Abstract
Noroviruses account for 96% of viral gastroenteritis cases worldwide, with GII.4 strains responsible >80% of norovirus outbreaks. Histo-blood group antigens (HBGAs) are norovirus binding ligands, and antigenic and preferential HBGA binding profiles vary over time as new GII.4 strains emerge. The capsid P2 subdomain facilitates HBGA binding, contains neutralizing antibody epitopes, and likely evolves in response to herd immunity. To identify amino acids regulating HBGA binding and antigenic differences over time, we created chimeric virus-like particles (VLPs) between the GII.4-1987 and GII.4-2006 strains by exchanging amino acids in putative epitopes and characterized their antigenic and HBGA binding profiles using anti-GII.4-1987 and -2006 mouse monoclonal antibodies (MAbs) and polyclonal sera, 1988 outbreak human sera, and synthetic HBGAs. The exchange of amino acids 393 to 395 between GII.4-1987 and GII.4-2006 resulted in altered synthetic HBGA binding compared to parental strains. Introduction of GII.4-1987 residues 294, 297 to 298, 368, and 372 (epitope A) into GII.4-2006 resulted in reactivity with three anti-GII.4-1987 MAbs and reduced reactivity with four anti-GII.4-2006 MAbs. The three anti-GII.4-1987 MAbs also blocked chimeric VLP-HBGA interaction, while an anti-GII.4-2006 blocking antibody did not, indicating that epitope A amino acids comprise a potential neutralizing epitope for GII.4-1987 and GII.4-2006. We also tested GII.4-1987-immunized mouse polyclonal sera and 1988 outbreak human sera for the ability to block chimeric VLP-HBGA interaction and found that epitope A amino acids contribute significantly to the GII.4-1987 blockade response. Our data provide insights that help explain the emergence of new GII.4 epidemic strains over time, may aid development of norovirus therapeutics, and may help predict the emergence of future epidemic strains.
INTRODUCTION
Noroviruses (NoVs) are members of the Caliciviridae family and represent the most significant cause of human acute viral gastroenteritis worldwide (3). Approximately 23 million norovirus infections occur each year in the United States alone (33), burdening retirement homes, day cares, the military, cruise ships, hospitals, educational institutions, and other community settings where close contact between humans is unavoidable. The elderly, very young, and immunocompromised are at the highest risk for severe complications and death (32, 34, 36), and economic costs of norovirus outbreaks are significant (23, 26). Although an estimated 200,000 deaths occur each year from NoV-induced gastroenteritis (37), there are no approved vaccines or antiviral therapies for the prevention or treatment of norovirus infections. However, current clinical trials are encouraging and support the use of virus-like particles (VLPs) of norovirus as a vaccine platform to ameliorate the human disease burden (17).
Noroviruses carry a 7.5-kb single-stranded, positive-sense RNA genome packaged within a 38-nm nonenveloped icosahedral capsid. The genome carries three open reading frames (ORFs). ORF1 encodes the replicase polyprotein, while ORF2 and ORF3 encode the major (VP1) and minor (VP2) structural proteins, respectively (13). Expression of VP1 from baculoviruses (22) or Venezuelan equine encephalitis (VEE) (4) virus replicon particles (VRPs) results in the production of NoV virus-like particles (VLPs). The capsid protein is divided into two distinct domains in the virion, the shell (S) and the surface protruding domain (P). The P domain is further subdivided into the P1 and P2 subdomains, with the P2 subdomain flanked by portions of P1 in the primary coding sequence (38). The shell forms the base of the capsid, while the P1 region forms a stalk protruding from the shell. The P2 subdomain is positioned atop the P1 stalk, where it is the most surface-exposed region, able to interact with both carbohydrates (CHOs) and antibodies (9, 31). Histo-blood group antigens (HBGAs) are a diverse family of CHOs expressed on mucosal surfaces. These CHOs are differentially expressed in humans and have been hypothesized to be receptors or coreceptors that allow NoVs to attach to and enter permissive cells. Conserved amino acids 343 to 345, 374, and 441 to 443 are important for HBGA binding (9), although it is unclear how nearby amino acid variation affects capsid surface topology and contributes to HBGA binding affinity differences noted in time-ordered GII.4 VLPs. Partly because there is no cell culture or small-animal model for human NoVs, the development and testing of vaccines and drug treatments for NoVs have only recently been evaluated in larger animal models of human disease (swine/primate models) (6, 44) and in humans (17). Importantly, VLPs, created by the expression of VP1 in a Venezuelan equine encephalitis (VEE) virus (4) or in baculovirus (22) expression vector, are both physically and antigenically similar to norovirus virions. These systems offer a promising strategy to study norovirus structure and function, which may lead to the development of effective vaccines and treatments (30).
Noroviruses are a genetically diverse group of viruses, divided into five genogroups (I to V) based on ORF2 sequence homology, in which protein sequence can differ by up to 60%. Each genogroup is further divided into several genoclusters, differing by 20% to 30% in VP1 sequence (46). Genogroups I and II predominantly encode strains that cause significant human disease (47). Among these, genocluster GII.4 noroviruses account for >80% of norovirus outbreaks (19). The emergence of new GII.4 NoV epidemic strains has been linked to changes in the HBGA binding profile using synthetic ABH and Lewis (Le) CHOs (29). The 1974 strain, currently the prototypic ancestral strain, binds O-type CHOs (H3 and Le-y) (5), the 1987 Camberwell strain binds O-type CHOs (H3 and Le-y), while the 1997 Grimsby strain, a pandemic strain, binds A trimer, B trimer, and O-type (H3 and Le-y) CHOs. The Farmington Hills 2002 strain binds B trimer and O-type (H3 and Le-y) CHOs (29). The most recent pandemic strain, 2006 Minerva, binds A trimer, B trimer, and O-type (H3, Le-b, and Le-y) CHOs (8). It is also clear that microvariation within the P2 domain between closely related strains influences CHO binding patterns (11, 14, 29, 41).
Among the GII.4 strains, the P2 region of ORF2 is evolving more rapidly than other domains and subdomains of the GII.4 norovirus capsid protein, with certain amino acids undergoing positive selection, likely in response to herd immunity (15, 29). Antigenic changes are also evident over time, culminating in the emergence of new GII.4 strains (15, 28, 42). On the capsid surface, highly variable amino acids surround the binding pocket, altering HBGA and antibody binding affinity. Multiple studies have identified specific residues within the P2 subdomain that are important for antibody and HBGA interactions (1, 2, 29, 45). Lindesmith et al. (29) showed that residues 393 to 395 are important for HBGA interactions and antigenicity in GII.4 noroviruses (29). A D393G change occurred between GII.4-1987 and GII.4-1997. Importantly, introduction of the mutation D393G into the 1987 strain converted GII.4-1987 into a B binding VLP (29). In addition, two groups identified amino acids 296 to 298 as potentially important for antigenicity and HBGA binding interactions (1, 15), and Allen et al. (1) demonstrated that these amino acids are involved in strain-specific antibody recognition (2). However, it was not clear whether antibodies targeting these sites blocked VLP-CHO binding interactions, as a surrogate neutralization assay was not performed (2). Both of these amino acid clusters (amino acids 296 to 298 and 393 to 395) comprise exposed loop regions of the P2 subdomain. In addition, Tan et al. identified amino acids 346 and 441 as important to A, B, and H binding and showed that several other amino acids in and around the binding pocket modulate HBGA specificity (45). A more recent study based on comparing crystal structures from the GII.4-1996 and GII.4-2004 strains suggests that temporal HBGA affinity differences exist among strains and that residues 393 to 395 directly impact Lewis HBGA but not ABH HBGA binding (41). However, the effects of amino acid differences at these positions may remodel the conformational and electrostatic landscape around the binding pocket, indirectly impacting ABH binding affinity. Together, the previous data demonstrate that clusters of mutations likely work independently or in concert to alter HBGA binding profiles and antibody binding. However, the specific interactions that take place to modulate HBGA binding and antigenicity are still not well understood.
In this work, we designed and produced a panel of recombinant and chimeric GII.4 NoV VLPs that exchanged surface-exposed predicted epitopes between an ancestral GII.4 strain (GII.4-1987, Camberwell) and the most recent pandemic GII.4 strain (GII.4-2006, Minerva). Our HBGA binding results provide further evidence for the hypothesis that amino acids 393 to 395 are important for HBGA A and B binding. Using a panel of mouse monoclonal antibodies (MAbs) derived against GII.4-1987 or GII.4-2006 (28) and human and mouse polyclonal sera, we employed gain- and loss-of-function assays to map varying antibody blockade epitopes, which are potentially critical sites of virus neutralization and protection from clinical disease (21, 29, 39). These data implicate amino acids 296 to 298 as well as other nearby amino acids that potentially contribute to antigenic differences among time-ordered GII.4 strains that are likely associated with escape from herd immunity. These data provide further support for the hypothesis that the GII.4 viruses are evolving by epochal evolution and antigenic variation in VP1 (15, 16, 29).
MATERIALS AND METHODS
Prediction of putative epitopes.
Multiple sequence alignments were generated for all full-length GII.4 norovirus capsid peptide sequences in GenBank using ClustalX, version 1.86 (12). A phylogenetic tree was generated by Bayesian inference using MrBayes (40), and 36 representative sequences were selected and analyzed based upon unique phylogenetic clustering characteristics. Representative strains of GII.4 noroviruses were selected from 1974, 1987, 1997, 2002, 2004, 2005, 2006, 2007, 2008, and 2009. These representative sequences were aligned using ClustalX, version 1.86, P2 subdomain variation was exported into Excel, and amino acid variation over time was analyzed. Homology models were generated using Modeller (Max Planck Institute bioinformatics toolkit), building structural models based upon the crystal structure of V387 (Protein Data Bank [PDB] accession no. 2OBT) (9) for each of the reference sequence P domains. Variable sites that were potentially exposed on the surface of the capsid were mapped and analyzed as these changes occurred over time from 1974 to 2009. Five discrete regions were identified as putative epitopes defined as local regions where clusters of amino acids map to similar structural regions on the capsid surface.
Production of VLPs.
Parental VLPs were expressed in a VEE expression system as previously described (4, 8, 29). Chimeric VLPs were created by replacing the P2 subdomain of ORF2 in the parental strains with the chimeric sequences (synthesized by BioBasic) (29). RNA was electroporated into 1 × 107 BHK cells using Bio-Rad GenePulser Xcell pulsed 3 times with a voltage of 850 V, capacitance of 25 μF, resistance of ∞ Ω, and a cuvette size of 4 mm. Cells were then incubated for 24 to 28 h at 37° in minimal essential medium (MEM)-alpha supplemented with 10% fetal calf serum (FCS), 10% tryptose phosphate broth (TPB), 1% antibiotic/antimycotic, and 1% nonessential amino acids (NEAAs). VLPs were harvested by spinning in 40% sucrose using a Ti-55 rotor at 31,500 rpm for 75 min. The sucrose cushion containing the VLPs was frozen at −80 degrees. VLPs were quantified by protein assay and confirmed by electron microscopy (EM).
ELISAs.
Enzyme-linked immunosorbent assays (ELISAs) were performed as previously described (28). Briefly, enzyme immunoassay (EIA) high-binding plates were coated with 100 μl of 2 μg/ml VLPs in phosphate-buffered saline (PBS) and incubated for 4 h at room temperature. Plates were washed 3 times with PBS-0.05% Tween 20, then 100 μl 5% Blotto was added to each well, and blocking was done overnight at 4°C. Plates were again washed 3 times with PBS-0.05% Tween 20, and 100 μl 2 μg/ml primary mouse MAb or a 1:500 dilution (1:500) of polyclonal sera in 5% Blotto was added to each well and incubated at 37° for 45 min. Following another wash as described above, 100 μl secondary anti-mouse AP antibody (Sigma, Jackson Laboratories) in 5% Blotto was added and allowed to incubate at room temperature (RT) for 30 min. Plates were again washed as described and detected using pNPP (Sigma). The values used in this paper indicate binding in the linear portion of the binding curve.
CHO binding assays.
CHO binding assays were performed as previously described (29). Briefly, avidin-coated plates were washed 3 times, and 100 μl 10 μg/ml biotinylated CHO (GlycoTech) in 5% Blotto was added to each well and incubated for 4 h. Plates were then washed 4 times with a 15-s soak in PBS-Tween, and 100 μl 2 μg/ml VLP was added to each well and incubated for 1.5 h at RT. Plates were washed 4 times with a 15-s soak, and 100 μl of 2 μg/ml primary MAb or 1:1,000 polyclonal sera was added and incubated for 45 min at RT. Plates were then washed 4 times with a 15-s soak, and 100 μl of 1:10,000 HRP-tagged anti-mouse antibody (GE Healthcare) was added and incubated for 30 min at RT. CHO binding was measured by addition of 100 μl Ultra TMB (3,3′,5,5′-tetramethylbenzidine)-ELISA solution (Thermo Scientific). Sera and mouse MAbs were produced as described previously (29).
Blockade assays.
Blockade assays were performed as described previously (28). Briefly, NeutriAvidin-coated plates (Pierce, Rockford, IL) were washed 3 times, and 100 μl of 10 μg/ml synthetic biotinyated-H3 CHO (GlycoTech, Gaithersburg, MD) in PBS was added to each well and incubated for 1 h. VLP-mouse MAb dilutions (1 μg/ml VLPs plus 2 μg/ml MAb) were prepared in a separate, nonbinding plate and incubated for 1 h. The avidin-coated plate was then washed 3 times, and VLP-Ab dilutions were added and incubated for 1.5 h. The plate was then washed 4 times with a 15-s soak, and 100 μl of 2 μg/ml anti-GII VLP rabbit polyclonal antisera in 5% Blotto was added to each well and incubated for 30 min. The plate was then washed 4 times with a 30-s soak, and 100 μl of a 1:10,000 dilution of horseradish peroxidase (HRP)-tagged anti-rabbit IgG antibody was added and incubated for 30 min. The plate was washed 4 times for 60 s and then developed by addition of 100 μl TMB-ELISA solution (Thermo Scientific) and 50 μl stop solution. The plate was read at an optical density of 450 (OD450). All incubations were done at room temperature. Sera and mouse MAbs were produced as described previously (29).
Statistical analysis.
Statistical analysis was performed using Prism GraphPad, and results with P values of <0.05 were considered significant. Two-way analysis of variance (ANOVA) was used to analyze CHO binding and MAb reactivity. One-way ANOVA was used to analyze the BT50s (dilutions at which 50% of the VLP-CHO interaction is blocked compared to the control) for blockade differences. Since GII.4-2006 was not blocked even at the highest mouse serum concentration, individual replicates not reaching the 50% blockade were assigned a BT50 value of 1:5 (2 times the highest serum concentration tested) for the purposes of statistical analysis.
RESULTS
Multiple sequence alignments were generated for all full-length GII.4 norovirus capsid genes in GenBank, and 36 representative sequences were selected and analyzed, based upon unique phylogenetic clustering characteristics. Representative GII.4 norovirus strains were selected from 1974, 1987, 1997, 2002, 2004, 2005, 2006, 2007, 2008, and 2009. These representative sequences were aligned, and P2 subdomain variation was mapped upon homology models of each protruding domain to identify variable sites that were potentially exposed on the surface of the capsid and therefore to the external environment. Five discrete regions (16, 19), two of which are discussed here, were identified as putative epitopes defined as local regions where variation appears to cluster on the surface (Fig. 1A and B). Putative epitope A is a conformational site comprised of amino acids 294, 296 to 298, 368, and 372, and epitope D is an epitope comprised of contiguous amino acids 393 to 395. Of interest, position 394 is the site of a single amino acid insertion that occurred in 2002 and has been fixed in the GII.4 NoV population since this time.
Fig 1.
Putative epitopes A and D. (A) Amino acid differences between GII.4-1987 Camberwell and GII.4-2006 Minerva in the P2 region of VP1. Green shading indicates amino acids within putative epitope A, and orange indicates amino acids within putative epitope D. Shared residue 296S was included because it falls within putative epitope A. (B) Epitope A is shown in green, consisting of 6 amino acids at positions 294, 296, 297, 298, 368, and 372. Epitope D is shown in orange, consisting of 3 amino acids at positions 393 to 395. Black shading indicates the HBGA binding pocket. (C) Epitope A chimeric VLPs. The parental GII.4-2006 and GII.4-1987 amino acid residues are shown in purple and yellow, respectively. GII.4-2006 Minerva residues (purple) were replaced in each chimera by GII.4-1987 Camberwell residues at the positions shown in yellow. A total of four epitope A chimeric VLPs were created. (D) Representative EM images of parental and chimeric VLPs. (a) GII.4-1987; (b) GII.4-2006; (c) GII.4-1987(+06D); (d) GII.4-2006(+87D); (e) GII.4-2006(+87A.1); (f) GII.4-2006(+87A.2); (g) GII.4-2006(+87A.3); (h) GII.4-2006(+87A.4).
Design of the GII.4-1987 and -2006 chimeric VLPs.
The P1 and P2 regions of the GII.4 NoV VP1 protein include residues that undergo high levels of variability over time. Using a panel of time-ordered mouse MAbs to GII.4-1987 and GII.4-2006 VLPs, we demonstrated clear antigenic differences among GII.4 time-ordered VLPs (28). To identify key varying residues that influence HBGA and antibody binding, we created a panel of chimeric VLPs by exchanging putative epitopes among strains that exhibit unique HBGA CHO binding and antigenic characteristics (29). GII.4-1987 (Camberwell) has previously been shown to bind synthetic HBGA H3 (29), while GII.4-2006 (Minerva) binds synthetic HBGAs A, B, and H3 (8). In this study, we focused on predicted epitopes A and D, which are surface exposed and likely play important roles in either HBGA binding or antigenicity, or both (Fig. 1B). We used synthetic cDNAs and created a panel of chimeric VLPs in which amino acids were exchanged between GII.4-1987 and GII.4-2006 within the identified putative epitopes (Fig. 1). Expression of the GII.4 parental and chimeric VP1 genes resulted in the production of robust levels of VLPs, as evidenced by SDS-PAGE gel and electron microscopy (EM) (Fig. 1D).
Epitope D affects HBGA binding but not MAb antigenicity.
Putative epitope D includes amino acids 393 to 395, which are surface exposed and vary greatly over time by either amino acid variation or insertion (Fig. 1A and B). In GII.4-2002, an amino acid insertion at position 394 evolved, which in later strains varied over time and is present in GII.4-2006 but not GII.4-1987. Previous work in our lab demonstrated that amino acid 393 influenced efficiency for HBGA B binding but not for A binding, suggesting that other amino acids likely coordinate this HBGA interaction (29). The Farmington Hills 2002a strain contains one change in the P2 region at position 395 compared to the Farmington Hills 2002 strain. Farmington Hills 2002a is able to interact with HBGA A, while Farmington Hills 2002 interactions are not robust, indicating that residue 395 likely influences A binding (29). GII.4-1987 has been shown to efficiently bind O-type CHOs but to only weakly bind (or not at all) synthetic HBGAs A and B. GII.4-2006, on the other hand, has been shown to efficiently bind O, A, and B CHOs. We created chimeric VLP GII.4-2006(+87D), in which we introduced residues 393 to 395 from GII.4-1987 into the parental GII.4-2006 strain and then built a reciprocal chimera, GII.4-1987(+06D), by inserting residues 393 to 395 from GII.4-2006 into the GII.4-1987 parental strain.
We performed a CHO binding assay to evaluate whether epitope D chimeric VLPs GII.4-2006(+87D) and GII.4-1987(+06D), in which amino acids 393 to 395 are exchanged, would recapitulate HBGA binding characteristics of the parental strains (Fig. 2A). As previously described, the GII.4-1987 parent did not efficiently bind A or B CHOs, while the GII.4-2006 parent bound both A and B in a time- and dose-dependent manner. However, GII.4-2006(+87D) did not bind to A or B, indicating that the 2006 residues at these positions are important for the ability of this strain to bind both HBGAs A and B. In addition, GII.4-1987(+06D) gained the ability to weakly bind HBGA A and strongly bind HBGA B. Despite this gain of function, the binding profile clearly differed from that of the GII.4-2006 parent, indicating there may be additional amino acids elsewhere in the capsid that contribute to subtle remodeling of the affinity of HBGA A and B interactions. These data are consistent with the idea that surface-exposed residues in and around the conserved HBGA binding sites influence HBGA binding efficiency (29, 41).
Fig 2.
Epitope D chimeric VLP reactivity with synthetic HBGAs and anti-GII MAbs. (A) HBGA reactivity. Synthetic CHO was diluted to 10 μg/ml and assayed by the CHO binding assay for interaction with GII.4-1987, GII.4-2006, GII.4-1987(+06D), and GII.4-2006(+87D). Amino acid residues from GII.4-1987 were exchanged with GII.4-2006 residues at positions 393 to 395. The dashed line represents 3 times the background level of binding and indicates a positive signal. Asterisks indicate significant differences. (B) Anti-GII.4 MAb reactivity. Anti-GII.4-1987 and -2006 MAbs were diluted to 2 μg/ml and assayed by EIA for reactivity with GII.4-1987, GII.4-2006, GII.4-1987(+06D), and GII.4-2006(+87D) VLPs. Amino acid residues from GII.4-1987 were exchanged with GII.4-2006 at positions 393 to 395. Bars represent the mean optical density values with standard errors of the means. The dashed line represents 3 times the background level of binding and indicates a positive signal.
We were also interested in whether epitope D amino acids are involved in antigenic differences between GII.4-1987 and GII.4-2006. Our group previously characterized a large panel of time-ordered mouse monoclonal antibodies that display varying interactions with time-ordered GII.4 VLPs (28). In particular, MAbs GII.4-1987-G1, GII.4-1987-G2, GII.4-1987-G3, GII.4-1987-G4, and GII.4-1987-G5 bound efficiently to GII.4-1987 but not to GII.4-2006 VLPs, while MAbs GII.4-2006-G3, GII.4-2006-G4, and GII.4-2006-G7 bound efficiently to GII.4-2006 but not to GII.4-1987 VLPs (28). GII.4-2006-G2 primarily bound GII.4-2006 but was somewhat cross-reactive with GII.4-1987 (28). The availability of MAbs that recognize distinct epitopes on the surfaces of GII.4-1987 and -2006 VLPs provides a means of mapping key residues that influence antibody escape from binding over time. To determine if amino acids 393 to 395 contribute to antigenic differences between GII.4-1987 and GII.4-2006, we tested the reactivity of these parents and chimeras with our panel of GII.4-1987 MAbs and GII.4-2006 MAbs. While subtle differences in overall binding were noted with anti-GII.4-1987 MAbs reacting with GII.4-1987 compared to GII.4-1987(+06D), we did not detect significant differences in antigenic phenotypes between GII.4-1987 and GII.4-1987(+06D) or between GII.4-2006 and GII.4-2006(+87D) (Fig. 2B) with these characterized MAbs. These data indicate that amino acids 393 to 395 do not explain the antigenic differences observed between GII.4-1987 and GII.4-2006 for any of the MAbs tested in our panel.
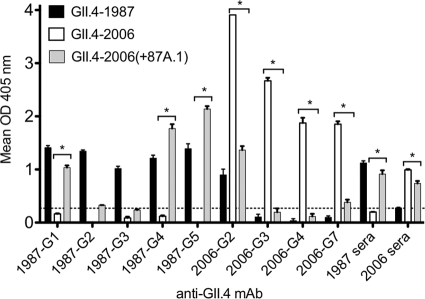
Putative epitope A amino acids contribute to antigenic differences between GII.4-1987 and GII.4-2006.
Putative epitope A consists of closely positioned surface-exposed amino acids 294, 296, 297, 298, 368, and 372 (Fig. 1A and B). These amino acids form a conformational cluster on the surface ridge of the capsid and display charge variation over time, suggesting that these residues may play a key role in the escape from herd immunity. We created chimeric VLPs in which combinations of amino acids 294, 297, 298, 368, and 372 from GII.4-1987 were introduced into the GII.4-2006 backbone (Fig. 1C). While we attempted to create the reciprocal exchange and produce VLPs comprised of GII.4-2006 epitope A amino acids in the GII.4-1987 backbone, after multiple tries, we were not able to yield VLPs that passed our quality control standards, which include robust protein production, as determined by SDS-PAGE gel and bicinchoninic acid (BCA) protein assay, visualization of particles by EM, and the ability to bind synthetic CHOs and one or more conformationally dependent MAbs. The ability of these chimeras to bind anti-GII.4-1987 MAbs and anti-GII.4-2006 MAbs was measured and compared to that of parental strains. As expected, GII.4-1987 VLPs demonstrated strong homotypic reactivity, binding to anti-GII.4-1987-G1, anti-GII.4-1987-G2, anti-GII.4-1987-G3, anti-GII.4-1987-G4, and anti-GII.4-1987-G5, while GII.4-2006 did not react with any of the GII.4-1987 MAbs tested (Fig. 3) (28). Chimeric VLP GII.4-2006(+87A.1), which contained all five of the epitope A variable residues from GII.4-1987, exhibited significant increases in reactivity with anti-GII.4-1987-G1, anti-GII.4-1987-G4, and anti-GII.4-1987-G5, and this resulted in 6-fold, 14-fold, and 11-fold increases in reactivity, respectively, compared to GII.4-2006 (Fig. 3) (P < 0.0001). However, GII.4-2006(+87A.1) did not significantly alter binding (P > 0.05) with the other two GII.4-1987 MAbs tested, anti-GII.4-1987-G2 or anti-GII.4-1987-G3, suggesting the presence of another epitope located elsewhere on the virion (Fig. 3). Anti-GII.4-1987-G1, -G4, and -G5 have previously been shown to be HBGA-VLP blocking antibodies (28), suggesting that epitope A amino acids may comprise a neutralizing antibody epitope for GII.4-1987.
Fig 3.
GII.4-2006(+87A.1) reactivity with anti-GII.4 MAbs. Anti-GII.4-1987 and -2006 IgG was diluted to 2 μg/ml and assayed by EIA for reactivity with GII.4-1987, GII.4-2006, and GII.4-2006(+87A.1) VLPs. Amino acid residues from GII.4-1987 were inserted into the GII.4-2006 backbone at positions 294, 297, 298, 368, and 372 to create GII.4-2006(+87A.1). Bars represent the mean optical density values with standard errors of the means. The dashed line represents 3 times the background level of binding and indicates a positive signal. Asterisks indicate significant differences.
Binding of anti-GII.4-2006 MAbs was also compared between GII.4-2006(+87A.1) and parental strains GII.4-1987 and GII.4-2006. GII.4-2006 had strong interactions with anti-GII.4-2006-G2, anti-GII.4-2006-G3, anti-GII.4-2006-G4, and anti-GII.4-2006-G7, while GII.4-1987 bound anti-GII.4-2006-G2 much less efficiently than GII.4-2006 but did not bind anti-GII.4-2006-G3, anti-GII.4-2006-G4, or anti-GII.4-2006-G7 (Fig. 3). Comparatively, GII.4-2006(+87A.1) more closely resembled the binding pattern seen for GII.4-1987. GII.4-2006(+87A.1) exhibited an intermediate binding profile for anti-GII.4-2006-G2 that was slightly greater than that seen for GII.4-1987 but greatly reduced compared to GII.4-2006 (Fig. 3). GII.4-2006(+87A.1) binding to anti-GII.4-2006-G3 and anti-GII.4-2006-G4 was negligible, and reactivity with anti-GII.4-2006-G7 was greatly reduced (Fig. 3). Overall, binding of GII.4-2006(+87A.1) was significantly reduced compared to that of GII.4-2006 for all anti-GII.4-2006 MAbs tested (P < 0.0001).
Next, we evaluated whether GII.4-2006(+87A.1) also had an increased reactivity to GII.4-1987 mouse polyclonal sera. We performed an ELISA comparing GII.4-2006(+87A.1) reactivity with both GII.4-1987 and GII.4-2006 polyclonal sera (Fig. 3). The GII.4-1987 and GII.4-2006 parental strains bound most efficiently to homotypic polyclonal sera, while GII.4-2006(+87A.1) had a significant, nearly 5-fold increase in binding to GII.4-1987 polyclonal sera (P < 0.0001) and a significant 20% decrease in binding to GII.4-2006 polyclonal sera (P < 0.01) compared to the GII.4-2006 parental VLP. However, GII.4-2006(+87A.1) did not bind to GII.4-1987 sera as strongly as the GII.4-1987 parental VLP. These results confirm that GII.4-2006(+87A.1) gained GII.4-1987 reactivity but that there are additional amino acid residues and/or epitopes (elsewhere on the capsid) that influence GII.4-1987 antigenic space.
Epitope A amino acids do not influence HBGA A and B binding.
To determine whether epitope A amino acids influence HBGA A and B binding, we performed CHO binding assays using GII.4-2006(+87A.1) and compared the binding profile to those of the parental GII.4-1987 and GII.4-2006 strains (Fig. 4). There were no significant differences in HBGA A and B binding between GII.4-2006(+87A.1) and the parental strain GII.4-2006, indicating that these amino acids do not influence differences in HBGA A and B binding between GII.4-1987 and GII.4-2006 under our treatment conditions.
Fig 4.

Epitope A chimeric VLP reactivity with synthetic HBGAs. Synthetic CHOs were diluted to 10 μg/ml and assayed by the CHO binding assay for interaction with GII.4-1987, GII.4-2006, and GII.4-2006(+87A.1). Amino acid residues from GII.4-1987 were inserted into the GII.4-2006 backbone at positions 294, 297 to 298, 368, and 372. The dashed line represents 3 times the background level of binding and indicates a positive signal.
Fine resolution mapping.
Putative epitope A consists of six surface-exposed amino acid residues that cluster on the surface of the virion. To further investigate the contribution of individual and/or combinations of amino acids to antibody reactivity, we characterized the antigenic profiles of additional chimeric VLPs in which only one or two of the residue changes within epitope A were switched between strains (Fig. 1C). Chimera GII.4-2006(+87A.2) contains two amino acid changes at positions R297H and N298D. By ELISA, GII.4-2006(+87A.2) did not react with any of the anti-GII.4-1987 MAbs, except for displaying minimal reactivity with anti-GII.4-1987-G1, which was not significantly different from the binding of the GII.4-2006 parent (Fig. 5). In contrast, GII.4-2006(+87A.2) reactivity with anti-GII.4-2006 was more similar to that of the GII.4-1987 parent, in that anti-GII.4-2006-G3 and anti-GII.4-2006-G4 binding was ablated and not significantly different from that of the GII.4-1987 parent. Under identical conditions, binding to anti-GII.4-2006-G7 was significantly reduced compared to the GII.4-2006 parent (Fig. 5) (P < 0.0001). This indicates that anti-GII.4-2006-G3, -G4, and -G7 may target similar or overlapping epitopes in GII.4-2006 comprising amino acids 297 and 298. However, GII.4-2006(+87A.2), like GII.4-2006, exhibited strong binding with anti-GII.4-2006-G2, indicating that GII.4-2006-G2 targets a different epitope than the other anti-GII.4-2006 MAbs in our panel; this is in agreement with previous data from our lab (28). This indicates that these amino acids modulate specificity to some anti-GII.2006 MAbs but do not account for the gain of reactivity seen with the additional amino acid substitutions in GII.4-2006(+87A.1).
Fig 5.
GII.4-2006(+87A.2) reactivity with anti-GII.4 MAbs. Anti-GII.4-1987 and -2006 IgG was diluted to 2 μg/ml and assayed by EIA for reactivity with GII.4-1987, GII.4-2006, and GII.4-2006(+87A.2) VLPs. Amino acid residues from GII.4-1987 were inserted into the GII.4-2006 backbone at positions 297 and 298 to create GII.4-2006(+87A.2). Bars represent the mean optical density values with standard errors of the means. The dashed line represents 3 times the background level of binding and indicates a positive signal. Asterisks indicate significant differences.
Both chimeras GII.4-2006(+87A.3) and GII.4-2006(+87A.4), which contain two amino acid changes at positions S368T and E372N and one amino acid change at position A294V, respectively, did not exhibit significantly different reactivity with anti-GII.4-1987 or anti-GII.4-2006 MAbs compared with the GII.4-2006 parent. GII.4-2006(+87A.3) and GII.4-2006(+87A.4) did not react to any of the anti-GII.4-1987 MAbs but showed reactivity with all anti-GII.4-2006 MAbs (Fig. 6 and 7). Taken together, our data indicate that all epitope A amino acids comprise a major antigenic site for GII.4-1987 and that epitope A residues 297 and 298 are important modulators of GII.4-2006 antigenicity (Table 1). Our data support the idea that several noncontiguous amino acids work in concert to account for the antigenic differences between these strains.
Fig 6.
GII.4-2006(+87A.3) reactivity with anti-GII.4 MAbs. Anti-GII.4-1987 and -2006 IgG was diluted to 2 μg/ml and assayed by EIA for reactivity with GII.4-1987, GII.4-2006, and GII.4-2006(+87A.3) VLPs. Amino acid residues from GII.4-1987 were inserted into the GII.4-2006 backbone at positions 368 and 372 to create GII.4-2006(+87A.3). Bars represent the mean optical density values with standard errors of the means. The dashed line represents 3 times the background level of binding and indicates a positive signal.
Fig 7.
GII.4-2006(+87A.4) reactivity with anti-GII.4 MAbs. Anti-GII.4-1987 and -2006 IgG was diluted to 2 μg/ml and assayed by EIA for reactivity with GII.4-1987, GII.4-2006, and GII.4-2006(+87A.4) VLPs. An amino acid residue from GII.4-1987 was inserted into the GII.4-2006 backbone at position 294 to create GII.4-2006(+87A.4). Bars represent the mean optical density values with standard errors of the means. The dashed line represents 3 times the background level of binding and indicates a positive signal.
Table 1.
Relative reactivity of chimeric versus parental homotypic VLPs
| MAb | Binding scorea |
|||||
|---|---|---|---|---|---|---|
| GII.4-1987 | GII.4-2006 | GII.4-2006(+87A.1) | GII.4-2006(+87A.2) | GII.4-2006(+87A.3) | GII.4-2006(+87A.4) | |
| GII.4-1987-G1 | 4 | 1 | 4 | 2 | 1 | 0 |
| GII.4-1987-G2 | 4 | 0 | 1 | 0 | 0 | 0 |
| GII.4-1987-G3 | 4 | 1 | 1 | 0 | 1 | 0 |
| GII.4-1987-G4 | 4 | 1 | 4 | 0 | 0 | 0 |
| GII.4-1987-G5 | 4 | 0 | 4 | 0 | 0 | 0 |
| GII.4-2006-G2 | 1 | 4 | 2 | 3 | 4 | 4 |
| GII.4-2006-G3 | 0 | 4 | 1 | 0 | 4 | 4 |
| GII.4-2006-G4 | 0 | 4 | 0 | 0 | 4 | 4 |
| GII.4-2006-G7 | 0 | 4 | 1 | 2 | 4 | 4 |
For anti-GII.4-1987 MAbs, binding of chimeric and parental GII.4-2006 VLPs was compared to that of GII.4-1987 VLPs. For anti-GII.4-2006 MAbs, binding of chimeric and parental GII.4-1987 VLPs was compared to that of GII.4-2006 VLPs. Scoring is as follows. VLPs that exhibited 0 to 5% of the binding level of the homotypic parental VLP (GII.4-1987 or GII.4-2006) received a score of 0, 6 to 25% received a score of 1, 26 to 50% received a score of 2, 51 to 75% received a score of 3, and 76% or higher received a score of 4.
Carbohydrate blockade.
Based on previous work by Lindesmith et al., anti-GII.4-1987-G1, anti-GII.4-1987-G4, and anti-GII.4-1987-G5 are blocking antibodies for the same epitope in the GII.4-1987 parent strain but do not block GII.4-2006 interaction with synthetic CHOs (28). While we determined that GII.4-2006(+87A.1) gained binding to the anti-GII.4-1987-G1, -G4, and -G5 MAbs (Fig. 3), we also determined whether these antibodies were able to block the interaction between GII.4-2006(+87A.1) and HBGAs and thus whether epitope A amino acids represented a possible neutralizing antibody epitope. To do this, we performed a CHO blockade assay comparing the ability of anti-GII.4-1987-G1, anti-GII.4-1987-G4, and anti-GII.4-1987-G5 to block HBGA interactions with GII.4-1987, GII.4-2006, and GII.4-2006(+87A.1). While anti-GII.4-1987-G1, -G4, and -G5 were not able to block GII.4-2006 interactions with H3 (28), all three MAbs were able to block H3 interactions with both GII.4-1987 and GII.4-2006(+87.1), indicating that these are blocking antibodies for GII.4-2006(+87A.1) and target epitope A. All three antibodies exhibited similar blocking patterns, and anti-GII.4-1987-G1 is shown as a representative (Fig. 8A). Since GII.4-2006(+87A.2), GII.4-2006(+87A.3), and GII.4-2006(+87A.4) do not bind anti-GII.4-1987-G1, -G4, or -G5, it stands to reason that these MAbs cannot be blocking antibodies for these chimeras. From these data, we determined that in addition to amino acids 296 to 298, which have previously been shown to modulate antigenicity, amino acids 294, 368, and 372 are important for the blockade response to GII.4-1987.
Fig 8.
Anti-GII.4 MAb blockade of VLP/synthetic HBGA interactions. (A, B) The ability of anti-GII.4-1987-G1 (A) and anti-GII.4-2006-G7 (B) to block VLP interaction with synthetic HBGA was measured by surrogate neutralization assay and expressed as a percentage of the control binding (100%). An antibody is considered a “blocking antibody” if it is able to block at least 50% of the control (dashed line). Anything below the dashed line represents potential neutralization. Error bars represent standard errors of the means from experiments run in triplicate.
We also determined whether a known GII.4-2006 blocking antibody was able to retain the ability to block our four epitope A chimeras. We performed a CHO blockade assay to determine if anti-GII.4-2006-G7, a mouse MAb identified by Lindesmith et al. (28) to block GII.4-2006 interaction but not GII.4-1987 interaction with HBGAs, could block HBGA interaction with GII.4-2006(+87A.1), GII.4-2006(+87A.2), GII.4-2006(+87A.3), and GII.4-2006(+87A.4). While anti-GII.4-2006-G7 was able to block H3 interaction with GII.4-2006, GII.4-2006(+87A.3), and GII.4-2006(+87A.4), it was unable to block HBGA binding to GII.4-1987, GII.4-2006(+87A.1), and GII.4-2006(+87A.2) (Fig. 8B). Taken together, this indicates that epitope A amino acids represent a blocking antibody epitope that is undergoing antigenic variation. The epitope encoded in both GII.4-1987 and GII.4-2006 is targeted by antibodies that block the homotypic VLP but not the heterotypic response.
GII.4-2006(+87A.1) exhibited increased reactivity with GII.4-1987 polyclonal mouse sera (Fig. 9A). To determine the degree to which epitope A amino acids contribute to the overall GII.4-1987 blockade response, we performed a blockade assay comparing the ability of GII.4-1987 polyclonal mouse sera to block H3 interaction with GII.4-1987, GII.4-2006, and GII.4-2006(+87A.1) VLPs (Fig. 9B). The GII.4-1987 polyclonal serum BT50 concentration for interaction between H3 and GII.4-1987 was 1:160 (0.7% sera), while the BT50 for GII.4-2006(+87A.1) was 1:40 (2.5% sera). GII.4-2006 did not have a mean BT50 even at the highest concentration of GII.4-1987 polyclonal sera tested (1:10); however, some individual replicates in the assays performed did have a BT50, and those that did not were assigned a BT50 of 1:5 (20% sera) (Fig. 9C). By one-way ANOVA, BT50 values for GII.4-1987 and GII.4-2006(+87A.1) were not significantly different. GII.4-2006 was significantly different from both GII.4-1987 and GII.4-2006(+87A.1) (P < 0.05). Overall, these results indicate that epitope A amino acids account for approximately 25% of the antibody blockade response encoded within GII.4-1987 mouse polyclonal sera.
Fig 9.

GII.4-1987 mouse polyclonal serum blockade. (A) GII.4-1987 polyclonal mouse sera was diluted 1:1,000 and assayed by EIA for reactivity with GII.4-1987, GII.4-2006, and GII.4-2006(+87A.1) VLPs. Bars represent the mean optical density values with standard errors of the means. The dashed line represents 3 times the background level of binding and indicates a positive signal. (B) GII.4-1987 polyclonal mouse serum blockade of GII.4 VLP/synthetic HBGA interactions. Ability of GII.4-1987 polyclonal mouse sera to block VLP/synthetic CHO was measured by surrogate neutralization assay and expressed as a percentage of the control binding (100%). An antibody is considered a “blocking antibody” if it is able to block at least 50% of the control (dashed line). Anything below the dashed line represents potential neutralization. Error bars represent standard errors of the means from experiments run in triplicate. (C) GII.4-1987 polyclonal mouse serum BT50 values for GII.4-1987, GII.4-2006, and GII.4-2006(+87A.1). Blocking titers of GII.4-1987 mouse polyclonal sera were determined as the serum concentration at which 50% of the VLP/synthetic HBGA was blocked. Box and whiskers plots represent the mean serum concentration at which BT50 was achieved for each VLP.
GII.4-2006(+87A.1) displayed increased reactivity with 1988 GII.4 NoV outbreak human sera compared to GII.4-2006 (Fig. 10A). To determine whether epitope A amino acids contribute to the GII.4-1987 blockade response in humans, we performed a blockade assay comparing the ability of 1988 GII.4 NoV outbreak human serum samples to block CHO interaction with GII.4-1987, GII.4-2006, and GII.4-2006(+87A.1). Since we were interested in the contribution of epitope A amino acids to the GII.4-1987 blockade response, we chose to utilize nine human serum samples that had the ability to block CHO interaction with the GII.4-1987 VLP (8, 29). Overall, the human serum samples were able to block GII.4-1987 most efficiently and, with the exception of one individual, were able to block GII.4-2006(+87A.1) more efficiently than GII.4-2006, although differences were not significant. When all nine serum samples were averaged at each time point, the same trend was seen (Fig. 10B). GII.4-1987 was blocked most efficiently at all dilutions. GII.4-2006(+87A.1) and GII.4-2006 were blocked to similar levels, although GII.4-2006(+87A.1) was blocked to a greater degree than GII.4-2006 at all sera dilutions. We then compared the BT50s (dilutions at which 50% of the VLP-CHO interaction is blocked compared to the control) for GII.4-1987, GII.4-2006(+87A.1), and GII.4-2006. The average BT50 values for GII.4-1987, GII.4-2006(+87A.1), and GII.4-2006 were 1:1,600, 1:800, and 1:400, respectively, indicating significant differences for BT50 values between GII.4-1987 and GII.4-2006 (P < 0.05) but not between GII.4-2006(+87A.1) and GII.4-1987 or GII.4-2006 (Fig. 10C). Together, these data suggest that epitope A contributes to the global GII.4-1987 blockade response in mice and humans. In vaccinated mice, epitope A accounts for approximately 25% of the total blockade response between GII.4-1987 and GII.4-2006. Using GII.4-1987 human antisera, we can detect clear differences in blockade responses using time-ordered VLPs. However, because of variation among human samples and unknown preexposure histories, additional human serum samples will be needed to detect significant differences in the blockade activity that targets individual epitopes.
Fig 10.
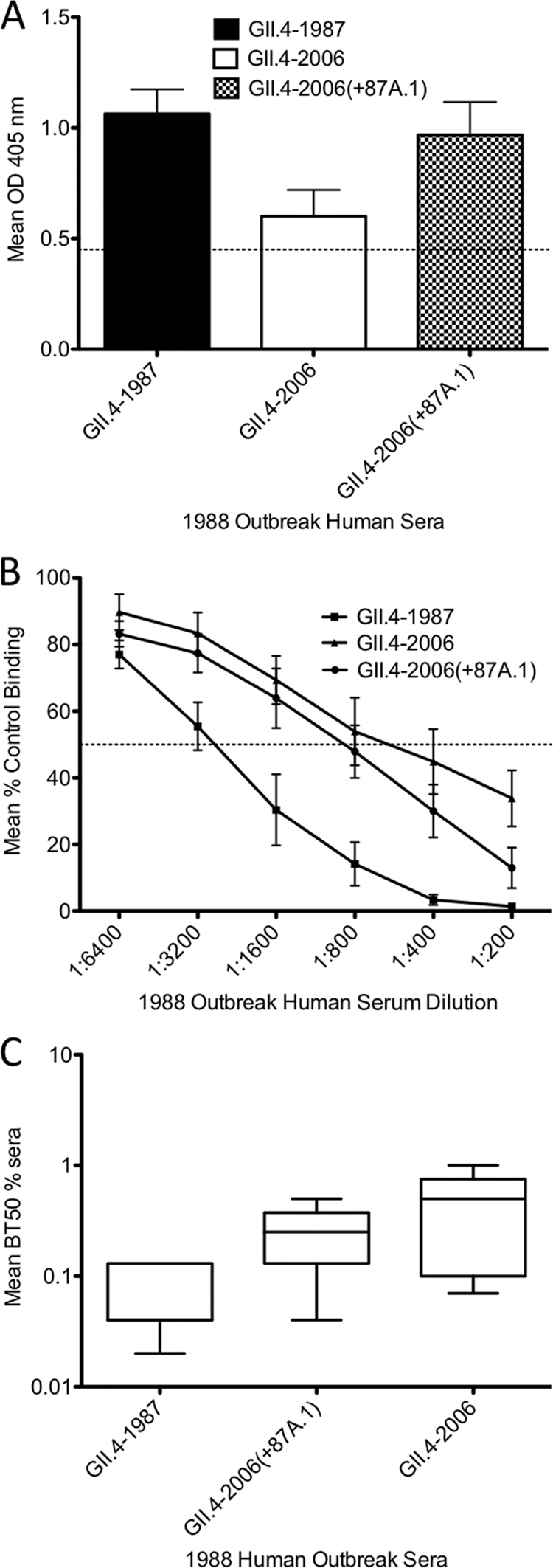
1988 human outbreak serum blockade. (A) 1988 human outbreak sera was diluted 1:500 and assayed by EIA for reactivity with GII.4-1987, GII.4-2006, and GII.4-2006(+87A.1) VLPs. Bars represent the mean optical density values with standard errors of the means. The dashed line represents 3 times the background level of binding and indicates a positive signal. (B) 1988 human outbreak serum blockade of GII.4 VLP/synthetic HBGA interactions. The ability of human sera to block VLP/synthetic CHO was measured by surrogate neutralization assay and expressed as a percentage of the control binding (100%). An antibody is considered a “blocking antibody” if it is able to block at least 50% of the control (dashed line). Anything below the dashed line represents potential neutralization. Error bars represent standard errors of the means from experiments run in triplicate. (C) 1988 human outbreak serum BT50 values for GII.4-1987, GII.4-2006, and GII.4-2006(+87A.1). Blocking titers of human sera were determined as the serum concentration at which 50% of the VLP/synthetic HBGA was blocked. Box and whiskers plots represent the mean serum concentration at which BT50 was achieved for each VLP.
DISCUSSION
GII.4 NoVs exhibit epochal evolution, whereby major circulating strains give rise to emergent strains, likely due to escape from herd immunity (15, 29, 43). Evidence supporting this includes greater positive selection in capsid P2 subdomain amino acids than in the rest of the capsid protein, robust data exhibiting altered antigenicity and HBGA binding over time in VLPs of major outbreak and pandemic GII.4 strains, and the identification of strain-specific potentially neutralizing antibodies in GII.4 NoVs (15, 28, 29). Similar to GII.4 NoVs, influenza A undergoes antigenic variation as mutations in the hemagglutinin 1 (HA1) domain of HA genes drive escape from the existing herd immunity (18, 24, 25). Additionally, amino acids in the GII.4 NoV P2 region, which interact with antibodies and binding ligands, undergo positive selection similar to the mechanism in influenza whereby HA interacts with antibodies and sialic acid and also contains many amino acids undergoing positive selection (7, 24). Individual epitopes in other viruses, like influenza, have been shown to lead to escape from herd immunity (18), providing the possibility that GII.4 NoVs undergo a similar mechanism of evolution. To provide further support for this idea, this study used a novel mapping strategy to identify a blockade epitope in GII.4-1987 and GII.4-2006 that changed over time in major GII.4 outbreak strains, and we identified a specific epitope involved in modulating HBGA binding patterns in different GII.4 strains. This is the first study to map an antibody blockade epitope in GII.4 NoVs. These findings will further contribute to the elucidation of mechanisms driving evolution in these viruses, provide insight into the emergence of new epidemic variants, and provide direction for broadening the antigenic design and cross-blockade activity of potential vaccines for NoVs.
Five putative epitopes were predicted via sequence analysis using 36 representative, time-ordered GII.4 stains from 1974 to 2009 to identify amino acid residues that varied consistently over time and were exposed on the capsid surface in the P2 subdomain (16). These varying residues were then mapped onto homology models of each respective representative, and the structural models were compared to find residues that map together in discrete regions of the capsid surface. We chose to build chimeras between GII.4-1987 and GII.4-2006 because of known differential HBGA binding and antigenicity profiles between these two strains (8, 28, 29). By examining the reactivity of our chimeric VLPs with synthetic HBGAs and mouse monoclonal antibodies against GII.4-1987 and GII.4-2006, we determined that epitopes A and D were important for antigenic and HBGA differences between these strains, respectively. Our results suggest that changes in surface residues drive antigenic change in GII.4 NoVs, as has been shown previously in influenza. Additionally, our results validate the utility of using bioinformatics approaches to identify important antigenic determinants.
Our data clearly show that amino acids 393 to 395 modulate HBGA A and B interactions, confirming earlier observations by our lab and others (2, 29). When we exchanged amino acids in positions 393 to 395 between GII.4-1987 and GII.4-2006, we observed a significant decrease in interaction between GII.4-2006(+87D) and HBGAs A and B compared to GII.4-2006 and a significant increase in interaction between these HBGAs and GII.4-1987(+06D) compared to GII.4-1987, indicating that these amino acids influence interaction with A and B CHOs. However, we also noticed that the interaction profile of GII.4-1987(+06D) was very different from that of the GII.4-2006 parent, suggesting that other amino acids contribute to A and B interactions as well. Teasing apart these interactions without a crystal structure can be complex. While modeling programs can be used to provide predictions about VLP-CHO interactions, these methods rely upon models based on previously solved structures. Studying the differences between GII.4 NoV strains has been impeded by the existence of only two GII.4 crystal structures, VA387, which models GII.4-1997 complexed with CHOs A and B, and Hunter 2004, bound to several CHOs. These structures were used to assemble homology models for GII.4-1987 and GII.4-2006. Recognizing the limitations inherent in this approach, our models identified potential mechanisms that explain increased GII.4-1987(+06D) interactions with HBGA B compared with GII.4-2006. Structural homology models for GII.4-1987, GII.4-2006, and GII.4-1987(+06D) predicted subtle differences in the positioning of residues 393 to 395 in a surface-exposed loop (Fig. 11A) and in polar interactions among binding pocket amino acids in sites 1 and 2 (residues 342 to 347, 374, 390 to 395, and 440 to 444) of GII.4-2006 and GII.4-1987(+06D) and in CHO B (Fig. 11B to F). Importantly, direct interactions between the CHO and residues 393 to 395 were not observed; however, rearrangement of polar interactions among residues 390, 393, and 395 (Fig. 11B) occurred, which may slightly juxtapose the interactions in the binding pocket. This suggests that these residues have long-range effects on polar interactions in the binding pocket. Variation in the loop programmed specific differences in the GII.4-1987(+06D) ability to (i) form a new hydrogen bond between the fucose of CHO B and amino acid 343 (Fig. 11C), (ii) terminate two hydrogen bonds from residue 345 and CHO B (Fig. 11D), and (iii) form new hydrogen bonds between residues 343 and 444 (Fig. 11E) and between residue 442 and CHO B (Fig. 11F). Together, these altered polar interactions are predicted to stabilize binding of CHOs within the binding site, allowing GII.4-1987(+06D) to exhibit stronger binding to CHO B and potentially to H3. However, we stress that the crystal structures of more GII.4 strains bound to CHOs are needed to further address these predictions. Other groups have suggested additional P2 amino acids that may modulate HBGA A and B binding. Tan et al. identified amino acid 346 as contributing to HBGA A and B interactions (45). Because this residue varies between GII.4-1987 and GII.4-2006, it is possible that amino acid 346 also modulates A and B binding; however, we did not test this possibility in this study, and this residue did not participate in direct polar interactions in our homology models. Yang et al. reported I389 as a single amino acid residue important for HBGA A binding (48). While this amino acid does not contribute to the differences in HBGA A and B binding between GII.4-1987 and GII.4-2006, as both strains carry I389, we cannot rule out the possibility that this residue contributes to HBGA binding in other GII.4 strains. However, Shanker et al. also found no evidence that residue 389 would contribute to HBGA A binding in GII.4-2004 (41).
Fig 11.
Homology models comparing GII.4-1987, GII.4-2006, and GII.4-1987(+06D). (A) Overlay of binding pocket of GII.4-1987 (yellow), GII.4-2006 (purple), and GII.4-1987(+06D) (gray). Side chains are shown for residues in sites 1 and 2 (residues 342 to 347, 374, 390 to 395, and 440 to 444). Residues 393 to 395 are shown in teal [GII.4-1987(+06D)], orange (GII.4-1987), and red (GII.4-2006). (B) Residue 390 (yellow) forms a hydrogen bond with residue 395 (yellow) in GII.4-1987(+06D) that is not found in GII.4-2006. GII.4-2006 residues 393 and 395 form hydrogen bonds not found in GII.4-1987(+06D). (C) Residue 343 (yellow) forms a hydrogen bond (red arrow) with the FUC of HBGA B (orange) in GII.4-1987(+06D) but not with that of GII.4-2006. (D) Three hydrogen bonds are formed between residue 345 (yellow) and the FUC of HBGA B in GII.4-2006, but only one is formed with GII.4-1987(+06D). (E) Residues 343 and 444 (both yellow) share a hydrogen bond in GII.4-1987(+06D) but not in GII.4-2006. (F) Residue 442 (yellow) participates in hydrogen bonding with the FUC of HBGA B in GII.4-1987(+06D) but not with that of GII.4-2006. In GII.4-1987(+06D), residue 442 (yellow) also forms a hydrogen bond with residue 443, which is not found in GII.4-2006.
Epitope A amino acids (294, 296 to 298, 368, and 372) change over time and map to the same area of the capsid surface, likely forming a noncontiguous conformational epitope. Influenza virus and HIV provide examples of viruses that have variable, noncontiguous conformational epitopes that change over time, resulting in viruses that are able to escape the immune response (10, 20, 49, 50). Our data demonstrate that epitope A amino acids collectively comprise a blocking antibody epitope for both GII.4-1987 and GII.4-2006, suggesting that the surrounding landscape and conformational changes associated with amino acid variation greatly impact antigenicity. Variation likely reshapes the surface of the P2 region, altering the affinity with which different MAbs bind and neutralize the virus; however, additional GII.4 crystal structures coupled with MAb are essential for understanding the molecular basis of antibody binding and blockade and likely essential for illuminating the mechanism of escape from herd immunity. While predicted epitope A also includes amino acid 296, this residue is conserved between GII.4-1987 and GII.4-2006, so our current work does not address the antigenic consequences of changes to residue 296. However, Allen et al. (2) demonstrated that amino acids 296 to 298 were important antigenic determinants between pre-2002 and post-2002 GII.4 strains, although they did not evaluate CHO blockade. Our data corroborate the importance of these amino acids in GII.4 antigenic changes over time. Introduction of smaller subsets of epitope A amino acids, as in GII.4-2006(+87A.2), GII.4-2006(+87A.3), and GII.4-2006(+87A.4), did not yield gains in binding to anti-GII.4-1987 MAbs for the chimeric VLPs tested, suggesting that all five epitope A amino acid changes in combination are needed to gain robust binding phenotypes to the anti-GII.4-1987 MAbs tested. While speculative, some flu epitopes are proposed to evolve by epochal evolution characterized by drift modified by mutations of large effect; similar mechanisms may be operating during GII.4 persistence in human populations (24). While it is possible that creating additional combinations of these five amino acids would yield a combination that accounts for the gain in binding to anti-GII.4-1987 MAbs, we argue that it is more likely that these noncontiguous amino acids constitute a conformational epitope in which they contribute, in concert, to the gain in binding to anti-GII.4-1987 MAbs. We do not exclude the possibility that additional amino acids may contribute to interactions with anti-GII.4-1987 and anti-GII.4-2006 MAbs. Importantly, based on sequence alignment data, epitope A is still evolving in the currently circulating Minerva variant, GII.4-2009 New Orleans. This suggests that this epitope may also constitute an important blocking epitope in future epidemic GII.4 strains, thus monoclonal antibodies against contemporary GII.4-2009 strains may provide greater resolution in understanding the potential mechanisms driving antigenic variation and antibody blockade escape over time. To our knowledge, this is the first study utilizing a panel of time-ordered chimeric viruses with exchanged epitopes to study these processes; however, this approach may be highly relevant to studying antigenic variation in other viruses, including influenza and HIV.
Because human NoVs lack a viable cell culture model, antibody neutralization potential is measured in a surrogate neutralization assay, the CHO blockade assay (21, 28). This assay measures an antibody's ability to block the interaction between immobilized HBGA and a particular VLP. Reeck et al. showed a correlation between the ability of antibodies to block VLP-HBGA interaction in this assay and protection from symptomatic Norwalk virus infection, demonstrating that the blockade assay is a relevant surrogate assay that measures a component of protective immunity (39). We showed that epitope A is a significant blockade epitope. Using anti-GII.4-1987-G1, -G4, and -G5 MAbs, we demonstrated that residues 294, 297 to 298, 368, and 372 work in concert to account for the GII.4-1987 blockade response for these antibodies. Using anti-GII.4-2006-G7, we demonstrated that amino acids 297 to 298 account for the GII.4-2006 blockade response for this antibody. GII.4-2006(+87A.1) and GII.4-2006(+87A.2), which both contain changes at these residues, were not blocked by anti-GII.4-2006-G7, but this antibody blocked VLP-HBGA interaction for chimeras GII.4-2006(+87A.3) and GII.4(+87A.4), which did not contain substitutions at residue 297 or 298. Approximately 25% of the total blockade response for GII.4-1987 polyclonal mouse sera was targeted against epitope A based on comparison of the CHO blockade potential of the parental GII.4-1987 to that of GII.4-2006. This result suggests that there are either additional unidentified blockade epitopes present for GII.4-1987 or additional unidentified residues that contribute to the overall antigenic variation in epitope A. This approach may be useful in future studies to predict the number of potentially neutralizing epitopes for each particular strain and the relative importance of each epitope in contributing to blocking antibody response over time.
Using GII.4 NoV human outbreak sera, we demonstrated that epitope A is a potential blockade epitope not only in mice but also in humans. Results gleaned from human data underscore the importance of individual variation and previous exposure history in the immune response to different GII.4 NoV strains. Mouse polyclonal serum data, produced in a background with no previous exposure history or genetic diversity, yielded clear differences in blockade among GII.4-1987, GII.4-2006(+87A.1), and GII.4-2006, in which GII.4-1987 and GII.4-2006(+87A.1) were blocked but GII.4-2006 was not. Human sera, on the other hand, was able to block VLP-CHO interactions with GII.4-1987, GII.4-2006(+87A.1), and GII.4-2006, though with differences in the overall BT50 values. While the human outbreak sera were able to detect significant differences in BT50 values between GII.4-1987 and GII.2006, this sera was unable to distinguish significant differences at the level of a single varying epitope. However, the overall trend showed that the human outbreak sera blocked GII.4-2006(+87A.1) more efficiently than GII.4-2006 at every dilution point. This suggests that epitope A likely contributes to the human blockade response in GII.4-1987 but that additional human serum samples are needed to dissect the overall contribution of an individual epitope. The differences in blockade between mouse and human sera may be due to varied NoV exposure histories from the human serum donors. Ogata et al. found that previous influenza vaccination resulted in greater levels of neutralizing antibodies against the newly emergent human H5N2 avian influenza virus (35). In addition, Lee et al. found that previous exposure to pre-1957 H1N1 strains was able to provide some cross-neutralizing protection from the 2009 H1N1 strain but not from a 2007 strain (27). This suggests that in some cases, previous exposure history may provide subtle to substantial levels of protection from related but distinct viral strains on an individual level. Since GII.4 NoVs and influenza exhibit similar evolutionary dynamics, previous exposure to NoV may provide protection from specific strains but not others. Our data provide evidence that epitope A accounts for up to 25% of the GII.4-1987 blockade response and suggests that there are additional blockade epitopes for GII.4-1987.
Our approach of using bioinformatics predictions to identify putative GII.4 NoV epitopes coupled with using our panel of chimeric and parental VLPs, time-ordered mouse MAbs, and synthetic CHOs in ELISA-based assays to validate our predictions offers a powerful method to elucidate the mechanisms driving GII.4 NoV evolution in the absence of a cell culture or small-animal model. Despite evidence that preferential HBGA binding patterns and antigenicity in GII.4 NoVs change over time, results among different labs vary and are difficult to directly compare due to differences that may exist among VLPs, P particles, and P dimers, differences in protein concentrations used in ELISA-based assays, and lot number differences in the reactivity of commercial biotinylated HBGAs. Variation in CHO binding levels among assays in this study is due to the differing reactivity among batches of commercially available synthetic biotinylated CHOs; however, due to the low supply of these reagents, this is unavoidable without altering the amount of CHO used in each experiment. However, it is clear that mutations outside the conserved CHO binding residues contribute to the antigenic and HBGA binding differences apparent over time. Future work will seek to validate our other predicted epitopes by expanding the epitopes to include surrounding amino acids and by building and characterizing additional chimeras in the GII.4-2002 Farmington Hills background.
GII.4 NoVs are significant human pathogens that cause considerable morbidity and mortality worldwide. The identification of highly significant, varying antigenic epitopes that influence VLP-ligand interaction provides important new insights into vaccine design and the development of therapeutics that target norovirus virions. For example, our data predict that vaccines derived from GII.4-1987(+06A) or GII.4-2006(+87A) should greatly broaden the breadth of the cross-antigenic and cross-blockade response against time-ordered GII.4 NoV. Our data predict that GII.4-2006(+09A) vaccines would induce stronger blockade responses against contemporary GII.4 strains than GII.4-2006 VLP vaccines. These experiments are under way. Finally, we anticipate that a full understanding of the varying antigenic and blockade epitopes of GII.4 NoV may not only help to predict the emergence of new epidemic strains but also simultaneously identify key reformulations in vaccine design that will protect the public health against contemporary and emerging epidemic strains of the future.
ACKNOWLEDGMENTS
We thank Victoria Madden and C. Robert Bagnell, Jr., of Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, University of North Carolina—Chapel Hill, for expert technical support.
This work was supported by grant AI056351 from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (to R.S.B.), and a Gillings Innovation Laboratory award from the UNC Gillings School of Global Public Health (to E.F.D.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Allen DJ, Gray JJ, Gallimore CI, Xerry J, Iturriza-Gomara M. 2008. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One 3:e1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allen DJ, et al. 2009. Characterisation of a GII-4 norovirus variant-specific surface-exposed site involved in antibody binding. Virol. J. 6:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atmar RL, Estes MK. 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. North Am. 35:275–290, viii [DOI] [PubMed] [Google Scholar]
- 4. Baric RS, et al. 2002. Expression and self-assembly of Norwalk virus capsid protein from Venezuelan equine encephalitis virus replicons. J. Virol. 76:3023–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bok K, et al. 2009. Evolutionary dynamics of GII. 4 noroviruses over a 34-year period. J. Virol. 83:11890–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bok K, et al. 2011. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc. Natl. Acad. Sci. U. S. A. 108:325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bush RM, Fitch WM, Bender CA, Cox NJ. 1999. Positive selection on the H3 hemagglutinin gene of human influenza virus A. Mol. Biol. Evol. 16:1457–1465 [DOI] [PubMed] [Google Scholar]
- 8. Cannon JL, et al. 2009. Herd immunity to GII.4 noroviruses is supported by outbreak patient sera. J. Virol. 83:5363–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao S, et al. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427 [DOI] [PubMed] [Google Scholar]
- 11. Chen Y, et al. 2011. Crystallography of a lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens. PLoS Pathog. 7:e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chenna R, et al. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clarke IN, Lambden PR. 1997. The molecular biology of caliciviruses. J. Gen. Virol. 78(Pt. 2):291–301 [DOI] [PubMed] [Google Scholar]
- 14. de Rougemont A, et al. 2011. Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens. J. Virol. 85:4057–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. 2008. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 225:190–211 [DOI] [PubMed] [Google Scholar]
- 16. Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. 2010. Viral shape-shifting: norovirus evasion of the human immune system. Nat. Rev. Microbiol. 8:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El-Kamary SS, et al. 2010. Adjuvanted intranasal Norwalk virus-like particle vaccine elicits antibodies and antibody-secreting cells that express homing receptors for mucosal and peripheral lymphoid tissues. J. Infect. Dis. 202:1649–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferguson NM, Galvani AP, Bush RM. 2003. Ecological and immunological determinants of influenza evolution. Nature 422:428–433 [DOI] [PubMed] [Google Scholar]
- 19. Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hager-Braun C, et al. 2010. Characterization of a discontinuous epitope of the HIV envelope protein gp120 recognized by a human monoclonal antibody using chemical modification and mass spectrometric analysis. J. Am. Soc. Mass Spectrom. 21:1687–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harrington PR, Lindesmith L, Yount B, Moe CL, Baric RS. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 76:12335–12343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang X, Wang M, Graham DY, Estes MK. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnston CP, et al. 2007. Outbreak management and implications of a nosocomial norovirus outbreak. Clin. Infect. Dis. 45:534–540 [DOI] [PubMed] [Google Scholar]
- 24. Koelle K, Cobey S, Grenfell B, Pascual M. 2006. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science 314:1898–1903 [DOI] [PubMed] [Google Scholar]
- 25. Koelle K, Ratmann O, Rasmussen DA, Pasour V, Mattingly J. 2011. A dimensionless number for understanding the evolutionary dynamics of antigenically variable RNA viruses. Proc. Biol. Sci. 278:3723–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee BY, et al. 2011. Economic value of norovirus outbreak control measures in healthcare settings. Clin. Microbiol. Infect. 17:640–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee VJ, et al. 2010. Inactivated trivalent seasonal influenza vaccine induces limited cross-reactive neutralizing antibody responses against 2009 pandemic and 1934 PR8 H1N1 strains. Vaccine 28:6852–6857 [DOI] [PubMed] [Google Scholar]
- 28. Lindesmith LC, Donaldson EF, Baric RS. 2011. Norovirus GII. 4 strain antigenic variation. J. Virol. 85:231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lindesmith LC, et al. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. LoBue AD, Lindesmith LC, Baric RS. 2010. Identification of cross-reactive norovirus CD4+ T cell epitopes. J. Virol. 84:8530–8538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lochridge VP, Jutila KL, Graff JW, Hardy ME. 2005. Epitopes in the P2 domain of norovirus VP1 recognized by monoclonal antibodies that block cell interactions. J. Gen. Virol. 86:2799–2806 [DOI] [PubMed] [Google Scholar]
- 32. Mattner F, et al. 2006. Risk groups for clinical complications of norovirus infections: an outbreak investigation. Clin. Microbiol. Infect. 12:69–74 [DOI] [PubMed] [Google Scholar]
- 33. Mead PS, et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murata T, et al. 2007. Prolonged norovirus shedding in infants ≤6 months of age with gastroenteritis. Pediatr. Infect. Dis. J. 26:46–49 [DOI] [PubMed] [Google Scholar]
- 35. Ogata T, et al. 2008. Human H5N2 avian influenza infection in Japan and the factors associated with high H5N2-neutralizing antibody titer. J. Epidemiol. 18:160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okada M, Tanaka T, Oseto M, Takeda N, Shinozaki K. 2006. Genetic analysis of noroviruses associated with fatalities in healthcare facilities. Arch. Virol. 151:1635–1641 [DOI] [PubMed] [Google Scholar]
- 37. Patel MM, et al. 2008. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14:1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prasad BV, et al. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290 [DOI] [PubMed] [Google Scholar]
- 39. Reeck A, et al. 2010. Serological correlate of protection against norovirus-induced gastroenteritis. J. Infect. Dis. 202:1212–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 41. Shanker S, et al. 2011. Structural analysis of histo-blood group antigen binding specificity in a norovirus GII.4 epidemic variant: implications for epochal evolution. J. Virol. 85:8635–8645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Siebenga JJ, et al. 2010. Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants. PLoS Pathog. 6:e1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siebenga JJ, et al. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932–9941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Souza M, Costantini V, Azevedo MS, Saif LJ. 2007. A human norovirus-like particle vaccine adjuvanted with ISCOM or mLT induces cytokine and antibody responses and protection to the homologous GII.4 human norovirus in a gnotobiotic pig disease model. Vaccine 25:8448–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan M, et al. 2008. Elucidation of strain-specific interaction of a GII-4 norovirus with HBGA receptors by site-directed mutagenesis study. Virology 379:324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vinje J, et al. 2000. Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses.” Arch. Virol. 145:223–241 [DOI] [PubMed] [Google Scholar]
- 47. Vinje J, Koopmans MP. 2000. Simultaneous detection and genotyping of “Norwalk-like viruses” by oligonucleotide array in a reverse line blot hybridization format. J. Clin. Microbiol. 38:2595–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang Y, et al. 2010. Genetic and phenotypic characterization of GII-4 noroviruses that circulated during 1987 to 2008. J. Virol. 84:9595–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yewdell JW. 2010. Monoclonal antibodies specific for discontinuous epitopes direct refolding of influenza A virus hemagglutinin. Mol. Immunol. 47:1132–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zolla-Pazner S. 2004. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 4:199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]