Abstract
Few studies have explored the role of neutralizing antibody (NAb) responses in controlling HIV-2 viremia and disease progression. Using a TZM-bl neutralization assay, we assessed heterologous and autologous NAb responses from a community cohort of HIV-2-infected individuals with a broad range of disease outcomes in rural Guinea-Bissau. All subjects (n = 40) displayed exceptionally high heterologous NAb titers (50% inhibitory plasma dilution or 50% inhibitory concentration [IC50], 1:7,000 to 1:1,000,000) against 5 novel primary HIV-2 envelopes and HIV-2 7312A, whereas ROD A and 3 primary envelopes were relatively resistant to neutralization. Most individuals also showed high autologous NAb against contemporaneous envelopes (78% of plasma-envelope combinations in 69 envelopes from 21 subjects), with IC50s above 1:10,000. No association between heterologous or autologous NAb titer and greater control of HIV-2 was found. A subset of envelopes was found to be more resistant to neutralization (by plasma and HIV-2 monoclonal antibodies). These envelopes were isolated from individuals with greater intrapatient sequence diversity and were associated with changes in potential N-linked glycosylation sites but not CD4 independence or CXCR4 use. Plasma collected from up to 15 years previously was able to potently neutralize recent autologous envelopes, suggesting a lack of escape from NAb and the persistence of neutralization-sensitive variants over time, despite significant NAb pressure. We conclude that despite the presence of broad and potent NAb responses in HIV-2-infected individuals, these are not the primary forces behind the dichotomous outcomes observed but reveal a limited capacity for adaptive selection and escape from host immunity in HIV-2 infection.
INTRODUCTION
Characterizing host and viral factors that contribute to protection from disease progression in natural HIV infection is vital to understanding HIV pathogenesis, and such studies can reveal novel prophylactic and therapeutic targets in the virus. Although HIV-2 proteins share up to 60% sequence identity with their HIV-1 counterparts (32), the outcomes of infection between these two closely related retroviruses are markedly different, with disease progression in HIV-2-infected individuals occurring at a much lower rate (reviewed in references 26 and 33). Although similar proviral loads are seen in HIV-1- and HIV-2-infected individuals matched for disease stage, the plasma viral load (VL) is low or undetectable in most HIV-2-infected individuals and approximately 30-fold lower than that in HIV-1-infected individuals at equivalent stages of infection (8, 12, 13, 59, 60). To describe HIV-2 as a less pathogenic virus, however, is not strictly accurate, as a proportion of infected individuals have high viremia and progress to AIDS which is clinically indistinguishable from that in HIV-1-infected subjects. Others, however, maintain plasma VLs below the limit of detection (elite controllers) and exhibit no higher mortality than uninfected individuals over almost 2 decades (68). This dichotomy in clinical outcomes makes HIV-2 an important human model of viral control and offers the opportunity to explore what features are required for natural containment of a potentially lethal retrovirus. Many studies of HIV-2 have rightly focused on characterizing the differences with HIV-1 and identified key distinguishing features. However, for understanding how humans can control a lethal lentivirus infection, it is perhaps more important to elucidate why infection with HIV-2 can lead to such contrasting outcomes in different individuals.
Potent and broadly neutralizing antibodies (NAbs) are crucial for defining an effective, sterilizing HIV vaccine, but induction of such NAbs has arguably represented the greatest challenge to investigators since the beginning of the HIV-1 epidemic. Significant advances in understanding the NAb response to HIV-1 infection have been made, including detailed description of appearance in acute infection, rapid ongoing viral escape resulting in low contemporaneous autologous NAb titers (i.e., the failure to neutralize the currently circulating virus), despite the development of NAb breadth in some individuals, and isolation of broadly neutralizing human monoclonal antibodies (MAbs) (reviewed in reference 6). Very few studies have explored the NAb response in HIV-2-infected individuals, and these studies have often been limited by small patient numbers and inclusion of HIV-2 progressors on antiretroviral therapy (ART), thus not allowing meaningful correlation with clinical status (14, 71). A cross-sectional evaluation of nine HIV-2-infected individuals suggested that subjects with AIDS had lower autologous NAb titers than those with asymptomatic infection (14) and that titers may be higher than those seen in HIV-1 infection. One of the only other descriptions of autologous NAb in four HIV-2-infected individuals indicated little change in longitudinal NAb titers, implying that env evolution and NAb escape may be limited (71). A key obstacle in interpreting studies on HIV neutralization has been the lack of a standardized and validated neutralization assay, as well as difficulties in culturing primary viral isolates for autologous NAb evaluation. The latter problem is even more pertinent in HIV-2, given the lower VLs and slower replication kinetics of some HIV-2 isolates (7, 15). Use of molecularly cloned envelopes in single-cycle TZM-bl cell luciferase reporter gene assays has overcome these barriers and is now commonplace in HIV-1 NAb studies. Only one published study to date has applied these techniques in HIV-2 NAb assessment (63), comparing heterologous NAb titers in HIV-1- and asymptomatic HIV-2-infected Senegalese subjects and concluding that greater breadth but lower potency distinguishes HIV-2 from HIV-1 infection. The lack of autologous NAb studies using cloned HIV-2 envelopes is likely due to the technical challenges in amplifying plasma RNA-derived envelopes from individuals with low or undetectable viremia.
It is important to clarify, using these newer NAb assays, whether in fact more potent and broadly neutralizing NAb responses are present in HIV-2 infection and to establish whether a potent autologous NAb response is a key distinguishing feature between HIV-2 controllers and those with higher viremia. Equally important is documenting how HIV-2 Env biology and evolution may differ from those of the HIV-1 envelope in relation to the NAb response and could provide crucial insight into HIV pathogenesis. We describe, to our knowledge for the first time, both autologous and heterologous NAb responses from a community cohort of ART-naïve HIV-2-infected individuals using a standardized neutralization assay, with the inclusion of subjects spanning the breadth of HIV-2 disease outcomes: from long-term nonprogressors with undetectable plasma VLs to those with higher viremia and advanced immunosuppression. A clinical correlation with NAb titers is made, as are key observations surrounding HIV-2 envelope susceptibility or resistance to NAb.
MATERIALS AND METHODS
Study population.
Study samples were obtained from the Caió community cohort, consisting of individuals from a rural village in northwestern Guinea-Bissau with community HIV serosurveys and ongoing follow-up of HIV-infected cases in place since 1989 (76). A number of studies on HIV-2 epidemiology, natural history, and immunological factors associated with HIV-2 viremic control have been reported previously from this cohort (42, 68, 76). Subjects with chronic HIV-2 infection and available plasma samples were chosen for env amplification and NAb studies. Twenty-one subjects covering a broad range of HIV-2 disease outcomes were chosen for autologous neutralization studies. All subjects had been infected for at least 6 years, were free of HIV-1 coinfection, and were antiretroviral therapy naïve at the time of sampling. Diagnosis of HIV-2 infection was determined using both serological and molecular diagnostics, CD4+ T-cell counts, and HIV-2 VLs as previously described (12, 13, 42). HIV-1-infected subject plasma samples used were also from the same cohort. Ethical approval for the study was provided by the joint MRC/Gambia Government Ethics Committee and the Guinea-Bissau Ministry of Health Ethics Committee.
HIV-2 env amplification and molecular cloning for neutralization studies.
Viral RNA was extracted from 200 μl plasma (and eluted in a 60-μl volume) using a QIAamp Ultrasens viral RNA extraction kit (Qiagen, United Kingdom) and used as template in a nested reverse transcription-PCR (RT-PCR) for HIV-2 env amplification. Initial primers were designed by selecting conserved regions flanking the HIV-2 envelope sequence from an alignment of all available HIV-2 isolates in the Los Alamos HIV Database, and subsequent primer modification was done on the basis of the initial local sequence data obtained, which resulted in improved PCR efficiency. The final optimized primer sequences used in the study are given below. HIV-2 env products amplified were dependent on downstream methods of pseudovirus or replication-competent virus generation (see below): either full-length gp160 envelopes (approximately 2.5 kb) or a fragment encoding the entire envelope ectodomain exposed to antibody recognition (gp140, approximately 2.1 kb), but excluding the signal peptide and the gp41 cytoplasmic tail. Reverse transcription and the first-round PCR were performed in a single reaction. Each 25 μl RT-PCR mixture contained the following: 1× PCR buffer Titan One Tube system (Roche Applied Science, Germany), 2.5 mM MgCl2, 400 nM deoxynucleoside triphosphate (dNTP) mix, 0.5 μM primers MO105 (GRGCMRGAGARCTCATTA) and MO133 (CCTACTWGGTCATCATCATCTGAATCT) for gp160 or primers MO148 (GTTTTTTAAACAAGGGGCTCGGGATA) and MO134 (GGTCATCATCATCTGAATCTACATCAT) for gp140, 0.208 U/μl RNase inhibitor, 1 μl of the Titan One Tube enzyme mix, and 5 μl of extracted RNA. Reverse transcription was performed at 45°C for 45 min, followed by 95°C for 3 min, 10 cycles of 94°C (15 s), 52°C (30 s), and 68°C (3 min), and then 30 cycles of 94°C (15 s), 52°C (30 s), and 68°C (3 min) plus a 5-s time extension at 68°C after each round and a final extension of 7 min at 68°C. The inner (nested) PCR used 1 μl of the first-round RT-PCR product in 50 μl containing 1× buffer (with 1.5 mM MgCl2, final concentration), 0.05 U/μl Expand HiFi Plus polymerase (Roche), 400 nM dNTP mix, and 0.5 μM primers MO108 (CTYCTRCAYCAGACAAGTGAG) and MO134 for gp160 or primers MO167 (CCAAATACGTAACTGTTTTYTATGG) and MO168 (GGGAAGAGAAAACCGGTCTATAGCC) for gp140. Primers MO167 and MO168 contained restriction recognition sites for SnaBI and AgeI (underlined), respectively, necessary for molecular cloning of these products into the 7312A-SNAG vector. For both gp160 and gp140 amplification, the second-round reaction was conducted at 95°C for 3 min and then with 40 cycles of 94°C (15 s), 56°C (30 s), and 72°C (3 min) and a final extension of 7 min at 72°C. The PCR products were purified using a MinElute gel extraction kit (Qiagen) according to the manufacturer's protocol. Purified full-length HIV-2 gp160 fragments were cloned into the pcDNA3.1/V5-His-TOPO vector according to the manufacturer's instructions (Invitrogen, United Kingdom), transformed into XL2-Blue MRF′ ultracompetent cells (Agilent Technologies, United Kingdom), and grown at 30°C for 24 h. Purified primary HIV-2 env gp140 fragments were cloned into a full-length HIV-2 provirus, 7312A-SNAG (in pBluescript SK+), constructed from parental HIV-2 plasmid pJK7312A (23, 30) and modified to contain unique restriction silent sites SnaBI and AgeI flanking gp140. Both primary gp140 env products and the 7312A-SNAG vector were first digested with SnaBI and AgeI (New England BioLabs, MA). Primary env fragments and the 7312A-SNAG env-deficient portion were purified using the MinElute gel extraction kit (Qiagen), mixed at a molar ratio of 5.5:1 (approximately 100 ng DNA of each) with 1× ligation buffer (New England BioLabs) and 400 cohesive end units of T4 DNA ligase (1 μl) in a 20-μl reaction mixture, and incubated at 16°C for 16 h. Approximately 10 ng of the ligation mix was then transformed into XL2-Blue MRF′ ultracompetent cells (Agilent Technologies) and grown at 30°C for 24 h. Between 96 and 192 colonies from each plate were screened by colony PCR for identification of colonies with HIV-2 env inserts (for both gp160 and gp140 cloning) using nested PCR primers and the conditions described above for gp140 amplification. Colonies containing inserts were inoculated in 5 ml of liquid lysogeny broth (LB) medium supplemented with 50 μg/ml ampicillin and incubated overnight at 30°C. Plasmid DNA was purified using a QIAprep Spin Miniprep kit (Qiagen) according to the manufacturer's protocol. The presence of inserts was confirmed by restriction enzyme digestion with SnaBI and AgeI for gp140 containing 7312A-SNAG constructs and with BstX1 (Promega, United Kingdom) for gp160 inserts in the pcDNA3.1/V5-His-TOPO vector (Invitrogen). Although the initial strategy utilized gp160-pcDNA3.1/V5-His-TOPO cloning, due to an appreciable improvement in cloning efficiency and availability of replication-competent chimeric virus, the majority of primary HIV-2 envelopes isolated for the study were gp140 fragments cloned into 7312A-SNAG.
HIV-2 and HIV-1 env amplification for comparison of sequence diversity and selective pressure.
Partial HIV-2 envelope sequences (C2 to gp41) from 76 Caió subjects were available as part of concurrent molecular epidemiological studies in the cohort (T. I. de Silva, unpublished data). These included the 21 subjects used for autologous neutralization studies in the current study. Methods used and sequences from 34 subjects (GenBank accession numbers GQ485517 to GQ485550) are described elsewhere (54). Partial HIV-1 envelope sequences (C2 to gp41) from 56 subjects infected with CRF02_AG were chosen from available Caió sequences. Amplification was carried out using the nested PCR techniques described above, but with first-round primers CAenvOF (CCAATTCCYATACATTATTGTGCYCC) and CAenvOR (TGGTGCARATGAGTTTTCCAGAGCA) and nested primers CAenvIF (AGYACRGTACAATGYACACATGGAAAT) and CAenvIR (CCCAAATTCCTAGGAGCTGTTGATC). Conditions were as follows: reverse transcription was performed at 50°C for 30 min, followed by 95°C for 3 min, 40 cycles of 94°C (15 s), 54°C (30 s), and 72°C (1 min), and a final extension at 72°C (10 min). The second-round reaction was conducted at 95°C for 3 min, followed by 40 cycles of 94°C (15 s), 56°C (30 s), and 72°C (1 min) and a final extension of 10 min at 72°C.
Sequencing of HIV-2 and HIV-1 env.
All HIV-1 and HIV-2 env genes were sequenced by Macrogen using a BigDye Terminator (version 3.1) cycle sequencing kit and an ABI 3730XL sequencer. The primers used for the nested PCRs described above, along with the following additional primers for HIV-2 env, were used in sequencing reactions: MO124 (CTAGTGCCATTAAAGCCAAACCA), MO72 (TCATGTGAYAAGCACTATTGGGA), MO122A (GTGGACTAACTGCAGAGGAGAATT), MO122.5 (CAACAGCTGTTAGACGTGGTCAA), and MO125A (AGAAAACCCAAGAACCCTAGCAC).
Virus propagation in 293T cells.
293T cells cultured in Dulbecco's modified Eagle medium (DMEM) plus GlutaMAX (Invitrogen) supplemented with 10% fetal calf serum (FCS) were plated in 6-well plates 24 h prior to transfection at a density of 4 × 105/well. The following day, 6 μl Fugene-HD (Roche) transfection reagent was premixed with 2 μg full-genome HIV-2 infectious molecular clone DNA (from either reference strains detailed below or primary gp140-7312A-SNAG chimeric proviral clones) in 100 μl serum-free Optimem medium (Invitrogen) for 15 min at room temperature and then added to each well. HIV-2 gp160 pcDNA3.1/V5-His-TOPO constructs were cotransfected with either pSG3Δenv or pNL4-3.Luc.R−E− (catalogue nos. 11051 and 3418, respectively; NIH AIDS Research and Reference Reagent Program) at a ratio of 1:2 to produce pseudovirus particles. After overnight incubation at 37°C, the medium was replaced and virus was harvested at 48 h and centrifuged at 250 × g for 15 min to remove cellular debris, prior to snap-freezing in liquid nitrogen for future use. Virus titers were determined on TZM-bl cells (77) (catalogue no. 8129; NIH AIDS Research and Reference Reagent Program), to obtain 200 50% tissue culture infectious doses (TCID50s).
Passage in PBMCs of 293T-grown virus.
Freshly isolated peripheral blood mononuclear cells (PBMCs) from healthy lab donors were resuspended at a concentration of 106/ml in R20 (RPMI 1640 medium [Invitrogen] with 20% heat-inactivated FCS, 2 mM l-glutamine [Sigma], 1% PenStrep [100 U/ml penicillin and 0.1 mg/ml streptomycin, final concentrations; Sigma]) supplemented with phytohemagglutinin (Remel Europe, United Kingdom) at a final concentration of 2.5 μg/ml. Following incubation at 37°C for 72 h, the cells were washed once in RPMI 1640 medium and resuspended at 106/ml in growth medium (R20 supplemented with 5 U/ml recombinant interleukin-2 [Peprotech, United Kingdom]). Cells were infected with virus following a further 1 to 3 days of growth at 37°C. Approximately 10 × 106 viable cells were resuspended in 1 ml growth medium and incubated with 500 μl to 1 ml of 293T-grown viral stock (multiplicity of infection, 0.005 to 0.05) for 2 h at 37°C, after which the cells were washed twice with RPMI 1640 medium and resuspended in 10 ml growth medium in a T25 flask. At 7 days after infection, virus was harvested and stored as described above.
HIV-2 human monoclonal antibodies.
Four anti-HIV-2 human MAbs, isolated from two Gambian HIV-2-seropositive donors, and the prototypic anti-HIV-2 envelope human MAb, 1.7a (provided by one of the authors [James Robinson]), were used in the study to further characterize the breadth and potency of the anti-HIV-2 humoral response against both laboratory-adapted and primary HIV-2 envelopes. Although the exact epitope specificities of these MAbs are currently being determined, preliminary data suggest that these antibodies target diverse regions of the HIV-2 envelope: 6.10B (CD4-binding site), 6.10F (V3), 1.4H (CD4-induced [CD4i]), 1.4B (overlaps V3), and 1.7A (V4) (35; James Robinson, unpublished data).
Neutralizing antibody assays.
Envelope clones that supported virus infection with adequate titers in TZM-bl cells were sequenced and used in neutralization assays. Briefly, 200 TCID50s of virus were added to 3-fold dilutions of complement-inactivated plasma or anti-HIV-2 human MAb in DMEM containing 10% FCS and incubated for 1 h at 37°C. TZM-bl cells (1 × 104) resuspended in DMEM containing 10% FCS and DEAE-dextran (Sigma, United Kingdom) to give a final concentration of 10 μg/ml were then added to each well, and the plate was incubated for a further 48 h. Cells were then lysed for 2 min using 100 μl of Bright-Glo luciferase assay substrate (Promega) and analyzed for luminescence activity on a luminometer. Neutralization titers were calculated as the plasma dilution or concentration of MAb causing a 50% reduction in luciferase activity compared to a plasma- or MAb-free virus-only condition (50% inhibitory concentration [IC50]), following subtraction of background luciferase activity from cell control wells. Nonspecific inhibition was assessed by testing all viruses against HIV-negative plasma from the same cohort of subjects and all HIV-positive plasma samples against the vesicular stomatitis virus (VSV) envelope (pseudotyped with pSG3Δenv). Nonspecific inhibition of the VSV envelope by HIV-2-infected subject plasma was observed at a median IC50 of 1:30 (range, 1:23 to 1:40). Pooled plasma from five HIV-negative Caió subjects showed minimal nonspecific inhibition of HIV-2 envelopes (all but one with an IC50 below the highest plasma concentration of 1:20 tested and one with an IC50 of 1:25). Neutralization was considered positive if the IC50 against the HIV-2 envelope was 3-fold above that seen against the VSV envelope.
Coreceptor tropism and CD4 dependence assays.
Human glioma (NP2) cell lines stably transfected with CD4, CCR5, and/or CXCR4 (73) were maintained in DMEM containing 10% FCS and used for coreceptor tropism and CD4 dependence assays, performed by using either 293T-grown chimeric gp140-7312-SNAG full-length HIV-2 molecular clones (and subsequent immunostaining of NP2 cells) or pseudovirus produced by cotransfection of gp160 envelopes in pcDNA3.1/V5-His-TOPO and pNL4-3.Luc.R−E− (and subsequent measurement of luminescence activity). NP2 immunostaining assays were performed as described previously (1). Briefly, cells were seeded at 2 × 104 per well in 48-well flat-bottom plates to yield semiconfluent layers after overnight incubation. Tenfold serially diluted viral stocks were then added in duplicate, and the plates were incubated for 2 h at 37°C, after which the cells were washed once and incubated with DMEM containing 10% FCS for 72 h at 37°C. The cells were then fixed and incubated with either heat-inactivated 1:4,000-diluted HIV-2 polyclonal sera (pooled from 10 HIV-2-infected donors not included in this study) or 1:40-diluted mouse anti-HIV-1 p24 monoclonal antibodies (ADP 365 and 266; NIBSC, Potters Bar, United Kingdom) for 1 h, followed by goat anti-human IgG or goat anti-mouse IgG monoclonal antibodies conjugated with β-galactosidase (Cambridge Biosciences, United Kingdom) at 2.5 μg/ml for 1 h. Following an incubation with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside substrate at 37°C, blue infected cells were counted as focus-forming units (FFU) using light microscopy and viral titers were calculated according to the formula FFU/(dilution factor × volume used for infection). Pseudovirus produced by cotransfection of gp160 in pcDNA3.1/V5-His-TOPO and pNL4-3.Luc.R−E− was diluted and plated in quadruplicate. NP2 cells were then added at 1 × 104 per well (96-well plate), and the plate was incubated at 37°C for 48 h. Luminescence measurement was carried out as described above, and the TCID50/ml was calculated.
Sequence analysis and tests for codon selection.
Sequences were aligned using ClustalW2 (40; http://www.ebi.ac.uk/Tools/clustalw2/index.html), all alignments were inspected and manually edited using the Se-Al (Sequence Alignment Editor, version 2.0a11) program (http://tree.bio.ed.ac.uk/software/seal/), and ambiguous regions were deleted prior to further analyses. Pairwise genetic distances were calculated using the PAUP* program (http://paup.csit.fsu.edu/paupfaq/faq.html) using the best-fit model of substitution for the relevant alignments selected by the jModel test (61): the HKY+I+G model of evolution for intrapatient HIV-2 gp140 alignments and the GTR+I+G model for both interpatient partial C2-gp41 HIV-1 and HIV-2 sets. HIV-1 and HIV-2 subtypes were determined using the REGAHIV subtyping algorithm (5), and for HIV-1, the subtype was confirmed with pol amplification, where ambiguous results were obtained (data not shown; de Silva, unpublished). Detection of N-linked glycosylation sites was performed using the N-GlycoSite tool (81). Diversity at each amino acid position in protein alignments was calculated using Shannon's entropy (37). A maximum likelihood (ML) phylogenetic tree of all subject HIV-2 gp140 sequences was constructed using PAUP* under the GTR+I+G model of nucleotide substitution. The statistical robustness of the ML topology was assessed by bootstrapping with 1,000 replicates using the neighbor-joining method.
Sites under positive and negative selection in HIV-1 and HIV-2 env alignments were identified by comparison of synonymous and nonsynonymous substitution rates using three different methods in the Datamonkey web server (57): single-likelihood ancestor counting (SLAC), fixed-effects likelihood (FEL), and random-effects likelihood (REL) (38, 58). Estimations were carried out using codon models of evolution selected by a genetic algorithm (25) on the Datamonkey suite, and all alignments were tested for recombination using the GARD tool (39). A single recombination breakpoint in each of the HIV-1 and HIV-2 partial C2-gp41 env alignments and two breakpoints in the HIV-2 gp140 set were identified, and therefore, multiple partition data sets were used for all analyses of selection. A P value cutoff of 0.05 (SLAC and FEL) or Bayes factor cutoff of 50 (REL) was used to define significant positive or negative selection at a codon.
Statistical analyses.
All statistical analyses were performed using Graphpad Prism (version 5.0) software for Macintosh (GraphPad Software, San Diego, CA). Comparisons between variables were performed using the Mann-Whitney U test (two-tailed) or the Kruskal-Wallis test with Dunn's posttest for multiple comparisons. Discrete data were compared by the chi-square test or Fisher's exact test. Correlations between nonnormally distributed data were made using the Spearman's rank correlation coefficient.
Nucleotide sequence accession numbers.
All env sequences (111 HIV-2 and 56 HIV-1 sequences) not previously described were submitted to GenBank under accession numbers JN863732 to JN863898.
RESULTS
Subject characteristics.
Heterologous NAb studies were conducted using plasma samples from 40 subjects with chronic HIV-2 infection known not to be closely epidemiologically linked to subjects from whom Env clones were derived. The median age of these subjects was 60 years (range, 35 to 84 years), and 28/40 (70%) were female. All plasma samples were taken in 2006, and (28/40) 70% of the cohort had been diagnosed with HIV-2 infection for at least 9 years. Autologous NAb studies were conducted with 69 plasma RNA-derived envelopes isolated from 21 HIV-2-infected subjects (subjects TD01 to TD21). Subjects had a median age of 56 years (range, 31 to 84 years) at the time of sampling, and 11/21 (52%) were female. Median CD4 count and VL (range) were 508 cells/μl (range, 100 to 1,754 cells/μl) and 11,675 copies/ml (range, <100 to 320,272 copies/ml), respectively; full characteristics are given in Table 1. The wide range in CD4 counts and VLs of these subjects reflects the variation in HIV-2 disease progression observed, including five long-term nonprogressors (LTNPs) with VLs of <100 copies/ml (TD01 to TD05) who had been infected with HIV-2 for at least 9 to 14 years. One further functional HIV-2 envelope was isolated but was used only in heterologous NAb studies due to insufficient autologous plasma (CA7205.8). Phylogenetic analysis confirmed that env isolates from each patient clustered together, as expected (data not shown). Seven individuals were included in both the autologous and heterologous NAb cohorts.
Table 1.
Details of HIV-2-infected individuals from whom 69 env variants for neutralization studies were derived
| Subject | Age (yr) at sample collection | Sexa | Yr of HIV-2 infection diagnosis | Yr of sample collection | No. (%) of CD4 cells/μlb | Plasma viral load (no. of copies/ml)b | No of env variants isolated |
|---|---|---|---|---|---|---|---|
| TD01 | 60 | M | 1997 | 2006 | 570 (35) | <100 | 1 |
| TD02 | 58 | F | 1989 | 2003 | 1,086 (44) | <100 | 3 |
| TD03 | 54 | M | 1989 | 2003 | 816 (40) | <100 | 2 |
| TD04 | 45 | F | 1989 | 2003 | 1,754 (45) | <100 | 4 |
| TD05 | 39 | F | 1997 | 2006 | 535 (35) | <100 | 4 |
| TD06 | 34 | M | 1997 | 2003 | 742 (32) | 667 | 5 |
| TD07 | 84 | F | 1989 | 2006 | 615 (28) | 1,343 | 1 |
| TD08 | 59 | M | 1989 | 2006 | 770 (29) | 2,078 | 1 |
| TD09 | 70 | M | 1989 | 2003 | 883 (30) | 3,482 | 4 |
| TD10 | 31 | M | 1997 | 2006 | 200 (18) | 6,431 | 1 |
| TD11c | 56 | M | 1997 | 2003 | 1,355 (32) | 7,260 | 3 |
| 59 | 2006 | 800 (30) | 14,104 | 1 | |||
| TD12 | 52 | F | 1989 | 2006 | 285 (28) | 9,979 | 1 |
| TD13 | 73 | F | 1997 | 2003 | 480 (29) | 13,371 | 3 |
| TD14 | 33 | F | 1989 | 2006 | 1,075 (31) | 23,889 | 1 |
| TD15 | 56 | F | 1997 | 2003 | 287 (11) | 43,396 | 5 |
| TD16c | 40 | F | 1997 | 2003 | 236 (12) | 76,493 | 5 |
| 43 | 2006 | 100 (9) | 148,593 | 4 | |||
| TD17 | 56 | M | 1997 | 2003 | 373 (20) | 109,221 | 3 |
| TD18 | 40 | F | 1997 | 2006 | 290 (13) | 146,284 | 1 |
| TD19c | 61 | F | 1997 | 2003 | 121 (15) | 223,924 | 3 |
| 64 | 2006 | 160 (8) | 37,503 | 3 | |||
| TD20 | 43 | M | 1997 | 2003 | 164 (12) | 313,188 | 5 |
| TD21 | 72 | M | 1996 | 2003 | 476 (33) | 320,272 | 5 |
F, female; M, male.
CD4 count and viral load are from the same sampling time point that env variants were derived.
Subjects TD11, TD16, and TD19 have clones from two different time points.
Broad and potent heterologous neutralizing antibody responses are found in chronically HIV-2-infected subjects.
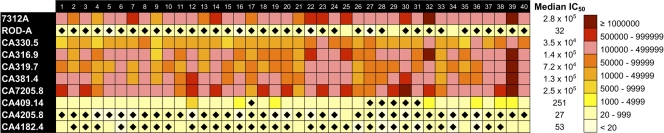
To assess the breadth and potency of the heterologous NAb response in chronic HIV-2 infection, 40 HIV-2-infected plasma samples were tested against eight newly isolated primary HIV-2 Env variants (from eight different subjects) and two reference HIV-2 strains, 7312A (a kind gift from Beatrice Hahn) and ROD A, a known neutralization-resistant laboratory-adapted HIV-2 isolate (74) (Fig. 1). Six primary envelopes were gp160 clones in pcDNA3.1/V5-His-TOPO (CA330.5, CA316.9, CA319.7, CA381.4, CA7205.8, CA409.14), and two were gp140 clones in 7312A-SNAG (CA4205.8 and CA4182.4). Two envelopes (CA381.4 and CA409.14), representing extremes of neutralization susceptibility phenotype, were compared in both gp140 and gp160 clone formats and as expected showed no appreciable difference in their neutralizability (data not shown). Potent heterologous neutralizing titers (IC50s, above 1:7,000) against five Caió Env variants and 7312A were seen with plasma from all (40/40) subjects, whereas ROD A and three Caió Envs appeared to be relatively resistant to neutralization (Fig. 1).
Fig 1.
Heat map depicting heterologous neutralization titers of 40 HIV-2-infected individuals against 10 HIV-2 envelopes. Values given are reciprocal IC50s. The panel clearly separates into envelopes with divergent neutralization phenotypes. Plasma-envelope combinations that were resistant to neutralization (based on a titer below a cutoff 3 times the VSV IC50) are additionally marked with a closed diamond.
A substantial breadth of heterologous neutralization was also observed, with 35/40 subjects neutralizing at least 7 of the 10 Env variants tested and 9 subjects being able to neutralize 8 or more Env variants. The magnitude of each subject's heterologous NAb response was defined as the median reciprocal IC50 titer against the 10 HIV-2 envelopes. Despite observing potent heterologous NAbs in most subjects, a significant positive association (two-tailed, Spearman's rho = 0.359, P = 0.02) was seen between the magnitude of the heterologous NAb response against the panel of HIV-2 envelopes tested and the plasma VL (Fig. 2). A nonsignificant trend toward a higher heterologous NAb magnitude in male subjects than female subjects was observed (median NAb IC50s, 1:85,557 versus 1:40,890, respectively; P = 0.07). This may, however, be related to a similar trend observed for higher VLs in male subjects (medians, 2,280 for male subjects versus 310 copies/ml for female subjects; P = 0.05). This relationship between higher HIV-2 VL and male sex has been noted previously in the Caió cohort (13). No gender difference in heterologous NAb breadth was observed.
Fig 2.
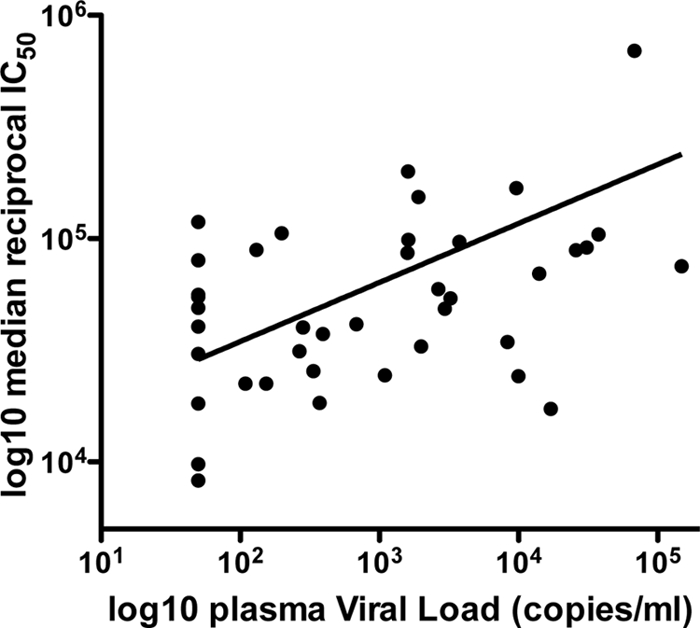
Relationship between plasma viral load and median reciprocal IC50 of heterologous neutralization titers against a panel of 10 HIV-2 envelopes from HIV-2-infected individuals (n = 40). A significant positive correlation between viremia and the magnitude of the heterologous neutralizing antibody response is found (Spearman's rho = 0.359, P = 0.02).
Evidence for highly potent contemporaneous autologous neutralizing antibody responses in chronic HIV-2 infection but no role in viremic control.
Similar to the high-magnitude heterologous NAb responses observed and described above, potent contemporaneous autologous NAb titers against 54/69 envelopes were observed, with IC50 titers being above 1:10,000 (Fig. 3). However, no significant association between the magnitude of contemporaneous autologous NAb titers and plasma viremia was found (Fig. 3; Kruskal-Wallis test with Dunn's posttest for multiple comparisons, P = 0.0938). Notably, autologous NAb titers in HIV-2 controllers with an undetectable plasma VL were equivalent to those in subjects with higher viremia and HIV-2 disease progression.
Fig 3.
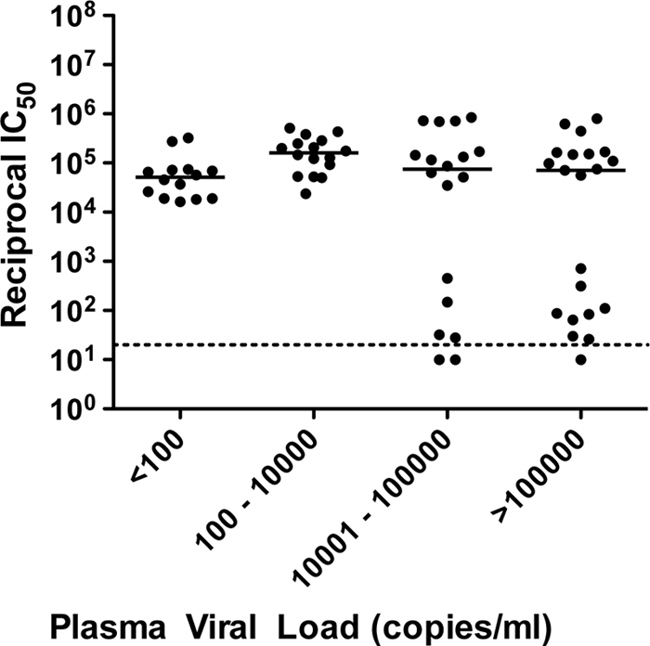
Contemporaneous autologous neutralization of HIV-2 envelopes stratified by plasma viral load. Of the 69 primary HIV-2 envelopes (from 21 subjects) used, 64 represented gp140 cloned into 7312A-SNAG. Values are means of two or three independent experiments. No significant difference in median IC50 is seen between viral load categories (P = 0.09).
A subset of envelopes (n = 15/69) was, however, found to be relatively resistant (IC50s < 1:1,000) to autologous contemporaneous neutralization, and these envelopes were isolated more frequently from patients with higher plasma viremia (P < 0.001, two-tailed Fisher's exact test for the proportion of resistant clones if viremia was <10,000 versus ≥10,000 copies/ml). Three of these envelopes were included in the panel used in heterologous NAb studies and found either to be resistant to neutralization by the majority of plasma samples tested (CA4205.8 and CA4182.4; Fig. 1) or to display only limited neutralization susceptibility (CA409.14; Fig. 1). In addition, five (of seven) subjects from whom more resistant envelopes were isolated (TD15, TD16, TD19, TD20, TD21) also had high-titer contemporaneous autologous NAbs against other variants in the quasispecies. Taken together, this indicates that the lower NAb titers seen in some cases reflect envelopes with a neutralization-resistant phenotype rather than subjects with poorer host humoral responses. Autologous NAb titers against six envelopes (of seven used in both autologous and heterologous NAb studies) were 1.5- to 8-fold higher than the median heterologous titer against the same envelope (data not shown), which would suggest that regardless of the neutralization phenotype of the envelope, HIV-2-infected subjects retain the ability to mount effective NAb responses specific to Env epitopes displayed on contemporaneous autologous isolates. No significant association between autologous and heterologous NAb titers in the same individual was seen (data not shown), although data from only 7 subjects were available for this analysis.
Neutralization by anti-HIV-2 Env human monoclonal antibodies reveals that diverse epitopes across the HIV-2 envelope are targeted in natural infection but are nevertheless susceptible to neutralization escape.
To further characterize the neutralization phenotype of the isolated HIV-2 Env variants, a subset was tested against five anti-HIV-2 Env human monoclonal antibodies (MAbs) with varied specificities. Although there was heterogeneity in susceptibility to MAbs among the 21 primary envelopes (from 13 subjects) and envelopes of the two reference strains tested (Table 2), those envelopes that were relatively resistant to autologous or heterologous plasma neutralization were globally resistant to neutralization by all HIV-2 human MAbs at the highest concentration tested (10 μg/ml). The anti-V3 MAb, 6.10F, showed the most potent neutralization, and MAb 1.4H, thought to be a CD4-induced antibody and therefore to potentially target the coreceptor binding site, neutralized many HIV-2 envelopes in the absence of soluble CD4 (sCD4). These data suggest that in HIV-2 natural infection, NAbs with a breadth of epitope specificities that can potently neutralize a variety of diverse envelopes are elicited, but also that neutralization-resistant HIV-2 envelopes are likely to represent variants that have accumulated mutations in multiple sites, resulting in a globally resistant phenotype.
Table 2.
Neutralization of HIV-2 envelopes with 5 human HIV-2 monoclonal antibodies
| Env | Mean IC50 (μg/ml)a |
Plasma neutralization titerb | ||||
|---|---|---|---|---|---|---|
| 1.7A (V4 dependent) | 6.10F (V3) | 6.10B (CD4 BSc) | 1.4B (overlaps V3) | 1.4H (CD4i) | ||
| 7312A | 0.08 | 0.004 | 0.1 | 0.038 | >10d | 2.8 × 105H |
| ROD ARe | >10 | >10 | >10 | >10 | >10 | 32Hf |
| CA316.9 | 0.18 | 0.01 | 0.19 | >10 | >10 | 7.3 × 105 |
| CA319.7 | >10 | 0.015 | 0.26 | 0.05 | 0.02 | 5.1 × 105 |
| CA330.5 | >10 | 0.01 | 0.165 | 0.18 | 0.175 | 5.7 × 104 |
| CA381.4 | 0.025 | 0.01 | 0.07 | >10 | 0.4 | 4.4 × 105 |
| CA4401.1 | 4.59 | 0.01 | 0.17 | 0.415 | 2.5 | 2.0 × 105 |
| CA4153.9 | >10 | 0.01 | 0.095 | 1.02 | 0.015 | 7.2 × 105 |
| CA7205.8 | 0.015 | 0.0075 | 0.18 | 0.02 | 0.015 | 2.5 × 105H |
| CA4145.13 | 0.17 | 0.006 | 0.2 | 0.18 | 0.02 | 3.8 × 105 |
| CA4291.2 | 0.54 | 0.006 | 0.155 | 0.115 | 0.025 | 1.6 × 104 |
| CA409.14R | >10 | >10 | >10 | >10 | >10 | 64 |
| CA4205.1R | >10 | >10 | >10 | >10 | >10 | 713 |
| CA4205.8R | >10 | >10 | >10 | >10 | >10 | 87 |
| CA4137.6 | >10 | 0.015 | 0.09 | 0.66 | 0.18 | 8.6 × 104 |
| CA4137.2 | >10 | 0.007 | 0.095 | 0.45 | 0.045 | 1.4 × 105 |
| CA4137.19R | >10 | >10 | >10 | >10 | >10 | <20 |
| CA4137.21 | >10 | 0.01 | 0.07 | 0.3 | 0.035 | 1.7 × 105 |
| CA4206.6 | >10 | 0.03 | 0.65 | 0.44 | 0.1 | 5.6 × 104 |
| CA4206.7 | >10 | 0.01 | 0.2 | 0.6 | 0.17 | 1.7 × 105 |
| CA4206.13 | >10 | 0.04 | 1.01 | 0.95 | 0.21 | 9.8 × 104 |
| CA4206.5R | >10 | >10 | >10 | >10 | >10 | 111 |
| CA4206.14R | >10 | >10 | >10 | >10 | >10 | 314 |
| VSV | >10 | >10 | >10 | >10 | >10 | |
Values given are the mean IC50s from two or three separate experiments.
Values for neutralizing antibody titers are reciprocal plasma autologous titers unless otherwise stated.
CD4 BS, anti-CD4 binding site.
Bold data indicate envelope-MAb combinations where no neutralization was observed.
R, envelope resistant to autologous and/or heterologous plasma neutralization.
H, median reciprocal heterologous neutralizing antibody titer (against 40 subject plasma samples).
Virus passage in peripheral blood mononuclear cells variably affects neutralization titers but not the overall HIV-2 envelope neutralization phenotype.
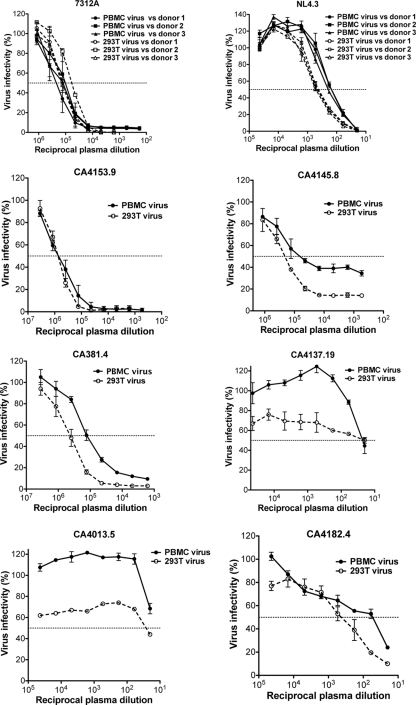
Differences in neutralization depending on the cell type used for viral propagation have previously been noted in HIV-1 isolates (44). The neutralization phenotype of a subset of HIV-2 Env isolates (gp140 in 7312A-SNAG) grown in 293T cells as described above was evaluated following minimal passage in mitogen-stimulated PBMCs for 7 days. Along with 7312A and HIV-1 NL4.3, six primary HIV-2 Env isolates (of 14 tested) produced PBMC-passaged viral titers high enough after 7 days to assess autologous neutralization in TZM-bl cells. Heterologous neutralization of 7312A and NL4.3 was assessed using plasma from three randomly chosen HIV-2- and HIV-1-infected donors, respectively. Consistent with previous observations (44), PBMC-passaged NL4.3 was less sensitive to neutralization by all three donor plasma samples (Fig. 4). However, no significant reduction in NAb titer of the highly sensitive HIV-2 Env isolate 7312A was observed. Likewise, no significant reduction in NAb titer of CA4153.9 was seen when PBMC virus was assessed against contemporaneous autologous plasma. PBMC CA381.4 and PBMC CA4145.8 showed only modest reductions in neutralization sensitivity. This indicates that the high NAb titers that we observed in HIV-2-infected subjects are unlikely to be due to an artifact of virus production in 293T cells. The neutralization-resistant isolates CA4137.19, CA4013.5, and CA4182.4 all remained resistant to (autologous contemporaneous) neutralization following PBMC passage, with a clear indication that the PBMC-passaged viruses were even more resistant than their 293T-grown counterparts.
Fig 4.
Comparison of neutralization of 239T-grown and PBMC-passaged virus with the same plasma sample. Shown are autologous neutralization of CA4153.9, CA4145.8, CA381.4, CA4137.19, CA4013.5, and CA4182.4 and heterologous neutralization of 7312A and NL4.3. Plotted values are means (and SDs) from three separate experiments. Virus infectivity (percent) is compared to a virus-only condition.
Neutralization resistance in HIV-2 is associated with greater env diversification and changes in N-linked glycosylation sites but not coreceptor tropism or CD4 dependence.
To explore the relationship between intrapatient viral diversity and the presence of neutralization-resistant envelopes, pairwise genetic distances between HIV-2 env isolates from each subject were calculated (Fig. 5), using only those with ≥3 functional env variants available (n = 14). Subjects from whom resistant envelopes were isolated had significantly higher intrapatient diversity than those in whom only neutralization-sensitive variants were found (medians, 0.034 versus 0.003 substitutions per site, respectively; P = 0.0007, two-tailed Mann-Whitney U test). Of note, within each subject, no env variants were 100% identical at the nucleotide level. These findings also provide indirect support for an association between higher viremia and viral diversification in HIV-2 infection, as the median plasma VL in the group with higher diversity was 166,576 copies/ml (range, 43,396 to 320,272 copies/ml), whereas that in the group with lower intrapatient diversity was 359 copies/ml (range, <100 to 13,371 copies/ml) (P = 0.002, two-tailed Mann-Whitney U test).
Fig 5.
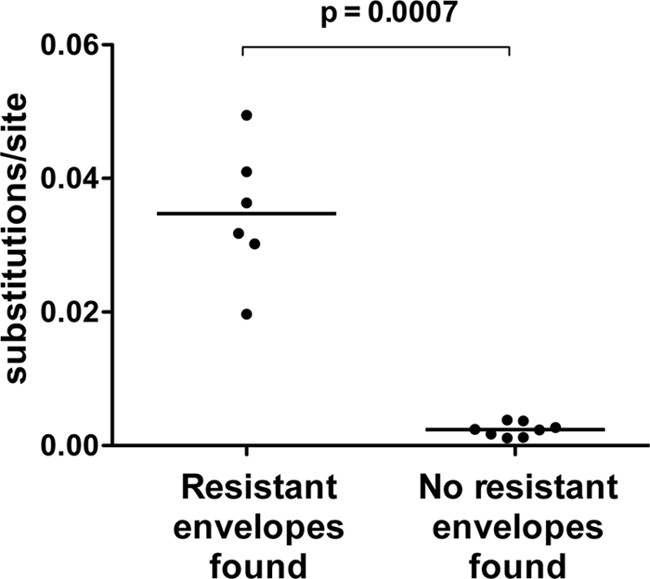
Association between greater intrapatient diversity and the presence of neutralization-resistant HIV-2 envelopes. Plotted values are mean pairwise genetic distances (substitutions/site) between env variants from each patient.
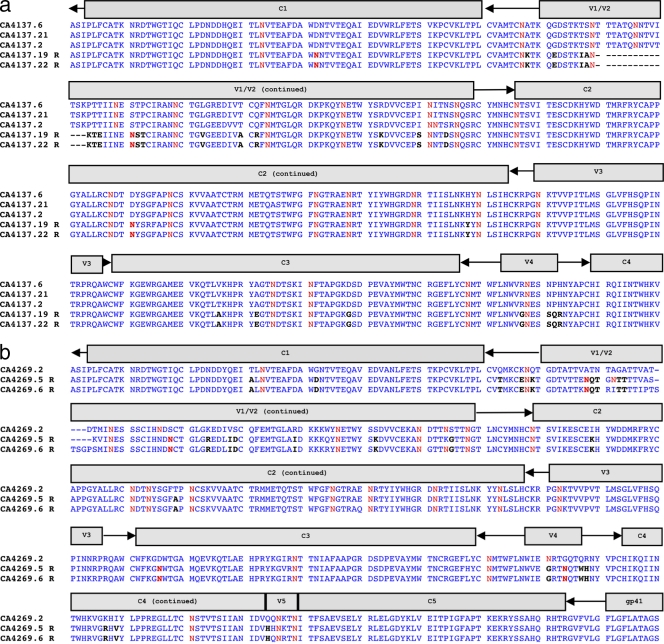
The concept of an evolving glycan shield as a mechanism of neutralization escape is well described in HIV-1 infection (78). Comparing sensitive and resistant Caió HIV-2 Envs used in this study, the numbers of potential N-linked glycosylation (PNLG) sites in any individual conserved or variable region or across the entire gp140 fragment (n = 26 median PNLG sites in both sets) were not significantly different between the two groups. It is likely, however, that akin to HIV-1, escape from NAb may occur via multiple pathways and could be via a unique route within each HIV-2-infected subject (65). We therefore compared PNLG sites in Envs from individuals where both resistant and sensitive variants had been isolated (n = 5). In all individuals, differences in either the number or position of PNLG sites were observed between sensitive and resistant envelopes (e.g., TD15 and TD19; Fig. 6a and b), although the extent of these differences varied from 1 to 4 additional or shifted sites. However, multiple additional amino acid substitutions unique to resistant variants can be seen throughout the envelope ectodomain (including deletions in V1/V2 common to resistant variants in 2/5 individuals), thus highlighting the complexity of neutralization escape in chronic HIV-2 infection.
Fig 6.
Alignments from two subjects (a and b) demonstrating amino acid changes specific to resistant envelopes (R). Either additional or shifted potential N-linked glycosylation sites are highlighted in bold red, and other changes are in black. Only regions with differences between resistant and sensitive envelopes are displayed for each subject.
Previous studies have also suggested an association between neutralization sensitivity and CD4 independence (74), as well as resistance to neutralization and CXCR4 tropism (47), in HIV-2. To explore whether the differences in neutralization phenotype seen in the current set of primary HIV-2 envelopes could be explained by similar mechanisms, their ability to infect NP2 cells expressing different combinations of CD4, CCR5, and CXCR4 was assessed (Table 3). Thirty-one HIV-2 envelopes (from 18 individuals) were tested, along with the envelopes of X4-tropic HIV-1 isolate NL4.3, the CD4-dependent HIV-2 isolate ROD A, and ROD B, a neutralization-sensitive and CD4-independent variant produced via passage of ROD A in C8166 cells (20). NL4.3, ROD A, and ROD B all displayed their reported coreceptor usage. Only one primary envelope (CA65331.3) showed a dual R5/X4-tropic phenotype, whereas all other envelopes were R5 tropic. Consistent with previous data, ROD B was able to infect CD4-negative cells efficiently using CXCR4, but a similar ability to infect cells expressing only the primary coreceptor was not observed in any of the primary HIV-2 envelopes cloned directly from plasma viral RNA and propagated in 293T cells. An association between sensitivity or resistance to neutralization and coreceptor tropism/CD4 dependence was therefore not evident in the HIV-2 envelope isolates described in this study.
Table 3.
Envelope coreceptor use and relationship to neutralization phenotype
| Vector and subject | Envelope | Infectivity on NP2 cells with use of the following coreceptor: |
Classificationa | Neutralization phenotypeb | ||||
|---|---|---|---|---|---|---|---|---|
| CD4/CCR5 | CD4/CXCR4 | CD4 alone | CCR5 alone | CXCR4 alone | ||||
| 7312A-SNAGc | ||||||||
| TD02 | CA4258.11 | 81.5 | NTd | 0.1 | NT | R5 | S | |
| TD05 | CA370.16 | 20.4 | NT | NT | R5 | S | ||
| TD06 | CA4401.1 | 53 | NT | NT | R5 | S | ||
| TD08 | CA381.4e | 255 | 0.1 | 1.2 | NT | R5 | S | |
| TD09 | CA4145.8 | 627 | NT | 0.8 | NT | R5 | S | |
| TD11 | CA4272.16 | 330 | 0.2 | 1.5 | NT | R5 | S | |
| CA65331.3 | 110 | 50.0 | 0.8 | NT | R5/X4 | S | ||
| TD12 | CA65332.1 | 42.5 | NT | 0.1 | NT | R5 | S | |
| TD13 | CA4153.9 | 2,500 | 2.0 | 1.3 | 0.9 | NT | R5 | S |
| TD15 | CA4137.2 | 1,328 | NT | NT | R5 | S | ||
| CA4137.19 | 7,125 | 1.4 | 1.7 | 1.2 | NT | R5 | R | |
| TD16 | CA4182.4 | 273 | NT | NT | R5 | R | ||
| CA4182.12 | 4,687 | 1.0 | 0.9 | 1.1 | NT | R5 | S | |
| CA65410.3 | 225 | 0.2 | 0.8 | NT | R5 | S | ||
| CA65410.5 | 134.5 | 0.1 | 1.5 | 0.5 | NT | R5 | S | |
| CA65410.7 | 300 | 0.4 | 2.5 | 0.9 | NT | R5 | S | |
| CA65410.13 | 59.5 | NT | NT | R5 | S | |||
| TD17 | CA4205.1 | 850 | 2.5 | 2.1 | 1.1 | NT | R5 | R |
| CA4205.8 | 1,100 | 5.5 | 3.0 | 1.4 | NT | R5 | R | |
| TD19 | CA4269.2 | 1,781 | 7.5 | 6.3 | 0.2 | NT | R5 | S |
| CA4269.6 | 2,625 | NT | 1.1 | NT | R5 | R | ||
| CA65339.3 | 32 | NT | NT | R5 | R | |||
| TD20 | CA4206.7 | 25 | NT | NT | NT | R5 | S | |
| TD21 | CA4013.5 | 3,187 | 2.4 | 1.9 | 0.9 | NT | R5 | R |
| CA4013.8 | 22.5 | NT | NT | NT | R5 | S | ||
| NAf | NL4.3 | 212.5 | NT | NT | NT | X4 | NA | |
| pcDNA3.1V5-His-TOPOg | ||||||||
| NA | ROD A | 69,877 | 31,250 | 250 | 112 | 112 | R5/X4 | R |
| NAh | ROD B | 1,250 | 31,250 | 50 | <50 | 13,975 | X4/CD4 independent | S |
| TD01 | CA330.5 | 69,877 | 559 | 559 | 250 | 112 | R5 | S |
| TD08 | CA381.4e | 69,877 | 250 | 112 | 250 | 112 | R5 | S |
| TD14 | CA316.9 | 349,386 | 1,250 | 559 | 559 | 559 | R5 | S |
| TD07 | CA319.7 | 349,386 | 2,795 | 559 | 559 | 559 | R5 | S |
| TD18 | CA409.14 | 156,250 | 1,250 | 1,250 | 1,250 | 1,250 | R5 | R |
| NA | CA7205.8 | 349,386 | 2,795 | 112 | 1,869 | 250 | R5 | S |
Viruses were classified as dual tropic if the titer on the alternative coreceptor was within 1.5 log10 units of the titer obtained from the dominant coreceptor cell line (4).
S, sensitive; R, resistant.
HIV-2 gp140 cloned into full-length HIV-2 7312A-SNAG vector. NL4.3 refers to virus derived from the full-length HIV-1 NL4.3 molecular clone. For infectivity on NP2 cells with the 7312A-SNAG vector, units are 103 FFU/ml, where FFU represents the number of focus-forming units measured by immunostaining of NP2 cells with or without CD4 and with or without coreceptors following infection with live virus.
NT, not tested.
Coreceptor tropism tested in both formats.
NA, subject identifier not assigned as envelope not used for autologous neutralization studies.
HIV-2 gp160 cloned into expression vector pcDNA3.1V5-His-TOPO. For infectivity on NP2 cells with the pcDNA3.1V5-His-TOPO vector, units are TCID50s, where TCID50s were calculated according to luciferase readout in NP2 cells following infection with pseudovirus produced by cotransfection of HIV-2 gp160 envelope plasmid and NL4.3LucR−E−.
Bold indicates ROD B, the only CD4-independent isolate of those tested.
Persistence of neutralization-sensitive envelope variants, despite the presence of high-magnitude autologous neutralizing antibodies.
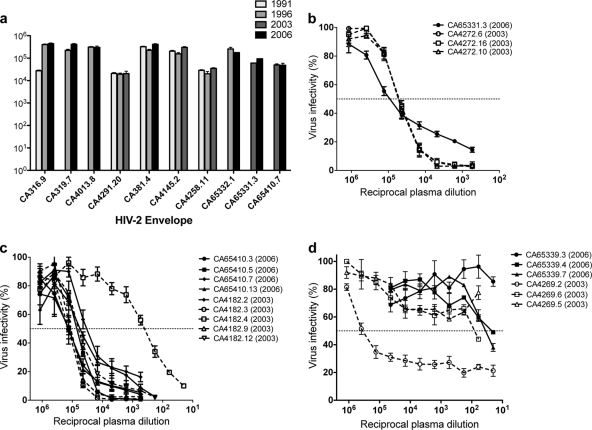
In HIV-1 infection, constant viral escape from the autologous NAb response usually results in not only poor contemporaneous autologous NAb titers but also even lower NAb titers when Envs are tested against plasma samples from older time points (78), reflecting the inability of the NAb repertoire to keep up with env evolution under NAb pressure. One previous publication (describing a study using four subjects) has suggested that, in contrast, escape from autologous NAbs is rarely seen in HIV-2 infection (71). To extend this observation with a larger data set, available archived plasma samples from previous Caió cohort bleeds were obtained for 10 subjects, and the ability of these historical samples to neutralize one of the most recent neutralization-sensitive envelopes available (from either 2003 or 2006) was tested. In almost all cases, the autologous NAb titers against HIV-2 envelopes from the most recent time point in plasma samples taken up to 15 years previously were as high as those seen in contemporaneous samples (Fig. 7a). The one exception was an approximate 1-log10 reduction in NAb titer against CA316.9 of a plasma sample collected in 1991, and in this case, a similar reduction was also seen in heterologous neutralization of 7312A (data not shown).
Fig 7.
Assessment of neutralization escape in HIV-2. (a) Neutralization of HIV-2 envelopes by autologous plasma from older time points in 10 subjects. The legend depicts the year of plasma sampling. HIV-2 envelopes were isolated from the most recent time point of plasma collection depicted for each clone (either 2003 or 2006). (b to d) Neutralization of envelopes from either 2003 or 2006 by contemporaneous plasma for subjects TD11 (b), TD16 (c), and TD19 (d), from whom variants were isolated at both time points. All values represent means (and SDs) of three independent experiments. Virus infectivity (percent) is compared to a virus-only condition.
Although the main objective of this study was not a longitudinal assessment of contemporaneous NAb responses, three individuals had adequate plasma available from two time points to isolate envelopes from both 2003 and 2006 (TD11, TD16, and TD19; Table 1). Two subjects (TD16 and TD19) had both sensitive and resistant envelopes isolated from 2003 plasma (Fig. 7c and d). For subjects TD11 and TD16 (Fig. 7b and c, respectively), no escape from neutralization was observed between envelopes isolated in 2003 and 2006. In fact, despite the presence of a relatively resistant envelope isolated in 2003 for subject TD16, all four 2006 envelopes were surprisingly sensitive to autologous neutralization (Fig. 7c). In contrast, subject TD19 had one sensitive and two resistant Env variants isolated from 2003 plasma, but all 2006 envelopes were neutralization resistant, indicating possible escape (Fig. 7d).
Selective pressure on HIV-2 envelope in the face of potent autologous neutralizing antibody responses.
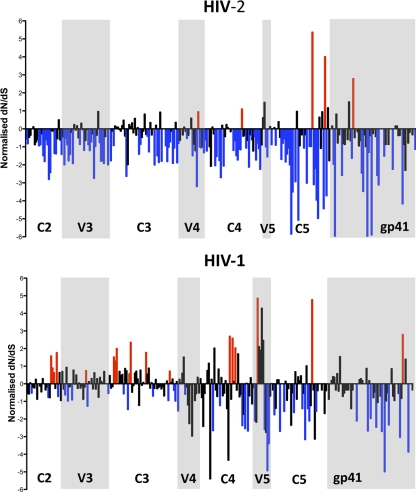
The neutralizing antibody response is well recognized to be the major driving force in env evolution and positive selective pressure in HIV-1 infection (29), with resulting continual escape from NAb. Previous studies focusing on the C2V3C3 region of the HIV-2 envelope have suggested that env evolution is slower in asymptomatic infection (45) and highlighted that this region in HIV-2 is largely under purifying or negative selective pressure, implying that possible functional constraints may play a role (10, 11). In view of the exceptionally high-magnitude autologous NAb observed in the current study, as well as the low neutralization escape seen, we examined selective pressure evident across the entire HIV-2 gp140 in the set of 70 functional envelopes that were isolated in this study. Using the three different algorithms (38, 58), the number of sites under significant positive selective pressure was remarkably low under the SLAC algorithm (11 sites) and FEL algorithm (19 sites), although it was higher under the FEL algorithm (44 sites), with the number of negatively selected sites being 175, 232, and 346, respectively (from a total of 641 codons). To compare how this differs from diversification of and selective pressure on the HIV-1 envelope in the same community, we used available partial C2 to gp41 ectodomain sequences from 75 HIV-2-infected and 56 HIV-1-infected individuals from Caió. The partial C2 to gp41 ectodomain is a fragment corresponding to nucleotides (nt) 6584 to 7490 of HIV-1 HXB2 (K03455) and nt 6984 to 7859 of HIV-2 ROD (M15390). Phylogenetic studies and the REGA HIV subtyping algorithm (5) showed that all HIV-2 sequences belonged to group A and all HIV-1 sequences were subtype CRF02_AG (data not shown). Surprisingly, pairwise genetic distances were significantly higher in the HIV-2 set than in the HIV-1 set, revealing a greater diversity in HIV-2 at the nucleotide level in this community (Table 4). This is likely to reflect the different ages of the two epidemics in Guinea-Bissau, with HIV-1 having been introduced more recently than HIV-2 (76). Despite this greater diversity in HIV-2, amino acid entropy and the number of sites under positive selection were significantly higher in the HIV-1 envelope, whereas the number of negatively selected sites was much higher in the HIV-2 envelope (Table 4 and Fig. 8). It appears, therefore, that at least at a community level there are constraints present on the HIV-2 envelope that restrict amino acid changes compared to the HIV-1 envelope, despite substantial pressure from neutralizing antibody responses.
Table 4.
Comparison of HIV-1 and HIV-2 envelope (partial C2 to gp41 ectodomain) diversity, amino acid entropy, and sites under positive and negative selective pressure
| Parameter | HIV-2 | HIV-1 | P value |
|---|---|---|---|
| Mean pairwise genetic distance (no. of substitutions/site) | 0.183 (0.181–0.185)a | 0.148 (0.146–0.151) | <0.0001 |
| Mean Shannon's entropy | 0.255 (0.207–0.303) | 0.347 (0.292–0.402) | 0.01 |
| Selection analysis (from a total of 277 codons)b | |||
| SLAC | |||
| No. of positively selected sites | 5 | 19 | <0.0001 |
| No. of negatively selected sites | 122 | 53 | |
| FEL | |||
| No. of positively selected sites | 11 | 22 | <0.0001 |
| No. of negatively selected sites | 140 | 79 | |
| REL | |||
| No. of positively selected sites | 21 | 51 | <0.0001 |
| No. of negatively selected sites | 195 | 143 |
Data in parentheses are 95% confidence intervals.
Codons with a P value of <0.05 (SLAC and FEL) or a Bayes factor of >50 (REL) considered significant.
Fig 8.
Comparison of the ratio of nonsynonymous (dN) to synonymous (dS) changes in HIV-2 (n = 75) and HIV-1 (n = 56) partial C2 to gp41 ectodomain fragments. Plotted values are those obtained under the SLAC algorithm with values normalized to alignment length. Positions found to be under significant positive selection (red) and negative selection (blue) under all three algorithms (SLAC, FEL and REL) are highlighted.
DISCUSSION
A major obstacle to the development of an antibody-based HIV-1 vaccine over the last 25 years has been identifying what defines protective humoral immunity in HIV infection: that is, lack of clarity over the role, if any, of NAbs elicited during natural HIV infection in limiting viral replication and the course of disease progression. Previous studies have reached contrasting conclusions. The constant antibody-driven viral evolution in HIV-1 env (62) indicates that the NAb response must be sufficiently functional to induce viral change. While some studies suggest a role in viral control (24, 51), most studies (in both acute and chronic HIV-1 infection) have failed to reveal a clear relationship between potent NAbs and lower VLs or slower disease progression (9, 31, 46, 55). With a high proportion of HIV-2-infected individuals with low or undetectable VLs, HIV-2 infection offers an important example of viral control with which to explore the relative importance of different host and viral factors implicated in limiting HIV pathogenesis. Only a few studies have attempted to establish the relationship between NAb in HIV-2-infected individuals and disease outcomes (14, 63, 71), in part due to the technical limitations in culturing primary virus or generating plasma RNA-derived env clones from individuals with low VLs with which to measure autologous NAbs. To our knowledge, the current study is the first to use cloned env plasma variants in a luciferase reporter gene assay to describe both autologous and heterologous NAb responses in HIV-2 infection, and with the inclusion of subjects ranging from LTNPs with undetectable plasma VLs to HIV-2 progressors with high VLs, it is also arguably the most comprehensive to date.
Heterologous NAb titers of strikingly high magnitude (IC50s, 1:7,000 to 1,000,000) and several log10 units higher than what is usually observed in HIV-1 infection using the same NAb assay (55) were found against primary HIV-2 envelopes in all 40 subject plasma samples tested. Yet despite the presence of potent heterologous NAbs, a significant positive association between these titers and plasma VL was found. This is consistent with findings from the one previous study on HIV-2 using a pseudotype reporter assay including only asymptomatic subjects (63) and confirms that, similar to HIV-1 infection (24), greater antigenic stimulation and/or abnormal polyclonal B-cell activation in HIV-2 progressors may result in greater heterologous NAb production. If NAbs do play a role in limiting HIV-2 replication, however, it is more likely to be reflected in contemporaneous autologous NAbs, which are conspicuously low or absent in most subjects with chronic HIV-1 infection (17, 24). Exhibiting a scenario clearly different from that in HIV-1-infected individuals, equally potent (IC50s > 1:10,000) contemporaneous autologous NAbs were found in 78% of plasma-envelope combinations tested, including in those with undetectable plasma VLs. HIV-1 elite controllers, in contrast, have very low autologous NAb titers (9, 46), leading to the argument that antigenic stimulation is required to maintain NAb. It is possible that this unique feature in HIV-2 elite controllers is due to factors such as intermittent viral blips (which are difficult to detect with our cohort design [68, 76]) providing a stimulus or ongoing viral replication in lymphoid tissue sufficient to maintain Env-specific memory B cells. It is worth noting that despite plasma viremia being below the limit of detection (100 copies/ml) in these individuals, our ability to amplify env using nested PCR in itself supports the existence of low-level viremia, potentially priming and in turn being controlled by adaptive immune responses. Previous work in the same cohort has also observed that a proportion of individuals with viremia below 100 copies/ml has detectable RNA by qualitative PCR (13). Interestingly, despite the absence of detectable viremia, HIV-2 elite controllers are also able to maintain a remarkably high proportion of HIV-2-specific CD8+ T cells in their circulation many decades after initial infection (41, 42). However, despite the presence of potent contemporaneous autologous NAb, we were unable to demonstrate a significant inverse relationship between NAb titers and plasma viremia. Given the 30-fold lower viremia in HIV-2-infected individuals than HIV-1-infected individuals in the asymptomatic stage (60), it is tempting to speculate that these high-magnitude NAbs could still play a role in containing HIV-2 replication, but what is clear from our study is that they are certainly not the predominant force responsible for the dichotomous outcomes observed in HIV-2 infection. In fact, if only NAb titers against sensitive envelopes are considered, individuals with detectable plasma VLs (≥100 copies/ml) do have significantly higher median autologous NAb titers than those with VLs of <100 copies/ml (1.47 × 105 versus 5.1 × 104, respectively; P = 0.0006). This suggests that although autologous NAb titers are unusually high in most HIV-2-infected subjects, a relationship between greater antigenic load and higher-magnitude autologous NAb may exist in HIV-2 infection, similar to our findings in heterologous NAbs, as well as previously reported findings in HIV-1 infection (46).
An interesting finding was the presence of HIV-2 envelopes that displayed a more resistant neutralization profile, notably, with contemporaneous autologous titers (IC50s < 1:1,000) closer to what is observed in HIV-1 infection. Of note, most of the isolated HIV-2 envelopes were either hypersusceptible or very resistant to neutralization, akin to HIV-1 tier 1A and tier 3 envelopes, respectively, whereas the majority of primary HIV-1 envelopes described tend to belong to the intermediate-susceptible tier 2 category (70). Although this finding requires confirmation with a greater number of individuals, the appearance of more resistant envelopes with higher viremia might suggest a scenario where HIV-2 replication to a level where neutralization escape is possible occurs more frequently with progressive HIV-2 infection. The association of greater intrapatient viral diversity with the presence of these envelopes would support this hypothesis, given that the capacity that HIV-1 has for diversification underlies its ability to escape readily from host immunity. The persistence of neutralization-sensitive envelopes despite the presence of NAb pressure up to 15 years previously also provides indirect support for the idea that neutralization escape occurs less often in HIV-2 infection (71). Certainly, a previous report has highlighted that in asymptomatic HIV-2 infection, long-term env evolution is remarkably low (45), although the authors suggest that lack of NAb pressure may account for this observation. Our current findings make an alternative explanation necessary. In some cases, these envelopes were found together with neutralization-sensitive variants in the same individual, an observation mirrored in some HIV-1 studies (2, 46, 72, 80). As evidence from acute HIV-1 infection shows that constant neutralization escape usually results in successive replacement of neutralization-sensitive variants with waves of variants resistant to the autologous antibody response (53, 78), this is a curious finding. One explanation might be that most neutralization-sensitive variants are derived from a long-lived latent viral reservoir, thought to be a prominent feature of HIV-2 infection and inferred due to a lack of phylogenetic separation between HIV-2 sequences over time (45). Nevertheless, in the face of greater HIV-2 replication, one would still expect the selective pressure exerted by such high NAb titers to result in elimination of neutralization-sensitive variants. In contrast, we observed in one subject (TD16) that a neutralization-resistant Env present (amid several sensitive variants) in 2003 failed to become the predominant population 3 years later. It is possible, therefore, that some sensitive HIV-2 Envs offer a selective advantage for alternative reasons. Extending previous studies focusing on the HIV-2 C2V3C3 (10, 11), we found limited evidence of positive selection across gp140 and significantly greater negative selective pressure than in HIV-1 in the region corresponding to C2-gp41. Together, these allude to the possibility that certain functional constraints unique to the HIV-2 envelope that limit amino acid changes may exist, exerting a greater selective pressure than that of autologous NAb. Whether this is related purely to replicative fitness or additional HIV-2 Env functions such as tetherin antagonism (43) requires further investigation.
It is difficult to conclude whether the exceptionally high NAb titers seen in most envelope-plasma combinations in our study are due to superior NAb production by HIV-2-infected individuals or features of most HIV-2 envelopes that result in greater susceptibility to NAb. Lack of env evolution and persistence of NAb epitopes over long periods of time may allow greater affinity maturation by B cells that is not possible in HIV-1-infected individuals, and CD4+ T-cell help is also better preserved in asymptomatic HIV-2 infection (27). A more open conformation and an exposed coreceptor site have been proposed for the HIV-2 envelope, as CD4-independent infection is related to global neutralization sensitivity in some HIV-2 isolates (74). We were, however, unable to find evidence of this phenotype in our current set of primary envelopes, although interestingly, the MAb 1.4H, thought to be a CD4-induced (CD4i) antibody (J. Robinson, unpublished data), was able to neutralize many isolates in the absence of soluble CD4 (sCD4). Previous work has shown that several HIV-2 strains (including 7312A) are neutralized by HIV-1 CD4i MAbs only in the presence of sCD4 (23), suggesting that an accessible coreceptor site is not a typical characteristic of the HIV-2 Env. Elegant work from the same group has also shown that transplantation of HIV-1 V3 in the HIV-2 envelope not only allows detection of potent anti-V3 NAb in HIV-1 sera but also results in a significant reduction in neutralization by HIV-2-infected sera (22), inferring that anti-HIV-2 V3 responses contribute to NAb in HIV-2-infected sera and that this region may be particularly exposed in the native HIV-2 envelope. A recent publication describing a study using structural modeling has claimed, however, that the HIV-2 V3 loop is much less exposed than in HIV-1 (10). Although our data showing a high degree of conservation in HIV-2 V3 are consistent with this hypothesis, the finding that the anti-V3 MAb 6.10F produced the most potent MAb neutralization would favor the conclusions drawn by Davis et al. (22) and imply that the lack of entropy in HIV-2 V3 may again be due to particular structural or functional constraints that limit its evolution. Although CXCR4-using HIV-2 isolates have been described in the literature (47, 69), a recent report suggests that an R5-to-X4 switch may occur less often in HIV-2 than in HIV-1 (18). Our observation that only 1 of 31 primary envelopes tested (including many from HIV-2 progressors) was able to use CXCR4 efficiently for cell entry would support this finding and may also relate to an evolutionary constraint on the V3 region, which is well-known to be the major determinant of coreceptor tropism.
Although we found no evidence that CD4 independence or CXCR4 usage was associated with neutralization resistance in our set of primary envelopes, comparison of resistant and sensitive envelopes found within the same individual revealed within the resistant variants clear changes in N-linked glycosylation sites, which are known to mediate escape from neutralization in HIV-1 infection (66, 72, 78). These changes in PNLG sites, as well as other amino acid mutations common to intrapatient resistant variants, were found across the entire ectodomain but were concentrated largely in V1/V2. This is in agreement with studies in both acute and chronic HIV-1 infection that suggest that V1/V2 may be a major target of NAbs and changes within this region are observed during neutralization escape (31, 52, 56, 64, 66). Despite the observation that all envelopes resistant to plasma neutralization were globally resistant to all MAbs tested, no clear patterns in PNLG site changes or other mutations that would reliably predict the resistant phenotype were observed across these isolates from different individuals. This is perhaps not surprising, given the complexity in neutralization escape described in HIV-1, with resistance to a single antibody possible through different pathways (65). Thus, this sequence-based comparison reveals parallels that exist between HIV-1 and HIV-2 when infection with the latter does result in env evolution and escape from NAb, but further detailed experimentation and inclusion of more individuals are required to expand on these findings.
Our study has certain limitations that must be mentioned. First, all our HIV-2 envelopes were derived through bulk PCR and molecular cloning, which would potentially have been at risk of creating recombinants that do not exist in vivo through Taq polymerase-driven template switching (50). Single-genome amplification (SGA) techniques are now increasingly used to avoid such issues (67) but at the time were beyond the capability or resources of the current study. More recent work, however, found no difference in sampling bias between standard bulk PCR/cloning and SGA, provided that an adequate number of templates is analyzed (34). Preliminary data (de Silva, unpublished) suggest that in subjects with low or undetectable plasma HIV-2 viremia, our amplification conditions are close to those that fulfill criteria for SGA (<30% PCR positivity [67]), but it may be important to confirm our findings through formal SGA analyses, particularly to demonstrate the presence of the HIV-2 envelopes with various neutralization sensitivities. Of note, recent work using env genes derived by SGA has indeed observed the presence of neutralization-sensitive and -resistant HIV-2 envelopes from a single individual (36). Second, for our heterologous NAb studies, we selected individuals who were known not to be epidemiologically linked to those from whom envelopes were derived, which could result in NAb titers appearing to be high purely through shared epitopes due to shared transmission. Given the cohort setting, it is still inevitable that many infected individuals have viruses that share a recent common ancestor. However, as similarly high titers against 7312A, an isolate originally from Côte d'Ivoire, in Caió plasma and against Caió HIV-2 envelopes in Gambian plasma samples have been observed (de Silva, unpublished), it is unlikely that the high antibody titers seen are unique to our cohort alone. Furthermore, recent findings from Kong et al. (36) using HIV-2-infected sera predominantly from Senegal and Côte d'Ivoire and an identical neutralization assay show equally potent NAb titers.
Despite the widespread use of luciferase reporter gene assays with TZM-bl cells as targets in HIV-1 studies, clear differences in assay sensitivity and performance compared to other available neutralization assays exist (28). Although we demonstrate that the high neutralization titers seen in HIV-2-infected sera are not an artifact of virus propagation in 293T cells, it would be important to validate our findings in the future using a range of different target cells and readouts. Encouragingly, recent work has shown similar anti-HIV-2 Env titers using a PBMC neutralization assay (36). Little is known regarding the degree of cell-cell spread of HIV-2, and previous work has shown that spread of HIV-1 in this manner can be inhibited by NAb (48). Future work should attempt to establish whether anti-HIV-2 NAbs can also prevent cellular HIV-2 transmission via the virological synapse and if the presence of these specific NAbs correlates with control of viremia.
Although our analyses of env diversity and selection pressure at a population level provide intriguing insight into HIV-2 pathogenesis, more detailed studies comparing intrapatient sequences in individuals with various disease outcomes can be considered. Of particular interest would be an analysis of whole-genome sequences to determine whether our findings are generalizable across the entire HIV-2 genome. Notably, escape within cytotoxic T-lymphocyte (CTL) epitopes in HIV-2 infection is also yet to be described, despite the presence of potent Gag-specific CTL responses (42), which is surprising, given the frequency with which this occurs in HIV-1 (19, 49). Finally, a study of NAb development and maintenance during and after acute HIV-2 infection would definitively resolve many unanswered questions about the role of NAb in defining the initial course of infection and whether escape from NAb is limited. However, the reducing incidence of HIV-2 infection on a background of substantially lower transmission than HIV-1 makes this incredibly difficult (21, 76).
In conclusion, our study reveals remarkably high autologous and heterologous NAb titers (and seemingly rare escape from NAbs) in natural HIV-2 infection, yet these NAbs do not appear to play a major role in determining the level of HIV-2 viremia. One could question what implication this has for predicting the benefit achievable through future NAb-based HIV vaccines, but of course, the nature and titer of NAb required for prophylaxis and those necessary for viremic control once infection is established may be very different. It is conceivable that the NAb titers seen in HIV-2 infection prevent further superinfection with other HIV-2 strains; this is not an uncommon occurrence in HIV-1 (reviewed in references 16 and 75) but has not been extensively described or investigated in HIV-2. The role of other antibody-dependent effector functions, such as complement-mediated lysis (3) or antibody-dependent cellular toxicity (79), is unknown in HIV-2 infection and warrants investigation within this model. Our findings also add to mounting evidence that the potential for adaptive selection and escape from host immunity may be limited in HIV-2 infection, which may underlie the lower VLs and slower disease progression observed in HIV-2-infected individuals. Determining whether this is due to a yet-unconfirmed potent host immune response or an intrinsic property of the virus deserves further investigation and has the potential to aid our understanding of HIV-1 pathogenesis and search for novel therapeutic targets considerably.
ACKNOWLEDGMENTS
We thank Tim Vincent and the Caió team, as well as the Caió cohort participants, for their invaluable contribution to this study; Maarten Schim van der Loeff, Aleksandra Leligdowicz, and Carla van Tienen for fieldwork and collection of samples used in the current study; Steve Kaye and Abraham Alabi for HIV-1 and HIV-2 VL assays; Anna Forsman, Suzanne Willey, Edward Wright, and Willey Koh for advice on experimental design; and Beatrice Hahn for the kind provision of HIV-2 7312A and 7312A-SNAG molecular clones.
T.I.D.S was supported by a Medical Research Council Clinical Research Training Fellowship (G0701313).
Footnotes
Published ahead of print 9 November 2011
REFERENCES
- 1. Aasa-Chapman MM, Aubin K, Williams I, McKnight A. 2006. Primary CCR5 only using HIV-1 isolates does not accurately represent the in vivo replicating quasi-species. Virology 351:489–496 [DOI] [PubMed] [Google Scholar]
- 2. Aasa-Chapman MM, et al. 2011. In vivo emergence of HIV-1 highly sensitive to neutralizing antibodies. PLoS One 6:e23961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aasa-Chapman MM, et al. 2005. Detection of antibody-dependent complement-mediated inactivation of both autologous and heterologous virus in primary human immunodeficiency virus type 1 infection. J. Virol. 79:2823–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aasa-Chapman MM, Seymour CR, Williams I, McKnight A. 2006. Novel envelope determinants for CCR3 use by human immunodeficiency virus. J. Virol. 80:10884–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abecasis AB, et al. 2010. Comparative performance of the REGA subtyping tool version 2 versus version 1. Infect. Genet. Evol. 10:380–385 [DOI] [PubMed] [Google Scholar]
- 6. Alter G, Moody MA. 2010. The humoral response to HIV-1: new insights, renewed focus. J. Infect. Dis. 20(Suppl 2):S315–S322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arien KK, et al. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 79:8979–8990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ariyoshi K, et al. 1996. A community-based study of human immunodeficiency virus type 2 provirus load in rural village in West Africa. J. Infect. Dis. 173:245–248 [DOI] [PubMed] [Google Scholar]
- 9. Bailey JR, et al. 2006. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J. Virol. 80:4758–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barroso H, et al. 2011. Evolutionary and structural features of the C2, V3 and C3 envelope regions underlying the differences in HIV-1 and HIV-2 biology and infection. PLoS One 6:e14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barroso H, Taveira N. 2005. Evidence for negative selective pressure in HIV-2 evolution in vivo. Infect. Genet. Evol. 5:239–246 [DOI] [PubMed] [Google Scholar]
- 12. Berry N, et al. 1998. Low peripheral blood viral HIV-2 RNA in individuals with high CD4 percentage differentiates HIV-2 from HIV-1 infection. J. Hum. Virol. 1:457–468 [PubMed] [Google Scholar]
- 13. Berry N, et al. 2002. Low level viremia and high CD4% predict normal survival in a cohort of HIV type-2-infected villagers. AIDS Res. Hum. Retroviruses 18:1167–1173 [DOI] [PubMed] [Google Scholar]
- 14. Bjorling E, et al. 1993. Autologous neutralizing antibodies prevail in HIV-2 but not in HIV-1 infection. Virology 193:528–530 [DOI] [PubMed] [Google Scholar]
- 15. Blaak H, van der Ende ME, Boers PH, Schuitemaker H, Osterhaus AD. 2006. In vitro replication capacity of HIV-2 variants from long-term aviremic individuals. Virology 353:144–154 [DOI] [PubMed] [Google Scholar]
- 16. Blish CA, Blay WM, Haigwood NL, Overbaugh J. 2007. Transmission of HIV-1 in the face of neutralizing antibodies. Curr. HIV Res. 5:578–587 [DOI] [PubMed] [Google Scholar]
- 17. Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calado M, et al. 2010. Coreceptor usage by HIV-1 and HIV-2 primary isolates: the relevance of CCR8 chemokine receptor as an alternative coreceptor. Virology 408:174–182 [DOI] [PubMed] [Google Scholar]
- 19. Carlson JM, Brumme ZL. 2008. HIV evolution in response to HLA-restricted CTL selection pressures: a population-based perspective. Microbes Infect. 10:455–461 [DOI] [PubMed] [Google Scholar]
- 20. Clapham PR, McKnight A, Weiss RA. 1992. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J. Virol. 66:3531–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. da Silva ZJ, et al. 2008. Changes in prevalence and incidence of HIV-1, HIV-2 and dual infections in urban areas of Bissau, Guinea-Bissau: is HIV-2 disappearing? AIDS 22:1195–1202 [DOI] [PubMed] [Google Scholar]
- 22. Davis KL, et al. 2009. Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J. Virol. 83:1240–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Decker JM, et al. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deeks SG, et al. 2006. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J. Virol. 80:6155–6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Delport W, et al. 2010. CodonTest: modeling amino acid substitution preferences in coding sequences. PLoS Comput. Biol. 6:e1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Silva TI, Cotten M, Rowland-Jones SL. 2008. HIV-2: the forgotten AIDS virus. Trends Microbiol. 16:588–595 [DOI] [PubMed] [Google Scholar]
- 27. Duvall MG, et al. 2006. Maintenance of HIV-specific CD4+ T cell help distinguishes HIV-2 from HIV-1 infection. J. Immunol. 176:6973–6981 [DOI] [PubMed] [Google Scholar]
- 28. Fenyo EM, et al. 2009. International network for comparison of HIV neutralization assays: the NeutNet report. PLoS One 4:e4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frost SD, et al. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. U. S. A. 102:18514–18519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao F, et al. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433–7447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gray ES, et al. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guyader M, et al. 1987. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature 326:662–669 [DOI] [PubMed] [Google Scholar]
- 33. Hodges-Mameletzis I, De Bree GJ, Rowland-Jones SL. 2011. An underestimated lentivirus model: what can HIV-2 research contribute to the development of an effective HIV-1 vaccine? Expert Rev. Anti Infect. Ther. 9:195–206 [DOI] [PubMed] [Google Scholar]
- 34. Jordan MR, et al. 2010. Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. J. Virol. Methods 168:114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kong R, et al. 2009. Epitope mapping of neutralizing antibody 1.7A on the HIV-2 gp160 glycoprotein, abstr 332. Abstr. 16th Conf. Retroviruses Opportunistic Infections, Montreal, Quebec, Canada [Google Scholar]
- 36. Kong R, et al. 2012. Broad and potent neutralizing antibody responses elicited in natural HIV-2 infection. J. Virol. 86:947–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Korber BT, et al. 1994. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J. Virol. 68:7467–7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kosakovsky Pond SL, Frost SD. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208–1222 [DOI] [PubMed] [Google Scholar]
- 39. Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23:1891–1901 [DOI] [PubMed] [Google Scholar]
- 40. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 41. Leligdowicz A, et al. 2010. Highly avid, oligoclonal, early-differentiated antigen-specific CD8(+) T-cells in chronic HIV-2 infection. Eur. J. Immunol. 40:1963–1972 [DOI] [PubMed] [Google Scholar]
- 42. Leligdowicz A, et al. 2007. Robust Gag-specific T cell responses characterize viremia control in HIV-2 infection. J. Clin. Invest. 117:3067–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Le Tortorec A, Neil SJ. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J. Virol. 83:11966–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Louder MK, et al. 2005. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology 339:226–238 [DOI] [PubMed] [Google Scholar]
- 45. MacNeil A, et al. 2007. Long-term intrapatient viral evolution during HIV-2 infection. J. Infect. Dis. 195:726–733 [DOI] [PubMed] [Google Scholar]
- 46. Mahalanabis M, et al. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J. Virol. 83:662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marcelino JM, et al. 2010. Potent and broadly reactive HIV-2 neutralizing antibodies elicited by a vaccinia virus vector prime-C2V3C3 polypeptide boost immunization strategy. J. Virol. 84:12429–12436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martin N, et al. 2010. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J. Virol. 84:3516–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McMichael AJ, Phillips RE. 1997. Escape of human immunodeficiency virus from immune control. Annu. Rev. Immunol. 15:271–296 [DOI] [PubMed] [Google Scholar]
- 50. Meyerhans A, Vartanian JP, Wain-Hobson S. 1990. DNA recombination during PCR. Nucleic Acids Res. 18:1687–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Montefiori DC, Hill TS, Vo HT, Walker BD, Rosenberg ES. 2001. Neutralizing antibodies associated with viremia control in a subset of individuals after treatment of acute human immunodeficiency virus type 1 infection. J. Virol. 75:10200–10207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moore PL, Gray ES, Morris L. 2009. Specificity of the autologous neutralizing antibody response. Curr. Opin. HIV AIDS 4:358–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moore PL, et al. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Onyango CO, et al. 2010. HIV-2 capsids distinguish high and low virus load patients in a West African community cohort. Vaccine 28(Suppl 2):B60–B67 [DOI] [PubMed] [Google Scholar]
- 55. Piantadosi A, et al. 2009. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J. Virol. 83:10269–10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pinter A, et al. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pond SL, Frost SD. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 58. Pond SL, et al. 2006. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput. Biol. 2:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Popper SJ, et al. 2000. Low plasma human immunodeficiency virus type 2 viral load is independent of proviral load: low virus production in vivo. J. Virol. 74:1554–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Popper SJ, et al. 1999. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 180:1116–1121 [DOI] [PubMed] [Google Scholar]
- 61. Posada D. 2006. ModelTest server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res. 34:W700–W703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rodriguez SK, et al. 2007. Comparison of heterologous neutralizing antibody responses between human immunodeficiency virus type 1 (HIV-1)- and HIV-2-infected Senegalese patients: distinct patterns of breadth and magnitude distinguish HIV-1 and HIV-2 infections. J. Virol. 81:5331–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rong R, et al. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 81:1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rong R, et al. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5:e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sagar M, Wu X, Lee S, Overbaugh J. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 80:9586–9598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Salazar-Gonzalez JF, et al. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schim van der Loeff MF, et al. 2010. Undetectable plasma viral load predicts normal survival in HIV-2-infected people in a West African village. Retrovirology 7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schramm B, et al. 2000. Cytopathicity of human immunodeficiency virus type 2 (HIV-2) in human lymphoid tissue is coreceptor dependent and comparable to that of HIV-1. J. Virol. 74:9594–9600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Seaman MS, et al. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shi Y, et al. 2005. Evolution of human immunodeficiency virus type 2 coreceptor usage, autologous neutralization, envelope sequence and glycosylation. J. Gen. Virol. 86:3385–3396 [DOI] [PubMed] [Google Scholar]
- 72. Skrabal K, et al. 2005. Human immunodeficiency virus type 1 variants isolated from single plasma samples display a wide spectrum of neutralization sensitivity. J. Virol. 79:11848–11857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Soda Y, et al. 1999. Establishment of a new system for determination of coreceptor usages of HIV based on the human glioma NP-2 cell line. Biochem. Biophys. Res. Commun. 258:313–321 [DOI] [PubMed] [Google Scholar]
- 74. Thomas ER, Shotton C, Weiss RA, Clapham PR, McKnight A. 2003. CD4-dependent and CD4-independent HIV-2: consequences for neutralization. AIDS 17:291–300 [DOI] [PubMed] [Google Scholar]
- 75. van der Kuyl AC, Cornelissen M. 2007. Identifying HIV-1 dual infections. Retrovirology 4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. van Tienen C, et al. 2010. Two distinct epidemics: the rise of HIV-1 and decline of HIV-2 infection between 1990 and 2007 in rural Guinea-Bissau. J. Acquir. Immune Defic. Syndr. 53:640–647 [DOI] [PubMed] [Google Scholar]
- 77. Wei X, et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 79. Wren L, Kent SJ. 2011. HIV vaccine efficacy trial: glimmers of hope and the potential role of antibody-dependent cellular cytotoxicity. Hum. Vaccin 7:466–473 [DOI] [PubMed] [Google Scholar]
- 80. Wu X, et al. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J. Virol. 80:835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang M, et al. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246 [DOI] [PubMed] [Google Scholar]