Abstract
Compared with human immunodeficiency virus type 1 (HIV-1), little is known about the susceptibility of HIV-2 to antibody neutralization. We characterized the potency and breadth of neutralizing antibody (NAb) responses in 64 subjects chronically infected with HIV-2 against three primary HIV-2 strains: HIV-27312A, HIV-2ST, and HIV-2UC1. Surprisingly, we observed in a single-cycle JC53bl-13/TZM-bl virus entry assay median reciprocal 50% inhibitory concentration (IC50) NAb titers of 1.7 × 105, 2.8 × 104, and 3.3 × 104, respectively. A subset of 5 patient plasma samples tested against a larger panel of 17 HIV-2 strains where the extracellular gp160 domain was substituted into the HIV-27312A proviral backbone showed potent neutralization of all but 4 viruses. The specificity of antibody neutralization was confirmed using IgG purified from patient plasma, HIV-2 Envs cloned by single-genome amplification, viruses grown in human CD4+ T cells and tested for neutralization sensitivity on human CD4+ T target cells, and, as negative controls, env-minus viruses pseudotyped with HIV-1, vesicular stomatitis virus, or murine leukemia virus Env glycoproteins. Human monoclonal antibodies (MAbs) specific for HIV-2 V3 (6.10F), V4 (1.7A), CD4 binding site (CD4bs; 6.10B), CD4 induced (CD4i; 1.4H), and membrane-proximal external region (MPER; 4E10) epitopes potently neutralized the majority of 32 HIV-2 strains bearing Envs from 13 subjects. Patient antibodies competed with V3, V4, and CD4bs MAbs for binding to monomeric HIV-2 gp120 at titers that correlated significantly with NAb titers. HIV-2 MPER antibodies did not contribute to neutralization breadth or potency. These findings indicate that HIV-2 Env is highly immunogenic in natural infection, that high-titer broadly neutralizing antibodies are commonly elicited, and that unlike HIV-1, native HIV-2 Env trimers expose multiple broadly cross-reactive epitopes readily accessible to NAbs.
INTRODUCTION
The contribution of neutralizing antibodies (NAbs) to virus containment in humans infected with human immunodeficiency virus type 1 (HIV-1) or HIV-2 remains an important question (31, 35, 38, 42, 45, 52, 58, 59, 61). Such information could inform our understanding of virus natural history and disease pathogenesis and contribute importantly to rational vaccine design. HIV-2, which resulted from cross-species transmission of simian immunodeficiency virus (SIV) from sooty mangabeys (SIVsmm) to humans (18, 25, 33), can cause AIDS but is generally far less pathogenic than HIV-1 (reviewed in references 16 and 40). Moreover, while HIV-1 continues to expand globally, the incidence of HIV-2 infection is declining (10, 26). Thus, while HIV-2 is dwarfed by HIV-1 in public health impact, the genetic and biological similarities of HIV-2 to HIV-1 and to SIVsmm and rhesus macaque SIV (SIVmac) make HIV-2 a potentially important virus for correlative studies of HIV natural history, pathogenesis, and prevention. In this respect, it is important to elucidate the immunogenicity and neutralization sensitivity of primary HIV-2 strains in order to better understand HIV-2 persistence and pathogenesis in humans and how HIV-2 relates to HIV-1 and to SIVsmm/SIVmac in the rhesus macaque model of virus transmission, pathogenesis, and vaccine protection (16, 40, 46).
Compared with HIV-1, less is known about the susceptibility of HIV-2 to antibody neutralization in natural human infection. Previous HIV-2 neutralization studies often employed laboratory-adapted virus strains and assay formats that differed from those commonly used to assess HIV-1 NAbs (4, 45, 52, 56, 59). For example, the single-cycle virus entry assay in JC53bl-13/TZM-bl cells (58) or U87 cells (42) has generally not been used in a comparable assay format to assess HIV-2 NAbs (4, 52, 56, 59). An exception is a study by Rodriguez and colleagues, which employed an HIV-2 Env-pseudotyped HIV-1 (NL4.3) backbone and U87 target cells (45). This study used bulk PCR amplification to generate a quasispecies mixture of Env glycoproteins for neutralization testing. Other studies employed syncytium formation (59), plaque formation (52), or peripheral blood mononuclear cell (PBMC) replication (4, 56) as the readout of virus entry. These widely varied approaches have made comparative evaluations of HIV-2 and HIV-1 NAb responses challenging. With this caveat, previous studies have suggested that HIV-2 infection elicited NAbs that were similar to or less than HIV-1 NAbs in their potency and equivalent or greater in their breadth of reactivity against primary, non-T-cell-line-adapted viruses (4, 45, 52, 56, 59). In addition, unlike HIV-1, where many of the epitopes recognized by autologous (strain-specific) and heterologous responses have been mapped (38, 50), the epitope regions that are targeted by NAbs in HIV-2 infection remain largely unknown.
In the present study, we employed a coordinated strategy for identifying accessible NAb epitopes on functional Env trimers of primary HIV-2 strains, for determining the immunogenicity of the HIV-2 Env glycoprotein in natural infection, and for assessing the neutralization sensitivity of primary HIV-2 strains to polyclonal and monoclonal antibodies from HIV-2-infected subjects. First, we used conventional amplification approaches to clone full-length and env-only viral DNA from patient PBMCs. We also performed single-genome amplification (SGA) and sequencing (27) to identify and clone env sequences from plasma viral RNA (vRNA). Second, we employed the JC53bl-13/TZM-bl cell single-cycle virus entry assay (58), in addition to more traditional PBMC (purified CD4+ T-cell) virus replication assays (64), to assess antibody neutralization of HIV-2 clones, env chimeras, and Env pseudotypes, in each case in the context of an HIV-2 backbone. Third, we created an HIV-1 gp160 env chimera into which we substituted the HIV-2 membrane-proximal external region (MPER) in order to test HIV-2-infected sera for HIV-2 MPER-specific NAbs, analogous to our previously described method for detecting HIV-1 MPER-specific NAbs (3, 11, 12, 23, 24). Fourth, we used a panel of human monoclonal antibodies (MAbs) specific for the V3, V4, CD4 binding site (CD4bs), CD4-induced (CD4i), and MPER epitopes of HIV-2 Env to probe the accessibility of these epitope regions to NAbs. Surprisingly, we observed potent and broad NAb responses to primary strains of HIV-2 in multiple assay formats and found that HIV-2 polyclonal and monoclonal antibodies target epitopes in V3, V4, CD4bs, and CD4i regions on the envelope glycoprotein. Interestingly, although HIV-2 MPER epitopes were accessible to monoclonal NAbs, naturally occurring anti-MPER NAbs in HIV-2-infected subjects were absent or of low titer. Potential implications of these findings for HIV-2 natural history and for interpreting antibody neutralization in the SIVsmm and SIVmac infection model are discussed.
MATERIALS AND METHODS
Study subjects.
Plasma or serum samples were obtained from 64 antiretrovirus therapy-naive subjects chronically infected with HIV-2 (see Table S1 in the supplemental material). These included samples from 52 Senegalese subjects enrolled between 1994 and 2004 (22, 63), 1 Ivory Coast subject (samples 7312Apl1992 and 7312Apl2003) (20), 6 source plasma donors whose country of origin was unknown (samples 8704Apl2006 and 8704Apl2007, 7810Apl1993, 7924Apl, 60667Kpl, 10849pl1995, and SLRHCNo.10pl1995), and 5 subjects from the NIH AIDS Research and Reference Reagent Program (1026se, Ivory Coast; 1030se, Senegal; 1032se, Ivory Coast; 1495se, Senegal; and 3660se, Guinea Bissau). HIV-1 clade B-infected plasma samples (SHROpl, BELIpl, FAROpl, PUMApl, and YOALpl) from chronically infected patients were obtained from the University of Alabama at Birmingham Center for AIDS Research HIV/AIDS tissue repository (39). HIV-1 clade C-infected plasma samples (8238Mpl, 5731Mpl, 7510Fpl, 5708Mpl, and 6765Mpl) were collected from chronically infected patients in Zambia. All samples were collected after obtaining informed consent and with regulatory approval and stored at −70°C. Before use, plasma and serum samples were heat inactivated at 56°C for 30 min.
Neutralization assays. (i) JC53bl-13/TZM-bl single-cycle virus entry assay.
Virus neutralization by plasma, sera, and MAbs was assessed on TZM-bl cells as described previously (11, 58). TZM-bl cells were seeded and cultured in 96-well plates for 24 h. The virus stocks were diluted in Dulbecco's modified Eagle medium containing 10% fetal bovine serum (FBS) and 80 μg/ml DEAE-dextran (Sigma-Aldrich, St. Louis, MO) to achieve 5 × 104 relative light units (RLU)/well. Equal-volume virus dilutions and 5-fold serially diluted plasma samples or MAbs were mixed and incubated at 37°C for 1 h. The supernatants were then removed from each well, and 100 μl virus-plasma mixture was added back. Luciferase activity was measured after 48 h of incubation at 37°C with 5% CO2. Medium-only control wells were measured as background, and virus-only control wells were included as 100% infection. For neutralization by plasma or serum samples, the concentrations of plasma or serum in all wells were normalized by the addition of plasma from healthy humans as described previously (11).
(ii) PBMC (purified CD4+ T cell) multicycle infectivity assay.
Human blood samples collected from healthy HIV-negative individuals (Research Blood Components, Boston, MA) were processed for PBMC isolation by gradient centrifugation by Ficoll-Hypaque Plus (GE Healthcare, Piscataway, NJ). We then purified CD4+ T cells from PBMCs using autoMACS and magnetic anti-human CD4 microbeads (Miltenyi Biotec, Auburn, CA). Purified CD4+ cells were activated by incubation in RPMI 1640 medium plus 15% FBS containing 3 μg/ml of staphylococcal enterotoxin B (SEB; Sigma-Aldridge) for 48 h at 37°C. Detailed protocols for cell isolation and activation were described previously (14). Activated CD4+ lymphocytes were cultured in complete medium (RPMI 1640 medium plus 15% FBS with 30 U/ml of interleukin-2; Roche Diagnostics). Neutralization assays were usually performed within 3 to 4 days after SEB activation. HIV-2-infected patient plasma samples were 5-fold serially diluted with RPMI 1640 medium and mixed with concentrated CD4+ lymphocyte-grown virus stocks (1 × 105 IU; multiplicity of infection [MOI] = 0.1) in a final volume of 25 μl. The starting dilution of patient plasma samples in the virus-plasma mixture was 1:100. Plasma samples from healthy humans were added into other wells, including a virus-only control (no patient plasma added), to normalize the concentration of human plasma samples. The virus-plasma mixtures were then incubated at 37°C with 5% CO2 for 1 h. After incubation, 1 × 106 SEB-activated CD4+ lymphocytes in 25 μl RPMI 1640 medium plus 15% FBS were combined with the virus-plasma mixture for an additional 2 h of incubation. After infection, cells were washed carefully 6 times to eliminate the viral inoculum, resuspended into 500 μl RPMI 1640 medium plus 15% FBS, equally separated into duplicate wells (250 μl/well) in a 96-well tissue culture plate, and incubated at 37°C with 5% CO2. Twenty-five microliters supernatants was taken for p27 measurement, and the same volume of RPMI 1640 medium plus 15% FBS was added back at 3, 4, and 5 days postinfection (dpi). The supernatants from virus-only control wells were first analyzed by SIV p27 antigen enzyme-linked immunosorbent assay (ELISA; Zeptometrix Corporation, Buffalo, NY) to determine the earliest time point with sufficient virus growth. Samples from that time point were then measured for p27 concentration to determine neutralization.
Viruses and Env targets of neutralization. (i) Infectious stocks of primary HIV-2 strains.
Infectious stocks of primary HIV-2 strains were produced in 293T cells by transfection with molecular clones of HIV-27312A, HIV-2ST, and HIV-2UC1 and by infection and productive replication in CD4+ T cells. To generate 293T-grown virus stocks, HIV-2 proviral clones pJK7312A and pSTsxb1 were transfected into 293T cells using Fugene 6 (Roche Applied Science, Indianapolis, IN) as described previously (13). The HIV-2UC1 env expression construct pSM-UC1 was cotransfected into 293T cells with HIV-2 backbone construct pJK7312AΔEnv. To generate PBMC-grown virus stocks, human CD4+ lymphocytes were isolated, activated, and cultured as described above. Within 3 to 4 days after SEB activation, 10 × 106 cells were incubated with 293T-grown HIV-27312A and HIV-2ST at a high MOI (0.5 or 1, based on β-galactosidase production-based titers on TZM-bl cells) in a small volume (<2 ml) for 2 h at 37°C, washed three times to remove inocula, and then cultured in complete medium at a concentration of 1 × 106 cells/ml. Starting from 3 dpi, medium was changed by low-speed centrifugation (1,200 rpm, 8 min) every 2 days until 12 dpi. The supernatants were collected, centrifuged at 3,000 rpm for 20 min, aliquoted, and stored at −80°C for future titration and infection. For CD4+ T-cell-based multicycle infectivity assay, the supernatants were concentrated up to 100-fold by sucrose gradient ultracentrifugation.
(ii) Chimeric viruses with HIV-2 Env ectodomain in an HIV-27312A backbone grown in 293T cells.
Vector p7/SNAG was created by introducing two restriction sites, SnaB1 and AgeI, into the proviral DNA clone pJK7312A through synonymous mutation using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The SnaB1 and AgeI restriction sites are located 72 bp and 2,187 bp downstream of the starting ATG of env, respectively. The ectodomains of 17 HIV-2 env genes were amplified from PBMC DNA or plasma from 9 HIV-2-infected patients using primers SnaB1-F (5′-CCAAATACGTAACTGTTTTTTATGGC-3′) and AgeI-R (5′-GAAAACCGGTCTATAGCCCTTTCTAAG-3′). The ectodomain of HIV-2MVP15132 (19) was amplified using the same primers. The PCR fragments were then cloned into p7/SNAG vector using restriction sites SnaB1 and AgeI. The resulting chimera clone, p7/SNAG-HIV2env, was transfected into 293T cells to generate virus stocks.
(iii) 293T-cell-grown pseudotyped viruses with SGA-derived HIV-2 Envs and HIV-27312A backbone.
Three plasma samples (7312Apl1992, 7312Apl2003, and 8704Apl2006) were selected for amplifying HIV-2 rev/env sequences by SGA (27). The following primer pairs were used for amplifying 7312A rev/env genes: for reverse transcription (RT; cDNA synthesis), 5′-TGGTTACAGCCCCTTCTGGAAAGTC-3′ (primer H2-NFKB-R); for first-round PCR, H2-NFKB-R and 5′-CGCTTGCTAACTGCGCTTGG-3′ (primer H2-Vpr-F1); and for second-round PCR, 5′-CGAGCCCTCTTCCTCCACTTCAGAG-3′ (primer H2-Tat-F1) and 5′-ACTGTAAAACATCCCTTCCAGTCCCCC-3′ (primer H2-U3-R). For amplifying 8704Apl2006 rev/env genes, a different forward primer (H2-Tat-F3, 5′-RTCCTRCAACGAGCCCTCTTC-3′) was used in the second-round PCR. The rev/env cassettes were then cloned into the pcDNA3.1D/V5-His-TOPO vector (pcDNA3.1 directional TOPO expression kit; Invitrogen, Carlsbad, CA) following the manufacturer's instructions. All the pcDNA3.1-rev/env expression vectors were cotransfected into 293T cells with backbone construct pJK7312AΔEnv.
(iv) 293T-grown primary HIV-1 strains 700010040 (CH40), ZM249, and TRJO4551.
HIV-1 transmitted/founder (T/F) proviral clones p700010040 and pTRJO4551 were obtained form John Kappes (University of Alabama at Birmingham). HIV-1 T/F proviral clone pZM249 has been described previously (49). HIV-1 stocks were generated by transfection of 293T cells.
Virus titration.
293T-grown and human CD4+ lymphocyte-grown viruses were titrated on TZM-bl reporter cells (8129; NIH AIDS Research and Reference Reagent Program), in which a Tat-inducible luciferase and β-galactosidase gene cassette are integrated. The infectious titers of virus stocks were measured on the basis of luciferase and β-galactosidase expression. The luciferase activity-based titers were measured on 96-well plates and presented as the number of RLU per μl virus stock, as described previously (11). The β-galactosidase production-based titers were measured on 24-well plates and presented as the number of infection events (β-galactosidase-expressing cell colonies) per μl virus stock (IU/μl), as described previously (15, 57).
Purification of plasma IgG.
IgG was purified from three HIV-2-infected patient plasma samples (7312Apl2003, SLRHCpl1995, and 10849pl1995) and three healthy human plasma samples (donors B, D, and J) using protein G columns (Pierce-Thermo Fisher Scientific, Rockford, IL). IgG purified from 200 μl of each plasma specimen was adjusted to a final volume of 200 μl. IgG concentration was determined using the bicinchoninic acid protein assay (Pierce-Thermo Fisher Scientific, Rockford, IL).
HIV-1/HIV-2 MPER Env chimeras.
The HIV-1/HIV-2 MPER chimeric clone pBG1168.1-C1env was made by exchanging the MPER of HIV-1BG1168.1 Env expression construct pBG1168.1env (D. C. Montefiori, Duke University) with the HIV-2ST MPER using strategies described previously (3, 11, 23). To generate virus stocks, pBG1168.1env and pBG1168.1-C1env were cotransfected into 293T cells with HIV-1 backbone construct pSG3ΔEnv (13).
HIV-2 and HIV-1 Env-specific monoclonal antibodies.
Human anti-HIV-2 MAbs 1.7A, 6.10B, 6.10F, 1.4H, and 2.6C have been described previously (8, 43, 44; R. Kong, J. E. Robinson, and G. M. Shaw, submitted for publication). MAbs 4E10 and 2F5 (contributed by Hermann Katinger) were obtained from the NIH AIDS Research and Reference Reagent Program.
Phylogenetic analysis.
Sequence alignments, diversity analysis, and neighbor-joining phylogenetic trees were made using the ClustalX (version 2.0.12) program.
Competition assay for HIV-2 MAb binding.
The competition ELISA was similar to assays described previously (13). Wells of ELISA plates were coated with 5 μg/ml human MAb 2.6C, a nonneutralizing MAb that binds to a conserved epitope of HIV-2 near the N terminus (8). Solubilized HIV-2ST or HIV-2MVP15132 (19) gp120 glycoproteins produced in 293T cells by a recombinant vaccinia virus expression vector were captured in 2.6C-coated wells. The plates were then washed and blocked with phosphate-buffered saline containing 4% whey and 0.5% Tween 20 at 25°C for 30 min. Serially diluted patient sera (starting at 1:50) and controls were incubated in the wells at 25°C for 1 h. HIV-2 MAbs were biotinylated as previously described (8), added to the wells at concentrations that gave less than maximal binding, and incubated for another hour. The color reaction was developed by sequential incubation with peroxidase-conjugated streptavidin and tetramethylbenzidine-H2O2. Color development was stopped by addition of 1 M phosphoric acid, and the absorbance was read at an optical density of 450 nm. The serum dilutions that gave half-maximum effective competition (reciprocal EC50) were calculated.
Plasma HIV-2 vRNA quantification.
Plasma HIV-2 vRNA levels were determined by a modified Amplicor Monitor test method previously described (22) and by a validated real-time PCR assay (9, 17).
Glycosylation site analysis.
The number of potential N-linked glycosylation sites in the V4 loop regions for a set of HIV-1 and HIV-2 strains was computed using the N-GlycoSite tool (62). The set of 181 HIV-1 strains was previously described (60), with 3 additional strains (BaL.26, R2, and Q769.d22) also being included. A total of 31 HIV-2 strains were used in the analysis (Table 1).
Table 1.
Neutralization titers of MAbs against HIV-2 molecularly cloned viruses
| HIV-2 strain | IC50 (μg/ml) |
||||
|---|---|---|---|---|---|
| 1.7A (V4) | 6.10F (V3) | 6.10B (CD4bs) | 1.4H (CD4i) | 4E10 (MPER) | |
| ST | 0.002 | 0.004 | 0.040 | 0.043 | 0.343 |
| UC1 | 0.001 | 0.011 | 0.655 | 0.028 | 0.591 |
| 7312A | 0.007 | 0.007 | 0.094 | 0.782 | >20 |
| 7312A-92-12a | 0.001 | 0.004 | 0.130 | 0.028 | NDb |
| 7312A-92-15a | 0.001 | 0.007 | 0.260 | 0.035 | ND |
| 7312A-92-17a | 0.001 | 0.003 | 0.131 | 0.022 | ND |
| 7312A-92-20a | 0.004 | 0.016 | 0.095 | 0.080 | ND |
| 7312A-92-21a | 0.001 | 0.005 | 0.267 | 0.141 | ND |
| 7312A-03-03a | 0.001 | 0.003 | 0.121 | 0.019 | ND |
| 7312A-03-04a | 0.001 | 0.004 | 0.029 | >10 | ND |
| 7312A-03-05a | 0.001 | 0.004 | 0.071 | 2.118 | ND |
| 7312A-03-07a | 0.002 | 0.007 | 0.181 | 0.021 | ND |
| 1370/4-5 | >10 | 0.014 | 0.169 | 0.117 | 6.703 |
| 1370/4-15 | >10 | 0.011 | 0.372 | 0.709 | >20 |
| 60415K-2 | 2.885 | 0.008 | 0.078 | 0.016 | >20 |
| SLRH4 | 0.009 | 0.011 | 0.063 | >10 | 0.364 |
| SLRH8 | 0.944 | 0.039 | >10 | >10 | 2.542 |
| 226711-11 | 0.038 | 0.014 | 0.104 | 0.034 | 0.439 |
| 226711-21 | 0.029 | 0.025 | 0.430 | 0.169 | 0.226 |
| 226711-33 | >10 | 0.055 | 1.885 | >10 | 9.226 |
| 1958/1-32 | 0.003 | 0.007 | 0.140 | 0.031 | 8.528 |
| 10849 | 0.015 | 0.006 | 0.273 | >10 | >20 |
| 227011-5 | >10 | 0.004 | 0.077 | 0.066 | 24.415 |
| 1871/5-5 | >10 | 0.012 | 0.183 | 0.038 | 1.499 |
| 1871/5-9 | 0.005 | 0.004 | 0.035 | 0.137 | 0.399 |
| 1871/5-10 | 0.008 | 0.003 | 0.053 | 0.043 | 0.076 |
| 8704A-06-01a | >10 | >10 | >10 | >10 | >20 |
| 8704A-06-06a | >10 | >10 | >10 | >10 | >20 |
| 8704A-06-16a | >10 | >10 | >10 | >10 | >20 |
| 8704A-06-23a | >10 | >10 | >10 | >10 | >20 |
| A2240-3 | >10 | >10 | >10 | >10 | >20 |
| A2240-12 | >10 | >10 | >10 | >10 | >20 |
| SIVmac239 | >10 | >10 | >10 | >10 | ND |
Env clone was generated by SGA.
ND, not determined.
Structure modeling.
Structural models of gp120s (without V1V2) from HIV-2 primary (7312A), HIV-1 primary (YU2), and HIV-1 laboratory-adapted (HXBc2) strains were obtained using the protein with Protein Data Bank accession number 3JWD (37) as a template. The GlyProt server (5) was used to model basic glycans at accessible potential N-linked glycosylation sites.
Statistical analysis.
Pearson correlation and linear regression analyses were performed to determine the correlation between plasma neutralization titers and competition titers of MAbs for HIV-2 gp120 glycoprotein binding. A P value of 0.05 was considered significant. A one-tailed paired t test was performed to compare the plasma neutralization titers (reciprocal 50% inhibitory concentration [IC50]) against HIV-27312A and HIV-2ST in the TZM-bl single-cycle virus entry assay and PBMC multientry cycle assay. A P value of 0.05 was considered significant.
RESULTS
Plasma or serum from subjects with chronic HIV-2 infection exhibits broad and potent virus neutralization in TZM-bl cells.
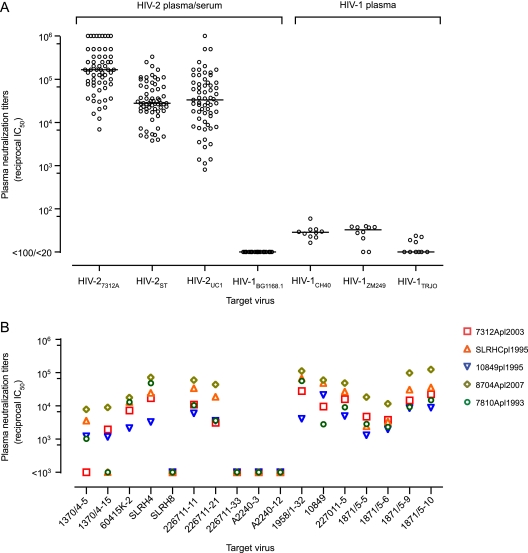
To evaluate the potency and breadth of naturally occurring NAbs in HIV-2-infected patients, we analyzed in the TZM-bl single-cycle virus entry assay (51, 58) the neutralizing activities of 64 plasma or serum samples from chronically infected subjects against three 293T-derived primary HIV-2 strains: HIV-27312A (group A), HIV-2ST (group A), and HIV-2UC1 (group B). HIV-27312A and HIV-2ST were generated from infectious molecular proviral clones and HIV-2UC1 was generated from Env-pseudotyped virus. Each of the three molecular clones was generated by conventional lambda phage cloning of viral DNA from short-term cultures of virus in human PBMCs or CD4+/CCR5+ T-cell lines (1, 18, 29). Previously, we showed that these viruses are CD4 and CCR5 dependent for entry and that their coreceptor binding surfaces are conformationally obscured from antibody recognition by coreceptor binding MAbs (17b, 21c, E51, 19e) that have molecularly defined binding specificities for the Env bridging sheet (13). After pretreatment with subinhibitory concentrations of soluble CD4 (sCD4), HIV-27312A, HIV-2ST, and HIV-2UC1 became sensitive to neutralization by the same CD4i MAbs (13). These are features typical of primary, non-T-cell-line-adapted HIV-2 strains. The maximum diversity of Env protein sequences among the three HIV-2 strains was 27%. Despite this, all 64 plasma and serum samples potently neutralized all three virus strains (Fig. 1A). The median and range (in parentheses) of reciprocal IC50 neutralization titers against HIV-27312A, HIV-2ST, and HIV-2UC1 were 1.7 × 105 (6.8 × 103 to 1 × 106), 2.8 × 104 (3.8 × 103 to 3.3 × 105), and 3.3 × 104 (8.1 × 102 to 1 × 106), respectively. These HIV-2 plasma and serum samples failed to neutralize HIV-1 strains, including HIV-1BG1168.1 (Fig. 1), or viruses pseudotyped with vesicular stomatitis virus (VSV) or murine leukemia virus (MuLV) Envs (data not shown), indicating the absence of antiretroviral drugs or nonspecific virus entry-inhibitory activities in these samples.
Fig 1.
Neutralization of primary HIV-2 and HIV-1 strains by plasma or serum from patients chronically infected by HIV-2 or HIV-1. (A) Neutralization titers (reciprocal IC50s) of HIV-2-infected patient plasma or serum (n = 64) against primary HIV-2 strains HIV-27312A (group A), HIV-2ST (group A), and HIV-2UC1 (group B) and HIV-1 strain HIV-1BG1168.1. IC50s not reached at a starting dilution of 1:100 or 1:20 are represented as <100 and <20, respectively. Neutralization titers of 10 clade B (n = 5) or clade C (n = 5) HIV-1-infected patient plasma samples against the primary HIV-1 strains HIV-1CH40 (subtype B, tier 2), HIV-1ZM249 (subtype C, tier 2), and HIV-1TRJO (subtype B, tier 3) are shown. Virus stocks were grown in 293T cells. Median values are represented by a horizontal line. (B) Neutralization titers of five HIV-2-infected patient plasma samples (7312Apl2003, SLRHCpl1995, 10849pl1995, 8704Apl2007, and 7810Apl1993) against 17 p7/SNAG-HIV2env chimeric viruses.
We also tested the neutralization of three primary HIV-1 strains representing subtype B (HIV-1CH40 and HIV-1TRJO) or C (HIV-1ZM249) (49) by plasma from 10 subjects chronically infected by HIV-1, and the results are shown in Fig. 1A. These assays were done in parallel with the HIV-2 neutralization assay so as to allow a direct comparison of the neutralization breadth and titers of HIV-2- and HIV-1-infected plasma samples. In contrast to the broad and potent neutralization of primary HIV-2 strains by HIV-2-infected patient plasma samples, none of the 10 HIV-1-infected patient plasma samples reached a reciprocal IC50 of 100 against HIV-1 strains, consistent with published reports showing that most HIV-1-infected sera lack neutralization breadth and potency against primary HIV-1 strains (51, 58).
To further evaluate the breadth of HIV-2 neutralizing responses, we tested plasma samples from five HIV-2-infected donors from whom greater volumes were available against a much larger panel of HIV-2 Env glycoproteins. For this analysis, 17 env gp160 genes from nine subjects were amplified from primary PBMC viral DNA and ligated into the HIV-2 proviral vector p7/SNAG to generate replication-competent provirus, as described in Materials and Methods. Virus stocks were generated by transfection of 293T cells. None of these viruses had been passaged in T-cell lines. All 17 HIV-2 Env protein sequences clustered within group A in phylogenetic analyses. The neutralization titers of plasma samples from five HIV-2-infected subjects tested against this virus panel are shown in Fig. 1B. All five plasma specimens potently neutralized most virus strains, although four strains (SLRH8, 226711-33, A2240-3, and A2240-12) were resistant to neutralization by all five HIV-2-infected plasma samples. Interestingly, both neutralization-sensitive and -resistant clones were found in subjects 226711 (226711-11 [sensitive], 226711-21 [sensitive], and 226711-33 [resistant]) and SLRH (SLRH4 [sensitive] and SLRH8 [resistant]). Collectively, the results depicted in Fig. 1A and B indicate that plasma or serum samples from HIV-2-infected subjects can generally neutralize a broad spectrum of HIV-2 strains at titers of 1:103 to 1:106, which are 100- to 10,000-fold higher than those for heterologous neutralization of HIV-1 by most HIV-1-infected patient plasma samples (51).
IgG mediates HIV-2 neutralization.
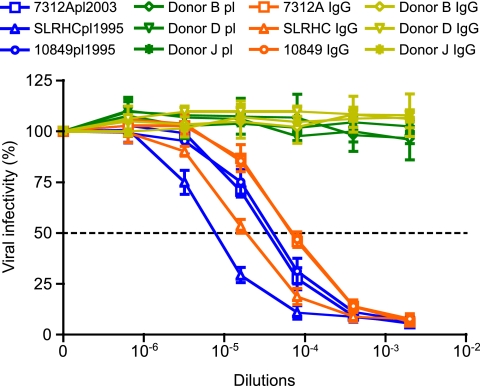
To determine if the observed neutralizing activity was mediated by IgG antibodies, IgG was purified from three HIV-2-infected patient plasma samples (7312Apl2003, SLRHCpl1995, and 10849pl1995) and from three non-HIV-2-infected control subjects (donors B, D, and J) and tested for neutralizing activity (Fig. 2). To compare the neutralizing activities of comparable concentrations of purified IgG from plasma IgG, purified IgG was reconstituted in buffer having the same volume as the original plasma, and the comparability of the respective IgG concentrations was confirmed. Neutralization curves of purified IgG preparations recapitulated those of the corresponding plasma samples (Fig. 2), indicating that the high-titer neutralizing responses in plasma were mediated by IgG. Control IgGs purified from healthy human plasma (donors B, D, and J) did not neutralize HIV-27312A.
Fig 2.
IgG-mediated neutralization of HIV-2. Neutralization of 293T-grown HIV-27312A by plasma from HIV-2-infected patients 7312Apl2003, SLRHCpl1995, and 10849pl1995 and healthy donors B, D, and J. IgG purified from these plasma samples are indicated 7312A IgG, SLRHC IgG, 10849 IgG, Donor B IgG, Donor D IgG, and Donor J IgG in the TZM-bl assay. Purified IgG was reconstituted in buffer having the same volume as the original plasma, and similarity in IgG concentrations was confirmed. Error bars represent three independent experiments. Dashed line indicates 50% reduction in virus infectivity.
Plasma from subjects with chronic HIV-2 infection exhibits broad and potent neutralization against HIV-2 propagated in primary human lymphocytes and tested in primary human CD4+ T target cells or TZM-bl target cells.
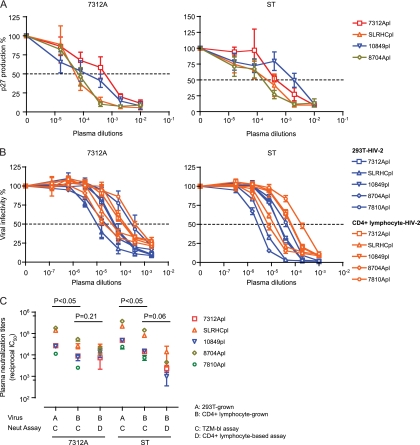
Because of the unanticipated breadth and potency of HIV-2 NAbs that we observed in the TZM-bl assay, we tested a subset of the HIV-2-infected patient plasma samples for which we had larger volumes in a human CD4+ T-cell multiple-infection-cycle assay. This was done using viruses (HIV-27312A and HIV-2ST) grown in activated CD4+ human T cells. Figure 3A shows potent neutralization by all four HIV-2-infected patient plasma samples (7312Apl2003, SLRHCpl1995, 10849pl1995, and 8704Apl2007). The median and range (in parentheses) of reciprocal IC50s against HIV-27312A and HIV-2ST were 1.5 × 104 (7.4 × 104 to 2.5 × 105) and 3.4 × 103 (9.7 × 102 to 1.4 × 104), respectively.
Fig 3.
Comparison of HIV-2 neutralization titrations using different assay formats. (A) Neutralization of human CD4+ lymphocyte-grown HIV-27312A and HIV-2ST stocks by four HIV-2-infected patient plasma samples in the human CD4+ T-cell multicycle infectivity assay. (B) Neutralization of human CD4+ T-cell-grown versus 293T-cell-grown HIV-27312A and HIV-2ST by five HIV-2-infected patient plasma samples in the TZM-bl assay. (C) Summary analysis of reciprocal IC50 neutralization shown in panels A and B. One-tailed paired t tests were performed, and P values are shown. Error bars were generated on the basis of at least three independent experiments. Dashed line indicates 50% reduction in virus infectivity.
Next we compared the neutralizing susceptibilities of viruses grown in 293T cells versus activated human CD4+ T cells in the TZM-bl assay. Figure 3B shows that all five HIV-2-infected patient plasma samples (7312Apl2003, SLRHCpl1995, 10849pl1995, 8704Apl2007, and 7810Apl1993) potently neutralized viruses produced in different cell types. The median and range (in parentheses) of reciprocal IC50s against 293T-grown HIV-27312A and HIV-2ST were 2.7 × 104 (1.1 × 104 to 1.8 × 105) and 4.9 × 104 (2.4 × 104 to 3.7 × 105), respectively, whereas the median and range of reciprocal IC50 against activated CD4+ T-cell-grown HIV-27312A and HIV-2ST were 9.0 × 103 (2.5 × 103 to 5.4 × 104) and 1.5 × 104 (7.4 × 103 to 1.4 × 105), respectively.
A statistical analysis of the results from different assays is shown in Fig. 3C. High-titer neutralization (reciprocal IC50 > 103) was observed in all assay formats. Viruses generated in activated human CD4+ T cells were modestly less sensitive than viruses generated in 293T cells when tested in TZM-bl cells (P < 0.05, one-tailed paired t test), which is consistent with previous reports on HIV-1 neutralization (34). Viruses grown in human CD4+ T cells and assayed in human CD4+ T cells were least sensitive to antibody neutralization, but even here, the median reciprocal IC50 was greater than 103 for all four HIV-2-infected patient plasma samples that were tested.
Autologous and heterologous neutralization of HIV-2 Envs generated by single-genome amplification.
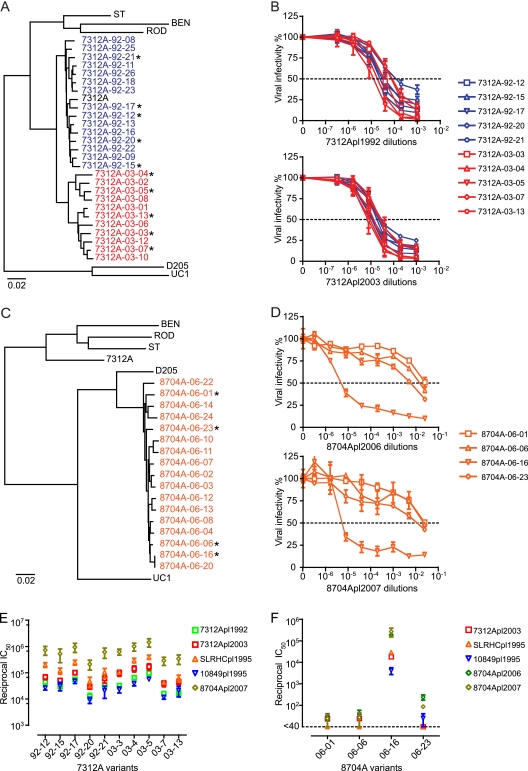
A recent innovation to identify, clone, and characterize HIV-1 env genes and full-length genomes that most closely approximate those viral sequences that exist in vivo utilizes single-genome amplification (SGA) and direct amplicon sequencing, followed by molecular cloning (27, 49). This approach avoids potential artifacts of in vitro Taq polymerase-mediated recombination, Taq-mediated base substitution errors in finished sequences, and founder effects resulting from biases in molecular cloning (27, 36, 48, 49, 53, 55). Because of the low plasma viral load and limited volumes of plasma from most HIV-2-infected patients, we focused on four plasma samples for SGA and neutralization analyses: 7312Apl1992 and 7312Apl2003 were collected from patient 7312A in 1992 and 2003, respectively, and 8704Apl2006 and 8704Apl2007 were collected from subject 8704A in 2006 and 2007, respectively. Fifteen env genes were amplified by SGA from the 7312Apl1992 sample, and 11 env genes were amplified from the 7312Apl2003 sample. The maximum nucleotide diversities were 2.46% among 7312Apl1992 env genes, 4.43% among 7312Apl2003 env genes, and 6.16% among genes from the two time points (Fig. 4A). Five genetically diverse env genes from each time point were cloned into the pcDNA3.1 vector and expressed and pseudotyped onto pJK7312AΔEnv, as described in Materials and Methods. As shown in Fig. 4B, all clones were highly sensitive to neutralization by autologous plasma samples 7312Apl1992 and 7312Apl2003 in the TZM-bl single-round entry assay. No neutralization escape mutants from the concurrent or later time points were observed. The median and range (in parentheses) of reciprocal IC50 neutralization titers of 7312Apl1992 and 7312Apl2003 were 3.3 × 104 (1.3 × 104 to 9.7 × 104) and 6.4 × 104 (2.9 × 104 to 1.7 × 105), respectively. The neutralization titers of 7312Apl2003 were significantly higher than those of 7312Apl1992 (P = 0.0002, one-tailed paired t test).
Fig 4.
Autologous and heterologous neutralization of single-genome amplification (SGA)-derived HIV-2 Env-pseudotyped virus. (A) Neighbor-joining tree of HIV-2 gp160 env gene sequences amplified from 7312Apl1992 (blue) and 7312Apl2003 (red) by SGA. HIV-2 group A (HIV-27312A, HIV-2ST, HIV-2BEN, and HIV-2ROD) and group B (HIV-2D205 and HIV-2UC1) reference sequences are included. The env genes tested for neutralization in panel B are labeled with asterisks. (B) 7312A-92 and 7312A-03 Envs were used to pseudotype the env-minus HIV-27312A backbone pJK7312AΔEnv in 293T cells. Autologous neutralization by plasma samples 7312Apl1992 (top) and 7312Apl2003 (bottom) in the TZM-bl assay is shown. (C) Neighbor-joining tree of HIV-2 gp160 env gene sequences amplified from 8704Apl2006 by SGA (orange). The env genes tested for neutralization in panel D are labeled with asterisks. (D) 8704A-06 Envs were used to pseudotype the env-minus HIV-27312A backbone pJK7312AΔEnv in 293T cells. Autologous neutralization by plasma samples 8704Apl2006 (top) and 8704Apl2007 (bottom) in the TZM-bl assay is shown. (E) Reciprocal IC50 of autologous plasma (7312Apl1992 and 7312Apl2003) and heterologous plasma (SLRHCpl1995, 10849pl1995, and 8704Apl2007) against 7312A-92 variants and 7312A-03 variants. (F) Reciprocal IC50 of autologous plasma (8704Apl2006 and 8704Apl2007) and heterologous plasma (7312Apl2003, SLRHCpl1995, and 10849pl1995) against 8704A-06 variants. Error bars reflect data from at least three independent experiments.
Seventeen env genes were amplified by SGA from 8704Apl2006. As shown in Fig. 4C, they were clustered within HIV-2 group B reference sequences. The maximum diversity of 8704Apl2006 env genes was 2.45%. Four env genes were cloned and tested for autologous neutralization. Clone 8704A-06-16 was potently neutralized by 8704Apl2006 and 8704Apl2007, with reciprocal IC50s of 1.9 × 105 and 2.7 × 105, respectively. The other three clones, 8704A-06-01, 8704A-06-06, and 8704A-06-23, were 1,000-fold more resistant.
We next tested the neutralization sensitivity of the 7312Apl1992, 7312Apl2003, and 8704Apl2006 variants to heterologous plasma samples. All 7312A variants and 8704A-06-16 were highly sensitive, with reciprocal IC50s ranging from 9.1 × 103 to 1.5 × 107. Variant 8704A-06-16 was potently neutralized by all three heterologous plasma samples tested, with reciprocal IC50s ranging from 4.1 × 103 to 2.9 × 105, whereas other 8704A variants were 1,000-fold more resistant (Fig. 4E and F). Thus, three of four 8704A variants were like four viral variants from subjects SLRH, 226711, and A2240 (Fig. 1B) in showing resistance to NAbs.
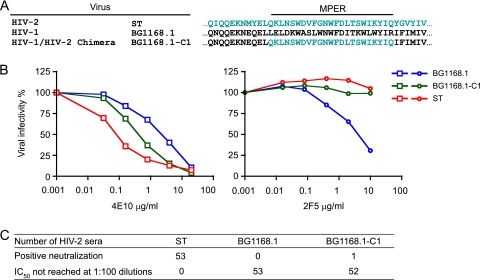
Antibodies directed at the HIV-2 MPER are not responsible for broad and potent neutralization.
The MPER in most HIV-2 Env glycoproteins is highly conserved, such that the HIV-1-elicited 4E10 neutralizing MAb, which neutralizes nearly all strains of HIV-1, also neutralizes many strains of HIV-2 (Table 1). Antibodies that bind the HIV-2 MPER peptide were detected in sera from multiple HIV-2-infected subjects (J. E. Robinson, unpublished data), but whether these antibodies were responsible for the neutralization observed in this study was unknown. We tested this by creating an HIV-1 gp160 Env backbone replaced by an HIV-2 MPER. This chimera is analogous to a reciprocal chimera that we previously developed and used to probe HIV-1-infected patient plasma samples for epitope-specific neutralization (3, 11, 12, 23, 24). For the current study, we used the HIV-1BG1168.1 gp160 backbone and replaced it with the HIV-2ST MPER sequence (Fig. 5A). As a control to demonstrate retained antigenicity and sensitivity to neutralization of the HIV-2 MPER, we tested the broadly reactive NAbs 4E10 and 2F5 against wild-type HIV-1, wild-type HIV-2, and the HIV-1/HIV-2 MPER chimera. As expected, the chimera BG1168.1-C1 and HIV-2ST were similarly sensitive to 4E10 but resistant to 2F5, whereas HIV-1BG1168.1 was sensitive to both 4E10 and 2F5 (Fig. 5B). We then assessed the neutralizing activities of 53 serum samples from HIV-2-infected patients against the chimeric virus BG1168.1-C1. As shown in Fig. 5C, only 1 out of 53 serum samples showed evidence of neutralization, and then only at a relatively low IC50 titer (1:252) compared with the titer of this sample against primary strain HIV-2ST (IC50 = 1:100,000). As a control, all 53 serum samples potently neutralized HIV-2ST with an average IC50 of 1:24,299 (Fig. 1A and Fig. 5C), and none of them neutralized HIV-1BG1168.1. This result strongly suggested that anti-MPER antibodies were not responsible for any of the broad and potent neutralizing responses in HIV-2 infection. Further evidence in support of this conclusion was the fact that HIV-2 MPER peptides could not adsorb neutralizing activity from HIV-2-infected patient plasma samples (Robinson, unpublished).
Fig 5.
Absence of MPER neutralizing antibodies in serum from HIV-2-infected patients. (A) Construction of the HIV-1/HIV-2 chimeric virus BG1168.1-C1 containing the complete MPER from HIV-2ST. (B) Neutralization of HIV-1BG1168.1, HIV-2ST, and BG1168.1-C1 by anti-MPER antibodies 4E10 and 2F5. (C) Neutralization of HIV-1BG1168.1, HIV-2ST, and BG1168.1-C1 by 53 HIV-2-infected patient serum samples. Positive neutralization was defined as a reciprocal IC50 of at least 100. One HIV-2-infected serum sample was positive for neutralization of the BG1168.1-C1 chimera with a reciprocal IC50 titer of 252. The median reciprocal IC50 titer of the 53 serum specimens against HIV-2ST was 2.8 × 104.
Epitopes involving V3, V4, CD4bs, CD4i, and MPER are accessible to neutralizing monoclonal antibodies on functional Env trimers of most primary HIV-2 strains.
To investigate what epitopes on the HIV-2 envelope glycoprotein might be targeted by HIV-2 NAbs in patient sera, we assessed the neutralization activities of human MAbs directed against the HIV-2 V3 (6.10F), V4 (1.7A), CD4bs (6.10B), CD4-inducible (CD4i) (1.4H), and MPER (4E10) epitopes (Table 1). Except for 4E10, which was obtained from an HIV-1-infected subject, these MAbs were obtained from Epstein-Barr virus-transformed B lymphocytes from subjects with HIV-2 infection (43; Kong et al., submitted). The binding epitopes or regions of these MAbs on the HIV-2 Env glycoprotein have been defined by peptide binding, site-directed mutagenesis, and antibody competition (8; Kong et al., submitted). The MAb 1.4H is defined as a CD4i antibody since its binding to monomeric HIV-2 gp120 is enhanced by preincubation of Env with sCD4 (Robinson, unpublished). However, 1.4H is not believed to recognize coreceptor binding surface epitopes since this region on the HIV-2 strains studied here (7312A, ST, and UC1) was previously shown to be inaccessible to binding by prototypic CD4i MAbs 17b, 21c, and 19e prior to Env engagement by sCD4 (13). All five MAbs—6.10F, 1.7A, 6.10B, 1.4H, and 4E10—potently neutralized HIV-2 Envs cloned from most infected subjects with IC50s of less than 0.1 μg/ml in most cases. Only Env clones from subjects 8704A and A2240 showed panresistance to all four MAbs. HIV-27312A Envs generated by SGA showed neutralizing sensitivities comparable to those of HIV-27312A Envs generated from PBMC proviral DNA through conventional amplification and cloning approaches. These results indicate that epitopes on V3, V4, CD4bs, and MPER on most HIV-2 strains were present and accessible to antibody neutralization and could potentially contribute to the potent and broadly reactive neutralizing responses observed with HIV-2-infected patient plasma samples.
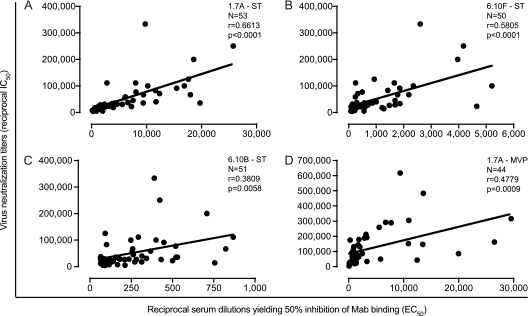
Antibodies in HIV-2-infected patient serum compete with MAbs 1.7A (V4), 6.10F (V3), and 6.10B (CD4bs) for binding HIV-2 gp120 Env glycoprotein.
Given that multiple epitope regions on HIV-2 Env trimers were accessible to neutralizing MAbs, we analyzed the panel of 54 HIV-2-infected patient serum samples for their ability to compete for MAb binding to the HIV-2 gp120. When tested for binding to the HIV-2ST gp120 glycoprotein, 53, 50, and 51 out of 54 total serum samples competed strongly with MAbs 1.7A, 6.10F, and 6.10B, respectively. The serum dilutions giving half-maximum effective competition (reciprocal EC50) are shown in Fig. 6 and Table S1 in the supplemental material. The median and range (in parentheses) of reciprocal EC50s against 1.7A, 6.10F, and 6.10B were 3,318 (148 to 25,767), 583 (78 to 5,209), and 212 (63 to 867), respectively. Comparable results were observed with the HIV-2MVP15132 gp120 glycoprotein, where 44 out of 51 serum samples competed effectively with 1.7A with a median and range (in parentheses) reciprocal EC50 of 1,707 (70 to 29,551) (Fig. 6D). There was a significant positive correlation between virus neutralization titers and MAb binding inhibition titers for MAb 1.7A against HIV-2ST Env (r = 0.6613, P < 0.0001) and HIV-2MVP15132 Env (r = 0.4779, P = 0.0009), for MAb 6.10F against HIV-2ST Env (r = 0.5805, P < 0.0001), and for MAb 6.10B against HIV-2ST Env (r = 0.3809, P = 0.0058). These results suggested that NAbs targeting all three epitope regions are associated with the potency and breadth of the neutralizing responses observed in HIV-2-infected patient plasma.
Fig 6.
Correlation between serum neutralization titers and MAb competition titers. (A) x axis, reciprocal serum dilutions yielding 50% inhibition of 1.7A binding to HIV-2ST gp120 (reciprocal EC50); y axis, virus neutralization titers (reciprocal IC50) of serum against HIV-2ST in the TZM-bl assay. Pearson correlation and linear regression analyses were performed. The correlation coefficient r, P value, and linear regression line are shown. Similar analyses are depicted for MAb 6.10F (B) and 6.10B (C) binding to HIV-2ST gp120 and 1.7A binding to HIV-2MVP15132 gp120 (D).
DISCUSSION
The present study investigated the potency and breadth of neutralizing antibody responses in natural HIV-2 infection and generated findings that significantly extend what has previously been reported (4, 45, 52, 56, 59). Surprisingly, we observed in 64 chronically infected subjects median reciprocal NAb titers of 1.7 × 105, 2.8 × 104, and 3.3 × 104 against three different primary HIV-2 strains (HIV-27312A, HIV-2ST, and HIV-2UC1, respectively) with a range of 8.1 × 102 to 1 × 106 when measured in the TZM-bl single-cycle virus entry assay (Fig. 1A). In the same assay, we tested plasma from 10 subjects chronically infected with HIV-1 for NAbs against three primary HIV-1 strains and observed the expected low titers of broadly reactive NAbs (reciprocal IC50 < 102). Previously, we and others have used the same TZM-bl assay to measure HIV-1 neutralization by plasma antibodies from hundreds of HIV-1-infected subjects (3, 21, 51, 58). Only a small fraction (<5%) exhibited reciprocal IC50 titers approaching 103 against a majority of genetically diverse tier 2 and tier 3 virus strains. Thus, the titer and breadth of HIV-2 neutralization described here far exceed, by orders of magnitude, what has been observed for HIV-1. They also exceed what has generally been reported for HIV-2 neutralization (4, 45, 52, 56, 59). The explanation for the latter differences likely involves differences in assay methods and design. Previous studies of HIV-2 neutralization commonly analyzed uncloned virus isolates and employed plaque reduction, syncytium inhibition, impairment in cell-to-cell virus propagation, reduction in proviral integration, and diverse cell targets as endpoints to assess virus neutralization (2, 4, 52, 56, 59). Given the enhanced sensitivity and reproducibility that recombinant-based single-cycle virus entry assays have brought to the analysis of HIV-1 neutralization (42, 51, 58), it is not surprising that findings made in the present study could differ from those of previous studies based on very different methodologies.
One prior study of HIV-2 neutralization that was more closely aligned with the present work was that by Rodriguez et al. (45). That study involved a single-cycle viral entry analysis on U87 cells but was different from the present study in that an env-minus HIV-1 pNL4-3.Luc backbone was pseudotyped with HIV-2 Envs that were in turn expressed from bulk-amplified HIV-2 env genes. This approach can result in Env heterotrimers derived from divergent or recombinant env genes where individual gp120 protomers having different primary amino acid sequences are mixed. This approach can also result in env gene products that contain in vitro-generated artificial recombinants and Taq polymerase nucleotide misincorporation errors (27, 48). In addition, pseudovirions with HIV-2 Envs paired with HIV-1 Gag and other HIV-1 virion components could have unpredictable effects on virus entry and neutralization. With these caveats, the study of Rodriguez et al. found expanded HIV-2 NAb breadth but titers of HIV-2 NAbs 1 to 4 orders of magnitude less than our results (45). Interestingly, Barnett and colleagues reported in an earlier study neutralization titers of 1:7,000 against HIV-2UC1 by heterologous patient plasma samples (1). Recently, using assay methods very similar to those in our study, de Silva and colleagues observed autologous and heterologous HIV-2 neutralization of primary HIV-2 strains by plasma from patients chronically infected by HIV-2 that is virtually indistinguishable in magnitude and breadth from our findings (16a). Thus, we conclude that when comparable assay methods are employed, the magnitude and breadth of HIV-2 NAbs far exceed those of HIV-1 NAbs from chronically infected humans, and such antibodies can be efficiently and reproducibly assayed.
Because of the remarkable breadth and potency of HIV-2 neutralization that was observed, we performed an extensive set of controls to ensure that neutralization was mediated by IgG antibody interacting with the HIV-2 Env and not some other surface molecule, that antibody neutralization was not confounded by surreptitious treatment with antiretroviral drugs or by the presence of chemokines or other entry inhibitors, and that no nonspecific inhibitory effect was present. To accomplish this, we first determined if virus entry inhibition specifically required virion-associated HIV-2 Env. Viruses pseudotyped with VSV, MuLV, or HIV-1 Env were infectious but were not neutralized by plasma from HIV-2-infected subjects that potently neutralized virions with HIV-2 Env. Second, we demonstrated that IgG purified from HIV-2-infected patient plasma, but not from uninfected control subjects, inhibited virus entry at the same concentrations as those found in plasma and that plasma-mediated inhibition could be completely accounted for by its IgG content. Third, we showed that broad and potent HIV-2 neutralization was not dependent on the cell source of virus or on the target cells used to assay virus entry inhibition. NAb titers were only moderately higher (P < 0.05, one-tailed paired t test; Fig. 3C) when virus was produced in 293T cells and tested on TZM-bl cells than when viruses were produced in activated primary human T cells and tested for entry inhibition on the same cells, consistent with previous findings with HIV-1 (34). Thus, we conclude that potent, broadly reactive HIV-2 IgG NAbs are a feature of HIV-2 infection and that these antibodies can be efficiently and reproducibly measured in the TZM-bl assay and the results corroborated in assays where virus is grown and tested on primary human CD4+ T cells.
We explored what molecular targets on the HIV-2 Env trimers might be recognized by broadly reactive antibodies in human plasma. We analyzed 32 different HIV-2 strains for neutralization sensitivity to the human MAbs 6.10F, 1.7A, 6.10B, 1.4H, and 4E10. Aside from the MPER-reactive MAb 4E10, whose binding specificity is precisely known (6, 65), the epitope specificities of the HIV-2-infected patient-derived MAbs are only partially known since antibody-protein crystal structures are not available. However, deletion mutation, site-directed mutation, and binding cross-competition studies have mapped 6.10F binding to V3, 1.7A binding to V4 and the base of V3, 6.10B binding to the CD4 binding site, and 1.4H binding to an unknown site inducible by sCD4 and competing for binding with 6.10B (8, 43; Kong et al., submitted). Each of these MAbs showed potent reactivity against most, but not all, HIV-2 Envs with IC50s frequently of 0.1 μg/ml or less (Table 1). Therefore, these epitopes are clearly accessible to antibody neutralization on the majority of HIV-2 Envs, including Envs derived by SGA from circulating plasma virions. It thus seems likely that these epitopes could contribute to the broad and potent neutralization observed with HIV-2-infected patient plasma. Evidence in support of this came from competition studies, where it was found that most patient sera competed with MAbs 6.10F, 1.7A, and 6.10B for binding to the HIV-2 gp120 glycoprotein. Importantly, competition titers correlated significantly with neutralization titers (Fig. 6). These results suggested that polyclonal antibodies targeting V3, V4, and CD4bs were elicited in most HIV-2-infected subjects and that these antibodies were likely to have contributed to neutralization of the primary virus strains. Conversely, antibodies targeting HIV-2 MPER did not appear to contribute appreciably to neutralization breadth or potency, since 52 of 53 HIV-2-infected patient serum samples did not neutralize an HIV-1 chimera into which we substituted the complete HIV-2 MPER (Fig. 5).
Initially, we tested the neutralization activity of plasma or sera from 64 HIV-2-infected patients against 20 HIV-2 group A and B viruses, Env chimeras, or Env pseudoviruses. These samples contained antibodies that neutralized all but 4 viruses (SLRH8, 226711-33, A2240-3, A2240-12). The latter viruses were also generally resistant to most neutralizing HIV-2 MAbs specific for V3, V4, CD4bs, and CD4i epitopes. The exceptions were SLHR8 and 226711-33, which were resistant to neutralization by antibodies in patient plasma (Fig. 1B) but were sensitive to neutralization by a subset of MAbs (Table 1). HIV-2-related viruses that exhibit a pan-neutralization-resistant phenotype include the SIVmac251 isolate and the SIVmac239 clone derived from it, but the observation that the vast majority of HIV-2 strains and Envs were actually highly sensitive to neutralization by both polyclonal and monoclonal human antibodies came as a surprise. To be certain that this was not due to short-term in vitro cultivation of these viruses prior to molecular cloning, we utilized recently described methods for single-genome amplification of viral sequences directly from plasma virions (27, 28) to more fully analyze env genomes and their expressed Env glycoproteins. These data are depicted for two subjects, 7312A and 8704A, in Fig. 4A and B and Table 1. What is evident is that all 10 7312A SGA-derived env genes derived from plasma specimens separated in time by 11 years are nearly indistinguishable in their exquisite sensitivity to autologous neutralization by contemporaneous, previous, or subsequent antibodies and to heterologous antibodies (Fig. 4E). These SGA-derived 7312A Envs were also nearly indistinguishable from lambda phage-derived 7312A Envs in their exquisite neutralization sensitivity to the panel of V4, V3, CD4bs, and CD4i MAbs (Table 1). SGA-derived Envs from subject 8704A (Fig. 4C) revealed a more complicated result: one clone (8704A-06-16) was exquisitely sensitive to autologous and heterologous patient antibodies (Fig. 4D and F) but not to MAbs (Table 1), whereas three other 8704A Env clones were resistant to polyclonal and monoclonal antibodies. SGA-derived HIV-2 Envs from 7312A and 8704A thus exhibit the broad range of neutralization sensitivities observed in the larger set of HIV-2 clones in the present study, and they confirm that such viral Envs exist in vivo. Of interest, we did not identify HIV-2 Envs that exhibited intermediate levels of neutralization sensitivity comparable to those of HIV-1 tier 2 strains (51). We do not have a mechanistic explanation for this dichotomy in neutralization sensitivities.
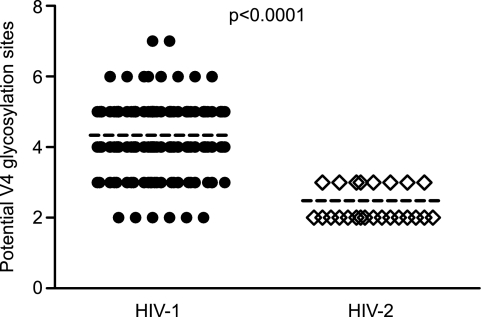
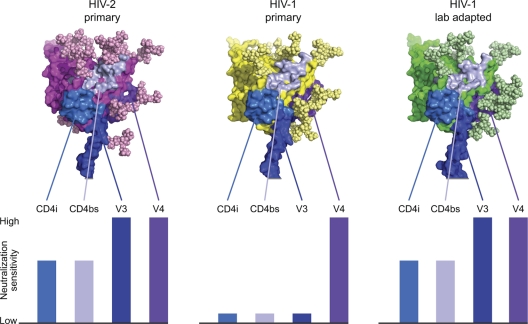
In light of the unexpected neutralization sensitivity of HIV-2 strains to antibodies elicited in chronically infected patients, we asked what molecular features of the HIV-2 Env might contribute to its enhanced antibody accessibility and sensitivity. Comparative analysis of Env sequences between many strains of HIV-1 and HIV-2 revealed a significantly higher number of potential V4 glycosylation sites for a diverse set of HIV-1 strains (mean = 4.34; median = 4; range = 2 to 7) compared to the panel of HIV-2 strains used in this study (mean = 2.48; median = 2; range = 2 to 3; P < 0.0001, Mann-Whitney test) (Fig. 7). Such a reduction in glycosylation in HIV-2 could conceivably serve to open the glycan shield surrounding V4, allowing better antibody access and neutralization. This is in agreement with previous studies in HIV-1, which showed that insertion of an epitope for a FLAG antibody into V4 resulted in potent virus neutralization by that antibody (41). Thus, with respect to V4-directed antibodies (1.7A and polyclonal NAbs in human plasma that compete with it for gp120 binding), HIV-2 appeared to have reduced glycan shielding. For V3 (6.10F), CD4bs (6.10B), and CD4i (1.4H) antibodies, HIV-2 neutralization sensitivities were similar to those of laboratory-adapted HIV-1 strains but very different from those of primary HIV-1 strains (Fig. 8). Different envelope conformations are required for recognition by these antibodies: CD4i antibodies recognize the CD4-bound conformation of HIV-1 gp120 with the bridging sheet assembled (32), CD4bs antibodies generally recognize the region under the bridging sheet (7), and V3 antibodies recognize any of a number of conformations in which the shielding V2 region is altered to expose the V3 loop (47). The potent neutralization of HIV-2 by V3 (6.10F), CD4bs (6.10B), and CD4i (1.4H) antibodies (Table 1) indicates that all these antibodies recognize the HIV-2 Env efficiently in its pre-CD4-bound state, suggesting that HIV-2 gp120 can more readily adopt a number of different conformations, similar to lab-adapted strains of HIV-1 but unlike primary strains of HIV-1. Consistent with this interpretation is the observation that the IC50s for sCD4 for HIV-27312A (9 nM), HIV-2UC1 (3 nM), and HIV-2ST (25 nM) (13) are all well below the mean and median values (both greater than 100 nM) for a large number of HIV-1 transmitted/founder and chronic Envs (27). The mechanism of immune evasion by HIV-1 that is related to fixation of the glycoprotein in a particular conformation—conformational masking (30)—thus distinguishes primary strains of HIV-1 from primary strains of HIV-2. Primary HIV-2 strains thus appear to have two mechanisms of immune evasion that are diminished in effectiveness relative to primary HIV-1 strains: glycan shielding (for V4-reactive antibodies) and conformational masking (for V3-, CD4bs-, and CD4i-reactive antibodies) (Fig. 8).
Fig 7.
Potential N-linked glycosylation sites in the V4 loops of HIV-1 and HIV-2 strains. Mean values are shown as dashed lines. For HIV-1, the mean was 4.34, the median was 4, and the range was 2 to 7. For HIV-2, the mean was 2.48, the median was 2, and the range was 2 to 3. The HIV-1 V4 sequences exhibited a significantly higher level of glycosylation than the HIV-2 V4 sequences (P < 0.0001, Mann-Whitney test).
Fig 8.
Structural models and relative neutralization sensitivities of primary and laboratory-adapted strains of HIV-1 compared with primary HIV-2 strains. Structural models of gp120s from HIV-2 primary (7312A), HIV-1 primary (YU2), and HIV-1 laboratory-adapted (HXBc2) strains are shown with modeled glycans and epitopes for V3, V4, CD4bs, and CD4i highlighted. Neutralization potencies of V3, V4, CD4bs, and CD4i antibodies are estimated schematically, with full bars corresponding to highly potent neutralization, medium bars to moderate neutralization, and small bars to very weak or nonneutralizing phenotypes. V4-directed antibodies are rarely observed in natural HIV-1 infection, but placement of antigenic tags into the V4 region reveals that antibodies directed here can neutralize HIV-1 potently (41). For V3-, CD4bs-, and CD4i-directed antibodies, epitopes are generally hidden on the assembled HIV-1 viral spike of primary HIV-1 strains by conformational masking. The increased flexibility of laboratory-adapted HIV-1 isolates allows exposure of these epitopes, which resembles the neutralization sensitivities observed with HIV-2 (Table 1).
Finally, given the extraordinary breadth and potency of NAbs observed in the 64 chronically infected subjects that we studied, we asked if NAb titers against any of the three HIV-2 virus strains (HIV-27312A, HIV-2ST, and HIV-2UC1) might correlate with either viral load or CD4 count. We found no significant associations (Table 2). Since this was a retrospective, cross-sectional study and not designed or powered to determine if there is a relationship between NAb responses and viral natural history and clinical outcome, little weight can be placed on such a negative result. We did, however, note that NAb titers in this study were generally very high (medians, 104 to 105), the HIV-2 vRNA loads were low (median, 121 vRNA copies/ml), and the CD4 counts were high (median, 482 cells/mm3) (see Table S1 in the supplemental material). It is possible that HIV-2 NAbs could exert a generally suppressive effect on virus replication that in concert with other elements of the immune system constrains virus replication and pathogenesis (16), but how such neutralization-sensitive HIV-2 strains could replicate and persist for years in chronically infected individuals in the face of potent, broadly reactive NAbs is a puzzle. Cell-to-cell virus transmission is a potential explanation (54).
Table 2.
Pearson correlation analysis of plasma viral load and CD4 count with neutralization titers
| Parameter and statistic | Neutralization titer (reciprocal IC50) |
||
|---|---|---|---|
| ST | UC1 | 7312A | |
| Viral load (n = 54) | |||
| r | −0.0100 | 0.1469 | 0.0425 |
| P | 0.9426 | 0.2891 | 0.7602 |
| CD4 count (n = 52) | |||
| r | −0.0516 | −0.2194 | −0.1681 |
| P | 0.7163 | 0.1182 | 0.2337 |
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Ferns (University College London, London, United Kingdom) for assistance with HIV-2 viral load assays, J. White for manuscript preparation, the clinical and DNA sequencing cores of the University of Alabama at Birmingham Center for AIDS Research, and the biostatistical core of the University of Pennsylvania Center for AIDS Research.
This work was supported by grants from the NIH (AI67854, AI88564, and AI87383).
Footnotes
Published ahead of print 26 October 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Barnett SW, Quiroga M, Werner A, Dina D, Levy JA. 1993. Distinguishing features of an infectious molecular clone of the highly divergent and noncytopathic human immunodeficiency virus type 2 UC1 strain. J. Virol. 67:1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Behrendt R, et al. 2009. A neutralization assay for HIV-2 based on measurement of provirus integration by duplex real-time PCR. J. Virol. Methods 159:40–46 [DOI] [PubMed] [Google Scholar]
- 3. Binley JM, et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bjorling E, et al. 1993. Autologous neutralizing antibodies prevail in HIV-2 but not in HIV-1 infection. Virology 193:528–530 [DOI] [PubMed] [Google Scholar]
- 5. Bohne-Lang A, von der Lieth CW. 2005. GlyProt: in silico glycosylation of proteins. Nucleic Acids Res. 33:W214–W219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cardoso RM, et al. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22:163–173 [DOI] [PubMed] [Google Scholar]
- 7. Chen L, et al. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cole KS, et al. 2001. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology 290:59–73 [DOI] [PubMed] [Google Scholar]
- 9. Damond F, et al. 2008. Quality control assessment of human immunodeficiency virus type 2 (HIV-2) viral load quantification assays: results from an international collaboration on HIV-2 infection in 2006. J. Clin. Microbiol. 46:2088–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. da Silva ZJ, et al. 2008. Changes in prevalence and incidence of HIV-1, HIV-2 and dual infections in urban areas of Bissau, Guinea-Bissau: is HIV-2 disappearing? AIDS 22:1195–1202 [DOI] [PubMed] [Google Scholar]
- 11. Davis KL, et al. 2009. Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J. Virol. 83:1240–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis KL, et al. 2009. High titer HIV-1 V3-specific antibodies with broad reactivity but low neutralizing potency in acute infection and following vaccination. Virology 387:414–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Decker JM, et al. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Decker JM, et al. 2009. Effective activation alleviates the replication block of CCR5-tropic HIV-1 in chimpanzee CD4+ lymphocytes. Virology 394:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Derdeyn CA, et al. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Silva TI, Cotten M, Rowland-Jones SL. 2008. HIV-2: the forgotten AIDS virus. Trends Microbiol. 16:588–595 [DOI] [PubMed] [Google Scholar]
- 16a. de Silva TI, et al. 2012. Potent autologous and heterologous neutralizing antibody responses occur in HIV-2 infection across a broad range of infection outcomes. J. Virol. 86:930–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferns RB, Garson JA. 2006. Development and evaluation of a real-time RT-PCR assay for quantification of cell-free human immunodeficiency virus type 2 using a brome mosaic virus internal control. J. Virol. Methods 135:102–108 [DOI] [PubMed] [Google Scholar]
- 18. Gao F, et al. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433–7447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao F, Yue L, Sharp PM, Hahn BH. 1993. Genetic typing of HIV-2 from a Senegalese/German heterosexual transmission. AIDS Res. Hum. Retroviruses 9:703–704 [DOI] [PubMed] [Google Scholar]
- 20. Gao F, et al. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in West Africa. Nature 358:495–499 [DOI] [PubMed] [Google Scholar]
- 21. Gnanakaran S, et al. 2010. Genetic signatures in the envelope glycoproteins of HIV-1 that associate with broadly neutralizing antibodies. PLoS Comput. Biol. 6:e1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gottlieb GS, et al. 2002. Equal plasma viral loads predict a similar rate of CD4+ T cell decline in human immunodeficiency virus (HIV) type 1- and HIV-2-infected individuals from Senegal, West Africa. J. Infect. Dis. 185:905–914 [DOI] [PubMed] [Google Scholar]
- 23. Gray ES, et al. 2009. Broad neutralization of human immunodeficiency virus type 1 mediated by plasma antibodies against the gp41 membrane proximal external region. J. Virol. 83:11265–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gray ES, et al. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hahn BH, Shaw GM, De Cock KM, Sharp PM. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607–614 [DOI] [PubMed] [Google Scholar]
- 26. Hamel DJ, et al. 2007. Twenty years of prospective molecular epidemiology in Senegal: changes in HIV diversity. AIDS Res. Hum. Retroviruses 23:1189–1196 [DOI] [PubMed] [Google Scholar]
- 27. Keele BF, et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keele BF, et al. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kong LI, et al. 1988. West African HIV-2-related human retrovirus with attenuated cytopathicity. Science 240:1525–1529 [DOI] [PubMed] [Google Scholar]
- 30. Kwong PD, et al. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 31. Kwong PD, Wilson IA. 2009. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat. Immunol. 10:573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kwong PD, et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lemey P, et al. 2003. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. U. S. A. 100:6588–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Louder MK, et al. 2005. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology 339:226–238 [DOI] [PubMed] [Google Scholar]
- 35. Mascola JR, Montefiori DC. 2010. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 28:413–444 [DOI] [PubMed] [Google Scholar]
- 36. Palmer S, et al. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pancera M, et al. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 107:1166–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pantophlet R, Burton DR. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24:739–769 [DOI] [PubMed] [Google Scholar]
- 39. Piatak M, Jr, et al. 1993. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science 259:1749–1754 [DOI] [PubMed] [Google Scholar]
- 40. Reeves JD, Doms RW. 2002. Human immunodeficiency virus type 2. J. Gen. Virol. 83:1253–1265 [DOI] [PubMed] [Google Scholar]
- 41. Ren X, Sodroski J, Yang X. 2005. An unrelated monoclonal antibody neutralizes human immunodeficiency virus type 1 by binding to an artificial epitope engineered in a functionally neutral region of the viral envelope glycoproteins. J. Virol. 79:5616–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robinson JE, et al. 1998. Production and characterization of SIV envelope-specific rhesus monoclonal antibodies from a macaque asymptomatically infected with a live SIV vaccine. AIDS Res. Hum. Retroviruses 14:1253–1262 [DOI] [PubMed] [Google Scholar]
- 44. Robinson JE, Holton D, Pacheco-Morell S, Liu J, McMurdo H. 1990. Identification of conserved and variant epitopes of human immunodeficiency virus type 1 (HIV-1) gp120 by human monoclonal antibodies produced by EBV-transformed cell lines. AIDS Res. Hum. Retroviruses 6:567–579 [DOI] [PubMed] [Google Scholar]
- 45. Rodriguez SK, et al. 2007. Comparison of heterologous neutralizing antibody responses of human immunodeficiency virus type 1 (HIV-1)- and HIV-2-infected Senegalese patients: distinct patterns of breadth and magnitude distinguish HIV-1 and HIV-2 infections. J. Virol. 81:5331–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rowland-Jones SL, Whittle HC. 2007. Out of Africa: what can we learn from HIV-2 about protective immunity to HIV-1? Nat. Immunol. 8:329–331 [DOI] [PubMed] [Google Scholar]
- 47. Rusert P, et al. 2011. Interaction of the gp120 V1V2 loop with a neighboring gp120 unit shields the HIV envelope trimer against cross-neutralizing antibodies. J. Exp. Med. 208:1419–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salazar-Gonzalez JF, et al. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salazar-Gonzalez JF, et al. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schief WR, Ban YE, Stamatatos L. 2009. Challenges for structure-based HIV vaccine design. Curr. Opin. HIV AIDS 4:431–440 [DOI] [PubMed] [Google Scholar]
- 51. Seaman MS, et al. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shi Y, et al. 2005. Evolution of human immunodeficiency virus type 2 coreceptor usage, autologous neutralization, envelope sequence and glycosylation. J. Gen. Virol. 86:3385–3396 [DOI] [PubMed] [Google Scholar]
- 53. Shriner D, Rodrigo AG, Nickle DC, Mullins JI. 2004. Pervasive genomic recombination of HIV-1 in vivo. Genetics 167:1573–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sigal A, et al. 2011. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 477:95–98 [DOI] [PubMed] [Google Scholar]
- 55. Simmonds P, Balfe P, Ludlam CA, Bishop JO, Brown AJ. 1990. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J. Virol. 64:5840–5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tamalet C, et al. 1995. Autologous neutralizing antibodies and viral load in HIV-2-infected individuals. AIDS 9:90–91 [PubMed] [Google Scholar]
- 57. Wei X, et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 59. Weiss RA, et al. 1988. HIV-2 antisera cross-neutralize HIV-1. AIDS 2:95–100 [DOI] [PubMed] [Google Scholar]
- 60. Wu X, et al. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888 [DOI] [PubMed] [Google Scholar]
- 62. Zhang M, et al. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246 [DOI] [PubMed] [Google Scholar]
- 63. Zheng NN, et al. 2007. Role of human immunodeficiency virus (HIV)-specific T-cell immunity in control of dual HIV-1 and HIV-2 infection. J. Virol. 81:9061–9071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhou JY, Montefiori DC. 1997. Antibody-mediated neutralization of primary isolates of human immunodeficiency virus type 1 in peripheral blood mononuclear cells is not affected by the initial activation state of the cells. J. Virol. 71:2512–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zwick MB, et al. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892–10905 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.