Abstract
Lambda-1 interferon (IFN-λ1) and cyclooxygenase-2 (COX-2) were reported to play an important role in host antiviral defense. However, the mechanism by which IFN-λ1 and COX2 are activated and modulated during viral infection remains unclear. In this study, we found that expression of both circulating IFN-λ1 and COX2-derived prostaglandin E2 (PGE2) was coordinately elevated in a cohort of influenza patients compared to healthy individuals. Expression of IFN-λ1 was blocked by a selective COX2 inhibitor during influenza A virus infection in A549 human lung epithelial cells but enhanced by overexpression of COX2, indicating that the production of IFN-λ1 is COX2 dependent. COX2 was able to increase IFN-λ1 expression by promoting NF-κB binding to the enhancer in the IFN-λ1 promoter. We found that epigenetic changes activate COX2 expression and PGE2 accumulation during viral infection. The expression of DNA methyltransferase 3a (DNMT3a) and DNMT3b, but not that of DNMT1, was downregulated following influenza A virus infection in both A549 cells and peripheral blood mononuclear cells (PBMCs). We showed that microRNA miR29 suppresses DNMT activity and thus induces expression of COX2 and PGE2. Furthermore, miR29 expression was elevated 50-fold in virally infected A549 cells and 10-fold in PBMCs from influenza patients, compared to expression after mock infection of A549 cells or in healthy individuals, respectively. Activation of the protein kinase A signaling pathway and phosphorylation of CREB1 also contributed to COX2 expression. Collectively, our work defines a novel proinflammatory cascade in the control of influenza A virus infection.
INTRODUCTION
Influenza virus (IV) causes a potentially serious infection with symptoms including headache, fever, pneumonia, and even death. The ease with which this pathogen can spread can cause widespread epidemics (38). The main antigenic determinants of influenza A viruses are the hemagglutinin (H) and neuraminidase (N) transmembrane glycoproteins. Based on the antigenicity of these glycoproteins, influenza A viruses are further subdivided into 16 H (H1 to H16) and 9 N (N1 to N9) subtypes. H1N1 and H3N2 subtypes are the main isolates currently circulating in the human population (38, 42). In recent years, H5N1 and H1N1 have also caused hundreds of deaths and billions of dollars in economic losses (21).
Cyclooxygenase-2 (COX2) expression is present in airway epithelial cells after influenza virus infection (27). COX2 catalyzes the first step in the biosynthesis of prostaglandins from arachidonic acid and is important in host responses to infection (7, 10, 33). The ability of COX2 products to modulate inflammation and immune responses is well documented (1, 4, 6, 22, 24, 29). COX2 is a main cause of inflammation during influenza virus infection and cooperates with other proinflammatory cytokines and interleukins (ILs), such as inducible nitric oxide, interferon (IFN), and IL-32. Furthermore, COX2 is the primary mediator of the body's protection from influenza virus infection (20). However, the acute and sometimes extensive inflammation in the respiratory system is also a common cause of death by influenza infection.
In mammalian cells, DNA methylation is performed by three members of the DNA methyltransferase (DNMT) family: DNMT1, DNMT3a, and DNMT3b. DNA methylation involves the formation of a covalent bond between a methyl group and a dinucleotide CpG (8). The majority of CpG dinucleotides in the genome, dispersed across retrotransposons or throughout coding regions and introns of genes, are methylated in normal cells. However, approximately 15% of CpGs are clustered in CpG islands in the promoter regions of genes and are normally unmethylated (8). Recent studies showed that tumorigenesis is coupled with aberrant methylation in the promoter of tumor suppressor genes and changes in expression of DNMT family members (9, 13). Additionally, some investigators have reported that the members of the DNMT family can be regulated by certain viral proteins such as the hepatitis B virus (HBV) X protein, which directly affects the expression of COX2, a gene critical to proinflammatory processes (17, 32, 48). The results of these studies indicate that similar aberrant epigenetic processes induced by certain viral proteins may occur in other viral infections and may constitute a potential new pathway leading to the expression of inflammation genes. However, the mechanism by which viral proteins can lead to epigenetic changes requires more investigation.
MicroRNAs (miRNAs) are small noncoding RNAs that naturally exist in mammalian cells and play a role similar to small interfering RNAs (siRNAs). Mature miRNAs are 19- to 25-nucleotide-long molecules cleaved from 70- to 100-nucleotide hairpin pre-miRNA precursor molecules. In animals, single-stranded miRNAs bind, through partial sequence homology, to the 3′ untranslated region (3′-UTR) of target mRNAs and block translation or, less frequently, induce mRNA degradation (3). The miRNA29 (miR29) family, which includes miR29a, miR29b, and miR29c, is involved in apoptosis, tumorigenesis, and chronic lymphocyte leukemia (12, 46). miRNAs of the miR29 family were identified bound to the 3′-UTR of DNMT3a and DNMT3b, indicating that they may have a role in regulating DNA methylation levels by regulating the expression of DNMT3a and 3b in lung cancer (11). However, little is known about the role of miR29 during viral infection.
The gene encoding lambda-1 IFN (IFN-λ1), also known as IL-29, is located on human chromosome 19. IFN-λ1, IL-28A (IFN-λ2), and IL-28B (IFN-λ3) were identified as type III IFNs in 2003 (16, 36). Type I and type III IFNs have similar biological functions, and their expression is induced in a number of viral infections including type IV (15, 44). Additionally, the antiviral activity of type 1 and type III INF on IV has also been reported (44). These studies indicate that IFN-λ1 may be an important member of the antiviral network to protect cells from IV infection.
As DNMT3a and DNMT3b have been reported to play a role during HBV infection, similar regulation may also occur in other viral infections. In this study, our aim was to discover how epigenetic modifications affect the expression of proinflammatory genes in response to influenza A virus infection. Our results demonstrate that cooperation between miR29-mediated epigenetic modifications and activation of the protein kinase A (PKA) signaling pathway mediate the upregulation of COX2 and consequent IFN-λ1 production. This study describes a previously unrecognized proinflammatory mechanism that occurs during influenza A virus infection.
MATERIALS AND METHODS
Clinical samples.
Peripheral blood samples were obtained from 32 patients (14 male, 18 female, aged 36.4 ± 18.2 years) confirmed to be infected by H1N1 influenza A virus. Blood samples from 32 healthy individuals (15 male, 17 female, aged 30.6 ± 11.5 years) were randomly selected as controls from the local blood donation center. Peripheral blood mononuclear cells (PBMCs) were isolated by standard Ficoll gradient centrifugation from the peripheral blood of either infected subjects or uninfected controls and used for miR29 determination by quantitative real-time reverse transcription-PCR (RT-PCR). Blood plasma was assayed for IFN-λ1 and prostaglandin E2 (PGE2) by enzyme-linked immunosorbent assay (ELISA). All research involving human participants was approved by the Institutional Review Board of the College of Life Sciences, Wuhan University, in accordance with the guidelines for the protection of human subjects. Written informed consent was obtained from each participant.
Virus and cell culture.
The IV strain A/Human/Hong Kong/498/97 (H3N2) was provided by the China Center for Type Culture Collection. Stock virus was propagated in 10-day-old embryonated chicken eggs for 36 to 48 h at 37°C. The allantoic fluid was then harvested, and aliquots were stored at −80°C until used. The final concentration of H3N2 virus infection used in this study was 1 multiplicity of infection (MOI).
A549 human lung epithelial cells were cultured in F12K medium with 10% heat-inactivated fetal bovine serum (FBS) and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) at 37°C and 5% CO2. PBMCs were isolated from blood of healthy donors by standard Ficoll gradient centrifugation and cultured in RPMI 1640 without FBS and antibiotics at 37°C and 5% CO2.
Antibodies and inhibitors.
The following antibodies were used in this study: goat anti-DNMT1 antibody (sc-10219; Santa Cruz Biotechnology), goat anti-DNMT3A antibody (sc-10231; Santa Cruz Biotechnology), goat anti-DNMT3B (sc-10236; Santa Cruz Biotechnology), rabbit anti-CREB1 (sc-20700; Santa Cruz), mouse anti-COX2 (Cayman), and rabbit anti-IL-29 (sc-130787; Santa Cruz Biotechnology).
The following inhibitors were used in this study: methylation inhibitor 5′-aza-CdR and signaling pathway inhibitors NS398 (iCOX2; 10 μM), H89 (iPKA; 10 μM), LY294002 (iPI3K; 10 μM), PD98059 (iERK; 10 μM), SP600125 (iJNK; 10 μM), SB203580 (iP38; 10 μM), U0126 (iMEK/ERK; 10 μM), GF109203 (iPKC; 10 μM), and Bay11-7082 (iNF-κB; 10 μM). All inhibitors were from Sigma-Aldrich.
Plasmids, siRNA, and miRNA.
Human DNMT1 full-length cDNA clone (pcDNA 3.1-DNMT1) was a gift form Dr. Moshe Szyf (Department of Pharmacology and Therapeutics, McGill University, Canada) (5, 39). Human DNMT3a and DNMT3b full-length cDNA clones (pcDNA3-DNMT3a, pcDNA3-DNMT3b) were gifts from Dr. Arthur D. Riggs (Beckman Research Institute of the City of Hope, Duarte, CA) (18, 45). Plasmids or miRNAs were transfected into the cells with Lipofectamine 2000 reagent (Invitrogen).
siRNAs, including siDNMT1, siDNMT3a, siDNMT3b, siP65, siP50, and siCREB1, were designed and then inserted into the pSilencer 2.1-U6 expression vector (Ambion); the interference efficiency was tested by transfection of siRNAs into A549 cells and RT-PCR analysis of the respective target. The sequences for the sense oligonucleotides for the most effective knockdown constructs are as follows: for siDNMT1, 5′-GATCCAA TGGCAGATGCCAACAGCTTCAA GAGAGCTGTTGGCATCTGCCATTCCTTTTTTGGAAA-3′; for siDNMT3a, 5′-GATCCACTACATCAGCAAGCGCAATTCAAGAGATTGCGC TTGCTGATGTAGTAGTTTTTTGGAAA-3′; for siDNMT3b, 5′-GATCCA TGACGGATGCCTAGAGTTCTCAAGAGAAACTCT AGGCAT CCGTCATCTTTTTTTGGAAA-3′; for siCREB1, 5′-GATCC GGAGTCAGTGGATAGTGTATTCAAGAGA TACACT ATCCACTGACTCCTTTTTTGGAAA-3′; for siP50,5′-GATCCGGGGCTATAATCCTGGACTTTCAAGAGAAGTCCAGGATTATAGCCCCTT TTTTGGAAA-3′; and for siP65,5′-GATCCGCCCCTTCCAAGTTCCTATATTCAAGAGATATAGGAACTTGGAAGGGGTTTTTTGGAAA-3′. miR29a, miR29b, and miR29c were synthesized as reported previously (11). siCOX2, siIRF3/7, and miR29 were designed and synthesized by Genephama (Shanghai).
Assays for DNMT activity and PGE2, IFN-λ1, and miR29 detection.
Total DNMT activity in nuclear extracts was tested with the EpiQuik DNA methyltransferase activity/inhibition assay kit (Epigentek). PGE2 expression was tested with the Biotrak prostaglandin E2 enzyme immunoassay system (R&D Systems). IFN-λ1 was detected by ELISA and Western blotting with IFN-λ1 antibody. miR29 family miRNAs were detected with Hairpin-it miRNA qPCR quantitation kit (GenePharma). The template used for miR29 detection was the RT-PCR product from RNA extracted from A549, PBMCs, or patient blood samples. The stem-loop structure RT primer was provided by the qPCR kit. Final results were read on the LC480 real-time PCR system. All experiments were carried out according to the manufacturer's instructions.
Quantitative real-time PCR.
Total RNA, isolated from A549 cells or PBMCs by using TRIzol reagent (Invitrogen), was treated with DNase I and reverse transcribed with murine leukemia virus (MLV) reverse transcriptase (Promega) and OligodT (Takara) to get total cDNA as templates.
PCR was performed with the following primer pairs: for COX2, 5′-ACAATGCTGACTATGGCTAC-3′ (sense) and 5′-ATGGGAGGAGTTCAGGGA-3′ (antisense); for DNMT1, 5′-CTGAGGCCTTCACGTTCAA-3′ (sense) and 5′-ACTTGTGGGTGTTCTCAGGC-3′ (antisense); for DNMT3a, 5′-CGTTGGCATCCACTGTGAATGA-3′ (sense) and 5′-TTACACACACGCAAAATACTCCTT-3′ (antisense); for DNMT3b, 5′-CCTGAGGCCTTCACGTTCAA-3′ (sense) and 5′-ACTTGTGGGTGTTCTCAGGC-3′ (antisense); and for β-actin, 5′-CCATCGAGCACGGCATCG TCACCA-3′ (sense) and 5′-CTCGGTGAGGATCTTC ATGAGGTAGT-3′ (antisense). The results of quantitative real-time PCR were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression.
Western blot analysis.
After sonification and centrifugation, protein extracts from cultured cells were prepared by suspending cells in lysis buffer (0.01% EDTA, 0.1% Triton X-100, and 10% proteinase inhibitor cocktail). Protein concentrations in supernatants were quantified using a protein assay kit (Bio-Rad). Western blot analysis was performed using antibodies specific for DNMT1, DNMT3a, DNMT3b, COX2, and IFN-λ1. Blots were developed using SuperSignal chemiluminescent reagent (Pierce, Rockford, IL).
ChIP assay.
A chromatin immunoprecipitation (ChIP) assay was performed according to the X-ChIP protocol (Abcam). Briefly, formaldehyde was added to the culture medium to a final concentration of 1%. The cells were then washed twice with phosphate-buffered saline (PBS), scraped, and lysed in lysis buffer (1% SDS, 10 mM Tris-HCl, pH 8.0, 1 mM phenylmethylsulfonyl fluoride [PMSF], 50 mg/ml of both aprotinin and leupeptin) for 10 min on ice. The lysates were sonicated on ice, and the debris was removed by centrifugation at 15,000 × g for 15 min at 4°C. One-fourth of the supernatant was used as DNA input control. The remaining supernatant was diluted 10-fold with dilution buffer (0.01% SDS, 1% Triton X-100, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0, and 150 mM NaCl) and incubated with antibody against CREB or DNMTs overnight at 4°C. Immunoprecipitated complexes were collected using protein A/G agarose beads. The pellets were washed with dialysis buffer (2 mM EDTA, 50 mM Tris-HCl, pH 8.0) and incubated at 67°C for 5 h to reverse formaldehyde cross-linking. DNA was precipitated with ethanol and extracted three times with phenol-chloroform. Finally, pellets were resuspended in TE buffer and subjected to PCR amplification using the following primers: for COX2, 5′-CCGATTTTCTCATTTCCGT GGG-3′ (sense) and 5′-CGAAATGACTGTTTCTT TCCGCC-3′ (antisense); for IFN-λ1 proximal, 5′-GCCAGTTGGCTGAAAGCTGCCCA-3′ (sense) and 5′-GGCAGGGCCAAGTGAGCTGGGA-3′ (antisense); and for distal, 5′-TTTAAGGGCAGGTGCAGGGTGTC-3′ (sense) and 5′-TTACCCAATGTGGTGGGCACCATC-3′ (antisense).
Bisulphate sequencing.
Bisulphate sequencing was performed as described previously (14). Briefly, 1 μg genomic DNA was denatured in 0.2 mol/liter NaOH. Sodium bisulphate and hydroquinone (Sigma-Aldrich) were added to final concentrations of 3.1 mol/liter and 0.5 mM, the pH was adjusted to 5.0 with NaOH, and the solution was incubated at 55°C for 16 h. Then, DNA samples were purified with a DNA clean-up system (Promega), precipitated with 75% ethanol, and resolved with 20 μl double-distilled water (ddH2O). The following primers were used for amplification of the COX2 promoter: 5′-TTTAAATTGGGGTAGTTTTT-3′ (sense) and 5′-CTACTAAAAAATTCCTAAA-3′ (antisense). PCR products were cloned to pGEM-T-easy vector system (Promega) and subjected to sequencing analysis.
RESULTS
Production of IFN-λ1 is enhanced by COX2 during influenza virus infection.
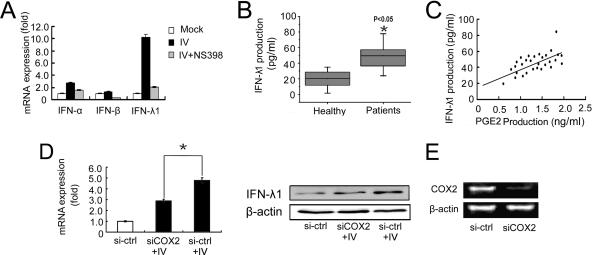
Although COX2 is a critical inflammatory gene induced by IV, it does not directly suppress viral replication in host cells. IFN-λ1 is a newly identified antiviral cytokine. The antiviral activity of IFN-λ1 in response to IV infection has been demonstrated in several recently published studies (15, 16, 30, 34, 36, 37, 44). We compared IFN-α, -β, and -λ1 expression in A549 human lung epithelial cells after infection with IV (MOI = 1) and treated with or without the selective COX2 inhibitor NS398 (10 μM): results showed that both IFN-α and -λ1 expression were upregulated 24 h after IV infection, but IFN-λ1 upregulation was more significant than that seen for IFN-α (Fig. 1A). It is reported that IFN-λ1 plays an important role in antiviral activity to protect cells form influenza virus infection in epithelial cells (31, 40). Our results indicated that both type I and type III interferons were suppressed by COX2 inhibitor (Fig. 1A). As a result, our work focused on clarifying the connection between IFN-λ1 and COX2. We thus measured the production of both IFN-λ1 and COX2-derived PGE2 in sera from a cohort of patients infected by influenza A virus (n = 32). The level of IFN-λ1 in influenza patients was elevated significantly compared to that in sera from healthy individuals (Fig. 1B). Interestingly, elevated IFN-λ1 in the sera of IV-infected patients was tightly correlated with the level of PGE2 (Fig. 1C). To further elucidate the relationship between COX2 and IFN-λ1, A549 cells were transfected with COX2-specific siRNA and infected with IV (MOI = 1). After a 24-h incubation, cells were harvested and IFN-λ1 expression was measured by both real-time RT-PCR and Western blotting. Our results indicated that IV infection-stimulated IFN-λ1 expression in A549 cells was suppressed by COX2-specific siRNA but not irrelevant control siRNA (Fig. 1D and E), which indicates that IFN-λ1 expression is enhanced by COX2 expression or activity. These results suggest that influenza A virus stimulated IFN-λ1 expression through the COX2 pathway.
Fig 1.
Expression of IFN-λ1 is enhanced by COX2 during influenza A virus infection. (A) A549 human lung epithelial cells were infected with IV (MOI = 1) and treated with or without the selective COX2 inhibitor NS398 (10 μM); 24 h after infection, cells were harvested and expressions of IFN-α, -β, and -λ1 were examined by quantitative real-time RT-PCR. (B) Analysis of IFN-λ1 level in sera collected from patients infected IV and healthy people. Thirty-two patients and 32 healthy individuals were enrolled in the experiments (*, P < 0.05). (C) IFN-λ1 and PGE2 levels in sera from 32 influenza patients were tested separately to show the correlation. The x axis shows COX2-derived PGE2 production, and the y axis shows IFN-λ1 production. (D) A549 cells were transfected with COX2-specific siRNA for 24 h and then infected with IV (MOI = 1). Cells were harvested 24 h after infection. IFN-λ1 expression was detected by real-time RT-PCR and Western blotting. An irrelevant siRNA was used as the control (*, P < 0.05). (E) The efficiency of COX2-specific siRNA (siCOX2) was tested by RT-PCR.
COX2 enhances IFN-λ1 expression through strengthening NF-κB binding activity to the distal NF-κB-rich cluster.
To further explore the signaling pathway by which COX2 enhances IFN-λ1 expression, different signaling pathway inhibitors were used (Fig. 2A). We found that the NF-κB inhibitor Bay11-7082 effectively suppressed IFN-λ1 expression in A549 cells transfected with pCMV-COX2. It is also reported that IFN-λ1 activation is regulated by IRF3/7 signaling pathway, and we thus designed specific siRNAs against IRF3, IRF7, and NF-κB P50 and P65 (siIRF3, siIRF7, siP50, and siP65) and cotransfected each of them along with pCMV-COX2 into A549 cells. Cells were collected 48 h after transfection, and both real-time RT-PCR and Western blotting were used to detect IFN-λ1 expression (Fig. 2B and C). The results showed that all four siRNAs (siIRF3, siIRF7, siP50, and siP65) suppressed IFN-λ1 expression induced by COX2 compared to what was seen for the irrelevant siRNA control, indicating that both IRF3/7 and NF-κB signaling pathways participate in IFN-λ1 expression during IV infection.
Fig 2.
COX2 enhances IFN-λ1 expression through strengthening the binding of NF-κB to the distal NF-κB-rich cluster. (A) A549 cells were incubated with eight signaling pathway inhibitors for 24 h and then infected with IV (MOI = 1). Cells were harvested 24 h after infection. IFN-λ1 mRNA expression was tested by real-time RT-PCR. (B) A549 cells were transfected with specific siRNAs against IRF3, IRF7, P50, and P65 for 24 h. An irrelevant siRNA served as the negative control. Cells were then infected with IV (MOI = 1) and harvested 24 h after infection for the detection of IFN-λ1 expression by real-time RT-PCR and Western blotting (*, P < 0.05). (C) The efficiency of specific siRNAs (siIRF3, siIRF7, siP50, and siP65) was tested by RT-PCR. (D) A549 cells were transfected with COX2-expressing plasmid pCMV-COX2 or infected with IV. The empty vector served as the negative control. Cells were harvested 48 h after transfection or 24 h after IV infection. P50, P65, IRF3, and IRF7 binding activities were tested by ChIP assay based on the sequence of the IFN-λ1 promoter.
Previous studies showed that IFN-λ1 gene expression is mediated by spatially separated promoter elements that independently interact with NF-κB and IRF3/7. Also, the distal cluster of NF-κB sites is required for maximal levels of lipopolysaccharide (LPS) induction (40). To verify the role of NF-κB and IRF3/7 during IV infection or after overexpression of COX2, we tested NF-κB binding activity to the distal and proximal NF-κB clusters and IRF3/7 binding activity to the IFN-λ1 promoter by ChIP assays (Fig. 2D). We found that the binding activities of NF-κB (P50, P65, and RelA), IRF3, and IRF7 were all strengthened after IV infection. However, after expression of COX2, the binding activity of NF-κB (P50, P65, and RelA) was strengthened mainly to the distal NF-κB cluster and slightly to the proximal NF-κB clusters. The binding activity of IRF3 and IRF7 to the IFN-λ1 promoter was slightly strengthened as well after expression of COX2. These results suggest that COX2 increases IFN-λ1 expression mainly through activating the distal NF-κB cluster enhancer. Although IRF3/7 do play an important role in IFN-λ1 induction during IV infection, they are relatively independent of COX2.
DNMT3a and DNMT3b downregulation induces COX2 expression.
To elucidate the relationship between DNMTs and COX2 expression, siRNA specific for each of three DNMTs (siDNMT1, siDNMT3a, and siDNMT3b) and 5′-aza-CdR, a specific DNMT inhibitor, were used to knock down DNMTs and to suppress DNMT activity, respectively. siDNMT3a and siDNMT3b significantly induced COX2-derived PGE2 production compared to irrelevant control siRNA. In contrast, siDNMT1 had only slight effects on PGE2 production (Fig. 3A). 5′-aza-CdR induced PGE2 production in a dose-dependent manner (Fig. 3B), indicating that downregulation of DNMTs leads to COX2 activation. To confirm this result, plasmids expressing each DNMT (DNMT1, DNMT3a, and DNMT3b) were transfected into A549 cells, and 8 h after transfection, cells were infected with IV (MOI = 1). Compared to what was seen for the vector control, transfection of these plasmids resulted in decreased expression of COX2 (Fig. 3C). But among three DNMTs, DNMT3b was more effective in suppressing COX2 expression, whereas DNMT1 and DNMT3a were less effective. Then, 5′-aza-CdR was used to suppress total DNMT activity. We found that the expression of COX2 was higher with the increased dose of DNMT inhibitor in PBMCs (Fig. 3D) and A549 cells (Fig. 3E). Thus, upregulation of COX2 and PGE2 is tightly correlated with decreased expression of DNMT3a and DNMT3b during IV infection.
Fig 3.
DNMTs suppress COX2 expression in both A549 cells and PBMCs. (A) PGE2 was tested in the culture supernatant of A549 cells 24 h after transfection with specific siRNA against each of three DNMTs. An irrelevant siRNA served as the negative control and was arbitrarily designated as 1.0. Data shown are mean ± standard error (SE); n = 3 (*, P < 0.05; **, P < 0.01). The efficiency of DNMT-specific siRNA (siDNMT1, siDNMT3a, and siDNMT3b) was tested by RT-PCR (right). (B) PGE2 was tested in the culture supernatant of A549 cells 24 h after treated with DNMT inhibitor 5′-aza-CdR in a dose-dependent manner. Data shown are mean ± SE; n = 3 (*, P < 0.05; **, P < 0.01). The negative control was designated as 1.0. (C) COX2 expression was tested by Western blot analysis in A549 cells transfected with each of three DNMTs expressing plasmids, and 8 h after transfection, cells were infected with IV (MOI = 1) for 24 h. The empty vector served as the negative control. (D and E) COX2 expression in PBMCs (D) and A549 cells (E) treated with DNMT inhibitor 5′-aza-CdR in a dose-dependent manner for 24 h. Western blots were used to analyze the COX2 expression.
Suppression of DNMTs triggers demethylation within the CREB binding site of the COX2 promoter.
To determine if IV infection affects DNA methylation in the COX2 promoter, we performed bisulfate DNA sequencing in A549 cells before and after IV infection. We found that CpG at the −57 position was dramatically demethylated following IV infection compared to mock infection (Fig. 4A). We then performed a chromatin immunoprecipitation (ChIP) assay to measure the binding activity for each of three DNMTs at the −57 CpG site of the COX2 promoter. Binding of both DNMT3a and DNMT3b was significantly reduced at this location after IV infection. In contrast, DNMT1 binding activity did not change significantly (Fig. 4B). These results indicate that DNMT3a and DNMT3b, but not DNMT1, are responsible for methylation at the −57 CpG, and DNMT3b may play the more important role in this process because the binding of DNMT3b was more significantly reduced over DNMT3a.
Fig 4.
Epigenetic modifications influence CREB binding activity at the CRE site in the COX2 promoter. (A) Bisulfite sequencing of the endogenous COX2 promoter region in A549 cells 24 h after infection with IV (MOI = 1). The sequencing region spanned from −227 to the +56 site in the COX2 promoter, including 11 CpGs in this area. Symbols: open circles, unmethylated; solid circles, methylated. (B) ChIP analysis using antibodies against DNMT1, DNMT3a, and DNMT3b in A549 cells 24 h after infection with IV (MOI = 1). IgG was used as the negative control. (C and D) ChIP analysis using antibodies against CREB1 in A549 cells 24 h after treatment with DNMT inhibitor 5′-aza-CdR (1 μM) (C) or 24 h after infection with IV (MOI = 1) (D). (E) A549 cells were transfected with CREB1-specific siRNA for 24 h. An irrelevant siRNA served as the negative control. Cells were then infected with IV (MOI = 1) and harvested 24 h after infection for the detection of COX2 expression by real-time RT-PCR and Western blotting. (F) The efficiency of CREB1-specific siRNA (siCREB1) was tested by RT-PCR.
We analyzed the sequence of the COX2 promoter and found that −57 CpG was located within the CREB binding motif, which indicates that CREB1 binding to the demethylated −57 position may be important for regulating COX2 expression. To determine whether there is a link between CREB1 binding and DNMT activity, we performed another ChIP to compare the CREB1 binding activities during treatment with the DNMT inhibitor 5′-aza-CdR and IV infection. We found that CREB1 binding activity increased after the treatment with 5′-aza-CdR (Fig. 4C) and IV infection (Fig. 4D). The results indicate that increased CREB1 binding activity is the direct consequence of demethylation at the CREB binding site. We can thus conclude that downregulation of DNMT3a and DNMT3b induces COX2 expression by demethylating CpG in CRE at the −57 position in the COX2 promoter, followed by an increase in CREB1 recruitment to the cognate binding site.
The next question is whether CREB1 binding induces COX2 expression. CREB1-specific siRNAs were transfected into A549 cells and infected with IV (MOI = 1). Results showed that COX2 expression was suppressed at both mRNA and protein levels in the presence of CREB1-specific siRNA but not the irrelevant siRNA control after IV infection (Fig. 4E and F). The results indicate that CREB1 binding is necessary for COX2 expression during IV infection.
DNMT3a and DNMT3b are downregulated in the early stages of influenza A virus infection.
To determine whether IV infection influences epigenetic modifications, A549 human lung epithelial cells were infected with IV (MOI = 1). Cells were harvested at various time points (0, 2, 4, 6, and 8 h), and the expression levels of three DNMTs were measured by quantitative real-time reverse transcription-PCR (RT-PCR). DNMT3a and DNMT3b, but not DNMT1, mRNA was markedly decreased as IV infection progressed over time (Fig. 5A). We then detected the level of DNMT family member proteins by Western blotting (Fig. 5B). Consistent with mRNA data, DNMT3a and DNMT3b proteins were downregulated, reaching the lowest levels at 6 and 8 h after IV infection. To determine the final DNMT catalyzing activity, we tested the total DNMT enzyme activity in both A549 cells and peripheral blood mononuclear cells (PBMCs) infected with IV (Fig. 5C). Total DNMT activity in the nuclei of infected A549 cells and PBMCs dropped sharply at 4 h after IV infection. These results indicate that the catalyzing activity of DNMTs decreases early after IV infection and should be a result of DNMT3a and DNMT3b, but not DNMT1, downregulation.
Fig 5.
Expression and activity of DNMT are reduced after IV infection. (A and B) A549 cells were infected with IV (MOI = 1). Cells were harvested at indicated time points (0, 2, 4, 6, and 8 h) and assayed for the expression levels of three DNMTs (DNMT3a, DNMT3b, and DNMT1) by quantitative real-time RT-PCR (A) and by Western blotting (B). (C) A549 cells and PBMCs were infected with IV (MOI = 1); 4 h after infection, cells were harvested for the preparation of nuclear extract. The total DNMT catalyzing activities in the nuclear extract from both A549 cells and PBMCs were measured. Data shown are mean ± SE; n = 3 (*, P < 0.05). OD, optical density.
miR29 plays an important role in DNMT downregulation during influenza virus infection.
Our data show that COX2 upregulation induced by IV infection was tightly correlated with downregulation of DNMT3a and DNMT3b. However, the mechanism by which the DNMTs were downregulated is unclear. As mentioned previously, DNMT downregulation occurred in the early stages of IV infection, and the rapid response time of the miRNA family molecules make them candidates for this mechanism. The miRNAs of the miR29 family have been shown to downregulate the expression of DNMT3a and DNMT3b (11, 43, 47).
To provide evidence supporting the relationship between the miR29 family and COX2 expression, the three members of miR29 family (miR29a, miR29b, and miR29c) were synthesized and transfected into A549 cells. COX2-derived PGE2 production was strongly induced by each of three miR29s compared to irrelevant control miRNA 24 h after transfection (Fig. 6A). We also measured total DNMT catalyzing activity in A549 cells transfected with the three miR29s. Following transfection with miR29s, total DNMT catalyzing activity significantly decreased in the nuclear extract of A549 cell lysates (Fig. 6B). Among the miR29 family members, miR29b most efficiently reduced DNMT catalyzing activity (Fig. 6B). Additionally, we measured the expression of COX2, DNMT3a, and DNMT3b in A549 cells transfected with miR29a, miR29b, and miR29c and found that COX2 expression was increased and DNMT3a and DNMT3b were decreased when the three miR29s were present (Fig. 6C). These results indicate that miRNAs of the miR29 family induce COX2 expression by regulating DNMT expression.
Fig 6.
COX2 upregulation caused by miR29 is tightly correlated with the decreased catalyzing activity of DNMTs. (A) A549 cells were transfected with each of three miR29 isoforms (miR29a, -b, and -c). PGE2 was tested in culture supernatant 24 h after transfection. Scramble miRNA served as the negative control and was designated as 1.0. Data shown are mean ± SE; n = 3 (*, P < 0.05). (B) A549 cells were transfected with each of three miR29 isoforms (miR29a, -b, and -c). Nuclear extracts were prepared 24 h after transfection. Total DNMT catalyzing activity was then tested. Scramble miRNA served as the negative control. Data shown are mean ± SE; n = 3 (*, P < 0.05). (C) Expressions of COX2, DNMT3a, and DNMT3b were detected by Western blotting 24 h after transfection with each of three miR29 isoforms (miR29a, -b, and -c) in A549 cells. Scramble miRNA served as the negative control. (D) A549 cells and PBMCs were infected with IV (MOI = 1). Cells were harvested at indicated time points (0, 2, 4, 6, and 8 h) and assayed for the expression level of miR29b. (E) Analysis of miR29b expression by quantitative real-time PCR in PBMCs from healthy individuals (n = 10) and IV-infected patients (n = 10; **, P < 0.01). (F) miR29 inhibitor was transfected into A549 cells for 24 h. Cells were then infected with IV (MOI = 1), and harvested 24 h after infection for the detection of DNMT3a, DNMT3b, and COX2 by both real-time RT-PCR and Western blotting. Scramble miRNA served as the negative control.
A critical question is whether the miR29 family miRNAs are upregulated during IV infection. We measured miR29b expression in both IV-infected A549 cells and PBMCs by quantitative real-time RT-PCR. Interestingly, we found that miR29b was upregulated in a time-dependent manner. Additionally, miR29b expression in both A549 cells and PBMCs was elevated at 2 h after infection and was increased 50-fold at 4 or 6 h after infection (Fig. 6D). We also tested miR29b expression in PBMCs of 10 patients infected with influenza A virus (H1N1) and 10 healthy individuals. miR29b expression in healthy people was relatively stable. In contrast, we found that miR29 was elevated an average of 10-fold in PBMCs from infected subjects compared to those from uninfected healthy controls (Fig. 6E). These results demonstrate that the miR29b upregulation occurs early after virus infection and influences DNMT expression in PBMCs. Through downregulating DNMT expression, the miR29 family induces expression of proinflammatory genes early in the course of a viral infection.
To verify the function of miR29 during IV infection, antago-miRNA29 was transfected into A549 cells as an inhibitor to block miR29 induced by IV (Fig. 6F). Results showed that downregulation of DNMT3a and DNMT3b induced by viral infection was reversed by the miR29 inhibitor but not by the scramble miRNA control at both mRNA and protein levels. COX2 expression induced by viral infection was also suppressed by the miRNA inhibitor. The results indicate that DNMT3a and DNMT3b downregulation induced by viral infection and consequent COX2 activation are dependent on miR29 expression. As we discussed before, COX2 expression is dependent on demethylation of the CREB1 site in the COX2 promoter. As a result, the miR29 inhibitor suppresses COX2 expression through reversing DNMT3a and DNMT3b downregulation induced by IV infection. Together, the results proved the important role of miR29 in DNMT downregulation, COX2 promoter demethylation, and COX2 expression during influenza virus infection.
Influenza virus infection activates COX2 through PKA signaling pathway.
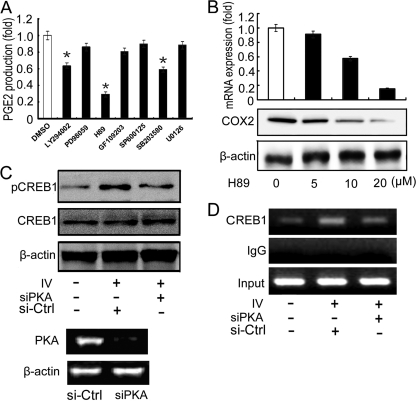
It has been reported that poly(I·C) mimics an influenza virus infection because the same double-stranded RNA (dsRNA) is produced during viral RNA replication (2, 19, 41). Other investigators have reported that dsRNA also activates PKA to phosphorylate CREB (25, 26), which is necessary for it to bind the COX2 promoter (35). To identify the signaling pathways activated during IV infection, A549 cells were infected with IV (MOI = 1) and treated with seven different signaling pathway inhibitors. At 24 h after infection, cells were harvested and PGE2 production was measured. The PKA inhibitor H89 reduced PGE2 production most effectively (Fig. 7A), indicating that PKA activation may play an important role in COX2 expression. Furthermore, administration of H89 suppressed IV-induced COX2 expression in a dose-dependent manner at both mRNA and protein levels (Fig. 7B). To determine the relationship between PKA and CREB1 phosphorylation, A549 cells were transfected with PKA-specific siRNA (Fig. 7C), an irrelevant siRNA was used as the negative control. Although no significant change in total CREB1 expression was observed, phosphorylated CREB1 was significantly elevated in response to IV infection and reduced in the presence of PKA-specific siRNA. We then performed a ChIP assay to examine the relationship between CREB1 phosphorylation and CREB1 binding to the COX2 promoter. We found that CREB1 binding activity is elevated upon IV infection and decreased in the presence of PKA-specific siRNA (Fig. 7D), indicating that only phospho-CREB1 is able to bind to the COX2 promoter. These results demonstrate that PKA activation and CREB1 phosphorylation are necessary for COX2 activation.
Fig 7.
The PKA-CREB pathway is involved in IV-induced COX2 expression. (A) A549 cells were infected with IV (MOI = 1) and treated with seven different signaling pathway inhibitors. After incubating for 24 h, cells were harvested and PGE2 was measured. Dimethyl sulfoxide (DMSO) served as the negative control. Data shown are mean ± SE; n = 3 (*, P < 0.05). (B) A549 cells were treated with the PKA inhibitor H89 at various doses (0, 5, 10, and 20 μM). After 24 h of incubation, cells were harvested and COX2 expression was measured by real-time RT-PCR and Western blotting. DMSO served as the negative control. (C and D) Specific siRNA against PKA was transfected into A549 cells for 24 h. Cells were then infected with IV (MOI = 1) and harvested 24 h after infection, and Western blotting was performed with antibody specific for CREB1 or phospho-CREB1 (C), or cells were harvested for the detection of CREB1 binding activity by ChIP assay (D). Irrelevant siRNA served as the negative control. The efficiency of PKA-specific siRNA (siPKA) was tested by RT-PCR (C, bottom).
DISCUSSION
Fatal influenza virus infections are often a result of acute inflammation induced by the viral infections, especially with regard to the highly pathogenic influenza viruses H5N1 and H1N1 epidemics of recent years. In this study, we provide a previously unrecognized mechanism by which both miR29-mediated epigenetic modifications and activation of the PKA signaling pathway play an important role in the regulation of COX2 and IFN-λ1 expression during infection with the influenza A virus.
By studying the blood samples from a cohort of subjects infected with influenza A virus and comparing to uninfected controls, we found that miR29, COX2-derived PGE2, and IFN-λ1 were all significantly increased in infected subjects, and levels of IFN-λ1 were tightly correlated with PGE2. To elucidate the mechanism behind this upregulation, we chose human lung epithelial cells A549 and PBMCs to mimic the viremic stage of influenza A virus infection in the body. We detected the changes of epigenetic modification status induced by the influenza virus infection. Aberrant epigenetic modification is currently being extensively studied, but most research has focused on the altered methylation pattern occurring in gametogenesis and early embryogenesis or chronic viral infections (9, 13). Here, we showed that in addition to epigenetic modifications, DNMT downregulation also occurs early in influenza virus infection. As a result, demethylation at the specific CREB binding sites leads to COX2 promoter activation. We then identified the miRNAs of the miR29 family as responsible for the rapidly decreased DNMT activity, thus explaining why DNMT expression downregulation occurs so soon following the onset of viral infection. Additionally, control by miR29 miRNAs also provides a way to restore DNMT expression when needed to avoid devastating consequences caused by DNMT downregulation. However, epigenetic modification changes induced by miR29 are not sufficient to activate COX2, which is further mediated by the activated PKA signaling pathway and CREB1 phosphorylation.
In uninfected cells, the proinflammatory genes such as COX2 and iNOS are silenced, and then activated during viral infection. A complex cell signaling network silences these genes, with DNA methylation regulation playing a critical role in this network (8). In this study, we provide evidence that DNMT3a and DNMT3b are involved in methylation regulation of COX2 promoter activation, whereas DNMT1 plays a less important role in this process. As we know, DNMT family has 5 main members, namely, DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3L, and their functions are quite different (8). DNMT1 maintains the methylation pattern of the newly synthesized DNA chain during DNA replication. Its maximum methylating activity needs semimethylated newly synthesized DNA double helix chain. DNMT2's methylating activity is very weak, and its function is not quite clear right now. DNBT3a and DNMT3b are involved in de novo acquisition of DNA methylation pattern, with the main methylation pattern happening during viral infection. DNMT3L is involved in maternal genomic imprinting and affects the activity of DNMT3a and 3b. In our research, we mainly focused on epigenetic modification caused by influenza virus infection, and DNMT3a and DNMT3b are the most functionally related DNMTs. We did not focus on DNMT1 because our results proved that DNMT1 expression was not affected by influenza virus infection, whereas DNMT3a and DNMT3b were downregulated. Furthermore, miR29 targeted on DNMT3a and DNMT3b but not DNMT1.
Interestingly, DNMT downregulation occurs very early after viral infection. As demethylation is an upstream event in proinflammatory gene expression, downregulation of DNMT3a and DNMT3b might trigger the entire inflammation signaling network. However, the induction of the miR29 family may happen even earlier. Previous research has mainly focused on the function and recognition site (12, 28, 46) of miR29, and little is yet known about its role during viral infection. In this study, we found that the expression of the three miR29 family miRNAs changed dramatically in the early stages of viral infection. This rapid response ensures that host cells activate an antiviral response early after infection with the virus. The expression of miR29 seems to be necessary for the early stage of inflammation signaling network activation. After this network has been activated, miR29 expression is rapidly reduced to a relatively low level. This reduction may avoid the gene expression disorder aroused by low-level methylation activity. The mechanism of miR29 regulation and the delicate process to keep perfect balance between methylation and demethylation levels during proinflammatory responses need further investigation.
The PKA-CREB signaling pathway has been reported to play an important role in influenza virus infection (23, 25, 26, 35, 41). However, this pathway can only partially explain how COX2 is induced during early viral amplification. It remains unclear how COX2 is initially activated after virus infection and how COX2 expression is rapidly suppressed after the infection is controlled. Our study emphasizes the role of cross talk between miR29-mediated epigenetic modifications and the PKA-CREB signaling pathway during viral infection. Both miR29-mediated demethylation at the specific CREB1 site in the COX2 promoter and activation of the PKA signaling pathway occur simultaneously and recruit CREB1 to the promoter, ultimately resulting in COX2 activation and consequent IFN-λ1 production.
To investigate the kinetic expressions of miR29, COX2, and IFN-λ1, A549 cells were infected with IV (MOI = 1), and cells were collected at different time points (0, 2, 4, 6, 8, 12, 24, and 48 h). The total miRNA and mRNA were extracted separately. Expressions of miR29, COX2, and IFN-λ1 were then tested by quantitative real-time RT-PCR. Results showed that the expression levels of IFN-λ1 were tightly correlated with COX2, which is consistent with the clinical sample analysis. Expression levels of miR29 increased more rapidly than those of IFN-λ1 and COX2 (Fig. 8A). A model of this mechanism, including miR29-mediated DNMTs, PKA activation, COX2, and IFN-λ1 production, is summarized in Fig. 8B. This model illustrates the cooperation and cross talk between the two pathways that ensures prompt activation of proinflammatory genes after virus infection and rapid suppression of these genes when the infection is under control.
Fig 8.
(A) The kinetic curves showing the expressions of miR29, COX2, and IFN-λ1 in a time-dependent manner. A549 cells were infected with IV (MOI = 1), and cells were collected at the indicated time points (0, 2, 4, 6, 8, 12, 24, and 48 h). The total miRNA and mRNA were extracted separately. Expressions of miR29, COX2, and IFN-λ1 were then tested by quantitative real-time RT-PCR, respectively. (B) A hypothetical model related to influenza A virus infection and host responses, showing simultaneous effects of both miR29-mediated demethylation at a specific CREB site in the COX2 promoter and activation of PKA signaling pathway-induced CREB recruitment to the promoter. These two pathways both contribute to the activation of COX2 and consequently to IFN-λ1 production.
It is well established that COX2 is important in activation of inflammation, although it does not directly suppress virus replication. IFN-λ1 is a newly discovered member of the IFN family, and its antiviral activity, especially with respect to the influenza virus, has been well studied in recent years (15, 16, 30, 34, 36, 37, 44). However, the genes that regulate IFN-λ1 have not been established. In this study, we investigated the relationship between COX2 and IFN-λ1 and found that the expression of IFN-λ1 was enhanced by COX2 through promoting NF-κB binding to the enhancer in the IFN-λ1 promoter. The potent antiviral activity of IFN-λ1 (15, 30, 34, 44) is enhanced by COX2 activation to protect host cells from IV infection. The data from this study provide details about the mechanism by which IFN-λ1 has both proinflammatory and antiviral functions. The promoter of IFN-λ1 has both typical IRF3/7 binding sites and two NF-κB clusters which may serve as bridges between the IFN family and the inflammation network, suggesting that the IFN family and the inflammation network may not work independently. The inflammatory signaling downstream of COX2 is a large and complex network, and the details of the mechanism require more investigation.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (30970144), by research grants from the Major State Basic Research Development Program of China (“973” project no. 2009CB522506 and no. 2012CB518900), by the National Key Scientific and Technological Project (2012ZX10004503-004), and by the Program for Changjiang Scholars and Innovative Research Team in University (no. IRT0745).
The funding agencies had no role in study design, data collection, or analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 9 November 2011
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Angeli V, et al. 2004. Activation of the D prostanoid receptor 1 regulates immune and skin allergic responses. J. Immunol. 172:3822–3829 [DOI] [PubMed] [Google Scholar]
- 2. Asahi-Ozaki Y, et al. 2006. Intranasal administration of adjuvant-combined recombinant influenza virus HA vaccine protects mice from the lethal H5N1 virus infection. Microbes Infect. 8:2706–2714 [DOI] [PubMed] [Google Scholar]
- 3. Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. Microbiol. 116:281–297 [DOI] [PubMed] [Google Scholar]
- 4. Betz M, Fox BS. 1991. Prostaglandin E2 inhibits production of Th1 lymphokines but not of Th2 lymphokines. J. Immunol. 146:108–113 [PubMed] [Google Scholar]
- 5. Campbell PM, Syzf M. 2003. Human DNA methyltransferase gene DNMT1 is regulated by the APC pathway. Carcinogenesis 24:17–24 [DOI] [PubMed] [Google Scholar]
- 6. Carey MA, et al. 2003. Accentuated T helper type 2 airway response after allergen challenge in cyclooxygenase-1-/- but not cyclooxygenase-2-/- mice. Am. J. Respir. Crit. Care Med. 167:1509–1515 [DOI] [PubMed] [Google Scholar]
- 7. Chen N, Restivo A, Reiss CS. 2002. Selective inhibition of COX-2 is beneficial to mice infected intranasally with VSV. Prostaglandins Other Lipid Mediat. 67:143–155 [DOI] [PubMed] [Google Scholar]
- 8. Chen T, Li E. 2004. Structure and function of eukaryotic DNA methyltransferases. Curr. Top. Dev. Biol. 60:55–89 [DOI] [PubMed] [Google Scholar]
- 9. de Maat MFG, et al. 2007. Epigenetic silencing of cyclooxygenase-2 affects clinical outcome in gastric cancer. J. Clin. Oncol. 25:4887–4894 [DOI] [PubMed] [Google Scholar]
- 10. Ejima K, et al. 2003. Cyclooxygenase- 2-deficient mice are resistant to endotoxin-induced inflammation and death. FASEB J. 17:1325–1327 [DOI] [PubMed] [Google Scholar]
- 11. Fabbri M, et al. 2007. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. U. S. A. 104:15805–15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garzon R, et al. 2009. MicroRNA 29b functions in acute myeloid leukemia. Blood 114:5331–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanson JA, et al. 2006. Gene promoter methylation in prostate tumor-associated stromal cells. J. Natl. Cancer Inst. 98:255–261 [DOI] [PubMed] [Google Scholar]
- 14. Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. 1996. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. U. S. A. 93:9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hui KP, et al. 2009. Induction of proinflammatory cytokines in primary human macrophages by influenza A virus (H5N1) is selectively regulated by IFN regulatory factor 3 and p38 MAPK. J. Immunol. 182:1088–1098 [DOI] [PubMed] [Google Scholar]
- 16. Kotenko SV, et al. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptorcomplex. Nat. Immunol. 4:69–77 [DOI] [PubMed] [Google Scholar]
- 17. Lee JO, Jung KHJK, Choi KH, Min DS, Jang KL. 2005. Hepatitis B virus X protein represses E-cadherin expression via activation of DNA methyltransferase 1. Oncogene 24:6617–6625 [DOI] [PubMed] [Google Scholar]
- 18. Li H, Chen RTZX, Szabó PE, Riggs AD, Pfeifer GP. 2006. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J. Biol. Chem. 281:19489–19500 [DOI] [PubMed] [Google Scholar]
- 19. Li W, et al. 2009. Negative feedback regulation of IL-32 production by iNOS activation in response to dsRNA or influenza virus infection. Eur. J. Immunol. 39:1019–1024 [DOI] [PubMed] [Google Scholar]
- 20. Li W, et al. 2008. Activation of interleukin-32 pro-inflammatory pathway in response to influenza A virus infection. PLoS One 3:e1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ludwig S, Pleschka S, Planz O, Wolff T. 2006. Ringing the alarm bells: signalling and apoptosisin influenza virus infected cells. Cell. Microbiol. 8:375–386 [DOI] [PubMed] [Google Scholar]
- 22. Luttmann W, et al. 1999. Modulation of cytokine release from mononuclear cells by prostacyclin, IL-4 and IL-13. Cytokine 11:127–133 [DOI] [PubMed] [Google Scholar]
- 23. Maggi LB, Jr, et al. 2002. Novel role for calcium-independent phospholipase A(2) in the macrophage antiviral response of inducible nitric-oxide synthase expression. J. Biol. Chem. 277:38449–38455 [DOI] [PubMed] [Google Scholar]
- 24. Martin JG, et al. 2002. The immunomodulatory actions of prostaglandin E2 on allergic airway responses in the rat. J. Immunol. 169:3963–3969 [DOI] [PubMed] [Google Scholar]
- 25. Martinson BD, Corbett ACJA, Wysolmerski RB, Ford DA. 2003. Calcium-independent phospholipase A2 mediates CREB phosphorylation in double-stranded RNA-stimulated endothelial cells. J. Lipid Res. 44:1686–1691 [DOI] [PubMed] [Google Scholar]
- 26. Misra UK, Pizzo SV. 2005. Coordinate regulation of forskolin-induced cellular proliferation in macrophages by protein kinase A/cAMP-response element-binding protein (CREB) and Epac1-Rap1 signaling: effects of silencing CREB gene expression on Akt activation. J. Biol. Chem. 280:38276–38289 [DOI] [PubMed] [Google Scholar]
- 27. Mizumura K, et al. 2003. Role of mitogen-activated protein kinases in influenza virus induction of prostaglandin E2 from arachidonic acid in bronchial epithelial cells. Clin. Exp. Allergy 33:1244–1251 [DOI] [PubMed] [Google Scholar]
- 28. Mott JL, Bronk KSSF, Gores GJ. 2007. Mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26:6133–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noguchi K, Endo H, Kondo H, Ishikawa I. 2001. Prostaglandin E2 upregulates interleukin-6 production in human gingival fibroblasts. J. Periodontal Res. 36:80–87 [DOI] [PubMed] [Google Scholar]
- 30. Osterlund P, et al. 2005. Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells. J. Virol. 79:9608–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. 2007. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-λ) genes. J. Immunol. 179:3434–3442 [DOI] [PubMed] [Google Scholar]
- 32. Park IY, et al. 2007. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology 132:1476–1494 [DOI] [PubMed] [Google Scholar]
- 33. Reddy RC, et al. 2001. Selective inhibition of COX-2 improves early survival in murine endotoxemia but not in bacterial peritonitis. Am. J. Physiol. 281:L537–L543 [DOI] [PubMed] [Google Scholar]
- 34. Rintahaka J, Kovanen WDPE, Alenius H, Matikainen S. 2008. Cytosolic antiviral RNA recognition pathway activates caspases 1 and 3. J. Immunol. 180:1749–1757 [DOI] [PubMed] [Google Scholar]
- 35. Scarfì S, et al. 2007. Ascorbic acid-pretreated quartz enhances cyclo-oxygenase-2 expression in RAW 264.7 murine macrophages. FEBS J. 274:60–73 [DOI] [PubMed] [Google Scholar]
- 36. Sheppard P, et al. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63–68 [DOI] [PubMed] [Google Scholar]
- 37. Sirén J, Pirhonen J, Julkunen I, Matikainen S. 2005. IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29. J. Immunol. 174:1932–1937 [DOI] [PubMed] [Google Scholar]
- 38. Stohr K. 2005. Avian influenza and pandemics-research needs and opportunities. N. Engl. J. Med. 352:405–407 [DOI] [PubMed] [Google Scholar]
- 39. Szyf M. 2002. Utilization of antisense oligonucleotides to study the role of 5-cytosine DNA methyltransferase in cellular transformation and oncogenesis. Methods 27:184–191 [DOI] [PubMed] [Google Scholar]
- 40. Thomson SJP, et al. 2009. The role of transposable elements in the regulation of IFN-λ1 gene expression. Proc. Natl. Acad. Sci. U. S. A. 106:11564–11569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Traynor TR, Bohnet MJSG, Krueger JM. 2004. Intratracheal double-stranded RNA plus interferon-gamma: a model for analysis of the acute phase response to respiratory viral infections. Life Sci. 74:2563–2576 [DOI] [PubMed] [Google Scholar]
- 42. Tumpey TM, et al. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77–80 [DOI] [PubMed] [Google Scholar]
- 43. Volinia S, et al. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U. S. A. 103:2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang J, et al. 2009. Differentiated human alveolar type II cells secrete antiviral IL-29 (IFN-lambda 1) in response to influenza A infection. J. Immunol. 182:1296–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xie ZH, et al. 2006. Mutations in DNA methyltransferase DNMT3B in ICF syndrome affect its regulation by DNMT3L. Hum. Mol. Genet. 15:1375–1385 [DOI] [PubMed] [Google Scholar]
- 46. Xiong Y, et al. 2010. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 51:836–845 [DOI] [PubMed] [Google Scholar]
- 47. Yanaihara N, et al. 2006. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9:189–198 [DOI] [PubMed] [Google Scholar]
- 48. Zheng DL, et al. 2009. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J. Hepatol. 50:377–387 [DOI] [PubMed] [Google Scholar]