Abstract
During V(D)J recombination, RAG targeting to correct sites versus off-target sites relies on both DNA sequence features and on chromatin marks. Kinetic analysis using the first highly active full-length purified RAG1/RAG2 complexes has now allowed us to define the important catalytic features of this complex. We found that the overall rate of nicking, but not hairpinning, is critical for the discrimination between correct (optimal) versus off-target (suboptimal) sites used in human T-cell lymphomas, and we show that the C-terminal portion of RAG2 is required for this. This type of kinetic analysis permits us to analyze only the catalytically active RAG complex, in contrast to all other methods, which are unavoidably confounded by mixture with inactive RAG complexes. Moreover, we can distinguish the two major features of any enzymatic catalysis: the binding constant (KD) and the catalytic turnover rate, kcat. Beyond a minimal essential threshold of heptamer quality, further suboptimal heptamer deviations primarily reduce the catalytic rate constant kcat for nicking. Suboptimal nonamers reduce not only the binding of the RAG complex to the recombination site (KD) but also the catalytic rate constant, consistent with a tight interaction between the RAG complex and substrate during catalysis. These features explain many aspects of RAG physiology and pathophysiology.
INTRODUCTION
The most fundamental aspect of any site-specific recombination process is how it achieves its site specificity, and this is also relevant for all nuclear sequence-specific proteins, including transcription factors. V(D)J recombination is the only truly site-specific recombination process in multicellular eukaryotes, because Ig class switch recombination is a regionally specific process (22). V(D)J recombination relies on RAG1 and RAG2 forming a complex that recognizes recombination signal sequences (RSSs) composed of a palindromic heptamer (5′-CACAGTG) and an AT-rich nonamer (5′-ACAAAAACA) (13, 33, 34). The heptamer and nonamer are separated by either a 12- or a 23-bp spacer, thereby specifying the 12RSS or the 23RSS. A single recombination event is initiated between one 12RSS and one 23RSS when double-strand breaks (DSBs) are created directly 5′ of the heptamer. The first step in creating these DSBs occurs when the RAG complex creates a nick at each of these RSS sites, followed by RAG use of the 3′-OH at that nick as a nucleophile to attack the strand opposite of the nick. This results in a hairpin at one DNA end (the coding end) and a blunt DNA end (5′-P and 3′-OH) at the RSS end (32, 35).
This DNA chemistry can be enzymatically catalyzed by segments of RAG1 and RAG2 that are called the core regions (designated with a c) (13, 33, 34). Enzymatic studies using both full-length (designated with an f) RAG1 and RAG2 have been rare because of the extremely poor solubility of these proteins (12, 37). The first report to examine the enzymatic properties of both fRAG1 and fRAG2 showed that this complex has poor cleavage activity at the RSS; however, it was not clear whether it was because of a lower specific activity of the protein itself or a lower percentage of active protein within the preparation (12). RAG complexes with fRAG1 and cRAG2 or with cRAG1 and fRAG2 have been successfully purified and used to investigate the properties of the noncore region. Studies of cRAG1 together with fRAG2 have nicely demonstrated that a plant homeodomain (PHD) finger in the noncore C-terminal region of RAG2 has a role in binding of the RAG complex to histone tails that have the H3K4me3 modification (24, 25, 30). This histone modification increases the kcat for nicking by the RAG complex, in addition to tethering the RAG complex to the region of active chromatin (36).
Human T-cell lymphomas are noteworthy because they illustrate the obvious danger of cell types that have evolved a site-specific recombination process in a multicellular organism (43). In human T-cell lymphomas, the most common initial genetic lesion is off-target action by the RAG complex at sites that are similar to RSSs, called cryptic RSSs (cRSSs). Previous work using replicating extrachromosomal substrates harboring cRSS sites demonstrated that recombination via those cRSS sites relies on the RAG complex in vivo (29). The choice of these cRSSs is likely reliant on the same H3K4me3 histone mark that is needed at physiologic RSS sites (36).
Although the open chromatin structure carrying the H3K4me3 mark and histone H3 and H4 hyperacetylation help narrow the fraction of the genome in which the RAG complex can act, the large fraction of the genome still included permits a large potential for off-target action (2–5, 17, 24, 25). It is clear that the RAG complex must somehow achieve sequence-specific discrimination between RSS and off-target (cRSS) sites, but the nature of such discrimination has been quite unclear.
Here, we report a set of major advances in our understanding of how the full-length (ff) RAG complex achieves sequence-dependent targeting or discrimination between RSS sites and off-target sites. This was possible for our study only because we have been able to generate the first highly active RAG complexes that are full length for both RAG1 and RAG2. We show here that fRAG2 is critical for the discrimination between optimal and suboptimal RSS sites. This function of the RAG2 C terminus is above and beyond its role in recognizing the H3K4me3 histone mark (24, 25). Hence, the RAG2 C terminus has three functions: (i) DNA sequence discrimination that determines the rate of nicking (this study), (ii) H3K4me3 binding (24, 25), and (iii) nick rate stimulation by H3K4me3 (15, 36). (It also has an important function in cell cycle regulation of RAG degradation [19, 23].) We used enzyme kinetic analysis of catalytically active RAG complexes to dissect the contributions of the heptamer and nonamer to the discrimination by the RAG complex between optimal RSS sites and suboptimal ones. This is the first opportunity in RAG biochemistry for assessment of binding of only the active RAG complexes, distinct from the catalytically inactive complexes. The overall rate of the nicking step can be formulated as a Michaelis-Menton reaction between active RAG complexes and RSS substrates, and this is reflected in the KM and the kcat values. We found that the nonamer quality affected both RAG complex binding (the KD, which we determine from the KM) and the catalytic rate constant kcat. However, the catalytic rate constant kcat for the nicking step is the major discriminating factor between RSS and off-target cRSS sites. These insights provide a unification of our understanding of the mechanism for how off-target events are suppressed normally and how such protective mechanisms fail on rare occasions and give rise to human T-cell lymphomas.
MATERIALS AND METHODS
Oligonucleotides.
Detailed information about the oligonucleotides used in this study is provided in the supplemental material.
Protein expression and purification.
Construction of the expression vectors for the maltose binding protein (MBP) fused to the core region or full-length form of mouse RAG1 and RAG2 have been described elsewhere (36). Proteins were purified over an amylose resin column (New England Biolab) exactly as described previously (1). Recombinant full-length HMGB1 protein was expressed in bacteria and purified as described previously (1). The success in obtaining active ff RAG complexes here was due to the higher expression of the EF1-α promoter and the simian virus 40 origin-driven replication of the vector in 293T cells.
Oligonucleotide cleavage.
An in vitro RAG nicking reaction was described previously (36). Briefly, the reaction mixture contained 20 nM 5′-end-labeled substrate, 40 nM each RAG complex, and 1 μM HMGB1 in a 5- or 10-μl final volume with a buffer composition of 25 mM K-morpholinepropanesulfonic acid (MOPS)–KOH (pH 7.0), 30 mM potassium glutamate, 30 mM potassium chloride (KCl), 5 mM MgCl2, 1 mM dithiothreitol, and 0.05 mg/ml bovine serum albumin and was incubated at 37°C for 30 min. Products were separated by sequencing gel and visualized using a Molecular Image FX apparatus (Bio-Rad). The intensity of autoradiography was quantified with the Quantity One software (Bio-Rad). In an in vitro hairpinning assay, the reaction mixture containing 20 nM labeled prenicked RSS, SIL, or SCL substrate was analyzed under the same conditions as those used in the in vitro nicking assay, but the mixture was further supplemented with 20 nM prenicked cold partner before addition of proteins. The reaction mixture was incubated at 37°C for 60 min.
Kinetic analysis of nicking.
A burst kinetic assay to determine the active fraction in a protein preparation was described previously (36). Briefly, various concentrations of RAG protein were tested in in vitro nicking assays with 12RSS oligonucleotide substrates, and the burst amount of nicked product was determined by extrapolating from the 2- to 6-min period back to time zero.
The active fraction was determined based on the slope of the plot of the burst products as a function of the concentrations of RAG proteins. An assay for initial rate of nicking reaction was described previously (36). Briefly, RAG proteins were tested in nicking assays with various concentrations of substrate. The reactions were incubated for periods of up to 2 to 6 min at 37°C. The constants of kinetic equations were determined by curve fitting using Delta Graph software (SPSS Inc. and Red Rock Software Inc.).
RESULTS
Experimental system.
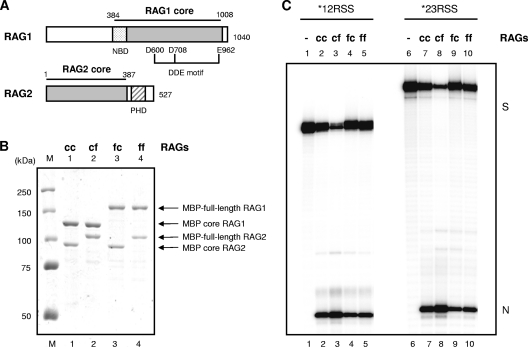
Nearly all RAG biochemistry reported to date has been obtained with one or both of the RAG1 and -2 proteins in a truncated configuration (RAG1 core, amino acids [aa] 384 to 1008 out of a total 1,040 aa; RAG2 core, aa 1 to 387 out of 527 aa) (Fig. 1A). Rare experiments describing full-length RAG1/2 complexes have reported very low enzyme activities, making conclusions very difficult (12, 37). However, recent studies have revealed that noncore regions of RAG proteins have important roles in regulating V(D)J recombination (20). The PHD finger of RAG2 located in the noncore region directly interacts with histone H3, with trimethylation on Lys4 (24, 25), and this interaction stimulates RAG cleavage activity, thereby resulting in improved efficiencies of V(D)J recombination (36). To explore further regulatory functions of the noncore regions, it was necessary to have both components in full-length form for a complete RAG complex. Here, we report full-length RAG1/2 complexes with enzyme activities that are similar to the activity of core RAG1/2 complexes (Fig. 1). Both RAG1 and RAG2 are coexpressed in 293T cells by using plasmid expression vectors with an N-terminal MBP tag for purification purposes are used, which increases the solubility of the RAG protein. The resulting proteins are shown in Fig. 1A. We prepared four different combinations of the RAG proteins: cRAG1/cRAG2 (cc), cRAG1/fRAG2 (cf), fRAG1/cRAG2 (fc), and fRAG1/fRAG2 (ff). All of the RAG complexes were catalytically active in in vitro nicking assays with single RSS substrates (Fig. 1C), as well as in coupled cleavage assays using 12RSS and 23RSS substrates together (data not shown).
Fig 1.
The full-length RAG1 and RAG2 complexes are as active as the truncated core RAG complex in vitro. (A) Schematic representation of RAG1 and RAG2. RAG1 core, aa 384 to 1008; NBD, nonamer-binding domain, aa 389 to 446; DDE motif, aa 600, 708, and 962. RAG2 core, aa 1 to 387; PHD finger, aa 414 to 481. (B) Purified RAG proteins analyzed by SDS-PAGE. Lane 1, MBP core RAG1/MBP core RAG2 (cc); lane 2, MBP core RAG1/MBP full-length RAG2 (cf); lane 3, MBP full-length RAG1/MBP core RAG2 (fc); lane 4, MBP full-length RAG1/MBP full-length RAG2 (ff). Each protein combination was expressed in 293T cells and purified by amylose resin column chromatography. Purified proteins were fractionated on SDS-PAGE with molecular mass standards and stained with Coomassie blue. (C) Purified RAG protein complexes were tested in in vitro nicking assays with 12RSS (left) or 23RSS (right) oligonucleotide substrate for 30 min at 37°C, as outlined in Materials and Methods. The products were analyzed on 10% denaturing PAGE gels, followed by autoradiography. The positions of substrate (S) and nicked (N) products are indicated in the right margin.
Recombinant RAG proteins include both catalytically active and inactive enzymes, as we found previously (36, 45). Therefore, it is necessary to determine the catalytically active fraction, in order to compare cleavage efficiencies of each protein preparation. Burst kinetic analysis was done with each RAG preparation across a range of substrate concentrations (7) (Table 1). Having determined the percentage of active fraction in each preparation, we calculated the concentration of active enzyme. The nicking efficiencies were defined as the number of nicked product molecules per active RAG complex (a RAG complex was taken to be a tetramer consisting of 2 RAG1 and 2 RAG2 proteins [13, 42]). As shown in Table 1, the number of nicked product molecules per active RAG complex for cc, cf, fc, and the two independent preparations of ff were between 1.1 and 1.9 catalytic events, indicating that both ff RAG complex preparations were at least as efficient as the cc RAG complex in an in vitro nicking assay.
Table 1.
Nicking efficiencies for optimal RSSs by full-length or truncated RAG complexesa
| Parameter | Value for RAG combination |
||||
|---|---|---|---|---|---|
| cRAG1/cRAG2 | cRAG1/fRAG2 | fRAG1/cRAG2 | fRAG1/fRAG21 | fRAG1/fRAG22 | |
| Nicked product concn (nM) | |||||
| 12RSS | 8.3 | 14.1 | 6.1 | 7.2 | 5.7 |
| 23RSS | 8.5 | 14.6 | 4.7 | 6.6 | 5.9 |
| Active RAG complex (%) | 17.6 | 33.4 | 10.3 | 9.3 | 12 |
| Active RAG complex concn (nM) | 7 | 13.4 | 4.1 | 3.7 | 4.8 |
| Product/active RAG ratio | |||||
| 12RSS | 1.2 | 1.1 | 1.5 | 1.9 | 1.2 |
| 23RSS | 1.2 | 1.1 | 1.1 | 1.8 | 1.2 |
Summary of the nicking assay with 12RSS and 23RSS with the cc, cf, fc, or two independent preparations (indicated by a superscript 1 or 2) of the ff RAG complex. The nicking assay mixture contained 20 nM DNA substrate and 40 nM RAG protein, and reaction mixtures were incubated for 30 min at 37°C. The concentration of nicked product (in nM) represents the mean of at least six independent experiments. The active RAG complex (as a percentage) of each RAG preparation was determined using burst kinetics. The concentration of active RAG complex (in nM) was calculated simply from the percentage of active RAG complex present in the 40 nM total RAG protein used in the reaction mixtures, assuming a RAG tetramer (2 RAG1 and 2 RAG2 monomers). Efficiencies of nicking at optimal RSSs by the RAG complex were defined as follows: (nicked product concentration)/(active RAG complex concentration).
The C terminus of RAG2 in discrimination between RSS and off-target sites used in human lymphomas.
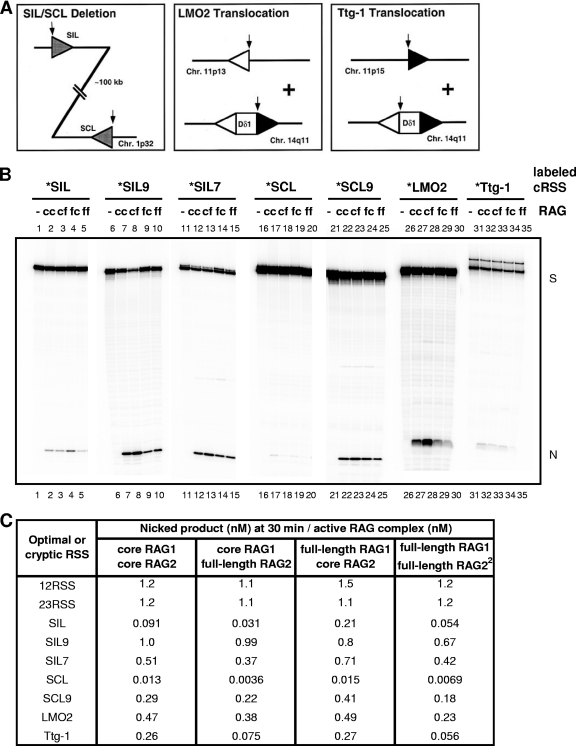
A fundamental question concerning RAG complexes is how they discriminate between optimal RSS sites and suboptimal ones, such as the ones commonly found in human T-cell lymphomas (e.g., SIL, SCL, LMO2, and Ttg1 sites) (Fig. 2A) (29, 46). Using the highly active full-length RAG complexes and combinations of truncated RAGs for comparison, we measured nicking at RSS sites and at suboptimal RSS sites (abbreviated cRSS for cryptic RSS) found in T-cell lymphomas as short oligonucleotide substrates (a representative gel image is shown in Fig. 2B). Figure 2C provides a summary of the nicking efficiencies defined by the nicked product (in nM) after 30 min for the suboptimal RSSs divided by the concentration of active RAG complex (in nM). Both full-length and truncated RAG complexes can variably nick cRSSs in vitro. Interestingly, the cf RAG complex had similar activity to the cc RAG complex at optimal RSS when the proteins were normalized for their active fractions; however, this complex was less active at many suboptimal sites (Fig. 2B and C). The signal sequence specificity of cc, cf, fc, and ff RAG complexes, defined by the nicking at 12RSS or 23RSS divided by the nicking at suboptimal RSSs (SIL, SCL, and Ttg-1), showed an interesting trend (Fig. 3A and B). The trend was that nicking was maximally efficient for RSS sites over cRSS sites when full-length RAG2 was used (Fig. 2C and 3). Core RAG2 makes a RAG complex that is less discriminating for RSS quality, a point suggested in a previous study (46). The full-length status of RAG1 is not relevant for this discrimination. Therefore, based on our in vitro studies here, a key role of the C-terminal (noncore) domain of RAG2 is in sequence discrimination among RSS sites, beyond its known role in H3K4me3 binding. Not surprisingly, as the quality of an RSS becomes increasingly close to optimal, even the ff RAG complex does not distinguish it (Fig. 3C and D).
Fig 2.
Nicking efficiencies by the full-length RAG1 and RAG2 complexes and truncated RAG complex at suboptimal RSS sites in vitro. (A) Diagram of the SIL/SCL, LMO2, and Ttg-1 events. The arrows in each panel indicate the proposed break region on the respective chromosome (29). The triangles represent the off-target RSS sites at the translocation junctions. The SIL/SCL deletion panel depicts the SIL/SCL interstitial deletion. Heptamer/nonamer-like sequences, indicated as triangles, are present adjacent to breakpoints at SIL and SCL genes. The triangles are intermediately shaded between those of 12RSS and 23RSS because of the uncertainty as to which RSS they are mimicking. The approximate length of the interstitial deletion is 100 kb. LMO2 translocation, the translocation breakpoint of the LMO2 gene at chromosome 11p13, is depicted. The breakpoint junction at chromosome 14q11 is located at the 23RSS of Dδ1. Ttg-1 translocation, the translocation breakpoint of the Ttg-1 gene at chromosome 11p15, is also depicted. The breakpoint junction at chromosome 14q11 is located at the12RSS of Dδ1. (B) Nicking of off-target T-cell lymphoma RSS sites by the full-length and truncated RAG complex in vitro. A representative image of in vitro nicking assay results is shown for cRSS oligonucleotide substrates by the cc, cf, fc, or ff RAG complex. All reactions mixtures, which included 20 nM DNA substrate and 40 nM RAG protein, were incubated for 30 min at 37°C as outlined in Materials and Methods. The products were analyzed on a 13% denaturing PAGE gel, followed by autoradiography. The positions of substrate (S) and nicked (N) products are indicated in the right margin. (C) The concentration of nicked product of optimal or suboptimal RSS site generated by cc, cf, fc, or ff RAG complex at the 30-min reaction time point. The table shows catalytic activities of cc, cf, fc, and ff RAG complexes at the optimal or suboptimal RSS site generated at the 30-min reaction time point. The nicked product concentration at 30 min generated by each RAG complex was divided by the active protein concentration for each preparation. Reactions were started with 20 nM DNA substrate and 40 nM RAG protein at 37°C. SIL9 represents the SIL sequence with a nonamer that was corrected to the optimal sequence (with a 23-bp spacer). SIL7 represents the SIL sequence with the heptamer corrected to the optimal sequence. SCL9 represents the SCL sequence with a nonamer corrected to the optimal sequence (with a 12-bp spacer). The results represent the means of at least three independent experiments.
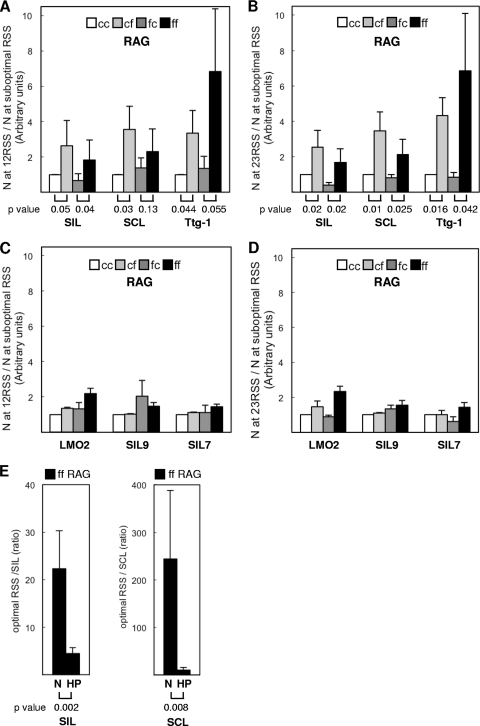
Fig 3.
RSS sequence discrimination by the RAG complex. (A) Histogram comparing the nicking efficiencies at an optimal RSS and suboptimal RSS by cc, cf, fc, or ff RAG complexes. Nicking at the 12RSS divided by nicking at the suboptimal RSS (y axis) was normalized to that of the cc RAG complex, which was arbitrarily set to 1. (There was wider variation in the yield of nicked product when Ttg-1 was used for the nicking assay, because the nicked products were very faint for Ttg-1 [Fig. 2B].) A one-tailed t test was carried out to test the hypothesis that the RAG complex with full-length RAG2 is capable of greater discrimination for signal sequence quality than is the complex containing core RAG2. The P values are listed under the histograms for the signal sequence discrimination by cc versus cf or by fc versus ff RAG complexes. (B) Histogram comparing the nicking efficiencies at an optimal RSS and suboptimal RSS. Nicking at the 23RSS divided by nicking at the suboptimal RSS (y axis) was normalized to that of the cc RAG complex, which was arbitrarily set to 1. As in panel A, the P values from the t test are listed under the histograms for signal sequence discrimination by cc versus cf or for fc versus ff RAG complexes. (C) Histogram comparing the nicking efficiencies at the optimal RSS and suboptimal RSS by cc, cf, fc, or ff RAG1 or RAG2 complexes. Nicking at 12RSS divided by nicking at the suboptimal RSS (y axis) was normalized to that of the cc RAG complex, which was arbitrarily set to 1. The P values from a t test for the data were substantially greater than 0.1. (D) Nicking at 23RSS divided by nicking at the suboptimal RSS (y axis) was normalized to that of the cc RAG complex, which was arbitrarily set to 1. The P values from the t test for the data were substantially greater than 0.1. (E) Histogram comparing sequence discrimination at nicking step and hairpin step by ff RAG complex. The amounts of nick (N) or hairpin (HP) product at optimal 12RSS and 23RSS sites were divided by those at SIL (left) or SCL (right) (y axes). The P values from the t test are listed under the histograms.
Most of the sequence discrimination by the RAG complex occurs at the nicking step rather than the hairpin step.
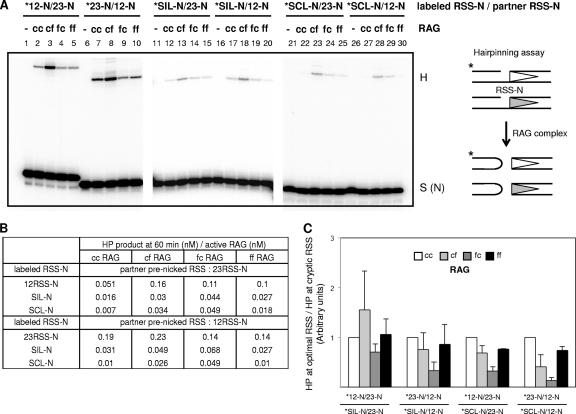
We were interested in whether sequence discrimination by full-length RAG complexes applies at the hairpinning step as well as the nicking step. To test this, we used prenicked RSS substrates to focus specifically on the hairpinning step and tested cc, cf, fc, and ff RAG complexes. Either prenicked optimal 12RSSs or prenicked optimal 23RSSs were used as partners of labeled prenicked RSS substrates. In the pathological chromosomal deletion events involving SCL and SIL, both of which are suboptimal RSS sites, and in our biochemical system, pairing SIL and SCL resulted in hairpin product at levels that were too low to be measured reliably, especially without H3K4me3; therefore, an optimal RSS was used here as the partner when assessing either of these two cryptic RSSs.
We found that none of the RAG complexes discriminated nearly as much at the hairpinning step as they did at the nicking step between optimal and suboptimal RSSs (merely ∼5-fold for hairpin formation versus ∼22-fold at the nicking step for SIL, and only ∼10-fold for hairpin versus ∼244-fold for nicking at the nicking step for SCL) (Fig. 2C, 3E, and 4B).
Fig 4.
Relatively limited discrimination in the signal sequence quality by the C terminus of RAG2 at the hairpin step. (A) On the left side is a representative gel image of an in vitro hairpin assay using the cc, cf, fc, or ff RAG complex. The scheme of the hairpin assay is depicted to the far right. Note that both the 12RSS and 23RSS substrates were prenicked. The substrates consisted of labeled (asterisks) prenicked RSSs (represented as RSS-N) and a prenicked cold partner, RSS-N. The substrate combinations used in each reaction mixture are indicated at the top of the gel image. The reaction mixtures, which included 20 nM prenicked RSS and 40 nM RAG proteins, were incubated for 60 min at 37°C as outlined in Materials and Methods. The products were analyzed on a 13% denaturing PAGE gel, followed by autoradiography. The positions of prenicked substrate [S (N)] and hairpin (H) products are indicated on the right margin of the gel image. (B) Summary of results of the experiment described for panel A. (C) Histogram comparing the hairpin efficiencies with optimal RSS and suboptimal RSS by cc, cf, fc, or ff RAG complex. The amount of hairpin product between 12RSS and 23RSS divided by hairpin product between cryptic RSS and 12/23 RSS (y axis) was normalized to that of the c/c RAG complex, which was arbitrarily set to 1. The P values from the t test were substantially greater than 0.1 in comparisons of the RAG complexes containing core RAG2 versus full-length RAG2, indicating that there is less discrimination for that hairpin step than the nicking step.
Interestingly, the C terminus of RAG2 is not necessary for the lower level of discrimination that is observed at the hairpin step (Fig. 4C). Therefore, most of the RSS quality discrimination by RAG complexes appears to occur at the nicking step, based on our in vitro studies here.
Our ff RAG complexes clearly showed no indication of autoinhibition caused by the C terminus of RAG2 (Fig. 4A and B), as was suggested in another recent study (15). Such autoinhibition would have shown up as reduced activity by the ff RAG complex relative to any of the core RAG-containing complexes. The earlier study's results (15) were complicated by the following: (i) all of the RAG1 that they used was N-terminally truncated, (ii) RAG2 was not full length, and (iii) no HMGB1 was present.
The overall rate of nicking is a critical factor in RAG complex discrimination between RSS and off-target sites.
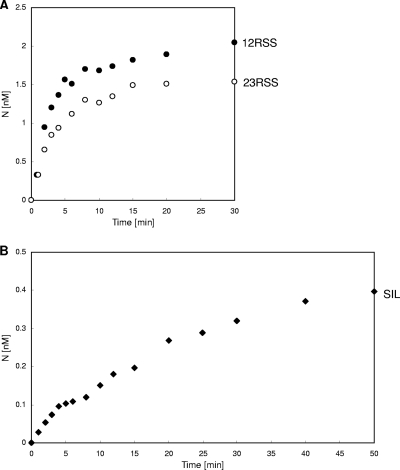
Full-length RAG complexes were tested in nicking time course assays with either RSSs or suboptimal RSSs (Fig. 5). We selected SIL as one example of a suboptimal RSS, because the nicking of SIL by the RAG complex is relatively more clear than for many other sites, even at the initial stage of nicking. Nicking at optimal 12RSS or 23RSS by the ff RAG complex reached plateaus in the first 10 min, whereas nicking at SIL continued to rise. The results indicated a slower nicking rate for the suboptimal RSS, SIL, by the ff RAG complex (Fig. 5).
Fig 5.
Kinetics of nicking by the full-length RAG complex. (A) Time course of nicking at 12RSS or 23RSS by the ff RAG complex. Reaction mixtures included 5 nM RSS and 20 nM RAG proteins, and an aliquot at each time point was analyzed. (B) Time course of nicking at SIL by the ff RAG complex. Reaction mixtures included 25 nM RSS and 40 nM RAG proteins, and an aliquot at each time point was analyzed.
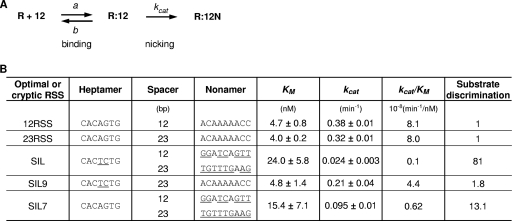
The nicking step is relatively simple, because it is a unireactant enzymatic reaction, and it allows us to determine kinetic constants (Fig. 6A). To investigate which kinetic factor(s) leads to slower nicking at suboptimal sites, we determined the Michaelis-Menten constants (KM) and catalytic rate constants (kcat) for the active RAG complexes. We utilized short oligonucleotide substrates to determine kinetic constants so that we could observe RAG binding reliant on only the RSS. The data indicated that optimal 12RSS and 23RSS sites have KM values of 4.7 nM and 4.0 nM, respectively, whereas the SIL site has a 5- to 6-fold-higher KM (∼24 nM). Optimal 12RSS and 23RSS sites had kcat values of 0.38 min−1 and 0.32 min−1, respectively, whereas the SIL site had a much lower kcat of 0.024 min−1 (Fig. 6B).
Fig 6.
Rate of nicking is critical in discriminating RSS sequence quality. (A) Kinetic model for binding and nicking steps mediated by the RAG complex. R, the active RAG complex; 12, the RSS substrate with a 12-bp spacer and an adjacent coding segment; 12N, the nicked form of the 12RSS. The nicking step has been schematized as a unireactant enzyme-catalyzed reaction. The forward and reverse kinetic constants for the active RAG complex in the binding step are designated a and b, respectively. The catalytic rate constant for the nicking step is kcat (also called c in the simulations in the supplemental material). (B) The kinetic constants for ff RAG complexes, determined by curve fitting for 12RSS, 23RSS, SIL, SIL with corrected nonamer as SIL9 (with a 23-bp spacer), and SIL with corrected heptamer as SIL7, respectively. Reactions were started with various concentrations of DNA substrate and 10 to 40 nM RAG proteins at 37°C and stopped at 2 to 6 min (well before the reaction reached its plateau). KM is the Michaelis-Menten constant, and kcat is the catalytic rate constant for nicking. Substrate discrimination was defined as follows: [(kcat/KM) for 12RSS]/[(kcat/KM) for RSS], where RSS refers to the 23RSS, SIL, SIL9, or SIL7. The results represent the means ± standard errors of the means of at least three independent experiments.
Modeling of the time courses (see Fig. S1 and Table S1 in the supplemental material) indicated that the values of kcat are lower than the rate of disassembly of the RAG:RSS complex back to the free RAG complex and RSS (Fig. 6A, kinetic constant b). This means that a KM of [(b + kcat)/a] is approximately equal to the KD, or b/a. Binding of only the enzymatically active fraction of RAG complexes has not been measurable previously. Previous estimates of KD were based on electrophoretic mobility shift assays (EMSA; reviewed in references 13, 33, and 42) or fluorescence anisotropy (47). Both methods include binding to the RSS by both catalytically inactive and active RAG complexes. The EMSA method gives KD values somewhat higher (∼25 to 32 nM) than the 4 to 5 nM that we observed. (Fluorescence anisotropy measurements have also been reported with RAG1 alone [6, 47].) Therefore, the determination of KD here is quite an improvement, because we can distinguish catalytically inactive from active RAG complexes.
In summary, kinetic analysis of the RSS variant, SIL, showed that KM and kcat both slowed the overall nicking rate. The KM, and hence KD, was 5- to 6-fold greater, reflecting looser binding. The kcat for nicking of the SIL site was 13- to 16-fold lower. Therefore, though both binding and the catalytic rate constant result in a slower overall nicking rate, the primary effect is on the catalytic rate constant (kcat).
Nonamer optimization improves both the KM (KD) and kcat, whereas heptamer optimization improves primarily the kcat.
The RSS variant SIL has deviations in both the heptamer and nonamer from the consensus sequence. The studies here permit the first opportunity to dissect the independent contribution of the heptamer deviations and the nonamer deviations to RAG catalysis. Based on studies using mutant RSSs, it has been established that an RSS with an intact heptamer but without a conserved nonamer sequence functions to direct the RAG complex to cleave it, although the presence of the nonamer stimulates cleavage efficiency (8, 16, 31). SIL has a heptamer-like sequence with variation at the fourth and fifth position (from the nick site), but it does not have an obvious nonamer. Our previous work demonstrated that SIL is more efficient in cellular V(D)J recombination activity when it is coupled with a 12RSS partner in an in vivo extrachromosomal V(D)J recombination than with 23RSS, suggesting that SIL functions as a 23RSS (29). Therefore, we designed a SIL mutant with an intact nonamer having a 23-bp spacer, designated SIL9. As expected, the overall nicking efficiency of SIL9 by the ff RAG complex was greatly improved compared to SIL but was still slightly weaker than the optimal RSS (Fig. 2B).
We were interested in determining which kinetic factor(s) of the ff RAG complex is responsible for the overall increased nicking of SIL9. We found that an optimized nonamer (SIL9) had a 5-fold-lower KM (4.8 nM), which was nearly as strong as for an optimal RSS, and a 9-fold faster kcat than the original SIL site (0.21 min−1) (Fig. 6B). Therefore, nonamer improvements affect both the KM and kcat. This is not surprising, as the enzyme must hold the substrate tightly during catalysis.
Changes in the SIL heptamer that converted it to the optimal heptamer sequence resulted in 1.55-fold-tighter binding (KM [also KD]). However, the kcat increased 4-fold. This is consistent with a primary role of the heptamer in directing the catalytic mechanism and a smaller role in the binding of the RAG complex to the RSS.
DISCUSSION
This is the first opportunity to study full-length RAG complexes in a context where the active fraction of each enzyme preparation is known. Kinetic analysis of these complexes resulted in several clear mechanistic insights of substantial biological and pathological relevance. First, the C terminus of RAG2 plays a key role in selecting optimal RSS sites at the DNA sequence level in mice (10) and in our in vitro studies (this study), beyond the role that it has in targeting active chromatin (Fig. 7). Second, kinetic analysis of the nicking step leads to a greater insight into the RAG complex genome-wide search process for RSS sites. Third, kinetic analysis indicates that the major barrier against RAG complex nicking at suboptimal sites is that the RAG complex falls off much faster than it nicks. This is a reflection of both the KD and the kcat differences between optimal RSS sites and suboptimal ones. These features reduce the risk of chromosomal translocations and provide a better mechanistic understanding of the rare translocations in human T-cell lymphomas that do arise.
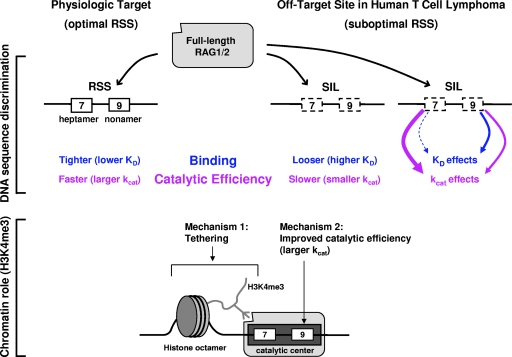
Fig 7.
Summary of DNA sequence factors and known chromatin factors affecting the binding and catalytic efficiencies of the full-length RAG complex. These diagrams depict the binding and nicking reactions of the RAG complex (large gray rectangle with rounded edges) at the optimal or suboptimal RSS site. Within the RSS, the heptamer is designated by a 7 and the nonamer is designated by a 9. The suboptimal heptamer and nonamer are indicated with hatched boxes. We found that the ff RAG complex had greater DNA sequence discrimination between optimal RSS sites and suboptimal (off-target) sites, such as SIL, used in human T-cell lymphomas. This was due primarily to the C terminus of RAG2. We found that this sequence discrimination applies to the nicking step but is much lower at the hairpin step. The overall rate difference between optimal and suboptimal RSSs was due primarily to differences in the catalytic rate constant (kcat), but there was a smaller contribution due to differences in binding efficiencies (KD). When the SIL heptamer was changed to the optimal sequence, then kcat increased substantially, with only a small improvement in the KD. When the SIL nonamer was changed to the optimal sequence, then both the KD and kcat were improved. The known contributions of chromatin (H3K4me3) to the tethering of the RAG complex and to increased kcat values are also shown. The catalytic center (black rectangle within the larger gray rectangle [the RAG complex]) refers to the portion that binds to the RSS and carries out the catalysis.
Why is there primary sequence discrimination at the nicking step?
Even a nick is a site of potential genetic instability. Therefore, it is best if this initial step is tightly controlled, which is what we observe here. Previous work showed that more than one V, D, or J segment RSS may be nicked within a single locus in vivo (9). It is not possible for such studies to be used to compare nick frequencies at optimal and off-target sites. Therefore, our studies here are key for understanding such discrimination.
It may be surprising that there is not equivalent sequence control at the hairpinning step, which includes synapsis under the physiologic reaction conditions used here (Fig. 3E). This may be because the hairpinning step involves substantial DNA structural distortion. Therefore, it may be difficult for primary DNA sequence information to be “read” or utilized, as it can be at the nicking step. We cannot formally rule out the possibility that there is more hairpin step discrimination between an optimal RSS and SIL (for example), if we could assess SIL when it is paired with SCL, but we do not favor this possibility.
Kinetic model of the RAG complex nicking step.
The full-length RAG complex studied here allowed an appraisal of how RAG catalysis relates to RAG biology. The nicking step conforms to a unireactant enzyme-catalyzed reaction. The steps of this reaction can be described by a series of differential equations that include parameters (or combinations of them) that we can measure, specifically, the kcat and KM. Additional parameters can be varied to determine the ranges of their values that permit fits of the full time courses for 12RSS or 23RSS nicking (Fig. 5). RAG binding to the RSS can be described in terms of a forward binding rate constant, a, and a dissociation rate constant, b. The ratio b/a = KD, and KM = (b + kcat)/a.
In the simulations, when we constrained kcat at or close to the measured kcat, there were only a narrow range of values for a and b that fit our observed time courses for both the 12RSS and the 23RSS (see the methods described in the supplemental material). We found that all values of a, b, and kcat that fit the observed time courses had b values that were ≫kcat. Thus far, we can only narrow the b value to a range of values, but even the lowest value for b that fit the observed full time courses was more than 170-fold greater than the kcat. This is important, because it means that KM is ≈KD = b/a. This allows us to know the equilibrium dissociation constant specifically for the active RAG complexes.
Previous studies of core RAG binding presumably represented a mixture of active and inactive RAG complexes (13, 42). All EMSA and other methods of binding measurements reflect the sum of both the active and inactive RAG complexes. Our RAG complex preparations ranged from 10% to 30% active (across ff, cf, fc, and cc forms), and the general natures of the time courses were similar. This suggests that the fraction of inactive RAG complexes does not fundamentally change any aspect of the kinetics of nicking (see the supplemental material).
Physical and biochemical model.
Nicking at optimal RSS sites has a kcat of ∼0.35 min−1. This corresponds to a half-time of 2 min. The dissociation rate of RAG complexes from optimal RSS sites is at least 128 min−1, or a half-time of 0.3 s. Therefore, RAG complex binding represents a rapid sampling of primary DNA sequence sites. Even when optimal sites are encountered, the rapid dissociation rate only permits a very small subset of encounters with optimal sites to result in a nick prior to their dissociation. It is possible that the relatively weak (KD, ∼10−5 M) secondary interactions between the RAG2 PHD finger and nearby H3K4me3 tails permit longer overall dwell times within regions of active chromatin (see below).
Interestingly, we found that sequence optimization at the heptamer primarily affected kcat, and these optimizations resulted in very little change in the KM (and KD). This suggests that nearly all of the binding interactions reside at the nonamer, and the role of the heptamer is in achieving a transition state for catalysis. This is consistent with the crystal structure information on a fragment of RAG1 and footprinting studies showing RAG (core or domain regions) binding contacts primarily at the nonamer (40–42, 44). Thus, nonamer contacts for RAG binding explain the KM (and KD) effect, and nonamer sequence effects on kcat are attributable to a requirement for the enzyme to tightly hold the substrate while it distorts the DNA for nicking catalysis. This study provides the first dynamic (kinetic) insights into which portion of the RSS is important for binding and which is important for catalysis for full-length RAG complexes.
A holistic view of RAG complex binding and catalysis includes both the primary sequence aspects examined here as well as chromatin effects (Fig. 7). The RAG complex catalyzes only a low level of nicking at suboptimal RSS sites relative to optimal ones. The lower level of nicking is due primarily to a low catalytic rate constant at suboptimal sites, as well as looser binding to those sites. When the SIL heptamer is changed to the optimal sequence, then kcat increases substantially, with only a small improvement in the KD. When the SIL nonamer is changed to the optimal sequence, then both KD and kcat are improved. Interaction of the RAG2 PHD finger with H3K4me3 at recombination sites or off-target sites provides up to two low-affinity (KD, ≈10−5 M) interactions for each RAG complex (assuming a RAG tetramer, as we have assumed here) and improves recombination efficiency (24, 25, 30). Our previous experiments showed that H3K4me3 stimulates the cleavage at RSS by the cf RAG complex (36). We found that this stimulation of RSS cleavage by H3K4me3 also applies when ff RAG complexes are used in in vitro cleavage assays (data not shown). When optimal and suboptimal RSSs both reside within H3K4me3 regions, then the DNA sequence effects described here provide the only discrimination between sites.
The RAG complex as a catalyst and RAG1 as a single catalysis subunit for the nicking step.
Our RAG complex preparations are very active relative to all others reported previously. Other than our earlier study of cc RAG complexes (45), no other studies have used formal burst kinetics to determine the active fraction. However, other studies typically report 30- or 60-min time point comparisons, and based on these, we did not see evidence that other cc, cf, or fc preparations were significantly more active than those that we have reported here for ff or previously for cc. Given this, it is interesting that we did not find more than two nicking events per RAG complex (range, 1.1 to 1.9). Because we assumed that each RAG complex included two RAG1 monomers (and two RAG2 monomers plus HMGB1) (14, 38, 39), it appears that each RAG1 monomeric subunit in such a tetrameric complex is capable of, at most, a single nicking event (range, 0.6 to 0.95 nicking events per RAG1 monomer). This suggests that RAG1 is a single-turnover subunit (sometimes called single catalysis enzymatic activity). The RAG tetrameric complex would then be capable of only two nicking events. Our analysis does not address whether these RAG1 subunits can proceed to carry out the hairpinning step, after their nicking action (see below).
If our inference applies to all other RAG preparations (cc, cf, fc, or ff) in future studies, then what is the potential biological significance of each RAG1 monomer being capable of only a single nicking event and what would be the evolutionary advantage of this? First, if each RAG1 monomer is only capable of one nicking event, then there must be mixed complexes that contain one expended monomer and one unexpended monomer. We infer this to be the case, given that the 12RSS and 23RSS nicking events can be (and, we suspect, probably typically are) independent events. Second, single catalysis enzyme action by each RAG1 monomer might provide tighter control between RAG1 production and RAG1 action.
In parallel with transposases, it seems likely that the same catalytic site in the RAG complex supports both nicking and hairpin formation in a sequential manner (26). Our studies suggest that the catalytic center of the RAG complex must change somewhat between the nicking step and the hairpin step. The nicking step is a hydrolysis reaction in which water is the nucleophile for attack on a phosphodiester linkage. The hairpin step involves nucleophilic attack of the 3′-OH on a phosphodiester linkage. If the active complex for both the nicking and hairpinning steps is a tetramer (14, 38, 39), and if each RAG1 monomer is capable of only one nicking event, then it must be true that one or both of the RAG1 monomers that are “expended” for nicking must still be capable of hairpin formation.
Relevance for the RAG genome-wide site search process and RAG complex distribution.
As mentioned in the previous section, the kinetic insights here indicate that the RAG complexes rapidly bind and dissociate even for optimal sites. For suboptimal sites, the dissociation by active RAG complexes is even faster and the nicking is much slower, thereby dramatically decreasing catalysis at such sites. Genome-wide binding studies have demonstrated that the H3K4me3 modification attracts RAG2 to most of the euchromatic regions, which could represent over 5% of the genome (18), whereas RAG1 and RAG2 complex binding to genomes relies on both an H3K4me3 mark and RSS. Within these H3K4me3 domains, the DNA sequence discrimination insights that we have described here would apply. It is difficult from genome-wide studies to infer the relative binding of the RAG complexes to off-target locations, because lower levels of binding may not be readily detectable, depending on the affinity of the antibody.
Extrachromosomal V(D)J substrates in murine pre-B cells provide insight regarding the contribution of suboptimal heptamers and nonamers to infrequent usage of cryptic sites in regions of active chromatin. Extrachromosomal substrates typically have high levels of H3K4me3 (27, 28) and thus would efficiently recruit the RAG complex via the RAG2 PHD domain (24, 25, 30, 36). Rarely, cryptic sites on such extrachromosomal substrates are used instead of the canonical 12RSS or 23RSS site (21). The frequency of these rare events has not been determined, but it appears to be well below 1 part in 1,000 relative to the level of canonical 12RSS and 23RSS site usage. These cryptic sites usually have no significant nonamer, and the heptamer consists only of CAC or CACA. This suggests that the low level of RAG complex binding via the PHD domain (since there is no nonamer) is sufficient to permit a very low level of nicking at the CAC or CACA sites. This then may represent the “noise” or nonspecific catalysis of the RAG complex within active chromatin. The nonamer would provide improved specificity of binding. Improved heptamers would also add to improved binding and faster catalysis to achieve the >1,000-fold improvement.
Relevance for the incidence of lymphomas in RAG mutant mice.
Our observations here provide a biochemical basis for understanding why a C-terminal RAG2 truncation contributes to dysregulation and lymphomas in mice. C-terminally truncated RAG2 would lead to less discrimination and presumably more action at the wide range of suboptimal sites (10). Moreover, without its C terminus, RAG2 is more likely to direct the RAG complex to the non-H3K4me3 regions (11). For these same reasons, the off-target action of the RAG complex without the C terminus of RAG2 is likely to explain the increase in lymphomas in mice (11).
Beyond these conclusions and inferences, our biochemical studies may provide an understanding of the differences between various RAG2 C terminus alterations. In particular, mice lacking the RAG2 C-terminal noncore region (11) differ considerably in phenotype from mice with a point mutation at the phosphorylation site at threonine 490 (45a), which is important for the destruction of RAG2 at the G1/S transition. Although mice in both models develop lymphomas (in the p53-deficient background), the RAG2 C terminus-truncated mice die more rapidly than the T490A mice. This may be because the RAG2-T490A mice have RAG activity throughout the cell cycle, whereas the RAG2 C-terminal truncation would not only cause this problem but would also cause reduced RSS site discrimination and presumably more action at a wide range of suboptimal sites. Therefore, the opportunity here to study highly active full-length RAG complexes provides important insights into both RAG biology and pathobiology.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants to M.R.L. N.S. was supported by JSPS.
We thank Irwin H. Segal (University of California—Davis for helpful comments regarding enzyme kinetics. We thank Nicholas Pannunzio and Xiaoping Cui for comments.
Footnotes
Published ahead of print 7 November 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Bergeron S, Anderson DK, Swanson PC. 2006. RAG and HMGB1 proteins: purification and biochemical analysis of recombination signal complexes. Methods Enzymol. 408:511–528 [DOI] [PubMed] [Google Scholar]
- 2. Chakraborty T, et al. 2007. Repeat organization and epigenetic regulation of the DH-Cμ domain of the immunoglobulin heavy-chain gene locus. Mol. Cell 27:842–850 [DOI] [PubMed] [Google Scholar]
- 3. Chowdhury D, Sen R. 2004. Mechanisms for feedback inhibition of the immunoglobulin heavy chain locus. Curr. Opin. Immunol. 16:235–240 [DOI] [PubMed] [Google Scholar]
- 4. Chowdhury D, Sen R. 2004. Regulation of immunoglobulin heavy-chain gene rearrangements. Immunol. Rev. 200:182–196 [DOI] [PubMed] [Google Scholar]
- 5. Chowdhury D, Sen R. 2001. Stepwise activation of the immunoglobulin mu heavy chain gene locus. EMBO J. 20:6394–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ciubotaru M, et al. 2003. RAG1-DNA binding in V(D)J recombination. Specificity and DNA-induced conformational changes revealed by fluorescence and CD spectroscopy. J. Biol. Chem. 278:5584–5596 [DOI] [PubMed] [Google Scholar]
- 7. Cornish-Bowden A. 1979. Fundamentals of enzyme kinetics. Butterworths, London, United Kingdom [Google Scholar]
- 8. Cuomo CA, Mundy CL, Oettinger MA. 1996. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol. 16:5683–5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Curry JD, Geier JK, Schlissel MS. 2005. Single-strand recombination signal sequence nicks in vivo: evidence for a capture model of synapsis. Nat. Immunol. 6:1272–1279 [DOI] [PubMed] [Google Scholar]
- 10. Curry JD, Schlissel MS. 2008. RAG2's non-core domain contributes to the ordered regulation of V(D)J recombination. Nucleic Acids Res. 36:5750–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deriano L, et al. 2011. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature 471:119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elkin SK, Matthews AG, Oettinger MA. 2003. The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J. 8:1931–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gellert M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101–132 [DOI] [PubMed] [Google Scholar]
- 14. Grundy GJ, et al. 2009. Initial stages of V(D)J recombination: the organization of RAG1/2 and RSS DNA in the postcleavage complex. Mol. Cell 35:217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grundy GJ, Yang W, Gellert M. 2010. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc. Natl. Acad. Sci. U. S. A. 107:22487–22492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hesse JE, Lieber MR, Mizuuchi K, Gellert M. 1989. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 3:1053–1067 [DOI] [PubMed] [Google Scholar]
- 17. Hesslein DG, et al. 2003. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 17:37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ji Y, et al. 2010. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell 141:419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang H, et al. 2005. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol. Cell 18:699–709 [DOI] [PubMed] [Google Scholar]
- 20. Jones JM, Simkus C. 2009. The roles of the RAG1 and RAG2 “non-core” regions in V(D)J recombination and lymphocyte development. Arch. Immunol. Ther. Exp. (Warsz.) 57:105–116 [DOI] [PubMed] [Google Scholar]
- 21. Lewis SM, Agard E, Suh S, Czyzyk L. 1997. Cryptic signals and the fidelity of V(D)J joining. Mol. Cell. Biol. 17:3125–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lieber MR. 1991. Site-specific recombination in the immune system. FASEB J. 5:2934–2944 [DOI] [PubMed] [Google Scholar]
- 23. Lin W-C, Desiderio S. 1994. Cell cycle regulation of RAG-2 V(D)J recombinase. Proc. Natl. Acad. Sci. U. S. A. 91:2733–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. 2007. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity 27:561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matthews AG, et al. 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450:1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Montano SP, Rice PA. 2011. Moving DNA around: DNA transposition and retroviral integration. Curr. Opin. Struct. Biol. 21:370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okitsu CY, Hsieh CL. 2007. DNA methylation dictates histone H3K4 methylation. Mol. Cell. Biol. 27:2746–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Okitsu CY, Hsieh JC, Hsieh CL. 2010. Transcriptional activity affects the H3K4me3 level and distribution in the coding region. Mol. Cell. Biol. 30:2933–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raghavan SC, Kirsch IR, Lieber MR. 2001. Analysis of the V(D)J recombination efficiency at lymphoid chromosomal translocation breakpoints. J. Biol. Chem. 276:29126–29133 [DOI] [PubMed] [Google Scholar]
- 30. Ramón-Maiques S, et al. 2007. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc. Natl. Acad. Sci. U. S. A. 104:18993–18998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramsden DA, McBlane JF, van Gent DC, Gellert M. 1996. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 15:3197–3206 [PMC free article] [PubMed] [Google Scholar]
- 32. Roth DB, Zhu C, Gellert M. 1993. Characterization of broken DNA molecules associated with V(D)J recombination. Proc. Natl. Acad. Sci. U. S. A. 90:10788–10792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schatz DG, Ji Y. 2011. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11:251–263 [DOI] [PubMed] [Google Scholar]
- 34. Schatz DG, Swanson PC. 2011. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 45:167–202 [DOI] [PubMed] [Google Scholar]
- 35. Schlissel M, Constantinescu A, Morrow T, Peng A. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 7:2520–2532 [DOI] [PubMed] [Google Scholar]
- 36. Shimazaki N, Tsai AG, Lieber MR. 2009. H3K4me3 stimulates V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol. Cell 34:535–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Swanson PC. 2004. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol. Rev. 200:90–114 [DOI] [PubMed] [Google Scholar]
- 38. Swanson PC. 2001. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21:449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swanson PC. 2002. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 22:7790–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swanson PC, Desiderio S. 1999. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 19:3674–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Swanson PC, Desiderio S. 1998. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity 9:115–125 [DOI] [PubMed] [Google Scholar]
- 42. Swanson PC, Volkmer D, Wang L. 2004. Full-length RAG-2, and not full-length RAG1, specifically suppress RAG-mediated transposition but not hybrid joint formation or disintegration. J. Biol. Chem. 279:4034–4044 [DOI] [PubMed] [Google Scholar]
- 43. Swerlow SH, et al. (ed.). 2008. WHO classification of tumours of haematopoietic and lymphoid tissues, 4th ed WHO Press, Lyon, France [Google Scholar]
- 44. Yin FF, et al. 2009. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat. Struct. Mol. Biol. 16:499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu K, Lieber MR. 2000. The nicking step of V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol. 20:7914–7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a. Zhang L, Reynolds TL, Shan X, Desiderio S. 2011. Coupling of V(D)J recombination to the cell cycle suppresses genomic instability and lymphoid tumorigenesis. Immunity 34:163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang M, Swanson PC. 2008. V(D)J recombinase binding and cleavage of cryptic recombination signal sequences identified from lymphoid malignancies. J. Biol. Chem. 283:6717–6727 [DOI] [PubMed] [Google Scholar]
- 47. Zhao S, Gwyn LM, De P, Rodgers KK. 2009. A non-sequence-specific DNA binding mode of RAG1 is inhibited by RAG2. J. Mol. Biol. 387:744–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.