Abstract
The innate immune response constitutes the first line of defense against viral infection and is extensively regulated through ubiquitination. The removal of ubiquitin from innate immunity signaling factors by deubiquitinating enzymes (DUBs) therefore provides a potential opportunity for viruses to evade this host defense system. It was previously found that specific proteases encoded by the unrelated arteri- and nairoviruses resemble the ovarian tumor domain-containing (OTU) family of DUBs. In arteriviruses, this domain has been characterized before as a papain-like protease (PLP2) that is also involved in replicase polyprotein processing. In nairoviruses, the DUB resides in the polymerase protein but is not essential for RNA replication. Using both in vitro and cell-based assays, we now show that PLP2 DUB activity is conserved in all members of the arterivirus family and that both arteri- and nairovirus DUBs inhibit RIG-I-mediated innate immune signaling when overexpressed. The potential relevance of RIG-I-like receptor (RLR) signaling for the innate immune response against arterivirus infection is supported by our finding that in mouse embryonic fibroblasts, the production of beta interferon primarily depends on the recognition of arterivirus RNA by the pattern-recognition receptor MDA5. Interestingly, we also found that both arteri- and nairovirus DUBs inhibit RIG-I ubiquitination upon overexpression, suggesting that both MDA5 and RIG-I have a role in countering infection by arteriviruses. Taken together, our results support the hypothesis that arteri- and nairoviruses employ their deubiquitinating potential to inactivate cellular proteins involved in RLR-mediated innate immune signaling, as exemplified by the deubiquitination of RIG-I.
INTRODUCTION
The first line of defense against viral infection is formed by the innate immune response. This system relies on a series of pattern-recognition receptors (PRRs) whose activation by pathogen-associated molecular patterns (PAMPs) ultimately leads to the transcription of genes encoding type I interferons (IFNs) and proinflammatory cytokines. In turn, these cytokines stimulate the expression of a myriad of effector molecules, the presence of which results in a cellular microenvironment that is hostile to viral replication. To date, four families of PRRs have been implicated in the response to viral infection: the membrane-bound Toll-like receptors (TLRs) and the cytosolic Nod-like receptors (NLRs), HIN-200 protein family members, and RIG-I-like receptors (RLRs) (27, 43).
Upon recognition of viral RNA by the RLRs RIG-I and MDA5, these sensors bind to the mitochondrially located adaptor protein MAVS, which is also known as IPS1, VISA, or Cardif. This interaction activates two divergent signaling pathways. The first involves the adaptor protein TRAF3 and triggers the activation of the TBK1-IκB kinase ε (IKKε) complex, which then phosphorylates IRF3. Upon subsequent dimerization, this transcription factor translocates to the nucleus to initiate transcription of the gene encoding beta IFN (IFN-β), a type I IFN. The second pathway downstream of MAVS involves the adaptor protein TRAF6 and the NEMO-IKKα/β complex, of which the latter is responsible for the phosphorylation event preceding the degradation of the NF-κB inhibitor IκB. With IκB no longer present, NF-κB is free to translocate to the nucleus and initiate the transcription of genes encoding proinflammatory cytokines and, together with activated IRF3, IFN-β. Through autocrine and paracrine signaling routes, IFN-β induces the transcription of numerous interferon-stimulated genes (ISGs), including those encoding the PRRs RIG-I and MDA5, and the antiviral effector molecules OAS, PKR, ISG15, and the Mx proteins, whose concerted action limits the spread of viral infection (6, 51).
In addition to phosphorylation, the signaling pathways involved in innate immunity are extensively regulated through ubiquitination (5, 63). Ubiquitin (Ub) is an 8-kDa protein moiety that can be covalently attached to a lysine in a target protein. Substrates can be tagged either by a single Ub moiety (monoubiquitination) or by a chain of Ub moieties that are linked by isopeptide bonds between their C-terminal glycine residue and an internal lysine residue (polyubiquitination). The configuration of the polyubiquitin chain can vary, depending on which one of the lysines present in the ubiquitin protein is used for conjugation. Of the possible configurations, those of Lys48- and Lys63-linked polyubiquitination have been studied most extensively. Lys48-linked ubiquitination induces translocation of the target protein to the proteasome, where degradation follows. In contrast, Lys63-linked ubiquitination results in activation or relocation of the target, which, for example, plays a role in signaling cascades.
Innate immune signaling is regulated through ubiquitination at multiple levels. For example, the IKKα/β-induced degradation of IκB relies on its Lys48-linked ubiquitination (28). In addition, a number of proteins involved in signal transduction were recently shown to be activated by Lys63-linked ubiquitination. This list includes RIG-I, MAVS, TBK1/IKKε, and TRAF3 and -6 (17, 45, 47, 65). Notably, this ubiquitin-mediated regulation poses an excellent opportunity for negative feedback and viral immune evasion since ubiquitination is a reversible process, with the deconjugation of ubiquitin being performed by deubiquitinating enzymes (DUBs). For example, the mammalian DUBs OTUB1 and OTUB2 downregulate the innate immune response through the deubiquitination of TRAF3 and -6 (35). In addition, Cezanne (11), DUBA (31), and A20 (45, 57) are mammalian DUBs that negatively regulate innate immune signaling.
DUBs can be grouped into five subclasses, based on their structural characteristics (42). Previously, members of two unrelated groups of RNA viruses, the arteri- and nairoviruses, were found to encode a protease that resembles the ovarian tumor domain-containing (OTU) subclass of DUBs (39). Interestingly, all of the above-mentioned mammalian DUBs that control the innate immune response also belong to this particular subclass. For this reason, Frias-Staheli et al. (2007) hypothesized that arteri- and nairoviral OTU DUBs are involved in the negative regulation of innate immune signaling in order to support viral replication (15).
The Arterivirus family, together with the Corona- and Ronivirus families, belongs to the order Nidovirales and currently comprises four species: Equine arteritis virus (EAV), Porcine respiratory and reproductive syndrome virus (PRRSV), Lactate dehydrogenase-elevating virus (LDV), and Simian hemorrhagic fever virus (SHFV). Of these pathogens, PRRSV poses the greatest burden on society, with regular outbreaks in pigs causing major economic losses, especially in North America and Southeast Asia (41, 75). PRRSV isolates are divided into two quite distant genotypes: European (type I) and North American (type II). Arteriviruses are positive-sense RNA viruses with a 13- to 16-kb nonsegmented genome. Their cytoplasmic replication starts with the translation of the genomic RNA to produce two large replicase polyproteins, pp1a and pp1ab. In the case of EAV, the proteolytic maturation of these polyproteins, which is directed by three internal proteinases, releases at least 13 nonstructural proteins, most of which assemble into a membrane-bound replication complex (66, 76). Eight structural proteins which are produced by synthesis and translation of a nested set of subgenomic mRNAs have been identified in arteriviruses (12, 46, 54).
The arterivirus DUB domain resides in the N-terminal region of nonstructural protein 2 (nsp2) and was previously shown to direct the proteolytic processing of the nsp2/3 site (22, 60). In addition to its resemblance to OTU DUBs, the nsp2 protease belongs to the papain-like protease (PLP) family and will therefore be referred to as PLP2-DUB in this paper. Interestingly, the (predicted) nsp2/3 junction in most arteriviruses only moderately resembles the LXGG↓ consensus motif commonly recognized by DUBs. In fact, only the EAV nsp2/3 site (LIGG) contains the consensus Leu residue at position P4, whereas PRRSV type I (PRRSV-I) (TTGG), PRRSV-II (PSGG), LDV (VSGG), and SHFV (VVGG) merely share the double glycine motif occupying the P1 and P2 positions of the cleavage site. This observation suggests that the arterivirus-encoded PLP2-DUBs constitute a unique protease subgroup that has evolved an intriguing specificity toward both viral and cellular substrates.
The Nairovirus genus of the Bunyavirus family currently consists of seven species, including the prototypic Dugbe virus (DUGV) and Crimean-Congo hemorrhagic fever virus (CCHFV), the causative agent of a severe hemorrhagic fever with case fatality rates of up to 30% in humans (70). CCHFV is one of the most widespread medically relevant arboviruses, being surpassed in geographical distribution only by dengue virus (18). CCHFV infections occur sporadically throughout Africa, Asia, the Middle East, and southeastern Europe and are correlated with the natural distribution of its preferred vector, ticks of the genus Hyalomma (70). Nairoviruses have a tripartite RNA genome of negative polarity, with the largest segment encoding the L protein, which harbors the RNA-dependent RNA polymerase (RdRp). The nairovirus DUB domain is situated in the N-terminal region of the L protein, but in contrast to its arteriviral counterpart, it is not involved in autoproteolysis and its catalytic activity is dispensable for RdRp activity (4).
In line with their hypothesis about a role in immune evasion, Frias-Staheli et al. (2007) showed that expression of DUBs from two arteriviruses (EAV and PRRSV) and two nairoviruses (DUGV and CCHFV) negatively affected the tumor necrosis factor alpha-induced activation of NF-κB. Furthermore, transgenic mice expressing the N-terminal part of the CCHFV L protein displayed an increased susceptibility to Sindbis virus infection (15). They also showed that these viral DUBs, unlike their mammalian counterparts, have the ability to target both ubiquitin and ISG15, a 15-kDa interferon-stimulated Ub-like modifier that plays an important role in antiviral defense (59). The recently solved crystal structure of the CCHFV DUB domain provided a structural basis for this observed catalytic promiscuity (1, 7, 26). In the arterivirus family, overexpression of the PRRSV PLP2-DUB was found to inhibit the Sendai virus-induced innate immune response (33, 64). Based on in vitro assays, Sun et al. (2010) proposed that this effect at least partially relies on the deubiquitination of IκBα, which would prevent the degradation of IκB and the subsequent nuclear translocation of NF-κB (64).
Here we report the characterization of the immune evasive properties of arteri- and nairovirus DUBs. We show for the first time that the DUB activity of the PLP2 domain is a conserved feature of all members of the arterivirus family and that it can inhibit the RLR-mediated immune response. We investigated the role of this pathway in the sensing of arteriviral RNA and found that in mouse embryonic fibroblasts (MEFs) the induction of IFN-β by EAV RNA is MAVS dependent and that the recognition of EAV RNA relies primarily on the PRR MDA5. Interestingly, we were able to show that overexpression of both arteri- and nairovirus DUBs inhibits RIG-I ubiquitination, suggesting that RIG-I is also likely involved in the response to arterivirus infection. Taken together, our data support the concept that the DUB-mediated immune evasion in arteri- and nairoviruses is, at least in part, the result of the removal of ubiquitin from RIG-I and likely also from other innate immune signaling factors in the RLR pathway.
MATERIALS AND METHODS
Plasmids.
nsp2 of the EAV Bucyrus strain (NCBI accession number NC_002532) comprises residues Gly261 to Gly832 of the EAV pp1a/pp1ab replicase polyproteins. Polyprotein amino acid numbers will be used throughout this paper. For bacterial expression of EAV nsp2(N), a cDNA fragment encoding amino acids (aa) 261 to 427 was cloned downstream of the maltose-binding protein (MBP) fusion partner into the pMalT vector (New England BioLabs). Active-site mutations C270A, H332A, and C270A/H332A, the last one of which is referred to from here on as C/H>A, were engineered by site-directed mutagenesis using Pfu DNA polymerase (Fermentas). Primer sequences are available upon request.
Mammalian expression constructs encoding the PLP2-DUB domains of all arteriviruses were made by cloning the following sequences encoding the indicated amino acids of the respective replicase polyproteins and placing them in frame with an N-terminal hemagglutinin (HA) tag in the pcDNA3.1 vector (Invitrogen): aa 261 to 1064 for EAV nsp2-3, aa 261 to 435 for EAV nsp2(N), aa 385 to 578 for PRRSV-I nsp2(N), aa 383 to 782 for PRRSV-II nsp2(N), aa 381 to 576 for LDV nsp2(N), and aa 480 to 829 for SHFV nsp2(N). The last two inserts were synthesized by the GeneArt gene synthesis service (Invitrogen) and were based on the sequences with NCBI accession numbers NC_001639 and NC_003092, respectively. For PRRSV-I nsp2(N) a sequence from the European SD-01-08 strain which was amplified from pCAGGS-nsp2 was used (64). The PRRSV-II nsp2(N) construct encodes the North American prototype VR-2332 strain. For the mammalian expression constructs of the CCHFV DUB domain, a fragment encoding aa 1 to 169 of the L protein was transferred from pCAGGS-HA-L(1-169) wild type and C40A/H151A (15) to the pcDNA3.1 vector. An insert containing the enhanced green fluorescent protein (EGFP) gene from pEGFP-N1 (Clontech) was placed into the multiple-cloning site of pcDNA3.1 to generate plasmid pcDNA-eGFP. All constructs were verified by sequence analysis.
The following expression plasmids were described elsewhere: pLuc-IFN-β (13), pRL-TK (Promega), pEBG-RIG-I(2CARD) (17), pcDNA-FLAG-MAVS (56), pEGFP-C1-IRF3(5D) (36), and pRBG4-CW7-myc-ubiquitin (69).
Viruses, cells, and antibodies.
The Bucyrus strain of EAV was propagated and concentrated by polyethylene glycol precipitation essentially as described previously (10). Propagation of mengovirus, derived from the pM16.1 full-length cDNA clone, was described before (23). Recombinant Newcastle disease virus (NDV), derived from the Hitchner B1 vaccine strain and containing a GFP insertion, was described elsewhere (49).
MEFs isolated from transgenic MAVS−/−, MDA5−/−, or RIG-I−/− mice and their respective wild-type littermate controls were described previously (29, 30, 62). MEFs, HEK293T cells, and Vero-E6 cells were all cultured in Dulbecco's modified Eagle medium (Lonza) supplemented with 10% fetal bovine serum (Bodinco BV), 100 units/ml each of penicillin/streptomycin (Lonza), and 2 mM l-glutamine (Lonza).
The antibodies used were anti-glutathione S-transferase (GST; sc459; Santa Cruz), anti-HA (ab18181; Abcam), anti-c-myc (9E10; Roche), anti-transferrin receptor (TFR; H68.4; Invitrogen), and anti-GFP from rabbits immunized with purified recombinant GFP.
In vitro DUB activity assay.
Escherichia coli strain BL21(DE3) transformed with wild-type or mutant pMalT-EAV-nsp2(N) was grown to an optical density at 600 nm of 0.6 in standard LB broth. Protein expression was then induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. After 3 h at 37°C, bacteria were resuspended in buffer A (20 mM Tris, 200 mM NaCl, 1 mM EDTA, pH 7.5) and lysed by sonication. After removal of cellular debris by centrifugation, the supernatant was incubated for 2 h with amylose resin (New England BioLabs). Resin was then washed 4 times with buffer A and protein was eluted using buffer B (20 mM Tris, 200 mM NaCl, 1 mM EDTA, 10 mM maltose, pH 7.5). Protein concentration and purity were determined using Bradford reagent (Bio-Rad) and SDS-PAGE analysis, respectively.
For an in vitro DUB activity assay, 1 μg of purified recombinant EAV nsp2(N) was incubated with 2.5 μg Lys48- or Lys63-linked polyubiquitin chains (Boston Biochem) in a final volume of 10 μl assay buffer (50 mM Tris, 5 mM MgCl2, 2 mM dithiothreitol [DTT], pH 7.5) for 2 h at 37°C. For nsp2(N) dilution series, a range of 2-fold dilutions starting at 0.5 μg per reaction was used. As a positive control, isopeptidase T (Boston Biochem) was included. The reaction was terminated by the addition of 3.5 μl 4× Laemmli sample buffer (4× LSB; 500 mM Tris, 4% SDS, 40% glycerol, 0.02% bromophenol blue, 2 mM DTT, pH 6.8). Proteins were separated in a 15% SDS-polyacrylamide gel and visualized using Coomassie brilliant blue staining. Gels were scanned using a GS-800 calibrated densitometer (Bio-Rad).
DUB activity assay in HEK293T cells.
HEK293T cells, grown to 80% confluence in 12-well plates, were transfected with a combination of plasmids encoding myc-ubiquitin (0.25 μg), GFP (0.25 μg), and the different viral proteins (1.5 μg) using the CaPO4 transfection method (19). Total DNA amounts were kept constant by the addition of empty vector. After 16 h at 37°C, cells were lysed in 250 μl 2× LSB (250 mM Tris, 2% SDS, 20% glycerol, 0.01% bromophenol blue, 1 mM DTT, pH 6.8). Samples were loaded on SDS-polyacrylamide gels, which were blotted to Hybond-P polyvinylidene difluoride membranes (GE Healthcare) using a semidry transfer cell (Bio-Rad). After incubation with the appropriate antibodies, protein bands were visualized using the Amersham ECL Plus detection reagent (GE Healthcare).
Quantitative real-time PCR.
Vero-E6 cells, grown to 90% confluence in 6-well plates, were infected at a multiplicity of infection of 10 with EAV, mengovirus, or NDV or were mock infected. Total RNA was isolated at 16 h postinfection using TRIzol reagent (Invitrogen). Subsequently, MEFs lacking MAVS, MDA5, or RIG-I and their respective wild-type controls, seeded at a density of 200,000 cells per well in 6-well plates on the previous day, were transfected with 4 μg of the aforementioned total RNA using Lipofectamine 2000 (Invitrogen). At 3 h posttransfection (p.t.), total RNA was isolated from these cells using a Nucleospin RNA II kit (Machery-Nagel) and used as a template for cDNA synthesis using Moloney murine leukemia virus reverse transcriptase (Fermentas) and oligo(dT)20 primer. Finally, samples were assayed by quantitative real-time PCR analysis (qRT-PCR) using commercially available gene expression assays for mouse IFN-β and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Mm00439552_s1 and Mm99999915_g1, respectively; Applied Biosystems). PCR was performed using a 7900HT Fast real-time PCR system and analyzed with SDS2.4 software (both from Applied Biosystems). PCR efficiencies were determined to be 86 and 93% for IFN-β and GAPDH, respectively, which allowed analysis using the comparative threshold cycle (CT) method (55). In addition, it was established that GAPDH expression did not differ between MEFs transfected with total RNA from infected or mock-infected Vero cells.
Luciferase-based IFN-β reporter assay.
HEK293T cells, grown to 80% confluence in 24-well plates, were transfected in quadruplicate with a combination of plasmids encoding firefly luciferase under the control of the IFN-β promoter (50 ng), Renilla luciferase (5 ng), RIG-I(2CARD), MAVS, or IRF3(5D) (25 ng) and the various viral proteins (1,000 ng) using Lipofectamine 2000. For dose range experiments, 2-fold serial dilutions of viral protein-encoding plasmids were used, starting at 1,000 ng of DNA. Total DNA amounts were kept equal by the addition of empty vector. At 12 h p.t., three out of four wells were lysed in 100 μl passive lysis buffer (Promega), and samples were assayed for luciferase activity using the Dual-Luciferase reporter assay system (Promega) on a Mithras LB 940 multimode reader (Berthold Technologies). The remaining well was lysed in 150 μl 2× LSB, and these samples were loaded onto SDS-polyacrylamide gels and analyzed by Western blotting as described above.
An unpaired two-tailed Student's t test was used to determine the statistical significance of the results obtained with the luciferase assays. P values of <0.05 were considered to be statistically significant.
RIG-I ubiquitination assay.
HEK293T cells, grown to 80% confluence in 6-well plates, were transfected with a combination of plasmids encoding myc-ubiquitin (0.5 μg), GFP (0.5 μg), GST–RIG-I(2CARD) (1.5 μg), and the various viral proteins (1.5 μg) using the CaPO4 transfection method. Total DNA amounts were kept constant by the addition of empty vector. At 16 h p.t., cells were lysed for 1 h in buffer C [50 mM Tris, 5 mM MgCl2, 0.5% NP-40, 25 mM N-ethylmaleimide, 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride, 5 μg/ml leupeptin, pH 7.5]. After removal of cellular debris by centrifugation, 1/10 of the lysate was mixed 1:1 with 2× LSB. The remaining lysate was incubated at 4°C for 3 h with glutathione Sepharose 4B resin (GE Healthcare). The resin was subsequently washed 3 times with buffer C, and proteins were eluted by addition of 2× LSB and incubation at 96°C for 5 min. Samples were then loaded on an SDS-polyacrylamide gel and analyzed by Western blotting as described above.
RESULTS
Recombinant EAV PLP2-DUB exhibits in vitro DUB activity, which is abolished by mutagenesis of active-site residues.
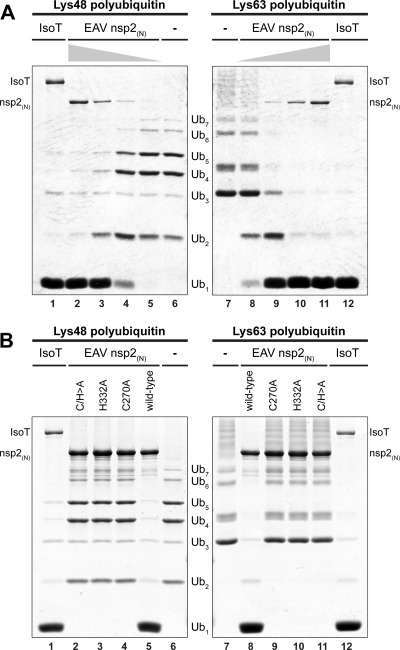
It was previously shown by Frias-Staheli et al. (2007) that overexpression of EAV and PRRSV nsp2 in mammalian cells resulted in a general decrease of the level of ubiquitinated proteins (15). In addition, Sun et al. (2010) showed that PRRSV-I PLP2, when immunoprecipitated from mammalian cells, cleaved Lys48-linked polyubiquitin chains (64). To confirm that the observed activities of nsp2 derive from the bona fide DUB activity of its protease and to exclude the possibility that these observations were caused by an indirect effect on cellular ubiquitination processes, a recombinant N-terminal fragment of EAV nsp2 containing the predicted PLP2-DUB domain [nsp2(N)] was purified from E. coli and tested in an in vitro activity assay. To this end, a dose range of purified recombinant protein was incubated with Lys48- or Lys63-linked polyubiquitin chains. Figure 1A shows that EAV nsp2(N) efficiently cleaved both types of substrates, while showing a slight preference for Lys63-linked chains in this assay. In addition, mutagenesis to Ala of the putative catalytic residues Cys270 and His332 (60) completely abolished the cleavage of ubiquitin chains of both linkage types (Fig. 1B). These results clearly establish that the PLP2-DUB domain present in EAV nsp2 possesses genuine DUB activity in the absence of other viral or cellular proteins.
Fig 1.
The EAV PLP2-DUB cleaves both Lys63- and Lys48-linked polyubiquitin chains. (A) To establish the bona fide DUB activity of the EAV/PLP2-DUB, an in vitro activity assay was developed using polyubiquitin chains as a substrate. To this end, a dose range consisting of 2-fold serial dilutions starting at 0.5 μg of purified recombinant wild-type EAV nsp2(N) per reaction was incubated with Lys48- or Lys63-linked polyubiquitin chains for 2 h at 37°C. The established DUB isopeptidase T (IsoT; 0.5 μg per reaction) was included as a positive control (21). (B) The effect of mutagenesis of the putative active-site residues (Cys270 and His332) of EAV PLP2-DUB was investigated by incubating Lys48- and Lys63-linked polyubiquitin chains with 1 μg of purified recombinant wild-type EAV nsp2(N) or mutant proteins in which one or both of these residues were replaced by Ala.
All arterivirus nsp2 proteins show DUB activity when overexpressed in HEK293T cells.
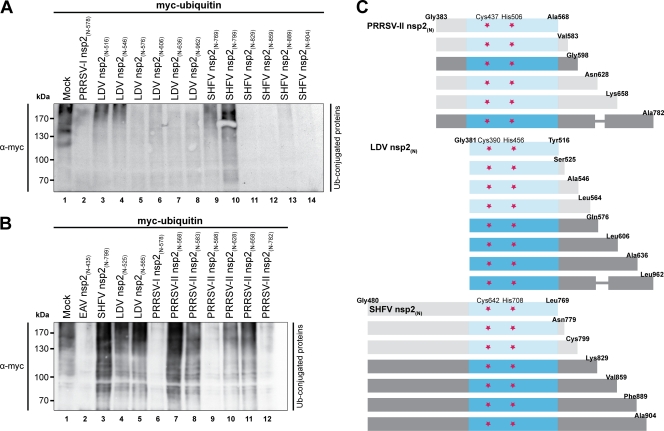
Comparative sequence analysis previously showed that all arterivirus nsp2 proteins contain sequences resembling the EAV PLP2-DUB domain (39, 60). To confirm that these proteins indeed possess DUB activity, mammalian expression constructs were made for the PLP2-DUB domains of all known arteriviruses. It soon became clear, however, that expression of the conserved core PLP2-DUB domain did not suffice for catalytic activity in the case of the PRRSV-II, LDV, and SHFV proteins. Therefore, expression constructs of variable length were designed, starting at the predicted N-terminal residue of nsp2 and gradually extending downstream of the C-terminal border of the PLP2-DUB core domain (Fig. 2C). The different nsp2(N) proteins were subsequently assayed for DUB activity by cotransfecting the expression plasmids encoding them together with a myc-ubiquitin expression vector into HEK293T cells. Figure 2A illustrates that in the case of LDV and SHFV, the nsp2(N) proteins displayed catalytic activity toward ubiquitinated cellular substrates only upon inclusion of a minimal stretch of residues flanking the core PLP2-DUB domain, while Fig. 2B shows that, at least for PRRSV-II, inclusion of the minimal required flanking sequence did not guarantee catalytic activity per se. Since all constructs expressed similar levels of protein, the observed lack of activity represented the expression of an inactive enzyme (data not shown).
Fig 2.
Delineation of the minimal flanking sequences required for LDV, SHFV, and PRRSV-II PLP2-DUB activity. (A and B) Since expression of the core PLP2-DUB domain did not suffice for catalytic activity in the case of the LDV, SHFV, and PRRSV-II proteins, constructs of variable length were designed, starting at the predicted N-terminal residues of the respective nsp2 proteins and gradually extending downstream of the predicted core PLP2-DUB domains. HEK293T cells were subsequently transfected with a combination of plasmids encoding myc-ubiquitin and the indicated viral proteins. Samples were subjected to SDS-PAGE and Western blot analysis and probed with anti-myc antibody to assess the levels of cellular protein ubiquitination. (C) Schematic representation of the constructs tested for PRRSV-II, LDV, and SHFV. Protein fragments were drawn to scale, except where a thinner line was used. Proteins that are represented in a lighter shade proved to be inactive. Indicated are the N- and C-terminal residues, the putative catalytic residues (red stars), and in blue the predicted PLP2-DUB domain. Numbering is based on amino acid positions of the respective replicase polyproteins.
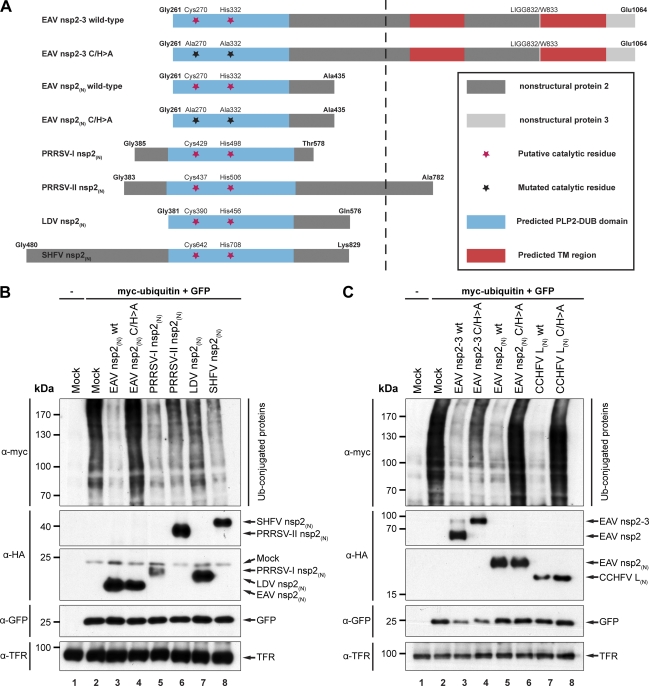
Figure 3A presents an overview of the arterivirus PLP2-DUB constructs used throughout the rest of the paper. For each arterivirus, overexpression of nsp2(N) proteins of sufficient length decreased the level of ubiquitinated proteins in transfected cell cultures (Fig. 3B lanes 3 and 5 to 8). It is clear from Fig. 3B that the PLP2-DUB domains of the various arteriviruses do not show the same level of DUB activity. Since these domains were taken out of their natural full-length-protein context for this experiment, this assay cannot be used to judge whether these differences are due to, for example, the use of suboptimal domain borders in these constructs or, alternatively, whether they might reflect an intrinsic property of the respective PLP2-DUBs. As expected, overexpression of a catalytically inactive double mutant (C/H>A) of EAV nsp2(N) did not result in a decrease in ubiquitinated protein levels (Fig. 3B, compare lanes 3 and 4). Although these experiments do not formally exclude the possibility that the observed effects are due to indirect effects on cellular ubiquitination events, they strongly suggest that all arterivirus nsp2 proteins are genuine DUBs, especially when taking the in vitro activity observed for recombinant EAV nsp2(N) into account (Fig. 1).
Fig 3.
All arterivirus nsp2 proteins possess deubiquitinating activity. (A) Schematic representation of arterivirus nsp2 constructs used. Indicated are the N- and C-terminal residues, putative catalytic residues and mutations thereof, and the EAV nsp2/3 junction (LIGG832/W833). Numbering is based on the amino acid positions of the respective replicase polyproteins. Protein fragments were drawn to scale, except for those parts of the depicted proteins that are to the right of the dashed line. TM, transmembrane. (B) To assess the effect of overexpression of arterivirus PLP2-DUBs on the level of cellular protein ubiquitination, HEK293T cells were transfected with a combination of plasmids encoding myc-ubiquitin, GFP, and the indicated viral proteins. Cells were lysed at 16 h p.t., and samples were subjected to SDS-PAGE and Western blot analysis, using the indicated antibodies. wt, wild type. (C) To further investigate the effect of overexpression of both arteri- and nairovirus DUBs on the level of cellular protein ubiquitination, HEK293T cells were transfected with plasmids encoding wild-type or catalytically inactive mutants (C/H>A) of EAV nsp2-3, EAV nsp2(N), or CCHFV L(N) in combination with plasmids encoding myc-ubiquitin and GFP. Results were analyzed as described for panel B.
In the infected cell, the arterivirus PLP2-DUB domain resides in a membrane-anchored viral replication and transcription complex which is associated with double-membrane structures in the perinuclear area of the cell (48, 67). However, the C-terminally truncated nsp2(N) proteins used as described above did not contain the transmembrane domain of nsp2 and therefore localized to the cytosol (data not shown). To mimic the natural situation more closely, the DUB activity assays described above were repeated using a self-cleaving, full-length EAV nsp2-3 polyprotein. A catalytically inactive derivative (C/H>A) of this protein, which does not process the nsp2/3 site, served as a negative control. Figure 3C shows that overexpression of both the wild-type and C/H>A forms of EAV nsp2-3 decreased the expression of the GFP transfection control. This suggested a general adverse effect on protein expression in this particular assay, which may also have affected the abundance of myc-ubiquitin. Nonetheless, a clear difference between the wild-type and C/H>A nsp2-3 proteins with regard to their effect on protein ubiquitination was observed (Fig. 3C, top; compare lanes 3 and 4), suggesting that at least part of the observed decrease in ubiquitination is due to the protease activity of EAV nsp2.
Finally, the DUB domain present in the L protein of the nairovirus CCHFV and a catalytically inactive mutant of this protein were included in this experiment (Fig. 3C). As was expected on the basis of previous work (15), overexpression of wild-type CCHFV L(N) decreased ubiquitinated protein levels, while overexpression of the C/H>A mutant did not (Fig. 3C, top, lanes 7 and 8).
The induction of IFN-β by EAV RNA in MEFs is MAVS dependent, and the recognition of EAV RNA relies primarily on MDA5.
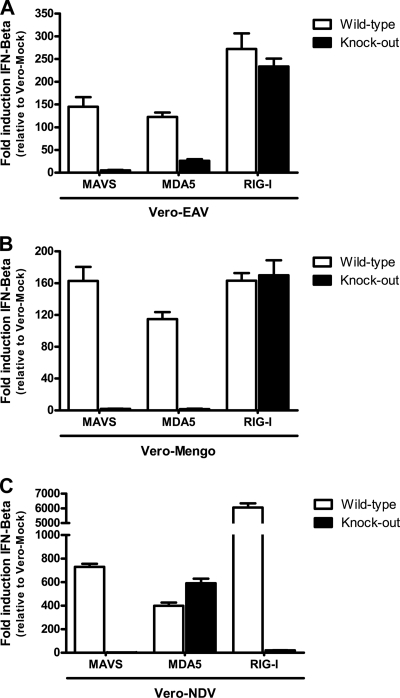
Since arteriviruses target a variety of tissues (61), it is likely that in vivo many of the infected cells have an operational RLR pathway that will trigger an antiviral response. To investigate the involvement of the RLR pathway in sensing EAV infection, we used MEFs isolated from transgenic mice lacking MAVS, MDA5, or RIG-I. Using qRT-PCR, we compared the IFN-β response in these MEFs with that in control cells derived from their respective wild-type littermates. MEFs are permissive to EAV infection, although the number of cells that can be infected is generally low, possibly due to differential expression of suitable receptors (data not shown). Moreover, low induction of IFN-β was observed upon infection, probably also due to innate immune-suppressing activities of the virus. Therefore, the IFN-β response was induced by transfecting these cells with total RNA isolated from EAV-infected Vero cells. As positive controls we employed total RNA isolated from Vero cells infected with mengovirus or NDV. Using the comparative CT method (55), IFN-β expression levels were normalized to those of GAPDH as an internal standard and compared to the IFN-β expression levels in MEFs transfected with total RNA from mock-infected Vero cells.
Our results show that transfecting wild-type MEFs with total RNA isolated from EAV-infected Vero cells induced a 100- to 300-fold increase of IFN-β expression compared to that by cells transfected with RNA from mock-infected cells (Fig. 4A). The absence of MAVS almost completely abolished this IFN-β response, indicating that, at least in MEFs, the RLR pathway is indeed involved in sensing EAV RNA. In addition, while a lack of RIG-I had no significant effect, the absence of MDA5 had a profound negative effect on the IFN-β response (Fig. 4A). However, in MDA5-knockout cells, the IFN-β response was not completely suppressed, unlike what was observed when RNA from mengovirus-infected cells was used (Fig. 4B), suggesting that either RIG-I or an as yet unidentified MAVS-dependent PRR can trigger an IFN-β response in the absence of MDA5. Consistent with the published literature, Fig. 4B and C clearly show that mengovirus RNA was exclusively recognized by MDA5, while NDV RNA was solely recognized by RIG-I (30).
Fig 4.
Induction of IFN-β by EAV RNA in MEFs is MAVS dependent, and the recognition of EAV RNA primarily relies on the PRR MDA5. To investigate the role of RLR sensors in the recognition of arteriviral RNA, MEFs lacking MAVS, MDA5, or RIG-I (black bars) and their respective wild-type controls (white bars) were transfected with total RNA isolated from Vero cells infected with EAV (A), mengovirus (B), or NDV (C). At 3 h p.t., total RNA was isolated from these cells and used as a template for cDNA synthesis. Gene expression levels of IFN-β and GAPDH were subsequently determined by means of qRT-PCR. Using the comparative CT method, the gene expression levels of IFN-β were normalized to those of GAPDH, which was used as an internal standard, and compared to the gene expression levels of IFN-β in MEFs transfected with total RNA from mock-infected Vero cells. This experiment was performed three times independently with similar outcomes. Bars represent the mean of PCR triplicates from one representative experiment ± standard deviation.
Arteri- and nairovirus DUBs inhibit the RIG-I-mediated IFN-β response.
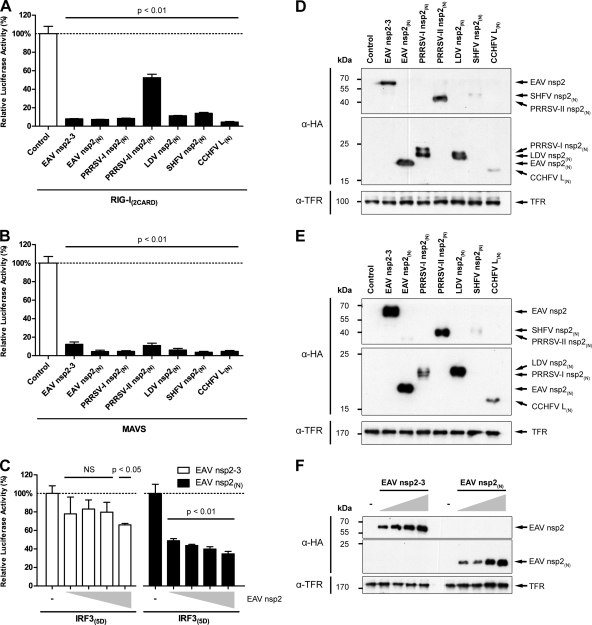
After establishing that the RLR pathway is indeed involved in the recognition of arteriviral RNA, we continued to investigate the immune evasive properties of the arteri- and nairoviral DUBs in more detail. To this end, we designed a luciferase-based reporter assay to investigate inhibition of RLR signaling by the various viral proteins. Plasmids encoding constitutively active RIG-I(2CARD), wild-type MAVS, or constitutively active IRF3(5D) were transfected to induce IFN-β expression. In addition, cells were cotransfected with a reporter plasmid encoding firefly luciferase under the control of the IFN-β promoter, and Renilla luciferase was included as a transfection control. As anticipated, expression of either RIG-I(2CARD), MAVS, or IRF3(5D) strongly induced reporter gene expression, which was set to 100% (Fig. 5A to C and 6A and B, leftmost bar of each graph). Western blots were included for all experiments to confirm correct transfection and expression of viral proteins (Fig. 5D to F and 6C and D).
Fig 5.
Overexpression of arteri- and nairoviral DUBs inhibits the RLR-mediated innate immune response. To investigate the effect of overexpression of the various viral DUBs on RLR-mediated signaling, a luciferase-based reporter assay was designed. To this end, HEK293T cells were transfected with a reporter plasmid encoding firefly luciferase under the control of the IFN-β promoter. A plasmid encoding Renilla luciferase was included as a transfection control, and RLR signaling was induced by the cotransfection of plasmids encoding RIG-I(2CARD) (A), MAVS (B), or IRF3(5D) (C). Dose ranges in panel C consist of 2-fold serial dilutions, where the largest amount of DUB-encoding plasmid DNA corresponds with the amounts used for the experiments whose results are presented in panels A and B. At 12 h p.t., three wells were used to perform a dual luciferase assay, while every fourth well was used for Western blot analysis to verify the correct expression of viral proteins by probing with the indicated antibodies (D to F). Comparable experiments were performed at least twice with similar results. Bars represent the mean of triplicates from one representative experiment ± standard deviation. NS, not statistically significant.
Fig 6.
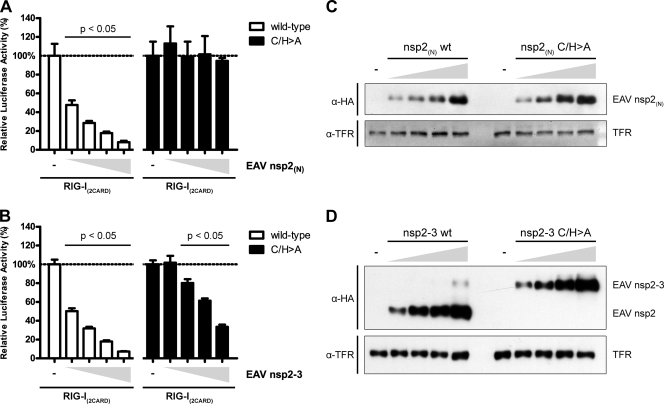
Inhibition of RLR-mediated signaling by EAV nsp2-3 depends only in part on its deubiquitinating activity. To further investigate the DUB dependency of the observed inhibitory activities of EAV nsp2(N) and nsp2-3, a luciferase-based reporter assay was performed using both wild-type and catalytically inactive forms of these proteins. To this end, HEK293T cells were transfected with a reporter plasmid encoding firefly luciferase under the control of the IFN-β promoter. A plasmid encoding Renilla luciferase was included as a transfection control, and RLR signaling was induced by the cotransfection of a plasmid encoding RIG-I(2CARD). In addition, cells were cotransfected in quadruple with 2-fold serial dilutions of plasmids encoding wild-type (white bars) or C/H>A (black bars) EAV nsp2(N) (A) or nsp2-3 (B). At 12 h p.t., three wells were used to perform a dual-luciferase assay, while every fourth well was used for Western blotting to control for correct expression of viral proteins by probing with the indicated antibodies (C and D). Comparable experiments were performed at least twice with similar results. Bars represent the mean of triplicates from one representative experiment ± standard deviation.
Figure 5A clearly shows that expression of each of the arterivirus nsp2(N) constructs, as well as the EAV nsp2-3 and CCHFV L(N) constructs, significantly decreased reporter gene activation upon induction by RIG-I(2CARD). This indicated that each of these proteins is able to inhibit the RLR-mediated immune response at the level of RIG-I or downstream of RIG-I. Consistent with its lower general DUB activity upon overexpression in HEK293T cells, PRRSV-II nsp2(N) showed a significant but relatively low inhibitory activity, which may be due to the use of suboptimal domain borders in this construct or may reflect an intrinsic property of this specific arterivirus PLP2-DUB. Figure 5B illustrates a comparable inhibition of MAVS-induced reporter gene activation by all DUB-containing proteins, which is suggestive of inhibition at the level of MAVS or downstream of MAVS.
To confirm that the observed effects on reporter gene expression indeed resulted from the specific inhibition of RLR signaling, a similar reporter assay in which constitutively active IRF3(5D) was used to activate the reporter gene was set up. Since the expression of constitutively active IRF3(5D) results in direct and presumably ubiquitin-independent reporter gene activation, one would not expect this induction to be affected by the expression of viral DUBs. Surprisingly, however, EAV nsp2(N) considerably inhibited reporter gene activation by IRF3(5D) (Fig. 5C), although the inhibition was less pronounced than that observed upon activation by RIG-I(2CARD) (Fig. 5A) or MAVS (Fig. 5B). Interestingly, EAV nsp2-3 only marginally affected reporter gene expression upon IRF3(5D)-mediated activation (Fig. 5C), while its effect on RIG-I or MAVS-induced IFN-β expression was comparable to that of EAV nsp2(N) (Fig. 5A and B). This supports the assumption that the subcellular localization of the PLP2-DUB is an important determinant of the natural range of substrates that is accessible to the enzyme in the infected cell.
To further investigate the DUB dependency of the observed inhibitory effects, the dose-dependent activities of wild-type and catalytically inactive (C/H>A) mutants of EAV nsp2(N) and nsp2-3 were compared (Fig. 6A and B). As expected, wild-type EAV nsp2(N) showed a clear dose-dependent increase in inhibition of reporter gene activity upon induction by RIG-I(2CARD), while EAV nsp2(N) C/H>A did not significantly inhibit with any of the tested amounts of transfected DNA. In contrast, although clearly decreased compared to the wild-type control, EAV nsp2-3 C/H>A still significantly inhibited RIG-I-mediated reporter gene activity, suggesting that this protein harbors one or more additional immune inhibitory functions that are not strictly dependent on its DUB activity.
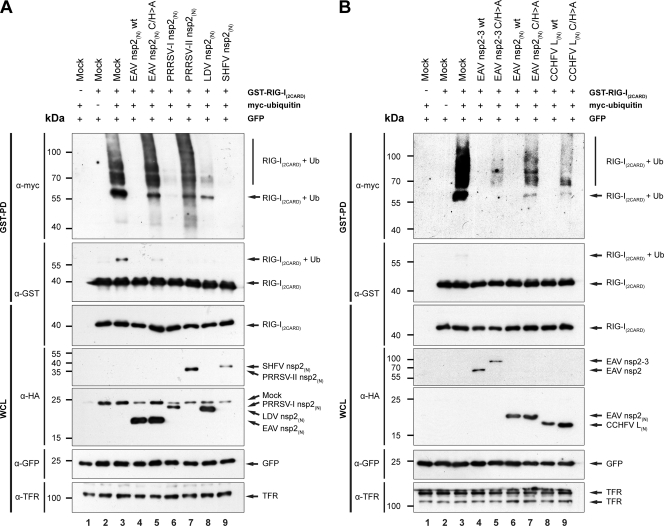
Arteri- and nairovirus DUBs can inhibit the ubiquitination of RIG-I.
RIG-I is a key PRR in the RLR pathway, and, unlike MDA5, its activation depends on ubiquitination (17). In addition, our data suggested a role for RIG-I in the recognition of arterivirus RNA and showed that arteri- and nairoviral DUBs can inhibit the RLR pathway at the level of RIG-I or downstream of RIG-I (Fig. 5A). This led us to examine whether these viral DUBs are capable of inhibiting RIG-I ubiquitination. We therefore cotransfected HEK293T cells with plasmids encoding GST–RIG-I(2CARD), myc-ubiquitin, GFP, and the indicated viral proteins and performed a GST pulldown experiment, followed by a Western blot analysis. In the absence of viral proteins, several bands representing (poly)ubiquitinated RIG-I(2CARD) could be observed upon overexpression of this protein in combination with myc-ubiquitin (Fig. 7A and B, top, lane 3). Notably, unlike others, our experiments were performed in the absence of proteasome inhibitors, which would merely have resulted in the accumulation of Lys48-linked substrates rather than the activating Lys63-linked form that we were interested in (68). Gack et al. (2007) previously used a similar approach to visualize ubiquitinated RIG-I(2CARD) and showed comparable band patterns, which they subsequently confirmed to be the Lys63-linked polyubiquitinated form using mass spectrometry analysis (17).
Fig 7.
Arteri- and nairovirus DUBs inhibit the ubiquitination of RIG-I upon overexpression. (A and B) The effect of overexpression of the arteri- and nairovirus DUBs on RIG-I ubiquitination was assessed by cotransfecting HEK293T cells with a combination of plasmids encoding GST–RIG-I(2CARD), myc-ubiquitin, GFP, and the indicated viral proteins. Following lysis at 16 h p.t., a GST pulldown experiment was performed and ubiquitination of RIG-I was subsequently analyzed by means of SDS-PAGE and Western blot analysis, using the indicated antibodies. PD, pull-down; WCL, whole-cell lysate.
Figure 7A shows that coexpression of wild-type EAV, PRRSV-I, LDV, or SHFV nsp2(N) resulted in the nearly complete disappearance of ubiquitinated RIG-I(2CARD) (Fig. 7A). PRRSV-II nsp2(N) exhibited only a modest effect (Fig. 7A, lane 7), in line with the other results obtained with this protein (Fig. 3B and 5A). The inhibitory effect of EAV nsp2(N) was clearly reduced upon mutagenesis of the catalytic residues (Fig. 7A, lane 5). Additionally, Fig. 7B shows that wild-type EAV nsp2-3 also completely inhibited RIG-I(2CARD) ubiquitination (Fig. 7B, top, lane 4). Expression of EAV nsp2-3 C/H>A resulted in an attenuated but nevertheless clear reduction of ubiquitinated RIG-I(2CARD) levels (Fig. 7B, top; compare lanes 3 to 5). This observation might, however, be explained by the fact that the overall amount of RIG-I(2CARD) appeared to be decreased in the presence of either wild-type or catalytically inactive EAV nsp2-3 (Fig. 7B, 2nd and 3rd panels from the top, lanes 4 and 5). Finally, the expression of wild-type CCHFV L(N) also resulted in the complete disappearance of ubiquitinated RIG-I, an effect that was clearly suppressed upon mutagenesis of the protease's catalytic residues (Fig. 7B, top; compare lanes 3, 8, and 9).
DISCUSSION
In this paper, using both in vitro and cell-based assays, we document for the first time that the nsp2 replicase subunits of all arteriviruses harbor a DUB activity that is likely used to counter RLR-mediated innate immune signaling. In addition, we established that the DUB domains of both arteri- and nairoviruses are capable of inhibiting RIG-I ubiquitination, suggesting that the deconjugation of ubiquitin from this PRR constitutes one of the mechanisms by which DUB-containing proteins from these viruses interfere with the innate immune response. The recent finding that a DUB encoded by Kaposi's sarcoma-associated herpesvirus, a DNA virus, also reduced IFN-β expression by inhibiting RIG-I ubiquitination highlights the potential benefits of this strategy for viruses at large (25). Since the viral DUBs studied here were also capable of inhibiting MAVS-mediated IFN-β induction, it stands to reason that the deubiquitination of MAVS or other signaling molecules downstream of this factor also plays a role in the immune evasion mediated by these viral proteins.
To date, our knowledge concerning the role of innate PRRs in the recognition of arterivirus infection is limited. There is some evidence to suggest that TLR3 is important (53), but our study yielded the first indication for the involvement of the RLRs in the recognition of arteriviral RNA. By transfecting MEFs with total RNA isolated from EAV-infected Vero cells, we have shown that the absence of MAVS, a central factor in RLR signaling, almost completely abrogated IFN-β expression, indicating that in these cells MAVS-mediated signaling is pivotal in the response to EAV infection (Fig. 4A). In addition, the absence of MDA5 strongly attenuated IFN-β induction compared to that in wild-type MEFs (Fig. 4A), suggesting that this sensor is the main RLR responsible for the recognition of arteriviral RNA. Since arteriviruses infect numerous different tissues and cell types during infection in vivo (61), it is likely that many of these indeed rely on the RLR-mediated antiviral response, supporting the relevance of our data acquired in MEFs. Considering our findings, it seems counterintuitive that an arteriviral protein would target RIG-I in order to evade host immune signaling. However, in contrast to what was observed for mengovirus, the absence of MDA5 reproducibly failed to cause a complete loss of EAV-induced IFN-β expression (compare Fig. 4A and B). This suggests that another PRR, likely RIG-I, can mount an immune response against EAV infection in the absence of MDA5. In fact, West Nile virus (14), dengue virus (40), and the coronavirus murine hepatitis virus (34) were previously found to be recognized by both MDA5 and RIG-I. Alternatively, not RIG-I but another PRR might be responsible for the observed residual level of IFN-β expression in the absence of MDA5. For example, recent work by Sabbah et al. (2009) showed that the NLR Nod2 detected viral RNA and that subsequent signaling by this molecule was also MAVS dependent (50). Nevertheless, the fact that both MDA5 and RIG-I are ISGs suggests a role for these proteins during later stages of infection which might be distinct from their initial activities as PRRs and would explain why a virus that is not primarily detected by RIG-I would still benefit from inhibiting its activation. This hypothesis is further supported by the finding that several picornaviruses induce RIG-I degradation, despite the fact that they are commonly believed to be recognized solely by MDA5 (2, 44).
As in the case of arteriviruses, little is known about the innate immunity signaling pathways involved in the recognition of nairoviruses. Nevertheless, Habjan et al. (2008) showed that CCHFV and the related bunyavirus Hantaan virus process the 5′ end of their genomes, likely to prevent recognition by RIG-I (20). In addition, there is recent evidence that infection with Hantaan virus can indeed be sensed by RIG-I (32). Since viruses belonging to the same family are often recognized by the same PRRs, these findings suggest that CCHFV is also recognized by RIG-I. In nairoviruses, the deubiquitination of RIG-I might therefore be an additional mechanism to further inhibit RLR-mediated signaling.
Notably, our results showed that inhibition of the innate immune response by EAV nsp2-3, in contrast to EAV nsp2(N), is not strictly dependent on its DUB activity. Although the inhibitory activity of EAV nsp2-3 is clearly reduced upon mutagenesis of the protease's active-site residues, it is not completely abolished (Fig. 6B). In contrast, mutagenesis of the same catalytic residues in EAV nsp2(N) almost completely abrogated its inhibitory activity (Fig. 6A).
These observations are consistent with results previously obtained by others for coronavirus DUBs (8, 9). Both arteri- and coronaviruses belong to the order Nidovirales and share a similar genome organization and expression strategy. Coronavirus nsp3, which can to a certain extent be considered the functional equivalent of arterivirus nsp2, contains two papain-like protease domains (PLP1 and PLP2). In some coronavirus species, however, PLP1 has lost its proteolytic activity or the entire PLP1 domain is missing. In these cases, PLP2 is the only active protease domain in nsp3 and is therefore referred to as PLpro. PLP2/PLpro of severe acute respiratory syndrome-associated coronavirus (SARS-CoV), murine hepatitis virus A59 (MHV-A59), human coronavirus NL63 (HCoV-NL63), and PLP1 of transmissible gastroenteritis virus (TGEV) were all shown to possess DUB activity, in addition to their role in the autoproteolytic maturation of the replicase polyproteins (3, 8, 37, 71, 74). Notably, the coronavirus DUBs are of the USP subclass instead of the OTU subclass of DUBs. Like the arterivirus PLP2-DUB, the PLP2/PLpro domains of various coronaviruses were found to inhibit the innate immune response (8, 9, 16, 74). While the immune inhibitory activity of MHV-A59 PLP2 was claimed to be completely dependent on its DUB activity, that of HCoV-NL63 and SARS-CoV PLP2/PLpro was shown to only partially depend on its proteolytic activity. These observations suggest that both arteri- and coronavirus replicase proteins harbor additional immune evasive features that may be linked to but are not strictly dependent on the DUB activity present in their nsp2 or nsp3 subunit, respectively. It is conceivable that a catalytically inactive DUB would still be able to bind ubiquitin or ubiquitinated proteins, thereby hindering interactions necessary for signal transduction. In addition, our results suggest that expression of both wild-type and catalytically inactive EAV nsp2-3 decreases the overall amount of RIG-I(2CARD), which may explain the mutant's inhibitory potential (Fig. 7B, 3rd panel from the top, lanes 4 and 5). Further research is needed to elucidate whether these DUB-containing viral proteins possess additional immune evasive properties.
Contrary to our expectations, we found that arterivirus nsp2(N), which localizes to the cytosol, inhibited the IRF3-mediated activation of IFN-β expression (Fig. 5C). In contrast, IRF3-mediated IFN-β induction was hardly affected by the expression of EAV nsp2-3, which is membrane anchored and localizes to the perinuclear region of the cell, while EAV nsp2(N) and nsp2-3 showed comparable inhibitory potential upon activation of signaling by RIG-I(2CARD) or MAVS (compare Fig. 5A to C). Similar results were previously obtained for SARS-CoV PLpro, of which a soluble form inhibited IRF3-mediated activation of IFN-β expression (16), while a membrane-anchored form did not (9). Taken together, these results suggest that the subcellular localization of these proteins is an important determinant of the range of substrates that is accessible to them. Since expression of the full-length nsp2 protein together with nsp3 more accurately reflects the situation in the infected cell, it is likely that during infection inhibition also takes place upstream of IRF3. This would be consistent with previous observations by Luo et al. (2008), who showed that inhibition of the RLR pathway during PRRSV infection occurred at the level of MAVS or upstream of this key factor (38).
Still, the question of the mechanism, albeit artificial, by which cytosolic EAV nsp2(N) is able to inhibit IRF3 signaling remains, since to date there is no evidence for activation of this transcription factor by Lys63-linked polyubiquitination. On the contrary, it is well established that IRF3 is negatively regulated by Lys48-linked polyubiquitination, the removal of which would appear to be counterproductive in view of controlling IFN signaling (24, 52, 72, 73). Interestingly, Shi et al. (2010) recently showed that ISG-ylation, i.e., the conjugation of ISG15 to a target protein, of IRF3 positively regulated its activation by preventing Lys48-linked polyubiquitination (58). In light of previous findings by Frias-Staheli et al. (2007) that arteri- and nairovirus DUBs can deconjugate ISG15 (15), it is tempting to speculate that cytosolic nsp2(N), in contrast to membrane-anchored full-length nsp2, is able to de-ISGylate IRF3. However, IRF3 ISGylation per se was shown not to be activating but merely increased the signaling potential of IRF3 by preventing its proteasomal degradation. Consequently, if a viral DUB would indeed de-ISGylate IRF3 to promote its degradation, it would at the same time need to refrain from removing any Lys48-linked polyubiquitin chains. This problem emphasizes that viral DUBs are likely able to distinguish between activating and inactivating Ub(-like) modifications. How viral DUBs make this distinction and how they would achieve a balance between the removal of the right and wrong modifiers pose interesting questions for future research.
Another interesting feature of viral DUBs is their apparent tendency to deubiquitinate all cellular proteins in a seemingly random fashion (Fig. 3B and C). In this respect, they differ from their mammalian counterparts, which generally have a more narrow specificity (15). A possible explanation for this observed promiscuity, which is seen for arteri-, nairo-, and coronavirus DUBs (15, 16), is the fact that the enzymes were often studied only as isolated domains taken out of their natural full-length-protein context. For this reason, we have also studied PLP2-DUB in the context of EAV nsp2-3, of which the membrane-associated subcellular localization is similar to that observed during EAV infection. Although EAV nsp2-3 did seem to be more restricted than nsp2(N) in the inhibition of IRF3-mediated IFN-β induction (Fig. 5C), PLP2-DUB also showed general DUB activity when overexpressed as part of full-length nsp2 (Fig. 3C). This suggests that the observed promiscuity is indeed an intrinsic property of these viral DUBs, although on the other hand, it should be noted that most of our experiments involved systems in which the DUBs were overexpressed. Future studies will aim to elucidate the role of viral DUB activity during the course of infection, for example, by using reverse genetics to engineer a virus that lacks this activity. Unfortunately, we have thus far been unable to create a viable mutant with this phenotype, mainly due to the intimate link between the DUB activity and polyprotein processing functions of PLP2.
Taken together, our results strongly suggest that arteriviruses as well as nairoviruses encode DUBs that are used to inactivate cellular proteins involved in innate immune signaling, as exemplified by the deubiquitination of RIG-I documented here. Strikingly, related DUBs from the OTU family seem to have been acquired and adapted for this purpose by apparently unrelated RNA viruses, the positive-stranded arteriviruses and the negative-stranded nairoviruses, thereby highlighting the selective advantages that must be linked to OTU DUB acquisition and the general plasticity of RNA virus genomes.
ACKNOWLEDGMENTS
We kindly thank the following people for providing us with reagents: Shizuo Akira (Osaka University), Zhijian Chen (University of Texas Southwestern Medical Center), Ying Fang (South Dakota State University), Michaela Gack (Harvard Medical School), John Hiscott (McGill University), Ron Kopito (Stanford University), Frank van Kuppeveld (Radboud University Nijmegen), Paul Moynagh (NUI Maynooth), and Herbert W. Virgin IV (Washington University School of Medicine). We thank Bart Tummers (Leiden University Medical Center) for his help with setting up the qRT-PCR assay. We thank Frank van Kuppeveld, Stanleyson Hato (Radboud University Nijmegen), Nadia Giannakopoulos, Herbert W. Virgin IV (Washington University School of Medicine), and Sasha Gorbalenya (Leiden University Medical Center) for helpful discussions and support.
This research was supported in part by the Division of Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW) through ECHO grant 700.59.008 to M.K. and E.J.S. and has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under SILVER grant agreement no. 260644. It was, furthermore, supported by NIH grants U54AI057158 and U19AI083025 to A.G.-S.
Footnotes
Published ahead of print 9 November 2011
REFERENCES
- 1. Akutsu M, Ye Y, Virdee S, Chin JW, Komander D. 2011. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc. Natl. Acad. Sci. U. S. A. 108:2228–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barral PM, Sarkar D, Fisher PB, Racaniello VR. 2009. RIG-I is cleaved during picornavirus infection. Virology 391:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barretto N, et al. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189–15198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bergeron E, Albarino CG, Khristova ML, Nichol ST. 2010. Crimean-Congo hemorrhagic fever virus-encoded ovarian tumor protease activity is dispensable for virus RNA polymerase function. J. Virol. 84:216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhoj VG, Chen ZJ. 2009. Ubiquitylation in innate and adaptive immunity. Nature 458:430–437 [DOI] [PubMed] [Google Scholar]
- 6. Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signaling. Nat. Rev. Immunol. 8:911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Capodagli GC, et al. 2011. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J. Virol. 85:3621–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clementz MA, et al. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84:4619–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Devaraj SG, et al. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 282:32208–32221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Vries AA, Chirnside ED, Horzinek MC, Rottier PJ. 1992. Structural proteins of equine arteritis virus. J. Virol. 66:6294–6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Enesa K, et al. 2008. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J. Biol. Chem. 283:7036–7045 [DOI] [PubMed] [Google Scholar]
- 12. Faaberg KS. 2008. Arterivirus structural proteins and assembly. In Perlman S, Gallagher T, Snijder EJ. (ed), Nidoviruses. ASM Press, Washington, DC [Google Scholar]
- 13. Fitzgerald KA, et al. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496 [DOI] [PubMed] [Google Scholar]
- 14. Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr 2008. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 82:609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frias-Staheli N, et al. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J. Virol. 83:6689–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gack MU, et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920 [DOI] [PubMed] [Google Scholar]
- 18. Gale P, et al. 2010. The feasibility of developing a risk assessment for the impact of climate change on the emergence of Crimean-Congo haemorrhagic fever in livestock in Europe: a review. J. Appl. Microbiol. 108:1859–1870 [DOI] [PubMed] [Google Scholar]
- 19. Graham FL, van der Eb AJ. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467 [DOI] [PubMed] [Google Scholar]
- 20. Habjan M, et al. 2008. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One 3:e2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hadari T, Warms JV, Rose IA, Hershko A. 1992. A ubiquitin C-terminal isopeptidase that acts on polyubiquitin chains. Role in protein degradation. J. Biol. Chem. 267:719–727 [PubMed] [Google Scholar]
- 22. Han J, Rutherford MS, Faaberg KS. 2009. The porcine reproductive and respiratory syndrome virus nsp2 cysteine protease domain possesses both trans- and cis-cleavage activities. J. Virol. 83:9449–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hato SV, et al. 2007. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell. Microbiol. 9:2921–2930 [DOI] [PubMed] [Google Scholar]
- 24. Higgs R, et al. 2008. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181:1780–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inn K-S, et al. 2011. Inhibition of RIG-I-mediated signaling by Kaposi's sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J. Virol. 85:10899–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. James TW, et al. 2011. Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc. Natl. Acad. Sci. U. S. A. 108:2222–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanneganti TD. 2010. Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol. 10:688–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karin M, Ben-Neriah Y. 2000. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18:621–663 [DOI] [PubMed] [Google Scholar]
- 29. Kato H, et al. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19–28 [DOI] [PubMed] [Google Scholar]
- 30. Kato H, et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 31. Kayagaki N, et al. 2007. DUBA: a deubiquitinase that regulates type I interferon production. Science 318:1628–1632 [DOI] [PubMed] [Google Scholar]
- 32. Lee MH, et al. 2011. RNA helicase retinoic acid-inducible gene I as a sensor of Hantaan virus (HTNV) replication. J. Gen. Virol. 92(Pt 9):2191–2200 [DOI] [PubMed] [Google Scholar]
- 33. Li H, et al. 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus non-structural protein 2 antagonizes interferon regulatory factor 3 activation. J. Gen. Virol. 91:2947–2958 [DOI] [PubMed] [Google Scholar]
- 34. Li J, Liu Y, Zhang X. 2010. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J. Virol. 84:6472–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li S, et al. 2010. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J. Biol. Chem. 285:4291–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin R, Heylbroeck C, Pitha PM, Hiscott J. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lindner HA, et al. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199–15208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luo R, et al. 2008. Porcine reproductive and respiratory syndrome virus (PRRSV) suppresses interferon-beta production by interfering with the RIG-I signaling pathway. Mol. Immunol. 45:2839–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Makarova KS, Aravind L, Koonin EV. 2000. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 25:50–52 [DOI] [PubMed] [Google Scholar]
- 40. Nasirudeen AM, et al. 2011. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl. Trop. Dis. 5:e926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Neumann EJ, et al. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 227:385–392 [DOI] [PubMed] [Google Scholar]
- 42. Nijman SM, et al. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773–786 [DOI] [PubMed] [Google Scholar]
- 43. O'Neill LA, Bowie AG. 2010. Sensing and signaling in antiviral innate immunity. Curr. Biol. 20:R328–R333 [DOI] [PubMed] [Google Scholar]
- 44. Papon L, et al. 2009. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology 393:311–318 [DOI] [PubMed] [Google Scholar]
- 45. Parvatiyar K, Barber GN, Harhaj EW. 2010. TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1-IKKi kinases. J. Biol. Chem. 285:14999–15009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pasternak AO, Spaan WJ, Snijder EJ. 2006. Nidovirus transcription: how to make sense? J. Gen. Virol. 87:1403–1421 [DOI] [PubMed] [Google Scholar]
- 47. Paz S, et al. 2009. Ubiquitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter. Mol. Cell. Biol. 29:3401–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pedersen KW, van der Meer Y, Roos N, Snijder EJ. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodriguez-Madoz JR, et al. 2010. Inhibition of the type I interferon response in human dendritic cells by dengue virus infection requires a catalytically active NS2B3 complex. J. Virol. 84:9760–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sabbah A, et al. 2009. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 10:1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saitoh T, et al. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7:598–605 [DOI] [PubMed] [Google Scholar]
- 53. Sang Y, Ross CR, Rowland RR, Blecha F. 2008. Toll-like receptor 3 activation decreases porcine arterivirus infection. Viral Immunol. 21:303–313 [DOI] [PubMed] [Google Scholar]
- 54. Sawicki SG, Sawicki DL, Siddell SG. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 56. Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 57. Shembade N, Ma A, Harhaj EW. 2010. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 327:1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shi HX, et al. 2010. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 30:2424–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Skaug B, Chen ZJ. 2010. Emerging role of ISG15 in antiviral immunity. Cell 143:187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Snijder EJ, Wassenaar AL, Spaan WJ, Gorbalenya AE. 1995. The arterivirus Nsp2 protease. An unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J. Biol. Chem. 270:16671–16676 [DOI] [PubMed] [Google Scholar]
- 61. Snijder EJ, Meulenberg JJ. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79(Pt 5):961–979 [DOI] [PubMed] [Google Scholar]
- 62. Sun Q, et al. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24:633–642 [DOI] [PubMed] [Google Scholar]
- 63. Sun SC. 2008. Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 8:501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun Z, Chen Z, Lawson SR, Fang Y. 2010. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J. Virol. 84:7832–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tseng PH, et al. 2010. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat. Immunol. 11:70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van Aken D, Zevenhoven-Dobbe J, Gorbalenya AE, Snijder EJ. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 87:3473–3482 [DOI] [PubMed] [Google Scholar]
- 67. van der Meer Y, van Tol H, Locker JK, Snijder EJ. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689–6698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang D, et al. 2011. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J. Virol. 85:3758–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ward CL, Omura S, Kopito RR. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83:121–127 [DOI] [PubMed] [Google Scholar]
- 70. Whitehouse CA. 2004. Crimean-Congo hemorrhagic fever. Antiviral Res. 64:145–160 [DOI] [PubMed] [Google Scholar]
- 71. Wojdyla JA, et al. 2010. Papain-like protease 1 from transmissible gastroenteritis virus: crystal structure and enzymatic activity toward viral and cellular substrates. J. Virol. 84:10063–10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yu Y, Hayward GS. 2010. The ubiquitin E3 ligase RAUL negatively regulates type I interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 33:863–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang M, et al. 2008. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 18:1096–1104 [DOI] [PubMed] [Google Scholar]
- 74. Zheng D, Chen G, Guo B, Cheng G, Tang H. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 18:1105–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhou L, Yang H. 2010. Porcine reproductive and respiratory syndrome in China. Virus Res. 154:31–37 [DOI] [PubMed] [Google Scholar]
- 76. Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853–879 [DOI] [PubMed] [Google Scholar]