Abstract
A subset of women in the Pumwani Sex Worker Cohort, established in 1985 in Nairobi, Kenya, remains uninfected despite repeated high-risk exposure (HIV-exposed, seronegative [HESN]) through active sex work. This HESN phenotype is associated with several alleles of human leukocyte antigens (HLAs) and specific CD8+ and CD4+ T cell responses to HIV-1. The associations of HLA alleles with differential HIV-1 infection are most likely due to their different abilities to present antigen and the different immune responses they induce. The characteristics of epitopes of HLA alleles associated with different outcomes of HIV-1 infection might therefore point to a vital clue for developing an effective vaccine. In this study, we systematically analyzed HIV-1 clade A and D Gag CD8+ T cell epitopes of two HLA class I alleles associated with different outcomes of HIV-1 infection. Binding affinity and off-rates of the identified epitopes were determined. Gamma interferon (IFN-γ) enzyme-linked immunospot (ELISpot) assays with patient peripheral blood mononuclear cells (PBMCs) validated the epitopes. Epitope-specific CD8+ T cells were further phenotyped for memory markers with tetramer staining. Our study showed that the protective allele A*01:01 recognizes only three Gag epitopes. By contrast, B*07:02, the allele associated with susceptibility, binds 30 epitope variants. These two alleles differ most importantly in the spectrum of Gag epitopes they can present and not in affinity, off-rates, the location of the epitopes, or epitope-specific Tem/Tcm frequencies. The binding of more epitopes and strong IFN-gamma ELISpot responses are associated with susceptibility to HIV-1 infection, while more focused antigen recognition of multiple subtypes is protective. Rational vaccine design should take these observations into account.
INTRODUCTION
Current candidate vaccines for HIV-1 have failed to provide protection and in some cases may have actually enhanced the likelihood of infection (3, 6–11, 44–46, 48) or produced only modest effects (15, 16, 51, 69, 70). This failure has been attributed to the difficulties of confronting a virus that targets cells which are a key component of the immune system and to the challenges posed by a pathogen that mutates rapidly and occurs with great genetic diversity. More critically, vaccines developed to date have been based on a conventional approach to control viral infection, which does not reflect a sufficient understanding of the correlates of protection against HIV-1. To develop a successful vaccine, we will need to understand which immunological parameters correlate with protection against HIV-1 and why.
Several cohort studies have documented that there is considerable heterogeneity in susceptibility to HIV-1 infection (21, 68). Despite repeated exposures, some individuals do not appear to become infected by HIV-1, and these individuals can be classified as HIV exposed, seronegative (HESN). Understanding why these individuals can escape HIV-1 infection and how their immune system works will help reveal parameters of protective immunity and aid in the development of effective vaccines and control strategies. A subset of women in the Pumwani Sex Worker Cohort (33, 65), established in 1985 in Nairobi, Kenya, remains HIV-1 seronegative and PCR negative despite repeated high-risk exposure through active sex work (21, 52). This resistance to HIV-1 infection is associated with several alleles of human leukocyte antigens (HLAs) and specific CD8+ and CD4+ T cell responses to HIV-1 (4, 5, 27, 28, 34, 43). HLAs constitute a group of host proteins that are central in regulating the immune response through the binding and presenting to T cells of peptides known as epitopes, which are derived from self and foreign proteins. The genes coding for HLAs are extremely polymorphic, resulting in a diversity of HLA alleles with variable recognition ability and affinity to the self and to the pathogenic proteins in the population. The contribution of different HLA alleles to virus control varies because of differences in antigenic recognition. The associations of specific HLA alleles with different outcomes of HIV-1 infection are most likely due to the differences in the antigenic peptides or epitopes of HIV being presented and the resulting immune responses following antigen recognition. Moreover, T cell responses to HIV-1 Gag correlate with viremia control and better outcomes of disease progression (25, 30, 37, 49, 54, 58, 62, 74). Finding out which differences in the Gag epitopes between HLA alleles are associated with different outcomes of HIV-1 infection might therefore offer a vital clue for developing an effective HIV-1 vaccine.
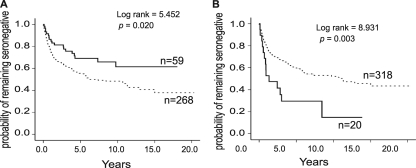
In this study, we systematically analyzed the HIV-1 clade A and D Gag epitope profiles of two HLA class I alleles, A*01:01 and B*07:02, which are independently associated with different outcomes of HIV-1 infection in order to elucidate the correlates of protective immunity to HIV-1. HLA-A*01 is significantly enriched in the HIV-1-resistant women (P = 0.016; odds ratio: 1.7; 95% confidence interval [95% CI]: 1.1 to 2.7). HIV-1-negative women with A*01 seroconvert significantly more slowly than women without this allele (Fig. 1A). By contrast, HLA-B*07:02 is associated with susceptibility to HIV-1 infection (P = 0.035; odds ratio: 0.38; 95% CI: 0.14 to 2.1) and rapid seroconversion (Fig. 1B) in the Pumwani cohort, as well as high viral loads and rapid disease progression in other populations (26, 67, 68). The peptide binding capacity of these two alleles was compared by screening an HIV-1 Gag overlapping peptide library with the iTopia epitope discovery system (Beckman Coulter). The identified epitopes were further characterized by affinity and off-rates and confirmed by gamma interferon (IFN-γ) ELISpot assays with patient peripheral blood mononuclear cells (PBMCs). Epitope-specific CD8+ T cells were further phenotyped for memory markers with tetramer staining. The broad epitope recognition and induced immune response were seen in the allele associated with susceptibility, while the more focused epitope recognition of multiple subtypes was observed in the allele associated with protection from HIV-1 infection. Thus, more epitopes and broad immune responses might not always be better for preventing HIV-1 infection. These observations should be considered when undertaking to design rational vaccines for HIV-1.
Fig 1.
Kaplan-Meier analysis of seroconversion in the Pumwani Sex Worker Cohort. (A) Women with A*01:01 were significantly less likely to seroconvert than individuals without A*01. Solid line, with A*01; dashed line, without A*01. (B) Women with B*07:02 were significantly more likely to seroconvert than individuals without B*07:02. Solid line, with B*07:02; dashed line, without B*07:02.
MATERIALS AND METHODS
Ethics statement.
All subjects gave informed consent to participate, and the study has been approved by the Institutional Review Boards at the Universities of Manitoba and Nairobi. Written informed consent has been obtained from all study participants.
Study cohort.
Subjects involved in this study were women enrolled in the Pumwani Sex Worker Cohort established in 1985 in Nairobi, Kenya (33, 65). The Pumwani Sex Worker Cohort is an open, prospective cohort to study factors influencing sexually transmitted diseases (33, 65). All enrollees have been followed biannually after enrollment, and a subgroup of women remain serologically and PCR negative for HIV-1 despite repeated exposure through high-risk sex work (21).
HLA class I typing.
DNA was isolated using QIAmp DNA mini kit and QIAgen EZ1 blood robot (QIAgen. Inc., Mississauga, ON, Canada). HLA-A, -B, and -C genes were amplified by PCR with gene specific primers (41, 42). The purified PCR products were sequenced with BigDye cycle sequencing kits (Applied Biosystems) using sequence specific primers and analyzed with an ABI3100 Prism genetic analyzer. Allele-specific primers were used to resolve ambiguous allele combinations. HLA-A, -B, and -C were typed using CodonExpress, a computer software program developed based on a taxonomy-based sequencing analysis (41, 42). Since exons 2 and 3 encode the peptide binding region of HLA class I molecules, analysis was restricted to these exons and all alleles were typed to 4-digit resolution. Homozygotes were classified based on the 4-digit typing for the peptide binding regions of the HLA genes. Between 1985 and 2001, 338 women who were seronegative at the cohort enrollment were included for the longitudinal seroconversion analysis. Of them, 327 were typed for HLA-A, 338 were typed for HLA-B, and 329 were typed for HLA-C.
Peptide synthesis and epitope screen using iTopia epitope discovery system.
Overlapping peptides spanning Gag of HIV-1 subtype A and D (predominant HIV subtypes in Kenyan) consensus clades were synthesized (JPT Peptides Technologies, Inc.). The peptide library is consisted of 632 peptides (9-mer overlapping by 8 amino acids) incorporating sequence variations of the subtype A and D consensus. The A*01:01 and B*07:02 kits of the iTopia epitope discovery system were purchased from Beckman Coulter, Inc. (San Diego, CA). The iTopia system consists of 3 assays to identify and characterize epitopes: peptide binding assay, off-rate assay, and affinity assay (72). The peptide binding assay evaluated the bindings of all 632 peptides to A*01:01 and B*07:02. Results are expressed as the percentage of the positive (POS) controls provided in the kits. Peptides showing binding of at least 30% of the positive control peptide were identified as binders and analyzed for binding affinity at the concentration range from 10−4 to 10−9 M. The binding affinity is expressed as 50% effective dose (ED50), which represents the concentration at which the peptide is bound to the major histocompatibility complex (MHC) at 50% maximum binding efficiency and is the midpoint of the sigmoidal dose-response curve. The identified peptides were analyzed for off-rate (complex stability) where t1/2 represents the time point when 50% of the peptide remains bound to the MHC. The off-rate was calculated as the time to the 50% of the highest binding (fluorescent intensity). Peptide binding, affinity, and off-rate values were calculated using the iTopia software and Graphpad Prism 4. ED50 was determined using a sigmoidal dose-response curve, and t1/2 was determined using a one-phase exponential decay curve (63, 72).
Validation of epitopes using ELISpot assays.
Peptides (9 amino acid) identified by the iTopia epitope discovery system were synthesized (Sigma Genosys, Oakville, ON) for confirmation with IFN-γ ELISpot assays using patient PBMCs. To confirm that peptides identified by the iTopia epitope discovery system are true epitopes capable of stimulating T cell responses, functional IFN-γ ELISpot assays were carried out as previously described (61). The peptide stocks were dissolved in dimethyl sulfoxide (DMSO), and the stocks were diluted to a final concentration of 10 mg/ml in RPMI medium for ELISpot assays. PBMCs were suspended in RPMI medium, and 105 cells were stimulated in duplicate for each peptide. Responses were considered positive if there were at least 50 spot-forming units (SFU)/million PBMCs after background subtraction and the positive control was successful (61).
Tetramer staining and phenotyping.
HIV-specific CD8+ T cells were detected with the following allophycocyanin (APC)-conjugated iTAg MHC class I (MHC-I) tetramers: HLA-A*01:01-NSSQVSQNY (HIV Gag 124 to 132), B*07:02-APRKKGCWK (HIV Gag 408 to 416), and B*07:02-SPRTLNAWV (HIV Gag 148 to 156) from Beckman Coulter (14). Fresh PBMCs isolated by Ficoll gradient centrifugation were incubated with antibodies: anti-CD3-Amcyan, anti-CD8-APC-Cy7, anti-CD4-PE-Cy5, anti-CCR7-PE-Cy7, anti-CD28-PE, and anti-CD27 Alexa Fluoro 700 from BD-Bioscience (San Diego, CA) and anti-CD45-RA-ECD (Beckman Coulter) for 30 min at room temperature. Cells were washed in phosphate-buffered saline with 2% fetal calf serum (FCS). After the wash, cells were fixed with 1% paraformaldehyde and analyzed on a BD LSRII system. Data were compensated and analyzed using Flowjo Software.
Statistical analysis.
Kaplan-Meier survival analysis was conducted among >300 women who enrolled in the Pumwani cohort between 1985 and 2001 and who were HIV seronegative at enrollment. Cox regression and Kaplan-Meier analysis were conducted for HLA alleles with genotype frequencies above 5% in this population. A total of 12 HLA-A, 14 HLA-B, and 11 HLA-C alleles with genotype frequencies greater than 5% in this population were included in the Cox regression analysis using a backward conditional method. The FACS data were analyzed with Graph Pad Prism 4.0 (Graph Pad Software, San Diego, CA). The nonparametric Kruskal-Wallis test was used to compare values between the independent groups. All differences were considered significant at P < 0.05.
RESULTS
A*01:01 and B*07:02 are independently associated with different outcomes of HIV-1 infection.
To identify HLA class I alleles associated with resistance or susceptibility to HIV-1 infection, we analyzed the effect of HLA class I alleles on the seroconversion rates of more than 300 women who were HIV uninfected at the cohort enrollment by Kaplan-Meier survival analysis. Kaplan-Meier survival analysis showed that women with A*01 seroconverted significantly slower than those without A*01 (Fig. 1A), whereas, women who have B*07:02 seroconvert significantly faster than those without B*07:02 (Fig. 1B). A total of 12 HLA-A, 14 HLA-B, and 11 HLA-C alleles with genotype frequencies greater than 5% in this population were included in the Cox regression analysis using a backward conditional method. Results showed that A*01, C*06:02, and C*07:01 are independently associated with slower seroconversion, while A*23:01, B*07:02, and B*42:01 are independently associated with rapid seroconversion (Table 1). The enrollment years and ages of women with and without these alleles are very similar.
Table 1.
Multivariate analysis of HLA class I alleles associated with seroconversion
| Allele | Cox regression result |
|||
|---|---|---|---|---|
| P valuea | Exp(B)b | 95% CIc for Exp(B) |
||
| Lower | Upper | |||
| Protective | ||||
| A*01 | 0.031 | 1.731 | 1.052 | 2.849 |
| C*06:02 | 0.015 | 1.679 | 1.105 | 2.551 |
| C*07:01 | 0.042 | 1.520 | 1.015 | 2.277 |
| Susceptible | ||||
| A*23:01 | 0.010 | 0.551 | 0.351 | 0.865 |
| B*07:02 | 0.011 | 0.483 | 0.276 | 0.846 |
| B*42:01 | 0.015 | 0.593 | 0.389 | 0.905 |
Significances are indicated.
Exp(B), Exponent of B, hazard ratio.
95% CI, confidence interval.
The spectrum and characteristics of HLA-A*01:01 HIV-1 Gag epitopes.
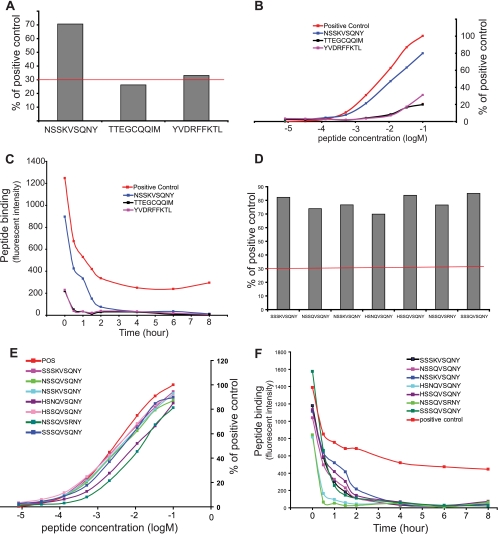
Epitope screening with the iTopia epitope discovery system showed that only 2 out of 632 peptides met the binding criteria (30% of the positive control peptide) for A*01:01 (Fig. 2A). The strongest binder was peptide NSSKVSQNY at 70.6% of the positive control, followed by two weaker peptides, YVDRFFKTL at 33.1% of the positive control and TTEGCQQIM at 26.2% of the positive control peptide. The binding affinity of peptide NSSKVSQNY displayed a typical sigmoidal binding curve over the concentrations tested, and the peptide has the highest affinity for A*01:01 with an ED50 at 1.21 × 10−5 M among the three positive peptides tested (Fig. 2B). The other two peptides have a relatively flat curve with ED50 values of 1.70 × 10−5 M for TTEGCQQIM and 4.91 × 10−5 M for YVDRFFKTL. The off-rate assay shows that the peptide NSSKVSQNY dissociated from the complex at a rate (t1/2) of 0.94 h. The t1/2 is 0.33 h for peptide TTEGCQQIM and 0.30 h for peptide YVDRFFKTL, both at a very low binding capacity (Fig. 2C). The peptide binding, affinity, and off-rate values are summarized in Table 2. We further tested 14 variants of peptide NSSKVSQNY representing major variants of HIV subtypes A, B, C, and D. A*01:01 binds 6 major variant peptides of HIV subtypes A, B, and D very well with comparable affinities and off-rates (Fig. 2D to F; Table 2).
Fig 2.
The binding, affinity, and off-rates of A*01:01 HIV-1 Gag epitopes. (A) The level of binding of Gag epitopes to A*01:01. (B) The binding affinity of A*01:01 Gag epitopes. (C) The off-rates of A*01:01 Gag epitopes. (D) The binding of variant peptides of A*01:01 epitope NSSKVSQNY. (E) The binding affinity of epitope variants to A*01:01. (F) The off-rates of A*01:01 epitope variants.
Table 2.
Summary of A*0101 and B*0702 iTopia and ELISpot data
| Allele | Peptide sequence | aab no. in Gag | % Peptide binding | Affinity (ED50) | Off-rate t1/2 (h) | Mean ELISpot responsec | Reported in HIV databasea |
|---|---|---|---|---|---|---|---|
| A*01:01 | NSSKVSQNY | 124–132 | 70.6 | 1.21 × 10−5 | 0.94 | 79 | Y |
| TTEGCQQIM | 53–61 | 26.2 | 1.70 × 10−5 | 0.33 | 77 | N | |
| YVDRFFKTL | 296–304 | 33.1 | 4.91 × 10−5 | 0.30 | 2,148 | Y | |
| SSSKVSQNY | 124–132 | 82.2 | 5.13 × 10−6 | 0.49 | 115 | N | |
| NSSQVSQNY | 124–132 | 73.9 | 5.02 × 10−6 | 0.48 | 96 | Y | |
| HSNQVSQNY | 124–132 | 69.9 | 1.43 × 10−5 | 0.31 | 115 | Y | |
| HSSQVSQNY | 124–132 | 83.7 | 4.30 × 10−6 | 0.79 | 74 | N | |
| NSSQVSRNY | 124–132 | 76.6 | 1.21 × 10−5 | 0.27 | 127 | N | |
| SSSQVSQNY | 124–132 | 85.1 | 5.18 × 10−6 | 0.43 | 130 | N | |
| B*07:02 | APPAEIFGM | 456–464 | 57.6 | 1.75 × 10−5 | 0.40 | 100 | N |
| APRKKGCWK | 406–414 | 145.3 | 8.96 E-07 | 0.78 | 148 | Y | |
| FPQSRPEPT | 447–455 | 55.6 | 1.50 × 10−5 | 0.70 | 55 | N | |
| GPGATLEEM | 338–346 | 35.1 | 1.78 × 10−5 | 0.81 | 80 | N | |
| GPGHKARVL | 355–363 | 93.1 | 4.80 × 10−6 | 0.42 | 84 | Y | |
| GPIPPGQMR | 221–229 | 45.9 | 4.84 × 10−4 | 0.78 | 215 | N | |
| GPSHKARVL | 355–363 | 91.6 | 3.14 × 10−5 | 0.44 | 95 | Y | |
| IVGGHQAAM | 190–198 | 46.4 | 4.49 × 10−5 | 0.47 | n/a | N | |
| IVQNAQGQM | 134–142 | 38.8 | 4.13 × 10−4 | 0.72 | n/a | N | |
| RALGPGATL | 335–343 | 52.2 | 1.49 × 10−5 | 0.45 | 115 | Y | |
| RLRPGGKKK | 20–28 | 32.4 | 7.83 × 10−5 | 0.71 | 135 | Y | |
| RPGNFPQSR | 443–451 | 37.3 | 5.37 × 10−5 | 0.73 | 80 | N | |
| SPRTLNAWV | 148–156 | 113.3 | 1.61 × 10−6 | 3.78 | 245 | Y | |
| TPQDLNMML | 180–188 | 36.9 | 8.12 × 10−5 | 0.83 | 905 | Y | |
| TPQEQIGWM | 242–250 | 44.8 | 2.60 × 10−5 | 0.67 | 185 | N | |
| TPQEQLGWM | 242–250 | 43.7 | 1.04 × 10−4 | 0.70 | <50 | N | |
| VRMYSPVSI | 274–282 | 41.6 | 1.69 × 10−4 | 0.83 | 435 | Y | |
| WPSSKGRPG | 437–445 | 81.0 | 2.71 × 10−5 | 0.40 | 115 | N | |
| YPLVSLKSL | 483–491 | 75.5 | 2.27 × 10−5 | 0.69 | 703 | N | |
| YVDRFFKTL | 296–304 | 60.0 | 4.37 × 10−5 | 0.50 | 1280 | Y | |
| KARVLAEAM | 359–367 | 66.5 | 8.08 × 10−6 | 0.44 | 78 | N | |
| QVQHTNIMM | 369–377 | 46.1 | 2.55 × 10−5 | 0.27 | 87 | N | |
| QAQQPNVMM | 369–377 | 46.2 | 1.92 × 10−5 | 0.35 | 96 | N | |
| QVNGNTAIM | 369–377 | 46.3 | 2.28 × 10−5 | 1.23 | 99 | N | |
| QANANTAIM | 369–377 | 46.0 | 1.36 × 10−5 | 1.35 | 109 | N | |
| ATNANAAIM | 370–378 | 37.4 | 2.08 × 10−5 | 0.25 | 95 | N | |
| NIMMQRGNF | 375–383 | 73.4 | 9.72 × 10−6 | 0.25 | 100 | N | |
| NIMMQRSNF | 375–383 | 73.0 | 9.31 × 10−6 | 0.25 | 121 | N | |
| NVMMQRSNF | 375–383 | 70.2 | 8.96 × 10−6 | 0.27 | 117 | N | |
| AIMMQRGNF | 375–383 | 91.6 | 3.45 × 10−6 | 0.38 | 113 | N |
Y, yes; N, no.
aa, amino acid.
n/a, not applicable.
The spectrum and characteristics of HIV Gag epitopes of HLA-B*07:02.
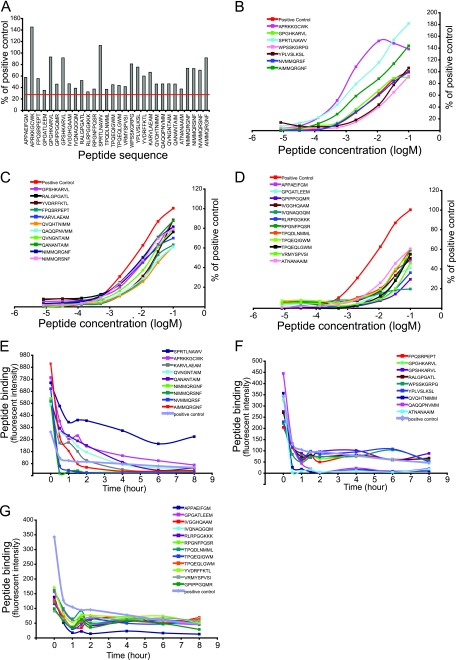
A total of 30 peptides bound to B*07:02 with greater than 30% of the positive control peptide (Fig. 3A; Table 2) as determined by the iTopia epitope discovery system screen. The peptides were divided into three groups based on their relative binding to the positive control peptide and analyzed their binding affinity to B*07:02 (Fig. 3B to D; Table 2). Group 1 (Fig. 3B) contained 7 peptides, three of which had higher affinity for B*07:02 than the positive control peptide. The binding of APRKKGCWK to B*07:02 was 145.3% of the positive control with an ED50 of 8.96 × 10−7 M, the binding of SPRTLNAWV to B*07:02 was 113.3% of the positive control with an ED50 of 1.61 × 10−6 M, and the binding of AIMMQRGNF to B*07:02 was 91.6% of the positive control with an ED50 of 3.45 × 10−6 M. Group 2 (Fig. 3C) contained 11 peptides with binding between 60% and 90% of the positive control peptide. These peptides were GPSHKARVL, RALGPGATL, YVDRFFKTL, FPQSRPEPT, KARVLAEAM, QVQHTNIMM, QAQQPNVMM, QVNGNTAIM, QANANTAIM, NIMMQRGNF, and NIMMQRSNF. Group 3 (Fig. 3D) contained 12 peptides with more than 30% but less than 60% binding of the positive control peptide. The peptides were APPAEIFGM, GPGATLEEM, GPIPPGQMR, IVGGHQAAM, IVQNAQGQM, RLRPGGKKK, RPGNFPQSR, TPQDLNMML, TPQEQIGWM, TPQEQLGWM, VRMYSPVSI, and ATNANAAIM. The detailed binding and ED50 values are shown in Table 2.
Fig 3.
The binding, affinity, and off-rates of B*07:02 HIV-1 Gag epitopes. (A) The level of binding of Gag epitopes to B*07:02. (B to D) The binding affinity of B*07:02 Gag epitopes. (E to G) The off-rates of B*07:02 Gag epitopes.
The off-rate assay was carried out on all 30 peptides to determine their rate of dissociation from B*07:02 (72). B*07:02 binds to several peptides with much higher capacity than the positive control peptide, and the binding persisted for a very long time; these peptides include SPRTLNAWV, APRKKGCWK, QANANTAIM, QVNGNTAIM, AIMMQRGNF, and KARVLAEAM (Fig. 3E). Most of the peptides dissociated from B*07:02 to the level of 50% of saturated binding within an hour, and their binding to B*07:02 is comparable to or lower than that of the positive control peptide (Fig. 3 E, F and G; Table 2).
Validation of A*01:01 and B*07:02 epitopes by IFN-γ ELISpot assay.
Using the iTopia epitope discovery system, we have determined HIV-1 Gag peptide binding capacity, affinity, and off-rates of A*01:01 and B*07:02. To confirm these peptides are true epitopes capable of stimulating T cell responses, we validated the epitopes with IFN-γ ELISpot assays with patient PBMCs. PBMCs isolated from 5 to 12 A*01:01+ women were tested by IFN-γ ELISpot assay for each of the 9 peptides (including the 6 variants of NSSKVSQNY) identified as A*01:01 epitopes (Table 3). Two or more A*01:01+ patients had positive ELISpot responses to the nine peptides tested (>50 SFU/million) (Table 3). We were able to detect responses for 3 epitopes and 6 variants of the epitope NSSKVSQNY for A*01:01 (Table 3). Four of the epitopes and epitope variants of A*01:01 are novel and have not been reported previously (Table 2). Among the 12 A*01:01+ subjects tested for the identified peptides, two are defined as resistant to HIV-1 infection (HESN) (enrolled in the cohort in 1987 and 1992, respectively), one is a seronegative new enrollee (NSN) and the rest are HIV-1 infected (POS). Positive ELISpot responses to the peptides were detected in 28% to 80% of the subjects tested (Tables 3 and 4). The magnitudes of the ELISpot responses among the tested subjects were comparable, except for one HIV+ patient whose response to peptide YVDRFFKTL is >10-fold higher than the average ELISpot responses to all peptides tested (Table 4). The HLA class I allele data for the subjects are listed below (see Table 6).
Table 3.
Summary of ELISpot responses to Gag peptides identified by iTopia epitope discovery system screening
| Peptide |
Patient sample |
Mean SFU/million | ||
|---|---|---|---|---|
| Allele | Sequence | Tested | Responded | |
| A*01:01 | NSSKVSQNY | 12 | 3 | 79 |
| TTEGCQQIM | 7 | 3 | 77 | |
| YVDRFFKTL | 7 | 2 | 2,148 | |
| SSSKVSQNY | 5 | 2 | 115 | |
| NSSQVSQNY | 5 | 4 | 96 | |
| HSNQVSQNY | 5 | 4 | 115 | |
| HSSQVSQNY | 5 | 4 | 74 | |
| NSSQVSRNY | 5 | 3 | 127 | |
| SSSQVSQNY | 5 | 4 | 130 | |
| B*07:02 | APPAEIFGM | 8 | 1 | 100 |
| APRKKGCWK | 8 | 2 | 148 | |
| FPQSRPEPT | 7 | 1 | 55 | |
| GPGATLEEM | 5 | 1 | 80 | |
| GPGHKARVL | 8 | 2 | 83 | |
| GPIPPGQMR | 5 | 1 | 215 | |
| GPSHKARVL | 8 | 3 | 62 | |
| RALGPGATL | 5 | 3 | 115 | |
| RLRPGGKKK | 5 | 1 | 135 | |
| RPGNFPQSR | 5 | 1 | 80 | |
| SPRTLNAWV | 8 | 2 | 245 | |
| TPQDLNMML | 5 | 1 | 905 | |
| TPQEQIGWM | 5 | 1 | 185 | |
| TPQEQLGWM | 4 | 0 | <50 | |
| VRMYSPVSI | 4 | 1 | 435 | |
| WPSSKGRPG | 8 | 3 | 115 | |
| YPLVSLKSL | 8 | 2 | 703 | |
| YVDRFFKTL | 8 | 2 | 1,280 | |
| GPGHKARVL | 8 | 4 | 84 | |
| GPSHKARVL | 8 | 6 | 95 | |
| KARVLAEAM | 8 | 5 | 78 | |
| QVQHTNIMM | 8 | 6 | 87 | |
| QAQQPNVMM | 8 | 5 | 96 | |
| QVNGNTAIM | 8 | 6 | 99 | |
| QANANTAIM | 8 | 6 | 109 | |
| ATNANAAIM | 8 | 5 | 95 | |
| NIMMQRGNF | 8 | 3 | 100 | |
| NIMMQRSNF | 8 | 6 | 121 | |
| NVMMQRSNF | 8 | 6 | 117 | |
| AIMMQRGNF | 8 | 4 | 113 | |
Table 4.
Detailed ELISpot data for tested HLA A*01:01 patientsb
| Peptide | Patient no., HIV statusa |
Responses/tested | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 893, RES (HESN) | 1589, RES (HESN) | 2137, POS | 1932, POS | 2003, POS | 2204, POS | 1811, POS | 2463, NSN | 2646, POS | 2297, POS | 2261, POS | 2178, POS | ||
| TTEGCQQIM | 15 | 15 | 85 | 65 | 10 | 80 | 40 | N/T | N/T | N/T | N/T | N/T | 3/7 |
| NSSKVSQNY | 55 | 80 | 15 | 65 | 25 | 15 | 30 | 60 | 35 | 95 | 75 | 120 | 7/12 |
| YVDRFFKTL | 0 | 10 | 30 | 50 | 4245 | 5 | 10 | N/T | N/T | N/T | N/T | N/T | 2/8 |
| SSSKVSQNY | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 0 | 45 | 30 | 90 | 140 | 2/5 |
| NSSQVSQNY | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 45 | 50 | 80 | 130 | 125 | 4/5 |
| HSNQVSQNY | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 55 | 15 | 50 | 115 | 240 | 4/5 |
| HSSQVSQNY | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 50 | 10 | 70 | 60 | 115 | 4/5 |
| NSSQVSRNY | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 10 | 125 | 45 | 155 | 100 | 3/5 |
| SSSQVSQNY | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 75 | 20 | 65 | 95 | 285 | 4/5 |
RES (HESN), HIV-1 resistant; POS, HIV-1 positive; NSN, newly enrolled seronegative; N/T, not tested.
Bold patient numbers indicate that fresh PBMCs were used. Lightface patient numbers indicate that PBMCs were previously frozen and stored in liquid nitrogen.
Table 6.
HLA class I alleles of all patients tested in ELISpot assays
| Allele | Patient no. | HLA class I allelea |
|||||
|---|---|---|---|---|---|---|---|
| HLA-A | HLA-B | HLA-C | |||||
| A*01:01 | 893 | 01:01 | 24:02 | 15:17 | 35:02 | 04:01 | 17:01 |
| 1589 | 01:01 | 30:02 | 58:02 | 58:02 | 06:02 | 06:06 | |
| 2137 | 01:01 | 23:01 | 58:02 | 44:15 | 03:02 | 04:07 | |
| 1932 | 01:01 | 01:01 | 81:01 | 37:01 | 06:02 | 18:01 | |
| 2003 | 01:01 | 68:02 | 81:01 | 15:10 | 03:04 | 18:01 | |
| 2204 | 01:01 | 68:02 | 42:01 | 49:01 | 07:01 | 17:01 | |
| 1811 | 01:01 | 23:01 | 07:02 | 45:01 | 16:01 | 07:02 | |
| 2463 | 01:01 | 02:01 | 41:01 | 42:01 | 07:01 | 17:01 | |
| 2646 | 01:01 | 29:02 | 57:01 | 45:01 | 07:01 | 06:02 | |
| 2297 | 01:01 | 30:01 | 57:03 | 08:01 | 18:01 | 07:04 | |
| 2261 | 01:01 | 49:01 | 27:26 | 02:02 | 02:02 | 07:01 | |
| 2178 | 01:01 | 29:02 | 81:01 | 81:01 | 07:04 | 08:04 | |
| B*07:02 | 2044 | 03:08 | 29:01 | 07:02 | 08:01 | 07:02 | 07:02 |
| 2230 | 01:01 | 03:01 | 07:02 | 58:02 | 06:06 | 07:02 | |
| 2181 | 30:01 | 31:01 | 07:02 | 53:01 | 07:14 | 03:04 | |
| 2127 | 01:01 | 03:01 | 07:02 | 41:01 | 07:01 | 07:23 | |
| 1961 | 03:01 | 23:01 | 07:02 | 42:01 | 17:01 | 04:01 | |
| 2035 | 23:01 | 30:01 | 07:02 | 15:10 | 07:14 | 03:04 | |
| 1394 | 02:01 | 68:02 | 07:02 | 13:02 | 06:02 | 04:01 | |
| 2257 | 30:02 | 23:01 | 07:02 | 18:01 | 07:02 | 15:05 | |
| 2427 | 68:02 | 29:02 | 07:02 | 39:10 | 07:27 | 12:03 | |
| 2320 | 33:01 | 02:01 | 07:02 | 49:01 | 07:01 | 07:02 | |
| 2400 | 68:02 | 02:01 | 07:02 | 42:01 | 08:02 | 17:01 | |
| 2545 | 68:02 | 30:02 | 07:02 | 18:01 | 07:04 | 07:02 | |
| 2619 | 02:01 | 68:01 | 07:02 | 53:01 | 07:02 | 04:01 | |
| 2423 | 68:02 | 68:02 | 07:02 | 07:02 | 07:02 | 07:02 | |
| 2425 | 29:02 | 68:02 | 07:02 | 39:10 | 12:03 | 07:02 | |
| 2644 | 36:01 | 68:02 | 07:02 | 53:01 | 17:01 | 06:02 | |
Bold data indicate A*01:01 or B*07:02 allele of the patients in ELISpot assays.
PBMCs from 4 to 10 patients were tested for each of the 28 peptides (peptide IVGGHQAAM and IVQNAQGQM failed to be synthesized for ELISpot assays) binding to B*07:02 (Table 3). All peptides except TPQEQLGQM had at least one positive ELISpot response (>50 SFU/million) from the tested patient samples. We were able to confirm 19 Gag epitopes and 8 epitope variants of B*07:02. Twenty-one epitopes or epitope variants are novel (Table 2). Among the 16 B*07:02 subjects tested for the identified peptide by using the iTopia epitope discovery system, one was defined as resistant to HIV-1 infection (HESN), nine subjects were HIV-1-negative new enrollees (NSN), and six subjects were HIV-1 infected (Table 5). Positive ELISpot responses to the peptides were detected in 12.5% to 75% of the subjects tested (Tables 3 and 5). The magnitudes of the ELISpot responses among the tested subjects were comparable, except for one HIV+ patient whose response to peptide YVDRFFKTL was much higher than the average ELISpot responses to all peptides tested (Table 5). The HLA class I allele data for the subjects are listed in Table 6. Peptide YVDRFFKTL can be recognized by both A*01:01 and B*07:02, and it induced the highest IFN-γ ELISpot responses in some patients. The higher magnitude of ELISpot responses to the peptide may be due to specific T cell receptor usage and need to be investigated further.
Table 5.
Detailed ELISpot data for tested HLA B*07:02 patientsb
| Peptide | Patient no., HIV statusa |
Responses/tested | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2044, NSN | 2230, NSN | 2181, POS | 2127, NSN | 1961, NSN | 2035, NSN | 1394, RES (HESN) | 2257, POS | 2427, POS | 2320, NSN | 2400, POS | 2545, POS | 2619, POS | 2423, NSN | 2425, NSN | 2644, NSN | ||
| APPAEIFGM | 0 | 0 | 100 | 0 | 5 | 0 | 10 | 20 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 1/8 |
| APRKKGCWK | 35 | 240 | 0 | 0 | 0 | 20 | 55 | 0 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 2/8 |
| FPQSRPEPT | 0 | N/T | 25 | 55 | 35 | 0 | 0 | 0 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 1/7 |
| GPGATLEEM | 0 | N/T | N/T | 80 | 25 | 35 | N/T | 45 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 1/5 |
| GPGHKARVL | 90 | 0 | 0 | 5 | 0 | 0 | 75 | 30 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 2/8 |
| GPIPPGQMR | 0 | N/T | N/T | 15 | 10 | 0 | N/T | 215 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 1/5 |
| GPSHKARVL | 55 | 0 | 5 | 65 | 20 | 65 | 0 | 10 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 3/8 |
| RALGPGATL | 0 | N/T | N/T | 175 | 80 | 0 | N/T | 90 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 3/5 |
| RLRPGGKKK | 0 | N/T | N/T | 135 | 10 | 35 | N/T | 20 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 1/5 |
| RPGNFPQSR | 0 | N/T | N/T | 0 | 0 | 0 | N/T | 80 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 1/5 |
| SPRTLNAWV | 115 | 375 | 0 | 0 | 35 | 0 | 25 | 20 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 2/8 |
| TPQDLNMML | 0 | N/T | N/T | 15 | 30 | 0 | N/T | 905 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 1/5 |
| TPQEQIGWM | 0 | N/T | N/T | 0 | 20 | 10 | N/T | 185 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 1/5 |
| TPQEQLGWM | 0 | N/T | N/T | 0 | 35 | 0 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 0/4 |
| VRMYSPVSI | 0 | N/T | N/T | 435 | 0 | 0 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 1/4 |
| WPSSKGRPG | 70 | 220 | 0 | 0 | 0 | 55 | 25 | 5 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 3/8 |
| YPLVSLKSL | 0 | 0 | 1320 | 85 | 30 | 0 | 10 | 40 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 2/8 |
| YVDRFFKTL | 0 | 0 | 2500 | 60 | 40 | 0 | 10 | 15 | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 2/8 |
| GPGHKARVL | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 55 | 75 | 70 | 45 | 5 | 5 | 15 | 135 | 4/8 |
| GPSHKARVL | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 70 | 67 | 70 | 55 | 15 | 70 | 10 | 235 | 6/8 |
| KARVLAEAM | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 45 | 50 | 80 | 75 | 0 | 115 | 10 | 70 | 5/8 |
| QVQHTNIMM | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 45 | 55 | 95 | 75 | 5 | 80 | 50 | 165 | 6/8 |
| QAQQPNVMM | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 65 | 30 | 70 | 70 | 0 | 10 | 50 | 225 | 5/8 |
| QVNGNTAIM | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 65 | 50 | 80 | 90 | 5 | 120 | 5 | 190 | 6/8 |
| QANANTAIM | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 75 | 25 | 65 | 80 | 5 | 115 | 55 | 265 | 6/8 |
| ATNANAAIM | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 60 | 60 | 85 | 75 | 15 | 25 | 5 | 195 | 5/8 |
| NIMMQRGNF | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 35 | 25 | 80 | 50 | 10 | 0 | 10 | 170 | 3/8 |
| NIMMQRSNF | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 65 | 70 | 125 | 50 | 5 | 105 | 15 | 310 | 6/8 |
| NVMMQRSNF | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 55 | 90 | 85 | 100 | 5 | 55 | 15 | 315 | 6/8 |
| AIMMQRGNF | N/T | N/T | N/T | N/T | N/T | N/T | N/T | N/T | 90 | 35 | 35 | 65 | 0 | 80 | 30 | 215 | 4/8 |
RES (HESN), HIV-1 resistant; POS, HIV-1 positive; NSN, newly enrolled seronegative; N/T, not tested.
Bold patient numbers indicate that fresh PBMCs were used. Lightface patient numbers indicate that PBMCs were previously frozen and stored in liquid nitrogen.
The binding motif of nine A*01:01 epitopes and epitope variants fits very well with the reported binding motif in the Immune Epitope database (IDEB; http://www.immuneepitope.org/MHCalleleId/142). However, among many novel B*07:02 Gag epitopes identified in this study (Table 2), 4 new amino acid residues at anchor positions P2 and P9 were identified (Table 7). Polar basic amino acids such as lysine and arginine are the surprising additions to the anchor positions. This suggests that the anchor residues of B*07:02 are more tolerant to different amino acid changes than previously reported (64).
Table 7.
B*07:02 Gag epitopes identified in this study and binding motifsa
| Binding motif | Amino acid at indicated anchor position |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | |
| Previously defined motif | X | PVA | X | X | X | X | X | X | FLM(AIVYW) |
| Epitopes identified in this study | A | P | P | A | E | I | F | G | M |
| A | P | R | K | K | G | C | W | K | |
| F | P | Q | S | R | P | E | P | T | |
| G | P | G | A | T | L | E | E | M | |
| G | P | G | H | K | A | R | V | L | |
| G | P | I | P | P | G | Q | M | R | |
| G | P | S | H | K | A | R | V | L | |
| R | A | L | G | P | G | A | T | L | |
| R | L | R | P | G | G | K | K | K | |
| R | P | G | N | F | P | Q | S | R | |
| S | P | R | T | L | N | A | W | V | |
| T | P | Q | D | L | N | M | M | L | |
| T | P | Q | E | Q | I | G | W | M | |
| T | P | Q | E | Q | L | G | W | M | |
| V | R | M | Y | S | P | V | S | I | |
| W | P | S | S | K | G | R | P | G | |
| Y | P | L | V | S | L | K | S | L | |
| Y | V | D | R | F | F | K | T | L | |
| G | P | G | H | K | A | R | V | L | |
| G | P | S | H | K | A | R | V | L | |
| K | A | R | V | L | A | E | A | M | |
| Q | V | Q | H | T | N | I | M | M | |
| Q | A | Q | Q | P | N | V | M | M | |
| Q | V | N | G | N | T | A | I | M | |
| Q | A | N | A | N | T | A | I | M | |
| A | T | N | A | N | A | A | I | M | |
| N | I | M | M | Q | R | G | N | F | |
| N | I | M | M | Q | R | S | N | F | |
| N | V | M | M | Q | R | S | N | F | |
| A | I | M | M | Q | R | G | N | F | |
Bolded amino acids are additional residues identified through iTopia peptide binding assays and confirmed by ELISpot assays. Previously described HLA binding motifs are summarized in reference 49.
Comparison of A*01:01 and B*07:02 Gag epitopes.
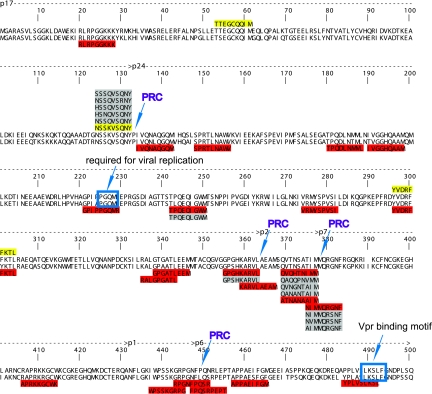
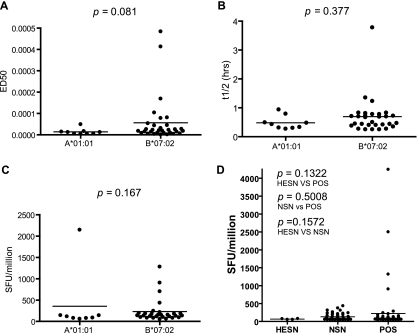
We compared the numbers, affinities, off-rates, and ELISpot responses, as well as the locations of the identified A*01:01 and B*07:02 Gag epitopes. As expected, only one epitope of A*01:01 overlaps with the epitopes of B*07:02. However, we were surprised to see the substantial difference in the numbers of Gag epitopes between the two alleles. B*07:02 Gag epitopes (19 epitopes) outnumber those of A*01:01 (3 epitopes) by 6-fold, excluding the epitope variants. B*07:02 epitopes are distributed along the entire Gag, while two of the three A*01:01 epitopes are located in p17 and one in p24 (Fig. 4). The locations of the A*01:01 Gag epitopes do not appear to be special, except for one, NSSKVSQNY, which is located right beside the p17/p24 protease cleavage site (Fig. 4). We further analyzed 15 variants of this peptide from HIV-1 subtypes A, B, C, and D. A*01:01 can bind to major variants of subtype A, B, and D and their recombinants (Fig. 2D to F; Table 1). However, with a few exceptions, A*01:01 does not bind the peptide variants of subtype C. There is no significant difference between the off-rates of the epitopes/epitope variants of A*01:01 and B*07:02 (P = 0.377) (Fig. 5B), and while B*07:02 binds to its epitopes with higher affinity than A*01:01 does to its epitopes, the difference is not significant (P = 0.081) (Fig. 5A). We observed no significant difference in the IFN-γ ELISpot SFU values between the Gag epitopes of A*01:01 and B*07:02 (Fig. 5C). There is no significant difference in magnitude of ELISpot responses to the individual peptides among HESN, NSN, and POS groups (Fig. 5D).
Fig 4.
The location of HIV-1 Gag epitopes of A*01:01 and B*07:02. A*01:01 epitopes are highlighted in yellow, and B*07:02 epitopes are highlighted in red. The epitope variants are shaded in gray. PRC, protease cleavage sites. The clade A Gag consensus is at the top of the alignment, and the clade D consensus is at the bottom.
Fig 5.
Comparison of the HIV-1 Gag epitopes of A*01:01 and B*07:02. (A) Binding affinities of HIV-1 Gag epitopes and epitope variants of A*01:01 and B*07:02. (B) Off-rates of HIV-1 Gag epitopes and epitope variants of A*01:01 and B*07:02. (C) IFN-γ ELISpot responses of HIV-1 Gag epitopes and epitope variants of A*01:01 and B*07:02. (D) Comparison of ELISpot responses to the A*01:01 and B*07:02 peptides among HESN, NSN, and POS.
A*01:01 and B*07:02 epitope-specific CD8+ T cells.
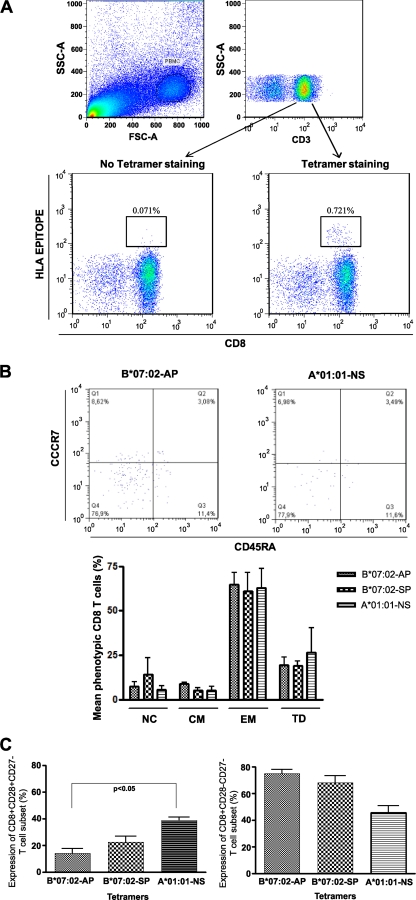
To determine if differences in potential epitope presentation between A*01:01 and B*07:02 epitopes may affect the functional phenotype of A*01:01- and B*07:02-responsive cells in proof-of-concept studies, we examined the memory phenotype using 3 epitope-specific tetramers and a phenotyping panel to define memory profiles among 6 chronic HIV-1 infected subjects (3 for tetramer A*01:01-NSSQVSQNY, and 3 for both B*07:02-APRKKGCWK and B*07:02-SPRTLNAWV) (Fig. 6A). The proportion of naïve (CD45RA+ CCR7+) and central memory (Tcm− CD45RA− CCR7+)-, effector memory (Tem− CD45RA− CCR7−)-, and terminal differentiated (Ttd− CD45RA+ CCR7)-specific CD8+ T cells was determined. We observed no statistically significant differences in the frequencies of memory phenotypes as defined by CD45RA and CCR7 of tetramer-specific HIV-1-specific CD8+ T cells (Fig. 6B). The majority responses classified as effector memory (Tem) and the proportion of terminal differentiated (Td) cells were very similar (Fig. 6B). We further examined the frequencies of different effector memory CD8+ T cell subsets with the costimulator markers CD27 and CD28. CCR7− CD45RA− CD28+ CD27− and CCR7− CD45RA− CD28− CD27− CD8+ T cells were identified within all of the tetramer-positive T cell populations. Higher frequencies of CCR7− CD45RA− CD28+ CD27− CD8+ T cells were identified with NSSKVSQNY-A*01:01 tetramer than with the B*07:02 tetramers (APRKKGCWK-B*07:02 and SPRTLNAWV-B*07:02), and the difference between NSSKVSQNY-A*01:01 and APRKKGCWK-B*07:02 is significant (Fig. 6C). Conversely, the B*07:02 tetramers identified more CCR7− CD45RA− CD28− CD27− CD8+ T cells, terminally differentiated effector memory T cells, than the NSSKVSQNY-A*01:01 tetramer, but the difference was not significant (Fig. 6C). This indicates that while the responses were phenotypically similar in Tem/Tcm frequencies, there may be subtle differences in the exhaustion and stimulatory potentials between the tetramer-specific T cell pools in A*01:01 and B*07:02 HIV-infected subjects, and perhaps this could be extrapolated to A*01:01 and B*07:02 responses in HESN subjects.
Fig 6.
Phenotype analysis of A*01:01 and B*07:02 epitope-specific CD8+ T cells. (A) Identification of circulating HIV-specific CD8+ T cells with Gag HLA peptide tetrameric complex. Results represent analyses of samples from representative HIV-1-seropositive subjects. (B) Visualization of virus-specific CD8+ T cells by staining ex vivo with MHC-I tetramers and surface expression of memory/effector phenotypic markers. All cells shown are a result of a gate on the specific CD8 subset to a tetramer. (Top) Representative figure showing surface expression of CCR7 and CD45-RA on specific HIV-1 CD8-T cells to A*01:01-NSSKVSQNY and B*07:02-APRKKGCWK tetramers. Numbers in the quadrants indicate percentages of cells. (Bottom) Comparison of percentages of markers expressed by CCR7 and CD45-RA (NC, naive cells; CM, central memory; EM, effector memory; TD: terminal differential) on specific HIV-1 CD8-T cells to A*01:01-NSSKVSQNY, B*07:02-APRKKGCWK, and B*07:02-SPRTLNAWV tetramers. Data are from the participants with HIV-1 tetramer reactivity (n = 3 for A*01:01-NSSKVSQNY and for B*07:02-APRKKGCWK and B*07:02-SPRTLNAWV). (C) Comparison of CD27 and CD28 expression in CD8+ T cells of A*01:01-NSSKVSQNY, B*07:02-APRKKGCWK, and B*07:02-SPRTLNAWV tetramers. Representative figure showing surface expression of CD28+ CD27− (left) and surface expression of CD28− CD27− (right) on specific HIV-1 CD8-T cells to each tetramer. Data are from participants with HIV-1 tetramer reactivity (n = 3 for A*01:01-NSSKVSQNY and for B*07:02-APRKKGCWK and B*07:02-SPRTLNAWV).
DISCUSSION
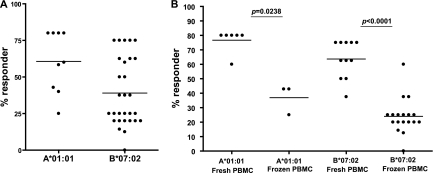
Numerous studies have documented the role of CD8+ T cells in controlling HIV-1 (18, 23, 24, 57, 66, 71). Specifically, studies have shown that CD8+ T cell responses to HIV-1 Gag are associated with viremia control (25, 37, 49, 54, 58, 62). Characterization of CD8+ T cell responses correlated with protection from HIV-1 infection needs to be conducted with PBMCs from individuals who are highly exposed but remain uninfected (HESN). Because such individuals represent a rare biological phenotype and identifying such individuals requires long-term careful observations, very few such studies have been reported. Many theoretical and empirical techniques have been used for epitope identification including predictive algorithms, defective cell lines, shotgun methods, and peptide elution techniques (63). These methods can be inaccurate and labor-intensive given the large number of antigens and their variations and the diversity of HLA (22, 63). Furthermore, screening a large number of HIV peptides with patient PBMCs is not practical because the numbers of PBMCs that can be obtained from patients are limited, and pooled peptides can obscure weak but real CD8+ T cell responses. In this study, we used an alternative approach to characterize CD8+ T cell epitopes that are more likely to be recognized by the HESN individuals. We first identified HLA class I alleles that are significantly associated with either protection from HIV-1 infection or susceptibility to HIV-1 infection. The epitopes of the identified alleles were then characterized by an in vitro, cell-free, peptide binding assay, the iTopia epitope discovery system. The identified epitopes were further confirmed by IFN-γ ELISpot assay with patient PBMCs. Our study showed that the iTopia epitope discovery system is a reliable in vitro system to rapidly identify epitopes of specific HLA alleles. Most peptides identified by this approach have been confirmed by IFN-γ ELISpot assays with fresh PBMCs from subjects with specific HLA alleles (Fig. 7) (60). The two alleles, A*01:01 and B*07:02, were chosen because they are independently associated with resistance or susceptibility to HIV-1 infection, and these two alleles are available in the iTopia epitope discovery system. Because studies have shown that CD8+ T cell response to HIV Gag is correlated with protection from disease progression and viremia control, in this study we focused on Gag of two common HIV subtypes in the Kenyan population. By comprehensively analyzing CD8+ T cell Gag epitopes of two HLA class I alleles associated with different outcomes of HIV-1 infection in a well-characterized high-risk sex worker cohort, we aim to characterize the CD8+ T cell responses that protect against HIV-1 infection.
Fig 7.
Confirmation of epitopes identified using the iTopia epitope discovery system by IFN-γ ELISpot assays. (A) Percent responders among all subjects with A*01:01 or B*07:02 tested using either fresh PBMCs or PBMCs that were stored in liquid nitrogen. (B) Percent responders among subjects with A*01:01 or B*07:02 tested using fresh PBMCs.
This study has shown that B*07:02, an allele associated with rapid seroconversion, can present a broad spectrum of Gag epitopes and induce strong immune responses, while the narrowly focused epitope presentation by A*01:01 correlates with protection. Because there is no significant difference in the magnitudes of IFN-γ ELISpot responses between A*01:01 and B*07:02 epitopes and between different groups of individuals (HESN, NSN, and POS), the major difference between these two alleles appears to be the spectrum of antigen presentation, although subtle differences may exist. As HLA class I alleles present viral peptides to activate CD8+ T cells, the ability to present more CD8+ T cell epitopes could mean more chances to induce CD8+ T cell responses. Indeed, most of the B*07:02 patients we examined have positive ELISpot responses to more than 3 different Gag epitopes and some have ELISpot responses to 8 different Gag epitopes. Thus, the ability to present more CD8+ T cell epitopes and induce more CD8+ T cell responses is not correlated with protection from HIV-1 infection. A*01:01, by contrast, can present only three Gag epitopes. The narrow spectrum of Gag epitope binding ability of the protective allele A*01:01 can explain previous observations that HIV-specific responses are generally lower in magnitude (4, 56) and narrower in breadth in HIV-1-resistant individuals than in HIV-infected donors (4). Although A*01:01 presents a very narrow spectrum of Gag epitopes, it appears to tolerate the variation of the presented epitope as seen in its binding to the epitope NSSKVSQNY and its variants of major subtypes A, B, and D. Thus, a more focused presentation of major HIV subtype variants appears to confer more-effective protection from infection. The ability of A*01:01 to present multiple major subtype variants of specific epitopes could be advantageous in dealing with multiple circulating subtypes in Kenya (35, 36, 50). To deal with the diversity of HIV-1 virus, a mosaic vaccine approach has been developed (13, 17, 20, 31, 59), and it appears to have enhanced coverage of diverse HIV strains (13, 59). In addition to the reports that Gag specific T cell responses are associated with lower viral loads (25, 30, 37, 49, 54, 58, 62, 74), studies have suggested that the failure of current vaccine approach may be due to targeting immunodominant variable epitopes and vaccine approaches targeting conserved regions of HIV-1 have been proposed and are under investigation (38, 55, 73). The narrowly focused antigen presentation of multiple variants by A*01:01 could be advantageous in dealing with viral diversity, in the meantime avoiding the negative effect of immunodominance of mutable epitopes (55).
Are the specific memory CD8+ T cells to A*01:01-Gag epitope complex phenotypically different from the B*07:02-epitope complex? Our preliminary study showed that although there is no difference in epitope-specific Tem/Tcm frequencies, the effector memory T cells (CCR7− CD45RA− CD28+ CD27−) induced by A*01:01-NSSKVSQNY are more numerous than those induced by B*07:02-APRKKGCWK and B*07:02-SPRTLNAWV. Thus, there appears to be a difference in the regulation of the costimulation marker, CD28 of the antigen-specific CD8+ T memory phenotype, between the allele associated with resistance to HIV-1 infection and those alleles associated with susceptibility. Further studies are needed to determine if these effects are independent of disease progression, as all the subjects in this analysis were in the chronic phase of HIV infection. We are conducting further studies to examine tetramer-specific responses in HESN subjects and chronically HIV-infected subjects. CD28 plays a key role in controlling the size and quality of pathogen-specific immune responses (53). CD28-mediated costimulation is needed for effective primary T cell expansion and for reactivating memory CD8+ T cells and secondary responses (46), and it helps generate and maintain virus-specific memory CD8+ T cells. Maintaining CD28 is important for proliferation and survival of CD8+ T cells; loss of these molecules is associated with reduced ability to respond to recurrent infection. The higher level of CCR7− CD45RA− CD28+ CD27− cells in the A*01:01 epitope NSSKVSQNY-specific T cell population could translate into a better ability to proliferate as well as enhanced cell survival. Future studies will address this and examine CD8+ T cell exhaustion and the functionality of these memory CD8+ T cells. Studying these parameters may help us better understand the advantage in chronic HIV-1 infection that this costimulator marker confers on memory CD8 T cells presented by this allele.
Recent studies have shown that in most cases mucosal transmission of HIV-1 is by a single or a few founder viruses (1, 12, 19, 29, 39, 40). Immune mechanisms preventing the establishment of a few founder viruses are likely different from the ones dealing with a full-blown viral infection after the virus has been well established in the host. It is possible that a lower-magnitude, narrowly focused, well-maintained virus-specific CD8+ T cell response to multiple subtypes is sufficient to destroy and eliminate a few founder viruses without inducing inflammatory responses that may activate more CD4+ T cells and provide more targets for HIV-1. More narrowly focused epitope recognition has also been reported for B*57:01, the allele associated with viral control in HIV-1-infected elite controllers (32, 47). There is also a common observation that the alleles with much broader capacity of peptide binding are not associated with viral control (2, 32). This study is the first to show that broader epitope recognition and induced immune response correlate to a detrimental outcome of HIV-1 infection, while the narrowly focused epitope presentation is correlated with protection from HIV-1 infection.
Generating strong immunological memory responses to infectious pathogens is a common vaccine approach. For many reasons, the traditional approaches have failed until recently to yield a successful AIDS vaccine. First, HIV-1 targets CD4+ T cells, and the activated CD4+ T cells are more readily infected. Second, HIV-1 virus exhibits a great diversity and evolves rapidly due to the high error rate of reverse transcriptase. In addressing the profound challenge these factors pose to developing an effective vaccine, we have shown here that the allele associated with protection from HIV-1 infection does not recognize a broad range of Gag epitopes but can present multiple variants of a few specific epitopes. By contrast, the allele associated with susceptibility to HIV-1 infection can recognize a broad spectrum of Gag epitopes and generate strong immune responses. Although we compared only Gag epitopes of two HLA class I alleles, and whether this observation can be extended to other alleles associated with different outcomes of HIV-1 infection remains to be determined, it is clear that recognizing more epitopes and generating stronger T cell responses are not always protective against HIV-1 infection. Since activated CD4+ T cells are more susceptible targets for HIV-1, a broad spectrum of antigen recognition may lead to broad T cell activation and thus provide more targets for HIV-1 and a greater probability of establishing infection. The much narrower epitope spectrum of the allele associated with a better HIV-1 outcome suggests that selectively targeting fewer and important locations of different HIV-1 subtypes might work better by destroying infected cells without overactivation of the immune system.
ACKNOWLEDGMENTS
This research project was supported by the Federal Intramural Genomics Research and Development Fund of Canada and the National Microbiology Laboratory, Public Health Agency of Canada. Part of the research was founded by grants from the Bill and Melinda Gates Foundation. F. Plummer is currently a Tier I CIHR Canada Research Chair.
There are no conflicts of interest.
We thank the nurses and staff working in the Pumwani Sex Worker Cohort (Jane Njoki, Jane Kamene, Elizabeth Bwibo, Edith Amatiwa, Ann Maingi), staff working at the University of Manitoba, the University of Nairobi, and the National Microbiology Laboratory of Canada for their dedication. Without their unstinting dedication, this research would not be possible. The women who participate in the Pumwani Sex Worker Cohort have made essential contributions to this research.
Footnotes
Published ahead of print 9 November 2011
REFERENCES
- 1. Abrahams MR, et al. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J. Virol. 83:3556–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahlers JD, Belyakov IM. Lessons learned from natural infection: focusing on the design of protective T cell vaccines for HIV/AIDS. Trends Immunol. 31:120–130 [DOI] [PubMed] [Google Scholar]
- 3. AIDS Vaccine Advocacy Coalition 2004. Support for the RV144 HIV vaccine trial. Science 305:177–180; author reply 177–180 [PubMed] [Google Scholar]
- 4. Alimonti JB, et al. 2006. Characterization of CD8 T-cell responses in HIV-1-exposed seronegative commercial sex workers from Nairobi, Kenya. Immunol. Cell Biol. 84:482–485 [DOI] [PubMed] [Google Scholar]
- 5. Alimonti JB, et al. 2005. CD4+ T cell responses in HIV-exposed seronegative women are qualitatively distinct from those in HIV-infected women. J. Infect. Dis. 191:20–24 [DOI] [PubMed] [Google Scholar]
- 6. Anonymous. 2003. AIDSVAX fails to prove efficacious in large-scale trial. Expert Rev. Vaccines 2:731. [PubMed] [Google Scholar]
- 7. Anonymous. 2007. HIV vaccine failure prompts Merck to halt trial. Nature 449:390. [DOI] [PubMed] [Google Scholar]
- 8. Anonymous. 2008. NIAID declines to proceed with HIV vaccine trial. AIDS Policy Law 23:1, 4 [PubMed] [Google Scholar]
- 9. Anonymous. 2003. Results in for first phase III vaccine trial. AIDS Patient Care STDS 17:607–608 [PubMed] [Google Scholar]
- 10. Anonymous. 2007. Therapeutic vaccine trial shows no benefit. IAVI Rep. 11:20. [PubMed] [Google Scholar]
- 11. Anonymous. 2007. Vaccine trial discontinued. AIDS Patient Care STDS 21:778–779 [PubMed] [Google Scholar]
- 12. Bar KJ, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J. Virol. 84:6241–6247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barouch DH, et al. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat. Med. 16:319–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bodinier M, et al. 2000. Efficient detection and immunomagnetic sorting of specific T cells using multimers of MHC class I and peptide with reduced CD8 binding. Nat. Med. 6:707–710 [DOI] [PubMed] [Google Scholar]
- 15. Cohen J. 2009. HIV/AIDS research. Beyond Thailand: making sense of a qualified AIDS vaccine “success.” Science 326:652–653 [DOI] [PubMed] [Google Scholar]
- 16. Cohen J. 2009. HIV/AIDS research. Surprising AIDS vaccine success praised and pondered. Science 326:26–27 [DOI] [PubMed] [Google Scholar]
- 17. Corey L, McElrath MJ. HIV vaccines: mosaic approach to virus diversity. Nat. Med. 16:268–270 [DOI] [PubMed] [Google Scholar]
- 18. Emu B, et al. 2008. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J. Virol. 82:5398–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fischer W, et al. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One 5:e12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fischer W, et al. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 13:100–106 [DOI] [PubMed] [Google Scholar]
- 21. Fowke KR, et al. 1996. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 348:1347–1351 [DOI] [PubMed] [Google Scholar]
- 22. Frahm N, et al. 2006. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat. Immunol. 7:173–178 [DOI] [PubMed] [Google Scholar]
- 23. Friedrich TC, et al. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 81:3465–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Friedrich TC, Watkins DI. 2008. Wanted: correlates of vaccine-induced protection against simian immunodeficiency virus. Curr. Opin. HIV AIDS 3:393–398 [DOI] [PubMed] [Google Scholar]
- 25. Geldmacher C, et al. 2007. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J. Virol. 81:2440–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goulder PJ, Watkins DI. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8:619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hardie RA, et al. 2008. A common human leucocyte antigen-DP genotype is associated with resistance to HIV-1 infection in Kenyan sex workers. AIDS 22:2038–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hardie RA, et al. 2008. Human leukocyte antigen-DQ alleles and haplotypes and their associations with resistance and susceptibility to HIV-1 infection. AIDS 22:807–816 [DOI] [PubMed] [Google Scholar]
- 29. Keele BF, et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kiepiela P, et al. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53 [DOI] [PubMed] [Google Scholar]
- 31. Kong WP, et al. 2009. Expanded breadth of the T-cell response to mosaic human immunodeficiency virus type 1 envelope DNA vaccination. J. Virol. 83:2201–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kosmrlj A, et al. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature 465:350–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kreiss JK, et al. 1986. AIDS virus infection in Nairobi prostitutes. Spread of the epidemic to East Africa. N. Engl. J. Med. 314:414–418 [DOI] [PubMed] [Google Scholar]
- 34. Lacap PA, et al. 2008. Associations of human leukocyte antigen DRB with resistance or susceptibility to HIV-1 infection in the Pumwani Sex Worker Cohort. AIDS 22:1029–1038 [DOI] [PubMed] [Google Scholar]
- 35. Land AM, et al. 2008. Full-length HIV type 1 proviral sequencing of 10 highly exposed women from Nairobi, Kenya reveals a high proportion of intersubtype recombinants. AIDS Res. Hum. Retroviruses 24:865–872 [DOI] [PubMed] [Google Scholar]
- 36. Land AM, et al. 2008. High prevalence of genetically similar HIV-1 recombinants among infected sex workers in Nairobi, Kenya. AIDS Res. Hum. Retroviruses 24:1455–1460 [DOI] [PubMed] [Google Scholar]
- 37. Leligdowicz A, et al. 2007. Robust Gag-specific T cell responses characterize viremia control in HIV-2 infection. J. Clin. Invest. 117:3067–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Letourneau S, et al. 2007. Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One 2:e984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H, et al. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6:e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu J, et al. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J. Virol. 84:10406–10412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo M, et al. 2001. Two-step high resolution sequence-based HLA-DRB typing of exon 2 DNA with taxonomy-based sequence analysis allele assignment. Hum. Immunol. 62:1294–1310 [DOI] [PubMed] [Google Scholar]
- 42. Luo M, Blanchard J, Pan Y, Brunham K, Brunham RC. 1999. High-resolution sequence typing of HLA-DQA1 and -DQB1 exon 2 DNA with taxonomy-based sequence analysis (TBSA) allele assignment. Tissue Antigens 54:69–82 [DOI] [PubMed] [Google Scholar]
- 43. MacDonald KS, et al. 2000. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J. Infect. Dis. 181:1581–1589 [DOI] [PubMed] [Google Scholar]
- 44. Maek ANW, et al. 2003. VaxGen vaccine trial fails the test but may offer insights. AIDS Alert 18:41–45 [PubMed] [Google Scholar]
- 45. McCarthy M. 2003. AIDS vaccine fails in Thai trial. Lancet 362:1728. [DOI] [PubMed] [Google Scholar]
- 46. McCarthy M. 2003. HIV vaccine fails in phase 3 trial. Lancet 361:755–756 [DOI] [PubMed] [Google Scholar]
- 47. Migueles SA, et al. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97:2709–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ojcius D. 2004. HIV vaccine trial halted. Nat. Rev. Microbiol. 2:776. [DOI] [PubMed] [Google Scholar]
- 49. Ondondo BO, et al. 2008. Comprehensive analysis of HIV Gag-specific IFN-gamma response in HIV-1- and HIV-2-infected asymptomatic patients from a clinical cohort in The Gambia. Eur. J. Immunol. 38:3549–3560 [DOI] [PubMed] [Google Scholar]
- 50. Peters HO, et al. 2008. An integrative bioinformatic approach for studying escape mutations in human immunodeficiency virus type 1 gag in the Pumwani Sex Worker Cohort. J. Virol. 82:1980–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Plotkin SA. 2010. The RV144 Thai HIV vaccine trial. Hum. Vaccin. 6:159. [PubMed] [Google Scholar]
- 52. Plummer FA, Ball TB, Kimani J, Fowke KR. 1999. Resistance to HIV-1 infection among highly exposed sex workers in Nairobi: what mediates protection and why does it develop? Immunol. Lett. 66:27–34 [DOI] [PubMed] [Google Scholar]
- 53. Riley JL, June CH. 2005. The CD28 family: a T-cell rheostat for therapeutic control of T-cell activation. Blood 105:13–21 [DOI] [PubMed] [Google Scholar]
- 54. Rolland M, et al. 2008. Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS One 3:e1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rolland M, Nickle DC, Mullins JI. 2007. HIV-1 group M conserved elements vaccine. PLoS Pathog. 3:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rowland-Jones SL, et al. 2001. How important is the ‘quality’ of the cytotoxic T lymphocyte (CTL) response in protection against HIV infection? Immunol. Lett. 79:15–20 [DOI] [PubMed] [Google Scholar]
- 57. Sacha JB, et al. 2007. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J. Immunol. 178:2746–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saez-Cirion A, et al. 2009. Heterogeneity in HIV suppression by CD8 T cells from HIV controllers: association with Gag-specific CD8 T cell responses. J. Immunol. 182:7828–7837 [DOI] [PubMed] [Google Scholar]
- 59. Santra S, et al. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat. Med. 16:324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Semeniuk CA, et al. 2010. Identification and characterization of HLA-A*0301 epitopes in HIV-1 gag proteins using a novel approach. J. Immunol. Methods 352:118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Semeniuk CA, et al. 2009. Multiple T-cell epitopes overlap positively-selected residues in the p1 spacer protein of HIV-1 gag. AIDS 23:771–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Serwanga J, et al. 2009. Host HLA B*allele-associated multi-clade Gag T-cell recognition correlates with slow HIV-1 disease progression in antiretroviral therapy-naive Ugandans. PLoS One 4:e4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shingler WH, Chikoti P, Kingsman SM, Harrop R. 2008. Identification and functional validation of MHC class I epitopes in the tumor-associated antigen 5T4. Int. Immunol. 20:1057–1066 [DOI] [PubMed] [Google Scholar]
- 64. Sidney J, et al. 2008. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Simonsen JN, et al. 1990. HIV infection among lower socioeconomic strata prostitutes in Nairobi. AIDS 4:139–144 [DOI] [PubMed] [Google Scholar]
- 66. Streeck H, et al. 2009. Human immunodeficiency virus type 1-specific CD8+ T-cell responses during primary infection are major determinants of the viral set point and loss of CD4+ T cells. J. Virol. 83:7641–7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Trachtenberg E, et al. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928–935 [DOI] [PubMed] [Google Scholar]
- 68. Trachtenberg EA, Erlich HA. 2001. A review of the role of the human leukocyte antigen (HLA) system as a host immunogenetic factor influencing HIV transmission and progression to AIDS, p I-43–I-60 In Korber BT, et al. (ed), HIV Molecular Immunology 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 69. Vaccari M, Poonam P, Franchini G. 2010. Phase III HIV vaccine trial in Thailand: a step toward a protective vaccine for HIV. Expert Rev. Vaccines 9:997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Weiner DB. 18 February 2010. RV144: old vs. new. Hum. Vaccin. 6:159–161 [PubMed] [Google Scholar]
- 71. Wilson NA, et al. 2009. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J. Virol. 83:6508–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wulf M, Hoehn P, Trinder P. 2009. Identification of human MHC class I binding peptides using the iTOPIA- epitope discovery system. Methods Mol. Biol. 524:361–367 [DOI] [PubMed] [Google Scholar]
- 73. Yang OO. 2009. Candidate vaccine sequences to represent intra- and inter-clade HIV-1 variation. PLoS One 4:e7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang OO, Daar ES, Ng HL, Shih R, Jamieson BD. Increasing CTL targeting of conserved sequences during early HIV-1 infection is correlated to decreasing viremia. AIDS Res. Hum. Retroviruses 27:391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]