Abstract
Noroviruses are the primary cause of epidemic gastroenteritis in humans, and GII.4 strains cause ∼80% of the overall disease burden. Surrogate neutralization assays using sera and mouse monoclonal antibodies (MAbs) suggest that antigenic variation maintains GII.4 persistence in the face of herd immunity, as the emergence of new pandemic strains is accompanied by newly evolved neutralization epitopes. To potentially identify specific blockade epitopes that are likely neutralizing and evolving between pandemic strains, mice were hyperimmunized with GII.4-2002 virus-like particles (VLPs) and the resulting MAbs were characterized by biochemical and immunologic assays. All of the MAbs but one recognized GII.4 VLPs representing strains circulating from 1987 to 2009. One MAb weakly recognized GII.4-1987 and -1997 while strongly interacting with 2002 VLPs. This antibody was highly selective and effective at blocking only GII.4-2002-ligand binding. Using bioinformatic analyses, we predicted an evolving GII.4 surface epitope composed of amino acids 407, 412, and 413 and subsequently built mutant VLPs to test the impact of the epitope on MAb binding and blockade potential. Replacement of the 2002 epitope with the epitopes found in 1987 or 2006 strains either reduced or ablated enzyme immunoassay recognition by the GII.4-2002-specific blockade MAb. These data identify a novel, evolving blockade epitope that may be associated with protective immunity, providing further support for the hypotheses that GII.4 norovirus evolution results in antigenic variation that allows the virus to escape from protective herd immunity, resulting in new epidemic strains.
INTRODUCTION
Noroviruses (NoVs) are the leading cause of severe viral gastroenteritis and are responsible for 50% of all acute gastroenteritis outbreaks in the United States and Europe (3). Although the severity of disease is usually moderate, lasting 1 to 3 days, infection can be especially virulent in young children, the elderly, and the immunocompromised (2, 18, 21, 22, 24, 35, 40). It is estimated that 200,000 people die each year from NoV infections, primarily children in the developing world (36). An effective vaccine would be particularly advantageous for young and aged populations, military personnel, food handlers, child and health care providers, cruise ship passengers, and citizens in the developing world. A central obstacle that blocks NoV vaccine development is the lack of understanding of the extensive antigenic diversity between the large number of NoV strains and the complex interrelationships between host protective immunity and virus antigenic heterogeneity.
NoVs are grouped by the major capsid protein amino acid sequence. Viruses with less than 14.3% difference are classified as belonging to the same strain, those with 14.3 to 43.8% difference are classified as having the same genotype, and those with 45 to 61.4% difference are classified as belonging to the same genogroup (49). Currently, NoVs are grouped into five genogroups (GI to GV). Viruses of GI and GII are responsible for most human infections. GI and GII are further subdivided into 8 and 21 different genotypes, respectively (3, 49). Over the past 2 decades, the majority of NoV outbreaks have been caused by strains within the GII.4 genotype. Between 1995 and 2006, four major NoV pandemics associated with evolving GII.4 strains have been documented based on molecular and clinical disease occurrences. During the mid-1990s (34), strain US95/96 was responsible for ∼55% of the NoV outbreaks in the United States and 85% of the outbreaks in the Netherlands (46). In 2002, the US95/96 strain was replaced by the Farmington Hills strain (47), which was associated with ∼80% of the NoV outbreaks (19) in the United States. In 2004, the Hunter GII.4 variant was detected in Australia, Europe, and Asia (8, 25, 37). This strain was subsequently replaced in 2006 by two new cocirculating GII.4 variants in the United States and Europe, Laurens (2006a) and Minerva (2006b) (2, 25, 42). In 2009, a new GII.4 variant emerged (GII.4 New Orleans), causing the majority of outbreaks in the United States, while the Minerva (2006b) strain also continues to circulate (3).
NoVs are ∼38-nm icosahedral viruses with an ∼7.5-kb single-stranded, positive-sense RNA genome that encodes three large open reading frames (ORFs). ORF1 encodes the replicase polyprotein, while ORF2 and ORF3 encode the major and minor capsid proteins, respectively. Expression of the major capsid protein (ORF2) in baculovirus (23) and Venezuelan equine encephalitis (VEE) virus (4) results in the formation of virus-like particles (VLPs) composed of 180 copies of the monomeric protein (38). The monomer is structurally divided into the shell domain (S), which forms the core of the particle, and the protruding domain (P) that extends away from the core. The P domain is further subdivided into the P1 subdomain (residues 226 to 278 and 406 to 520) and the P2 subdomain (residues 279 to 405) (38). P2 represents the most exposed polypeptide on the surface of the viral particle and determines its interactions with potential neutralizing antibodies and with histo-blood group antigens (HBGAs) (10, 12, 28, 29, 32). The P2 domain of the major capsid protein of GII.4 strains is evolving rapidly, resulting in new epidemic strains with altered ligand binding properties and antigenicity (1, 5, 7, 15, 29, 41, 43).
To further study GII.4 antigenic variation, we prepared panels of both time-ordered GII.4 VLPs and mouse monoclonal antibodies (MAbs) to compare epitopes between VLPs that represent various stages of NoV evolution. VLPs were constructed representing each of the pandemic strains (GII.4-1997 [US95/96], -2002 [Farmington Hills], -2004 [Hunter], and -2006 [Minerva]), an ancestral strain that circulated prior to the first pandemic outbreak (GII.4-1987), a strain circulating during the brief period between the 2004 and 2006 pandemic strains (GII.4-2005), and a newly emergent GII.4 New Orleans strain (GII.4-2009). The lack of cell culture or small-animal models for NoV cultivation has restricted the study of neutralization antibodies and epitopes to in vitro assays that measure the ability of an antibody to “block“ the binding of a VLP to an HBGA ligand. This in vitro assay has proven to be highly sensitive, as it differentiates between NoV strains too similar to be distinguished by enzyme immunoassay (EIA). The clinical relevance of the blockade assay, as a surrogate neutralization assay, has been confirmed in both infected chimpanzees (6) and Norwalk virus-infected humans (39).
Using EIAs and a surrogate neutralization assay based on VLP-HBGA interactions, GII.4-1987 and -1997 were antigenically indistinguishable from each other as measured by human NoV outbreak sera, VLP-immunized mouse sera, and mouse MAbs (9, 28, 29). The VLPs of strains circulating post-2002 had significantly less reactivity with sera directed against earlier strain VLPs and minimal reactivity to mouse MAbs directed against GII.4-1987. Conversely, selected mouse MAbs generated against GII.4-2006 reacted only with VLPs that circulated from 2002 or later. These data suggested that the pandemic GII.4-2002 was an important antigenic hub, while it retained important antigenic features (blockade epitopes) that characterized the first GII.4 pandemic strain, GII.4-1997, it also had new antigenic features (blockade epitopes) that were retained in the 2006 pandemic strain, GII.4-2006. No blockade antibodies were found to be in common between GII.4-1987 and GII.4-2006.
To further investigate the complex antigenic profile of the pandemic GII.4-2002 strain, relative to other GII.4 outbreak strains, we created a panel of mouse MAbs directed against GII.4-2002 and compared the reactivity of these MAbs to that of a panel of time-ordered GII.4 VLPs using EIAs and surrogate neutralization assays. Combining these empirical data with bioinformatic analyses, a GII.4-2002-specific neutralizing epitope was predicted and genetic approaches were used to construct mutant VLPs with chimeric epitopes. EIA and blockade assays were then used to test the mutant VLPs to confirm the unique GII.4-2002 blockade epitope. These data further support the hypothesis that GII.4 NoV evolution results in antigenic variation of “neutralizing” epitopes defined by antibody blockade assays and thus confirms that protective herd immunity is a driving force in NoV evolution.
MATERIALS AND METHODS
VLPs.
A diverse panel of VLPs representing GI and GII NoV strains and epitope E mutants was assembled. Briefly, for all constructs except GII.4 New Orleans, ORF2 constructs were inserted directly into the VEE virus replicon vector for the production of virus replicon particles (VRPs), and VLPs were expressed in VRP-infected BHK cells and purified by velocity sedimentation in sucrose. New clones were verified as approximately 35-nm particles visualized by negative-staining electron microscopy (4, 30). The GII.4 New Orleans (2009) VLPs were expressed in the baculovirus system and purified by cesium chloride gradient centrifugation. VLP protein concentrations were determined using the BCA protein assay (Pierce).
Mouse immunization, hybridoma production, and IgG purification.
MAbs were produced and purified by the University of North Carolina—Chapel Hill Immunology Core (http://mabs.unc.edu). Briefly, to develop the anti-GII.4-2002 MAbs, Swiss Webster mice were immunized on days 0, 20, 40, and 80 with 50 μg of VLP in PBS plus 50 μl GERBU adjuvant (Fisher Scientific) for a total of 100 μl and splenocytes were fused on day 84. The resulting hybridomas were subcloned by limiting dilution, isotyped (Roche), and purified by protein G chromatography (GE Healthcare).
EIAs.
MAb reactivity was determined by EIA as reported previously (26, 28). Briefly, plates were coated with 1 μg/ml VLP in phosphate-buffered saline (PBS) before the addition 2 μg/ml purified IgG and incubation, followed by anti-mouse–alkaline phosphatase and color development with pNPP substrate solution (Sigma Chemicals). Each step was followed by washing with PBS–0.05% Tween 20, and all antibodies were diluted in 5% dry milk in PBS–0.05% Tween 20. The EIA data shown in the figures represent the average of at least three replicates, and these data are representative of similar data from at least two independent trials. We established these EIAs using the new MAbs and included PBS and genogroup I VLP-coated wells as negative controls and polyclonal anti-VLP mouse sera as positive controls. Antibodies were considered positive for reactivity if the mean optical density, after background subtraction, of the VLP-coated wells was greater than three times the mean optical density of the PBS-coated wells. A two-way analysis of variance (ANOVA) with Bonferroni's multiple-comparison test was used to compare BT50 values of an antibody with different VLPs. A difference was considered significant if the P value was <0.05.
HBGA expression in commercial mucins.
To characterize the HBGAs present in commercial pig gastric mucin (PGM) type III (Sigma Chemicals), mucin was solvated in PBS at 5 mg/ml and used at 10 μg/ml in PBS to coat EIA plates, which were incubated for 4 h at room temperature, and blocked by incubation overnight at 4°C in 5% dry milk in PBS–0.05% Tween 20 before the addition of anti-HBGA MAbs for 1 h of incubation at 37°C. Antibodies to blood group antigen A or B (Ortho Clinical Diagnostics, Rochester, NY) were used at a 1/20 dilution, and antibodies to H, Lewis A, Lewis B, Lewis X (Santa Cruz Biotechnology, Santa Cruz, CA), or Lewis Y (Calbiochem, Gibbstown, NJ) were used at 1 μg/ml. Primary antibody incubations were 1 h at 37°C and were followed by a cocktail of anti-mouse IgM/IgG-horseradish peroxidase (HRP; Sigma Chemicals/GE Healthcare) for 30 min at 37°C and color development with 1-Step Ultra TMB (3,3′,5,5′-tetramethylbenzidine) ELISA (enzyme-linked immunosorbent assay) HRP substrate solution. Each step was followed by a washing step (PBS–0.05% Tween 20), and all of the antibodies were diluted in 5% dry milk in PBS–0.05% Tween 20. Samples were assayed in triplicate. HBGA expression was considered positive if the mean optical density of coated wells, after background subtraction, was greater than three times the mean optical density of PBS-coated wells. As PGM is a crude preparation, the HBGA phenotype of each lot should be verified.
VLP binding to synthetic carbohydrates.
Biotinylated synthetic HBGAs (Bi-HBGA; Glycotech, Gaithersburg, MD) were bound to Neutri-avidin-coated plates (Pierce) at 10 μg/ml for 1 h prior to the addition of 1 μg/ml VLP for 1.5 h. VLP binding was detected with a mouse MAb, followed by rabbit anti-mouse IgG-HRP and 1-Step Ultra TMB ELISA HRP substrate (Fisher Scientific). All of the incubations were done at room temperature. Each step was followed by washing with PBS–0.05% Tween 20, and all reagents were diluted in 5% dry milk in PBS–0.05% Tween 20. VLP binding to synthetic HBGAs was considered positive if the mean optical density, after background subtraction (no HBGA coating), of HBGA-coated wells was greater than three times the mean optical density of PBS-coated wells.
VLP binding to PGM.
To evaluate VLP binding to PGM, EIA plates were coated with PGM as described above for HBGA expression of PGM. VLPs (1 μg/ml) were added to the coated plates for 1 h of incubation, followed by a cocktail of rabbit anti-GI and anti-GII NoV polyclonal sera and mouse MAb anti-GII.4-2006-G6 (28) for 45 min of incubation. Subsequently, a mixture of anti-rabbit/anti-mouse IgG-HRP was added for 30 min of incubation and color was developed with 1-Step Ultra TMB ELISA HRP substrate solution. Each step was followed by washing with PBS–0.05% Tween 20, and all VLPs and antibodies were diluted in 5% dry milk in PBS–0.05% Tween 20. Samples were assayed in triplicate. VLP binding to mucin was considered positive if the mean optical density, after background subtraction, of mucin-coated wells was greater than three times the mean optical density of PBS-coated wells.
Synthetic carbohydrate binding blockade.
VLP-HBGA binding blockade experiments were conducted as previously reported by our group (28, 29), with minor modifications. Synthetic HBGAs were bound to Neutri-avidin-coated plates at 10 μg/ml for 1 h prior to the addition of MAb-pretreated VLP at 1 μg/ml for 1.5 h of incubation. VLP binding was detected with rabbit anti-NoV antiserum, followed by anti-rabbit-IgG-HRP and TMB substrate. The percent control binding was defined as the binding level in the presence of antibody pretreatment compared to the binding level in the absence of antibody pretreatment multiplied by 100. The HBGA ligands used were H type 3 for GII.4-1987 and B trimer for GII.4-1997, -2002, -2005, -2006, and -2009 (9, 29). All of the incubations were done at room temperature. Each step was followed by washing with PBS–0.05% Tween 20, and all of the reagents were diluted in 5% dry milk in PBS–0.05% Tween 20. Two criteria were used to designate an antibody “blockade competent” (28) (i) a positive dose-response relationship between antibody treatment and mean percent control binding, as indicated by a negative slope significantly different from zero by linear regression analysis, and (ii) at least 50% blockade of the VLP-HBGA interaction within the dilution series tested. The 50% blockade titer (BT50) was defined as the lowest MAb concentration tested that blocked at least 50% of VLP binding compared to levels determined in the absence of antibody pretreatment. Samples that did not reach a BT50 by the antibody concentration were assigned a BT50 value equal to 2× the maximum concentration tested for statistical analysis. Serum samples that blocked >50% binding at the lowest concentration tested were assigned a BT50 value equal to 1/2× the minimum concentration tested for statistical analysis (29). A one-way ANOVA with Bonferroni's multiple-comparison test was used to compare the BT50 values of an antibody with different VLPs. A difference was considered significant if the P value was <0.05.
Mucin binding blockade.
VLP-mucin binding blockade experiments were conducted as described for VLP-mucin binding and synthetic carbohydrate binding blockade assays, except that antibody-pretreated VLPs were used at 0.5 μg/ml and incubated with PGM for 1 h. A blockade antibody was defined as described above.
Western blot analysis.
GII.4-2002 VLP was suspended in Laemmli loading buffer, and 9 μg of protein was loaded onto a prep-well 7.5% SDS-polyacrylamide gel (Bio-Rad, Hercules, CA). After electrophoresis, proteins were transferred to Immuno-Blot polyvinylidene difluoride (PVDF) membrane (Bio-Rad) and blocked. Immobilized proteins were probed with 1 and 5 μg/ml MAb. Antibody-reactive protein was visualized with goat anti-mouse IgG-HRP secondary antibody (GE Healthcare) and the ECL Western blotting detection kit (GE Healthcare). All of the incubations were done at room temperature. Each step was followed by washing with PBS–0.05% Tween 20, and all reagents were diluted in 5% dry milk in PBS–0.05% Tween 20.
Structural models of VLP P domains.
The amino acid sequences of GII.4.1987, GII.4.2002, and GII.4.2006 VLPs were individually aligned with the VA387 P domain sequence using ClustalX 1.86 (14), and the VA387 P domain dimer X-ray crystal structure (PDB accession no. 2OBT) (10) was used as a template for generating homology models. Homology models were generated using the program Modeler available via the Max Planck Institute Bioinformatics Toolkit (http://toolkit.tuebingen.mpg.de/). The structural models were analyzed and compared, and figures were generated using Mac Pymol (Delano Scientific).
RESULTS
Anti-GII.4-2002 MAb reactivity.
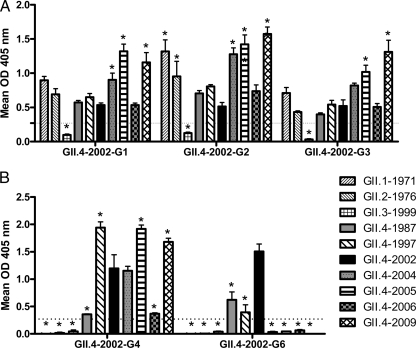
The capsid genes of the GII.4 NoVs are undergoing epochal evolution resulting in phenotypic changes at the level of antigenicity and ligand binding affinity (9, 28, 29). Our comprehensive study of GII.4 antigenicity over time, utilizing mouse MAbs to the ancestral strain GII.4-1987 and a more contemporary strain, GII.4-2006, illustrated that the pandemic strain GII.4-2002 was divergent from both GII.4-1987 and GII.4-2006 but retained some epitopes of each strain. These findings suggest that GII.4-2002 may be an antigenic switching point that set the stage for significant GII.4 evolution and antigenic change. To test this hypothesis, we developed a panel of mouse MAbs to GII.4-2002 (Table 1) and evaluated MAb reactivity across a large panel of NoV VLPs (Fig. 1). As observed with the MAbs directed against other GII.4 VLPs, none of the anti-GII.4-2002 MAbs recognized any of the GI VLPs tested (data not shown). Three of the five MAbs (G1, G2, and G3) reacted across the panel of 1987 to 2009 time-ordered GII.4 VLPs and with additional GII VLPs representing GII.1-1971 and GII.2-1976. All three of these MAbs had similar reactivity patterns to each VLP, indicating that they may share an epitope. Two MAbs (G4 and G6) recognized only the time-ordered panel of GII.4 VLPs. Anti-2002-G4 reactivity was variable between the GII.4 VLPs. Anti-GII.4-2002-G6 detected only a subset of GII.4 VLPs representing early strains GII.4-1987, GII.4-1997, and GII.4-2002, the same pattern identified for antibodies generated against GII.4-1987. These EIA data suggest that our panel of anti-GII.4-2002 MAbs recognize at least two different epitopes on GII.4 NoV capsids.
Table 1.
MAbs developed in this study
| Clone | Species | Clone no. | Immunogen | Isotype |
|---|---|---|---|---|
| GII.4-2002G1 | Mouse | 66.72.12.12 | GII.4-2002 | IgG2b |
| GII.4-2002G2 | Mouse | 66.691.7.2 | GII.4-2002 | IgG2a |
| GII.4-2002G3 | Mouse | 66.757.1.7 | GII.4-2002 | IgG2b |
| GII.4-2002G4 | Mouse | 66.71.4.2 | GII.4-2002 | IgG2b |
| GII.4-2002G6 | Mouse | 66.573.13.2 | GII.4-2002 | IgG1 |
Fig 1.
EIA cross-reactivity of anti-GII.4-2002 MAbs to NoV VLPs. Purified MAbs were assayed by EIA for reactivity to GI and GII NoV VLPs. (A) MAbs with cross-genogroup reactivity. (B) MAbs with cross-genotype reactivity. Bars represent means with standard errors. Positive reactivity was defined as a mean optical density (OD) at 405 nm ≥3 time that of the background (dashed line). Asterisks indicate VLPs with reactivity significantly different from that of GII.4-2002.
Characterization of a commercially available PGM as a substrate for NoV surrogate neutralization assays.
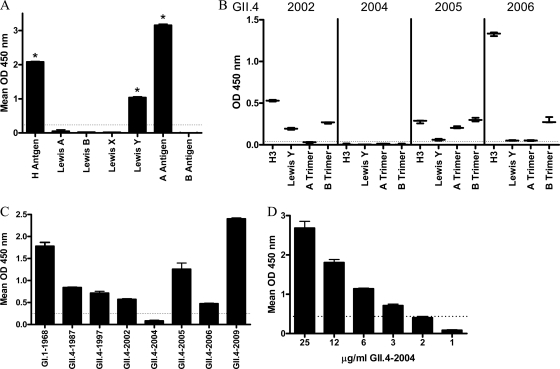
EIAs provide information on the presence or absence of an epitope on a VLP. Neutralization assays provide information on the function of an antibody in protection from infection. In the absence of a cell culture model, blockade of VLP binding to Bi-HBGAs has been used extensively as a NoV surrogate neutralization assay (20, 27, 28). However, the high cost, limited availability, and limited diversity of commercial synthetic HBGAs have hampered progress in the field. Robust human NoV VLP binding to PGM has been reported (44, 45), reflecting the native HBGA milieu in biological samples. To determine if commercial mucin preparations could serve as a biochemically semicharacterized HBGA ligand source in surrogate neutralization assays, we first characterized the HBGA distribution of semipurified PGM type III using MAbs to H antigen, Lewis A antigen, Lewis B antigen, Lewis X antigen, Lewis Y antigen, A antigen, and B antigen. In agreement with previous reports of HBGA distribution in individual animals (11), PGM was positive for relatively high levels of H and A antigens and more moderate levels of Lewis Y antigen (Fig. 2A). Lewis A, Lewis B, Lewis X, and B antigens were not detected (11). These data suggest that PGM may have broad NoV VLP binding properties because of abundantly available α-1,2-fucose (H antigen) and α-1,4-fucose (Lewis antigen) moieties.
Fig 2.
Characterization of PGM HBGA expression and VLP binding. (A) The HBGA phenotype of commercial PGM was determined by reactivity to a panel of anti-HBGA MAbs by EIA. Asterisks indicate HBGAs with reactivity significantly different from that of B antigen. (B) Binding to synthetic HBGA found in PGM of the GII.4 VLPs used in this study and not previously reported. The mean OD is indicated by the line. Whiskers indicate the maximum and minimum values of replicates. (C) PGM binding ability of NoV VLPs determined by incubating GII.4 and GI.1-1968 VLPs with PGM-coated plates, followed by VLP detection with an anti-NoV antibody cocktail. (D) GII.4-2004 binds to PGM at higher VLP concentrations. Bars represent means with standard errors. Positive reactivity was defined as a mean optical density (OD) at 450 nm ≥3 time that of the background (dashed line).
GII.4 VLPs from 1987 to 2002 have all been reported to bind to at least one of the HBGAs abundant in PGM (29). However, synthetic HBGA binding ligands for GII.4-2004, -2005, and -2009 VLPs have not been reported. Therefore, the carbohydrate binding profile of these VLPs was determined (Fig. 2B). GII.4-2002 bound to H type 3, Lewis Y, and B trimer (29, 48). Consistent with our previous report (29), a ligand binding partner for GII.4-2004 VLP was not identified in this assay, perhaps reflecting residue microvariation occurring within the different GII.4-2004 Hunter strains used by different laboratories or the difference between VLP binding compared to P particle binding (29, 48). MAb based detection reagents identified GII.4-2005 VLP binding to H type 3, Lewis Y(weakly), A, and B trimer; as reported for GII.4-2005 P particles (48). Consistent with GII.4-2006 VLP binding profiles, GII.4-2009 bound preferentially to H type 3, Lewis Y, and B trimer and modestly to A trimer. These results indicate that the panel of GII.4 VLPs, except GII.4-2004, bind to the HBGA found in PGM.
To confirm this suggestion, the panel of GII.4 time-ordered VLPs was tested directly for PGM binding. VLP binding to PGM was consistent with synthetic HBGA binding profiles. All of the VLPs tested, except GII.4-2004, bound to H, A, and/or Lewis antigens in the synthetic HBGA assays (Fig. 2B) (9, 29), and all VLPs tested, except GII.4-2004, bound to PGM (Fig. 2C) at a 1 μg/ml VLP concentration. The HBGA binding characteristics of GII.4-2004 have been reported as either less robust than other GII.4 strains using P particles (48) or below the limit of detection using VLPs (Fig. 2B) (29). To determine if GII.4-2004 VLPs were structurally incapable of binding PGM or if they had a relatively weak ligand binding affinity, the PGM binding assay was repeated with increasing concentrations of GII.4-2004 VLP and binding was detected with a cocktail of MAbs and polyclonal antibodies. Under these conditions, dose-dependent binding of GII.4-2004 to PGM was detected, confirming that GII.4-2004 VLPs do bind mucin ligand but with lower affinity than other GII.4 VLPs (Fig. 2D). Threefold more VLP (3 μg/ml) than the other GII.4 VLPs (1 μg/ml) was required to detect binding of GII.4-2004 to PGM. The broad ability of PGMs to bind to NoV VLPs indicates that PGM could be a suitable ligand for NoV surrogate neutralization assays.
Neutralization potential of anti-GII.4-2002 MAbs.
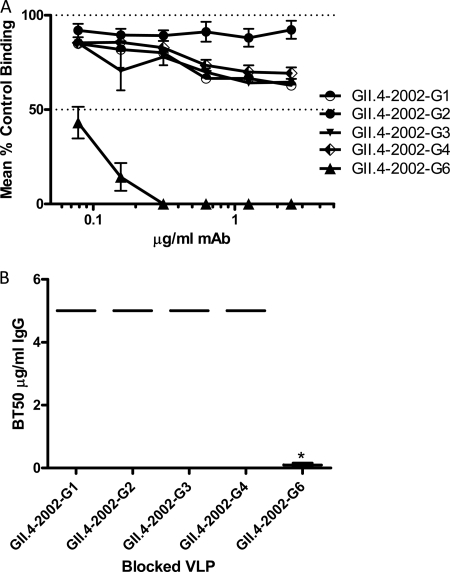
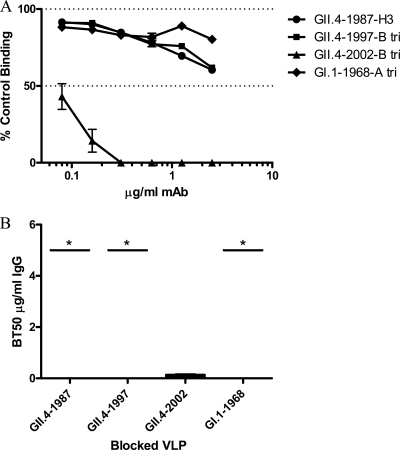
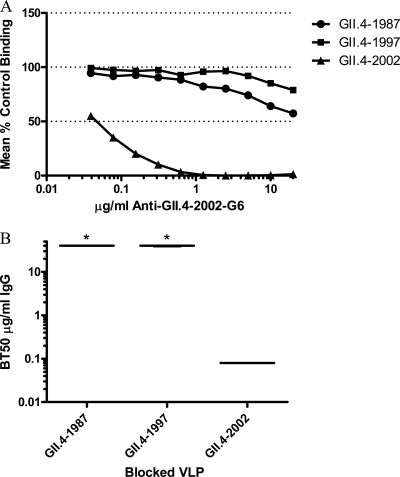
To evaluate the VLP-mucin blockade assay, the blockade phenotype of our panel of anti-GII.4-2002 MAbs was compared using both the traditional VLP-synthetic HBGA and VLP-PGM blockade assays. The ability of the anti-GII.4-2002 MAbs to block the interaction of GII.4-2002 with Bi-B trimer was determined first. The three antibodies (G1, G2, and G3) with broad reactivity across GII VLPs by EIA and anti-GII.4-2002-G4 were unable to block homotypic VLP binding to Bi-B trimer (Fig. 3). Anti-GII.4-2002-G6 did block homotypic VLP interactions (BT50, 0.08 μg/ml) (Fig. 3) but was unable to block any other GII.4 VLP interaction with synthetic HBGAs (Fig. 4). In agreement with Bi-HBGA blockade results, anti-GII.4-2002-G6 efficiently blocked the GII.4-2002 VLP-PGM interaction (BT50, 0.08 μg/ml) (Fig. 5). This blockade was selective, as the antibody had no effect on the GII.4-1987 or GII.4-1997 VLP interactions with PGM or Bi-HBGA (Fig. 4 and 5), despite limited EIA reactivity with both VLPs. Anti-GII.4-2002-G6 did not recognize any VLPs post-2002 by EIA; therefore, no additional VLPs were assessed for blockade. The synthetic HBGA and the PGM-based surrogate neutralization assays provided similar measures of the antibody's ability to block VLP-ligand interaction and indicate that MAb GII.4-2002-G6 recognizes a GII.4-2002-specific blockade epitope.
Fig 3.
Homotypic blockade of GII.4-2002 VLP-B trimer interaction by anti-GII.4-2002 MAbs. (A) GII.4-2002 VLPs were incubated with increasing concentrations of anti-GII.4-2002 MAbs, and the mean percent control binding was calculated by comparing the amount of VLP bound in the presence of antibody pretreatment to the amount of VLP bound in the absence of antibody pretreatment. Error bars represent the standard errors of the means. (B) Mean MAb concentration (μg/ml) needed to block 50% of GII.4-2002 ligand binding. The mean titer is indicated by the line in the graph. The upper and lower broken lines in the graph represent the maximum and minimum values. The asterisk indicates an antibody with a BT50 significantly different from that of GII.4-2002-G1.
Fig 4.
Heterotypic blockade of GII.4 VLP-Bi-HBGA interactions by anti-GII.4-2002-G6. (A) Increasing concentrations of anti-GII.4-2002-G6 antibody were incubated with additional GII.4 VLPs positive for EIA reactivity and GI.1-1968 as a negative control. The mean percent control binding was calculated by comparing the amount of VLP bound to Bi-HBGA in the presence of antibody pretreatment to the amount of VLP bound in the absence of antibody pretreatment. Error bars represent the standard errors of the means. (B) The Mean MAb concentration (μg/ml) needed to block 50% of GII.4 VLP ligand binding is indicated by the line in the graph. The upper and lower broken lines in the graph represent the maximum and minimum values. Asterisks indicate VLPs with BT50s significantly different from that of GII.4-2002.
Fig 5.
Blockade of GII.4 VLP-PGM interactions by GII.4-2002-G6. Increasing concentrations of anti-GII.4-2002-G6 were incubated with GII.4 VLPs positive for EIA reactivity, and the mean percent control binding was calculated by comparing the amount of VLP bound to PGM in the presence of antibody pretreatment to the amount of VLP bound in the absence of antibody pretreatment. Error bars represent the standard errors of the means. (B) The Mean MAb concentration (μg/ml) needed to block 50% of GII.4 VLP ligand binding is indicated by the line in the graph. The upper and lower broken lines in the graph represent the maximum and minimum values. Asterisks indicate VLPs with BT50s significantly different from that of GII.4-2002.
Epitope characterization by Western blot analysis.
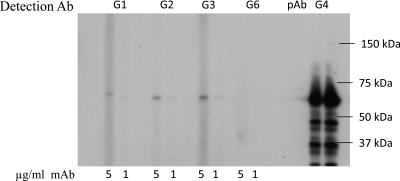
Initial epitope characterization was done via Western blot analysis to determine if the epitopes recognized by the panel of MAbs were conformational or linear sequences. As expected, MAbs without VLP-ligand binding blockade activity (GII.4-2002-G1 to -G4) recognized denatured VLP and the MAb with VLP-ligand binding blockade activity (GII.4-2002-G6) did not recognize a linear epitope, as measured by Western blot reactivity (Fig. 6). These data, combined with EIA data demonstrating broad GII.4 reactivity of GII.4-2002-G1, -G2, -G3, and -G4, suggest that these antibodies may recognize highly conserved linear epitopes and may be suitable for diagnostics.
Fig 6.
Detection of GII.4-2002 by Western blot analysis with anti-GII.4-2002 MAbs. GII.4-2002 VLPs were denatured, electrophoresed, and transferred to PVDF before being probed with 5 μg/ml and 1 μg/ml MAb. Mouse polyclonal anti-GII.4-2002 serum was used as a positive control for detection.
Epitope mapping of anti-GII.4-2002-G6.
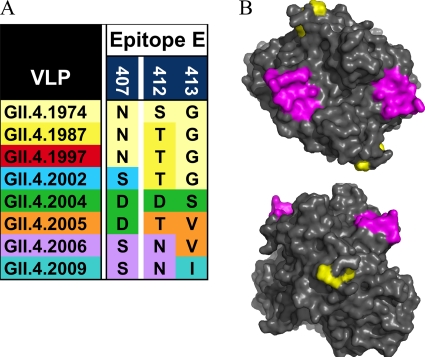
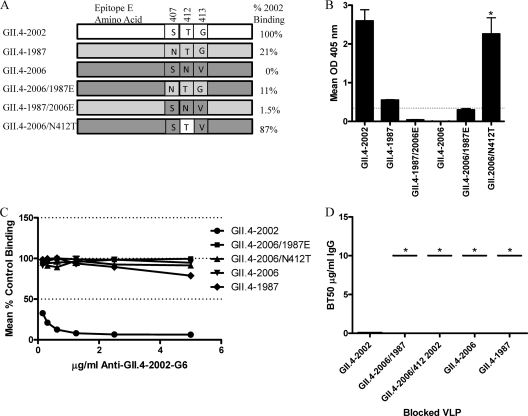
Previous work has indicated that blockade epitopes are located in the P2 subdomain and are likely dependent on the appropriate confirmation (17, 28, 31). Bioinformatic analyses identified five putative epitopes based upon variation that maps to the P2 subdomain surface of the NoV capsid (16). Epitope E, composed of surface-exposed residues 407, 412, and 413, is located in the P2 subdomain lateral to the receptor binding sites on the top of the surface. Because epitope E is located distal to the other epitopes, encodes significant amino acid change over time, and varies among 1987, 2002, and 2006; this putative epitope was selected for further characterization (Fig. 7). Using the GII.4.1987, GII.4.2002, and GII.4.2006 VLPs and capsid sequences as a guide, mutants were engineered that contained chimeric combinations of epitope E in the GII.4-1987 and GII.4-2006 wild-type backgrounds (Fig. 8). To test if any of the anti-GII.4-2002 MAbs interacted with epitope E, exchange mutant VLPs were constructed (Fig. 8A) and reactivity to anti-GII.4-2002 MAbs was evaluated by EIA. Anti-GII.4-2002-G6 recognized the ancestral GII.4-1987 VLP but not the contemporary GII.4-2006 VLP by EIA (Fig. 1B). Epitope E of the nonreactive GII.4-2006 VLP was put into the reactive GII.4-1987 backbone, resulting in VLP GII.4-1987/2006 E. This exchange resulted in complete loss of GII.4-2002-G6 binding (Fig. 8B). Conversely, when epitope E of the reactive GII.4-1987 VLP was put into the nonreactive GII.4-2006 backbone, creating VLP GII.4-2006/1987 E, binding of MAb GII.4-2002-G6 was gained, although at low levels near the 3-fold background. To test the impact of amino acid 412 within epitope E, N412 of GII.4-2006 was replaced with T412 of GII.4-1987, resulting in VLP GII.4-2006/N412T (Fig. 9A). This amino acid change restored the original 2002 sequence at 407 and 412 and converted a GII.4-2002-G6 nonbinding backbone into a binding VLP. MAb binding was restored to approximately 87% of GII.4-2002 levels (Fig. 8B). Consistent with a lack of blockade of GII.4-1987, epitope E exchange mutations into the 2006 backbone did not restore the GII.4-2002 blockade phenotype (Fig. 8C and D). However, exchanging the entire epitope E sequence and amino acid 412 alone between the reactive and nonreactive GII.4 backbones allowed us to determine that the E epitope itself is impacting GII.4-2002-G6 binding and indicates that epitope E is a potential GII.4-2002 neutralization epitope that varies over time. These exchanges did not impact the binding of any of the other anti-GII.4-2002 MAbs (data not shown). Further, VLPs with changes in other predicted epitopes (16) were tested and did not show differential reactivity to any of the anti-GII.4-2002 MAbs (data not shown). These data identify epitope E as a GII.4-2002-specific blockade epitope.
Fig 7.
Epitope E was defined by variable residues 407, 412, and 413. (A) Variation in epitope E within GII.4 strains. (B) Epitope E (yellow) and HBGA binding sites (magenta) mapped onto the P domain dimer of GII.4-2002.
Fig 8.
Characterization of epitope E, a GII.4-2002-specific potential neutralization epitope. (A) VLPs engineered to contain variations of epitope E were assembled to test the impact of the epitope and the nonepitope backbone on anti-GII.4-2002-G6 binding and neutralization. (B) Anti-GII.4-2002-G6 EIA reactivity to epitope E constructs. Asterisks indicate chimeric VLPs with reactivity significantly different from that of the parental VLPs. (C) Blockade of epitope E constructs binding to PGM by anti-GII.4-2002-G6. (D) The mean MAb concentration (μg/ml) needed to block 50% of GII.4 VLP ligand binding is indicated by the line in the graph. The upper and lower broken lines in the graph represent the maximum and minimum values. Asterisks indicate VLPs with BT50s significantly different from that of GII.4-2002.
Fig 9.
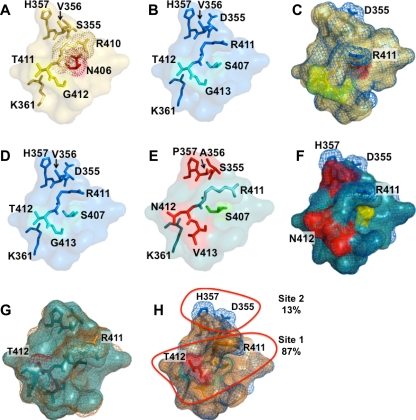
Variation in epitope E alters capsid structure. Epitope E was defined by variable residues 407, 412, and 413 (406, 411, and 412 for GII.4.1987) and all residues within 8 Å of these sites, as these additional residues are likely to be impacted by the structural differences (driven by the variable sites) that contribute to MAb recognition differences. GII.4.1987 epitope E (A) differs from GII.4.2002 epitope E (B) at positions 406/407 and 355. The N-to-S difference at 406/407 likely alters the rotameric position of R411 in the GII.4.2002 VLP, allowing it to extend further from the surface. (C) GII.4.1987 (yellow) and GII.4.2002 (blue) superimposed. Because R411 is more buried in 1987, the MAb likely cannot interact as strongly with this residue in the GII.4.1987 VLP. GII.4-2002 (D) differs from GII.4-2006 (E) at five positions in the expanded epitope (positions 355 to 357, 412, and 413). (F) Superimposition suggests that R411 of GII.4.2006 (teal) is buried, and variation at 355 to 357 may alter key interactions involving H357 and D355. Red, differences between epitopes; yellow, key residues. (G) GII.4-2006/N412T (orange) superimposed upon GII.4.2006. Two potentially important residues are R411, which is more surface exposed in GII.4-2006/N412T, and T412, which is buried. (H) GII.4-2002 (blue) superimposed on GII.4-2006/N412T (orange). R411 is nearly identical, suggesting that N412T frees the R411 side chain to extend away from the surface, where it likely interacts with the MAb. Resides that regulate R411 make up site 1, and 87% of binding can be recovered with structural modifications to R411. A second site, site 2, is composed of residues 355 to 357, and particularly D355, which adds negative potential to the second site.
Modeling of amino acid variation in epitope E over time.
Residues 407, 412, and 413 were identified as key amino acids that were exposed to the surface and that changed frequently over time, suggesting a role in escape from herd immunity. Because a potential MAb binding site is larger than three amino acids, residues within 8 Å of the key amino acids were analyzed to determine the impact of amino acid replacements at positions 407, 412, and 413 on the local structural neighborhood. This distance was selected because it accounted for most of the residues that were localized around the E epitope and included all of the residues that would likely be directly impacted by changes at positions 407, 412, and 413. By expanding from these three sites to include residues within 8 Å of those sites, additional variations at positions 355 to 357 were noted (Fig. 9).
Modeling of the predicted impacts of amino acid variation provides potential mechanistic insights into antibody-VLP interactions at a specific site. Structural modeling predicted that the primary interaction between GII.4.2002 and MAb GII.4-2002-G6 appears to be amino acid R411, which is invariant among GII.4 strains after 1977 (Fig. 9). Importantly, R411 appears to extend from the surface of the GII.4-2002 capsid, where it is well positioned to directly interact with potential MAb binding partners. R411 has variant rotameric positions that are likely regulated by S407 and T412 of GII.4-2002, which are uncharged and hydrophilic and do not directly interact with R411 in the GII.4-2002 homology model (Fig. 9). In contrast, GII.4-1987 epitope E differs from GII.4-2002 epitope E at two positions, 407 (position 406 in GII.4-1987) and 355. GII.4-2002 encodes S407 and D355, while GII.4-1987 encodes N406 and S355. The N-to-S difference at 406/407 likely alters the rotameric position of R411 in the GII.4.1987 capsid, where N406 likely interacts with R411, preventing the side chain from extending to the surface and changing the surface topology. This likely reduces the ability of MAb GII.4-2002-G6 to recognize and bind to the GII.4-1987 VLP. In addition, D355 of GII.4-2002 is likely an additional residue that is targeted by MAb GII.4-2002-G6, as the negative charge of this residue extends from the VLP surface (Fig. 9). Structural studies on cocrystals are needed to validate these predictions.
The GII.4-2006 VLP is different from the GII.4-2002 VLP at five positions, 355 to 357, 412, and 413 (Fig. 9). Interestingly, MAb GII.4-2002-G6 does not bind to GII.4-2006. This is likely due to at least three structural modifications. (i) Changing 355DVH357 of GII.4-2002 to 355SAP357 of GII.4-2006 removes the negative potential of D355 from the VLP surface, which may be a target of MAb GII.4-2002-G6. (ii) Changing T412 of GII.4-2002 to N412 of GII.4-2006 likely alters the rotameric position of R411, causing it to be more buried and therefore less exposed to the surface, where it can interact with the MAb. (iii) The combination of N412 and V413, as encoded in the GII.4-2006 VLP, alters the structure of the putative MAb GII.4-2002-G6 binding site, adding bulky side chain modifications that are predicted to interfere with antibody binding (Fig. 9D, E, and F).
DISCUSSION
Understanding the molecular mechanisms governing GII.4 antigenic variation is key to designing immunotherapeutics and vaccines and developing diagnostic predictions regarding the epidemic potential of newly emergent strains. The study of antigenicity among related GII.4 NoV strains using mouse MAbs developed against a time-ordered panel of GII.4 VLPs has demonstrated two important findings relative to NoV vaccine design. First, the major antigenic determinant of GII.4 NoVs (the major capsid protein) is undergoing antigenic variation over time. Second, antibodies can be developed that recognize and potentially neutralize a broad range of GII.4 strains. Detailed studies utilizing our panel of time-ordered GII.4 VLPs and mouse MAbs revealed that antibodies that similarly blocked 1987/1997 VLP-ligand interactions required higher concentrations to block GII.4-2002 ligand interactions, suggesting that from a neutralization standpoint, the pandemic GII.4-2002 strain is antigenically divergent from the earlier 1987/1997 strains. Divergence at this epitope, and potentially others, may have provided a herd immunity “escape hatch” for GII.4-2002, leading to the second NoV pandemic. This hypothesis is supported by the results of blockade assays utilizing mouse MAbs derived from GII.4-2006 immunizations. MAbs that effectively blocked GII.4-2006-ligand binding either required more MAb for blockade or were completely unable to block GII.4-2002 and these were universally unable to block GII.4-1987 or 1997-HBGA interactions. These data suggest that the divergence of neutralizing epitopes as defined by carbohydrate blockade assays corresponds to new GII.4 pandemic strain emergence and that GII.4-2002 represents an antigenic switching point in GII.4 evolution.
These data indicate that a successful NoV vaccine regimen will require periodic sampling to identify future strains for inclusion in subsequent vaccine formulations. The identification of evolving GII.4 antigenic epitopes provides targeted sites that may be useful for surveillance and prediction of new strain emergence. Therefore, we produced a panel of MAbs to the GII.4-2002 VLP with the goal of identifying epitopes that are changing over time within GII.4 genotypes. Surprisingly, unlike MAbs generated to GII.4-1987 and 2006, anti-GII.4-2002 MAbs demonstrated little difference in EIA reactivity among the time-ordered panel of GII.4 VLPs. Four out of five MAbs recognized the entire panel of time-ordered VLPs. For three MAbs, the reactivity extended to other GII VLPs. The four broadly reactive MAbs detected denatured capsid protein by Western blot analysis, suggesting that these antibodies recognize linear epitopes that are conserved within the genotype. These data suggest that these antibodies (anti-GII.4-2002-G1, -G2, -G3, and -G4) do not recognize neutralizing epitopes, and although not informative about viral antigenic evolution, they do provide potential diagnostic reagents for detection of broader groups of NoV strains. These antibody reactivity patterns are reminiscent of those generated by immunizing mice with P particles (48), as the antibodies react with denatured capsid protein by Western blot analysis but did not block VLP-ligand interactions. These data underscore an important complication of using Western blot analysis to determine the antigenic relatedness of GII.4 strains. As all of the blockade antibodies we have identified to date (Fig. 4 to 6) (28) recognize conformational epitopes and Western blot analysis is restricted to denatured epitope evaluation, this technique confounds any rational interpretation of antigenic relatedness between strains and explains why Western blot analyses have failed to demonstrate antigenic differences between GII.4 strains (48).
Anti-GII.4-2002-G6 is the only MAb identified to be specific for blockade of GII.4-2002. This blockade antibody was also the only GII.4-2002 MAb that did not cross-react with the entire panel of time-ordered GII.4 VLPs but instead detected only additional VLPs GII.4-1987 and GII.4-1997. Using genetic approaches and chimeric VLPs, we identified epitope E, composed of amino acids 407, 412, and 413; as the binding site for MAb GII.4-2002-G6. Our findings characterize the first novel blockade epitope specific for the Farmington Hills pandemic GII.4 strain (GII.4-2002). An interesting feature of epitope E is its location on the surface of the NoV capsid. Unlike other predicted GII.4 epitopes, this epitope resides lateral to the HBGA binding site. In contrast to the carbohydrate binding pockets that occur directly on top of the P2 subdomain; residues 407, 412, and 413 are positioned on the side of the P2 subdomain extending away from the surface. However, expanding the epitope to include all amino acid residues within 8 Å of residues 407, 412, and 413 revealed a second region proximal to these sites that includes residues 355 to 357, which are more exposed to the top of the P2 subdomain. We hypothesize that these residues are part of the same epitope as residues 407, 412, and 413. The inclusion of these residues in an expanded epitope E suggests that MAb GII.4-2002-G6 blocks the ability of the VLP to interact with its ligand by direct hindrance of the binding pocket. Mutants have been designed to determine if amino acid positions 355 to 357 and 407, 412, and 413 make up a single epitope.
Other GII.4 epitopes have been predicted using bioinformatic methods (16) and mapped using molecular biological approaches (14a). Allan et al. (1) recently compared the reactivities of five MAbs against a pre- and post-2002 epidemic GII.4 strain and identified a conformational epitope(s) composed of residues 294 to 296 and 393 to 395. Interestingly, both of these regions were previously predicted to be epitopes using bioinformatic methods (1, 16, 28) and were later verified by molecular analyses (1, 29). The carbohydrate blockade potential of these antibodies was never tested, so the role of this site in escape from herd immunity is uncertain. Unfortunately, none of our VLPs allowed a direct comparison between the two studies but the finding that residues 393 to 395 were antigenically important supported our previously published work identifying amino acid 395 as an antigenic determinant in the GII.4-2002 Farmington Hills strain (29).
Blockade of VLP binding to biotinylated synthetic HBGAs has been used extensively as a NoV surrogate neutralization assay by our group and more recently by others. The key limitation of this assay is the commercial availability of biotinylated HBGAs, which occasionally suffer from inconsistent product availability, variable quality control, and limited offerings compared to the diversity of the biological molecules. Human salivary samples are a potential source of biologically relevant HBGAs, but collection is hampered by the need for institutional review board approval and donor collection (26). Further, each human salivary sample contains variable components in variable quantities/time, depending on an individual's genetics and behaviors, making it nearly impossible to develop a standardized sample pool for the NoV field. To address these limitations, we devised an antibody-mediated VLP-PGM blockade assay based on the ability of NoV VLPs to bind to commercially available, semipurified PGM (45). By nature, the composition of gastric mucin is complex and each lot may require HBGA phenotyping before use as a substrate, but it is inexpensive and available in semipurified lots, providing a platform for the evaluation of VLP-carbohydrate interactions. We used anti-human HBGA MAbs to phenotype the HBGA content of semipurified PGM. These antibodies confirmed the presence of α-1,2-fucose (H antigen), α-1,4-fucose (Lewis antigen), and A antigen for VLP binding. B antigen was not detected, confirming previous reports of porcine HBGA expression (11). Clearly, the lack of expressed B antigen limits the depth of the assay, but it does not limit the functionality of the assay, as all of the VLPs tested by our group and others bind to H, Lewis, or A antigen. Previously, binding of NoV VLPs to HBGAs has been shown to be dependent upon the valency of the carbohydrate (33). Our PGM binding data support this observation. VLP binding to PGM is stronger than binding to synthetic HBGAs, allowing us to use half as much VLP per binding assay and still maintain robust binding signal/noise ratios. For example, GII.4-2004 binding to PGM was detected at 3 μg/ml but binding to synthetic HBGA was not detected at 5 μg/ml. To establish the VLP-PGM blockade assay, the blockade potential of anti-GII.4-2002-G6 was characterized using both the traditional biotinylated HBGAs and the PGM blockade assay. These finding have been replicated for additional panels of anti-NoV MAbs not raised against GII.4-2002. The response curves of both assays were similar for all of the MAbs and polyclonal sera tested (data not shown), indicating that PGM provides a blockade assay platform that is affordable and readily available for use in the increasing numbers of labs interested in NoV surrogate neutralization assays.
The lack of availability of quality-controlled, diverse, biotinylated HBGAs not only impacts the ability of the research community to access potential neutralization of anti-NoV antibodies, it also impacts the key assay used to characterize HBGA binding profiles of NoV strains. Conflicting data on GII.4 ligand binding patterns produced from different labs has been controversial in the NoV field. Even though a consensus is forming around the acknowledgment that altered ligand recognition occurs within the GII.4 strains over time (29), it is important to also acknowledge that GII.4 ligand recognition varies by assay as no two labs have exactly the same set of assay reagents, reagent concentrations, or conditions. Even within our own set of reagents, we have discovered that the choice of detection antibody can impact ligand binding results. Utilizing MAbs rather than polyclonal sera for detection increased the assay sensitivity enough to detect lower levels of biotinylated-HBGA binding. The enhanced detection was apparent only for lower binding signals, such as GII.4-2002-B trimer and GII.4-2005-H type 3, -Le Y, -A, and -B trimer interactions. These refined HBGA binding profile results agree with those published by others and emphasize the complexity of the NoV-HBGA binding assays. A final complicating factor in comparing HBGA binding data between labs is the microvariation within each strain. As has been demonstrated repeatedly, one amino acid difference within the P2 domain can significantly alter VLP-HBGA affinity (13, 15, 29); therefore, making comparisons between data sets is misleading without the accompanying P2 subdomain sequence information.
The data presented here on GII.4-2002-HBGA binding and antigenicity support our hypothesis that GII.4 NoV strains persist in human populations by altering their affinity for HBGA ligands in response to antibody-driven antigenic variations. Both of these features are mediated by epochal evolution of the major capsid protein. Use of MAbs and blockade assays allowed us to demonstrate that the pandemic GII.4-2002 strain is antigenically distinct from the previous pandemic strain GII.4-1997 and the following pandemic strain GII.4-2006, indicating that new pandemic strain emergence is associated with changes in neutralizing epitopes. Identification of a novel GII.4 blockade epitope provides a target for monitoring new strain emergence and potential therapeutics. Finally, the identification of these evolving epitopes will provide key targets for effective vaccine redesign, allowing the rational reformulation of existing vaccine components to target newly emerged contemporary strains over time.
ACKNOWLEDGMENTS
We thank Victoria Madden and C. Robert Bagnell, Jr., of the Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, University of North Carolina—Chapel Hill for expert technical support.
This work was supported by a grant from the National Institutes of Health National Institute of Allergy and Infectious Diseases (AI056351) and a Gillings Innovation Laboratory award from the University of North Carolina Gillings School of Global Public Health.
The findings and conclusions in this article are ours and do not necessarily represent the views of the Centers for Disease Control and Prevention (CDC). This article did receive clearance through the appropriate channels at the CDC prior to submission. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Allen DJ, Gray JJ, Gallimore CI, Xerry J, Iturriza-Gomara M. 2008. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One 3:e1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anonymous. 2007. Norovirus activity–United States, 2006–2007. MMWR Morb. Mortal. Wkly. Rep. 56:842–846 [PubMed] [Google Scholar]
- 3. Anonymous. 2011. Updated norovirus outbreak management and disease prevention guidelines. MMWR Recommend. Rep. 60(RR-3):1–18 [PubMed] [Google Scholar]
- 4. Baric RS, et al. 2002. Expression and self-assembly of Norwalk virus capsid protein from Venezuelan equine encephalitis virus replicons. J. Virol. 76:3023–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bok K, et al. 2009. Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 83:11890–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bok K, et al. 2011. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc. Natl. Acad. Sci. U. S. A. 108:325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bull RA, Eden JS, Rawlinson WD, White PA. 2010. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6:e1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bull RA, Tu ET, McIver CJ, Rawlinson WD, White PA. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44:327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cannon JL, et al. 2009. Herd immunity to GII.4 noroviruses is supported by outbreak patient sera. J. Virol. 83:5363–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao S, et al. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheetham S, et al. 2006. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 80:10372–10381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen R, Neill JD, Estes MK, Prasad BV. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. U. S. A. 103:8048–8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y., et al. 2011. Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens. PLoS Pathog. 7(7):e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chenna R, et al. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a. Debbink K, Donaldson EF, Lindesmith LC, Baric RS. 2012. Genetic mapping of a highly variable norovirus GII.4 blockade epitope: potential role in escape from human herd immunity. J. Virol. 86:1214–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Rougemont A, et al. 2011. Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens. J. Virol. 85:4057–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. 2008. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 225:190–211 [DOI] [PubMed] [Google Scholar]
- 17. Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. 2010. Viral shape-shifting: norovirus evasion of the human immune system. Nat. Rev. Microbiol. 8:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Estes MK, Prasad BV, Atmar RL. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19:467–474 [DOI] [PubMed] [Google Scholar]
- 19. Fankhauser RL, et al. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1–7 [DOI] [PubMed] [Google Scholar]
- 20. Harrington PR, Lindesmith L, Yount B, Moe CL, Baric RS. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 76:12335–12343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harris JP, Edmunds WJ, Pebody R, Brown DW, Lopman BA. 2008. Deaths from norovirus among the elderly, England and Wales. Emerg. Infect. Dis. 14:1546–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hutson AM, Atmar RL, Estes MK. 2004. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. 12:279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang X, Wang M, Graham D, Estes M. 1992. Expression, self-assembly and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koopmans M, et al. 2000. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181(Suppl. 2):S262–S269 [DOI] [PubMed] [Google Scholar]
- 25. Kroneman A, et al. 2006. Increase in norovirus activity reported in Europe. Euro Surveill. 11:E061214.1. [DOI] [PubMed] [Google Scholar]
- 26. Lindesmith L, et al. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9:548–553 [DOI] [PubMed] [Google Scholar]
- 27. Lindesmith LC, et al. 2010. Heterotypic humoral and cellular immune responses following Norwalk virus infection. J. Virol. 84:1800–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lindesmith LC, Donaldson EF, Baric RS. 2011. Norovirus GII.4 strain antigenic variation. J. Virol. 85:231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lindesmith LC, et al. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. LoBue AD, et al. 2006. Multivalent norovirus vaccines induce strong mucosal and systemic blocking antibodies against multiple strains. Vaccine 24:5220–5234 [DOI] [PubMed] [Google Scholar]
- 31. Lochridge VP, Hardy ME. 2007. A single-amino-acid substitution in the P2 domain of VP1 of murine norovirus is sufficient for escape from antibody neutralization. J. Virol. 81:12316–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lochridge VP, Jutila KL, Graff JW, Hardy ME. 2005. Epitopes in the P2 domain of norovirus VP1 recognized by monoclonal antibodies that block cell interactions. J. Gen. Virol. 86:2799–2806 [DOI] [PubMed] [Google Scholar]
- 33. Marionneau S, Airaud F, Bovin NV, Le Pendu J, Ruvoen-Clouet N. 2005. Influence of the combined ABO, FUT2, and FUT3 polymorphism on susceptibility to Norwalk virus attachment. J. Infect. Dis. 192:1071–1077 [DOI] [PubMed] [Google Scholar]
- 34. Noel JS, Fankhauser RL, Ando T, Monroe SS, Glass RI. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334–1344 [DOI] [PubMed] [Google Scholar]
- 35. Okada M, Tanaka T, Oseto M, Takeda N, Shinozaki K. 2006. Genetic analysis of noroviruses associated with fatalities in healthcare facilities. Arch. Virol. 151:1635–1641 [DOI] [PubMed] [Google Scholar]
- 36. Patel MM, et al. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Phan TG, et al. 2006. Changing distribution of norovirus genotypes and genetic analysis of recombinant GIIb among infants and children with diarrhea in Japan. J. Med. Virol. 78:971–978 [DOI] [PubMed] [Google Scholar]
- 38. Prasad BV, et al. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290 [DOI] [PubMed] [Google Scholar]
- 39. Reeck A, et al. 2010. Serological correlate of protection against norovirus-induced gastroenteritis. J. Infect. Dis. 202:1212–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schorn R, et al. 2010. Chronic norovirus infection after kidney transplantation: molecular evidence for immune-driven viral evolution. Clin. Infect. Dis. 51:307–314 [DOI] [PubMed] [Google Scholar]
- 41. Shanker S, et al. 2011. Structural analysis of HBGA binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J. Virol. 85:8635–8645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Siebenga J, Kroneman A, Vennema H, Duizer E, Koopmans M. 2008. Food-borne viruses in Europe network report: the norovirus GII.4 2006b (for US named Minerva-like, for Japan Kobe034-like, for UK V6) variant now dominant in early seasonal surveillance. Euro Surveill. 13(2):8009. [PubMed] [Google Scholar]
- 43. Siebenga JJ, et al. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tian P, et al. 2007. Binding of recombinant norovirus like particle to histo-blood group antigen on cells in the lumen of pig duodenum. Res. Vet. Sci. 83(3):410–418 [DOI] [PubMed] [Google Scholar]
- 45. Tian P, et al. 2010. Specificity and kinetics of norovirus binding to magnetic bead-conjugated histo-blood group antigens. J. Appl. Microbiol. 109:1753–1762 [DOI] [PubMed] [Google Scholar]
- 46. Vinjé J, Altena S, Koopmans M. 1997. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 176:1374–1378 [DOI] [PubMed] [Google Scholar]
- 47. Widdowson MA, et al. 2004. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus—United States, 2002. J. Infect. Dis. 190:27–36 [DOI] [PubMed] [Google Scholar]
- 48. Yang Y, et al. 2010. Genetic and phenotypic characterization of GII-4 noroviruses that circulated during 1987 to 2008. J. Virol. 84:9595–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng DP, et al. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323 [DOI] [PubMed] [Google Scholar]