Abstract
Mitochondrial respiratory complexes of the electron transport chain (CI, CIII, and CIV) can be assembled into larger structures forming supercomplexes. We analyzed the assembly/stability of respiratory complexes in mouse lung fibroblasts lacking the Rieske iron-sulfur protein (RISP knockout [KO]cells), one of the catalytic subunits of CIII. In the absence of RISP, most of the remaining CIII subunits were able to assemble into a large precomplex that lacked enzymatic activity. CI, CIV, and supercomplexes were decreased in the RISP-deficient cells. Reintroduction of RISP into KO cells restored CIII activity and increased the levels of active CI, CIV, and supercomplexes. We found that hypoxia (1% O2) resulted in increased levels of CI, CIV, and supercomplex assembly in RISP KO cells. In addition, treatment of control cells with different oxidative phosphorylation (OXPHOS) inhibitors showed that compounds known to generate reactive oxygen species (ROS) (e.g., antimycin A and oligomycin) had a negative impact on CI and supercomplex levels. Accordingly, a superoxide dismutase (SOD) mimetic compound and SOD2 overexpression provided a partial increase in supercomplex levels in the RISP KO cells. Our data suggest that the stability of CI, CIV, and supercomplexes is regulated by ROS in the context of defective oxidative phosphorylation.
INTRODUCTION
The Rieske iron-sulfur protein (RISP) is one of the catalytic subunits of ubiquinol-cytochrome c oxidoreductase, also known as complex III (CIII), from the electron transport chain (ETC). CIII contains two other catalytic subunits, cytochrome b and cytochrome c1. The active enzyme is a homodimer that catalyzes the transfer of electrons from ubiquinol (coenzyme Q) to cytochrome c in a bifurcated mechanism mediated by RISP (62).
In the last few years, structural evidence indicates that the mitochondrial complexes of the oxidative phosphorylation (OXPHOS) system interact with each other to form supramolecular structures in the inner mitochondrial membrane named supercomplexes. Their association into megacomplexes was proposed to form a functional unit known as the “respirasome” (7, 60, 69). Because the electron transport chain is one of the major contributors to free radicals in the cell, respirasomes could minimize the generation of reactive oxygen species (ROS) by allowing a more efficient electron transfer and substrate channeling among the complexes, therefore avoiding the diffusion of reactive intermediates (29, 42).
Supercomplex assemblies have been observed in a wide variety of organisms, including bacteria, plants, fungi, and mammals, although their composition might vary from organism to organism. Mammalian supercomplexes are composed mainly by CI, CIII, and CIV in different stoichiometries (I/III, I/III2, I2III2, I/III/IV, I/III2/IV, I/III/IV2, III/IV, and III2/IV1-2) (58, 69). Recently, Acin-Perez et al. (2) investigated the composition and functionality of the different mammalian supercomplexes. The authors showed that some of these structures contained coenzyme Q and cytochrome c. Moreover, they demonstrated that some supercomplexes were able to “respire” by transferring electrons from NADH to oxygen (2).
Ultimately, the capacity to form supercomplexes arrangements relies on the stability of its components. Respiratory complex interdependence has been observed in numerous cases. Generally, CI appears to be the most labile complex of the electron transport chain. Cells with defects in CIII or CIV assembly have decreased levels of CI (1, 17, 20, 43), whereas cells lacking cytochrome c showed defects in CIV and CI (64). Likewise, defects in CV have been associated with defects in CIII and CIV in both yeast and mammalian cells (55, 59).
In the last few years, studies have attempted to elucidate the role of deranged mitochondrial supercomplexes and their pathophysiological significance in disease conditions. Alterations in the supramolecular architecture of OXPHOS complexes have been observed in the rat cortex during aging, and decreased respirasome levels have been reported in animal models of severe heart failure (26, 54). Whether these alterations result in significant physiological changes is still unclear.
Here, we analyzed CIII assembly and supercomplex formation in mouse fibroblasts deficient in RISP and uncovered a ROS-dependent mechanism mediating decreases in CI, CIV, and supercomplexes.
MATERIALS AND METHODS
Cell culture.
A primary culture lung fibroblast line was produced from a knock-in mouse homozygous for the floxed UQCRFS1 gene (exon 2) encoding the Rieske iron-sulfur protein (RISP) (28). The introduction of loxP sites into the gene did not interfere with RISP function, since the CIII activities from tissues derived from wild-type or floxed mice were comparable. Cells were grown at 37°C in a 5% CO2 atmosphere in high-glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, gentamicin, fungizone, 1 mM pyruvate, and 50 μg/ml uridine. The derived cell line was immortalized by transduction with a G418-resistant retrovirus expressing E6/E7 oncogenes of type 6 papillomavirus (kindly provided by E. Shoubridge, McGill University) (45), and G418-resistant clones were obtained by ring cloning. These immortalized cell lines containing the floxed gene were used as control cell lines from which knockout (KO) cells were derived. To ablate the floxed UQCRFS1 gene, one of the immortalized clones was transfected with a hygromycin-resistant plasmid expressing P1 Cre recombinase (kindly provided by J. Marth, University of California San Diego). The control cell lines used in this study, referred to as clones 8 and 14, contained the floxed UQCRFS1 gene, whereas the knockout cells deficient in RISP were derived from clone 8 and are referred to as clones 8.2, 8.4, 8.5, and 8.17.
Multiplex PCR.
Cre-mediated recombination was determined by multiplex PCR using 3 primers allowing the simultaneous amplification of the floxed, wild-type, and deletion alleles, as shown in Fig. 1A. The primer sequences were as follows: (i) forward, 5′TTCCCTCCTCAGGCTTCACTTGAC3′; (ii) reverse, 5′GATTGGGAAGACAATAGCAGGCATG3′; and (iii) reverse, 5′TTGGCTAGAGAGTAAAATTCAGTCTT3′. The relative positions and orientations of the primers are depicted in Fig. 1A by arrows.
Fig 1.
Ablation of RISP leads to CIII deficiency. (A) Diagram shows the floxed allele containing 3 loxP sites (triangles) flanking exon 2 of the UQCRFS1 gene (encoding RISP) and a selection cassette (TK/Neo) present in the knock-in mouse, the deletion allele obtained after Cre recombination, and the wild-type allele. (B) Efficiency of recombination of the floxed gene. Knockout clones only showed the band corresponding to the deletion allele by multiplex PCR (primers shown in panel A by arrows), indicating complete recombination. The wild-type (WT) allele was amplified from skin fibroblasts from a wild-type mouse. (C) Enzymatic activities of CIII, CIV, and citrate synthase (CS) determined spectrophotometrically. Values represent the means and standard deviations of specific activities. Statistical significance is shown by an asterisk (P ≤ 0.05). (D) Western blot of mitochondrial proteins and OXPHOS complex subunits (CI, NDUFA9; CIII, Core1, Core2, and RISP; and CIV, Cox1). VDAC1 and Tim23 were used as mitochondrial loading controls.
Mitochondrial preparation and enzyme activities.
Mitochondria were obtained from 8 confluent T-75 flasks by nitrogen cavitation as described previously (20) and used for determination of the enzyme activities of different respiratory complexes and citrate synthase as described previously (3). Total cell respiration was determined by polarography with a Clark oxygen electrode (Hansatech Instruments, Norfolk, United Kingdom) as described in reference 20.
Protein determination.
Protein concentrations were determined using bovine serum albumin as the standard and either the Bradford or the bicinchoninic acid (BCA) (detergent-compatible) protein assay kit from Bio-Rad and Pierce, respectively.
BNGE.
To identify individual respiratory complexes, mitochondrial preparations were treated with lauryl maltoside (Sigma) and separated by blue native gel electrophoresis (BNGE) in 4 to 13% acrylamide gradient gels (8, 19). For Western blot analysis, amounts of about 10 to 25 μg of proteins were separated, transferred to polyvinylidene difluoride (PVDF) membrane (Bio-Rad), and immunodecorated sequentially with antibodies against several subunits of the different mitochondrial respiratory complexes. For in-gel activity assays, amounts of about 40 μg of proteins were separated by BNGE and gels were incubated with solutions containing the respective substrates for complex I or IV as described before (19).
For supercomplex analysis, mitochondrial preparations were treated with digitonin (Calbiochem) at a detergent-to-protein ratio of 8:1 (wt/wt) as described previously (20). Proteins (25 μg) were separated by BNGE in 3 to 12% precast blue native gels (Invitrogen).
For analysis of respiratory complexes by 2 dimensional (2D) BNGE, BN gel strips from the first dimension corresponding to each lane were excised and treated with 1% SDS, 1% β-mercaptoethanol for 10 min, then with 1% SDS for 5 min, and overlaid on a 10% SDS-acrylamide gel with a 4% stacking gel (20). Proteins were separated in the second dimension, transferred to PVDF membranes, and blotted sequentially with different antibodies.
For experiments that included different cell treatments (pharmacological inhibition or normoxia/hypoxia conditions), a digitonin-enriched mitochondrial fraction was obtained by resuspending 2 × 106 cells in 200 μl phosphate-buffered saline (PBS) containing protease inhibitors and adding 70 μl of 8 mg/ml digitonin (Sigma) for 10 min on ice. Subsequently, samples were diluted with 1 ml PBS and centrifuged at 21,000 × g for 5 min. Membranes were washed with PBS, and pellets were either stored at −80°C until needed or resuspended in 100 μl of 1.5 M aminocaproic acid, 50 mM Bis-Tris (pH 7.0) buffer (8). Supercomplexes were extracted from these preparations with highly purified digitonin (Calbiochem) in an 8:1 ratio (detergent to protein), and proteins were separated by BNGE (20).
Western blotting.
Mitochondrial samples were separated by either SDS-PAGE in 4 to 20% acrylamide gradient gels (Bio-Rad) or by BNGE and 2D-BNGE and transferred to PVDF membranes. Membranes were blocked with 5% nonfat dry milk in PBS–0.1% Tween 20 and then blotted with specific primary antibodies. Secondary antibodies conjugated to horseradish peroxidase (Cell Signaling) were used, and the reaction was detected by chemiluminescence using SuperSignal West reagent (Pierce, Rockford, IL) or RapidStep ECL reagent (EMD).
The following antibodies were used to identify individual respiratory complexes: complex I, NDUFA9 and NDUFS3 (Mitosciences); complex II, SDHA (succinate dehydrogenase subunit A) flavoprotein (Molecular Probes); complex III, Core1, Core2 (Mitosciences), and Rieske iron-sulfur protein (Molecular Probes); complex IV, Cox1 (Mitosciences); and complex V, ATP synthase β (Molecular Probes). Additionally, premade antibody cocktails from Mitosciences specific for BNGE (NDUFA9, SDHA, Core2, Cox IV, and ATPase α) or for SDS-PAGE (NDUFB8, SDHB [succinate dehydrogenase subunit B], Core2, Cox1, and ATPase α) were used. Antibodies against VDAC1 (voltage dependent anion-selective channel protein 1; Mitosciences) or Tim23 (protein import component of the inner mitochondrial membrane; Molecular Probes) were used as mitochondrial loading controls.
To detect HIF1-α, cells were immediately lysed after hypoxia in ice using 2× Laemmli buffer without bromophenol blue. Cells were scraped off the tissue culture plate, and lysate heated at 96°C for 20 min and sonicated. Amounts of about 50 to 60 μg of proteins were separated in 6% acrylamide gels and transferred to PVDF membranes, and HIF1-α was detected with a specific antibody obtained from Novus Biologicals.
Signal intensities were quantified using the free NIH software Image J available for download at http://rsb.info.nih.gov/ij/. Fold changes in protein levels annotated in the text were calculated as the ratio between the protein signal and the corresponding loading control (in the same membrane).
Recombinant lentivirus.
Wild-type RISP was amplified from mouse cDNA and cloned into the pLenti-MP2-MCS-IRES-EGFP lentiviral vector containing an internal ribosome entry site (IRES) site allowing for the expression of both RISP and green fluorescent protein. A mutant form of the RISP was created by site-directed mutagenesis. The mutant RISP had two point mutations in positions 716 and 717 to change the wild-type CAT codon to a CGG codon. These point mutations resulted in an amino acid change from histidine to arginine that corresponded to amino acid 239 in the murine sequence. Lentiviruses expressing green fluorescent protein (GFP) and wild-type or mutant RISP were produced by the Viral Core Facility at the University of Miami.
RISP KO fibroblasts were transduced with 107 pg of viral particles with lentiviruses expressing GFP, wild-type RISP-GFP, or mutant RISP-GFP. High-GFP-expressing cells were sorted out by fluorescence-activated cell sorting (FACS) in an Aria-IIu cell sorter (Becton Dickinson) and maintained as a cell line for subsequent experiments.
Lentiviral particles expressing superoxide dismutase 2 (SOD2; enzyme cloned in the pWXIRESpuro vector) were used to transduce RISP-deficient cells as described above. Lentivirus expressing SOD2 contained puromycin resistance. Antibiotic-resistant stable cell lines were used in the experiments.
Pharmacological inhibition of mitochondrial respiratory complexes.
The pharmacological inhibition of CIII was achieved by growing cells in the presence of 2 nM antimycin A or 2 μM myxothiazol in complete medium for 3 days. The inhibition of CI, CIV, and CV was achieved by growing cells in the presence of 100 nM rotenone, 400 μM potassium cyanide (KCN), or 5 μg/ml oligomycin, respectively, for 3 days in complete medium. Fresh medium containing the respective drug was changed daily to avoid highly acidified medium. All OXPHOS inhibitors were obtained from Sigma.
Hypoxia treatment.
Cells (∼3 × 106) were plated in 10-cm dishes and immediately placed under conditions of normoxia or hypoxia. A hypoxic chamber with a Proox-110 oxygen controller (BioSpherix; Reming Bioinstruments) directly connected to a nitrogen-CO2 mixture tank (Mediblend clinical blood gas mixture, 5% CO2 plus nitrogen) was placed in a 37°C incubator. Cells were exposed for 4 h or 24 h to hypoxic (1% O2, 5% CO2) conditions, and control replicates were placed under normoxic conditions (21% O2, 5% CO2) at 37°C. In some experiments, cells were treated with different concentrations of MitoQ or decyltriphenylphosphonium bromide (dTPP; control compound to account for any nonspecific effect of lipophilic cations) (38) for 4 h prior to the exposure to either normoxia or hypoxia for 4 h.
Assessment of oxidative stress.
To measure superoxide levels, cells (3 × 105/well in 6-well plates) were plated, and the next day, the medium was replaced with fresh medium containing 10 μm oligomycin (positive control) or dimethyl sulfoxide (DMSO; vehicle) for 1.5 h at 37°C. Then, either 2.5 μM MitoSox (Invitrogen) or 2.5 μM dihydroethidium (DHE, Calbiochem) was added and the mixture incubated for 30 min. Cells were harvested by trypsinization, washed, and resuspended in Hanks balanced solution. Fluorescence was detected by FACS in an Aria-IIu flow cytometry cell sorter (Becton Dickinson) using excitation/emission wavelengths of 396/580 for MitoSox and 518/605 for DHE.
To measure hydrogen peroxide production in normoxia/hypoxia experiments, cells were plated in 96-well plates at a density of 104 cells/100 μl/well and incubated with 8 μM H2DCF-DA (2′,7′-dichlorodihydrofluorescein diacetate) in 200 μl medium (56) prior to the treatment. Then, cells were placed under normoxic or hypoxic conditions for 24 h. At the end of the incubation period, DCF fluorescence was measured immediately with a plate reader (Victor II; Perkin Elmer). The fluorescence values were corrected for the fluorescence of the medium alone. Cells were incubated with 100 μM tert-butyl hydroperoxide (tBOOH) and 2 mM n-acetyl cysteine (NAC) as positive controls for the assay. Values represent means and standard deviations of the results of 3 to 6 wells of a representative experiment of at least 3 individual determinations.
Analysis of cellular antioxidant system during normoxia/hypoxia treatment.
The levels and enzyme activities of Mn- and CuZn-superoxide dismutases (SOD1 and SOD2) were determined by Western blot analysis and by in-gel activity assay as described previously (68). To determine steady-state levels of these enzymes by Western blotting, we used an anti-SOD1 antibody from Calbiochem and an anti-SOD2 antibody from Upstate. Glutathione (GSH) levels were determined using the GSH-Glo glutathione assay kit from Promega, and glutathione peroxidase (GPx) activity was measured with the GPx assay kit from Cayman Chemicals, following the manufacturer's instructions.
Statistical analysis.
Values represent the means and standard deviations of at least 3 independent measurements. Statistical significance was determined by using the two-tailed unpaired Student t test. A P value of ≤0.05 was considered statistically significant.
RESULTS
Creation and characterization of RISP KO fibroblasts.
Primary cultures of lung fibroblasts from a knock-in mouse (28) homozygous for the floxed UQCRFS1 gene were immortalized by E6/E7 expression. The knock-in mouse contained loxP sites flanking exon 2 of the UQCRFS1 gene and a selection cassette at the 3′ untranslated region (UTR) (Fig. 1A) (28). The UQCRFS1 gene encodes the Rieske iron-sulfur protein (RISP), one of the catalytic subunits of complex III (CIII). Two immortalized clones harboring the floxed UQCRFS1 (clones 8 and 14) were used as control cell lines in this study.
To obtain CIII-deficient cells, the floxed clone 8 was transfected with a plasmid expressing the Cre recombinase. The efficiency of gene ablation in several clones was determined by a PCR detecting the floxed, deletion, and wild-type alleles simultaneously (Fig. 1A). PCR of the RISP knockout (KO) clones (clones 8.2, 8.4, 8.5, and 8.17) only amplified the band corresponding to the deletion allele. The band corresponding to the floxed allele was only amplified in the control cells, clones 8 and 14. These results indicate a complete recombination of the floxed gene (Fig. 1B).
The RISP KO clones were unable to consume oxygen and quickly acidified the medium, indicating increased production of lactate due to impaired respiration (not shown). Biochemical analysis of the enzymatic activities of the OXPHOS complex measured spectrophotometrically showed, as expected, that deletion of RISP completely abolished CIII enzymatic activity (Fig. 1C). RISP deletion was also associated with a decrease in complex IV (CIV) activity in most of the KO clones compared to the activity in the parental cell line (Fig. 1C). Citrate synthase (CS) activity was increased in all the KO fibroblasts compared to the activity in the parental clone 8. We found a marked variation in CIV activity among the KO clones, with a clear reduction compared with their parental cell line. Control cell lines 8 and 14 showed variation in CIV and CS (Fig. 1C), possibly related to genome instability in cultured cells.
Analysis of the steady-state levels of mitochondrial proteins by Western blotting (Fig. 1D) showed no detectable levels of RISP in the KO clones, confirming the deletion of the gene. Other CIII subunits (UQCRC1 and UQCRC2, also known as Core1 and Core2) were decreased in the KO clones (1.6- to 2-fold and 7- to 10-fold lower than control levels, respectively). The levels of CI subunits NDUFA9 and NDUFS3 were also decreased (1.9- to 2.7-fold and 7- to 9.6-fold lower than control levels, respectively). Likewise, the levels of the CIV subunit Cox1 were decreased in those clones with decreased CIV activity (2- to 7-fold lower than control levels). In contrast, CII and CV subunits (SDHA and ATPase-β, respectively) were increased in the RISP KO clones compared to the levels in controls (2- to 4-fold higher). The levels of other mitochondrial proteins, such as cytochrome c, VDAC1 (voltage-dependent anion-selective channel protein 1), and Tim23 (inner mitochondrial protein import complex component) were comparable in all samples analyzed (Fig. 1D).
Deletion of RISP did not alter mitochondrial protein synthesis, and the levels of newly synthesized Cox1 in the KO cells were comparable to control levels (not shown). These results suggest that Cox1 was destabilized posttranslationally.
Assembly of OXPHOS complexes is impaired in the absence of RISP.
We investigated the effect of deletion of RISP on the levels of assembled OXPHOS complexes using blue native gel electrophoresis (BNGE). Proteins were extracted with lauryl maltoside to detect individual complexes. The deletion of RISP significantly reduced the levels of assembled CIII in all KO clones. The absence of RISP also had a profound effect on the assembly/stability of CI, which could only be detected upon overexposure of the blots. The levels of CIV were decreased in those KO clones with lower CIV activity, whereas the levels of CV were not altered in the KO clones (not shown).
Extraction with milder detergents, such as digitonin, preserves OXPHOS supramolecular interactions, and we were able to detect the presence of isolated CI, CIII, and supercomplexes in the control cell line when analyzed by 2-dimensional blue native gel electrophoresis (2D-BNGE) (Fig. 2A). Still, only by overexposing or loading more protein into the gels were we able to observe low levels of CI; however, we were unable to detect supercomplexes in the RISP KO clones (Fig. 2A and B).
Fig 2.
Ablation of RISP reduced the levels of CI, CIII, and supercomplexes. (A) Two-dimensional BNGE (2D-BNGE) and Western blot of mitochondrial proteins. Membranes were sequentially blotted with different antibodies, and signals obtained are indicated with an arrow in each panel. NDUFS3 antibody showed a strong unknown signal (?). Cox1 signal is circled with a white dotted line to distinguish it from the unknown signal from NDUFS3. Positions of respiratory complexes and supercomplexes (CIII/CI or CIII/CIV) are marked with dotted lines. (B) Presence of a subassembly/degradation product (sub) in RISP KO clones. (C) Western blot of VDAC1 as protein loading control.
Although all the steps for CIII assembly in mammals are unknown, the final steps in yeast appear to be the incorporation of RISP and the Qcr10 subunits. RISP incorporation is mediated by the chaperone Bcs1. In mammals, the homologue subunits are termed RISP and UQCRC11, respectively, and the homologue chaperon is BCS1L. The fully assembled CIII is a dimer with a molecular mass of about 550 kDa (16, 70). The CIII observed in the RISP KO clones was actually an intermediate precomplex that lacks RISP and UQCR11 (pre-CIII). Under the conditions used, we were unable to detect gel mobility differences between the fully assembled CIII and pre-CIII from control and RISP KO cells, respectively, by BNGE. The pre-CIII lacking RISP can be observed in Fig. 3A (GFP lanes, probed with Core2-specific antibody). These results suggest that at this point in assembly, a dimerization of assembly intermediates already occurred to form the pre-CIII prior to the addition of the last two subunits. We also found a CIII subassembly/degradation intermediate containing Core2 but not Core1 in the KO clones (Fig. 2B).
Fig 3.
Expression of wild-type RISP in KO cells reconstituted the levels of fully assembled CIII and CIV. (A) BNGE and Western blot of KO fibroblasts transduced with GFP, wild-type (WT) RISP, or mutant (MUT) RISP lentivirus. WT RISP restored levels of fully assembled CI and increased levels of CIII and CIV in KO cells. (B) BNGE and in-gel activity stain for CI and CIV. (C) Steady-state levels of OXPHOS complex subunits by Western blotting.
Respiratory complex and supercomplex instability has been attributed, in some instances, to low levels of phospholipids, such as cardiolipin (47, 52, 67). However, analysis of phospholipids by thin-layer chromatography did not reveal major differences between control and RISP KO cells (data not shown).
To determine whether the pleiotropic effects on CI and CIV in the KO clones were directly related to the ablation of RISP, reconstitution experiments introducing RISP back into the KO clones were performed. We infected KO clones with lentiviruses containing GFP (control infection), mouse wild-type RISP (WT-RISP), or the mutant RISP (MUT-RISP). We mutated the mouse histidine 219 residue to an arginine that corresponded to the histidine 181 in yeast. We chose this particular mutation because the yeast H181R mutant had the highest levels of RISP protein among the collection of no-activity mutants (32).
Wild-type RISP was incorporated into CIII in RISP KO cells (Fig. 3A, RISP panel) and restored the levels of the fully assembled CIII to control levels (Core2 panel), although the amount of RISP protein expressed was lower than in the control cells (Fig. 3C). The expression of either GFP or MUT-RISP did not increase the levels of CIII in KO clones. In fact, we were unable to detect the mutant RISP protein (Fig. 3C). It is possible that the mouse mutant protein, unlike the yeast mutant H181R, was not stable and it was degraded. The levels of other CIII subunits, Core1 and Core2, were increased in KO clones expressing WT-RISP compared to the levels in the clones expressing either GFP or MUT-RISP (Fig. 3C). Not surprisingly, these results suggest that the fully assembled CIII containing wild-type RISP is more stable than the pre-CIII.
WT-RISP was able to restore CIII activity in KO clones only partially (perhaps due to lower-than-normal expression from the integrated transgene). The improvement in CIII activity ranged between 14% and 32% of that of the parental cell line. The expression of GFP or MUT-RISP did not have any rescuing effect on CIII activity (not shown).
Wild-type RISP is required for CI, CIV, and supercomplex assembly/stability.
The pleiotropic CI defect due to ablation of RISP was abrogated by expressing the wild-type protein in the KO cells. Fully assembled CI levels were increased to control levels in KO clones transduced with the WT-RISP (Fig. 3A, NDUFA9 panel). Moreover, analysis by BNGE followed by in-gel activity staining showed that the restored CI was enzymatically active (Fig. 3B). However, CI levels and activities remained very low in the KO cells expressing GFP or mutant RISP. Similarly, only WT-RISP improved the levels and activity of CIV (Fig. 3B). The increase in CIV in-gel activity was only observed in those clones that initially had low levels of CIV (clones 8.2, 8.5, and 8.17), in agreement with spectrophotometric measurements (not shown). The increase in CIV was accompanied by higher steady-state levels of the CIV subunits Cox1 and Cox4 (Fig. 3C). The levels of other subunits, such as ATPase-α from CV or SDHA from CII, were unchanged upon the expression of GFP, wild-type, or mutant RISP (Fig. 3C).
Analysis of reconstituted KO cells by 2D-BNGE showed that WT-RISP restored supercomplexes. Figure 4 shows color-coded OXPHOS complexes and their organization into supercomplexes. Images of individual antibodies added sequentially to the same membrane were superimposed. Each color reflects an antibody. For identification of CIII, we used antibodies against Core2 (red), RISP (pink), and the assembly factor Bcs1L (blue); for CI, we used NDUFA9 (yellow); and for CIV, we used Cox1 (green). RISP KO clones transduced with the WT-RISP lentivirus contained higher levels of CI compared to those observed in cells transduced with GFP lentivirus (Fig. 4, yellow spots). Moreover, CI was also detected in supercomplex assemblies (CI/CIII/CIV dotted line). Detection of CIV with Cox1 antibody (green spots) showed that its levels were increased in those cells expressing WT-RISP.
Fig 4.

Expression of wild-type RISP increased levels of CI and CIV and restored supercomplex assemblies. Two-dimensional BNGE and Western blotting were performed to assess supercomplex assembly. Antibodies were added sequentially to the same membrane. Pseudocolors identifying each antibody are used for clarity. For identification of CIII, we used antibodies against Core2, RISP, and the assembly factor Bcs1L; for CI, NDUFA9; and for CIV, Cox1. The positions of complexes and supercomplexes are marked by dotted lines. Arrows indicate molecular mass of RISP (32 kDa).
RISP KO cells are able to induce HIF1-α in hypoxia.
Free radicals generated by CIII through RISP have been proposed to stabilize the hypoxic transcription factor HIF1-α during hypoxia by inhibiting the prolyl hydroxylase that constantly targets HIF1-α for degradation. Evidence supporting this hypothesis was provided by RISP knockdown experiments (6, 33).
The availability of our bona fide RISP knockout cells allows us to examine the levels of HIF1-α in hypoxic conditions. We found marked differences in the levels of HIF1-α depending on the time in hypoxia. Figure 5A shows that, when exposed to hypoxia (1% O2) for 24 h, the RISP KO cells were able to stabilize HIF1-α. Moreover, the levels of the transcription factor were higher in the RISP KO cells than in control cells (clones 8.4 and 8.5 had higher levels than control cells, with a 4- and 2.7-fold increase, respectively). Additionally, HIF1-α stabilization was also induced when cells were exposed to CoCl2, a prolyl hydroxylase inhibitor, although the chemical stabilization of HIF1-α was less robust than with hypoxia (Fig. 5A).
Fig 5.
RISP is not required for HIF1-α stability during hypoxia. (A) Steady-state levels of HIF1-α in cells exposed to either hypoxia (1% O2) or CoCl2 (50 μM) for 24 h. (B) Levels of HIF1-α in cells preincubated with 2.5 μM MitoQ (MQ) or 2.5 μM dTPP for 4 h and then subjected to either normoxia (N; 21% O2) or hypoxia (H; 1% O2) for 4 h. (C) Levels of HIF1-α in reconstituted RISP KO cells expressing wild-type RISP. Tubulin was used as loading control.
On the other hand, we found that after 4 h of hypoxia, the RISP KO clones' ability to stabilize HIF1-α was severalfold lower (2.9 to 19.7 times depending on the clone) than that of the control cells (Fig. 5B), in agreement with previous observations (4, 6, 12, 33). However, unlike previous observations (4, 57), MitoQ, a mitochondrial antioxidant (38), when used at 1 μM concentration, did not alter the levels of HIF1-α in any of the cell lines compared with the control compound dTPP (not shown). At 2.5 μM MitoQ, we observed a slight decrease in the levels of HIF1-α in some of the cell lines (Fig. 5B) but still not as robust as previously described (4, 57). The levels of HIF1-α were still detectable and only decreased 1.4- and 5.7-fold in the control and KO clone 8.4, whereas it remained unchanged in the other clones (Fig. 5B). Similar results were obtained with the reconstituted clones expressing WT-RISP (Fig. 5C). In these cells, HIF1-α levels only decreased 1.8- and 2.9-fold in the presence of 2.5 μM MitoQ but did not change in the presence of 5 μM MitoQ (Fig. 5C).
Hypoxia increased the levels of OXPHOS complexes and supercomplexes in RISP-deficient cells.
In the course of the hypoxia experiments, we also tested whether the decrease in oxygen levels affected the assembly of OXPHOS complexes. We subjected cells to 1% O2 for 24 h since that is the time frame required to observe fully assembled CI and supercomplexes (2, 40, 51). To our surprise, exposure of RISP KO cells to hypoxia for 24 h resulted in a significant increase in the levels of OXPHOS complexes in all cell lines (Fig. 6A). In particular, the levels of CI and CIV were markedly increased in the KO clone 8.5 under low-oxygen conditions. Assessment of the enzymatic activity of fully assembled complexes by in-gel activity assay revealed an increase in CI and CIV activities after 24 h of hypoxia in both control and KO cells compared to the normoxic activity (not shown).
Fig 6.
Hypoxia increased the levels of OXPHOS complex and supercomplex assemblies. (A) Cells were subjected to either normoxia or hypoxia (1% O2) for 24 h, and supercomplexes (SC) analyzed by BNGE. (B) Steady-state levels of OXPHOS complex subunits and other proteins in either normoxia or hypoxia. VDAC1 was used as loading control. LDH, lactate dehydrogenase.
Analysis of the steady-state levels of OXPHOS subunits showed that they were slightly increased in the hypoxia-treated samples (Fig. 6B). NDUFAB8 levels in KO clones 8.4 and 8.5 were increased 1.5- and 1.6-fold, respectively, during hypoxia, whereas its levels were unchanged in the control cell line. The highest changes were observed for Core2, with 4.3- and 3.5-fold the normoxia levels in the KO clones 8.4 and 8.5, respectively, and a 1.9-fold increase for control cells. Likewise, the levels of SDHB in KO clones 8.4 and 8.5 were 2.4- and 1.3-fold higher than in normoxia. The levels of ATPase-α increased 1.7-fold after hypoxia exposure in both the control and KO clone 8.4 but not in KO clone 8.5. The levels of other mitochondrial proteins, such as Grp75 and VDAC1, remained unaltered (Fig. 6B).
An increase in reactive oxygen species affects supercomplex stability.
From the results described above, we inferred that the stability/assembly of respiratory complexes might be affected by the production of free radicals. We reasoned that under low oxygen levels (hypoxia), fewer free radicals should be produced, and therefore, the levels of respiratory complexes and supercomplexes should improve. To test this hypothesis, we treated control cells with different OXPHOS inhibitors (many known to produce ROS) and determined their effects on the assembly/stability of OXPHOS complexes and supercomplexes.
Control cells were grown in the presence of either rotenone, antimycin A, KCN, or oligomycin (CI, CIII, CIV, and CV inhibitors, respectively) for 3 days, and OXPHOS complexes analyzed by BNGE after digitonin extraction. Treatment with oligomycin had a profound effect on CI assembly/stability and completely disrupted the supercomplexes containing CI (Fig. 7A, NDUFA9 panel). Analysis of steady-state levels of the respiratory complex subunits showed that oligomycin treatment strongly affected the levels of the CI subunit NDUFB8, whereas and to a lesser extent, it also affected the levels of Core2 and Cox1 from CIII and CIV, respectively (Fig. 7B, Cocktail panel).
Fig 7.
Pharmacological inhibition of OXPHOS complexes affected supercomplex assembly/stability. (A) BNGE and Western blot of control fibroblasts treated with rotenone (Rot; 100 nM), antimycin A (AA; 20 nM), KCN (400 μM), and oligomycin (Olig; 5 μg/ml). Ctrl, untreated control. (B) Steady-state levels of OXPHOS complex subunits in control cells treated with the different OXPHOS inhibitors. (C) BNGE and Western blot of control cells treated with CIII-specific inhibitors antimycin A (20 nM) and myxothiazol (Myx; 2 μM). (D) Steady-state levels of mitochondrial proteins of cells treated with CIII inhibitors. VDAC1 and Tim23 signals were used as loading controls.
Treatment with antimycin A had an effect on CI stability/assembly that was similar to that of the oligomycin treatment, albeit milder (Fig. 7A). Conversely, CI was not affected when cells were treated with either rotenone or KCN. In fact, KCN treatment mainly affected CIV subunit levels and, in turn, also affected supercomplexes containing CIV (Fig. 7A and B, respectively). Previous studies have shown that oligomycin and antimycin A markedly increase ROS generation, whereas KCN does not (44).
We further tested the effects of antimycin A and myxothiazol on supercomplex assembly and stability since the results of pharmacological inhibition of CIII presumably should resemble more closely the data for our RISP KO cells. Antimycin A and myxothiazol bind to different sites in CIII (62). Pharmacological inhibition of CIII decreased the levels of supercomplexes containing CI, CIII, and CIV compared to the levels in untreated cells (Fig. 7C). Similarly, the levels of CI were also reduced (NDUFA9 panel). Most of CIII appeared to be in supercomplexes in untreated cells, but with the pharmacological inhibition, this association was disrupted and most of the complex appeared in its dimeric form (Fig. 7C, Core2 panel). Likewise, the association of CIV with other complexes was also disrupted by both CIII inhibitors (Fig. 7C, Cox1 panel). The instability of CI upon CIII inhibition was also reflected by the lower steady-state levels of the NDUFB8 subunit (3- to 6-fold lower levels than in untreated cells) (Fig. 7D, Cocktail panel). Other subunits of the electron transport chain complexes (ATPase α, Cox1, Core2, and SDHB) or mitochondrial membrane proteins (Tim23 and VDAC1) were not significantly altered (Fig. 7D). Incubation with antimycin A and myxothiazol inhibited CIII activity and did not have any significant effect on CIV or citrate synthase activity measured spectrophotometrically (not shown).
RISP-deficient cells had increased levels of free radicals.
Because antimycin A and oligomycin are potent generators of reactive oxygen species (ROS), we investigated whether instability/assembly of CI, CIV, and supercomplexes in the RISP-deficient cells was related to increased levels of free radicals in these cells.
We measured the levels of superoxide by flow cytometry with the fluorescent probe MitoSox. Figure 8A shows that clone 8.4 contained higher levels of superoxide and clone 8.5 had only a slight increase in the levels of mitochondrial superoxide, presumably due to its lower mitochondrial membrane potential (not shown). Oligomycin increased ROS production in all cell lines, although it had a more pronounced increase in clone 8.5 (Fig. 8A). The reason for this marked difference in response to oligomycin is unknown, but it suggests that clone 8.5 produced larger amounts of ROS when CV was disturbed. In addition, we also measured the levels of total cellular superoxide using another dye, dihydroethidium (DHE), confirming that the RISP KO clones produced more ROS than the parental cell line (Fig. 8B).
Fig 8.
Deletion of RISP resulted in increased levels of ROS. (A) Mitochondrial superoxide radicals were measured by MitoSox fluorescence using flow cytometry. Cells were preincubated with oligomycin (Oligo) to increase ROS production. (B) Determination of cellular ROS using dihydroethidium (DHE) fluorescence by flow cytometry as described for panel A. Values from representative experiments are shown. Vertical lines indicate the mean fluorescence values for untreated control cells.
Antioxidants stabilized OXPHOS complexes/supercomplexes.
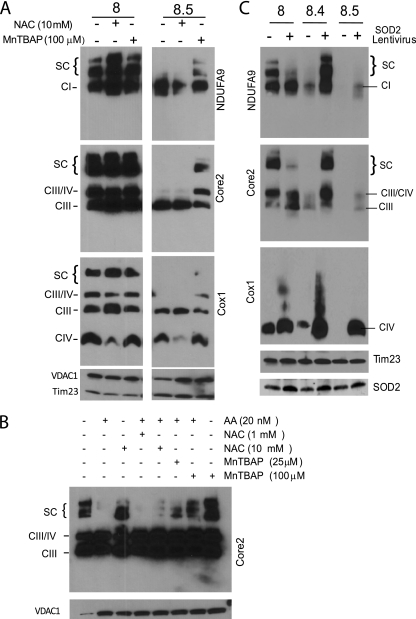
To test whether the stability of OXPHOS complexes, particularly CI and supercomplexes, is affected by increased free radicals, we treated cells with different antioxidant compounds. Cells were grown for 3 days in the presence of either a hydrogen peroxide scavenger, n-acetylcysteine (NAC), or with a superoxide dismutase mimetic compound, MnTBAP. The results in Fig. 9A show that NAC had a beneficial effect in CI and supercomplex stability in control cells (4.9-fold increase in supercomplex levels) but did not stabilize supercomplexes in the RISP-deficient fibroblasts. In contrast, when MnTBAP was used, supercomplex stability was preserved in the RISP KO (Fig. 9A).
Fig 9.
Scavenging superoxide radicals increases CI and supercomplex stability. (A) BNGE and Western blot of cells treated with either NAC (10 mM) or MnTBAP (100 μM). (B) BNGE of control cells treated with antimycin A (20 nM) and the effects of different concentrations of either NAC or MnTBAP on supercomplex (SC) stability. (C) BNGE of control and RISP KO fibroblasts infected with lentivirus expressing mitochondrial SOD2. Antibodies used are indicated beside each panel. The levels of SOD2 were determined by SDS-PAGE. VDAC1 and Tim23 were used as protein loading controls.
We also tested the effects of both antioxidants in control cells treated with a CIII inhibitor. Low NAC concentrations (1 mM) could not block the detrimental effect of antimycin A on CI and supercomplex stability. At higher concentrations (10 mM), NAC slightly protected supercomplex stability from antimycin A. Similarly, higher concentrations of MnTBAP improved the supercomplex levels in cells treated with antimycin A (Fig. 9B).
Taken together, these results showed that OXPHOS components, especially CI and supercomplexes, are vulnerable to increased levels of ROS. It also showed that by scavenging superoxide (but not hydrogen peroxide), we were able to partially rescue this instability. To confirm these results, we transduced the fibroblasts with lentivirus expressing mitochondrial SOD2 and obtained stable cell lines. Figure 9C shows that the SOD2 expression increased about 2-fold for both the control and KO clone 8.4 after lentiviral transduction. This increase in SOD2 levels completely rescued the instability of CI and supercomplexes in the RISP-deficient clone. An improvement of CI and supercomplex stability/assembly was also observed in clone 8.5 but to a much lesser extent, since SOD2 was only increased 1.3-fold after lentiviral transduction (Fig. 9C). We also analyzed cells with a more-than-20-fold increase in SOD2 levels, but the high levels of expression were detrimental for cell growth and OXPHOS complexes (not shown).
Antioxidant defenses were increased in RISP KO cells.
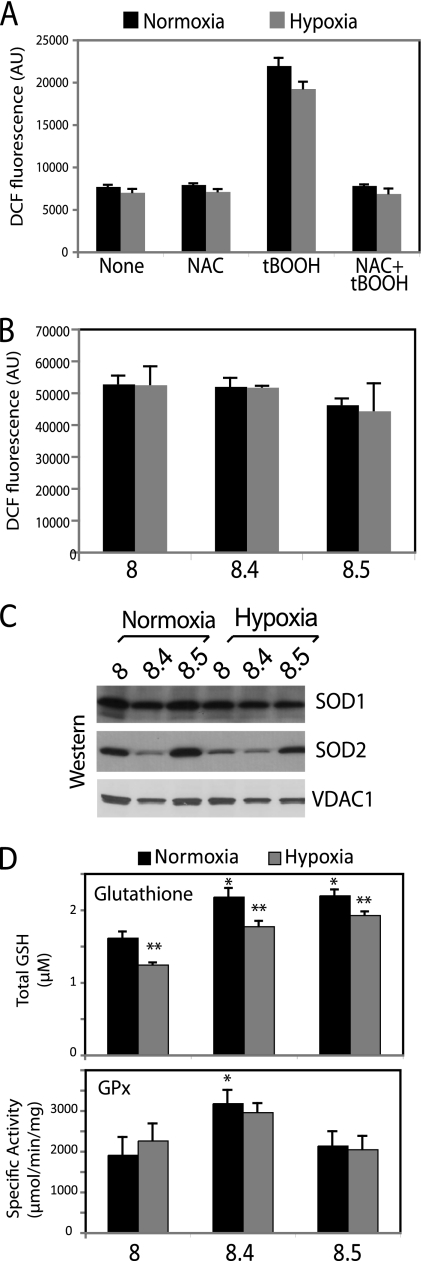
The production of free radicals under hypoxia remains controversial and unresolved. Therefore, we tested whether exposure of cells to low levels of oxygen resulted in increased free radical production in our cells. We measured hydrogen peroxide levels in control cells exposed to normoxia or hypoxia (1% O2) for 24 h. We were unable to observe any difference in H2O2 levels in control cells under hypoxia compared to the levels under normoxia (Fig. 10A). In contrast, in the positive control of the assay, cells that were treated with tert-butyl hydroperoxide (tBOOH) produced a significant increase in DCF fluorescence which was abrogated by the presence of NAC under both normoxic and hypoxic conditions (Fig. 10A). Using the same assay, we were unable to detect changes in the levels of H2O2 in RISP KO cells during hypoxia compared to the normoxic levels, indicating that our cells did not produce ROS when exposed to hypoxia (Fig. 10B).
Fig 10.
Ablation of RISP leads to an increase in cellular antioxidant defenses. (A) H2O2 levels were determined by DCF-DA fluorescence in control cells exposed to normoxia (21% O2) or hypoxia (1% O2) for 24 h. Cells were treated with either 100 μM tBOOH or a mixture of 2 mM NAC and 100 μM tBOOH to increase or suppress ROS production. AU, arbitrary units. (B) DCF-DA fluorescence of cells exposed to normoxia or hypoxia for 24 h. Values in panels A and B represent the means and standard deviations of fluorescence in triplicates of a representative experiment of at least 3 independent determinations. (C) Steady-state levels of the superoxide scavenger enzymes SOD1 and SOD2 in cells exposed to either normoxia or hypoxia for 24 h. (D) Total glutathione levels and glutathione peroxidase (GPx) activities in cells exposed to normoxia and hypoxia for 24 h. Values represent means and standard deviations. *, P ≤ 0.05 for KO cells compared to control in normoxia; **, P ≤ 0.05 for hypoxia compared to normoxia.
We also examined the antioxidant defense system under normoxia and hypoxia. The results in Fig. 10C show that the RISP clone 8.5 had higher steady-state levels of both SOD1 and SOD2 than the control and clone 8.4. The increased steady-state levels of these two proteins also correlated with increased levels in their enzymatic activity as assessed by in-gel activity assays (not shown). When cells were exposed to hypoxia (1% O2) for 24 h, the levels of SOD2 slightly decreased in the control and clone 8.5 (0.4- and 0.6-fold decrease, respectively), although in clone 8.4, the levels remained unchanged (Fig. 10C). After hypoxia, the levels of SOD1 in the control and clone 8.4 were unchanged, whereas clone 8.5 had a slight decrease of SOD1 levels of about 0.6-fold (Fig. 10C). Reduction of the levels of superoxide scavenger enzymes during hypoxia suggests less production of free radicals. Since SOD2 is an inducible enzyme, its high levels suggest increased levels of superoxide, particularly in the RISP KO clone 8.5. This could explain why we could not detect higher levels of mitochondrial superoxide with MitoSox in this clone (Fig. 8A). Higher levels of superoxide dismutase could counterbalance the mitochondrial ROS that is being produced.
The levels of GPx were significantly increased in the RISP KO clone 8.4 compared to levels in the control cell line (Fig. 10D). Exposure of cells to hypoxia did not alter the levels of GPx in any of the cells tested. However, the levels of total glutathione were significantly increased in RISP KO cells compared to the levels in the control in normoxia, in agreement with alterations in oxidative status. When cells were exposed to hypoxia, glutathione levels diminished significantly in all cell lines, suggesting a reduction in free radical production (Fig. 10D).
DISCUSSION
The present study characterized the effect of the deletion of the Rieske iron-sulfur protein on CIII assembly and showed that the interdependence and supramolecular organization of OXPHOS complexes are associated with the increased levels of ROS.
The complete assembly process of CIII is still under investigation, but work in yeast suggested that it starts with the formation of 3 subassembly intermediates composed by subunits cytb/Qcr7/Qcr8, cytc1/Qcr6/Qcr9, and Core1/Core2. By an unknown assembly process, these three intermediates form a precomplex, pre-CIII, to which only the last two subunits (RISP and Qcr10) need to be inserted (70, 71). In yeast, three chaperones, Cbp3, Cbp4, and Bcs1, aid in the assembly process (16, 39). The addition of RISP is catalyzed by Bcs1 in an ATP-dependent manner, and it appears that the structural dimerization of the complex occurs before RISP is incorporated (16). The mammalian CIII contains an additional subunit (UQCR9) that is derived by the proteolytic cleavage of the amino-terminal portion of RISP (5). It is not clear when the cleavage of RISP to produce UQCR9 occurs, but presumably it is after its addition into the complex by BCS1L (human homolog of Bcs1). Interestingly, the Core1 and Core2 subunits display mitochondrion-processing peptidase activity (18).
We did not detect major differences in the electrophoretic mobility of the fully assembled CIII and that of the one lacking RISP. Therefore, we have confirmed that in mammals, the dimerization and the formation of the pre-CIII assembly intermediate take place prior to RISP addition. Moreover, the pre-CIII appears to be structurally unstable, as we observed low levels of pre-CIII and subassembly intermediates/degradation products in the RISP KO clones. These results are in agreement with observations made in Bcs1L patients, where little incorporation of RISP into the pre-CIII occurs (25, 31, 36, 49).
The absence of RISP affected not only the levels of pre-CIII but also the levels of CI, CIV, and supercomplexes. Several possibilities that could contribute to the interdependence of respiratory complexes include the existence of specific threshold levels of respiratory complexes (17), specific subunits and lipid interactions (47, 52), or as we showed in this study, increased levels of ROS.
Mitochondria, in particular the electron transport chain (ETC), are major generators of ROS (reviewed in reference 41). The established sites for mitochondrial ROS production are CI, CII, and CIII. Complexes I and II produce superoxide within the mitochondrial matrix, whereas CIII releases the radicals into either the matrix or the mitochondrial intermembrane space (30, 35, 41, 72).
Analysis of RISP KO clones showed increased levels of ROS associated with destabilization of CI and supercomplexes. Complex I instability has been shown previously in both human and mouse cell lines with mutations in the cytochrome b gene that impeded the assembly of CIII (1). Studies in NDUFS4 KO mice showed that CI was stabilized by the formation of supercomplexes containing CIII in the absence of the CI subunit NDUFS4 (9).
Our results suggest that CI stability is affected by factors other than the “physical support” conferred by CIII to assemble into supercomplex structures. Here, we show that the presence of RISP per se is not required for CI stability, since the exposure of RISP KO fibroblasts to hypoxia and, presumably, low ROS was sufficient to increase CI to the levels observed in control cells. This indicates that the partial assembly of CIII into the pre-CIII in the RISP KO cells was enough to stabilize CI and supercomplex formation. Unfortunately, the previous studies on CI stability mentioned above did not investigate the effect of increased free radicals in stability/assembly (1, 9).
To test whether increased levels of ROS affected OXPHOS complex stability, we exposed control cells to different OXPHOS inhibitors known to increase free radicals. Studies in submitochondrial particles suggested that rotenone inhibits the binding of coenzyme Q to its reduction site in CI, permitting the release of electrons directly to oxygen at the N2 center and forming superoxide radicals (24). The generation of free radicals by CIII is intimately linked to its catalytic mechanism. During the Q cycle, ubiquinol is oxidized and two electrons enter CIII in a bifurcate fashion. According to this model, inhibition of CIII with antimycin A and myxothiazol, which bind to center N or center P, respectively, can generate superoxide anions. Myxothiazol binds close to heme bL and does not interfere with ubiquinol binding; in this way, ubiquinol electrons can access RISP but not heme bH, allowing for the formation of superoxide (61). Among the drugs that we used, CIII and CV inhibitors known to be ROS generators displayed the most detrimental effect on CI and supercomplex stability. Acin-Perez et al. (1) showed that antimycin A decreased the levels of CI in mouse control cells derived from the L929 line (subcutaneous aerolar and adipose tissue) (1). However, the decrease that they observed in CI levels was not as severe as the one we observed in this study. This difference could be attributed to variations in cell type or drug treatment time. It is possible that during their long treatment period with the drug (2 weeks), cells in culture adapted to the inhibition or that the “less fit” cells were selected against in growing cultures.
RISP is thought to be required for the production of reactive oxygen species by CIII, indicating that the increased levels of ROS observed in our RISP KO fibroblasts are probably generated by other sources (i.e., CI or CII). Along these lines, Hinson et al. (36) showed that the levels of free radicals produced by mitochondria from patients with Björnstad syndrome (Bcs1L mutations) and other CIII deficiencies were increased by 50 to 80%. The authors determined that the increased levels of H2O2 detected in the patients were produced by CI and not by CIII (36). More recently, Moran et al. confirmed increased levels of H2O2 and upregulation of ROS-scavenging enzymes in fibroblasts derived from patients with different mutations in the BCS1L gene. Remarkably, the levels of ROS in these patients correlated with the severity of the disease (49). These results support our findings of increased ROS in the RISP KO cells.
The production of free radicals under hypoxia is still controversial. In this paradox, researchers supporting hypoxia-induced ROS contend that at low oxygen levels, the flow of electrons through the electron transport chain slows down, increasing the likelihood for the electrons to escape and produce free radicals. In contrast, other reports showed decreased ROS during hypoxia (23, 48, 63). This controversy might rely on the nature of the free radical species measured. As pointed out by Poyton's group, not only reactive oxygen species but also reactive nitrogen species (RNS) can be generated during hypoxia (10, 53). Low levels of both ROS and RNS act as signaling molecules during physiological conditions, and only excessive amounts, usually generated during a pathological state, produce oxidative stress (34). A potential reconciliation of this controversy came from the studies measuring the oxidation state of redox-sensitive GFP (roGFP) probes in different cellular compartments. Waypa et al. (66) found that hypoxia increased the oxidation of roGFP in the cytosol and intermembrane space, whereas it decreased oxidation in the mitochondrial matrix (66). Decreased mitochondrial matrix ROS supports our observations of increased OXPHOS complex stability/assembly during hypoxia.
It has been proposed that CIII is the mitochondrial oxygen sensor that regulates cellular responses during hypoxia and that the ROS generated by CIII is responsible for stabilizing the transcription factor HIF1-α that initiates the hypoxic signaling cascade (reviewed in reference 11). This hypothesis is based in numerous studies performed in different cell types, including rho0 cells (cells devoid of mitochondrial DNA [mtDNA]), cytochrome b and cytochrome c mutants, specific OXPHOS inhibitors, mitochondrially targeted antioxidants, and knockdown studies (4, 6, 12, 13, 33, 46). Of particular interest are the studies on the effect of RISP knockdown on HIF1-α stability. Brunelle et al. (6) showed that transient transfection of two RISP small interfering RNA sequences led to a reduction in the stabilization of HIF1-α in Hek293 cells exposed to hypoxia (1.5% O2) for 2 h (6). Stable knockdown of RISP in 143B cells showed similar results, and when exogenous H2O2 was added, hypoxic HIF1-α stabilization was restored, suggesting that ROS is required for stability of the transcription factor (33). Hypoxic abrogation of HIF1-α stabilization upon knockdown of RISP (15, 50) and the role of ROS (46) have been reported in other cell types also. Interestingly, recent studies implicate another CIII subunit (UQCRB) in the oxygen-sensing role during hypoxia (37).
The use of MitoQ, a mitochondrially targeted antioxidant, supported the hypothesis that mitochondrial ROS was required for hypoxic signaling. Control and cytochrome b mutant cybrids pretreated with MitoQ prior to hypoxic exposure failed to stabilize HIF1-α (4). This concept has been recently challenged by Chua et al. (14). The authors observed that pharmacological inhibition of the electron transport chain decreased the HIF1-α half-life under hypoxia to the same extent independent of the complex inhibited and that there was no increase in ROS production during hypoxia (14). The authors proposed that, rather than requiring ROS produced by CIII, HIF1-α stability during hypoxia is related to the intracellular concentrations of oxygen, which are determined by the rate of mitochondrial respiration.
Similar to results of the RISP knockdown studies described above, we were able to detect a marked decrease in the levels of HIF1-α after exposing our RISP KO cells to hypoxia (1% O2) for 4 h. However, after 24 h of hypoxia, the levels of HIF1-α in KO cells increased, reaching levels that were even higher than the ones in control fibroblasts. Moreover, reducing mitochondrial ROS with different concentrations of MitoQ did not have a significant effect on HIF1-α stability. This observation suggests that HIF1-α stability in the RISP KO cells during hypoxia may be independent of ROS production, as has been suggested (14). The RISP KO cells had increased levels of superoxide, and antioxidant defenses were upregulated. Presumably, in our cells, ROS is generated by means other than CIII. However, we did not observe an increase in hydrogen peroxide during hypoxia.
We were surprised to find that hypoxia also increased the stability/assembly of CI, CIV, and supercomplexes. Although the results of the experiments described above do not rule out the participation of HIF1-α in this process, they showed that ROS is an active mediator of complex/supercomplex stability. Interestingly, there are two forms of CI: one form, called the A-form, is catalytically active, and the other, called the D-form, is catalytically inactive (65). Deactivation occurs when all reactive redox centers of CI are in the reduced state. During hypoxia, a deactivation of CI was observed in epithelial kidney cells (27). The authors proposed that the deactivation of CI could be a protective mechanism for a potential burst of free radicals during reoxygenation (27). Unfortunately, they did not investigate the stability of the CI and supercomplexes during hypoxia.
The fact that we observed that conditions producing high levels of free radicals (antimycin A, myxothiazol, and oligomycin) led to CI instability and affected the levels of supercomplexes and, conversely, that conditions of lower oxygen and, presumably, lower levels of free radicals preserved supercomplexes even in OXPHOS-defective cells prompts us to propose a possible model for regulation of OXPHOS complex interactions into supercomplexes. Figure 11 illustrates this model: in wild-type cells, OXPHOS complexes are able to form stable supercomplexes. Alterations in OXPHOS function can produce increased free radicals, which are potentially dangerous. To avoid this, CI is degraded and supercomplexes disassembled. Under conditions of low oxygen or increased superoxide scavengers (SOD2 or MnTBAP), less ROS is produced, resulting in the restoration of a safe environment for the respiratory complexes and supercomplexes to reassemble. In addition, supercomplex stability/assembly could be further regulated by the expression of hypoxia-specific isoforms of subunits of the respiratory chain and could be mediated by HIF1-α.
Fig 11.
Model of hypoxic-induced stability of OXPHOS complexes and supercomplexes. The mitochondrial OXPHOS complexes are able to associate, forming stable supercomplexes in wild-type (WT) cells. Defects in OXPHOS function, such as the absence of RISP, can cause increased levels of potentially dangerous superoxide. To avoid this, CI is degraded and supercomplexes disassembled. CI stability was particularly compromised by the absence of RISP. Under conditions of low oxygen or increased superoxide scavengers, less ROS is produced, resulting in the restoration of a safe environment for reassembly of OXPHOS complexes and supercomplexes. HIF1-α might mediate assembly/stability by expressing hypoxia-specific isoforms. Supercomplexes may confer protection against increased ROS upon future changes in oxygen levels.
Our results also addressed a previously puzzling observation. Mouse tissues defective in CIV (21, 22) or CIII (F. Diaz, unpublished observations) did not show a decrease in CI, as consistently observed in cultured cells. However, the oxygen concentration in tissues is 3 to 6%, which is markedly lower than the 21% concentration observed in cultured cells. Therefore, we feel compelled to speculate that increases in tissue oxygenation (physiological or pathological) may lead to a signaling pathway that controls the levels of CI and supercomplexes to avoid further exacerbation of the oxidative stress, as postulated in our model.
In conclusion, our data suggest that localized increases in ROS levels at the mitochondrial inner membrane affect the assembly/stability of CI and supercomplexes. CI appears to be particularly sensitive to this oxidant environment effect. This system may be physiologically relevant for the control of respiration and ROS levels.
ACKNOWLEDGMENTS
We express our gratitude to Alejandro Ocampo, Xiao Wang, Sofia Garcia, and Ana LaTorre for their help with flow cytometry and technical assistance, to T. Wenz for insightful discussions, and to A. Barrientos and E. Perales-Clemente for critical review of the manuscript. We also thank Mike Murphy and Rob Smith for providing MitoQ and dTPP.
We are indebted to the following funding agencies for supporting our work: the James and Esther King Biomedical Research Program, Florida Department of Health, for grant 08KN-01 (F.D.), PHS for grants NS041777, CA085700, and EY10804 (C.T.M.), and the Spanish Ministry of Education for DGA-B55 grants SAF2009-08007 and CSD2007-00020 (J.A.E.). The CNIC is supported by the Spanish Ministry of Science and Innovation and the Pro-CNIC Foundation.
Footnotes
Published ahead of print 21 November 2011
REFERENCES
- 1. Acin-Perez R, et al. 2004. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Acín-pérez R, Fernández-Silva P, Peleato ML, Pérez-Martos A, Enriquez JA. 2008. Respiratory active mitochondrial supercomplexes. Mol. Cell 32: 529–539 [DOI] [PubMed] [Google Scholar]
- 3. Barrientos A, Fontanesi F, Diaz F. 2009. Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr. Protoc. Hum. Genet. Chapter 19:Unit19.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bell EL, et al. 2007. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J. Cell Biol. 177: 1029–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brandt U, Yu L, Yu CA, Trumpower BL. 1993. The mitochondrial targeting presequence of the Rieske iron-sulfur protein is processed in a single step after insertion into the cytochrome bc1 complex in mammals and retained as a subunit in the complex. J. Biol. Chem. 268: 8387–8390 [PubMed] [Google Scholar]
- 6. Brunelle JK, et al. 2005. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 1: 409–414 [DOI] [PubMed] [Google Scholar]
- 7. Bultema JB, Braun HP, Boekema EJ, Kouril R. 2009. Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim. Biophys. Acta 1787: 60–67 [DOI] [PubMed] [Google Scholar]
- 8. Calvaruso MA, Smeitink J, Nijtmans L. 2008. Electrophoresis techniques to investigate defects in oxidative phosphorylation. Methods 46: 281–287 [DOI] [PubMed] [Google Scholar]
- 9. Calvaruso MA, et al. 7 October 2011. Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum. Mol. Gen. [Epub ahead of print [DOI] [PubMed] [Google Scholar]
- 10. Castello PR, David PS, McClure T, Crook Z, Poyton RO. 2006. Mitochondrial cytochrome oxidase produces nitric oxide under hypoxic conditions: implications for oxygen sensing and hypoxic signaling in eukaryotes. Cell metabolism 3: 277–287 [DOI] [PubMed] [Google Scholar]
- 11. Chandel NS. 2010. Mitochondrial complex III: an essential component of universal oxygen sensing machinery? Respir. Physiol. Neurobiol. 174: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chandel NS, et al. 1998. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. U. S. A. 95: 11715–11720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chandel NS, et al. 2000. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem/ 275: 25130–25138 [DOI] [PubMed] [Google Scholar]
- 14. Chua YL, et al. 2010. Stabilization of hypoxia-inducible factor-1alpha protein in hypoxia occurs independently of mitochondrial reactive oxygen species production. J. Biol. Chem. 285: 31277–31284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Comito G, et al. 2011. HIF-1alpha stabilization by mitochondrial ROS promotes Met-dependent invasive growth and vasculogenic mimicry in melanoma cells. Free Rad. Biol. Med. 51: 893–904 [DOI] [PubMed] [Google Scholar]
- 16. Cruciat CM, Hell K, Folsch H, Neupert W, Stuart RA. 1999. Bcs1p, an AAA-family member, is a chaperone for the assembly of the cytochrome bc(1) complex. EMBO J. 18: 5226–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. D'Aurelio M, Gajewski CD, Lenaz G, Manfredi G. 2006. Respiratory chain supercomplexes set the threshold for respiration defects in human mtDNA mutant cybrids. Hum. Mol. Genet. 15: 2157–2169 [DOI] [PubMed] [Google Scholar]
- 18. Deng K, Shenoy SK, Tso SC, Yu L, Yu CA. 2001. Reconstitution of mitochondrial processing peptidase from the core proteins (subunits I and II) of bovine heart mitochondrial cytochrome bc(1) complex. J. Biol. Chem. 276: 6499–6505 [DOI] [PubMed] [Google Scholar]
- 19. Diaz F, Barrientos A, Fontanesi F. 2009. Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using blue native gel electrophoresis. Curr. Protoc. Hum. Genet. Chapter 19:Unit19.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diaz F, Fukui H, Garcia S, Moraes CT. 2006. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 26: 4872–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diaz F, et al. 2008. Pathophysiology and fate of hepatocytes in a mouse model of mitochondrial hepatopathies. Gut 57: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Diaz F, Thomas CK, Garcia S, Hernandez D, Moraes CT. 2005. Mice lacking COX10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum. Mol. Genet. 14: 2737–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fandrey J, Frede S, Jelkmann W. 1994. Role of hydrogen peroxide in hypoxia-induced erythropoietin production. Biochem. J. 303(Pt 2): 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fato R, et al. 2009. Differential effects of mitochondrial complex I inhibitors on production of reactive oxygen species. Biochim. Biophys. Acta 1787: 384–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fernandez-Vizarra E, et al. 2007. Impaired complex III assembly associated with BCS1L gene mutations in isolated mitochondrial encephalopathy. Hum. Mol. Genet. 16: 1241–1252 [DOI] [PubMed] [Google Scholar]
- 26. Frenzel M, Rommelspacher H, Sugawa MD, Dencher NA. 2010. Ageing alters the supramolecular architecture of OxPhos complexes in rat brain cortex. Exp. Gerontol. 45: 563–572 [DOI] [PubMed] [Google Scholar]
- 27. Galkin A, Abramov AY, Frakich N, Duchen MR, Moncada S. 2009. Lack of oxygen deactivates mitochondrial complex I: implications for ischemic injury? J. Biol. Chem. 284: 36055–36061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garcia S, Diaz F, Moraes CT. 2008. A 3′ UTR modification of the mitochondrial Rieske iron sulfur protein in mice produces a specific skin pigmentation phenotype. J. Investig. Dermatol. 128: 2343–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Genova ML, et al. 2008. Is supercomplex organization of the respiratory chain required for optimal electron transfer activity? Biochim. Biophys. Acta 1777: 740–746 [DOI] [PubMed] [Google Scholar]
- 30. Genova ML, et al. 2001. The site of production of superoxide radical in mitochondrial complex I is not a bound ubisemiquinone but presumably iron-sulfur cluster N2. FEBS Lett. 505: 364–368 [DOI] [PubMed] [Google Scholar]
- 31. Gil-Borlado MC, et al. 2009. Pathogenic mutations in the 5′ untranslated region of BCS1L mRNA in mitochondrial complex III deficiency. Mitochondrion 9: 299–305 [DOI] [PubMed] [Google Scholar]
- 32. Graham LA, Trumpower BL. 1991. Mutational analysis of the mitochondrial Rieske iron-sulfur protein of Saccharomyces cerevisiae. III. Import, protease processing, and assembly into the cytochrome bc1 complex of iron-sulfur protein lacking the iron-sulfur cluster. J. Biol. Chem. 266: 22485–22492 [PubMed] [Google Scholar]
- 33. Guzy RD, et al. 2005. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 1: 401–408 [DOI] [PubMed] [Google Scholar]
- 34. Hamanaka RB, Chandel NS. 2010. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 35: 505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Han D, Williams E, Cadenas E. 2001. Mitochondrial respiratory chain-dependent generation of superoxide anion and its release into the intermembrane space. Biochem. J. 353: 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hinson JT, et al. 2007. Missense mutations in the BCS1L gene as a cause of the Björnstad syndrome. N. Engl. J. Med. 356: 809–819 [DOI] [PubMed] [Google Scholar]
- 37. Jung HJ, et al. 2010. Terpestacin inhibits tumor angiogenesis by targeting UQCRB of mitochondrial complex III and suppressing hypoxia-induced reactive oxygen species production and cellular oxygen sensing. J. Biol. Chem. 285: 11584–11595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kelso GF, et al. 2001. Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. J. Biol. Chem. 276: 4588–4596 [DOI] [PubMed] [Google Scholar]
- 39. Kronekova Z, Rodel G. 2005. Organization of assembly factors Cbp3p and Cbp4p and their effect on bc(1) complex assembly in Saccharomyces cerevisiae. Curr. Genet. 47: 203–212 [DOI] [PubMed] [Google Scholar]
- 40. Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. 2007. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol. Cell. Biol. 27: 4228–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lenaz G. 2001. The mitochondrial production of reactive oxygen species: mechanisms and implications in human pathology. IUBMB Life 52: 159–164 [DOI] [PubMed] [Google Scholar]
- 42. Lenaz G, Genova ML. 2010. Structure and organization of mitochondrial respiratory complexes: a new understanding of an old subject. Antioxid. Redox Signal. 12: 961–1008 [DOI] [PubMed] [Google Scholar]
- 43. Li Y, et al. 2007. An assembled complex IV maintains the stability and activity of complex I in mammalian mitochondria. J. Biol. Chem. 282: 17557–17562 [DOI] [PubMed] [Google Scholar]
- 44. Liu Y, Schubert DR. 2009. The specificity of neuroprotection by antioxidants. J. Biomed. Sci. 16: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lochmuller H, Johns T, Shoubridge EA. 1999. Expression of the E6 and E7 genes of human papillomavirus (HPV16) extends the life span of human myoblasts. Exp. Cell Res. 248: 186–193 [DOI] [PubMed] [Google Scholar]
- 46. Mansfield KD, et al. 2005. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 1: 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McKenzie M, Lazarou M, Thorburn DR, Ryan MT. 2006. Mitochondrial respiratory chain supercomplexes are destabilized in Barth syndrome patients. J. Mol. Biol. 361: 462–469 [DOI] [PubMed] [Google Scholar]
- 48. Mehta JP, et al. 2008. Generation of oxidants by hypoxic human pulmonary and coronary smooth-muscle cells. Chest 133: 1410–1414 [DOI] [PubMed] [Google Scholar]
- 49. Moran M, et al. 2010. Cellular pathophysiological consequences of BCS1L mutations in mitochondrial complex III enzyme deficiency. Hum. Mutat. 31: 930–941 [DOI] [PubMed] [Google Scholar]
- 50. Patten DA, et al. 2010. Hypoxia-inducible factor-1 activation in nonhypoxic conditions: the essential role of mitochondrial-derived reactive oxygen species. Mol. Biol. Cell 21: 3247–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Perales-Clemente E, et al. 2010. Five entry points of the mitochondrially encoded subunits in mammalian complex I assembly. Mol. Cell. Biol. 30: 3038–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pfeiffer K, et al. 2003. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278: 52873–52880 [DOI] [PubMed] [Google Scholar]
- 53. Poyton RO, Ball KA, Castello PR. 2009. Mitochondrial generation of free radicals and hypoxic signaling. Trends Endocrinol. Metab. 20: 332–340 [DOI] [PubMed] [Google Scholar]
- 54. Rosca MG, et al. 2008. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovasc. Res. 80: 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saddar S, Dienhart MK, Stuart RA. 2008. The F1F0-ATP synthase complex influences the assembly state of the cytochrome bc1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J. Biol. Chem. 283: 6677–6686 [DOI] [PubMed] [Google Scholar]
- 56. Salnikow K, Su W, Blagosklonny MV, Costa M. 2000. Carcinogenic metals induce hypoxia-inducible factor-stimulated transcription by reactive oxygen species-independent mechanism. Cancer Res. 60: 3375–3378 [PubMed] [Google Scholar]
- 57. Sanjuan-Pla A, et al. 2005. A targeted antioxidant reveals the importance of mitochondrial reactive oxygen species in the hypoxic signaling of HIF-1alpha. FEBS Lett. 579: 2669–2674 [DOI] [PubMed] [Google Scholar]
- 58. Schagger H, Pfeiffer K. 2001. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J. Biol. Chem. 276: 37861–37867 [DOI] [PubMed] [Google Scholar]
- 59. Soto IC, et al. 2009. Synthesis of cytochrome c oxidase subunit 1 is translationally downregulated in the absence of functional F1F0-ATP synthase. Biochim. Biophys. Acta 1793: 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Strecker V, Wumaier Z, Wittig I, Schagger H. 2010. Large pore gels to separate mega protein complexes larger than 10MDa by blue native electrophoresis: isolation of putative respiratory strings or patches. Proteomics 10: 3379–3387 [DOI] [PubMed] [Google Scholar]
- 61. Sun J, Trumpower BL. 2003. Superoxide anion generation by the cytochrome bc1 complex. Arch. Biochem. Biophys. 419: 198–206 [DOI] [PubMed] [Google Scholar]
- 62. Trumpower BL. 1990. The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex. J. Biol. Chem. 265: 11409–11412 [PubMed] [Google Scholar]
- 63. Vaux EC, Metzen E, Yeates KM, Ratcliffe PJ. 2001. Regulation of hypoxia-inducible factor is preserved in the absence of a functioning mitochondrial respiratory chain. Blood 98: 296–302 [DOI] [PubMed] [Google Scholar]
- 64. Vempati UD, Han X, Moraes CT. 2009. Lack of cytochrome c in mouse fibroblasts disrupts assembly/stability of respiratory complexes I and IV. J. Biol. Chem. 284: 4383–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vinogradov AD. 1998. Catalytic properties of the mitochondrial NADH-ubiquinone oxidoreductase (complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim. Biophys. Acta 1364: 169–185 [DOI] [PubMed] [Google Scholar]
- 66. Waypa GB, et al. 2010. Hypoxia triggers subcellular compartmental redox signaling in vascular smooth muscle cells. Circ. Res. 106: 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wenz T, et al. 2009. Role of phospholipids in respiratory cytochrome bc(1) complex catalysis and supercomplex formation. Biochim. Biophys. Acta 1787: 609–616 [DOI] [PubMed] [Google Scholar]
- 68. Weydert CJ, Cullen JJ. 2010. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 5: 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wittig I, Carrozzo R, Santorelli FM, Schagger H. 2006. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim. Biophys. Acta 1757: 1066–1072 [DOI] [PubMed] [Google Scholar]
- 70. Zara V, Conte L, Trumpower BL. 2009. Evidence that the assembly of the yeast cytochrome bc1 complex involves the formation of a large core structure in the inner mitochondrial membrane. FEBS J. 276: 1900–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zara V, Conte L, Trumpower BL. 2007. Identification and characterization of cytochrome bc(1) subcomplexes in mitochondria from yeast with single and double deletions of genes encoding cytochrome bc(1) subunits. FEBS J. 274: 4526–4539 [DOI] [PubMed] [Google Scholar]
- 72. Zhang L, Yu L, Yu CA. 1998. Generation of superoxide anion by succinate-cytochrome c reductase from bovine heart mitochondria. J. Biol. Chem. 273: 33972–33976 [DOI] [PubMed] [Google Scholar]