identified components of this bridge and studied their interactions.
SR and SR-related proteins contribute to multiple steps of pre-mRNA splicing, functioning as activators/repressors in splicing regulation, affecting splice site recognition, spliceosome assembly, and catalysis (18). SR proteins usually consist of an RS domain, enriched in Arg-Ser dipeptides, and an RNA-binding motif or other functional domain. The RS domain mediates protein-protein interactions or contacts the pre-mRNA and can also be a signal for nuclear import/export and subcellular localization (6, 34). SpPrp5 contains an RS-like domain and a U2 snRNP-binding domain within its N terminus and a conserved ATPase/helicase domain within its C terminus (Fig. 1A) (42). Therefore, as a central protein in the bridge between U1 and U2 snRNPs, the RS-like and U2-binding domains in SpPrp5 could provide binding sites for U1 and U2 components.
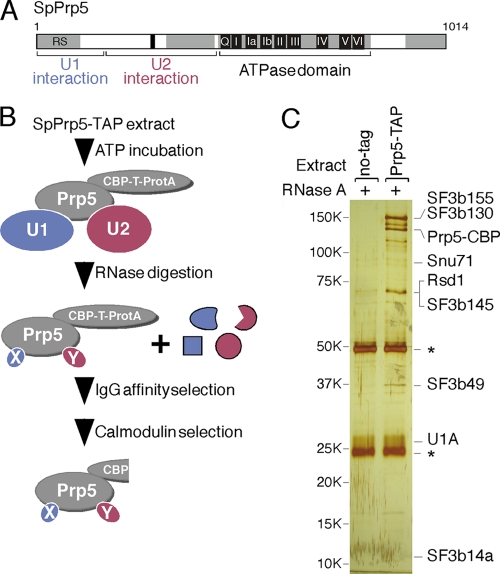
Fig 1.
Identification of SpPrp5-associated proteins. (A) Schematic of S. pombe Prp5 protein, indicating regions previously shown to be sufficient for interaction with U1 and U2 snRNPs (42). (B) Strategy for identification of protein interaction partners of SpPrp5. ProtA, protein A. (C) Silver-stained gel to visualize proteins that copurified with SpPrp5. An untagged extract was used as a negative control. The two large bands in each lane indicated by an asterisk to the right of the gel are the heavy and light chains of IgG, derived from the resin. The positions of molecular weight markers (in thousands) are indicated to the left of the gel.
Here, we purified and characterized SpPrp5-containing complexes and investigated protein-protein interactions between the components. We identify the SR-related protein Rsd1 as a mediator of the interaction between SpPrp5 and U1 snRNP and the SF3b complex as the binding target of SpPrp5 in U2 snRNP. We also describe a phylogenetically conserved DPLD motif in Prp5, mutation of which disrupts SpPrp5-SF3b and SpPrp5-U2 snRNP interactions and the formation of prespliceosomes in vitro, yielding intron definition defects in vivo. These findings elucidate a network of interactions between U1 and U2 snRNPs that contributes to intron definition.
MATERIALS AND METHODS
Yeast strains and extracts.
Yeast strains are described in Table 1. Schizosaccharomyces pombe strain 972 h− was used for C-terminal tagging of each protein of interest by PCR-based gene targeting (2, 23); tagged strains were selected by G418 resistance and confirmed by Western and PCR analyses. Intact and tagged-protein-depleted extracts from S. pombe strains were prepared as described previously (42). Strains containing mutations of the DPLD motif within the endogenous SpPrp5 gene were generated using targeted homologous recombination in SP286 (h+/h+) diploid strain, followed by switching the mating type to h+/h− using plasmid pON177 that contains the S. pombe MAT1-1 M locus, and then by dissection of the sporulated tetrads. PCR products for targeting DPLD mutations contained the KanMX6 cassette, an upstream silent mutation of a SacI site (within SpPrp5) and individual DPLD motif mutations; mutant strains were confirmed by sequencing of the SpPrp5 gene. S. pombe 972 h− and its derivative tagged strains were cultured in YE medium (0.5% yeast extract, 3.0% glucose); the SpPrp5 DPLD mutant strains were cultured in YES medium (YE medium plus amino acid supplements). Saccharomyces cerevisiae strains were derived from yYZX02 with ACT1-CUP1 reporters (43). The S. cerevisiae prp5-DPLD mutants were constructed using in vivo gap repair cloning in pRS314 plasmids.
Table 1.
Yeast strains used in this study
| Yeast strain | Relevant genotype or phenotype or descriptiona |
|---|---|
| S. pombe strains | |
| 972 h− | h− 972 |
| 972 h−(Prp5-TAP) | h−prp5::prp5-TAP-KanMx |
| 972 h−(Rsd1-TAP) | h−rsd1::rsd1-TAP-KanMx |
| 972 h−(SF3b145-TAP) | h−sap145::sap145-TAP-KanMx |
| 972 h−(SF3b49-TAP) | h−sap49::sap49-TAP-KanMx |
| 972 h−(SF3b130-flag) | h−prp12::KanMx-FLAG-sap49 |
| SP286 (h+/h+) | h+/h+ade6-M210/ade6-M216 ura4-D18/ura4-D18 leu1-32/leu1-32 |
| SpPrp5-WT | prp5::KanMx-prp5-WT |
| SpPrp5-APLD | prp5::KanMx-prp5-D303A |
| SpPrp5-DALD | prp5::KanMx-prp5-P304A |
| SpPrp5-DPAD | prp5::KanMx-prp5-L305A |
| S. cerevisiae strains | |
| yYZX02 | MATa ade2 cup1Δ::ura3 his3 leu2 lys2 prp5Δ::loxP trp1 pRS314- PRP5(PRP5 URA3 CEN ARS) |
| ScPrp5-DPLD mutants | Constructed from yYZX02; Prp5 alleles were in pRS316-TRP, and the WT allele in pRS314 was removed using 5-FOA |
WT, wild type; 5-FOA, 5-fluoroorotic acid.
Copurification and mass spectrometry.
S. pombe extracts were incubated at 30°C for 30 min to assemble SpPrp5-associated complexes, followed by RNase A digestion (40 ng/μl) for 30 min. The mixture was applied to an IgG-Sepharose column (Amersham Pharmacia) and washed using IPP100 buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.1% Nonidet P40). Bound proteins were released by tobacco etch virus (TEV) protease (Invitrogen) cleavage at 16°C for 4 h and applied to calmodulin resin (Stratagene) in the presence of 2 mM CaCl2. Copurified proteins were eluted with buffer containing 2 mM EGTA, separated by SDS-PAGE, and analyzed using a Dionex capillary/nano-high-performance liquid chromatography (nano-HPLC) system and tandem mass spectrometry (MS-MS) by a Finnigan LCQ mass spectrometer. The MS-MS data set was searched using Sonar MS-MS and MASCOT for protein identification.
Mutagenesis, recombinant protein expression, and purification.
All mutations in the chromosomal SpPrp5 gene and the constructs for in vitro protein expression were generated by two- or three-step overlapping PCR and confirmed by sequencing. We used pGEX-4T-1 vector for glutathione S-transferase (GST) tag cloning and pET-33b for His6 tag cloning. Recombinant proteins were purified by either glutathione-Sepharose or Ni-agarose (Qiagen) chromatography under standard conditions, except that the lysis and binding buffers contained 500 mM NaCl. The purified recombinant proteins were dialyzed against buffer D (20 mM HEPES-KOH [pH 7.9], 0.2 mM EDTA, 100 mM KCl, 0.5 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], and 20% glycerol) after purification.
In vitro transcription and translation for [35S]methionine-labeled proteins.
cDNAs of S. pombe genes were generated by Qiagen Sensiscript reverse transcriptase from total RNA using an oligo(dT) primer, and the coding sequence of each gene of interest was amplified by PCR adding a T7 promoter and a Kozak sequence (AGCCACC) for optimal protein translation. The PCR products were confirmed by sequencing and used directly as templates for [35S]methionine incorporated protein synthesis by TNT T7 quick coupled transcription/translation system (Promega). Twenty-five microliters of 35S-labeled protein was used for each protein-protein interaction assay and visualized by autoradiography.
Western and Northern blots.
Antiserum against SF3b155 was generated by immunizing rabbits with 331EKELPAALPTEIPGVC peptide (Genemed Synthesis Inc.). Western blots were probed using monoclonal antibody MAb 12CA5 (Roche) and sheep anti-mouse antibody conjugated to horseradish peroxidase (HRP) (Amersham) for hemagglutinin (HA) or using rabbit anti-chicken antibody conjugated to HRP (Pierce) to detect the protein A component of the tandem affinity purification (TAP) tag. Northern analyses were performed by transferring RNA from urea gels to Hybond N membranes (Amersham), followed by probing with 32P-labeled antisense DNA oligonucleotides.
Reverse transcription-PCR (RT-PCR) analysis for in vivo mutants.
Mutant strains were cultured at 30°C to an optical density (OD) of 0.8 measured at 600 nm; cells were harvested, and total RNA was extracted as described previously (43). Reverse transcription was performed using an oligo(dT) primer (ReverTra Ace-a; Toyobo), and PCR used the primers listed in Table 2. PCRs were carried out for 30 cycles using primers listed in Table 2; products were semiquantified using a Tanon-2500 gel analysis system.
Table 2.
Primers for RT-PCR
| Gene | Sense primer |
Antisense primer |
||
|---|---|---|---|---|
| Location | Sequence | Location | Sequence | |
| Intron-containing genes | ||||
| pyp3a | Exon 1 | 5′-CTCGTGTTCGTTTAGATCCAATG | Exon 2 | 5′-GGCGTTCCCTCAATTTGGTAAGC |
| erf1a | Exon 2 | 5′-GGAGATCAGCTGAAGGCTTCTAC | Exon 3 | 5′-TACAAGCAGCATCTACACGGTCC |
| Exon 1 | 5′-CCTGAAGAGCCAGAAGATATGTG | Exon 5 | 5′-TGCTGAATATGTAGTCATACAAG | |
| sF3b155 | Exon 1 | 5′-CTTCGCCATCAACATGTCAACTG | Exon 3 | 5′-GGTCATAACTTTCATCAGCGTATTCATT |
| cdc2 | Exon 1 | 5′-ATGGAGAATTATCAAAAAGTCGAAA | Exon 3 | 5′-CTTCTAGAATGGCAAAAATTTACAC |
| Exon 2 | 5′-CTTGAAGATGAATCTGAGGGAGTTC | Exon 4 | 5′-CAGGAGCACGATACCATAAAGTGAC | |
| Exon 3 | 5′-ACATGGACCGAATTTCAGAAACTG | Exon 5 | 5′-CACTAATGCGATGGGCAGGGTC | |
| nda3 | Exon 1 | 5′-CATAAATTAGTCTAGATGCGTGAG | Exon 3 | 5′-ATTCCAGCTGAATCCAAACCATG |
| Exon 5 | 5′-TAAAGCATCACATGCTTCAGCTTC | |||
| Exon 6 | 5′-CTTGGATAGAAGTACTGTTACC | |||
| cgs2 | Exon 1 | 5′-GCATGCAGCACTCGAAATCAAAG | Exon 3 | 5′-TCAGAAATCAAATGACTGCTACAG |
| Exon 4 | 5′-TGCAGACGACTTTAGCTCTTGAC | |||
| Intron-less genes | ||||
| alp16 | 5′-GTTTCAGCGTTGGCTTCGTCTT | 5′-ATTCAAGTTAAGCGGACAGACG | ||
| prp5 | 5′-GACCGCGGTATGCTAGGAAATGAGCAAGC | 5′-TTTATTTTTGAGTGATCAACTGTGATTACGTCTTTC | ||
| sF3b145 | 5′-ATGATGCCTTCTTTCGCTACCA | 5′-TCATAGCCTGCTCAGACACCAACT | ||
Data on primers for these two genes are taken from reference 40.
RESULTS
Identification of Prp5 interaction partners.
To characterize the Prp5-mediated connection between U1 and U2 snRNPs, U1-(Prp5-TAP)-U2 complexes from S. pombe extract were assembled and then digested with RNase A to degrade the snRNAs. Proteins bound to SpPrp5-TAP were purified by two-step affinity chromatography and identified by mass spectrometry (Fig. 1B and C). A similar purification without RNase A treatment and a mock purification from an untagged strain were used as controls.
Three groups of proteins were found associated with SpPrp5 (Fig. 1C and Table 3). The first group contained two U1 snRNP proteins: U1A and S. pombe Snu71 (SpSnu71). The second group contained five U2 snRNP SF3b subunits: SF3b155, SF3b130, SF3b145, SF3b49, and SF3b14a. A third group, non-U1 or non-U2 proteins, contained one member: Rsd1, an SR-like protein (15) whose human homolog, CAPER (also known as HCC1, CC1.3, RNPC2, and RBM39), was originally identified as a nuclear autoantigen in hepatocellular carcinoma (12) and has been reported to coactivate AP1 and ER (estrogen receptor) transcription factors (9, 13), is overexpressed in breast cancer (19), and was recently found in purified exon definition complexes (27, 29). All of the identified proteins were also found in highly purified S. pombe prespliceosomes (T. Huang and C. C. Query, unpublished observations) and in human complex A (4, 14). Several additional proteins (data not shown) were found with low abundance in the SpPrp5-purified material and are not further pursued here.
Table 3.
Results of mass spectrometry
| S. pombe protein | MMa (kDa) | No. of peptides | Human protein | S. cerevisiae protein |
|---|---|---|---|---|
| U1 snRNP | ||||
| U1A/Usp102/Mud1 (SPBC4B4.07c) | 28.3 | 2 | U1A | Mud1p |
| SpSnu71p/Usp107 (SPBC839.10) | 81.0 | 3 | Snu71p | |
| U2 snRNP | ||||
| SF3b155/Sap155/Prp10 (SPAC27F1.09c) | 135.8 | 16 | SF3B1/SF3b155 | Hsh155p |
| SF3b130/Sap130/Prp12 (SPAPJ698.03c) | 135.8 | 20 | SF3B3/SF3b130 | Rse1p |
| SF3b145/Sap145 (SPAC22F8.10c) | 69.4 | 5 | SF3B2/SF3b145 | Cus1p |
| SF3b49/Sap49 (SPAC31G5.01) | 36.0 | 3 | SF3B4/SF3b49 | Hsh49p |
| SF3b14a/Sab14 (SPBC29A3.07c) | 15.7 | 5 | SF3b14a | Ist3p/Snu17p |
| SR protein | ||||
| Rsd1 (SPAC19G12.07c) | 69.7 | 4 | HCC1/CAPER |
MM, molecular mass.
Rsd1 mediates Prp5 interaction with U1 snRNP.
To investigate direct protein-protein interactions, we used GST pulldown assays with a series of GST fusion proteins and [35S]methionine-labeled in vitro translation products expressed in reticulocyte lysate. GST-SpPrp5 did not copurify U1A or SpSnu71; however, it did efficiently copurify the non-U1/U2 protein, Rsd1 (Fig. 2A). Furthermore, GST-Rsd1 copurified U1A, but not SpSnu71, and GST-SpSnu71 copurified U1A, but not Rsd1 (Fig. 2B). These data suggest that the interaction between SpPrp5 and U1 snRNP involves a mediator, Rsd1, that binds directly to SpPrp5 and U1A (Fig. 2C).
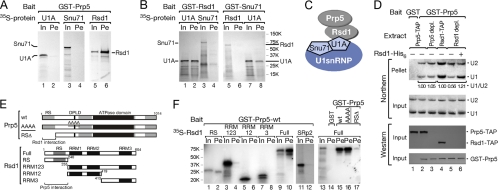
Fig 2.
Rsd1 mediates SpPrp5 interaction with U1 snRNP. (A) In vitro protein-protein interactions indicate that SpPrp5 binds directly to Rsd1, but not to core U1 proteins. GST-tagged proteins were incubated with 35S-labeled in vitro translation products and purified using glutathione-Sepharose. Lanes: In, 1/6 of total input; Pe, copurifying proteins. (B) GST-Rsd1 binds directly to U1A, but not to SpSnu71 (left); GST-Snu71 binds to U1A, but not to Rsd1 (right). (C) Schematic of proposed interaction network between SpPrp5 and U1 snRNP mediated by Rsd1. (D) Rsd1 facilitates SpPrp5 interaction with U1 snRNP. SpPrp5 or Rsd1 was depleted from extract by IgG-Sepharose binding under high-salt conditions (Western blotting), and then the extract was incubated with ATP and GST-SpPrp5 followed by affinity selection. Copurifying snRNAs were analyzed by Northern blotting. The U1-to-U2 snRNA ratio was normalized to the amount of U1 and U2 snRNAs in each TAP-tagged extract. (E and F) SpPrp5 and Rsd1 interact through their RS and RS-like domains. GST pulldowns were performed as described above fir panel A. WT-SpPrp5, SpPrp5-AAAA306, and SpPrp5-RSΔ (amino acids [aa] 1 to 172 deleted) were expressed as GST-tagged proteins (bait); Rsd1 full-length and truncated proteins, RS domain (aa 1 to 240), RRM123 (aa 235 to 604), RRM12 (aa 235 to 419), RRM3 (aa 413 to 604), and SRp2 were translated in vitro with 35S labeling.
To confirm that Rsd1 contributes to SpPrp5-U1 interaction, we depleted Rsd1 from Rsd1-TAP-tagged extracts and reconstituted it using purified Rsd1-His6 protein. When Rsd1-TAP was depleted, GST-SpPrp5 copurified much less U1 snRNA, indicated by a reduction in the ratio of Prp5-bound U1 to U2 from 1 to 0.56 (Fig. 2D, cf. lane 4 to 5); the copurification of U1 snRNA was restored after adding back Rsd1-His6 protein, indicated by an increased ratio of SpPrp5-bound U1 to U2 from 0.56 to 1.21(cf. lane 5 to 6). As a control, depleting endogenous SpPrp5-TAP did not decrease the GST-SpPrp5 affinity selection of U1 snRNA (cf. lane 3 to 2). The remaining level of U1 bound to GST-SpPrp5 after Rsd1-TAP depletion may be due to the presence of some Rsd1 that was cleaved from its TAP tag prior to depletion or to additional contacts between SpPrp5 and U1 snRNP components. Together, these data suggest that Rsd1 mediates the SpPrp5-U1 snRNP interaction.
Prp5 and Rsd1 interact through their RS domains.
To define domains required for SpPrp5-Rsd1 binding, we constructed mutants of both SpPrp5 and Rsd1 for in vitro protein interaction assays (Fig. 2E). GST-tagged full-length SpPrp5 and SpPrp5-AAAA306, which contains a mutation (underlined) in the U2-binding region and is defective in binding to SF3b (described below), efficiently copurified 35S-labeled full-length Rsd1; however, GST-SpPrp5ΔRS, in which the N-terminal RS-like domain was deleted, did not copurify Rsd1 (Fig. 2F, lanes 15 to 17), indicating that the RS-like domain of SpPrp5 is required for binding to Rsd1. This is consistent with our previous observation that the RS-like domain of SpPrp5 alone copurified U1 snRNP from S. pombe extract (42).
To address which domain in Rsd1 interacts with SpPrp5, we divided Rsd1 into its RS domain, RRM123 (all three RRMs), RRM12 (RRM1 and RRM2), or RRM3 only. None of the RRMs within Rsd1 copurified with GST-SpPrp5 (Fig. 2F, lanes 4, 6, and 8), whereas the RS domain of Rsd1 alone was sufficient to bind to GST-SpPrp5 (lane 2). Another S. pombe SR protein, SRp2, which is also involved in splicing (40), was tested as a control for RS domain specificity and showed no detectable binding to GST-SpPrp5 (lane 12). Thus, SpPrp5 and Rsd1 interact primarily through their RS/RS-like domains.
SF3b mediates Prp5 interaction with U2 snRNP.
The U2 components identified by mass spectrometry were five of the SF3b subunits; the remaining two components of the heptameric SF3b complex are less than 12 kDa and were likely too small to be detected. To confirm that SpPrp5 binds to the SF3b complex, we asked whether recombinant GST-SpPrp5 could affinity select SF3b. We partially purified SF3b from SF3b145-TAP extract; subsequently, GST-SpPrp5, but not GST alone, affinity selected SF3b, indicated by the SF3b155 signal (Fig. 3A). Consistent with the purification shown in Fig. 1, this SpPrp5-SF3b interaction did not depend on RNA (Fig. 3B), nor did it require ATP hydrolysis by SpPrp5 (Fig. 3C, lane 2, and 3E, lane 4), indicated by unaffected SF3b binding by an ATPase domain motif III mutant SAA, which is defective in ATP hydrolysis and cannot form prespliceosomes (42). We tested three other SF3b subunits, SF3b49, SF3b130, and SF3b145 (Fig. 3C), which were also associated. We attempted to test individual protein-protein interactions using in vitro-translated SF3b subunits, as we did above for U1 proteins, but most SF3b proteins were poorly translated or did not interact in this assay.
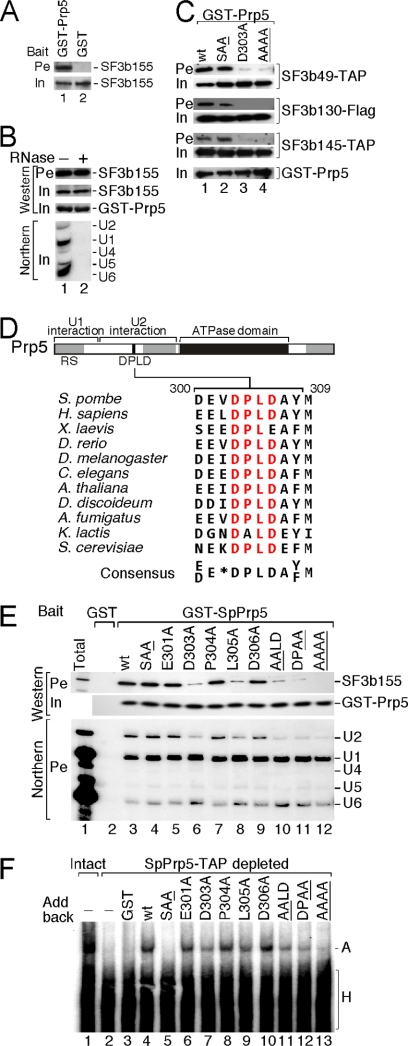
Fig 3.
The DPLD motif and SF3b mediate SpPrp5 association with U2 snRNP. (A) Recombinant GST-SpPrp5 interacts with the SF3b complex partially purified from SF3b145-TAP extract. Signal of SF3b155, detected by Western blotting using anti-SF3b155 antibody, is shown as a representative of the SF3b complex. Pulling down with GST alone was used as a negative control. (B) Interaction between SpPrp5 and SF3b is RNA independent. Recombinant GST-SpPrp5 can pull down the SF3b complex both in the presence (+) and absence (−) of snRNAs, which were detected by Northern blotting. (C) Alanine mutations in the DPLD motif disrupt SpPrp5 interaction with SF3b49, SF3b130, and SF3b145, whereas mutation of the SAT motif in the ATPase domain has no effect on interaction with SF3b proteins. SF3b subunits were either TAP or Flag tagged as indicated and detected by Western blotting. (D) Phylogenetic comparison of the conserved DPLD motif in Prp5. Sequences from Schizosaccharomyces pombe, Homo sapiens, Xenopus laevis, Danio rerio, Drosophila melanogaster, Arabidopsis thaliana, Dictyostelium discoideum, Aspergillus fumigatus, Kluyveromyces lactis, and Saccharomyces cerevisiae are shown. (E) Alanine mutations at D303 and L305 inhibit SpPrp5 interaction with SF3b155 and U2 snRNP. Recombinant GST-tagged proteins containing mutations in the DPLD motif of SpPrp5 were incubated with S. pombe extract, then selected, and assayed for interaction with SF3b proteins (indicated by Western blotting for SF3b155) and U2 snRNP (indicated by Northern blotting for snRNAs). (F) Mutations in the DPLD motif inhibit assembly of prespliceosomes. Recombinant SpPrp5 proteins and 32P-labeled pre-mRNA substrate were incubated with S. pombe extract depleted of endogenous SpPrp5; their abilities to form prespliceosomes (complex A) were analyzed by 4% native gel.
A conserved DPLD motif of Prp5 interacts with the U2-SF3b complex.
Previously, we defined a U2-binding domain of SpPrp5 encompassing amino acids 171 to 426, which alone was sufficient to affinity select U2 snRNP from S. pombe extract (42). However, an antibody prepared against a human Prp5 (hPrp5) peptide within this domain, EELDPLDAYMEEV, did not coimmunoprecipitate U2 snRNP from HeLa cells, whereas other anti-hPrp5 antibodies did (42). These two findings suggested this peptide as a candidate for the Prp5-SF3b interaction site. This region is conserved from yeasts to human, the consensus (E/D)EXDPLDA(Y/F)M having a nearly invariant core motif, DPLD (Fig. 3D).
To investigate whether the DPLD motif contributes to Prp5-SF3b interaction, we tested the effects of single, double, and tetra-alanine substitutions. In comparison with wild-type SpPrp5 (wt-SpPrp5), D303A and L305A mutants (but not E301A, P304A, and D306A mutants) reduced SpPrp5 interaction with the SF3b155 subunit; consistent with this, the amount of copurified U2 snRNA, but not U1 snRNA, was also decreased (Fig. 3E, cf. lanes 5 to 9 to lane 3). Two mutants with double-alanine mutations in the DPLD motif, AALD and DPAA mutations, exhibited similarly reduced levels of copurified SF3b155 and U2 snRNA as did the D303A and L305A mutants. The tetra-alanine mutant exhibited even stronger defects, consistent with cumulative effects of mutations at positions D303 and L305 (Fig. 3E, lanes 10 to 12). Furthermore, in a depletion/reconstitution system in which endogenous SpPrp5-TAP protein was depleted from extract and recombinant wt-SpPrp5 or mutants were supplemented, the SpPrp5 mutants described above assembled prespliceosomes less efficiently than wt-SpPrp5 did (Fig. 3F, cf. lanes 7, 9, and 11 to 13 to lane 4). In both the pulldown and prespliceosome assembly assays, the behavior of E301, P304, and D306 mutants was indistinguishable from wt-SpPrp5 (Fig. 3E and F). Other SF3b subunits were also tested: mutations at position D303 or L305 decreased SpPrp5 interaction with SF3b49, SF3b130, and SF3b145 (Fig. 3C), consistent with the DPLD motif being critical for Prp5 interaction with the intact SF3b particle.
Prp5-DPLD motif mutants yield intron retention and exon skipping defects.
To investigate the effects of the DPLD motif on splicing in vivo, we generated alanine substitution mutants using homologous recombination to replace the endogenous SpPrp5 gene in an S. pombe diploid strain. After sporulation and tetrad dissections, haploids containing the alanine mutation were confirmed by DNA sequencing. The strains containing D303A and L305A single mutations showed growth defects, especially at lower temperatures. The defects of the tetra-alanine substitution AAAA were stronger, as the haploid was nonviable, but the P304A mutant had no observed defects (Fig. 4A). These data parallel the relative effects observed in the in vitro analyses.
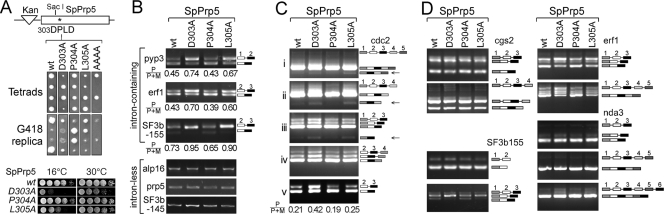
Fig 4.
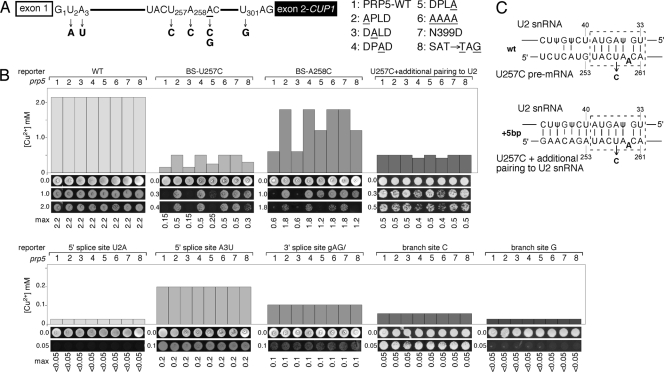
Prp5-DPLD motif mutants yield intron retention and exon skipping in S. pombe. (A) In vivo, mutations at D303 and L305 in the DPLD motif of SpPrp5 affect the growth of fission yeast. (Top) Strategy; (middle) tetrad dissection and growth on G418 (only Kan+ cells grow; wild-type Kan− cells leave a faint background of dead cells; for the AAAA mutant, two of the spots are blank, because this mutant is inviable); (bottom) temperature growth assay. (B) RT-PCR analysis reveals that D303A and L305A mutants of Prp5 inhibit pre-mRNA splicing of intron-containing genes, yielding intron retention. Expression levels of intronless genes were not affected. P/P+M, precursor/(precursor + mature), is an estimate of the fraction of unspliced RNA for various transcripts. (C) D303A and L305A mutants of Prp5 trigger skipping of exon 2 of the cdc2 gene. RT-PCRs were tested by various sets of primers across five exons of cdc2, and the PCR products were confirmed by sequencing. (D) D303A and L305A (but not P304A) mutants of Prp5 inhibit pre-mRNA splicing of multiple-intron-containing genes, yielding intron retention, as shown here by increased levels of pre-mRNA and decreased levels of mRNA (most notably for erf1 and SF3b155, but decreased effects for other genes like cgs2 and nda3). D303A and L305A mutants of Prp5 result in skipping of exon 2 of the cdc2 gene (panel C), but no detectable exon skipping for four other multiple-intron-containing genes. RT-PCRs were performed using primers listed in Table 2.
We tested the levels of mRNA isoforms by RT-PCR for a number of genes to ask whether the DPLD mutations affected splicing activity. For several intron-containing genes, such as pyp3, erf1 (characterized for U2AF dependence in reference 39), SF3b155 (prp10), cdc2, nda3, and cgs2, mRNA levels were reduced in the D303A and L305A mutant strains, with a concomitant increase in the levels of pre-mRNA or of intron-containing mRNA (intron retention) relative to the wild-type strain (Fig. 4B, C, and D). In contrast, for intron-less genes, including alp16 (tubulin), SpPrp5, and SF3b145, mRNA levels were not detectably different from the wild-type strain (Fig. 4B). These results are consistent with the growth defect of mutant strains being due to a widespread inhibition of pre-mRNA splicing. To detect possible exon skipping, we tested five multi-intron-containing genes: exon skipping was observed in the D303A and L305A mutant strains for exon 2 of the cdc2 gene (Fig. 4C, panels i to iii, indicated by the black arrows). We did not detect exon skipping in DPLD mutant strains for the other cdc2 exons (Fig. 4C and D), nor for other multiple intron-containing genes tested (prp10, erf1, nda3, and cgs2; Fig. 4D), consistent with the notion that intron/exon specification in fission yeast is mostly via an intron definition pathway (26). As a caveat, exon-skipped products may be underrepresented due to degradation by nonsense-mediated decay. We conclude that Prp5-DPLD mutations result in defects in intron/exon definition.
Functional contribution of the Prp5-DPLD motif is conserved in S. cerevisiae.
We also tested the effects of DPLD motif mutations in S. cerevisiae, using characterized strains with intron mutations in the ACT1-CUP1 reporter gene. The APLD, DPAD, and AAAA mutants strongly improved splicing of branch region mutants U257C and A258C, which reduced the pairing with U2 snRNA (Fig. 5B, cf. lanes 2, 4, and 6 to lanes 1), but did not detectably alter splicing of the wt reporter or of 5′SS, 3′SS, or branch nucleophile mutants, whereas DALD and DPLA mutants did not improve or only slightly improved the splicing of branch region mutants (Fig. 5B, lanes 3 and 5). This pattern of altered splicing is identical to that observed with prp5-ATPase domain mutants (43) (N399D and TAG448; Fig. 5B, lanes 7 and 8), suggesting that the DPLD-SF3b interaction is integrally linked with Prp5 ATPase activity.
Fig 5.
Prp5-DPLD motif mutants modulate substrate selectivity of suboptimal branch regions in S. cerevisiae. (A) Schematic of ACT1-CUP1 pre-mRNA, indicating intron mutations at 5′SS, BS, and 3′SS used in panel B. (B) Analysis of prp5-DPLD mutant alleles that alter splicing of branch region mutants. Graphs of maximum copper concentration tolerated (top) and growth on selected copper plates (bottom) are shown. Previously described prp5-N399D and -TAG448 alleles (25) were tested for comparison. prp5-DPLD mutants improved the copper tolerance of branch region mutants U257C and A258C that decrease pairing with U2 snRNA but do not alter splicing and growth on copper of 5′SS, 3′SS, or branch site C or G mutants. The presence of additional base pairs between the branch region and U2 snRNA (25) abrogates the effects of both DPLD mutants and ATPase mutants on the U257C branch region mutation. max, maximum. (C) Additional potential base pairs between U2 snRNA and the intron branch region partially suppress the U257C defect; prp5 alleles provide no additional improvement. (Top) Schematic of base pairing interactions between U2 snRNA and the intron branch region, indicating BS-U257C ACT1-CUP1 reporter mutation, which is improved by prp5 alleles, and (bottom) BS-U257C plus five additional base pairs to U2 snRNA, which is not improved by prp5 alleles, shown in panel B.
DISCUSSION
U1 and U2 snRNP binding to pre-mRNA are critical steps in spliceosome assembly, specifying the intron-exon structure. Communication between U1 and U2 is necessary for this process, both in exon and intron definition phases, but little has been known about direct U1-U2 interaction. Here, we present an interaction network for U1 and U2 snRNPs, centered on the ATPase Prp5.
Rsd1 bridges Prp5 interaction with U1 snRNP.
U1 snRNP is composed of U1 snRNA and 10 tightly bound proteins, the core of which has been described structurally (22). Several lines of evidence support a role for Rsd1 in the early stage of pre-mRNA splicing. The results of analysis of mass spectrometry and individual protein-protein interactions demonstrate that SpPrp5 does not interact directly with U1 snRNP, but instead binds strongly to Rsd1, which binds to the U1 core protein U1A. Biochemical depletion of Rsd1 in vitro reduces SpPrp5-U1 snRNP interaction and adding Rsd1 back restores it. Rsd1 is also found in human complex A (4, 14). These data support a model of Prp5 interaction with U1 snRNP mediated by Rsd1.
Fission yeast Rsd1 is an SR-related protein, containing an N-terminal RS domain with 30 RS/SR/RD dipeptides, and three RRMs. Its human homolog, CAPER, has been proposed to couple transcription with splicing, influencing alternative splicing of the steroid hormone receptor (9, 13). Because both the presence of a 5′SS and the binding of snRNPs have been shown to enhance transcription (8), the mechanism by which CAPER can stimulate transcription may be related to its role in U1 binding.
Conventional SR proteins typically have one RS domain and one to three RNA-binding motifs (RRM or PWI). The RS domain is believed to mediate interaction with other proteins; the RNA-binding motif typically binds to RNA targets, although RRMs can also contribute to protein-protein interactions (e.g., reference 7 and references therein). Here, we demonstrate that the RS domain of Rsd1 is sufficient for binding to SpPrp5; likewise, the RS-like domain of SpPrp5 is required for binding to Rsd1. Because RS domains are known to be phosphorylated and dephosphorylated, an intriguing possibility is that this interaction may be modulated by phosphorylation states. These two domains are conserved from S. pombe to human, but there is no identifiable Rsd1 homolog in the budding yeast S. cerevisiae, consistent with the lack of conventional SR proteins (with the possible exception of Npl3 [16]); this suggests that other U1-Prp5 or U1-U2 connections may exist, which have allowed for the loss of the Rsd1-mediated interaction in S. cerevisiae.
SF3b bridges Prp5 interaction with U2 snRNP.
U2 snRNP is an approximately 17S particle, composed of U2 snRNA and ∼20 proteins. Although U2 snRNP has not been specifically purified from S. pombe, all of the orthologous proteins are found in the S. pombe genome. Our data indicate that SpPrp5 binds directly and stably to the SF3b protein complex, but not to other U2 snRNP proteins.
We present several lines of evidence that a phylogenetically conserved DPLD motif in Prp5 is critical for the interaction with U2-SF3b. Mutations at D303 or L305 (DPLD) disrupt the interaction with SF3b in vitro, the interaction with U2 snRNP, and the formation of prespliceosomes. In vivo, mutations within the SpPrp5-DPLD motif yield significant defects: mutants with the DPLD-to-AAAA mutation were inviable and D303 and L305 mutants showed growth and splicing defects (discussed more below), whereas the P304 mutant was indistinguishable from the strain with wt-Prp5. This is consistent with the conservation of the DPLD motif across species, where the first Asp and Leu residues are invariant, but the Pro and second Asp residues are occasionally divergent (e.g., DALD in Kluyveromyces lactis and DPLE in Xenopus laevis) (Fig. 3D), suggesting that D303 and L305 residues play critical roles for Prp5 interaction with SF3b.
Although we have not yet determined which subunit(s) of SF3b mediates the interaction with Prp5, two lines of evidence suggest that Prp5 may bind directly to SF3b155. (i) S. cerevisiae Prp5 can interact with Hsh155, the SF3b155 homolog, in yeast two-hybrid assays (36). (ii) In a cocrystal structure of chloroplast signal recognition particle protein cpSRP43 with a light-harvesting chlorophyll-binding protein (LHCP), a DPLG peptide in LHCP was bound by a helix-turn-helix motif in cpSRP43 that is similar to those found in the heat repeats of SF3b155 (32), suggesting that Prp5-DPLD could interact similarly with SF3b155. In this structure, the D and L residues were engaged in specific contacts, consistent with the functional importance of these two residues observed in Prp5.
Communication between U1 and U2 snRNPs in the prespliceosome.
Prior to a requirement for ATP, cross-exon interactions bridge from a 3′SS to the next 5′SS or to downstream exon enhancers (5, 27), and cross-intron bridging interactions connect from U1 snRNP at the 5′SS to SF1/BBP at the branch site in S. cerevisiae or to U2AF at the PPT in mammals (1, 41). In the first ATP-dependent transition of spliceosome assembly, the branch site-SF1/BBP interaction, or the PPT-U2AF interaction, is disrupted and replaced by branch site-U2 snRNP interactions (28). We have proposed that the 5′SS-branch site connection is maintained at this stage by a Prp5-mediated U1-U2 interaction (42).
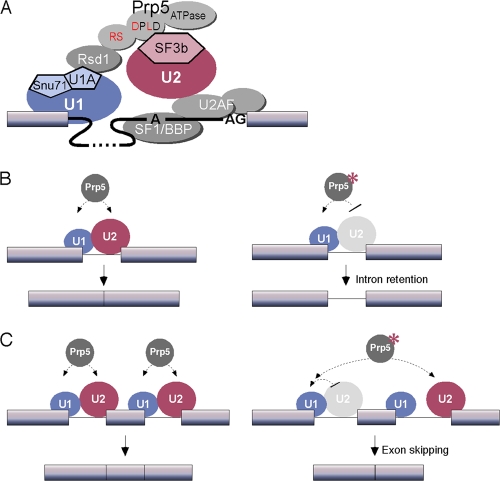
Several lines of evidence have long argued for communication between U1 and U2 snRNPs, including the stimulation of U2-branch site binding by the presence of a 5′SS (25) and the decrease in U2 binding upon U1 snRNP depletion (3). Such U1-U2 communication has been proposed to contribute to both intron and exon definition in spliceosome assembly models (5), and both snRNPs have been found in purified intron- and exon-defined complexes (11, 27, 29). Further hints at such an interaction came from the U12-dependent spliceosome, in which U11 and U12 snRNPs (analogs of U1 and U2) were found stably associated as a di-snRNP (38). However, how U1 and U2 snRNPs might interact has remained unknown. Here, we show that Prp5 interacts directly with U2 snRNP SF3b proteins and that Prp5-Rsd1 and Rsd1-U1A interactions mediate the connection between Prp5 and U1 snRNP. These findings elucidate a network of interactions between U1 and U2 snRNPs (Fig. 6A).
Fig 6.
Protein interaction network for U1-U2 snRNP communication during intron specification and prespliceosome assembly. (A) During intron definition, the U1 snRNP core protein U1A binds to an SR-like protein, Rsd1. Rsd1 also contacts SpPrp5, mediated by SR-like domains in both proteins. SpPrp5 then contacts U2 snRNP through SF3b, mediated by a conserved DPLD motif in Prp5. The ATPase domain of Prp5 is not required for these protein-protein interactions but instead is required for the remodeling of U2 snRNP for its stable binding with the branch site. (B) Prp5 contributes to communication between the 5′SS and branch region, helping to define the intron (left). Loss of Prp5-SF3b interaction results in failure of U2 snRNP to engage the intron, leading to intron retention (right). (C) In multi-intron genes, loss of Prp5-SF3b interaction at a weak PPT can result in a U1 connection to U2 snRNP at a downstream branch region, resulting in exon skipping (right). The asterisk indicates a prp5 mutant that is impaired or slow in interactions with U2 snRNP.
Disruption of the Prp5-SF3b interaction by mutation of the DPLD motif causes splicing defects that result in both intron retention and exon skipping in S. pombe (Fig. 4 and 6B and C). Does the observed exon skipping imply participation of the network in an exon definition bridge, or does it represent alternative 3′SS selection in an intron-defined mode of splicing? A defect in intron definition would be predicted to yield mostly intron inclusion and occasionally exon skipping, whereas an exon definition defect should mostly yield exon skipping. Although S. pombe does have genes with multiple introns, and thus might exhibit exon definition, the behavior of these multi-intron genes suggests that they are intron defined (26), and our data are consistent with this. The intron retention observed here is general, in that many intron-containing genes are affected, whereas the exon skipping defect was observed for one example (exon 2 of cdc2), and only weakly. Some introns/exons were more sensitive to inclusion/skipping than others, and this presumably reflects sequence differences: the observation of exon skipping only for exon 2 of cdc2 pre-mRNA correlates with the absence of a strong PPT in the preceding intron 1 3′SS (UAAUGC) and the presence of a strong PPT in the following intron 2 3′SS (CCUUUUUU). Thus, a failure to engage intron 1 3′SS in prp5-DPLD mutant strains could result in recognition of the next strong 3′SS and pairing of it with the intron 1 5′SS (Fig. 6C). Thus, all of the effects observed in vivo in S. pombe are consistent with a role of the Prp5-SF3b interaction in intron definition. This does not exclude the possibility of a role in exon definition in other organisms with long introns.
One model to explain the mechanism of these effects is that a weak Prp5-SF3b interaction allows a longer time (stochastically) for U2 to bind stably to the branch site. A strong PPT allows for stable U2AF binding and thereby a stable tethering (long dwell time) of U2 snRNP (10), whereas a weak PPT would not. This model is supported by data from analogous prp5-DPLD mutants tested in vivo in S. cerevisiae, which resulted in increased use of suboptimal branch regions. We have argued previously that such mutants are slow at pairing with U2 snRNA and that prp5-ATPase domain mutants allow a longer time for stable U2 binding to weak branch sites (43). Increasing the pairing potential between U2 snRNA and the branch region abrogates the effects of both ATPase and DPLD prp5 mutants (Fig. 5C) (43). Thus, in both S. pombe and S. cerevisiae, the phenotype of prp5-DPLD mutants is consistent with an intron definition defect that allows a longer time for stable engagement at weak branch sites.
That the same alanine point mutations in Prp5 alter intron engagement in both S. pombe and S. cerevisiae indicates that the interaction with SF3b components has been conserved since the last common predecessor of these two yeasts (∼380 million years ago [mya] [30]; by comparison, the last common predecessor of the entire mammalian class existed ∼165 mya [20]), and we expect to find analogous Prp5-SF3b interactions in mammalian cells. It has been argued that the ATPase Prp2 results in removal of SF3b proteins from the branch region (17, 37); based on the requirement of Prp5 in complex A formation (42), the appearance of SF3b proteins around the branch at this time (10, 42), and the physical interaction of Prp5 with SF3b proteins described here (disruption of which leads to defects in complex A formation in vitro and in intron definition in vivo), it is likely that one consequence of Prp5 ATPase action is the deposition of SF3b proteins around the branch site, resulting in stable U2 snRNP binding.
ACKNOWLEDGMENTS
We thank Haiteng Deng in the protein facility of the Rockefeller University for mass spectrometry identification and A. Moldón and M. Konarska for helpful discussions and critical reading of the manuscript.
This work was supported by NIH grant GM57829 to C.C.Q., by the National Natural Science Foundation of China grant 30972622 and Science and Technology Commission of Shanghai Municipality grant 09PJ1411500 to Y.-Z.X., and by a Cancer Center Support (core) grant from the NCI to the Albert Einstein College of Medicine. C.C.Q. is a scholar of the Irma T. Hirschl Trust.
Footnotes
Published ahead of print 7 November 2011
REFERENCES
- 1. Abovich N, Rosbash M. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89:403–412 [DOI] [PubMed] [Google Scholar]
- 2. Bahler J, et al. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951. [DOI] [PubMed] [Google Scholar]
- 3. Barabino SM, Blencowe BJ, Ryder U, Sproat BS, Lamond AI. 1990. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell 63:293–302 [DOI] [PubMed] [Google Scholar]
- 4. Behzadnia N, et al. 2007. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26:1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berget SM. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270:2411–2414 [DOI] [PubMed] [Google Scholar]
- 6. Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138:225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho S, et al. 2011. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. U. S. A. 108:8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Damgaard CK, et al. 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell 29:271–278 [DOI] [PubMed] [Google Scholar]
- 9. Dowhan DH, et al. 2005. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol. Cell 17:429–439 [DOI] [PubMed] [Google Scholar]
- 10. Gozani O, Potashkin J, Reed R. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18:4752–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. House AE, Lynch KW. 2006. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat. Struct. Mol. Biol. 13:937–944 [DOI] [PubMed] [Google Scholar]
- 12. Imai H, Chan EK, Kiyosawa K, Fu XD, Tan EM. 1993. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J. Clin. Invest. 92:2419–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jung DJ, Na SY, Na DS, Lee JW. 2002. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J. Biol. Chem. 277:1229–1234 [DOI] [PubMed] [Google Scholar]
- 14. Jurica MS, Moore MJ. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5–14 [DOI] [PubMed] [Google Scholar]
- 15. Kaüfer NF, Potashkin J. 2000. Analysis of the splicing machinery in fission yeast: a comparison with budding yeast and mammals. Nucleic Acids Res. 28:3003–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kress TL, Krogan NJ, Guthrie C. 2008. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol. Cell 32:727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lardelli RM, Thompson JX, Yates JR, III, Stevens SW. 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16:516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Long JC, Caceres JF. 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417:15–27 [DOI] [PubMed] [Google Scholar]
- 19. Mercier I, et al. 2009. Genetic ablation of caveolin-1 drives estrogen-hypersensitivity and the development of DCIS-like mammary lesions. Am. J. Pathol. 174:1172–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O'Brien SJ, et al. 1999. The promise of comparative genomics in mammals. Science 286:458–481 [DOI] [PubMed] [Google Scholar]
- 21. Perriman R, Ares M., Jr 2010. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol. Cell 38:416–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458:475–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Puig O, et al. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218–229 [DOI] [PubMed] [Google Scholar]
- 24. Reed R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340–345 [DOI] [PubMed] [Google Scholar]
- 25. Robberson BL, Cote GJ, Berget SM. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 10:84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Romfo CM, Alvarez CJ, van Heeckeren WJ, Webb CJ, Wise JA. 2000. Evidence for splice site pairing via intron definition in Schizosaccharomyces pombe. Mol. Cell. Biol. 20:7955–7970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schneider M, et al. 2010. Exon definition complexes contain the tri-snRNP and can be directly converted into B-like precatalytic splicing complexes. Mol. Cell 38:223–235 [DOI] [PubMed] [Google Scholar]
- 28. Schwer B. 2001. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 8:113–116 [DOI] [PubMed] [Google Scholar]
- 29. Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. 2008. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol. 15:183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sipiczki M. 2000. Where does fission yeast sit on the tree of life? Genome Biol. 1:REVIEWS1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith DJ, Query CC, Konarska MM. 2008. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol. Cell 30:657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stengel KF, et al. 2008. Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science 321:253–256 [DOI] [PubMed] [Google Scholar]
- 33. Tanner NK, Linder P. 2001. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell 8:251–262 [DOI] [PubMed] [Google Scholar]
- 34. Wagner S, Chiosea S, Nickerson JA. 2003. The spatial targeting and nuclear matrix binding domains of SRm160. Proc. Natl. Acad. Sci. U. S. A. 100:3269–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136:701–718 [DOI] [PubMed] [Google Scholar]
- 36. Wang Q, He J, Lynn B, Rymond BC. 2005. Interactions of the yeast SF3b splicing factor. Mol. Cell. Biol. 25:10745–10754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Warkocki Z, et al. 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 16:1237–1243 [DOI] [PubMed] [Google Scholar]
- 38. Wassarman KM, Steitz JA. 1992. The low-abundance U11 and U12 small nuclear ribonucleoproteins (snRNPs) interact to form a two-snRNP complex. Mol. Cell. Biol. 12:1276–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Webb CJ, Lakhe-Reddy S, Romfo CM, Wise JA. 2005. Analysis of mutant phenotypes and splicing defects demonstrates functional collaboration between the large and small subunits of the essential splicing factor U2AF in vivo. Mol. Biol. Cell 16:584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Webb CJ, Romfo CM, van Heeckeren WJ, Wise JA. 2005. Exonic splicing enhancers in fission yeast: functional conservation demonstrates an early evolutionary origin. Genes Dev. 19:242–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu JY, Maniatis T. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061–1070 [DOI] [PubMed] [Google Scholar]
- 42. Xu YZ, et al. 2004. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 23:376–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu YZ, Query CC. 2007. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol. Cell 28:838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]