Abstract
MicroRNA-122 (miR-122) enhances hepatitis C virus (HCV) fitness via targeting two sites in the 5′-untranslated region (UTR) of HCV. We used selective 2′-hydroxyl acylation analyzed by primer extension to resolve the HCV 5′-UTR's RNA secondary structure in the presence of miR-122. Nearly all nucleotides in miR-122 are involved in targeting the second site, beyond classic seed base pairings. These additional interactions enhance HCV replication in cell culture. To our knowledge, this is the first biophysical study of this complex to reveal the importance of ‘tail’ miR-122 nucleotide interactions.
TEXT
MicroRNAs (miRNAs) are small endogenous RNAs that alter cellular protein expression by targeting messenger RNAs (mRNAs) (1, 21). The selective targeting of mRNAs by miRNAs is thought to be partially mediated by canonical base pairings between the first 2 to 8 nucleotides (nt) in the miRNA (the “seed site”) and complementary nucleotides in the targeted mRNA (1, 9).
Hepatitis C virus (HCV) has been found to coopt miR-122. Based on genetic data, binding of miR-122 at these sites enhances viral replication, and to a lesser extent, viral translation (7, 8). Two genetically validated miR-122 target sites (S1 and S2) within the HCV 5′-untranslated region (UTR) have been identified. No structural studies to date have described the details of these miR-122:HCV complexes.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) analysis is one of the most robust and well-characterized chemical probing methods for mapping RNA secondary structure (17, 22, 24, 25). SHAPE quantifies the flexibility (base-paired versus single-stranded RNA) of every nucleotide in an RNA structure with striking accuracy and speed compared to traditional enzymatic mapping methods (4). Here, we used SHAPE technology to resolve the structure of a miR-122:HCV complex.
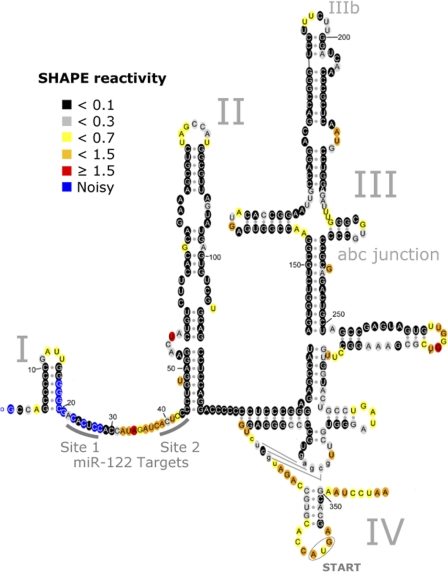
HCV 5′-UTR structure with and without miR-122.
In vitro-transcribed HCV 5′-UTR RNA (genotype [GN] 1b/con1; nt 1 to 377) was folded as previously described (10) (except the buffer was 100 mM HEPES [pH 8]), and this folded structure was interrogated using N-methylisotoic anhydride (NMIA; 10 mM final concentration; Invitrogen) (24). NMIA distinguishes single-stranded from double-stranded nucleotides by covalently attaching only to single-stranded nucleotides. NMIA-labeled nucleotides are then identified by reverse transcription (RT) primer extension (with SuperScript III; Invitrogen) (18). SuperScript III pauses at NMIA-labeled nucleotides, resulting in a series of DNA fragments whose lengths (relative to the primer site) correspond to the location of single-stranded nucleotides in the target RNA. In this study, these fragments can be resolved by capillary electrophoresis (17) by using an ABI 3100 Genetic Analyzer.
The ABI 3100 Genetic Analyzer (50-cm capillaries filled with POP6 matrix; voltage, 15 kV; temperature, 60°C; injection time, 15 s) and GeneScan (ABI) were used to acquire the data for each sample, which consisted of the purified RT DNA fragments resuspended in 9.75 μl of Hi-Di formamide, to which 0.25 μl of DNA internal sizing standard (ROX 500; ABI catalog number 602912) was added. Peaks in the resulting electrophoretic traces were integrated, and their fragment lengths were determined using PeakScanner (ABI Biosystems).
The data from PeakScanner were then processed into SHAPE data by using FAST (fast analysis of SHAPE traces), a custom program that we built. FAST automatically corrects for signal differences due to handling errors, adjusts for signal decay (11), and converts fragment length to nucleotide position, using a ddGTP ladder as an external sizing standard and the local Southern method (20). This software is freely available at glennlab.stanford.edu.
SHAPE-determined reactivities were then incorporated as a pseudo-nearest-neighbor free-energy change parameter (4, 15) in the program RNAstructure (14), resulting in the predicted 5′-UTR structure (Fig. 1). Generally, SHAPE reactivities below ∼0.3 are associated with nucleotides constrained by base pairing or other interactions, while higher reactivities are associated with flexible, single-stranded nucleotides (4, 23). RNA structures were colored to reflect SHAPE reactivities by using RNAViz 2 (3) (Fig. 1). Notably, this proposed secondary structure model is in concordance with all the subdomains of the HCV 5′-UTR that have previously been resolved by nuclear magnetic resonance (NMR) or crystallography studies (12). The benefit of having SHAPE-derived information is also evident from a comparison of the secondary structures predicted using RNAstructure with (Fig. 1) and without (see Fig. S2b in the supplemental material) SHAPE-determined reactivities. We were unable to resolve the state of nt 17 to 28, which contain the S1 miR-122 binding site, due to a high background signal.
Fig 1.
Secondary structure of the HCV 5′-UTR. The 5′-UTR consists of 4 domains, labeled I to IV. The proposed secondary structure is consistent with all available crystallographic and/or NMR structural data (see Fig. S2a in the supplemental material). Colors denote SHAPE reactivity, as indicated, which is proportional to the probability that a nucleotide is single stranded.
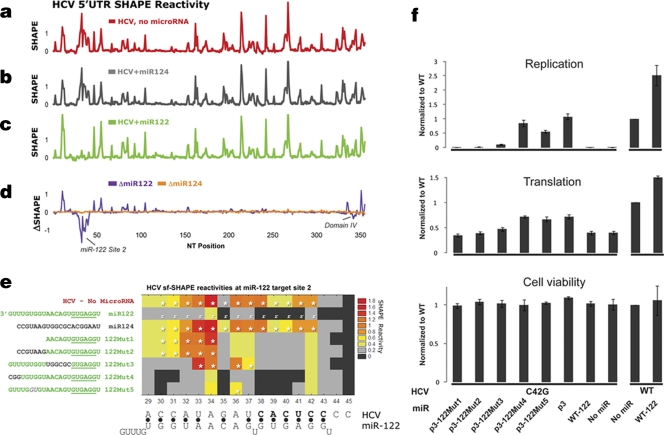
To determine the effect of miR-122 on this structure, miR-122 was added to the folded HCV RNA at 37°C for 25 min. We used miR-124, a microRNA shown to have no effect on HCV (7, 8), as a control. As expected, miR-124 did not alter the structure of the HCV 5′-UTR (red [panel a] − gray [panel b] = orange trace [panel d]) (Fig. 2a). In contrast, miR-122 induced significant changes in the reactivity of the 5′-UTR (red [panel a] − green [panel c] = purple trace [panel d]).
Fig 2.
MicroRNA-122-induced changes in the HCV 5′-UTR. (a to c) HCV 5′-UTR SHAPE reactivities in the presence of no microRNA (a, red), control miR-124 (b, gray), or miR-122 (c, green) are shown. (d) The change in SHAPE reactivities [ΔSHAPE = (with miR) − (without miR)] induced by control miR-124 (d, orange) and miR-122 (d, purple). (e) Heat map of SHAPE reactivities for HCV nt 29 to 45 in the presence of various miR-122 mutants. Boxes with an asterisk indicate values were statistically different (P < 0.001) from the SHAPE reactivity determined in the presence of WT miR-122 (reference [r]). (f) Effects of the miR-122 mutants on HCV translation and replication.
The most prominent decreases in SHAPE reactivities induced by miR-122 occurred at the region of nt 29 to 42, which contains the second miR-122 target site (S2). No reactivity changes were seen in domain III (D-III) of the internal ribosome entry site (D-III; nt 125 to 323), suggesting that binding of miR-122 to the region between domains I and II does not affect D-III. Changes in flexibility were observed for nucleotides in domain IV and are discussed below.
Base paring between HCV nt 38 and 42 and the seed nucleotides of miR-122 (nt 2 to 7) is likely responsible for the decreased reactivity of HCV nt 38 to 42. The decrease in SHAPE reactivity for HCV nt 29 to 37, however, was unexpected, since these nucleotides fall outside the complementary seed region.
To further investigate these changes, SHAPE reactivities of the 5′-UTR were determined in the presence of a series of miR-122 (3′) tail mutants (Mut1 to -5) (Fig. 2e). Each experiment was performed in triplicate. Statistically significant differences in SHAPE reactivity, calculated using the Tukey-Kramer multiple comparisons test (InStat 3) are denoted with asterisks. Subsequently, to test the effects of these miR-122 tail mutants on HCV replication, we constructed these miR-122 mutants and also mutated the guanosine at position 3 (p3) to cytidine (p3miR-122), similar to the approach taken by Machlin et al. (13). In parallel, an HCV RNA mutant (HCV C42G) was constructed by mutating position 42 from a C to a G (C42G), which disrupted its binding to miR-122 at position 3 of site 2 (HCV site 2). This HCV mutant failed to replicate in cell culture. Exogenous wild-type (WT) miR-122 failed to rescue its replication defects. However, exogenous complementary p3miR-122 allowed this HCV mutant to replicate to WT levels (Fig. 2f). We then assessed the ability of all the p3miR-122 tail mutants to rescue replication of the HCV C42G viral RNA. Cell culture experiments were performed using the J6/JFH1 strain and the Renilla luciferase reporter (5). The miR duplexes were transfected into Huh7 cells 1 day prior to transfection of in vitro-transcribed HCV C42G at time zero and 1 day after. The luciferase signal was measured after 4 h for translation and 48 h for replication. Alamar blue was used to assess cell viability (5). In general, these miR-122 mutants had modest effects on HCV translation (Fig. 2f) but more significant effects on replication, as described next.
122Mut1 lacks the last 9 nt of miR-122. SHAPE analysis revealed that 122Mut1 exhibits base pairing only with HCV nt 38 to 42 but not with HCV nt 29 to 34, suggesting that the terminal 9 nt of miR-122 are required to decrease the flexibility of HCV nt 29 to 34. In complementary cell culture experiments, exogenous p3-122Mut1 failed to rescue the replication defect of HCV site 2, suggesting that binding of the terminal 9 nt of miR-122 is important for HCV replication.
122Mut2 is a substitution mutation in which the tail of miR-124 has replaced the last 9 nt. This mutant resulted in a nearly identical SHAPE reactivity pattern as for 122Mut1. This result suggests that the specific sequence found in the tail of miR-122 is required to decrease the flexibility of HCV nt 29 to 34. Together, these data suggest that a direct interaction between the tail of miR-122 and HCV nt 29 to 34 is likely. In corresponding cell culture experiments, exogenous p3-122Mut2 failed to rescue the replication defect of site 2 in HCV C42G, further confirming that binding of the tail of miR-122 to HCV nt 29 to 34 is required for HCV replication.
122Mut3 is a substitution mutant in which the middle 6 nt of miR-122 have been swapped for the middle 6 nt of miR-124. If, as speculated, these middle 6 nucleotides form a bulge when bound to the miR-122 binding site (13), then the resulting miR-122:HCV complex should not be affected by this swap. Instead, we found the identity of these nucleotides to be relevant. The data obtained with 122Mut3 suggest that miR-122 nucleotides 8 to 13 bind to HCV nucleotides 33 to 37, resulting in the decreased flexibility observed for these HCV nucleotides. In complementary cell culture experiments, exogenous p3-122Mut3 partially rescued the replication defect of HCV site 2, although the replication efficiency was only one-fifth that of the WT level. This suggests that binding of miR-122 nt 8 to 13 to HCV nt 33 to 37 is not as critical for HCV replication, although it may play an enhancing role.
122Mut4 is a substitution mutant in which the last 3 nt of miR122 have been swapped for the last 3 nt of miR-124. The lack of any difference in SHAPE reactivity between this mutant and wild-type miR-122 suggests that these nucleotides are not important for binding. Indeed, as expected, exogenous p3-122Mut4 rescued the replication defect of HCV site 2 up to the WT level.
Together, this structural information is most consistent with the proposed miR-122:HCV complex shown in Fig. 3a. To further validate this structure, we made the 122Mut 5 in which miR-122 nt 16 and 17 were swapped. The SHAPE reactivity for 122Mut5 demonstrated both seed and tail binding, but it revealed a slight increase in flexibility for HCV nt 34 and 36. Perhaps decreasing the stability of miR-122 tail–HCV interactions increases the flexibility of the A-A mismatches found at nt 34 and 36. In cell culture, exogenous p3-122Mut5 can significantly rescue the replication defect of HCV site 2 to 60% of the WT level. This result further demonstrates that binding at nt 34 and 36 (which fall between nt 33 and 37) are not critical for HCV replication and may have a modest enhancement effect. Taken together, our SHAPE data can accurately predict the effects of the structural changes on HCV replication and show that direct binding of HCV RNA and the tail of miR-122 is important for HCV replication.
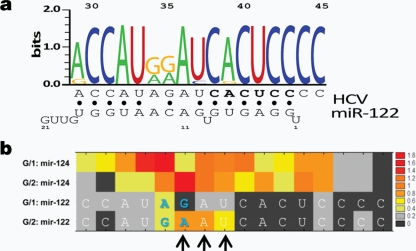
Fig 3.
MicroRNA-122:HCV complex strain specificity. (a) Naturally occurring HCV S2 target site polymorphisms, depicted as a WebLogo. Nucleotides 1 to 22 of miR-122 are depicted at the bottom, from right to left. The secondary structure of the proposed HCV:miR-122 complex is shown below, with HCV nucleotides complementary to the seed region shown in bold and Watson-Crick base pairs indicated by filled circles. (b) HCV SHAPE reactivities as a function of HCV genotype, as determined in the presence of either miR-124 (control) or miR-122. In the G/1 strain, nt 35 is a G. As predicted, in the J6 strain, a base pair did not form between HCV nt 35 and miR-122 nt 11; this mismatch destabilized other nearby interactions (arrows).
Effects of HCV natural polymorphisms.
We next sought to understand the effects of naturally occurring HCV mutations on this miR-122:HCV complex. The conservation pattern of this region is depicted in Fig. 3a as a WebLogo (2). Naturally occurring variations at HCV nt 29, 37, and 39 preserve the proposed base pairing.
The natural variation that occurs at HCV nt 35, however, does not. In the GN1b/con1 construct, HCV nt 35 is a G. In other strains, such as the GN2/J6 strain, HCV nt 35 is an A. This led us to predict that the miR-122:HCV complex at site 2, for strains with the A35 polymorphism, have greater flexibility in this region. To test this hypothesis, we used SHAPE to determine the structure of the GN2/J6 miR-122:HCV complex (Fig. 3b). As predicted, this region of the complex, specifically HCV nt 35 to 37, was significantly more flexible when the A35 polymorphism was present. Most importantly, despite this increase in flexibility in the middle of the complex, the identified tail interactions between miR-122 and HCV were preserved.
Further demonstration of the importance of tail interactions in HCV replication.
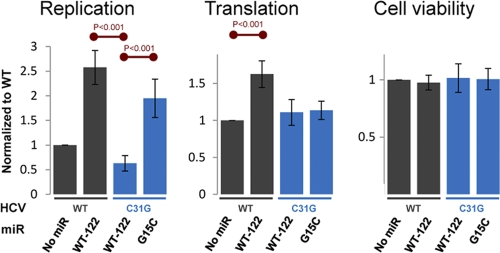
To further confirm the importance of tail interactions in HCV replication, we first mutated HCV nucleotide 31 from a C to a G in order to disrupt its proposed interaction with miR-122 tail nucleotide G15. As expected, HCV mutant C31G replicated less well than did WT HCV (Fig. 4) in the presence of WT miR-122. However, the exogenous, complementary miR-122 mutant, G15C, was able to enhance the replication of the HCV mutant C31G (Fig. 4, left panel) over that achieved in the presence of WT miR-122, which lacks the compensatory mutation. This experiment further confirms that the interaction of the tail of miR-122 with HCV's second target site is important for HCV replication.
Fig 4.
Effect of the miR-122 3′-tail interaction on HCV replication. Values were normalized to the signal obtained for WT HCV in the absence of additional microRNA (addition of miR-124 had no effect). Error bars denote 1 standard deviation, as determined from 5 separate experiments, with each experiment consisting of four replicates, which were averaged together. Red lines denote statistically significant differences (P < 0.001) between the indicated conditions.
Also consistent with prior experiments, ectopic supplementation of WT HCV with WT miR-122 did enhance WT HCV translation (7) (Fig. 4, middle panel), but it enhanced replication to an even greater degree (8) (Fig. 4, left panel). Supplementation of WT HCV with either the miR-122 mutant G15C or miR-124 had no effect on translation or replication (data not shown).
Finally, changes in SHAPE reactivity in D-IV in the presence of miR-122 were also observed. To explore how these D-IV changes relate to the known target sites (S1 and S2), we mutated S1 and S2 and also generated an HCV deletion mutation that lacked both S1 and S2 target sites (ΔNT1-44) to prevent seed pairing. In the presence of miR-122, these HCV mutants were found to have the same D-IV SHAPE reactivity patterns as WT HCV. This suggested that the observed D-IV changes were independent of the S1 and S2 target sites. We therefore explored the possibility that these changes were the result of direct binding of miR-122 to HCV RNA (i.e., to a third miR-122 target site). We found that the base-pairing scheme between miR-122 and D-IV illustrated in Fig. S1 of the supplemental material would be consistent with the SHAPE reactivities we observed. The ability of miR-122 to directly bind to this region was subsequently confirmed by UV cross-linking experiments (19). Preliminary studies, however, have not shown that miR-122 binding at this site enhances translation or RNA replication. Additional experiments aimed at identifying a potential role for this putative third binding site in the HCV life cycle are ongoing.
In conclusion, this is the first use of structure-based mapping to characterize a miR-122:HCV complex. Moreover, we were able to exploit this structural technique to predict the importance of miR-122 tail nucleotides for miR-122's effect on HCV. While the manuscript was in preparation, Machlin et al. (13) demonstrated, using genetic techniques, that this same tail interaction at site 2 (as well as site 1) is of functional importance to HCV RNA stability.
More generally, microRNA tail interactions with mRNA targets, termed 3′ supplemental sites, have previously been observed (1, 6). However, such sites are thought to play only a limited role in mRNA target recognition (1, 6). Nevertheless, we speculate that the lack of structural tools to detect such interactions have limited the ability to accurately characterize the importance of interactions between microRNA tails and their mRNA targets. SHAPE provides a structural means of overcoming this important limitation and enables the direct mapping of microRNA:target complexes in a high-throughput manner.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jody Puglisi and Valaiporn Rusmintratip for feedback and Peter De Rijk for a custom version of RNAviz.
P.S.P., J.S.G., S.G.P., and M.R.E. designed SHAPE. P.S.P. designed FAST; P.S.P. and S.G.P. ran fragment analyses. E.A.P. performed tissue culture experiments. E.A.P. and S.G.P. performed quantitative reverse transcription-PCR. P.S.P. and E.A.P. wrote the manuscript; P.S.P., M.E., J.S.G., S.G.P., M.R.E., and E.A.P. edited the manuscript.
This work was supported by RO1AI087917 and a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research (J.S.G.), NRSA grant F32AI082930 (P.S.P.), and a Soros, MedScholars and HHMI Fellowship (E.A.P.).
A patent application describing SHAPE and its uses for structural studies and drug discovery has been filed by Stanford University.
Footnotes
Published ahead of print 9 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Rijk P, Wuyts J, De Wachter R. 2003. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics (Oxford) 19:299–300 [DOI] [PubMed] [Google Scholar]
- 4. Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U. S. A. 106:97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Einav S, et al. 2008. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nature Biotechnol. 26:1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grimson A, et al. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27:91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henke JI, et al. 2008. MicroRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27:3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 9. Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. 2007. The role of site accessibility in microRNA target recognition. Nat. Genet. 39:1278–1284 [DOI] [PubMed] [Google Scholar]
- 10. Kieft JS, et al. 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292:513–529 [DOI] [PubMed] [Google Scholar]
- 11. Low JT, Weeks KM. 2010. SHAPE-directed RNA secondary structure prediction. Methods 52:150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lukavsky PJ. 2009. Structure and function of HCV IRES domains. Virus Res. 139:166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Machlin ES, Sarnow P, Sagan SM. 2011. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. U. S. A. 108:3193–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mathews DH. 2005. RNA secondary structure analysis using RNAstructure. Curr. Prot. Bioinformatics 2005:12.14.11. [DOI] [PubMed] [Google Scholar]
- 15. Mathews DH, et al. 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 101:7287–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127:4223–4231 [DOI] [PubMed] [Google Scholar]
- 17. Mitchelson KR, Cheng J. (ed.). 2001. Capillary electrophoresis of nucleic acids, vol. 1 Humana Press, Totowa, NJ [Google Scholar]
- 18. Mortimer SA, Weeks KM. 2009. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat. Protoc. 4:1413–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sontheimer EJ. 1994. Site-specific RNA crosslinking with 4-thiouridine. Mol. Biol. Rep. 20:35–44 [DOI] [PubMed] [Google Scholar]
- 20. Southern EM. 1979. Measurement of DNA length by gel electrophoresis. Anal. Biochem. 100:319–323 [DOI] [PubMed] [Google Scholar]
- 21. Thomas M, Lieberman J, Lal A. 2010. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 17:1169–1174 [DOI] [PubMed] [Google Scholar]
- 22. Weeks KM. 2010. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 20:295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilkinson KA, et al. 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 6:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1:1610–1616 [DOI] [PubMed] [Google Scholar]
- 25. Wilkinson KA, et al. 2009. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA 15:1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.