Abstract
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) replicates in the nucleus of insect cells to produce nucleocapsids, which are transported from the nucleus to the plasma membrane for budding through GP64-enriched areas to form budded viruses. However, little is known about the anterograde trafficking of baculovirus nucleocapsids in insect cells. Preliminary confocal scanning laser microscopy studies showed that enhanced green fluorescent protein (EGFP)-tagged nucleocapsids and capsid proteins aligned and colocalized with the peripheral microtubules of virus-infected insect cells. A colchicine inhibition assay of virus-infected insect cells showed a significant reduction in budded virus production, providing further evidence for the involvement of microtubules and suggesting a possible role of kinesin in baculovirus anterograde trafficking. We investigated the interaction between AcMNPV nucleocapsids and kinesin-1 with fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy (FRET-FLIM) and show for the first time that AcMNPV capsid proteins VP39 and EXON0, but not Orf1629, interact with the tetratricopeptide repeat (TPR) domain of kinesin. The excited-state fluorescence lifetime of EGFP fused to VP39 or EXON0 was quenched from 2.4 ± 1 ns to 2.1 ± 1 ns by monomeric fluorescent protein (mDsRed) fused to TPR (mDsRed-TPR). However, the excited-state fluorescence lifetime of an EGFP fusion of Orf1629 remained unquenched by mDsRed-TPR. These data indicate that kinesin-1 plays an important role in the anterograde trafficking of baculovirus in insect cells.
INTRODUCTION
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is a member of the Baculoviridae family with a large, closed circular DNA genome packaged into a rod-shaped capsid. Baculoviruses have a biphasic replication cycle, which results in two virion phenotypes, namely, the occluded derived virus (ODV) and budded virus (BV). ODV is involved in the primary infection of insect caterpillars, whereas BV is required for cell-to-cell transmission and is mainly used in baculovirus research.
Many viruses exploit host surface receptors and macromolecules for entry and the cytoskeleton and motor proteins for intracellular trafficking for replication and cell-to-cell spread (19, 42). In a similar manner, AcMNPV particles enter insect cells via clathrin-mediated endocytosis (28) when AcMNPV GP64 interacts with unknown host surface receptors (31). Naked nucleocapsids, which are released from endosomes due to a low endosomal pH (3), induce actin polymerization through capsid protein Orf1629 (p78/83) (14, 29). It has been suggested that actin polymerization is crucial for efficient translocation of nucleocapsids toward the nucleus (7) and through the nuclear pores (33). Myosins have also been suggested to play a role in the transport of naked nucleocapsids toward the nucleus (26). Inside the nucleus, replicated DNA and synthesized capsid proteins are assembled into nucleocapsids (45) and are transported toward the plasma membrane, the so-called anterograde trafficking, before budding through GP64-enriched areas to form budded viruses (34).
In comparison to the retrograde trafficking detailed by Volkman and colleagues (7, 33), very little is known about anterograde trafficking of baculovirus. An earlier study by Volkman and Zaal (46) suggested that microtubules are not required for transport of AcMNPV nucleocapsids from the nucleus to the cell periphery after an insignificant reduction in budded virus in colchicine-treated, virus-infected Sf9 cells was observed. However, a more recent study has suggested that microtubules may be involved in the anterograde trafficking of baculovirus when virus-infected cells are treated with colchicine at a different time point (12). This adds baculovirus to other DNA viruses such as vaccinia virus (18), African swine fever virus (21), and herpes simplex virus (27), which employ microtubules for anterograde trafficking of virions. However, unlike actin polymerization, which leads to the transport of cargoes, anterograde microtubule-based trafficking is mediated by kinesins, which use microtubules as “tracks or superhighways” to transport cargoes to the desired destinations (44).
A conventional kinesin (kinesin-1) is a tetramer consisting of two kinesin heavy chains (KHCs), the globular head, which are linked to the neck coil (the stalk) via a neck linker. The neck coil is connected to the two kinesin light chains (KLCs). The motor domain of the globular heads binds to and moves along microtubules when ATP is hydrolyzed in the motor catalytic core (22). The KLCs consist of at least two structurally unique regions, an N-terminal region of heptad repeats and a C-terminal region that has a six-tetratricopeptide repeat (TPR). The TPR is the cargo binding domain of kinesin (2).
One of the most sensitive methods available for investigating the interaction of kinesins with their cargoes is fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy (FRET-FLIM). FRET interactions in live cells can be detected by tagging proteins with suitable fluorophores such as an enhanced green fluorescence protein (EGFP) and a monomeric fluorescent protein (mDsRed). The EGFP acts as an excited-state donor, whereas mDsRed functions as an acceptor. FLIM is the most effective approach available to detect the steady-state FRET (43), and quenching of the donor's fluorescence decay lifetime in the presence of the acceptor fluorophore indicates a direct physical interaction between the paired fluorophores (35). The FRET-FLIM technique is not only highly sensitive, but also the decay time of a fluorophore does not change upon intensity variations and is unaffected by donor bleed-through (47).
Two-photon-induced FRET-FLIM (2P-FRET-FLIM) offers several advantages over single-photon FRET-FLIM. 2P-FRET-FLIM offers a reduced phototoxicity by the use of near-infrared excitation light, which is not absorbed by cellular components (9), and also reduces bleaching of the donor fluorophore (40). FRET-FLIM has been employed in examining a direct interaction between nonviral proteins, including β1 integrin and the receptor for epidermal growth factor (30), and also to determine a direct interaction between fowlpox proteins and kinesin-1 (20).
In this study, using 2P-FRET-FLIM, we investigated the direct interaction between an EGFP fusion of AcMNPV capsid proteins (VP39, EXON0, and Orf1629) and a mDsRed fusion of TPR. The excited-state fluorescence decay lifetimes of both EGFP fusions of VP39 and EXON0 were quenched by mDsRed-TPR; however, the decay lifetime of the EGFP-Orf1629 remained unquenched in the presence of mDsRed-TPR. For the first time, we provide evidence that AcMNPV nucleocapsids interact with TPR of kinesin-1. This study further confirms our preliminary work that microtubules are indeed involved in the anterograde transport of baculovirus, as evidenced by both colocalization studies and colchicine inhibition assays.
MATERIALS AND METHODS
Cells and viruses.
Trichoplusia ni 368 (TN-368) cells (16) were maintained in TC100 medium supplemented with 10% fetal calf serum (Gibco, United Kingdom), and IPLB-Sf9 cells (Invitrogen, United Kingdom) were maintained in serum-free Ex-cell 420 medium (Sigma, United Kingdom) at 28°C. Both wild-type virus AcMNPV clone 6 (C6) (1) and recombinant baculoviruses generated for this study were propagated in IPLB-Sf9 cells at 28°C by using standard procedures (23).
Antibodies.
Monoclonal antibodies against VP39 (a kind gift from Taro Ohkawa, University of California, Berkeley, CA) and anti α-tubulin (Sigma, United Kingdom) were used in staining AcMNPV capsid protein VP39 and microtubules, respectively. Anti-mouse immunoglobulin G (IgG) secondary antibodies conjugated with Alexa Fluor 488/568 was obtained from Invitrogen.
Construction of recombinant baculoviruses.
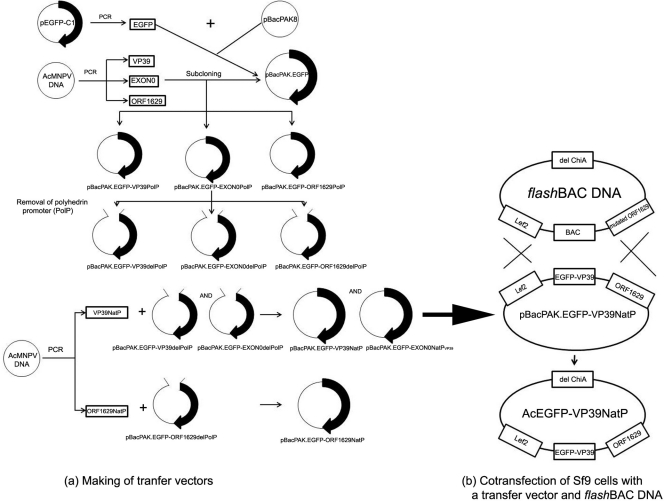
Recombinant AcMNPV baculoviruses (rAcMNPVs) were generated in a two-step approach (17) by cotransfecting Sf9 cells with both a recombinant transfer vector and a flashBAC DNA (Oxford Expression Technologies, United Kingdom) as illustrated in Fig. 1.
Fig 1.
A schematic representation of recombinant baculovirus generation via homologous recombination. The first step is the generation of a transfer vector-pBacPAK.EGFP-VP39NatP (a), followed by a second step, which is the production of AcEGFP-VP39NatP via homologous recombination (b). VP39NatP, VP39 native promoter; ORF1629NatP, ORF1629 native promoter; EGFP, enhanced greed fluorescent protein; BAC, bacterial artificial chromosome; ChiA, chitinase A; PolP, polyhedrin promoter; del, deletion.
Briefly, rAcMNPV transfer vectors pBacPAK.EGFP-VP39NatP, pBacPAK.EGFP-EXON0NatPVP39, and pBacPAK.EGFP-ORF1629 were generated. An EGFP gene fragment was PCR amplified from pEGFP-C1 vector (Molecular Probes, United Kingdom) by using the JD-FP-EGFP/JD-RP-EGFP primer pair (Table 1). The resultant PCR products were subcloned into pCR 2.1-TOPO TA vector (Invitrogen, United Kingdom), and the integrity of the inserts was confirmed by DNA sequencing. The PstI/XhoI EGFP fragment released from the pCR 2.1-TOPO TA vector was ligated into PstI/XhoI-digested pBacPAK8 (Invitrogen, United Kingdom), which contains the polyhedrin promoter (PolP), to produce the pBacPAK8-EGFP-PolP vector. The coding DNA sequence of the capsid proteins, vp39, exon0, or orf1629, was amplified from AcMNPV C6 DNA template by using FP-VP39/FP-VP39, AJ_Ac141_FP/AJ_Ac141_RP, and JD_FP_ORF1629/JD_RP_ORF1629 primers, respectively (Table 1). The resultant PCR fragments were TOPO subcloned. The XhoI/XbaI fragments containing vp39 and exon0 and the KpnI/EcoRI fragment of orf1629 were released from their respective pCR 2.1-TOPO TA vectors and ligated into pBacPAK8.EGFP-PolP in frame with the N-terminal EGFP. The resultant vectors were named pBacPAK8-EGFP-VP39PolP, pBacPAK.EGFP-EXON0PolP, and pBacPAK.EGFP-ORF1629PolP.
Table 1.
Primers used for making recombinant transfer vectors
| Gene | Primer name | Primer sequence (5′–3′)a | Restriction enzyme |
|---|---|---|---|
| egfp | JD-FP-EGFP | aaaCTG CAG AAG CTA GCG CTA CCG GTC | PstI |
| JD-RP-EGFP | aaC TCG AGA TCT AGT CCG GAC | XhoI | |
| vp39 | FP-VP39 | aaC TCG AGA TGG CGC TAG TGC CCG | XhoI |
| RP-VP39 | aaT CTA GAG ACG GCT ATT CCT CCA CCT GC | XbaI | |
| orf1629 | JD_FP_ORF1629 | aaG GTA CCA CGA ATC GTA GAT ATG AAT CTG | KpnI |
| JD_RP_ORF1629 | aGG AAT TCT TAA GCG CTA GAT TCT GTG CG | EcoRI | |
| exon0 | AJ_Ac141_FP | CC CTC GAG ATA AGA ACC AGC AGT CAC GTG | XbaI |
| AJ_Ac141_RP | CC TCT AGA TTA TTT ATA CGA TGT CCT G | XhoI | |
| vp39natp | VP39P-EcoRV-FP | GAT ATC TTG TTC GCC ATC | EcoRV |
| VP39P-BamH-RP | GGA TCC TTG TTG CCG TTA TAA ATA TG | BamHI | |
| orf1629natp | ORF1629P-EcoRV-FP | GAT ATC GAA TTT TTT CCG GAC TAA AAT AC | EcoRV |
| ORF1629P-PstI-RP | CTG CAG GGC CAC CAC AAA TGC TAC | PstI |
The bold lowercase a, aa, and aaa are enzyme spaces. The bold underlined sequences are the restriction enzyme sequences, with the names of the restriction enzymes listed under restriction enzymes.
To express the fusion EGFP-capsid genes under the control of their respective native promoters, except egfp-exon0, which was placed under the control of AcMNPV VP39 promoter, the polyhedrin promoter of both pBacPAK.EGFP-VP39PolP and pBacPAK.EGFP-EXON0PolP was replaced with the AcMNPV VP39 promoter; in pBacPAK.EGFP-ORF1629PolP, the polyhedrin promoter was replaced with the AcMNPV ORF1629 promoter. pBacPAK.EGFP-VP39PolP and pBacPAK.EGFP-EXON0PolP were digested with EcoRV and BamHI and pBacPAK.EGFP-ORF1629PolP was digested with EcoRV and PstI to yield pBacPAK.EGFP-VP39ΔPolP, pBacPAK.EGFP-EXON0ΔPolP, and pBacPAK.EGFP-ORF1629ΔPolP, respectively. Natural promoters of AcMNPV VP39 (VP39NatP) and ORF1629 (ORF1629NatP) were PCR amplified from AcMNPV C6 DNA by using VP39P-EcoRV-FP/VP39P-BamHI-RP and ORF1629P-EcoRV-FP/ORF1629P-PstI-RP primer pairs (Table 1), respectively. The PCR products were TOPO subcloned and sequenced. The EcoRV/BamHI fragment of VP39NatP was ligated to EcoRV/BamHI-digested pBacPAK.EGFP-VP39ΔPolP or pBacPAK.EGFP-EXON0ΔPolP, and the resultant vectors were named pBacPAK.EGFP-VP39NatP and pBacPAK.EGFP-EXON0NatPVP39, respectively. Likewise, the EcoRV/PstI fragment of ORF1629NatP was subcloned into the EcoRV/PstI site of pBacPAK.EGFP-ORF1629ΔPolP to produce pBacPAK.EGFP-ORF1629NatP.
Homologous recombination between pBacPAK.EGFP-VP39NatP, pBacPAK.EGFP-EXON0NatPVP39, or pBacPAK.EGFP-ORF1629NatP and flashBAC DNA in Sf9 cells produced recombinant baculovirus AcEGFP-VP39NatP, AcEGFP-EXON0NatPVP39, or AcEGFP- ORF1629NatP, respectively (Fig. 1). To confirm the presence of fusion genes in the recombinant AcMNPV genome, viral DNA was extracted and analyzed by PCR using the primers listed in the Table 1.
Construction of mDsRed-kinesin vector.
A plasmid vector expressing the tetratricopeptide repeat (TPR) of kinesin-1 light chain (KLC) tagged with monomeric Discosoma sp. red fluorescent protein (mDsRed) under the control of a constitutive OpMNPV immediate early (IE) promoter, pIZ.KLC-TPR-mDsRed, was generated. Vector pEL.KLC-TPR-mDsRed (20) was digested with NcoI and blunt ended with T4 polymerase. The linearized blunt-ended pEL.KLC-TPR-mDsRed vector was digested with XbaI to release the KLC-TPR-mDsRed fragment. A pIZ-mcherry-Actin vector (a kind gift from Matthew Welch, University of California, Berkeley, CA) was first digested with SacI and blunt ended with T4 polymerase. The mcherry-Actin fragment was released by XbaI digestion, and the KLC-TPR-mDsRed fragment was then subcloned into the pIZ backbone to produce pIZ.KLC-TPR-mDsRed.
Confocal immunofluorescence microscopy.
TN-368 cells (0.25 × 106 cells/dish) were seeded onto sterile glass coverslips in 35-mm dishes and incubated at 28°C overnight. Cells were infected with 100 μl of AcEGFP-ORF16291629NatP, AcEGFP-VP39NatP, AcEGF-EXON0NatPVP39, or AcMNPV at a multiplicity of infection (MOI) of 5 PFU/cell and incubated at room temperature. After 1 h of virus absorption, the virus inoculum was replaced with growth medium and incubated at 28°C for the required time. Virus-infected cells were washed with phosphate-buffered saline (PBS) and fixed with freshly prepared 4% paraformaldehyde (PFA) in PBS for 45 min at room temperature. Cells were washed three times with PBS, permeabilized with 0.1% (vol/vol) Triton X-100 (Sigma, United Kingdom) in PBS for 10 min, and blocked with 1% bovine serum albumin (BSA) in PBS for 30 min. The capsid protein VP39 was immunostained using mouse α-VP39 and secondary goat anti-mouse IgG conjugated with Alexa Fluor 488. Microtubules were stained using mouse anti α-tubulin and secondary goat anti-mouse IgG conjugated with Alexa Fluor 568. Cells on the coverslips were mounted in Vectashield fluorescence microscopy mountant with DAPI (Vector Laboratories). Confocal images were acquired using Zeiss LSM 510 META and processed with LSM Image Browser software and Adobe Photoshop (CS2).
Effect of colchicine on BV production.
To determine the effect of colchicine, which inhibits microtubule polymerization, on budded virus (BV) production, TN-368 cells were treated with colchicine at different hours postinfection (hpi). Cells seeded at 0.5 × 106 cells/dish onto sterile glass coverslips in 35-mm dishes were allowed to double at 28°C overnight. The cells were infected with AcEGFP-VP39NatP at an MOI of 5 to 15 PFU/cell and incubated for 1 h at room temperature to allow virus absorption. The virus inoculum was removed, and the cells were washed three times with growth medium and treated with fresh growth medium containing 3 μg/ml colchicine (colchicine concentration as described in references 6 and 46) at 6 hpi or 12 hpi. Control samples were washed and treated with fresh medium containing no colchicine. At 24 hpi, BV of both control and colchicine-treated samples were harvested. The colchicine-treated cells were then washed three times with fresh medium containing 3 μg/ml colchicine and then incubated with fresh medium containing 3 μg/ml colchicine. The control cells were washed with only fresh medium, and the last wash was replaced with fresh medium without any colchicine. BV was harvested at 5, 15, 30, 60, and 120 min thereafter and titrated by plaque assay as described previously (23). Cells were fixed, stained for microtubules, and processed for confocal laser scanning microscopy (CLSM) as previously described. The experimental data were analyzed by a two-tailed Student t test using a statistical program, SPSS (version 15).
Two-photon fluorescence lifetime imaging analysis of transfected insect cells.
Two-photon-induced fluorescence lifetime images were obtained with multiphoton microscopy apparatus and an Eclipse TE2000 (Nikon) with confocal scanning capability FLIM (4, 5). Briefly, a high-powered titanium sapphire laser (MIRA 900; Coherent Lasers) was pumped by a frequency-doubled neodynium:vanadate laser (Verdi V18; Coherent Lasers) to produce a 920 ± 5-nm laser light of 180-fs pulses at 75 MHz. Specimens on a Nikon TE2000U microscope stage were excited by focusing the near-infrared laser beam to a diffraction-limited spot through a 60× water immersion objective (numerical aperture [NA], 1.2). Fluorescence emissions from specimens were collected, bypassing the scanning system, through a band-pass filter (BG39, Comar) using a non-descanned port of the confocal microscope. Single-photon pulses were detected by an external fast microchannel plate photomultiplier tube (Hammamatsu R3809U). The scan, which was operated in a normal mode and line, frame, and pixel clock signals were synchronized and collected with a time-correlated single-photon-counting (TCSPC) PC module (SPC-830; Becker and Hickl, Germany).
Nikon confocal microscopy coupled with argon 488-nm and HeNe 543-nm lasers to excite EGFP fusion and mDsRed fusion proteins, respectively, was used to confirm the expression of fluorescently labeled proteins prior to FLIM acquisition. Cells expressing fluorescence of either EGFP fusion proteins alone (as controls) or both EGFP fusion and mDsRed fusion proteins were selected for FRET-FLIM. The fluorescence lifetimes of EGFP were analyzed with SPCIamge 2.92 analysis software (Becker and Hickl, Germany) as previously detailed (41). The average fluorescence lifetime was statistically analyzed using SPSS version 15. FRET-FLIM images were processed by Adobe Photoshop software (CS2).
RESULTS
Characterization of recombinant baculoviruses expressing tagged capsid proteins.
In order to study the possible interaction between AcMNPV capsid proteins and kinesin, a number of recombinant baculovirus were prepared that expressed EGFP-tagged capsid proteins (VP39, EXON0 or Orf1629) under the control of the VP39 promoter (VP39 and EXON0) or the Orf1629 promoter (Orf1629), at the polyhedrin gene locus, as described in Materials and Methods. A native copy of the same gene was present in all constructs. All gene constructions were confirmed by DNA sequencing, and analysis of recombinant baculoviruses by PCR demonstrated that DNA fragments of the correct size had been amplified according to the predicted sizes of the gene fusions, indicating that the correct recombinant baculoviruses had been made (data not shown).
Analysis of recombinant virus growth curves prepared for each recombinant virus to determine whether additional copy of gene (fusion gene) compromised recombinant virus replication suggested that fusion genes did not affect the replication of the recombinant baculoviruses.
CLSM fluorescence imaging of the interaction of baculovirus and microtubules.
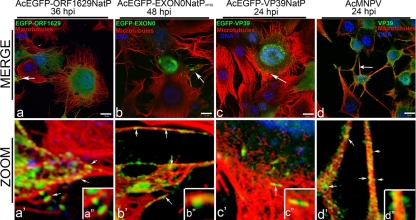
Having confirmed that recombinant viruses were expressing the correct fusion proteins, confocal laser scanning microscopy (CLSM) was used to determine the localization of the three EGFP fusion capsid proteins (EGFP-VP39, EGFP-Orf1629, and EGFP-EXON0) in relation to cellular microtubules. Insect TN-368 cells were infected with recombinant virus expressing the fusion protein under the control of VP39 or Orf1629 native promoters. Nucleocapsid-like structures containing EGFP-Orf1629 were observed aligned and colocalized along peripheral microtubules at 36 hpi (Fig. 2a, a′, and a″). Similarly, alignment and colocalization of nucleocapsids containing EGFP-EXON0 with microtubules were observed at 48 hpi (Fig. 2, b, b′, and b″), which support a similar observation by Fang et al. (12). Cells infected with AcEGFP-VP39NatP produced green fluorescent rod-shaped structures at 24 hpi, which colocalized with microtubules at the cell periphery (Fig. 2 c, c′, and c″). Further, wild-type AcMNPV-infected cells in which VP39 was immunostained showed that the native/untagged VP39 aligned and colocalized with microtubules (Fig. 2 d, d′, and d″), providing stronger evidence that baculovirus may utilize microtubules during anterograde trafficking.
Fig 2.
Alignment and colocalization of EGFP-tagged or immunolabeled capsid proteins with microtubules in virus-infected TN-368 cells. TN-368 cells infected with individual recombinant AcMNPV expressing EGFP fusion (green) as EGFP-Orf1629 (a, a′, and a″), EGFP-EXON0 (b, b′, and b″), or EGFP-VP39 (c, c′, and c″) or infected AcMNPV and the VP39 immunolabeled (d, d′, and d″) using mouse α-VP39 antibody/Alexa Fluor 488 goat anti-mouse IgG (green). Cells were stained for microtubules with anti-α-tubulin/Alexa Fluor 568 goat anti-mouse IgG (red) and DNA with Vectashield DAPI (4′,6-diamidino-2-phenylindole; blue). Merged channels are shown in panels a, b, c, and d; zoomed sections of merged channels are shown in panels a′, b′, c′, and d′ and insets a″, b″, c″, and d″. White solid arrows indicate probably nucleocapsids. Bar, 10 μm.
Colchicine treatment halts the release of budded virus in infected insect cells.
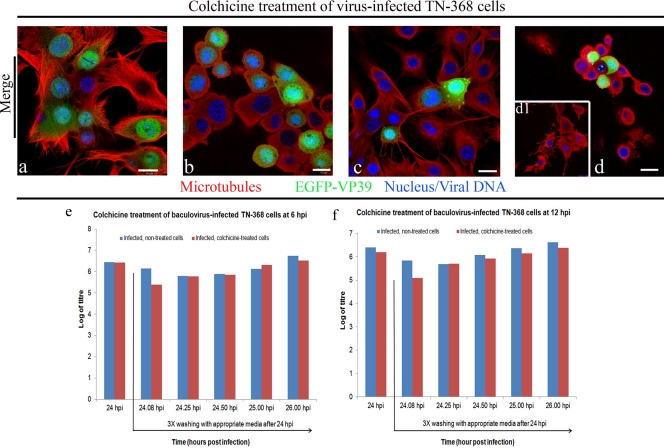
The association of the EGFP-tagged capsid proteins VP39, EXON0, and Orf1629 with microtubules (Fig. 2) suggests that microtubules may be involved in baculovirus anterograde transport. To confirm whether microtubules are required for baculovirus anterograde trafficking, recombinant virus-infected cells were treated with the microtubule depolymerizing drug, colchicine. The microtubules of TN-368 cells infected at 6 hpi and 12 hpi appear the same. The microtubules of these virus-infected TN-368 cells emanate from the nuclear periphery and crisscrossed at the cell periphery (Fig. 3a and c). The microtubules of virus-infected TN-368 cells treated with colchicine from 6 to 24 hpi showed a complete disappearance of microtubule filaments rather patchy microtubules—an indication of a complete depolymerization of microtubules (Fig. 3b). However, a partial depolymerization of microtubules of virus-infected TN-368 cells colchicine treated from 12 to 24 hpi was observed. These cells showed some remnant peripheral microtubule filaments (Fig. 3d and d1). The levels of expression of EGFP-VP39 in both the control (Fig. 3a and c) and colchicine-treated cells (Fig. 3c and d) were similar, indicating that colchicine did not affect the expression of capsid proteins and probably suggesting that replication is not compromised by colchicine. Treatment of virus-infected cells with 3 μg/ml colchicine at 6 hpi produced a 1-log decrease in budded virus production at 24 hpi, which is very similar to that previously reported by Volkman and Zaal (46).
Fig 3.
Colchicine treatment of virus-infected TN-368 cells: TN-368 cells infected with AcEGFP-VP39NatP (green) were fixed at 6 h (a) or 12 h (c), and additional AcEGFP-VP39NatP-infected cells were treated with 3 μg/ml colchicine at 6 hpi (b) or 12 hpi (d and d1). At 24 hpi, budded virus of both virus-infected and treated and virus-infected, nontreated cells was harvested. The virus-infected, colchicine-treated cells were washed colchicine containing medium, whereas virus-infected, nontreated cells were washed with only medium. Both virus-infected, colchicine-treated and virus-infected, nontreated cells were incubated in the appropriate medium and budded viruses were harvested 5, 15, 30, 60, and 120 min following the last wash. Fixed cells were stained for microtubules with anti-α-tubulin/Alexa Fluor 568 goat anti-mouse IgG (red) and DNA with Vectashield DAPI (blue). (b) The image shows a complete depolymerization of microtubules. (d) The image (middle plane) shows a complete microtubule depolymerization. Image d1, which shows a partial microtubule depolymerization, is taken from the surface plane of image d. Budded viruses harvested from cells colchicine-treated at 6 hpi (e) or 12 hpi (f) were titrated by plaque assay. Bar, 10 μm. Error bars are indicated by mean ± SD. n = 5.
There was approximately a 1.03-log decrease in budded virus production at 120 min, following 24 hpi after cells had been washed three times and replaced with fresh medium. The average reduction in budded virus production in virus-infected cells which were colchicine treated at 6 hpi in comparison to nontreated virus-infected cells was statistically insignificant (P = 0.3), similar to results reported by Volkman and Zaal (46). In a similar experiment, virus-infected cells, which were colchicine treated at 12 hpi, produced a 1.03-log reduction in budded virus production at 24 hpi. A 1.04-log reduction in budded virus production at 120 min, following 24 hpi after cells had been washed three times and replaced with fresh medium, was observed. Importantly, the average decrease in budded virus production of virus-infected cells treated with colchicine at 12 hpi gave a statistical significance with a P value of 0.03.
FLIM analysis indicates a direct interaction between AcMNPV VP39 and EXON0 with the TPR of kinesin.
Previous experiments indicated that EGFP-tagged baculovirus capsid proteins (EGFP-VP39, EGFP-EXON0, and EGFP-Orf1629) and immunolabeled VP39 (wild-type VP39) aligned and colocalized with peripheral microtubules, suggesting that baculovirus nucleocapsids indirectly interact with microtubules via kinesin during anterograde trafficking of baculovirus. To test this hypothesis, TN-368 cells were transfected with either recombinant viral DNA (AcEGFP-Orf1629NatP, AcEGFP-VP39NatP, or AcEGFP-EXON0NatPVP39) or recombinant viral DNA and pIZ.mDsRed-TPR. Direct interaction of nucleocapsids (EGFP fusion capsid proteins) and kinesin (mDsRed-TPR) was detected with the FRET-FLIM technique.
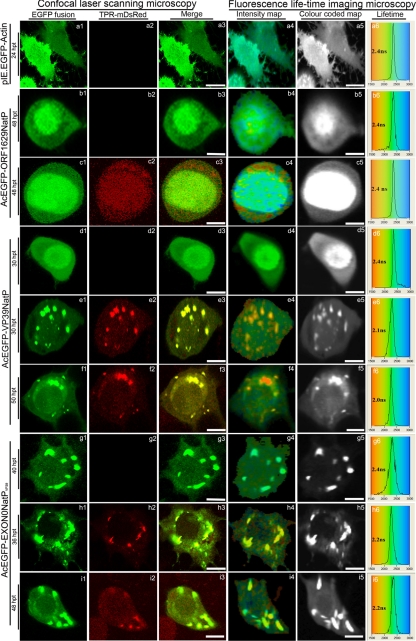
CLSM of virus-infected TN-368 showed a strong colocalization of EGFP-ORF1629 proteins with microtubules (Fig. 2a′). At 48 h posttransfection (hpt) EGFP-Orf1629 was localized in the entirety of the transfected-TN-368 cells but heavily concentrated in the nucleus (Fig. 4b1 to b3) and gave an average lifetime of 2.4 ± 0.1 ns (Fig. 4b4 to b6; Table 2), similar to the average lifetime of EGFP-actin, 2.4 ± 0.1 ns (Fig. 4a4 to a6; Table 2). When both EGFP-Orf1629 and KLC-TPR-mDsRed were expressed in cells at 48 hpt (Fig. 4c1 to c3) the average lifetimes remained at 2.4 ± 0.1 ns (Fig. 4c4 to c6; Table 2), suggesting that AcMNPV Orf1629 does not interact with the KLC-TPR. However, using the FLIM technique, it has been shown that the colocalization between EGFP-Orf1629 and microtubules (Fig. 2a′) may not be a true indirect interaction of EGFP-Orf1629 and microtubules.
Fig 4.
Confocal microscopy and fluorescence lifetime imaging microscopy analysis of transfected TN-368 cells: (a1 to a6) Expression of only EGFP-actin (pIE.EGFP-Actin) and the expression of EGFP-Orf1629 (AcEGFP-Orf1629NatP), EGFP-VP39 (AcEGFP-VP39NatP), and EGFP-EXON (AcEGFP-VP39NatPVP39) are shown in b1 to b6, d1 to d6, and g1 to g6, respectively, as unquenched, negative controls. Transfected cells show expressions EGFP-ORF1629 and KLC-TPR-mDsRed (c1 to c6), EGFP-VP39 and KLC-TPR-mDsRed (e1 to e6 at 36 hpt and f1 to f6 at 50 hpt), and EGFP-EXON0 and KLC-TPR-mDsRed (h1 to h6 at 36 hpt and i1 to i6 at 48 hpt). Colocalization of EGFP fusions and KLC-TPR-mDsRed (yellow) are shown in e3, f3, h3, and i3 and interaction of dual proteins in e5, f5, h5, and i5. The lifetime panels show the distribution curves of the relative occurrence frequency of the lifetimes. The distribution curves of a6, b6, d6, and g6 show the lifetimes of only EGFP fusions with observed lifetimes of ∼ 2.4 ns. Transfected cells (c6) showing expression of both EGFP-Orf1629 and KLC-TPR-mDsRed indicated an observed lifetime of ∼2.4 ns, whereas lower values of interaction are distributed from 1.5 to 2.2 ns. All the images were processed with Adobe Photoshop (CS2). Bar, 15 μm.
Table 2.
FRET-FLIM analysis of interactions of baculovirus capsid proteins and kinesina
| Virus/vector | Donor | Acceptor | Sample size (n) | Lifetimes [ns (mean ± SD)] | P valueb |
|---|---|---|---|---|---|
| pIE.EGFP-Actin | EGFP-actin | None | 5 | 2.4 ± 0.1 | NA |
| AcEGFP-ORF1629NatP | EGFP-ORF1629 | None | 5 | 2.4 ± 0.1 | NA |
| AcEGFP-VP39NatP | EGFP-VP39 | None | 5 | 2.4 ± 0.1 | NA |
| AcEGFP-EXON0-NatPVP39 | EGFP-EXON0 | None | 5 | 2.4 ± 0.1 | NA |
| AcEGFP-ORF1629NatP | EGFP-ORF1629 | KLC-TPR-mDsRed | 5 | 2.4 ± 0.1 | 0.4 |
| AcEGFP-VP39NatP | EGFP-VP39 | KLC-TPR-mDsRed | 13 | 2.1 ± 0.1 | 0.0001 |
| AcEGFP-EXON0-NatPVP39 | EGFP-EXON0 | KLC-TPR-mDsRed | 10 | 2.15 ± 0.1 | 0.0009 |
FRET-FLIM data were collected at different time points as shown in Fig. 5 and analyzed statistically using a two-tailed Student t test. In all the experiments, 40 cells (in duplicate) expressing EGFP fusions and KLC-TPR-mDsRed were selected for FRET-FLIM. The number of cells (n) that indicated a true FLIM from all the cells imaged is presented in Table 2.
NA, not available.
The expression of EGFP-VP39 alone in cells was localized evenly; however, it was heavily concentrated in the nucleus at 30 hpt (Fig. 4d1 to d3). The average excited-state lifetime of EGFP-VP39 in the absence of KLC-TPR-mDsRed was 2.4 ± 0.1 ns (Fig. 4d4 to d6). Cells expressing both EGFP-VP39 and KLC-TPR-mDsRed at 30 hpt and 50 hpt showed a strong interaction between both proteins (Fig. 4e1 to e4 and f1 to f4). The average lifetime of EGFP-VP39 in live cells was reduced from 2.4 ± 0.1 ns to 2.1 ± 0.1 ns at 30 hpt (Fig. 4e4 to e6; Table 2) and from 2.4 ± 0.1 ns to 2.1 ± 0.1 ns at 50 hpt (Fig. 4f4 to f6; Table 2) in the presence of KLC-TPR-mDsRed. Reduction in the fluorescence lifetimes of EGFP-VP39 at various time points suggest that EGFP-VP39 proteins were in a close proximity of 1 to 10 nm with mDsRed-TPR. This finding indicates that there is a direct interaction between EGFP-VP39 and KLC-TPR-mDsRed, as evidenced by energy transfer from EGFP to mDsRed. We have previously shown that a reduction of as little as ∼200 ps (0.2 ns) in the excited-state lifetime of the EGFP-labeled protein represents quenching a protein-protein interaction (20, 35, 38, 39). The P values, statistically significant indicators (Table 2), clearly show that AcMNPV capsid protein VP39 interacts with the KLC-TPR.
Expression of EGFP-EXON0 was localized in the cytoplasm at 40 hpi (Fig. 4g1 to g3). The average lifetime of EGFP-EXON0 alone was observed to be 2.4 ± 0.1 ns (Fig. 4g4 to g6; Table 2). Cells expressing both EGFP-EXON0 and KLC-TPR-mDsRed showed interaction between the two proteins (Fig. 4h1 to h3 and i1 to i3). The fluorescence of the EGFP-EXON0 was quenched by mDsRed attached to KLC-TPR (KLC-TPR-mDsRed) at 36 hpt (Fig. 4h4 to h6) and at 48 hpt (Fig. 4i4 to i6), reducing the average lifetimes from 2.4 ± 0.1 to 2.1 ± 0.1 ns (Table 2). This also strongly indicates that AcMNPV capsid protein EXON0 also interacts with the KLC-TPR. This finding adds more support to the proposal of Fang et al. (12) that EXON0 may interact with kinesin.
DISCUSSION
Some viruses exploit the host actin and/or microtubule cytoskeleton and their associated motor proteins, including myosins, kinesins, and dyneins, for intracellular transport to the sites of replication and for egress and cell-to-cell spread (15). Baculovirus in particular induces actin polymerization for the transportation of nucleocapsids into the nucleus (7, 33); however, little is known about anterograde transport of baculovirus in insect cells. In the present study, we examined the roles of both microtubules and kinesin in baculovirus anterograde trafficking. Recombinant baculoviruses that have capsid protein Orf1629 (AcEGFP-Orf1629NatP), VP39 (AcEGFP-VP39NatP), and EXON0 (AcEGFP-EXON0NatPVP39) fluorescently tagged with EGFP were made and characterized to probe the interaction of baculovirus with the host microtubule cytoskeleton and how kinesins are recruited during anterograde trafficking of nucleocapsids.
CLSM examination of individual recombinant virus-infected TN-368 cells showed that EGFP-Orf1629, EGFP-VP39, and EGFP-EXON0 aligned and colocalized with the peripheral microtubules of virus-infected TN-368 cells (Fig. 2). Some of the EGFP-Orf1629 proteins appeared to have DNA associated with them, probably suggesting nucleocapsids. AcEGFP-VP39NatP infected cells showed rod-like structures of EGFP-VP39 which colocalized with the peripheral microtubules. It is uncertain whether these rod-like structures are just proteins or nucleocapsids, as no viral DNA was found to associate with the EGFP-VP39. Immunolabeling of VP39 of AcMNPV-infected TN-368 cells, which showed colocalization of VP39 and peripheral microtubules, was used to confirm that the EGFP-VP39 colocalization was a true interaction and not fluorescence saturation. However, a lack of antibodies against capsid protein Orf1629 and EXON0 meant that the observed colocalization between capsid proteins Orf1629 and EXON0 with microtubules could not be confirmed in AcMNPV-infected TN-368 cells. The report that FLAG-EXON0 also colocalized with microtubules in infected Sf9 and TN-5b cells (12) provides evidence that the interaction of EGFP-EXON0 and microtubules is not fluorescence saturation.
Colocalization of two proteins in formaldehyde-fixed cells does not necessarily indicate an interaction, because formaldehyde has been shown to cross-link proteins, making nonassociating proteins appear associated or interacting (24). To further confirm that the association of EGFP fusion capsid proteins with microtubules was not due to paraformaldehyde, virus-infected cells were treated with colchicine and the budded virus production was monitored. Microtubules have been shown to physically interact with F-actin (37), and since F-actin is explicitly involved in baculovirus replication (8), it was important to determine whether colchicine treatment of virus-infected TN-368 cells will disrupt baculovirus replication and perhaps nucleocapsid assembly. The expression of EGFP-VP39 in AcEGFP-VP39NatP-infected cells (Fig. 3a to d) confirmed a previous study that colchicine does not affect baculovirus replication (46) and that colchicine may be used to investigate the role of microtubules in baculovirus trafficking. It is also presumed that colchicine did not have any structural defect on budded virus production, as normal plaque sizes were obtained from both virus-infected control cells and virus-infected, colchicine-treated cells.
At present, there are differing views on the anterograde transport of AcMNPV nucleocapsids from colchicine inhibition assay. Fang et al. (12) reported a 90% reduction in budded virus production and suggested that microtubules are utilized in baculovirus anterograde trafficking. In contrast, an earlier study by Volkman and Zaal (46) concluded that microtubules were not required for AcMNPV anterograde transport. A repeat of these authors' colchicine inhibition assay in both Sf9 and TN-368 cells, in which budded viruses were titrated by plaque assay instead of the endpoint dilution used by Fang et al. (12), showed insignificant budded virus reductions of 25%, 32%, and 50% at 24, 48, and 72 hpi, respectively (data not shown). In addition, treatment of virus-infected TN-368 cells at 6 hpi resulted in reduction of budded virus production (Fig. 3E) as reported by Volkman and Zaal (46), and both studies produced statistically insignificant results. However, treatment of virus-infected TN-368 cells with colchicine at 12 hpi (Fig. 3F) resulted in a statistically significant reduction of budded virus (P = 0.03, P < 0.05), suggesting that the colchicine effect may be pronounced only at the time when nucleocapsids are most likely transported to the cell periphery.
It is proposed that the infected cells may contain different forms of nucleocapsids during anterograde trafficking, and this may explain why colchicine treatment at 12 hpi, but not at 6 hpi, produced a significant reduction in budded virus production. For example, if nucleocapsids should bud though the nuclear envelope as proposed in an earlier study (25), there will be enveloped nucleocapsids that have to de-envelope before budding through plasma membrane. In that instance, enveloped and naked nucleocapsids may presumably interact differently with microtubules or motor proteins. It is therefore possible that naked nucleocapsids which are closer to the cell periphery may use microtubules, which would explain the significant reduction of budded virus when infected cells were treated with colchicine at 12 hpi.
The association of fluorescently tagged nucleocasipds or capsid proteins with microtubules indicated that an indirect interaction between the two proteins was possibly mediated by N-kinesins, which are responsible for transporting cargoes on microtubules from the nucleus to the plasma membrane (44). To provide further evidence for the role of microtubules and kinesin in virus trafficking, the direct physical interaction of kinesins with AcMNPV nucleocapsids was examined using the very sensitive protein-protein interaction technique of FRET-FLIM. Because kinesins bind to cargoes by using the TPR domain of kinesin light chain region (44), we analyzed the physical interaction between capsid proteins VP39, EXON0, and Orf1629 (which showed colocalization with microtubules using CLSM) and the TPR domain. The average lifetime of the EGFP fusion protein (in the absence of the acceptor, mDsRed) was determined to be 2.4 to 2.5 ns (Fig. 4), which is consistent with our previous reports (20, 35, 38, 39). However, the average lifetimes of EGFP-EXON0 were reduced in the presence of KLC-TPR-mDsRed at 36 and 48 hpt from 2.4 ± 0.1 to 2.2 ± 0.1 ns (Table 2). These data confirm that EXON0 may likely interact with kinesin. Similarly, the average lifetime of EGFP-VP39 was reduced from 2.4 ± 0.1 to 2.1 ± 0.1 ns in the presence of KLC-TPR-mDsRed. However, the average lifetimes of EGFP-Orf1629 remained the same, indicating that unlike VP39 and EXON0, Orf1629 does not appear to interact with kinesin (Table 2). This finding also indicates that all the known domains and motifs of Orf1629 protein are not involved in interacting with the TPR of kinesin.
The FRET-FLIM data provide evidence that baculovirus nucleocapsids trafficking is likely to be mediated by kinesin-1 through interaction with the capsid proteins EXON0 and/or VP39. There are several DNA viruses, which have been demonstrated to recruit or interact with kinesins during the anterograde trafficking of their virions. For example, vaccinia virus (36), African swine fever virus (21), herpes simplex virus 1 (11), and fowlpox virus (20) use both microtubules and kinesin-1 for anterograde trafficking. However, it is not clear if there is any evolutionary pressure on how DNA viruses exit infected cells. All the interactions between EGFP-EXON0 and EGFP-VP39 with KLC-TPR-mDsRed occurred at the cell periphery, where colocalization of EGFP fusion capsid proteins with microtubules was most prominent. Interesting also, no FRET interactions between EGFP fusion capsid proteins and KLC-TPR-mDsRed were observed in the nucleus or at the perinuclear regions. This may suggest either that kinesin-1 is not involved in intranuclear trafficking of newly formed nucleocapsids or that an alternative intranuclear transport exists that does not involve microtubules. One possible explanation is that newly formed nucleocapsids may have different structural forms (for example, nucleocapsids enwrapped with nuclear envelope) which may not interact with either microtubules and/or kinesin at the subnuclear peripheral regions, but as the nucleocapsids move toward the cell periphery, where microtubules and kinesin are recruited, they may become de-enveloped and thus able to interact with both microtubules and kinesin.
The interaction of both VP39 and EXON0 with KLC-TPR raises several further research questions, including why both capsid proteins interact with KLC-TPR and whether EXON0 and VP39 interact with KLC-TPR consequentially or one capsid protein activates the other. The vaccinia virus structural proteins A36 and F12 have been demonstrated to also interact with KLC-TPR (32, 48). In addition, the surface protein Fpv140 and type II membrane protein Fpv198 of fowlpox virus have been shown to interact with the KLC-TPR (20), suggesting that it is not uncommon for two or more viral structural proteins to interact with kinesin. The AcMNPV capsid protein EXON0 has the RING finger (10), which is a structural domain of zinc finger type and an essential component of the cellular ubiquitin-proteosome system and also involved in protein-protein interactions (13). It is uncertain whether the RING finger is responsible for or involved in binding or interacting with the TPR of kinesin-1. A bioinformatics analysis of AcMNPV VP39 protein sequence did not reveal any functional domains or motifs, which would have suggested or explained the interaction between AcMNPV VP39 and the TPR of kinesin-1. It is likely that the large surface area provided by the VP39 capsid protein makes it well exposed to interact with kinesin-1. Future work will examine whether a mutation in the RING finger of EXON0 would hamper the interaction of AcMNPV EXON0 and TPR of kinesin-1. Furthermore, mutation analysis of the terminal domains of VP39 may explain which domain interacts with TPR of kinesin-1.
ACKNOWLEDGMENTS
We thank Chris Hawes and John Runions for expert advice on the use of confocal laser scanning microscopy and Alasdair McKendzie, Pierre Burgos, and Rahul Yadav for their expert assistance with the FLIM data collection.
This project was funded by an Oxford Brookes University Ph.D. studentship and a Science and Technology Facilities Council-funded access grant to the Central Laser Facility and Multiphoton Laboratory, Harwell.
Footnotes
Published ahead of print 9 November 2011
REFERENCES
- 1. Ayres MD, Howard SC, Kuzio J, Lopez-Ferber M, Possee RD. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586–605 [DOI] [PubMed] [Google Scholar]
- 2. Blatch GL, Lassle M. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932–939 [DOI] [PubMed] [Google Scholar]
- 3. Blissard GW, Wenz JR. 1992. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 66:6829–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Botchway SW, et al. 2005. A novel method for observing proteins in vivo using a small fluorescent label and multiphoton imaging. Biochem. J. 390:787–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Botchway SW, Parker AW, Bisby RH, Crisostomo AG. 2008. Real-time cellular uptake of serotonin using fluorescence lifetime imaging with two-photon excitation. Microsc. Res. Tech. 71:267–273 [DOI] [PubMed] [Google Scholar]
- 6. Carpentier DC, Griffiths CM, King LA. 2008. The baculovirus P10 protein of Autographa californica nucleopolyhedrovirus forms two distinct cytoskeletal-like structures and associates with polyhedral occlusion bodies during infection. Virology 371:278–291 [DOI] [PubMed] [Google Scholar]
- 7. Charlton CA, Volkman LE. 1993. Penetration of Autographa californica nuclear polyhedrosis virus nucleocapsids into IPLB Sf 21 cells induces actin cable formation. Virology 197:245–254 [DOI] [PubMed] [Google Scholar]
- 8. Charlton CA, Volkman LE. 1991. Sequential rearrangement and nuclear polymerization of actin in baculovirus-infected Spodoptera frugiperda cells. J. Virol. 65:1219–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Y, Periasamy A. 2004. Characterization of two-photon excitation fluorescence lifetime imaging microscopy for protein localization. Microsc. Res. Tech. 63:72–80 [DOI] [PubMed] [Google Scholar]
- 10. Dai X, Stewart TM, Pathakamuri JA, Li Q, Theilmann DA. 2004. Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus. J. Virol. 78:9633–9644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diefenbach RJ, et al. 2002. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 76:3282–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fang M, Nie Y, Theilmann DA. 2009. AcMNPV EXON0 (AC141) which is required for the efficient egress of budded virus nucleocapsids interacts with beta-tubulin. Virology 385:496–504 [DOI] [PubMed] [Google Scholar]
- 13. Freemont PS. 2000. RING for destruction? Curr. Biol. 10:R84–R87 [DOI] [PubMed] [Google Scholar]
- 14. Goley ED, et al. 2006. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science 314:464–467 [DOI] [PubMed] [Google Scholar]
- 15. Greber UF, Way M. 2006. A superhighway to virus infection. Cell 124:741–754 [DOI] [PubMed] [Google Scholar]
- 16. Hink WF. 1970. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature 226:466–467 [DOI] [PubMed] [Google Scholar]
- 17. Hitchman RB, et al. 2010. Improved expression of secreted and membrane-targeted proteins in insect cells. Biotechnol. Appl. Biochem. 56:85–93 [DOI] [PubMed] [Google Scholar]
- 18. Hollinshead M, et al. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsieh MJ, White PJ, Pouton CW. 2010. Interaction of viruses with host cell molecular motors. Curr. Opin. Biotechnol. 21:633–639 [DOI] [PubMed] [Google Scholar]
- 20. Jeshtadi A, et al. 2010. Interaction of poxvirus intracellular mature virion proteins with the TPR domain of kinesin light chain in live infected cells revealed by two-photon-induced fluorescence resonance energy transfer fluorescence lifetime imaging microscopy. J. Virol. 84:12886–12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jouvenet N, Monaghan P, Way M, Wileman T. 2004. Transport of African swine fever virus from assembly sites to the plasma membrane is dependent on microtubules and conventional kinesin. J. Virol. 78:7990–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kasprzak AA, Hajdo L. 2002. Directionality of kinesin motors. Acta Biochim. Pol. 49:813–821 [PubMed] [Google Scholar]
- 23. King LA, Possee RD. 1992. The baculovirus expression system: a laboratory guide. Chapman & Hall, New York, NY [Google Scholar]
- 24. Klockenbusch C, Kast J. 2010. Optimization of formaldehyde cross-linking for protein interaction analysis of non-tagged integrin beta1. J. Biomed. Biotechnol. 2010:927585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knudson DL, Harrap KA. 1975. Replication of nuclear polyhedrosis virus in a continuous cell culture of Spodoptera frugiperda: microscopy study of the sequence of events of the virus infection. J. Virol. 17:254–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lanier LM, Volkman LE. 1998. Actin binding and nucleation by Autographa california M nucleopolyhedrovirus. Virology 243:167–177 [DOI] [PubMed] [Google Scholar]
- 27. Lee GE, Murray JW, Wolkoff AW, Wilson DW. 2006. Reconstitution of herpes simplex virus microtubule-dependent trafficking in vitro. J. Virol. 80:4264–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Long G, Pan X, Kormelink R, Vlak JM. 2006. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 80:8830–8833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Machesky LM, Insall RH, Volkman LE. 2001. WASP homology sequences in baculoviruses. Trends Cell Biol. 11:286–287 [DOI] [PubMed] [Google Scholar]
- 30. Masi A, Cicchi R, Carloni A, Pavone FS, Arcangeli A. 2010. Optical methods in the study of protein-protein interactions. Adv. Exp. Med. Biol. 674:33–42 [DOI] [PubMed] [Google Scholar]
- 31. Monsma SA, Oomens AG, Blissard GW. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 70:4607–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morgan GW, et al. 2010. Vaccinia protein F12 has structural similarity to kinesin light chain and contains a motor binding motif required for virion export. PLoS Pathog. 6:e1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohkawa T, Volkman LE, Welch MD. 2010. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J. Cell Biol. 190:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oomens AG, Blissard GW. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254:297–314 [DOI] [PubMed] [Google Scholar]
- 35. Osterrieder A, et al. 2009. Fluorescence lifetime imaging of interactions between Golgi tethering factors and small GTPases in plants. Traffic 10:1034–1046 [DOI] [PubMed] [Google Scholar]
- 36. Rietdorf J, et al. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992–1000 [DOI] [PubMed] [Google Scholar]
- 37. Rodriguez OC, et al. 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5:599–609 [DOI] [PubMed] [Google Scholar]
- 38. Sparkes I, et al. 2010. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22:1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stubbs CD, Botchway SW, Slater SJ, Parker AW. 2005. The use of time-resolved fluorescence imaging in the study of protein kinase C localisation in cells. BMC Cell Biol. 6:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suhling K, French PM, Phillips D. 2005. Time-resolved fluorescence microscopy. Photochem. Photobiol. Sci. 4:13–22 [DOI] [PubMed] [Google Scholar]
- 41. Sun Y, Day RN, Periasamy A. 2011. Investigating protein-protein interactions in living cells using fluorescence lifetime imaging microscopy. Nat. Protoc. 6:1324–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taylor MP, Koyuncu OO, Enquist LW. 2011. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 9:427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tramier M, Zahid M, Mevel JC, Masse MJ, Coppey-Moisan M. 2006. Sensitivity of CFP/YFP and GFP/mCherry pairs to donor photobleaching on FRET determination by fluorescence lifetime imaging microscopy in living cells. Microsc. Res. Tech. 69:933–939 [DOI] [PubMed] [Google Scholar]
- 44. Verhey KJ, Hammond JW. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777 [DOI] [PubMed] [Google Scholar]
- 45. Volkman LE. 1988. Autographa californica MNPV nucleocapsid assembly: inhibition by cytochalasin D. Virology 163:547–553 [DOI] [PubMed] [Google Scholar]
- 46. Volkman LE, Zaal KJ. 1990. Autographa californica M nuclear polyhedrosis virus: microtubules and replication. Virology 175:292–302 [DOI] [PubMed] [Google Scholar]
- 47. Wallrabe H, Periasamy A. 2005. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16:19–27 [DOI] [PubMed] [Google Scholar]
- 48. Ward BM, Moss B. 2004. Vaccinia virus A36R membrane protein provides a direct link between intracellular enveloped virions and the microtubule motor kinesin. J. Virol. 78:2486–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]