Abstract
Drosophila melanogaster Pumilio is an RNA-binding protein that potently represses specific mRNAs. In developing embryos, Pumilio regulates a key morphogen, Hunchback, in collaboration with the cofactor Nanos. To investigate repression by Pumilio and Nanos, we created cell-based assays and found that Pumilio inhibits translation and enhances mRNA decay independent of Nanos. Nanos robustly stimulates repression through interactions with the Pumilio RNA-binding domain. We programmed Pumilio to recognize a new binding site, which garners repression of new target mRNAs. We show that cofactors Brain Tumor and eIF4E Homologous Protein are not obligatory for Pumilio and Nanos activity. The conserved RNA-binding domain of Pumilio was thought to be sufficient for its function. Instead, we demonstrate that three unique domains in the N terminus of Pumilio possess the major repressive activity and can function autonomously. The N termini of insect and vertebrate Pumilio and Fem-3 binding factors (PUFs) are related, and we show that corresponding regions of human PUM1 and PUM2 have repressive activity. Other PUF proteins lack these repression domains. Our findings suggest that PUF proteins have evolved new regulatory functions through protein sequences appended to their conserved PUF repeat RNA-binding domains.
INTRODUCTION
Precise regulation is required for expression of the appropriate quantity of proteins at the proper time and location. Posttranscriptional regulation of mRNAs is an integral control point mediated by cis-acting sequences and trans-acting regulators (18). PUF (Pumilio and Fem-3 binding factor) proteins are a family of mRNA regulators defined by a conserved RNA-binding domain (59). PUFs exert their function by selectively binding to single-stranded RNA sequences with high affinity and specificity (33, 55, 64, 65). Drosophila melanogaster Pumilio, the focus of this study, binds the consensus sequence 5′-UGUANAUA (19, 38, 57, 64).
Pumilio controls diverse processes, including stem cell proliferation (2, 15, 32, 42), motor neuron function, and memory formation (12, 35, 36, 46). Pumilio was initially identified by its requirement for embryonic development. Mutations in Pumilio disrupt abdominal segmentation (30). Early embryonic development is regulated through intricate expression patterns of maternally derived mRNA transcripts, while the genome is transcriptionally silent (60). During this stage, Pumilio regulates the mRNA encoding Hunchback (3, 29, 51). For proper development, Hunchback protein must be restricted to the embryonic anterior but not the posterior, yet the mRNA is distributed throughout the embryo (51, 52). The zinc finger protein Nanos spatially restricts Hunchback expression (31, 40, 58). A gradient of Nanos emanates from the posterior pole, opposing the Hunchback gradient (54). Two RNA sequences located in the 3′ untranslated region (3′ UTR) of Hunchback mRNA, Nanos response elements (NREs), are necessary for repression of Hunchback (58). The NREs are, in fact, binding sites for Pumilio, which is evenly dispersed throughout the embryo (3, 34, 38). In the posterior, Nanos partners with Pumilio on the NREs to form a trimeric Nanos-Pumilio-NRE complex that represses Hunchback (48).
Pumilio repression correlates with shortening of the 3′ poly(A) tail of target mRNAs (i.e., deadenylation) (5, 16, 58, 61). Yeast and Caenorhabditis elegans PUFs also enhance deadenylation (20, 21, 24, 25, 41, 49). However, multiple lines of evidence indicate that additional repression mechanisms exist. Pumilio inhibits mRNAs that lack a poly(A) tail, implicating a poly(A)-independent mechanism (5). Pumilio repression was shown to be dependent on the 5′ 7-methyl guanosine cap (5); however, in the Drosophila eye, Pumilio inhibited a reporter whose translation was driven by an internal ribosome entry site, suggesting a cap-independent mechanism (57). Therefore, Pumilio likely uses multiple means to repress mRNAs, though the precise mechanism(s) remains unknown.
To inhibit Hunchback, Pumilio is thought to recruit corepressors. Assembly of the Pumilio-Nanos complex on the Hunchback NRE recruits the Brain Tumor protein (47). Like Pumilio and Nanos, Brain Tumor promotes formation of the correct Hunchback gradient and abdominal segments (47). A lack of Brain Tumor shifts the Hunchback gradient and limits segmentation. A similar phenotype is produced by mutations in the eIF4E homologous protein, 4EHP, which partners with Brain Tumor (8). 4EHP competes with eIF4E for binding to the 5′ cap structure and inhibits translation (9). Inactivation of 4EHP or mutations that abrogate its recruitment shift the Hunchback gradient toward the posterior and reduce abdominal segmentation, though the effect is not fully penetrant (8). Therefore, recruitment of 4EHP by Brain Tumor is proposed to interfere with cap-dependent translation to refine the Hunchback protein gradient (8).
Nanos may be an obligatory cofactor for Pumilio repression. Supporting this notion, Nanos is necessary for Pumilio repression of Hunchback, cyclin B, and Paralytic in the embryonic posterior, primordial germ cells, and larval neurons, respectively (26, 37, 58). One study suggested that the main function of Pumilio is to recruit Nanos (26). Orthologs of Nanos serve as cofactors for PUFs in C. elegans and Xenopus (28, 39). However, PUFs in yeast repress, though no Nanos ortholog is present. Evidence in Drosophila also hints that Nanos might not be essential in all contexts. For instance, Pumilio regulates Bicoid and cyclin B in the anterior of the embryo, where Nanos is not detected (16, 53). Therefore, the universal necessity of Nanos in Pumilio repression remains questionable.
The C-terminal RNA-binding domain (RBD) of Pumilio binds Nanos and Brain Tumor, which in turn recruits 4EHP (8, 14, 47, 48). Because overexpression of the Pumilio RBD partially rescued embryonic segmentation defects in pumilio mutant embryos, this region was thought to be sufficient for function (57). Biochemical studies focused on the 336-amino-acid (336-aa) RBD because it is amenable to purification, whereas full-length Pumilio (1,533 aa) has been recalcitrant (63, 64). The functions of regions outside the RBD are obscure, though evidence hints at their importance (3, 36, 37, 57). Analysis of the molecular functions of these sequences has awaited development of new approaches to measure their activities.
In the research presented here, we develop assays that measure repression by Pumilio and Nanos. We uncover two modes of Pumilio-mediated repression: a Nanos-dependent mode and Pumilio repression that is independent of Nanos. We examine the roles of corepressors Brain Tumor and 4EHP and find that they are dispensable. Furthermore, we engineered a new Pumilio protein with altered RNA-binding specificity to direct repression of a new target mRNA. A key discovery was that full-length Pumilio mediates robust repression, whereas the RNA binding domain displays weak activity. The major repressive activity of Pumilio resides within three unique repression domains in the protein's N terminus. We show that equivalent regions of human PUFs also exhibit repressive activity.
MATERIALS AND METHODS
Plasmids.
To create the pAc5.1 FF control, Firefly luciferase was PCR amplified from pGL4.13 (Promega) and inserted into plasmid pAc5.1/V5-His A (Invitrogen). Reporter plasmids were created by inserting the Renilla luciferase open reading frame (ORF) with a minimal 3′ UTR into pAc5.1. This 3′ UTR contains a multiple cloning site and cleavage and polyadenylation signal from psiCHECK1 (Promega). To create the reporters, oligonucleotides encoding wild-type NREs (Rn1xNRE and Rn3xNRE) or mutant NREs (Rn3xNREmut) were inserted into XhoI and NotI sites in multiple cloning site of pAc5.1 Renilla luciferase. The NRE sequences, derived from Drosophila hunchback (NM_169234), are as follows (with the Pum site underlined and mutations in boldface): NRE, 5′-UUGUUGUCGAAAAUUGUACAUAAGCCAA; NRE mutant, 5′-UUCAUCACGAAAAUACAACAUAAGCCAA; and NRE UGG, 5′-UUGGUGGCGAAAAUUGGACAUAAGCCAA.
The RnMS2 reporter plasmid for the tethered-function assays was created by inserting oligonucleotides containing two MS2 binding sites into the XhoI and NotI sites in the 3′ UTR of pAc5.1 Renilla luciferase. The sequence of the tandem MS2 binding sites is 5′-AAAACATGAGGATCACCCATGTCTGCAGGTCGACTCTAGAAAACATGAGGATCACCCATGTC. (The stem-loops are underlined.) Drosophila Pumilio (NP_731315.1) and the Pumilio RBD (aa 1091 to 1426) were amplified by (RT-PCR) from oligo(dT)-primed cDNA from Schneider 2 cells and inserted into pIZ/V5-His vector (Invitrogen). Nanos (NP_476658.1) was cloned from whole fly cDNA and also inserted into the pIZ/V5-His vector. Mutations in Pumilio and the RBD were created by QuikChange site-directed mutagenesis (Stratagene). RNA-binding-defective Pumilio and RBD were created by mutations S1342A, N1343A, and E1346A of the seventh PUF repeat. The Pumilio R6SYE mutant was created by mutations N1306S and Q1310E of the sixth PUF repeat. The Pumilio and RBD mutants F1367S or G1330D were also created by site-directed mutagenesis. To create an RNA-binding-defective Nanos mutant, amino acid C354Y was mutated by site-directed mutagenesis. For the tethered-function expression vectors, DNA encoding MS2 coat protein was amplified from the pLexA N55K three-hybrid vector and fused in-frame to the N terminus of Pumilio. Control plasmid pIZ MS2 was created by inserting MS2 coding sequence into the pIZ plasmid. The control pIZ-HT plasmid was created by inserting the HaloTag (Promega) open reading frame (ORF) into pIZ plasmid. Brat (NP_476945.1) or 4EHP (NP_788729.1) coding sequences were amplified from S2 cell cDNA and inserted into pIZ-HT to create HT-Brat and HT-4EHP.
Cell culture.
D.mel-2 cells (Invitrogen) were cultured in Sf-900 III serum-free medium (Invitrogen) with 5 ml/liter penicillin-streptomycin using standard cell culture techniques. Cells were grown at 28°C.
Transfections.
D.mel-2 cells were transfected with plasmid DNA using Effectene (Qiagen) according to the manufacturer's specifications. Unless otherwise noted, standard transfection conditions of 2.2 ml per well of a 6-well plate are as follows: 1.6 ml D.mel-2 cells (1.5 × 106cells/ml), 600 μl Sf-900 III media, 10 ng of Renilla reporter plasmid DNA, 5 ng firefly control plasmid DNA, and Effectene. For transfection of Pumilio expression vectors, 400 ng of DNA was used unless otherwise noted in the figures. For Nanos expression, 10 ng was the standard amount, unless otherwise noted. Cells were harvested for dual-luciferase assay, Western blotting, TMR labeling and fluorescence detection, and quantitative RT-PCR (qRT-PCR) after 2 days of growth following transfection. Where necessary, the total DNA transfected in each sample was held constant by balancing transfection with an empty pIZ vector.
RNAi.
For RNA interference (RNAi), double-stranded RNAs corresponding to each target gene were generated by in vitro transcription from DNA templates. The templates were created by PCR amplification of regions of 250 to 600 bp of open reading frame from either plasmid vectors or cDNA from D.mel-2 cells. Both forward and reverse PCR primers had T7 promoters appended. The oligonucleotides used are listed as follows, with the T7 sequence underlined and the gene-specific sequence in boldface: LacZ control, forward primer, 5′-dGGATCCTAATACGACTCACTATAGGGTGACGTCTCGTTGCTGCATAAAC, and reverse primer, 5′-dGGATCCTAATACGACTCACTATAGGGGGCGTTAAAGTTGTTCTGCTTCATC; Pumilio, forward primer, 5′-dGGATCCTAATACGACTCACTATAGGGGTCAAGGATCAGAATGGCAATCATGT, and reverse primer, 5′-dGGATCCTAATACGACTCACTATAGGGCTTCTCCAACTTGGCATTGATGTGC; Nanos, forward primer, 5′-dGGATCCTAATACGACTCACTATAGGGCATTCCACTCGCCACCCACTGG, and reverse primer, 5′dGGATCCTAATACGACTCACTATAGGGCTAAACCTTCATCTGTTGCTTGTAGTAAC; Brain Tumor-1, forward primer, , and reverse primer, 5′-dGGATCCTAATACGACTCACTATAGGGCATACCCACTGGCGCCAGTTGG; Brain Tumor-2, forward primer, , and reverse primer, 5′-dGGATCCTAATACGACTCACTATAGGGGGTGTGACTGTTGGTGGTGGCC; 4EHP-1, forward primer, , and reverse primer, 5′-dGGATCCTAATACGACTCACTATAGGGCAATGGGCCTTTATTAATTGAAACATA; and 4EHP-2, forward primer, , and reverse primer, 5′-dGGATCCTAATACGACTCACTATAGGGCGACCATGTGCAGCGACTTGC.
From each PCR template, double-stranded RNA (dsRNA) was transcribed in vitro with the T7 RiboMAX large-scale RNA production system (Promega), treated with Turbo DNase (Ambion) for 3 h, and purified using the SV total RNA isolation system (Promega). For knockdown of each gene's expression, 6 μg of dsRNA per well of a 6-well plate was added to cells 10 min before transfection of reporters and expression vectors.
Luciferase assays.
Luciferase assays were performed 2 days posttransfection. To do so, 100 μl of transfected D.mel-2 cells was plated into three or four wells of a 96-well plate. Firefly expression and Renilla luciferase expression were measured using the Dual-Glo luciferase assay (Promega) according to the manufacturer's specifications and the GloMax Multi+ detection system (Promega) luminometer. The measured relative light units (RLU) were used to calculate a relative response ratio (RRR) using the equation RRR = Renilla RLU/firefly RLU. A percent repression value was then calculated as percent repression = 100 × (1 − RRRvariable/RRRmutant). For Nanos-stimulated repression, RRRmutant corresponds to the Nanos C354Y mutant. For Pumilio repression, Pumilio mutR7 (S1342A N1343A E1346A) was used as the mutant control. For RNA interference (RNAi)-treated samples, percent repression was calculated for each sample relative to the negative control Pum mutR7 treated with LacZ control dsRNA using the equation percent repression = 100 × (1 − RRRvariable/RRRcontrol). To measure activation of the Pumilio RBD R6SYE by Nanos, the RRRmutant control was measured from cells expressing RBD mutR7. Percent repression in the tethered-function assay was determined relative to the control samples expressing MS2CP from the pIZ MS2CP plasmid, using the equation percent repression = 100 × (1 − RRRvariable/RRRMS2CP). To measure experimental error, we calculated standard error of the mean (SEM) from triplicate or quadruplicate samples in each experiment. The reported SEMs are from technical replicates and are representative of multiple biological replicates performed at different times from different cell populations. Data were analyzed using Microsoft Excel. A graph of Pumilio repression relative to fold overexpression was created using the GraphPad Prism software.
Western blotting.
For Western blotting, 1-ml aliquots were taken from the same transfected D.mel-2 samples used for dual luciferase expression analysis. Two days posttransfection, cells were centrifuged at 1,000 × g for 3 min and pellets were lysed for 1 h on ice in lysis buffer (0.5% Igepal CA-630 [USB], 50 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 2 mM MgCl2, 150 mM NaCl, 20 nM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 μg/ml leupeptin). Lysates were cleared by centrifugation at 16,000 × g for 2 min, and supernatants were saved as whole-cell protein extracts. Extracts were separated via SDS-polyacrylamide (12%) gel electrophoresis (Tris-glycine running buffer), and proteins were transferred onto Immobilon-FL polyvinylidene difluoride (PVDF) membranes (Millipore). Membranes were blocked in blocking buffer (phosphate-buffered saline [PBS], 5% milk, 0.01% Tween 20), probed with V5 antibody (Invitrogen), washed in buffer, probed with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Thermo Scientific), and washed again, and then ECL (enhanced chemiluminescence) Western blotting reagent (Pierce) was added, and luminescence was imaged on autoradiography film.
Fluorescent labeling and visualization of HaloTag protein constructs.
Protein extracts from HaloTag expression cells were harvested as above and mixed with HaloTag TMR Ligand (900 nM final) for 30 min on ice in the dark. After labeling, extracts were separated via SDS-polyacrylamide (4 to 20%) gel electrophoresis (Tris-glycine running buffer), and protein fluorescence (532 excitation [Ex]/580 emission [Em]) was measured with a Typhoon Trio+ imager (GE Healthcare). Relative fluorescence was quantified using ImageQuant TL software (GE Healthcare).
RNA isolation and cDNA preparation.
For isolation of RNA, 1 ml of transfected D.mel-2 cells was centrifuged at 1,000 × g for 3 min, washed twice in PBS, and lysed with QIAzol reagent (Qiagen) according to the manufacturer's specifications. Upon ethanol precipitation and resuspension, whole-cell RNA was treated with Turbo DNase (Ambion) for 3 h before adding stop solution (Promega) (20 mM EGTA [pH 8.0]). For isolates prepared from the tethered-function assay, RNA was purified from cell pellets using Maxwell LEV simplyRNA cells (Promega). RNAs were primed with random hexamers (IDT) for synthesis of cDNAs using GoScript reverse transcriptase (Promega). The final concentration of RNA in RT reactions was 60 ng/μl.
qPCR.
To measure endogenous mRNA levels, quantitative PCR (qPCR) was performed on 5 μl of cDNA product in a 50-μl reaction using 100 nM specific primers and GoTaq qPCR master mix (Promega). To measure firefly and Renilla luciferase mRNAs, multiplexed qPCR was performed in 25-μl reactions with 200 nM fluorescent primers (Biosearch Technologies) and Plexor master mix (Promega). Reactions were performed with a C1000 thermal cycler equipped with the CFX96 real-time system (Bio-Rad). Standard control reactions were performed without reverse transcriptase or without RNA template. For GoTaq reactions, cycling conditions were performed using the following sequence of steps: (i) step 1 was 95°C for 3 min, (ii) step 2 was 95°C for 10 s, (iii) step 3 was 65°C for 30 s, and (iv) step 4 was 72°C for 40 s, with steps 2 to 4 repeated for 40 cycles. For Plexor reactions, (i) step 1 was 95°C for 2 min, (ii) step 2 was 95°C for 5 s, and (iii) step 3 was 60°C for 35 s, with steps 2 and 3 repeated for 40 cycles. In the case of Fig. 1B, GoTaq qRT-PCR was performed for 30 cycles and products were visualized by 0.8% agarose gel electrophoresis. Each qPCR was analyzed via a thermal melting curve and gave a single peak with the expected melting temperature. Amplification efficiencies of each primer set were optimized. Plexor primers were optimized at 100% for the Plexor qPCR protocol, while all other primers had efficiencies between 90 and 110% with 65°C elongation steps.
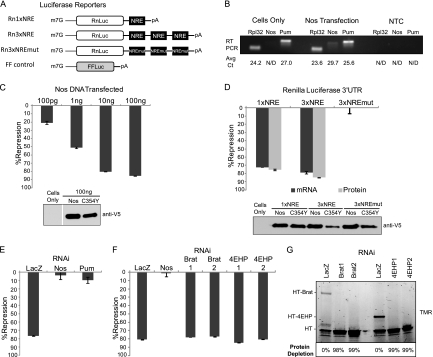
Fig 1.
Nanos stimulates Pumilio-mediated repression. (A) Diagrams of Renilla (Rn) and firefly (FF) luciferase reporters with wild-type (NRE) or mutant (NREmut) Nanos response elements are depicted. (B) qRT-PCR analysis of Nanos (Nos), Pumilio (Pum), or positive-control Rpl32 mRNAs from D.mel-2 cells (cells only) or cells transfected with Nos expression plasmid. Average cycle threshold (Avg Ct) values are indicated below corresponding samples. NTC, no-template control reactions; N/D, not detected. (C) Percent repression by Nos, reported for the indicated mass of transfected plasmid, was calculated relative to mutant Nanos C354Y. See Fig. S2A in the supplemental material for raw data. (D) Percent repression of luciferase protein (dual-luciferase assay) and mRNA (qRT-PCR). See Table S1 in the supplemental material for the corresponding data. (E) Percent repression by Nanos in cells treated with dsRNA for LacZ (negative control), Nos, Pum, or (in panel F) Brain Tumor (Brat) and 4EHP. In each figure, Western blotting confirmed expression of V5 epitope-tagged proteins. Data for panels E and F are reported in Table S2 in the supplemental material. (G) Fluorescent imaging of SDS-polyacrylamide (4 to 20%)-separated TMR-labeled HaloTag (HT) Brat or 4EHP proteins. Protein depletion by dsRNA to Brat or 4EHP was calculated relative to control LacZ RNAi.
Cycle thresholds (CTs) were measured using CFX Manager software (Bio-Rad) for GoTaq reactions, while the raw data were imported into Plexor Analysis Desktop (Promega) for Plexor reactions. Differences in mRNA levels were calculated using the ΔΔCT method. For analysis of RNAi depletion of endogenous mRNAs (i.e., Pumilio, Nanos, Brain Tumor, and 4EHP), CT values were measured and normalized to the internal control Rpl32 mRNA for each sample using the equation ΔCT target RNAi = CT target − CT control. A normalized ΔCT control RNAi was also calculated for each mRNA in the LacZ dsRNA-treated samples. To measure relative changes in each mRNA level, ΔΔCT was calculated for each gene as ΔΔCT = ΔCT target RNAi − ΔCT control RNAi. The fold change in mRNA level was then calculated as 2−ΔΔCT. For the measurement of reporter mRNA levels, the same method was used, but the normalizations were calculated relative to the internal control firefly mRNA (FF control). The ΔCT for each sample was calculated as ΔCT = CTRenilla − CT firefly. To measure changes in reporter mRNA levels induced by Pumilio or Nanos, ΔΔCT was calculated as ΔCT WT − ΔCT mutant, where “mutant” refers to samples expressing RNA-binding-defective Pumilio (mutR7) or Nanos (C354Y), as indicated in the figure legends. The fold change was then calculated from 2−ΔΔCT. Percent repression values were derived using the equation percent repression = 100 × 1-fold change.
qPCR primer sequences.
The qPCR primer sequences are as follows: firefly luciferase reporter, forward primer, 5′-dGATCCTCAACGTGCAAAAGAAGC, and reverse primer, 5′-d carboxyfluorescein (FAM)-isoC-TCACGAAGGTGTACATGCTTTGG; Renilla luciferase reporter, forward primer, 5′-d CAL Fluor Orange 560-isoC-CGCAACTACAACGCCTACCTTC, and reverse primer, 5′-dCCCTCGACAATAGCGTTGGAAAA; Rpl32, forward primer, 5′-dGCCCAAGGGTATCGACAACAG, and reverse primer, 5′-dGCACGTTGTGCACCAGGAAC; Pumilio, forward primer, 5′-dGCCTGATGACCGATGTCTTTGG, and reverse primer, 5′-dCGATTTCCTGCTGCTGCTCC; Nanos, forward primer, 5′-dCTGGCTCGATGCAGGATGTG, and reverse primer, 5′-dGTCTGCAGCTGGGCAGGATT; Brain Tumor, forward primer, 5′-dCAACTACAGACGGGCATTCAGG, and reverse primer, 5′-dGCCCGAATGTAACCAAAGGTG; and 4EHP, forward primer, 5′-dCCAGCGTGCAGCAGTGGTGG, and reverse primer, 5′-dCAAACGTTCTCCCAGGCCCG.
RESULTS
A cell-based, reporter mRNA assay recapitulates Nanos-dependent repression.
To dissect repression by Pumilio and Nanos, we used the D.mel-2 cell line, derived from Drosophila embryo Schneider 2 cells (45). A reporter mRNA encoding Renilla luciferase was created with efficient cleavage and poly-adenylation signals within a minimal 3′ UTR. To study regulation by Pumilio and Nanos, one or three NRE sequences (58) were inserted into the 3′ UTR to create Rn1×NRE and Rn3×NRE, respectively (Fig. 1A). As a control, the NRE sequences were mutated by changing the U1G2U3 trinucleotide, crucial for Pumilio binding, to ACA in Rn3×NREmut (Fig. 1A). Reporter plasmids were individually transfected into D.mel-2 cells. As an internal control, a firefly luciferase plasmid was cotransfected (FF control) (Fig. 1A). To measure protein expression, the enzymatic activities of Renilla and firefly luciferases were assayed. The transfection efficiency of each sample was normalized by calculating a relative response ratio of Renilla activity divided by firefly activity. Relative response ratios and standard errors for all experiments are reported in Tables S1 to S17 in the supplemental material. We observed that Renilla expression from reporters with three NREs was nearly equivalent to that in reporters lacking NREs or with mutant NREs (see Fig. S1A in the supplemental material). Titration of the Renilla reporters from 1 to 100 ng did not alter this result (data not shown). However, RNAi-mediated depletion of endogenous Pumilio caused a 1.8-fold increase in Rn3×NRE expression, but did not affect Rn3×NREmut (see Fig. S1B), indicating that endogenous Pumilio inhibits the mRNA with Pumilio binding sites. In the same experiment, dsRNA against Nanos had no effect (see Fig. S1B).
A possible explanation for the minimal NRE-dependent regulation is that a key regulator—Pumilio or Nanos—may be limiting. Therefore, we measured expression of Pumilio and Nanos in D.mel-2 cells. Reverse transcription followed by quantitative PCR (qRT-PCR) revealed that D.mel-2 cells express Pumilio mRNA (cycle threshold [CT] of 27.0); however, Nanos mRNA was not detected. A constitutively expressed ribosomal subunit Rpl32 was detected at a CT of 24.2 (Fig. 1B). Nanos mRNA was detectable when cells were transfected with a Nanos expression plasmid, demonstrating that the qRT-PCR assay is valid (Fig. 1B). Nanos, Pumilio, and Rpl32 were not detected in control reaction mixtures lacking template (no-template control [NTC]) (Fig. 1B) or reverse transcriptase (data not shown). These results demonstrate that D.mel-2 cells express Pumilio but not detectable amounts of Nanos, supported by a recent microarray study of mRNA expression in Schneider 2 cells (7).
The effect of Nanos was tested by transfecting increasing amounts of a Nanos-expressing plasmid into D.mel-2 cells along with the Rn3×NRE reporter (Fig. 1C) (Raw data are reported in Fig. S2A in the supplemental material). As a control, equivalent samples were prepared using an inactive Nanos mutant wherein the cysteine at position 354 of the RNA-binding zinc finger was changed to a tyrosine (C354Y) (11). Western blotting confirmed expression of both wild-type and mutant Nanos proteins (Fig. 1C). To measure Nanos-mediated repression, luciferase activities were measured and normalized. Next, a percent repression value was determined for each amount of transfected Nanos, relative to the equivalent amount of Nanos C354Y. Nanos inhibited expression of the Rn3×NRE reporter mRNA in a dose-dependent manner (Fig. 1C). Transfection of 100 ng of Nanos expression plasmid caused 85.6% repression of Renilla expression (Fig. 1C). Titration of the Nanos expression plasmid over a 2,000-fold range demonstrated that Nanos repression plateaus at 10 ng and continues to repress greater than 80% up to 200 ng with no observed squelching effect (see Fig. S2B). Therefore, this assay recapitulates the ability of Nanos to repress NRE-bearing target mRNA.
Nanos inhibits protein expression and reduces levels of target mRNAs.
Nanos targets mRNAs through direct interactions with its partner, Pumilio, and the NRE (48). To address the necessity of the NRE for Nanos-directed regulation, the effect of Nanos on various Renilla reporters was tested (Fig. 1A). Reporters were transfected into cells with plasmid expressing either wild-type or mutant Nanos C354Y. Each sample was split into three portions: luciferase activity assays were performed on one, qRT-PCR was performed on RNA from another, and Western blots were performed on the final portion (Fig. 1D). Wild-type but not mutant Nanos repressed luciferase expression from both Rn1×NRE (76% repression) and Rn3×NRE (85% repression) reporters, whereas the mutant Rn3×NREmut reporter was not affected (Fig. 1D). Repression of mRNA level corresponded to the observed change in luciferase protein. Rn1×NRE and Rn3×NRE mRNAs were reduced by 72% and 79%, respectively, whereas the Rn3×NREmut mRNA was not affected (Fig. 1D). Western blot analysis confirmed expression of wild-type and mutant Nanos proteins (Fig. 1D). These data show that the NRE is necessary for Nanos-directed regulation, in agreement with data from embryos (58), and that Nanos potently decreases reporter protein and mRNA levels.
Depletion of Pumilio and Nanos, but not Brain Tumor or 4EHP, abrogates Nanos-dependent repression.
Pumilio and Nanos assemble on the NRE and are thought to recruit Brain Tumor and 4EHP to inhibit translation (8, 47). We sought to examine the role of each corepressor in Nanos-dependent repression. Using qRT-PCR, we confirmed expression of endogenous Brain Tumor and 4EHP mRNAs in D.mel-2 cells with specific CT values of 23.3 and 24.2, respectively. We then depleted each protein by RNA interference using specific double-stranded RNAs (dsRNAs). As a negative control, cells were treated with dsRNA corresponding to the bacterial LacZ gene. Rn3×NRE reporter, and Nanos expression plasmids were subsequently transfected into these dsRNA-treated cells. As before, Nanos dependent repression was calculated relative to mutant Nanos C354Y. Depletion of exogenously expressed Nanos or endogenously expressed Pumilio almost completely abolished repression, whereas nontargeting LacZ dsRNA had no effect (Fig. 1E). These results demonstrate that Pumilio is essential for Nanos-dependent repression of the NRE-containing mRNA and validate the RNAi efficacy. Surprisingly, depletion of Brain Tumor or 4EHP had no effect on Nanos-dependent repression (Fig. 1F). This result was corroborated using two different dsRNAs that targeted separate regions of the Brain Tumor or 4EHP coding sequences (Fig. 1F). We confirmed depletion of each mRNA by qRT-PCR. Exogenously expressed Nanos mRNA was depleted by up to 95% by treatment with Nanos dsRNA (data not shown). Across multiple experiments, specific dsRNAs depleted Pumilio mRNA by up to 67%, Brain Tumor mRNA by up to 80%, and 4EHP mRNA by up to 84% (data not shown). To further verify the RNAi efficacy, we tested the ability to deplete Brain Tumor or 4EHP proteins by overexpressing fluorescently labeled HaloTag (HT) fusions of Brain Tumor or 4EHP (Fig. 1G). Both dsRNAs for Brain Tumor and 4EHP completely ablated expression (98 to 99% depletion) of HT-Brain Tumor and HT-4EHP, respectively, as measured by fluorescence detection (Fig. 1G). As an internal control, Halotag was also overexpressed and was not depleted by the dsRNAs (Fig. 1G). We conclude that Nanos and Pumilio collaborate to repress. Moreover, Nanos-dependent repression remains effective when Brain Tumor and 4EHP are significantly depleted.
Pumilio represses mRNAs independent of Nanos.
The observation that Pumilio represses mRNAs in the anterior of the developing embryo, where Nanos is not detected, suggests that Nanos may not be absolutely required (16, 53). While depletion of endogenous Pumilio in D.mel-2 cells increased RnLuc3×NRE expression, RNAi of Nanos had no effect (see Fig. S1B in the supplemental material). Since Nanos is undetectable in D.mel-2 cells, this indicates that Pumilio can repress independently of Nanos (Fig. 1B). We tested the ability of Pumilio to repress NRE-containing reporters by transfecting a Pumilio expression plasmid, which caused dose-dependent repression of Rn3×NRE (Fig. 2A). (Raw data are reported in Fig. S3A in the supplemental material.) At the highest expression level, Pumilio repressed expression by 66% (Fig. 2A). A broader titration range of Pumilio is shown in Figure S3B. Repression increased as the dosage of Pumilio increased, with no indication of squelching (see Fig. S3B). The assay is very responsive to modest increases in Pumilio. Overexpression at maximum repression, measured by qRT-PCR, was 1.34-fold above that of endogenous Pumilio (see Fig. S3C). Pumilio repression values were calculated relative to equivalent amounts of an inactive mutant Pumilio (Pum mutR7) that is incapable of binding to RNA by way of alanine substitutions in the RNA recognition amino acids in the seventh PUF repeat (Fig. 2A). Both wild-type and mutant Pumilio proteins were expressed (Fig. 2A) and increased in response to the mass of transfected expression plasmid (see Fig. S3D). This data indicates that Pumilio can indeed repress in a Nanos-independent fashion.
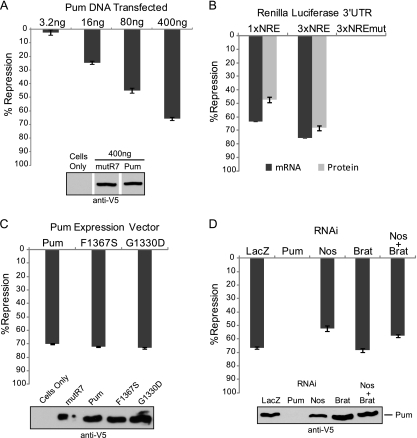
Fig 2.
Pumilio represses independent of Nanos and Brain Tumor. (A) Percent repression of Rn3×NRE reporter by the indicated mass of transfected Pumilio (Pum) plasmid, relative to mutant Pum mutR7. Data are reported in Fig. S3A in the supplemental material. (B) Percent repression by Pum of the Renilla luciferase protein and mRNA expression for the indicated reporters. (C) Percent repression of the Rn3×NRE reporter by wild-type, mutant F1367S, or mutant G1330D Pumilio. Repression was calculated relative to Pum mutR7. (D) Percent repression by wild-type Pum (400 ng) in cells treated with dsRNA to LacZ control, Pum, Nanos (Nos), Brain Tumor (Brat), or both Nos and Brat. Efficacy of RNAi was demonstrated in Fig. 1F. All data points represent mean values with standard errors of the mean indicated. Western blotting confirmed expression of V5 epitope-tagged proteins. The data for panels B, C, and D are reported in Tables S3 to S5 in the supplemental material.
Pumilio reduces protein and mRNA levels in an NRE-dependent manner.
To verify that Nanos-independent Pumilio repression is mediated by the NRE, we examined the effect of Pumilio on reporter protein and mRNA levels using Rn1×NRE, Rn3×NRE, and control Rn3×NREmut reporters. Pumilio potently decreased NRE-containing reporter mRNA levels and caused a corresponding reduction in Renilla luciferase protein activity (Fig. 2B). Rn1×NRE and Rn3×NRE mRNAs were reduced 63% and 76%, respectively (Fig. 2B). Luciferase expression was repressed by 47% for Rn1×NRE and 68% for Rn3×NRE (Fig. 2B). Mutation of the NRE alleviates repression entirely (Rn3×NREmut) (Fig. 2B). These results demonstrate that Pumilio overexpression elicits Nanos-independent repression of NRE-containing reporters by reducing protein and mRNA expression.
Nanos and Brain Tumor are not necessary for Pumilio-mediated repression.
We further examined the potential involvement of Nanos and Brain Tumor in Pumilio repression. Although Nanos expression could not be detected in D.mel-2 cells, we wished to use multiple strategies to rule out the possibility that trace amounts of Nanos, below the limit of detection, might be sufficient to aid Pumilio repression. First, we utilized previously identified Pumilio mutants that are inactive for interaction with either Nanos or Brain Tumor. The F1367S mutation in Pumilio blocks interaction with Nanos (13, 14). The G1330D mutation in Pumilio, originally identified as the pum680 allele that eliminates abdominal segmentation (30, 57), binds the NRE but is unable to recruit Brain Tumor into the Pumilio-Nanos-NRE complex (13, 47). We tested the ability of these mutants to repress Rn3×NRE reporter relative to wild-type Pumilio and mutant Pumilio mutR7. Neither F1367S nor G1330D affected Pumilio repression (Fig. 2C).
As an additional means of assessing participation of Nanos and Brain Tumor in Pumilio repression, cells were treated with corresponding dsRNAs to induce RNA interference, and the resulting impact on repression by overexpressed Pumilio was measured. As a positive control, RNAi depletion of Pumilio completely alleviated repression (Fig. 2D). Treatment with LacZ dsRNA had no effect on Pumilio repression (Fig. 2D). RNAi of Nanos, Brain Tumor, or simultaneous knockdown of Nanos and Brain Tumor had negligible effects on Pumilio repression (Fig. 2D). Therefore, using three approaches, we have shown that Pumilio can repress by a mechanism that is independent of Nanos and Brain tumor: (i) Pumilio represses in Nanos-deficient cells, (ii) mutations in Pumilio that inhibit Nanos and Brain Tumor ternary complex formation do not affect repression, and (iii) depletion of Nanos, Brain Tumor, or both does not alleviate Pumilio repression. These findings provide strong evidence indicative of a previously uncharacterized regulatory function of Pumilio, which we now explore.
The N terminus of Pumilio is necessary for optimal repression.
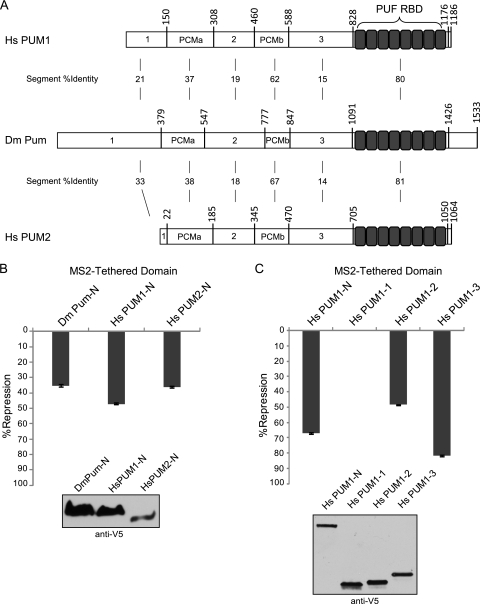
To characterize the domains of Pumilio that are necessary for repression, we examined the activities of the RNA-binding domain and full-length Pumilio. The RBD is composed of eight PUF repeats located at the C terminus of Pumilio protein (amino acids 1091 to 1426) (Fig. 3A) and is necessary and sufficient for high-affinity binding to the NRE RNA and interaction with corepressors Nanos and Brain Tumor (13, 14, 47, 48, 57, 63, 64). Outside of the RBD, no domains or motifs have been documented. However, within the large N-terminal region of Pumilio (aa 1 to 1090), we identified two regions conserved in PUF proteins from insects to vertebrates, designated Pumilio conserved motifs a and b (PCMa and PCMb, respectively) (Fig. 3A).
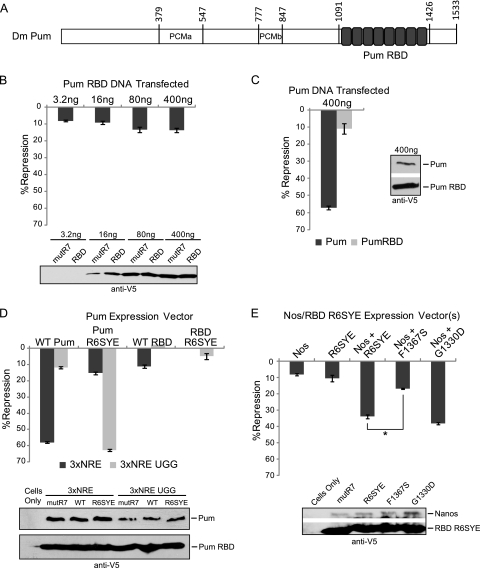
Fig 3.
Nanos stimulates repression through the RNA-binding domain of Pumilio. (A) Diagram of Pumilio (Pum) domains, including Pumilio conserved motifs PCMa and PCMb and the RNA binding domain (RBD) with PUF repeats indicated by gray boxes. (B) Percent repression of Rn3×NRE by Pum RBD, calculated relative to mutant Pum RBD mutR7. (C) Percent repression of Rn3×NRE by full-length Pum or RBD. (D) Percent repression of the Rn3×NRE with wild-type or UGG NREs by full-length Pum (wild type [WT Pum] and programmed mutant [Pum R6SYE]) or by Pum RBD (wild type [WT RBD] or programmed mutant [RBD R6SYE]). (E) Percent repression by Nanos (Nos) and Pum RBD R6SYE using the Rn3×NRE UGG reporter. Mutations of F1367S or G1330D in Pum RBD R6SYE block binding to Nanos or Brain Tumor, respectively. Mean values with standard errors are indicated in each graph. Statistical significance is indicated with P < 0.001 in a two-tailed t test. Western blotting confirmed expression of V5 epitope-tagged proteins. Data for panels B to E are reported in Tables S6 to S9 in the supplemental material.
Repression by the Pumilio RBD was measured relative to an inactive, RNA-binding defective mutant RBD mutR7. The RBD repressed the Rn3×NRE reporter by 8 to 15% (Fig. 3B). While repression increased slightly over a gradient of transfected RBD, the maximum level of repression did not exceed 15% (Fig. 3B). Under identical conditions, the full-length Pumilio protein repressed by 57%, while the RBD repressed by 11% (Fig. 3C). This difference cannot be attributed to poor protein expression of the RBD because Western blotting revealed that the RBD expressed to a higher level than full-length Pumilio (Fig. 3C). We scrutinized repression by Pumilio and the RBD over a 250-fold range of transfected plasmids (see Fig. S3B in the supplemental material). At each transfected amount, full-length Pumilio repressed greater than the RBD. Repression by the RBD never exceeded 15%, whereas repression by full-length Pumilio continued to increase up to 61% at the maximum amount of transfected plasmid (see Fig. S3B). The RBD mRNA was maximally overexpressed by 3.31-fold and full-length Pumilio by 1.34-fold, relative to endogenous Pumilio mRNA (see Fig. S3C). Comparison of conditions in which the mRNAs and proteins were overexpressed at similar levels (e.g., 400 ng Pumilio compared to 80 ng RBD) shows that Pumilio repressed 49%, while the RBD only repressed 11% (see Fig. S3D and E). Therefore, differential repression does not result from disparate expression levels. These results indicate that the N-terminal 1,090 amino acids of Pumilio contain the major repressive activity, illuminating a previously unknown function.
Programming Pumilio RNA-binding specificity confers repression of a new target mRNA.
We engineered a Pumilio protein with altered RNA-binding specificity that recognizes a new binding site, allowing examination of the activity of exogenously introduced Pumilio mutants without potential interference by the endogenous protein. Previous studies deciphered an RNA-binding code for PUF repeats (6, 33, 55). Three RNA recognition amino acids of each PUF repeat bind one nucleotide (55). The third base of the Pumilio binding site is a uracil highlighted in boldface: U1G2U3, which is recognized by amino acids N1306, Y1307, and Q1310 of the sixth PUF repeat. The two flanking RNA recognition amino acids, N1306 and Q1310, make hydrogen bonds, while Y1307 mediates base stacking interactions between uracil and the following nucleotide base, adenine (55). By changing the RNA recognition amino acids of repeat 6 (N1306S and Q1310E) while leaving intact the stacking residue (Y1307), we programmed the mutant Pumilio R6SYE to bind an NRE sequence with the U1G2G3 trinucleotide, instead of U1G2U3, in the Rn3×NRE UGG reporter.
We tested the ability of wild-type Pumilio to repress the Rn3×NRE and Rn3×NRE UGG reporter mRNAs. As expected, Pumilio repressed the wild-type NRE reporter by 58% but only slightly affected the Rn3×NRE UGG reporter by 12% (Fig. 3D). We then measured the activity of Pumilio R6SYE, which minimally repressed the wild-type NRE by 15% (Rn3×NRE) (Fig. 3D). In contrast, Pumilio R6SYE dramatically repressed the Rn3×NRE UGG reporter by 63%. This result demonstrates that PUF proteins can be programmed to repress new target mRNAs.
We then used Pumilio R6SYE to examine the activity of the RNA-binding domain relative to full-length protein. We considered that repression by the exogenous Pumilio RBD tested in Fig. 3B and C might be antagonized by endogenous Pumilio. This concern could be eliminated by the altered specificity approach. First, we confirmed that the wild-type Pumilio RBD repressed the Rn3×NRE reporter by 11% but was incapable of repressing the Rn3×NRE UGG reporter (Fig. 3D). Next, repression by RBD R6SYE was examined. RBD R6SYE repressed the UGG reporter weakly (5%) but had no effect on the reporter bearing wild-type NREs (Fig. 3D). Expression of Pumilio RBD and derivatives was confirmed by Western blotting (Fig. 3D). These results reaffirm that repression by the RBD is substantially deficient relative to that by full-length Pumilio.
The RBD is sufficient for Nanos stimulation of repression.
The effectiveness of Pumilio R6SYE created the opportunity to further examine Nanos-dependent repression. We hypothesized that inefficient repression by the RBD might be caused by the lack of Nanos in D.mel-2 cells. Because Nanos enhances repression by endogenous Pumilio of the wild-type NRE reporter (Fig. 1), we circumvented this issue using the altered-specificity approach. Using the Rn3×NRE UGG reporter, the ability of Pumilio RBD R6SYE to respond to Nanos was tested. While Nanos and the RBD R6SYE mutant have low activity when tested individually (Fig. 3E) (8% and 11% repression, respectively), when expressed together, they synergize to repress by 34% (Fig. 3E). To confirm that Nanos-dependent repression is mediated by interaction with the RBD, we tested the ability of Nanos to affect the RBD R6SYE mutant, F1367S, which disrupts the Pumilio-Nanos interaction (13). RBD R6SYE F1367S with Nanos repressed by 17%—less than additive compared to Nanos and RBD R6SYE controls alone (Fig. 3E); therefore, Nanos cannot synergize with the Pumilio RBD in the absence of a direct protein interaction. Using an R6SYE RBD with the G1330D mutation, we tested the requirement of interaction with Brain Tumor (47). Nanos stimulated repression by RBD R6SYE G1330D to 38% (Fig. 3E). We confirmed expression of Nanos and RBD R6SYE proteins by Western blotting (Fig. 3E). We conclude that repression by the RBD is enhanced by interaction with Nanos but does not require binding to Brain Tumor.
The N-terminal portion of Pumilio contains the major repressive activity.
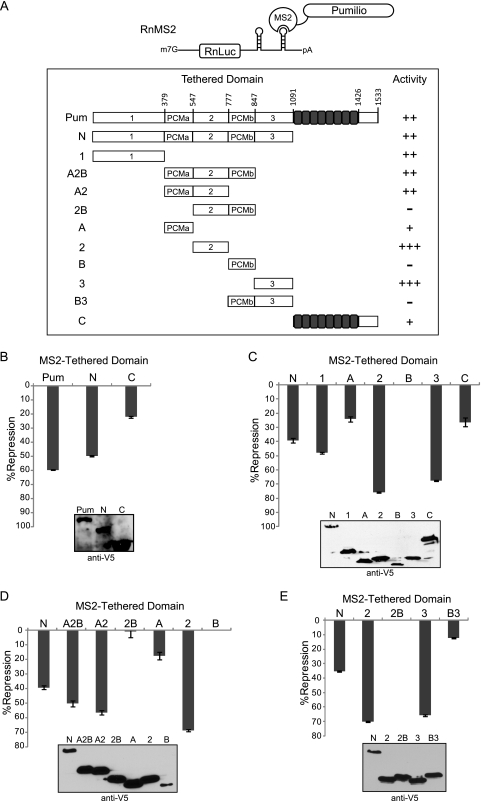
Full-length Pumilio exhibits greater repression than the RBD, indicating that the major repression domain resides outside the PUF repeats. To separate Pumilio repression and RNA-binding activities, we utilized a tethered-function approach. Pumilio or portions thereof were fused to the MS2 coat protein, which binds a specific RNA stem-loop. A Renilla luciferase reporter was constructed with two MS2 binding sites in a minimal 3′ UTR (RnMS2) (Fig. 4A). If the test protein represses when tethered by MS2, then reporter expression will be reduced. As a control, firefly luciferase was coexpressed. Luciferase activities were normalized by dividing Renilla signal by that of firefly to calculate a relative response ratio. Percent repression values of each test protein, summarized in Fig. 4A, were calculated relative to control MS2 protein.
Fig 4.
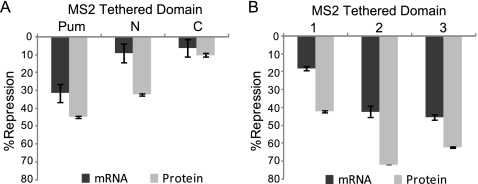
The N terminus of Pumilio contains multiple autonomous repression domains. (A) Diagram of tethered-function Renilla luciferase reporter with tandem stem-loop binding sites for MS2 coat protein in the 3′ UTR (RnMS2). Shown is a schematic of test proteins fused to MS2 coat protein (MS2) including full-length Pumilio (Pum) and segments with the indicated amino acid positions. Relative repression by Pum segments is based on the following scale: −, <10% repression; +, 10 to 30% repression; ++, 31 to 60% repression; and +++, >60% repression. (B) Percent repression of RnMS2 by tethered Pum, the N terminus (N), and C terminus (C), calculated relative to the MS2 control. (C) Percent repression of RnMS2 reporter by tethered Pum segments, with names corresponding to the diagrams in panel A. (D) Percent repression of RnMS2 by the indicated MS2-tethered Pum segments. (E) Percent repression by Pum regions 2 and 3 alone or fused to PCMb (2B or B3). Western blotting confirmed expression of V5 epitope-tagged proteins. Data from panels B to E are reported in Tables S10 to S13 in the supplemental material.
When tethered, full-length Pumilio repressed by 60% (Fig. 4B), a magnitude similar to that observed for repression of Rn3×NRE (Fig. 2A). The N-terminal two-thirds of Pumilio (N, aa 1 to 1090) repressed by 50%, whereas the C-terminal RBD (C, aa 1091 to 1533) repressed by 22% (Fig. 4B). Repression is dependent on tethering because, when not fused to MS2, full-length Pumilio and RBD had no effect on RnMS2 (data not shown). Consistent with results in Fig. 3, the C-terminal RBD represses inefficiently. Therefore, the N-terminal 1,090 amino acids contain the major repressive activity of Pumilio.
Multiple domains within the Pumilio N terminus have autonomous repressive activity.
We further dissected repression by the Pumilio N terminus using the tethered-function assay (Fig. 4C). Internal deletions did not cause loss of repression, thus we reasoned that the N terminus harbors multiple repression domains (see Fig. S4 in the supplemental material). Six segments of Pumilio were then separately fused to MS2 including amino acids 1 to 378 (region 1), 379 to 547 (PCMa; region A), 548 to 776 (region 2), 777 to 847 (PCMb; region B), 848 to 1090 (region 3), and 1091 to 1533 (region C) (Fig. 4A). When tethered, three segments repressed more efficiently than the N terminus: region 1, 48% repression; region 2, 76% repression; and region 3, 68% repression (Fig. 4C). PCMb (region B) did not repress while PCMa (region A) and region C repressed to lesser degree (24% and 26% repression) (Fig. 4C). Therefore, multiple domains of Pumilio can independently repress an mRNA.
Conserved motifs may regulate activity of an autonomous Pumilio repressor domain.
The role of PCMa and PCMb remained unclear. We reasoned that these domains may regulate Pumilio function. To investigate this idea, we created tethered constructs with one or both conserved motifs connected to a Pumilio repression domain (Region 2, aa 548 to 776) (Fig. 4D). By itself, region 2 had maximal activity (69% repression) (Fig. 4D). When PCMa was fused to this repression domain (region A2, aa 379 to 776), the activity remained robust (56% repression) (Fig. 4D). Strikingly, when PCMb was fused to this repression domain (region 2B) (aa 548 to 847), the activity was completely lost (Fig. 4D) (1% repression). In agreement with Fig. 4C, PCMa and PCMb exhibited weak or no repression on their own (Fig. 4D). When PCMb was fused to another repression domain (region B3, aa 777 to 1090), activity was also severely reduced (12% repression) (Fig. 4E). Western blots confirmed protein expression (Fig. 4D and E). This data indicates that PCMb inhibits repression domains in region 2 and region 3.
We measured the activity of a segment containing PCMa, region 2, and PCMb; this construct (region A2B, aa 379 to 847) repressed the mRNA by 50% (Fig. 4D). Deletion of PCMa from the tethered N terminus also leads to a small but significant decrease in repression (see Fig. S4A in the supplemental material). We speculate that PCMa may antagonize the negative regulatory function of PCMb, perhaps via autoinhibitory interactions.
The N-terminal Pumilio repression domains reduce protein and mRNA levels.
Full-length Pumilio reduces protein expression and mRNA levels with comparable efficiency (Fig. 1D). We next tested the ability of individual Pumilio repression domains to do the same. Using the tethered-function approach, we measured the effect of the N terminus and C terminus on luciferase protein and mRNA expression. Reduction of protein levels correlated with reduction of mRNA levels for both the full-length protein (Pum, 45% protein and 32% mRNA) and the C terminus (region C, 10% protein and 6% mRNA) (Fig. 5A). Surprisingly, the N terminus had a substantially greater effect on protein expression than mRNA (region N, 32% protein and 9% mRNA) (Fig. 5A). When each N-terminal repression domain was tested, similar differences were observed for regions 1 (18% versus 42%), 2 (42% versus 72%), and 3 (45% versus 62%) (Fig. 5B). This suggests that the N-terminal repression domains inhibit translation to a greater extent than they enhance mRNA degradation.
Fig 5.
The N-terminal repression domains inhibit protein expression and, to a lesser extent, reduce mRNA levels. (A) Percent repression of RnMS2 mRNA and protein by tethered full-length Pumilio (Pum), Pum N terminus, or Pum C terminus. (B) Percent repression of RnMS2 mRNA and protein by tethered Pum N-terminal repression domains. In panels A and B, percent repression of RnMS2 was normalized to the FF control and calculated relative to control MS2CP alone for both qRT-PCR (mRNA) and dual-luciferase (protein) assays. Data are reported in Tables S14 and S15 in the supplemental material.
The N termini of human PUF proteins PUM1 and PUM2 possess repression domains.
We hypothesized that the N-terminal repression domains of Pumilio may be conserved by other PUF proteins. Insect and vertebrate PUF proteins share a similar architecture, including a highly conserved C-terminal RNA-binding domain (>80 identical) and N-terminal domains, including PCMa and PCMb motifs (Fig. 6A). We compared repression by the N termini of human PUM1 (Homo sapiens PUM1-N [HsPUM1-N]; aa 1 to 827) and PUM2 (HsPUM2-N; aa 1 to 704) to that of Drosophila Pumilio (D. melanogaster Pum-N [DmPum-N]) using the tethered-function assay (Fig. 6B). All three proteins were expressed, as confirmed by Western blotting (Fig. 6B). Human PUM1-N and PUM2-N repressed RnMS2 reporter by 47% and 36%, respectively, comparable to the 35% repression caused by the N terminus of Pumilio (Fig. 6B). Next, we tested whether the regions of human PUM1 corresponding to Pumilio repression domains possessed autonomous repressive activity. When tethered, region 1 of human PUM1 (HsPUM1-1; aa 1 to 150) lacked repressive function (Fig. 6C). However, PUM1 region 2 (HsPUM1-2, aa 309 to 459) and region 3 (HsPUM1-3, aa 589 to 827) robustly repressed 48% and 82%, respectively (Fig. 6C). These results show that the N termini of human PUFs contain potent repressive domains, indicating a conserved regulatory function.
Fig 6.
Conservation of repression activity by the N termini of human PUFs PUM1 and PUM2. (A) Comparison of human PUM1 and PUM2 to Drosophila Pumilio. The percentage of identical amino acids between segments is indicated. (B) Percent repression by the N terminus of each protein, fused to MS2 coat protein, was determined relative to control MS2 alone. Test proteins include the N termini of Drosophila Pumilio (DmPum-N) and human PUM1 (HsPUM1-N) and PUM2 (HsPUM2-N). (C) Percent repression by MS2-tethered regions of human PUM1 corresponding to Pumilio repression domains (HsPUM1-1, HsPUM1-2, and HsPUM1-3). All assays used RnMS2 reporter and the FF control. Western blot analysis confirmed expression of V5 epitope-tagged proteins. Data are provided for panels B and C in Tables S16 and S17 in the supplemental material, respectively.
DISCUSSION
Pumilio and Nanos control important functions, including development (30, 31, 54), stem cell proliferation (2, 15, 32, 42), and learning (12). Previous analyses of Pumilio and Nanos function were restricted to mutant or transgenic Drosophila. The experiments presented in this work build upon these elegant studies to elucidate the mechanism of regulation. We developed a reporter assay that recapitulates Nanos-dependent repression. Nanos is not detectable in D.mel-2 cells (Fig. 1B) (7), but expression of Nanos confers potent repression of an mRNA bearing Hunchback NREs (Fig. 1C and D). Pumilio is essential for Nanos repression (Fig. 1E) (3, 31). Therefore, Nanos activates Pumilio. Acting together, Nanos and Pumilio inhibit protein expression and cause a corresponding decrease in mRNA level (Fig. 1D). This data is consistent with Nanos and Pumilio collaborating to repress Hunchback mRNA in the Drosophila embryo (3, 30, 31, 38, 48, 51, 58).
Pumilio also represses independently of Nanos (Fig. 2). Without Nanos, endogenous Pumilio in D.mel-2 cells minimally represses the NRE-bearing reporter (see Fig. S1B in the supplemental material); however, efficient repression was elicited by increasing the concentration of Pumilio (Fig. 2A; see Fig. S3 in the supplemental material). A likely explanation is that the amount of endogenous Pumilio is insufficient to efficiently repress. Like Nanos-dependent repression, Pumilio potently decreased reporter protein and mRNA levels (Fig. 2B). Several facts support the conclusion that Nanos was not necessary for repression by Pumilio. First, Nanos is not detectable in D.mel-2 cells. Also, RNAi of Nanos did not affect Pumilio repression (Fig. 2D). Furthermore, a mutation that blocks Nanos binding to Pumilio (F1367S) did not alleviate repression (Fig. 2C). Nanos-inde-pendent Pumilio repression is supported by observations that Pumilio regulates Bicoid and cyclin B mRNAs in the anterior of Drosophila embryos, where Nanos is below the limit of detection (16, 53).
The finding that Pumilio represses independently of Nanos raises the question: what is the function of Nanos? One logical answer is that Nanos strengthens Pumilio repression. The observation that Nanos activates endogenous Pumilio supports a model wherein Nanos enhances the RNA-binding activity of Pumilio. Previous work strengthens this hypothesis: Nanos and Pumilio interact with each other and both contact the NRE RNA through a network of protein-protein and protein-RNA interactions that may cooperate to enhance binding (47, 48). The necessity of Nanos could be obviated by increasing the level of Pumilio (Fig. 2), likely resulting from increased occupancy of the NRE reporter. Another hypothesis is that binding of Nanos to Pumilio might displace a negative regulatory factor, resulting in activation of endogenous Pumilio. Nanos may also collaborate with Pumilio to recruit corepressors. The Nanos-Pumilio-NRE complex is thought to recruit Brain Tumor and 4EHP to refine regulation of the Hunchback gradient (8, 47). However, RNAi depletion of Brain Tumor and 4EHP did not abrogate Nanos-dependent repression (Fig. 1). We interpret this as evidence that Nanos and Pumilio repress mRNAs through additional mechanisms (see below), with the caveat that residual Brain Tumor and 4EHP might be sufficient to support Nanos-dependent repression. As an alternative model, Nanos and Pumilio may collaborate to recruit the Ccr4-Not deadenylase complex through interactions with Not4 and Pop2 subunits, respectively (22, 26). Future research is necessary to address these models.
Potent Pumilio repression in the absence of Nanos indicates that Pumilio independently inhibits protein expression and enhances mRNA decay. Involvement of known corepressors Brain Tumor and 4EHP is improbable because recruitment of these proteins depends on Nanos (8, 47). Furthermore, a mutant Pumilio (G1330D) that cannot bind Brain Tumor is fully active for Nanos-independent repression (Fig. 2C). In addition, depletion of Brain Tumor by RNAi did not affect Pumilio repression (Fig. 2D). These findings reveal that mechanisms other than Brain Tumor-4EHP-mediated inhibition of 5′-cap-dependent translation are utilized by Pumilio. Previous studies concluded that Brain Tumor, and therefore 4EHP, are dispensable for Pumilio repression in certain contexts. For instance, Pumilio repression of cyclin B in embryonic pole cells does not require Brain Tumor (26, 47). Furthermore, while Brain Tumor is necessary for Pumilio repression in motor neurons, it is not essential in other neurons (37). Finally, the G1330D mutant Pumilio, which is deficient for recruitment of Brain Tumor, is functional for regulation of dendritic morphology (62). We do not dismiss the importance of Brain Tumor and 4EHP in embryonic development. Instead, these findings illustrate that while Brain Tumor and 4EHP facilitate repression of Hunchback in the embryo, in other contexts, Pumilio represses by other means.
We identified Pumilio domains that mediate Nanos-inde-pendent repression. The Pumilio RBD has modest repressive activity compared to the full-length protein (Fig. 3C and D; see Fig. S3 in the supplemental material), indicating that regions outside the RBD must confer repressive activity. Previous analysis of the ability of Pumilio transgenes to rescue abdominal segmentation defects in a pumilio mutant embryo support this conclusion. Whereas overexpression of the Pumilio RBD partially rescued segmentation defects, the full-length Pumilio fully restored proper embryonic development (3, 57). Indeed, we discovered three repression domains within the N-terminal two-thirds of Pumilio that provide the major repressive activity (Fig. 4). These unique repression domains (i.e., aa 1 to 378, 548 to 776, and 848 to 1090) do not share sequence homology. Each functions autonomously when tethered to mRNA (Fig. 4). Because all known Pumilio cofactors (i.e., Nanos, Brain Tumor, 4EHP, and Pop2) interact with the RBD (8, 26, 47, 48), the N-terminal repression domains likely function through novel mechanisms. While full-length Pumilio affects both mRNA and protein levels almost equally (Fig. 1D and 2B), the individual repression domains affect protein expression more than mRNA levels (Fig. 5A and B). This suggests translational inhibition may be their predominant function.
The repressive function of the Pumilio N terminus may be evolutionarily conserved. Sequence alignments indicated that the N terminus of vertebrate PUMs, including human PUM1 and PUM2, are related to the Pumilio N terminus (Fig. 6A). When tethered, the N-terminal portions of human PUM1 and PUM2 repressed, providing evidence that human PUFs are repressors (Fig. 6B). Two regions in human PUM1 are autonomous repression domains (Fig. 6C). These regions are small (region 2, 152 amino acids; region 3, 240 amino acids), share 19% and 15% identity with Pumilio, and do not contain previously identified motifs. We propose that they may contact novel corepressors, which remain to be identified.
We compared the Pumilio N terminus to other PUF proteins; no detectable relationship could be found with six Saccharomyces cerevisiae PUFs or 12 C. elegans PUFs. Instead, these PUFs have evolved unique sequences, appended to their RNA-binding domains, whose function remains unknown. We also searched the nonredundant protein sequence database (www.ncbi.nlm.nih.gov) using the BLAST algorithm (1) to identify protein sequences similar to the Pumilio N terminus: no proteins, other than PUF family members, share homology. The broad implication is that members of the PUF family have evolved unique domains, appended to the evolutionarily conserved PUF repeat RNA-binding module, which may confer unique regulatory activities to individual PUFs. Consistent with this idea, specific PUFs have been shown to affect translation, mRNA degradation, mRNA localization, and, for one PUF, activation of target mRNAs (10, 17, 20, 23, 27, 41, 43, 44, 50).
We identified two sequence motifs in the N terminus of Pumilio, designated PCMa and PCMb, which are conserved between insects (e.g., Drosophila) and vertebrates (e.g., humans) (Fig. 6A). PCMb encompasses a motif in Xenopus PUM2 proposed to interfere with cap-dependent translation (4). However, when tethered, PCMb does not repress (Fig. 4), nor does deletion of PCMb diminish Pumilio repression (see Fig. S4A in the supplemental material). In addition, mutation of a PCMb tryptophan residue (W783) proposed to contact the 5′ cap (4) had no effect (data not shown). Therefore, we find no evidence that the putative cap binding motif of PCMb is important for Pumilio repression. Instead, PCMb negatively affects repression domains aa 548 to 776 (region 2) and aa 848 to 1090 (region 3) (Fig. 4E). While PCMa had weak repressive activity on its own, it could counteract the inhibitory effect of PCMb (Fig. 4D), and PCMa deletion of PCMa caused a minor but significant drop in repression (Fig. see S4A in the supplemental material). The precise roles of PCMa and PCMb remain to be determined; we speculate that they may have autoregulatory functions.
We successfully programmed Pumilio to repress a new target mRNA. By changing the RNA recognition amino acids in the sixth PUF repeat of Pumilio from NYQ to SYE, the specificity was altered from uridine to guanosine, thereby conferring repression to an mRNA with an altered binding site (Rn3×NRE UGG) (Fig. 3D). This experiment provides the proof of principle that PUF proteins with programmed RNA-binding specificity can be engineered to repress new mRNAs. While programmed Pumilio fully represses its new target, a similarly programmed RBD lacks substantial activity (Fig. 3), further emphasizing the importance of the N-terminal repression domains. This finding has important implications for future engineering of PUFs. The Pumilio RBD provides a protein module with low intrinsic regulatory activity that can be programmed to bind new RNA sequences. Functional domains—either repression or activation domains—can be attached to this module to create novel RNA regulators. Consistent with this idea, a recent study reported that addition of splicing effector domains and a nuclear localization signal transformed a PUF RBD into a splicing regulator (56).
An important question for future research is how do the Pumilio repression domains function? A probable hypothesis is that the repression domains inhibit the translation machinery. Alternatively, the repression domains may activate enzymes that degrade mRNAs. In future experiments, we seek to identify corepressors that interact with these domains. Also worth consideration is why Pumilio possesses multiple repression domains. These domains may recruit the same corepressor, either acting redundantly or collaboratively. Alternatively, each repression domain could bind to a different corepressor, perhaps affecting different steps in the gene expression pathway (e.g., translation initiation or mRNA degradation). In this case, their individual repressive activities would collaborate to increase the efficiency of repression. Addressing these crucial questions will help reveal how Pumilio regulates mRNAs to control diverse biological functions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Wagner for reagents and comments on the manuscript. We are grateful to Trista Schagat for comments on the manuscript and design of reporter mRNAs and multiplexed qRT-PCR assays. We appreciate comments and advice from Jamie Van Etten, Nathan Blewett, Joel Hrit, and Nathan Raynard.
This research was supported by a Faculty Research Grant from the Rackham Graduate School at the University of Michigan to A.C.G. C.A.W. is supported by the Michigan Predoctoral Training Program in Genetics through NIH National Research Service Award 5T32GM007544-33.
Footnotes
Published ahead of print 7 November 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. 1999. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1:431–437 [DOI] [PubMed] [Google Scholar]
- 3. Barker DD, Wang C, Moore J, Dickinson LK, Lehmann R. 1992. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 6:2312–2326 [DOI] [PubMed] [Google Scholar]
- 4. Cao Q, Padmanabhan K, Richter JD. 2010. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA 16:221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chagnovich D, Lehmann R. 2001. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A. 98:11359–11364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheong CG, Hall TM. 2006. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. U. S. A. 103:13635–13639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cherbas L, et al. 2011. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 21:301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cho PF, et al. 2006. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr. Biol. 16:2035–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cho PF, et al. 2005. A new paradigm for translational control: inhibition via 5′–3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121:411–423 [DOI] [PubMed] [Google Scholar]
- 10. Chritton JJ, Wickens M. 2010. Translational repression by PUF proteins in vitro. RNA 16:1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Curtis D, et al. 1997. A CCHC metal-binding domain in Nanos is essential for translational regulation. EMBO J. 16:834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dubnau J, et al. 2003. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 13:286–296 [DOI] [PubMed] [Google Scholar]
- 13. Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. 2001. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105:281–289 [DOI] [PubMed] [Google Scholar]
- 14. Edwards TA, Wilkinson BD, Wharton RP, Aggarwal AK. 2003. Model of the brain tumor-Pumilio translation repressor complex. Genes Dev. 17:2508–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forbes A, Lehmann R. 1998. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125:679–690 [DOI] [PubMed] [Google Scholar]
- 16. Gamberi C, Peterson DS, He L, Gottlieb E. 2002. An anterior function for the Drosophila posterior determinant Pumilio. Development 129:2699–2710 [DOI] [PubMed] [Google Scholar]
- 17. Garcia-Rodriguez LJ, Gay AC, Pon LA. 2007. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J. Cell Biol. 176:197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gebauer F, Hentze MW. 2004. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5:827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 103:4487–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldstrohm AC, Hook BA, Seay DJ, Wickens M. 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13:533–539 [DOI] [PubMed] [Google Scholar]
- 21. Goldstrohm AC, Seay DJ, Hook BA, Wickens M. 2007. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 282:109–114 [DOI] [PubMed] [Google Scholar]
- 22. Goldstrohm AC, Wickens M. 2008. Multifunctional mRNA deadenylase complexes. Nat. Rev. Mol. Cell Biol. 9:337–344 [DOI] [PubMed] [Google Scholar]
- 23. Gu W, Deng Y, Zenklusen D, Singer RH. 2004. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 18:1452–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hook BA, Goldstrohm AC, Seay DJ, Wickens M. 2007. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 282:15430–15438 [DOI] [PubMed] [Google Scholar]
- 25. Jackson JS, Jr, Houshmandi SS, Lopez Leban F, Olivas WM. 2004. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA 10:1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kadyrova LY, Habara Y, Lee TH, Wharton RP. 2007. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134:1519–1527 [DOI] [PubMed] [Google Scholar]
- 27. Kaye JA, Rose NC, Goldsworthy B, Goga A, L'Etoile ND. 2009. A 3′UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron 61:57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kraemer B, et al. 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9:1009–1018 [DOI] [PubMed] [Google Scholar]
- 29. Lehmann R, Nusslein-Volhard C. 1987. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev. Biol. 119:402–417 [DOI] [PubMed] [Google Scholar]
- 30. Lehmann R, Nusslein-Volhard C. 1987. Involvement of the pumilio gene in the transport of an abdominal signal in the Drosophila embyro. Nature 329:167–170 [Google Scholar]
- 31. Lehmann R, Nusslein-Volhard C. 1991. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development 112:679–691 [DOI] [PubMed] [Google Scholar]
- 32. Lin H, Spradling AC. 1997. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124:2463–2476 [DOI] [PubMed] [Google Scholar]
- 33. Lu G, Dolgner SJ, Hall TM. 2009. Understanding and engineering RNA sequence specificity of PUF proteins. Curr. Opin. Struct. Biol. 19:110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Macdonald PM. 1992. The Drosophila pumilio gene: an unusually long transcription unit and an unusual protein. Development 114:221–232 [DOI] [PubMed] [Google Scholar]
- 35. Mee CJ, Pym EC, Moffat KG, Baines RA. 2004. Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J. Neurosci. 24:8695–8703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Menon KP, et al. 2004. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron 44:663–676 [DOI] [PubMed] [Google Scholar]
- 37. Muraro NI, et al. 2008. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J. Neurosci. 28:2099–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murata Y, Wharton RP. 1995. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80:747–756 [DOI] [PubMed] [Google Scholar]
- 39. Nakahata S, et al. 2001. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 276:20945–20953 [DOI] [PubMed] [Google Scholar]
- 40. Nusslein-Volhard C, Frohnhofer HG, Lehmann R. 1987. Determination of anteroposterior polarity in Drosophila. Science 238:1675–1681 [DOI] [PubMed] [Google Scholar]
- 41. Olivas W, Parker R. 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19:6602–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parisi M, Lin H. 2000. Translational repression: a duet of Nanos and Pumilio. Curr. Biol. 10:R81–R83 [DOI] [PubMed] [Google Scholar]
- 43. Quenault T, Lithgow T, Traven A. 2011. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 21:104–112 [DOI] [PubMed] [Google Scholar]
- 44. Saint-Georges Y, et al. 2008. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One 3:e2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schneider I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27:353–365 [PubMed] [Google Scholar]
- 46. Schweers BA, Walters KJ, Stern M. 2002. The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics 161:1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sonoda J, Wharton RP. 2001. Drosophila Brain Tumor is a translational repressor. Genes Dev. 15:762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sonoda J, Wharton RP. 1999. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 13:2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Suh N, et al. 2009. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics 181:1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tadauchi T, Matsumoto K, Herskowitz I, Irie K. 2001. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20:552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tautz D. 1988. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature 332:281–284 [DOI] [PubMed] [Google Scholar]
- 52. Tautz D, Pfeifle C. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81–85 [DOI] [PubMed] [Google Scholar]
- 53. Vardy L, Orr-Weaver TL. 2007. The Drosophila PNG kinase complex regulates the translation of cyclin B. Dev. Cell 12:157–166 [DOI] [PubMed] [Google Scholar]
- 54. Wang C, Lehmann R. 1991. Nanos is the localized posterior determinant in Drosophila. Cell 66:637–647 [DOI] [PubMed] [Google Scholar]
- 55. Wang X, McLachlan J, Zamore PD, Hall TM. 2002. Modular recognition of RNA by a human pumilio-homology domain. Cell 110:501–512 [DOI] [PubMed] [Google Scholar]
- 56. Wang Y, Cheong CG, Hall TM, Wang Z. 2009. Engineering splicing factors with designed specificities. Nat. Methods 6:825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y. 1998. The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1:863–872 [DOI] [PubMed] [Google Scholar]
- 58. Wharton RP, Struhl G. 1991. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67:955–967 [DOI] [PubMed] [Google Scholar]
- 59. Wickens M, Bernstein DS, Kimble J, Parker R. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18:150–157 [DOI] [PubMed] [Google Scholar]
- 60. Wickens M, Goodwin EB, Kimble J, Strickland S, Hentze MW. 2000. Translational control in developmental decisions, p 295–370 In Mathews M. (ed.), Translational control, 2nd ed Cold Spring Harbor Press, New York, NY [Google Scholar]
- 61. Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. 1997. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development 124:3015–3023 [DOI] [PubMed] [Google Scholar]
- 62. Ye B, et al. 2004. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 14:314–321 [DOI] [PubMed] [Google Scholar]
- 63. Zamore PD, Bartel DP, Lehmann R, Williamson JR. 1999. The PUMILIO-RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry 38:596–604 [DOI] [PubMed] [Google Scholar]
- 64. Zamore PD, Williamson JR, Lehmann R. 1997. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 3:1421–1433 [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang B, et al. 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390:477–484 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.