Abstract
Long interspersed elements (LINEs) are mobile elements that comprise a large proportion of many eukaryotic genomes. Although some LINE-encoded open reading frame 1 proteins (ORF1ps) were suggested to be required for LINE mobilization through binding to their RNA, their general role is not known. The ZfL2-1 ORF1p, which belongs to the esterase-type ORF1p, is especially interesting because it has no known RNA-binding domain. Here we demonstrate that ZfL2-1 ORF1p has all the canonical activities associated with known ORF1ps, including self-interaction, nucleic acid binding, and nucleic acid chaperone activities. In particular, we showed that its chaperone activity is reversible, suggesting that the chaperone activities of many other ORF1ps are also reversible. From this discovery, we propose that LINE ORF1ps play a general role in LINE integration by forming a complex with LINE RNA and rearranging its conformation.
INTRODUCTION
Long interspersed elements (LINEs), or non-long terminal repeat (non-LTR) retrotransposons, are transposable elements that comprise a large proportion of many eukaryotic genomes. Mobilization or amplification of LINEs causes various alterations in their host genomes, thus having profound effects on eukaryotic genome evolution (7, 12). LINEs mobilize by a mechanism called retrotransposition. In LINE retrotransposition, a LINE-encoded endonuclease (EN) nicks a target site of the host genomic DNA by which a 3′ hydroxyl group is generated. The 3′ hydroxyl group is then used as a primer from which a LINE-encoded reverse transcriptase (RT) initiates reverse transcription of the LINE RNA. This reaction, which is called target-primed reverse transcription (TPRT), is characteristic of LINE retrotransposition (1, 16). After TPRT, the newly synthesized LINE DNA is integrated into the genomic DNA with the help of the host DNA repair system(s), although the mechanism of this integration is not well understood (32).
LINEs are divided into 12 or more clades based on phylogenetic analysis of the RTs they encode (18). The clades are classified into two major groups that differ in structure. One group encodes a single multidomain protein that is responsible for TPRT and contains a restriction-like endonuclease (RLE) and an RT domain. LINEs of this group are all integrated into a specific site of the host genome DNA defined by their RLE (4). The other group encodes two proteins, called open reading frame 1 and 2 proteins (ORF1p and ORF2p). ORF2p, which is responsible for TPRT, contains an apurinic/apyrimidinic endonuclease (APE) and an RT domain (6). APE-type LINEs are usually dispersed in the host genomes because most APEs do not have strict specificity for their target sequence. ORF1p is also required for retrotransposition, although its role is not well understood (24, 29, 33).
ORF1ps in APE-type LINEs do not contain any domains conserved in common, and their amino acid sequences are frequently quite different among LINEs of different clades and even among those classified into one clade. This contrasts with the fact that the APE and RT domains of ORF2p are well conserved among APE-type LINEs. Research regarding the function of ORF1p is predominantly conducted using ORF1p encoded by the mammalian LINE, L1, which is classified into the L1 clade. L1 ORF1p is composed of three distinct structural domains, an N-terminal coiled-coil (CC), a middle noncanonical RNA recognition motif (RRM), and a C-terminal domain (CTD) (10, 13, 22) (Fig. 1A). Biochemical analyses showed that L1 ORF1p forms a trimer via the CC domain and requires the RRM and CTD for nucleic acid binding (13, 19, 22). Consistent with these findings, L1 ORF1p forms a large ribonucleoprotein particle (RNP) with L1 RNA in cultured cells (3, 14). These data suggest that L1 ORF1p is necessary for RNP formation, although the role of the RNP in retrotransposition remains unclear. The RRM domain is present in many other ORF1ps encoded by LINEs of various clades, suggesting that ORF1p having the RRM domain has a common feature for RNP formation (13). Consistent with this notion, the ORF1p encoded by the fruit fly LINE, I factor (which encodes the RRM domain in the I clade), forms a multimer and binds nucleic acids in vitro, and the ORF1p encoded by the silkworm LINE, SART1 (which encodes the RRM domain in the R1 clade), forms an RNP in cultured cells (2, 23) (Fig. 1A). Besides the RRM domain, intriguingly, a zinc knuckle motif at the C terminus of SART1 ORF1p is essential for packaging of the SART1 RNA into the RNP (23). Zinc knuckles are also found in many other ORF1ps in which they are frequently located downstream of the RRM, indicating that the zinc knuckle and RRM may cooperatively bind LINE RNA.
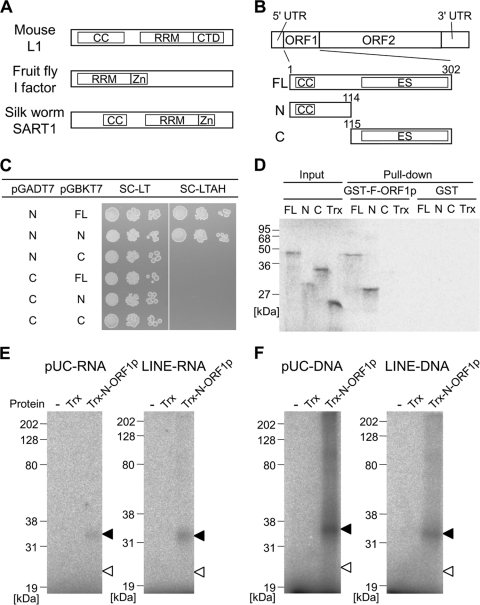
Fig 1.
ZfL2-1 ORF1p interacts with itself and binds to nucleic acids through the N-terminal region. (A) Schematic diagrams of LINE ORF1ps biochemically studied so far. The domain structures were predicted by HHpred (30). CC, coiled-coil motif; RRM, RNA recognition motif; CTD, C-terminal domain; Zn, zinc knuckle motif. (B) Schematic diagram of the zebrafish LINE, ZfL2-1. ZfL2-1 is composed of a 5′ untranslated region (UTR), two open reading frames (ORF1 and ORF2), and a 3′ UTR. The full-length ZfL2-1 ORF1p (FL) and its N- and C-terminal portions (N and C) were used in this study. Numbers above the boxes indicate the amino acid positions from the first methionine. ES, esterase domain. (C) Yeast two-hybrid assay. The pGADT7 derivatives express the activation domain of GAL4 fused with the N- or C-terminal portion of ZfL2-1 ORF1p. The pGBKT7 derivatives express the binding domain of GAL4 fused with the full-length ORF1p or its N- or C-terminal portion. AH109 cells that have the two derivatives were spotted in 10-fold serial dilutions on synthetic complete medium depleted of leucine and tryptophan (SC-LT) or that depleted of leucine, tryptophan, adenine, and histidine (SC-LTAH) and grown at 30°C for 3 days. (D) Autoradiogram of GST pulldown assay. Trx and the Trx fusions of the full-length, N-terminal portion, and C-terminal portion of ZfL2-1 ORF1p were synthesized in vitro as 35S-labeled proteins and incubated with the GST fusion of full-length ZfL2-1 ORF1p (GST-F-ORF1p) or GST. Equal amounts of the 35S-labeled proteins before the incubation were subjected to SDS-PAGE (Input). 35S-labeled proteins bound to GST-F-ORF1p or GST were purified and electrophoresed on the gel (Pull-down). Sizes of standard marker proteins are indicated at the left of the autoradiogram. (E and F) UV cross-linking assays. Trx or the Trx fusion of the N-terminal portion of ZfL2-1 ORF1p (Trx-N-ORF1p) was incubated with 32P-labeled RNA (E) derived from the pUC19 vector (pUC-RNA) or the 3′ tail of ZfL2-1 (LINE-RNA) and 32P-labeled DNAs (F) derived from the pUC19 vector (pUC-DNA) or the 3′ tail of ZfL2-1 (LINE-DNA). Black and white arrowheads indicate the positions of Trx-N-ORF1p and Trx, respectively. The left lane of each autoradiogram shows the results with no protein (−). Sizes of standard marker proteins are indicated at the left of each autoradiogram. The RNA and DNA sequences are shown in Table 1.
Nucleic acid chaperones are defined as proteins that catalyze rearrangement of nucleic acids into a conformation that has the maximum number of base pairs (15). Nucleic acid chaperones can catalyze two reactions in vitro: nucleic acid annealing and strand exchange. Nucleic acid annealing is the reaction in which two single-stranded nucleic acids complementary to each other are annealed, whereas strand exchange is the reaction in which the replacement of strands occurs between a double-stranded nucleic acid and a single-stranded nucleic acid. Recently, nucleic acid chaperones that can catalyze only one of these reactions were found, suggesting that their catalytic mechanism can be dissected into at least two distinct manners (28). However, their molecular basis is not well understood. The mouse L1 ORF1p catalyzes both reactions, and mutations abolishing its nucleic acid chaperone activity dramatically decrease the frequency of L1 retrotransposition (20, 21). This indicates that the nucleic acid chaperone activity is required for L1 retrotransposition, although its precise role has not been elucidated (21). It has not been determined, however, whether the ORF1ps other than the one encoded by L1 have nucleic acid chaperone activity, except for the I factor-encoded ORF1p, which catalyzes nucleic acid annealing but has not been tested for strand exchange (2).
We previously isolated two retrotransposition-competent LINEs from the zebrafish genome, one of which is ZfL2-1 (31). ZfL2-1 is an APE-type LINE and belongs to the L2 clade. ORF1ps encoded by the L2-clade LINEs can be divided into at least three types based on their structures. One type comprises the RRM and zinc knuckle domains such as fly I factor and silkworm SART1 (Fig. 1A). The combination of these domains is most typical among ORF1ps. The second type comprises the RRM and CTD domains such as mouse L1 (Fig. 1A), resembling the ORF1ps encoded by L1 clade LINEs. The third type does not contain any known RNA-binding domains and instead contains an esterase (ES) domain of unknown function (11). Thus, it is not clear whether ORF1ps of the third type have biochemical properties similar to those with RNA-binding domains. The ORF1p encoded by ZfL2-1, typical of the third type, comprises a CC motif at its N terminus and an ES domain in its C-terminal half, but it does not contain any known domains responsible for RNA binding (Fig. 1B). In this study, we show that ZfL2-1 ORF1p has all the canonical activities, that is, self-interaction, nucleic acid binding, nucleic acid annealing, and strand exchange, in its N-terminal region, suggesting that this ES-type ORF1p has a “typical” function in retrotransposition. In addition, we gained insight into the molecular mechanism by which ZfL2-1 ORF1p rearranges the conformation of nucleic acids.
MATERIALS AND METHODS
Oligonucleotides.
DNA oligonucleotides were purchased from Sigma Genosys. Oligonucleotides used in UV cross-linking, annealing, melting, and strand exchange assays were PAGE-purified grade. RNA for the UV cross-linking assay was synthesized in vitro as follows. PCR was performed using ZF1-stemT7F (5′-CTAATACGACTCACTATAGTGTGAAGCGCTTTGACACAATCTAC-3′) and ZF1-stemR (5′-TTATTTGTATAGCGCTTTTACAATGTAG-3′) in the presence of ZL08 vector (31) as a template for LINE-RNA (26) or using pUC-T7F (5′-CTAATACGACTCACTATAGTCGTGCCAGCTGCATTAATGAATC-3′) and pUC-R (5′-AACCGCCTCTCCCCGC-3′) in the presence of pUC19 (Invitrogen) as a template for pUC-RNA, and then the PCR products were transcribed in vitro using the AmpliScribe T7 transcription kit (Epicentre Biotechnologies). The transcribed RNA oligonucleotides were gel purified from 10% denaturing polyacrylamide gels. 32P labels were introduced using [γ-32P]ATP (Perkin Elmer) and T4 polynucleotide kinase (Epicentre Biotechnologies), and the unincorporated isotope was removed using the QIAquick nucleotide removal kit (Qiagen). Preannealed duplexes used in this study were prepared as follows. More than 3 μM 32P-labeled oligonucleotides was combined with equimolar amounts of the nonlabeled complementary oligonucleotides in 50 mM NaCl. After incubation at 95°C for 5 min, the mixtures were cooled to 20°C at −1°C/min and then stored frozen until use. The names and sequences of the oligonucleotides used in UV cross-linking, annealing, melting, and strand exchange assays are indicated in Tables 1 and 2.
Table 1.
Oligonucleotides used in the UV cross-linking assaya
| Name | Sequence (5′ to 3′) |
|---|---|
| pUC-DNA | GTCGTGCCAGCTGCATTAATGAATCGGCCAACGCGCGGGGAGAGGCGGTT |
| pUC-RNA | GUCGUGCCAGCUGCAUUAAUGAAUCGGCCAACGCGCGGGGAGAGGCGGUU |
| LINE-DNA | GTGTGAAGCGCTTTGACACAATCTACATTGTAAAAGCGCTATACAAATAA |
| LINE-RNA | GUGUGAAGCGCUUUGACACAAUCUACAUUGUAAAAGCGCUAUACAAAUAA |
pUC oligonucleotide sequences were derived from pUC19; LINE oligonucleotide sequences were derived from the 3′ UTR of ZfL2-1 and can form stem-loop structures (26). The stem regions are underlined.
Table 2.
Oligonucleotides used in the annealing, melting, and strand exchange assaysa
| Name | Sequence (5′ to 3′) |
|---|---|
| co25 | GCGAGTTGATGTTAGACTGTGTACT |
| o29 | AAAAAGTACACAGTCTAACATCAACTCGC |
| co29 | GCGAGTTGATGTTAGACTGTGTACTTTTT |
| co294m | GCGAGTTGACGTCAGACCGTGCACTTTTT |
| o293′ | AGTACACAGTCTAACATCAACTCGCAAAA |
| o33 | AAAAAGTACACAGTCTAACATCAACTCGCAAAA |
| co33 | TTTTGCGAGTTGATGTTAGACTGTGTACTTTTT |
| o29T4 | AAAAAGTACACAGTCTAACATCAACTCGCTTTT |
| o293′T4 | TTTTAGTACACAGTCTAACATCAACTCGCAAAA |
Oligonucleotides are named for their length except for o29T4 and o293′T4, which are 33 nucleotides long. “T4” indicates that four T's were added to one end, and “c” indicates the complementary strand. The nucleotides in co294m that form mismatches when hybridized to o29 are underlined.
Vectors.
The entire ORF1 sequence of ZfL2-1 was amplified by PCR using primers ZF1F2 (5′-ACACCATATGTCGCTTCCGTCTCTGTCC-3′) and ZF1R2 (5′-ACACGGTACCATGTCGCTTCCGTCTCTGTCCTTG-3′) with genomic DNA from zebrafish (Danio rerio) as a template. The PCR product was digested with KpnI and HindIII and then cloned into KpnI-HindIII digested pET-32c(+) (Novagen), generating the p32F vector. To create the N-terminal or C-terminal region of the ORF1 fusion protein expression vector (p32N or p32C), PCR was performed using ZF1F2 and ZF1R6 (5′-ACACAAGCTTCACGTTCTGCCCCGGGATC-3′) or ZF1F6 (5′-ACACGGTACCTCTCCTCCTGTGTTCGAGATC-3′) and ZF1R2 in the presence of p32F as a template, and then the PCR products were subcloned into the KpnI-HindIII site of pET-32c(+). To create activation domain (AD) fusion protein expression vectors (pAD-N or pAD-C), PCR was performed using ZF1F7 (5′-ACACGGATCCTTATGTCGCTTCCGTCTCTGTCC-3′) and ZF1R8 (5′-ACACCTCGAGTCACGTTCTGCCCCGGGATC-3′) or ZF1F8 (5′-ACACGGATCCTTTCTCCTCCTGTGTTCGAGATC-3′) and ZF1R7 (5′-ACACCTCGAGTCAGATGGTGCGAAGTAGTCTGGAG-3′) in the presence of p32F as a template, and then the PCR products were subcloned into the BamHI-XhoI site of pGADT7 (Clontech). To create binding domain (BD) fusion protein expression vectors (pBD-FL, pBD-N, or pBD-C), PCR was performed using ZF1F7 and ZF1R7, ZF1F7 and ZF1R8, or ZF1F8 and ZF1R7, and then the PCR products were subcloned into the BamHI-SalI site of pGBKT7 (Clontech). To create a glutathione S-transferase (GST) fusion protein expression vector (pGF), PCR was performed using ZF1F7 and ZF1R7 in the presence of p32F as a template, and then the PCR product was subcloned into the BamHI-XhoI site of pGEX-5X-2 (GE Healthcare). Cloning products were all verified by sequencing.
Sequence analyses of ZfL2-1 ORF1p.
The ORF1p sequence contains a single conservative amino acid substitution (A45V) relative to the ORF1p sequence of ZfL2-1 that was shown to retrotranspose in HeLa cells (31). The boundary between the N-terminal and C-terminal portions of ORF1p was selected based on computational analyses. An esterase-like domain was predicted by Kapitonov and Jurka (11). The secondary structure of ORF1p was predicted by the new joint method (9, 25), and then the boundary was determined at a nonstructured region upstream of the esterase-like domain. The prediction of a coiled-coil domain was established by COILS (17).
Protein production.
For the GST pulldown assay, a GST fusion of the full-length ORF1p (GST-F-ORF1p) and GST protein was produced as follows. pGEX-5X-2 or pGF was transformed in Escherichia coli Rosetta(DE3)pLysS (Novagen). The transformants were grown to an optical density at 600 nm (OD600) of 0.6 at 37°C in LB broth containing 50 μg/ml ampicillin and 35 μg/ml chloramphenicol, and then the expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h at 37°C. Cells from 500-ml cultures were resuspended in 20 ml NET-N+ buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 0.5% [wt/vol] Triton X-100 [pH 7.5]) and lysed by sonication. The soluble fraction was separated by centrifugation at 7,200 × g for 5 min at 4°C. To produce the thioredoxin (Trx) fusion of the full-length ORF1p and its N- and C-terminal portions and Trx protein, p32F, p32N, p32C, and pET-32c(+) were transcribed in vitro and translated using the TNT T7 quick-coupled transcription/translation kit (Promega) in the presence of [35S]methionine.
For UV cross-linking, annealing, melting, and strand exchange assays, the thioredoxin-tag protein and the N-terminal portion of ZfL2-1 ORF1p fused with the thioredoxin-tag protein (Trx-N-ORF1p) were produced as follows. pET-32c(+) or p32N was transformed in E. coli Rosetta(DE3)pLysS for production of Trx or Trx-N-ORF1p, respectively. The transformants were grown to an OD600 of 0.5 at 37°C in LB broth containing 50 μg/ml ampicillin and 35 μg/ml chloramphenicol, and then the expression was induced by the addition of 0.8 mM IPTG for 3 h at 37°C. Cells from 200-ml cultures were resuspended in 10 ml binding buffer (20 mM sodium phosphate, 500 mM NaCl, 10 mM imidazole [pH 7.4]) and lysed by sonication. The insoluble fractions were washed three times with the binding buffer and resuspended in eB50 with 8 M urea buffer (20 mM sodium phosphate, 500 mM NaCl, 50 mM imidazole, 8 M urea [pH 7.4]) by pipetting and incubation for 15 min at 50°C, for 15 min at 60°C, and overnight at room temperature with shaking. After centrifugation at 7,200 × g for 30 min and 17,200 × g for 15 min at room temperature, the supernatants were purified by HiTrap Chelating HP (GE Healthcare) with Ni2+ because the proteins contained 6×His tags. To refold the purified proteins, 2 ml of the protein solutions was diluted in 50 ml refolding buffer (50 mM Tris-HCl, 9.6 mM NaCl, 0.4 mM KCl, 500 mM arginine hydrochloride, 1 mM dithiothreitol [DTT] [pH 8.2]) at 4°C. The refolded proteins were concentrated by Amicon Ultra-15 nominal molecular weight limit (NMWL) 10 kDa (Millipore) and diluted in ref-DTT+gly buffer (50 mM Tris-HCl, 9.6 mM NaCl, 0.4 mM KCl, 500 mM arginine hydrochloride, 40% glycerol [pH 8.2]) at 4°C. The concentration and dilution were repeated up to more than a 100-fold dilution, and then the refolded soluble proteins were recovered in the supernatant after centrifugation at 17,200 × g for 30 min at 4°C. The soluble fractions were stored at −20°C. The purity of the protein at each step was analyzed by 12% SDS-PAGE and Quick-CBB (Wako). Western blot analysis was performed using the final protein preparation using anti-His6-peroxidase (Roche Applied Science) and Immobilon Western (Millipore). The protein concentrations were measured by Coomassie Plus protein assay (Pierce).
Yeast two-hybrid assay.
All vectors, yeast strains, reagents, and methods were adapted from Matchmaker two-hybrid system 3 (Clontech). The constructs derived from pGADT7 and pGBKT7 were cotransformed into the yeast strain, AH109 (Clontech). The transformants with both vectors were selected on synthetic complete medium depleted of leucine and tryptophan (SC-LT) after 3 days of growth at 30°C. At least five independent colonies were picked and grown to an OD600 of 0.4 to 0.6 in SC-LT liquid cultures at 30°C. After equalization of concentration, 5 μl of 1:10, 1:100, and 1:1,000 diluted solutions was spotted onto SC-LT and synthetic complete medium depleted of leucine, tryptophan, adenine, and histidine (SC-LTAH) plates. These plates were incubated at 30°C for 3 days.
GST pulldown assay.
GST-F-ORF1p or GST protein (20 ml) was incubated for 2 h at 4°C with 400 μl glutathione Sepharose 4B beads (Amersham Biosciences) that had been washed three times with 1 ml of NET-N+ buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100 [pH 7.5]). The beads bound with GST or GST fusion protein (25 μl) were incubated with 10 μl of in vitro translated proteins in 300 μl of NET-N+ buffer. After incubating overnight at 4°C with rotation, the beads were washed five times with NET-N+ buffer. After removal of the last wash, 25 μl SDS-PAGE loading buffer (62.5 mM Tris-HCl, 2% SDS, 10% glycerol, 0.015% bromophenol blue [pH 6.8]) was added and boiled for 5 min. The proteins were separated by 12% SDS-PAGE and analyzed by autoradiography (BAS 2000 and Image Gauge; Fujifilm).
UV cross-linking assay.
The labeled oligonucleotides were incubated with or without 1 μM purified proteins in 10 μl of GS buffer (20 mM HEPES-NaOH, 10 mM NaCl, 2 mM MgCl2, 2 mM DTT, 15% glycerol [pH 7.5]) at 4°C for 30 min, and then the solutions were irradiated with UV light at 254 nm for 20 min using Funa-UV-Linker (Funakoshi). The DNA and RNA solutions were incubated at 37°C for 30 min with 2.5 U of DNase I (Epicentre Biotechnologies) and 0.5 μg of RNase A (Sigma-Aldrich), respectively. SDS-PAGE loading buffer (10 μl) was added and boiled for 5 min. The proteins were separated by 12% SDS-PAGE and analyzed by autoradiography.
Annealing assay.
The 32P-labeled oligonucleotide ([32P]o29) (2 nM) was added to HET buffer (20 mM HEPES-NaOH, 1 mM EDTA, 1 mM DTT, 0.1% Triton X-100 [pH 7.1]) containing 2 nM nonlabeled oligonucleotide (co29) and the indicated amount of protein. Reaction mixtures were incubated for 3 min at 37°C and then mixed with a half volume of the stop solution (0.2% SDS, 15% glycerol, 0.2% bromophenol blue) on ice to stop the reactions. [32P]o29 and [32P]o29-co29 were separated by 15% native PAGE in TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA). The gel was dried and analyzed by autoradiography. The amount of each oligonucleotide was quantified by Image Gauge.
Melting assay.
Preannealed 32P-labeled duplex (2 nM) was added to the HET buffer in the presence or absence of protein. The temperature of the mixture was increased in a stepwise manner. After incubation at the indicated temperature for 5 min, nine-tenths volume of the stop solution was added to stop melting. The oligonucleotides were separated by 15% native PAGE in TBE, followed by autoradiography to quantify the amount of labeled oligonucleotide.
Strand exchange assay.
Strand exchange assays were set up on ice in HET buffer containing 1 mM DTT, 2 nM preannealed 32P-labeled duplex, 100 nM indicated oligonucleotide, and the indicated amount of protein. Strand exchange assays were initiated by placing reaction tubes at 37°C and stopped on ice by addition of nine-tenths volume of the stop solution at the indicated time points. The oligonucleotides were separated by 15% native PAGE in TBE, and then the gels were analyzed by autoradiography. The amount of each oligonucleotide was quantified by Image Gauge. Initial rates were calculated from the amount of labeled single strand at 0.25, 0.5, 1, 1.5, and 2 min.
RESULTS
ZfL2-1 ORF1p interacts with itself through the N-terminal region.
To examine whether the ZfL2-1 ORF1p interacts with itself, the yeast two-hybrid assay was performed using the yeast strain, AH109, and two kinds of vectors, pGBKT7 and pGADT7. In this system, AH109 cells that contain the two vectors grow on synthetic complete medium depleted of leucine and tryptophan (SC-LT), whereas cells in which the two ORF1ps from these vectors interact with each other grow on synthetic complete medium depleted of leucine, tryptophan, adenine, and histidine (SC-LTAH). As shown in Fig. 1C, AH109 cells that carry pGADT7 expressing the N-terminal portion of the ZfL2-1 ORF1p grew on SC-LTAH only when they concomitantly carried pGBKT7 expressing full-length ORF1p or the N-terminal portion but not the C-terminal portion, suggesting that ZfL2-1 ORF1p interacts with itself through the N-terminal portion. Consistent with this result, pGADT7 expressing the C-terminal portion of the ZfL2-1 ORF1p did not allow AH109 cells to grow on SC-LTAH in all cases with pGBKT7 vectors (Fig. 1C).
Self-interaction of ZfL2-1 ORF1p was also examined by GST pulldown assays in vitro (Fig. 1D). Full-length ORF1p or the N- or C-terminal portion, each of which was produced as a Trx fusion protein by in vitro translation in the presence of [35S]methionine, was mixed with full-length ORF1p, which was fused with GST (GST-F-ORF1p). Full-length ORF1p or the N-terminal portion interacted with GST-F-ORF1p, whereas the C-terminal portion or Trx alone did not (Fig. 1D). Together with the data from the yeast two-hybrid assay, these results indicated that ZfL2-1 ORF1p interacts with itself through the N-terminal region.
The N-terminal portion of ZfL2-1 ORF1p interacts with nucleic acids.
Trx and the N-terminal portion of ZfL2-1 ORF1p fused with Trx (here denoted as Trx-N-ORF1p) were expressed in Escherichia coli, purified, and used in all the experiments reported below. To examine whether ZfL2-1 ORF1p could interact with nucleic acids, we conducted UV cross-linking assays in vitro (Fig. 1E and F). UV irradiation creates covalent bonds between proteins and nucleic acids that are in close contact. Trx-N-ORF1p or Trx was mixed with 32P-labeled nucleic acid followed by UV irradiation, and then the interaction was analyzed by SDS-PAGE. In this assay, the protein directly interacting with the nucleic acid was labeled by 32P derived from the nucleic acid. First, two RNA species, one derived from the pUC19 vector and the other from the 3′ tail of ZfL2-1, were subjected to the assay (Fig. 1E). In both cases, 32P signals were detected on Trx-N-ORF1p but not on Trx, indicating that the N-terminal portion of the ORF1p interacts with RNA in a non-sequence-specific manner. Two DNA species of the same sequence were then subjected to this assay, and the N-terminal portion also nonspecifically interacted with DNA (Fig. 1F). These results indicated that ZfL2-1 ORF1p bears non-sequence-specific nucleic acid binding ability in its N-terminal region.
The N-terminal portion of ZfL2-1 ORF1p accelerates nucleic acid annealing.
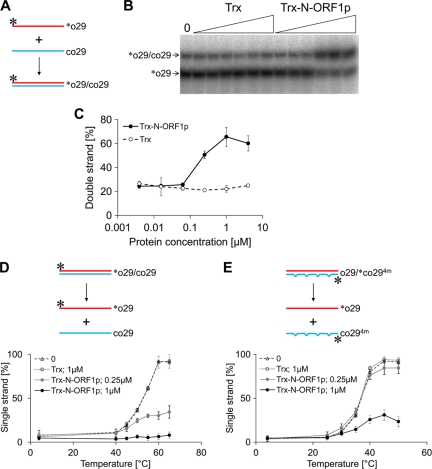
We performed a nucleic acid annealing assay to examine whether ZfL2-1 ORF1p can act as a nucleic acid chaperone (Fig. 2A to C). Equal amounts of 32P-labeled single-stranded DNA ([32P]o29) and its full complement (co29) were incubated together in the presence of Trx-N-ORF1p or Trx (0.0039 to 4 μM), and the ratio of the single- to double-stranded form was determined by native PAGE (Fig. 2A to C). The proportion of the double-stranded form (∼25%) was not altered when Trx was added at any concentration. On the other hand, the addition of Trx-N-ORF1p at ≥0.25 μM increased the proportion of double-stranded DNA to up to ∼75%, indicating that the N-terminal portion of ZfL2-1 ORF1p accelerated the annealing of the single-stranded DNAs, thus serving as a nucleic acid chaperone. In addition, we examined the effect of ZfL2-1 ORF1p on the melting temperature of double-stranded DNAs using duplexes without mismatches ([32P]o29-co29) and with four mismatches (o29-[32P]co294m). The addition of Trx-N-ORF1p increased the melting temperatures of both duplexes (Fig. 2D and E), suggesting that ZfL2-1 ORF1p functions not only to rearrange the conformation of nucleic acids into a stable form but also to maintain it.
Fig 2.
The N-terminal portion of ZfL2-1 ORF1p accelerates nucleic acid annealing and keeps double-stranded DNA stable. (A) Schematic diagram of the nucleic acid annealing assay. The two complementary oligonucleotides, o29 and co29, are indicated by red and blue lines, respectively. Their sequences are shown in Table 2. o29 was labeled by 32P at its 5′ end (asterisks). (B) A representative autoradiogram of [32P]o29 separated by native PAGE. The two oligonucleotides were incubated with 4-fold serial dilutions of Trx or the Trx fusion of the N-terminal portion of ZfL2-1 ORF1p (Trx-N-ORF1p) (triangles, 0.0039 to 4 μM). The left lane indicates the result with no protein (0). (C) Acceleration of annealing. Five independent experiments were performed, and the averages and standard deviations are shown. (D and E) Schematic diagrams and the fraction of single-stranded labeled oligonucleotides in the melting assays. The 32P-labeled duplex ([32P]o29/co29 in panel D or o29/[32P]co294m in panel E) was mixed with no protein (0), 1.0 μM Trx, 0.25 μM Trx fusion of the N-terminal portion of ZfL2-1 ORF1p (Trx-N-ORF1p), or 1 μM Trx-N-ORF1p and then incubated at the indicated temperatures for 5 min. The oligonucleotide sequences are shown in Table 2. One strand of each initial duplex was labeled by 32P at its 5′ end (asterisks). Five independent experiments were performed, and the averages and standard deviations are shown.
The N-terminal portion of ZfL2-1 ORF1p exchanges strands between a double-stranded DNA and a single-stranded DNA.
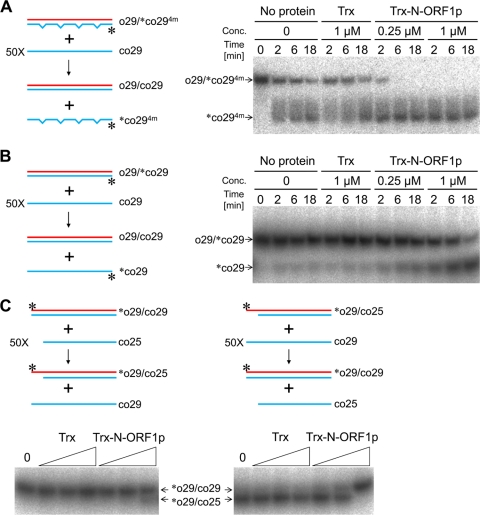
We examined whether the nucleic acid chaperone activity of ZfL2-1 ORF1p can also carry out the strand exchange reaction between a double-stranded DNA and a single-stranded DNA. A duplex, one strand of which was labeled by [32P]phosphate, was mixed with a 50-fold excess of a single-stranded DNA that was complementary to the nonlabeled strand of the duplex (Fig. 3A and B). When strand exchange occurred between them, the 32P-labeled oligonucleotide was released from the duplex as a single strand. Thus, strand exchange was detected as the emergence of the labeled single strand on native PAGE. First, the duplex composed of the 29-base oligonucleotide (o29) and its complement containing four mismatches ([32P]co294m) was mixed with a 50-fold excess of the 29-base oligonucleotide (co29) that was fully complementary to o29 (Fig. 3A). In this case, the duplex became more stable if strand exchange occurred. Because of the instability of the duplex, the labeled oligonucleotide was released from the duplex in the absence of any proteins. The addition of Trx did not alter the ratio of the single- to double-stranded DNA. However, the addition of Trx-N-ORF1p promoted the release of the labeled strand, indicating that the N-terminal portion of ORF1p facilitated strand exchange in the direction that increased the stability of the duplex. Next, the perfectly matched duplex of 29 bp (o29-[32P]co29) was mixed with a 50-fold excess of the unlabeled co29 (Fig. 3B). In this case, the stability of the duplex did not change even if strand exchange occurred. Although only a small fraction of the labeled oligonucleotide was released from the stable duplex with the addition of no protein or Trx, the addition of Trx-N-ORF1p increased its release remarkably, indicating that the N-terminal portion of the ORF1p caused strand exchange without any stability changes in the duplex. These results prompted us to examine whether the ZfL2-1 ORF1p could cause strand exchange in the direction that decreased the stability of a target duplex, although it is believed that the mouse L1 ORF1p cannot (20). We performed a strand exchange assay using the duplex of 29 bp ([32P]o29-co29) and a 50-fold excess of the 25-base oligonucleotide (co25) (Fig. 3C, left). In this case, the duplex of 25 bp with a 4-base overhang ([32P]o29-co25) was produced if strand exchange occurred, decreasing the stability of the initial duplex. It should be noted that in this assay, the strand of the duplex that was not replaced by strand exchange was labeled by [32P]phosphate because the 29- and 25-bp duplexes could be distinguished by their distinct mobilities on native PAGE (Fig. 3C). Surprisingly, Trx-N-ORF1p caused strand exchange between these oligonucleotides, although the efficiency was very low compared with that for the reverse combination (Fig. 3C). Taken together, these results revealed that the N-terminal portion of ZfL2-1 ORF1p could cause strand exchange in all the directions that increase, keep, or decrease the stability of a target duplex.
Fig 3.
The N-terminal portion of ZfL2-1 ORF1p exchanges strands between double- and single-stranded DNAs. Strand exchange assays were performed in the presence of Trx or the Trx fusion of the N-terminal portion of ZfL2-1 ORF1p (Trx-N-ORF1p). Schematic diagrams of the strand exchange assays and the resulting autoradiograms are shown. In the diagrams, oligonucleotides are indicated by blue and red lines. Their sequences are shown in Table 2. One strand of each initial duplex was labeled by 32P at its 5′ end (asterisks). (A, B) Left, schematic diagram of the strand exchange proceeding in the direction increasing the stability of the duplex (A) or causing no stability change (B). Mismatches in the duplex are shown by notches in the line. Right, a representative autoradiogram of the strand exchange with the proteins of interest. (C) Top, schematic diagrams of the strand exchange proceeding in the direction decreasing (left) or increasing (right) the stability of the duplex. Bottom, representative autoradiograms of the strand exchanges. The strand exchanges were conducted for 30 min with increasing amounts of Trx or Trx-N-ORF1p (triangles; 0.063, 0.25, and 1 μM each protein). The left lane of each autoradiogram shows the result with no protein (0).
The strand exchange reaction caused by ZfL2-1 ORF1p is reversible.
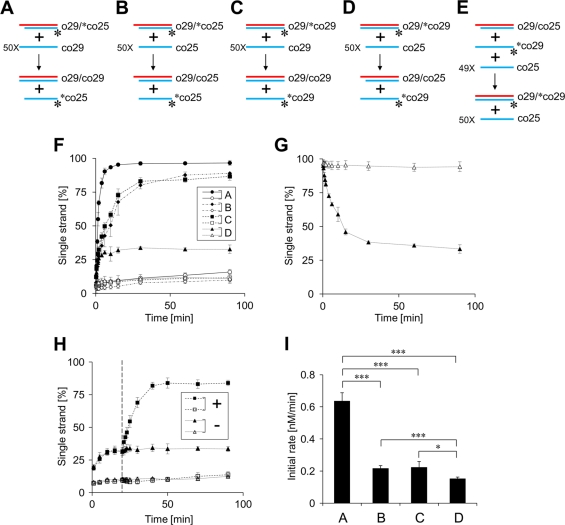
To gain insight into the molecular basis of the ORF1p-dependent strand exchange, we further conducted strand exchange assays using several combinations of double-stranded DNA and single-stranded DNA (Fig. 4). In all cases, 32P-labeled oligonucleotides in the duplexes were released by strand exchange (Fig. 4A to D), and the time course of their release was measured. First, the duplex of 25 bp (o29-[32P]co25) was mixed with a 50-fold excess of the 29-base oligonucleotide (co29) in the presence of Trx-N-ORF1p (Fig. 4A). In this case, the initial duplex was converted to the more stable duplex of 29 bp (o29-co29). Most of the 32P-labeled molecules were immediately released from the initial duplex in a few minutes and reached a state in which ∼97% of the labeled molecules were present as a single strand (Fig. 4F). Next, the duplex of 25 bp (o29-[32P]co25) was mixed with a 50-fold excess of co25, the oligonucleotide whose length is the same as the duplex (Fig. 4B). Also, the duplex of 29 bp (o29-[32P]co29) was mixed with a 50-fold excess of co29 (Fig. 4C). In these cases, the stability of the initial duplexes did not change even if strand exchange occurred (here we call these reactions “equivalent strand exchanges”). Each labeled strand was released at a similar rate and reached states in which 87 to 89% of the labeled molecules were present as a single strand. Their release rates were somewhat slow compared with that of the strand exchange increasing the stability of the duplex (Fig. 4F). Finally, when the duplex of 29 bp (o29-[32P]co29) was mixed with a 50-fold excess of the 25-base oligonucleotide (co25), the less-stable duplex of 25 bp (o29-co25) was produced by strand exchange (Fig. 4D). The labeled molecules were also released from the initial duplex with a maximum of ∼33% single-stranded oligonucleotide (Fig. 4F). These results indicated that the N-terminal portion of ZfL2-1 ORF1p could cause strand exchange regardless of stability changes in a target duplex and suggested that the ORF1p-dependent strand exchange is reversible. Thus, the obvious difference in the states observed in the strand exchange assays most likely represented the distinct equilibrium of each reaction. Accordingly, we designed an experiment to directly examine whether the strand exchanges were in equilibrium in which the forward reaction (Fig. 4D) and the reverse reaction (Fig. 4E) were balanced. In the assay for the reverse reaction (Fig. 4E), we used the combination of double-stranded DNA (o29-co25) and single-stranded DNA ([32P]co29 and a 49-fold excess of co25), which is theoretically identical to the concentration of the DNAs present in the state where the forward reaction (Fig. 4D) had proceeded to completion. As shown in Fig. 4G, the “released” oligonucleotide ([32P]co29) was incorporated into the duplex and reached the state nearly identical to that of the “forward” reaction (∼33% of [32P]co29 were single strands) (Fig. 4G). Furthermore, the addition of another 50-fold excess oligonucleotide (co29) in the forward reaction (Fig. 4D) changed the final state (from ∼33% to ∼84%) (Fig. 4H). These results indicated that the strand exchange was in equilibrium.
Fig 4.
The strand exchange caused by ZfL2-1 ORF1p is reversible. Strand exchange assays were performed in the presence of Trx or the Trx fusion of the N-terminal portion of ZfL2-1 ORF1p (Trx-N-ORF1p). (A to E) Schematic diagrams of the strand exchange assays are shown. Oligonucleotides used in the assays are indicated by blue and red lines. Their sequences are shown in Table 2. One strand of each initial duplex was labeled by 32P at its 5′ end (asterisks). (F) Time course for the single-stranded fraction of the labeled oligonucleotides in the strand exchange assays (A to D). Filled symbols indicate values with Trx-N-ORF1p; open symbols indicate values with Trx. The strand exchanges were conducted with 1 μM proteins at 37°C. Five independent experiments were performed, and the averages and standard deviations are shown. (G) Time course for the single-stranded fraction of the labeled oligonucleotide in the strand exchange assay (E). Filled and open symbols are as described for panel F. (H) Time course for the single-stranded fraction of the labeled oligonucleotide in strand exchange assays. The strand exchange assay (D) was conducted with (+) or without (−) addition of a 50-fold excess co29. co29 was added at 20 min after the initiation (dashed line). Filled and open symbols are as described for panel F. (I) The initial rates of the strand exchange reactions (A to D). The averages of the initial rates and standard deviations are indicated. Statistical analysis was performed using Student's t test; *, P < 0.05; ***, P < 0.001.
To compare the efficiency of the strand exchange reactions without the effects of the reverse reactions, their initial rates were calculated using five time points within 2 min after the initiation of the reaction, during which the labeled strands were released linearly with time (Fig. 4I). The initial rate of the strand exchange that increased the stability of the duplex was approximately 3 times higher than those of the equivalent strand exchanges (compare A with B and C in Fig. 4I), whereas the initial rate of the strand exchange that decreased the stability of the duplex was similar to those of the equivalent strand exchanges (compare B and C with D in Fig. 4I). These results indicated that the strand exchange reactions caused by ZfL2-1 ORF1p proceeded with the different efficiencies.
Short annealed segment between target DNAs accelerates strand exchange.
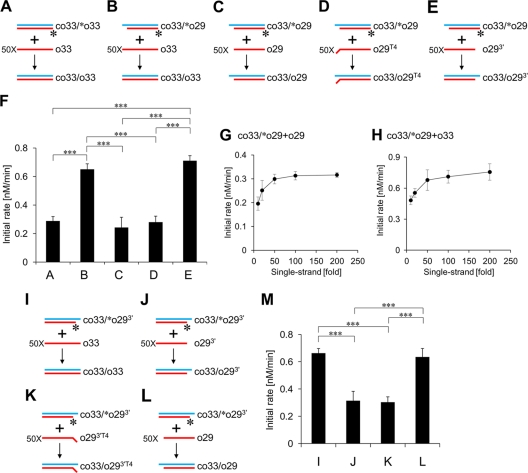
To determine what causes the difference in their initial rates, we conducted further strand exchange assays (Fig. 5). As shown in Fig. 5A, strand exchange between the duplex of 33 bp (co33-[32P]o33) and a 50-fold excess of the 33-base oligonucleotide (o33) was induced by the presence of Trx-N-ORF1p, and its initial rate was measured (Fig. 5F). Generation of a 5′ overhang on the duplex enhanced the initial rate 2-fold (Fig. 5B and F). Because the overhang could form a short annealing with the single-stranded DNA, it was possible that this short annealed segment enhanced the initial rate. Disruption of the short annealed segment by deletion or mutation in the single-stranded DNA abolished the rate enhancement (Fig. 5C, D, and F), supporting this notion. These data prompted us to conduct a further strand exchange assay to examine whether the annealing but not the stability of the duplex was responsible for the observed acceleration of the strand exchange. In this experiment, we used the double-stranded DNA (co33-[32P]o29) and single-stranded DNA (o293′), which could form the short annealed segment but did not change the length and GC content of the duplex during strand exchange (Fig. 5E). The rate enhancement was still observed, indicating that the annealing itself accelerated the strand exchange (Fig. 5F).
Fig 5.
Short annealed segment between target DNAs accelerates strand exchange. Strand exchange assays were performed with 1 μM Trx fusion of the N-terminal portion of ZfL2-1 ORF1p (Trx-N-ORF1p) at 37°C. (A to E and I to L) Schematic diagrams of the strand exchange assays. Oligonucleotides used in the assays are indicated by blue and red lines. Their sequences are shown in Table 2. One strand of each initial duplex was labeled by 32P at its 5′ end (asterisks). (F and M) The initial rates of the strand exchange reactions (A to E and I to L, respectively). Five independent experiments of each strand exchange assay were performed, and the averages and standard deviations are shown. Statistical analysis was performed using Student's t test; ***, P < 0.001. (G and H) Strand exchange reactions between the duplex (co33-[32P]o29) and increasing excess amounts of the single-stranded DNA (o29 in panel G or o33 in panel H) were induced by Trx-N-ORF1p, and their initial rates were calculated. Five independent experiments were performed for each concentration, and the averages and standard deviations are shown.
Next, in place of the duplex with a 5′ overhang, we used a duplex with a 3′ overhang and examined whether annealing between the duplex with a 3′ overhang and a single strand accelerated strand exchange (Fig. 5I to M). Similar to the results obtained with the 5′-overhang duplex (Fig. 5A to F), the initial rates were enhanced when the 3′ overhang could form a short annealed segment with the single strand. These results indicated that the annealing accelerated strand exchanges regardless of the position of the overhang. Furthermore, we showed that the initial rates measured in these experiments were not affected by the concentration of the single-stranded DNA in the range between 100 nM and 400 nM (a 50-fold and 200-fold excess, respectively) (Fig. 5G and H), indicating that the concentration of the single-stranded DNA is saturated at above 50-fold. Thus, the acceleration was not due to the promotion of the association between the target DNAs.
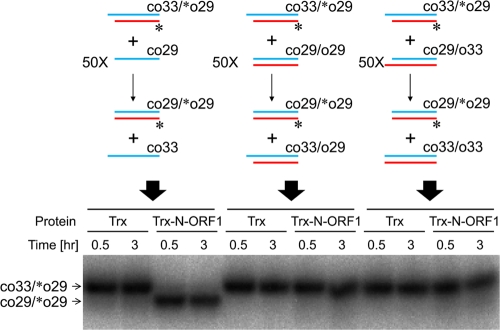
The N-terminal portion of ZfL2-1 ORF1p does not induce strand exchange between two duplexes.
We examined whether ZfL2-1 ORF1p could facilitate strand exchange between two duplexes (Fig. 6). In the presence of Trx-N-ORF1p, strand exchange between the 29-bp duplex (co33-[32P]o29) and a 50-fold excess of the 29-base single strand (co29) was efficiently induced (Fig. 6, left). In contrast, strand exchange between the duplex (co33-[32P]o29) and another duplex (co29-o29 or co29-o33) was not observed even if a short annealed segment could form between the two duplexes (co33-[32P]o29 and co29-o33) (Fig. 6, middle and right). These results indicated that ZfL2-1 ORF1p does not induce strand exchange between duplexes.
Fig 6.
The N-terminal portion of ZfL2-1 ORF1p does not induce strand exchange between two duplexes. Strand exchange assays were performed with 1 μM Trx or the Trx fusion of the N-terminal portion of ZfL2-1 ORF1p (Trx-N-ORF1p) at 37°C. Schematic diagrams of the strand exchange assays (top) and the resulting autoradiogram (bottom) are shown. In the diagrams, oligonucleotides are indicated by blue and red lines. Their sequences are shown in Table 2. One strand of each initial duplex was labeled by 32P at its 5′ end (asterisks). The duplex (co33-[32P]o29) was incubated for the indicated times with a 50-fold excess of the single-stranded oligonucleotide co29 (left), the duplex co29-o29 (middle), or the duplex co29-o33 (right).
DISCUSSION
Many ORF1ps are likely to bind their own RNAs through the RRM domain; the CCHC-type zinc knuckle or CTD, one of which is frequently present downstream of the RRM, also appears to be important for this binding (13). However, not all ORF1ps have these RNA binding domains. Typical examples are the ORF1ps that contain an ES domain of unknown function (11). We have shown here that ZfL2-1 ORF1p, an ES-containing ORF1p, has self-interaction and nucleic acid binding activities. This is the first experimental evidence suggesting that the ES-type ORF1ps can form an RNP during retrotransposition.
The ES-type ORF1ps are encoded by LINEs of the L2, CR1, and RTEX clades (11, 27) and share a conserved structure that can be divided into two parts: the N-terminal half containing a CC motif and the C-terminal half composed of an ES domain. The self-interaction and nucleic acid binding activities of ZfL2-1 ORF1p are both present in its N-terminal region exclusive of the ES domain. Most ES-type ORF1ps retain the CC motif in their N-terminal regions, suggesting that the CC confers the self-interaction ability. However, the N-terminal regions do not contain any other conserved domains except for the CC and are highly divergent. This markedly contrasts with the high conservation of the C-terminal regions containing the ES domains. Thus, it was originally thought that the N-terminal regions had no function except for self-interaction. We demonstrated, however, that ZfL2-1 ORF1p bound RNAs in vitro (Fig. 1E) and, furthermore, formed an RNP together with the ZfL2-1 RNA when it was heterogeneously expressed in HeLa cells (M. Kajikawa and N. Okada, unpublished data), suggesting that the N-terminal region is involved in RNP formation through its RNA-binding activity. This raises the intriguing question of why the N-terminal regions of the ES-type ORF1ps are so divergent. Identification of a domain(s) responsible for RNA binding in each ES-type ORF1p and comparative analysis of the domain(s) will shed light on the molecular basis underlying the RNP formation of the ES-type LINEs. This may be distinct from that of the RNP formation of RRM-type LINEs, which have widely conserved RNA-binding domains.
In addition to the unexpected RNA-binding activity of the ORF1p of ZfL2-1, the discovery that it has nucleic acid chaperone activity in its N-terminal region is surprising. So far, there are two examples in which ORF1ps retain this activity, namely, mouse L1 and fruit fly I factor, both of which are RRM-type LINEs (2, 20). The present discovery and these two examples suggest that the nucleic acid chaperone activity is an essential property of all ORF1ps regardless of their structures and that it probably has a role in LINE retrotransposition. The nucleic acid chaperone activity was also found in the C-terminal region adjacent to the RT domain in Trypanosoma LINE (8), which does not encode a canonical ORF1p, further suggesting a general role in retrotransposition.
Initial strand exchange assays were designed to examine properties of the mouse L1 ORF1p and the Trypanosoma LINE protein; these proteins exchange strands between a single-stranded DNA and a double-stranded DNA to increase, but not decrease, the stability of the duplex (8, 20). This result suggests that the increase in the stability is a key factor for the strand exchange. However, this idea remains controversial because the mouse L1 ORF1p was recently shown to exchange strands without any stability changes (5). In the present study, we clearly demonstrated the detailed properties of strand exchange by nucleic acid chaperone activity as follows. First, ZfL2-1 ORF1p accelerated the annealing of two complementary single-stranded DNAs (Fig. 2). Second, ZfL2-1 ORF1p exchanged strands between a single-stranded DNA and a double-stranded DNA regardless of the stability of the duplex formed (Fig. 3 and 4), showing that the strand exchange was reversible. Third, a short annealed segment between target DNAs accelerated their strand exchange in the presence of ZfL2-1 ORF1p (Fig. 5). Fourth, ZfL2-1 ORF1p did not exchange strands between two duplexes (Fig. 6). From these results, we propose that the function of ZfL2-1 ORF1p is to convert a single-stranded nucleic acid to a double-stranded form using its complement existing as a single or double strand. This reaction will continue as long as single-stranded nucleic acids are present.
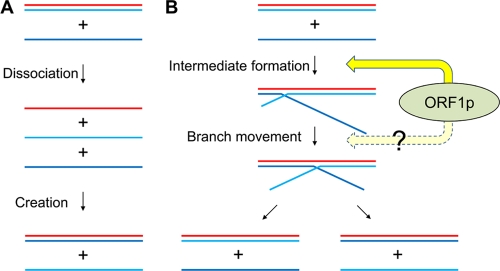
The rearrangement of nucleic acids in strand exchange can be dissected into two reactions: the dissociation of preexisting base pairs and the creation of new base pairs. However, it is not clear whether these two reactions are related to or independent of each other, and the molecular basis underlying strand exchange is not well understood (15). Our strand exchange assays provide novel insight into this mechanism. The initial rate of strand exchange between the 33-bp duplex (co33-[32P]o33) and the 33-base oligonucleotide (o33) was enhanced by the deletion of four nucleotides at the end of [32P]o33 in the duplex (Fig. 5A and B). If the reactions of dissociation and creation are independent, the deletion affects only the dissociation step because the deleted strand is involved only in the dissociation reaction in the two-step model (Fig. 7A). However, the rate enhancement was abolished by deletion or mutation at the end of o33 (Fig. 5C and D), which should be involved only in the creation step of the two-step model. These data indicate that the dissociation and creation reactions are related to and not independent of each other. Remarkably, we showed here that a short annealed segment between target DNAs increased the initial rate of strand exchange (Fig. 5). This suggests that a branched structure formed through the short annealed segment is an intermediate of strand exchange, and its formation is the rate-limiting step (Fig. 7B). Once the branched structure is formed, its branch point will move along the duplex because the preexisting base pairs and the newly formed base pairs are equivalent thermodynamically. The movement seems to be bidirectional because the short annealed segment accelerated strand exchange regardless of its position (Fig. 5). There is little doubt that the branched structure by itself is highly unstable, as it is formed only through a few base pairs. The ZfL2-1 ORF1p may function to keep it stable, which then facilitates branch point movement, resulting in strand exchange. It is possible that ORF1p may also promote branch point movement (Fig. 7B). Currently, we cannot distinguish these two steps in our experiments; precise characterization of these steps will help us to understand the role of ORF1p in strand exchange.
Fig 7.
Models of strand exchange caused by ZfL2-1 ORF1p. (A) Strand exchange model with independent base-pair dissociation and creation. If the dissociation and the creation are independent steps in strand exchange, dissociation is followed by creation. (B) Strand exchange model with interdependent base-pair dissociation and creation. If the dissociation is related to the creation, a branched structure is formed as a reaction intermediate. ZfL2-1 ORF1p facilitates formation of the intermediate. The branch point of the intermediate can move in both directions in a ZfL2-1 ORF1p-dependent or -independent manner. When the branch point moves back to the origin, the incoming single strand is released and the original double strand returns to the double-stranded form (left). On the other hand, when the branch point reaches the opposite end, the new double-stranded DNA forms and one strand of the original double strand is released (right).
In retrotransposition, the LINE RNA functions not only as the mRNA for the LINE-encoded proteins but also as the template for TPRT. Thus, it must be protected from endogenous RNA degradation processes. The major role of ORF1ps may be to protect LINE RNA from degradation because ORF1ps interact with LINE RNA to form a complex. In the complex, ORF1ps plausibly rearrange the RNA into a stable conformation via the nucleic acid chaperone activity. This conformational change of the RNA, by which the double-stranded region of the RNA increases and the single-stranded region decreases, may serve to protect it. On the other hand, the RNA must be unfolded during reverse transcription, which is inhibited by the presence of the double-stranded region. Considering that the double-stranded region in the LINE RNA of the complex is subject to an exchange of one strand by another single-stranded region that must transiently exist in the RNA, reverse transcription might be facilitated by its rearrangement, which probably continues via the nucleic acid chaperone activity. That is, LINE RNA likely is dynamic in its conformation in the presence of the ZfL2-1 ORF1p. This dynamic nature may reconcile those two different requirements, namely, the protection of RNA and the facilitation of reverse transcription.
Another zebrafish LINE, ZfL2-2, is closely related to ZfL2-1. The structures of these two LINEs are highly similar except for the presence of ORF1p. It should be noted, however, that ZfL2-2 is also capable of retrotransposition (31). It will be interesting to explore what new component in ZfL2-2 compensates for the function of ORF1p elucidated in the present study.
ACKNOWLEDGMENT
This work was supported by a grant-in-aid to M.K. and N.O. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print 21 November 2011
REFERENCES
- 1. Cost GJ, Feng Q, Jacquier A, Boeke JD. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21:5899–5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dawson A, Hartswood E, Paterson T, Finnegan DJ. 1997. A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids. EMBO J. 16:4448–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doucet AJ, et al. 2010. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 6:e1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eickbush TH, Malik HS. 2002. Origins and evolution of retrotransposons, p 1111–1144 In Craig NL, Craigie R, Gellert M, Lambowitz AM. (ed), Mobile DNA II. American Society for Microbiology, Washington, DC [Google Scholar]
- 5. Evans JD, Peddigari S, Chaurasiya KR, Williams MC, Martin SL. 2011. Paired mutations abolish and restore the balanced annealing and melting activities of ORF1p that are required for LINE-1 retrotransposition. Nucleic Acids Res. 39:5611–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905–916 [DOI] [PubMed] [Google Scholar]
- 7. Goodier JL, Kazazian HH., Jr 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135:23–35 [DOI] [PubMed] [Google Scholar]
- 8. Heras SR, Lopez MC, Garcia-Perez JL, Martin SL, Thomas MC. 2005. The L1Tc C-terminal domain from Trypanosoma cruzi non-long terminal repeat retrotransposon codes for a protein that bears two C2H2 zinc finger motifs and is endowed with nucleic acid chaperone activity. Mol. Cell. Biol. 25:9209–9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ito M, Matsuo Y, Nishikawa K. 1997. Prediction of protein secondary structure using the 3D-1D compatibility algorithm. Comput. Appl. Biosci. 13:415–424 [DOI] [PubMed] [Google Scholar]
- 10. Januszyk K, et al. 2007. Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1. J. Biol. Chem. 282:24893–24904 [DOI] [PubMed] [Google Scholar]
- 11. Kapitonov VV, Jurka J. 2003. The esterase and PHD domains in CR1-like non-LTR retrotransposons. Mol. Biol. Evol. 20:38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kazazian HH., Jr 2004. Mobile elements: drivers of genome evolution. Science 303:1626–1632 [DOI] [PubMed] [Google Scholar]
- 13. Khazina E, Weichenrieder O. 2009. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc. Natl. Acad. Sci. U. S. A. 106:731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kulpa DA, Moran JV. 2005. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 14:3237–3248 [DOI] [PubMed] [Google Scholar]
- 15. Levin JG, Guo J, Rouzina I, Musier-Forsyth K. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 80:217–286 [DOI] [PubMed] [Google Scholar]
- 16. Luan DD, Korman MH, Jakubczak JL, Eickbush TH. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595–605 [DOI] [PubMed] [Google Scholar]
- 17. Lupas A, Van Dyke M, Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164 [DOI] [PubMed] [Google Scholar]
- 18. Malik HS, Burke WD, Eickbush TH. 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16:793–805 [DOI] [PubMed] [Google Scholar]
- 19. Martin SL, Branciforte D, Keller D, Bain DL. 2003. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 100:13815–13820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin SL, Bushman FD. 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21:467–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin SL, et al. 2005. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J. Mol. Biol. 348:549–561 [DOI] [PubMed] [Google Scholar]
- 22. Martin SL, Li J, Weisz JA. 2000. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J. Mol. Biol. 304:11–20 [DOI] [PubMed] [Google Scholar]
- 23. Matsumoto T, Hamada M, Osanai M, Fujiwara H. 2006. Essential domains for ribonucleoprotein complex formation required for retrotransposition of telomere-specific non-long terminal repeat retrotransposon SART1. Mol. Cell. Biol. 26:5168–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moran JV, et al. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927 [DOI] [PubMed] [Google Scholar]
- 25. Nishikawa K, Noguchi T. 1991. Predicting protein secondary structure based on amino acid sequence. Methods Enzymol. 202:31–44 [DOI] [PubMed] [Google Scholar]
- 26. Nomura Y, et al. 2006. Solution structure and functional importance of a conserved RNA hairpin of eel LINE UnaL2. Nucleic Acids Res. 34:5184–5193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Putnam NH, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317:86–94 [DOI] [PubMed] [Google Scholar]
- 28. Rajkowitsch L, Schroeder R. 2007. Dissecting RNA chaperone activity. RNA. 13:2053–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seleme MC, Busseau I, Malinsky S, Bucheton A, Teninges D. 1999. High-frequency retrotransposition of a marked I factor in Drosophila melanogaster correlates with a dynamic expression pattern of the ORF1 protein in the cytoplasm of oocytes. Genetics 151:761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sugano T, Kajikawa M, Okada N. 2006. Isolation and characterization of retrotransposition-competent LINEs from zebrafish. Gene 365:74–82 [DOI] [PubMed] [Google Scholar]
- 32. Suzuki J, et al. 2009. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 5:e1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takahashi H, Fujiwara H. 2002. Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. EMBO J. 21:408–417 [DOI] [PMC free article] [PubMed] [Google Scholar]