Abstract
The P9-1 protein of Rice black streaked dwarf virus accumulates in viroplasm inclusions, which are structures that appear to play an important role in viral morphogenesis and are commonly found in viruses in the family Reoviridae. Crystallographic analysis of P9-1 revealed structural features that allow the protein to form dimers via hydrophobic interactions. Each dimer has carboxy-terminal regions, resembling arms, that extend to neighboring dimers, thereby uniting sets of four dimers via lateral hydrophobic interactions, to yield cylindrical octamers. The importance of these regions for the formation of viroplasm-like inclusions was confirmed by the absence of such inclusions when P9-1 was expressed without its carboxy-terminal arm. The octamers are vertically elongated cylinders resembling the structures formed by NSP2 of rotavirus, even though there are no significant similarities between the respective primary and secondary structures of the two proteins. Our results suggest that an octameric structure with an internal pore might be important for the functioning of the respective proteins in the events that occur in the viroplasm, which might include viral morphogenesis.
INTRODUCTION
Rice black streaked dwarf virus (RBSDV) is a member of the genus Fijivirus in the family Reoviridae, multiplying both in plants and in an invertebrate insect vector. RBSDV is transmitted propagatively to rice, maize, barley, and wheat by the small brown planthopper (Laodelphax striatellus Fallén). RBSDV has a double-shelled, icosahedral capsid of approximately 75 to 80 nm in diameter and contains 10 segments of double-stranded RNA (dsRNA) (27). It carries six putative structural proteins, namely, P1, P2, P3, P4, P8, and P10. The capsid of RBSDV, which encloses a putative RNA-dependent RNA polymerase (P1) and 10 segments of dsRNA, appears to consist of at least the following five different proteins: P2 and P4, which are a putative capsid shell protein and B spike protein, respectively (52); P3, which is a putative capping enzyme (37); and P8 and P10, which are core and major outer capsid proteins, respectively (17, 23). The protein encoded by S5 has no significant homology to any known protein (52). Other segments of dsRNA encode putative nonstructural proteins, namely, P6, P7-1, P7-2, P9-1, and P9-2. P6 functions as a viral RNA-silencing suppressor (53). The gene encoding RBSDV S7 is 2,193 nucleotides (nt) long and contains two long, nonoverlapping open reading frames (ORFs) (S7-1 and S7-2), which encode polypeptides of 41.2 kDa (P7-1) and 36.4 kDa (P7-2), respectively (3). P7-2 has not been detected in RBSDV-infected plants and insects, while P7-1 has been characterized as a putative tubular protein (17). The S9 gene is approximately 1,900 nt long and contains two long, nonoverlapping ORFs (S9-1 and S9-2), which encode polypeptides of 39.9 kDa (P9-1) and 24.2 kDa (P9-2), respectively. There is no apparent similarity between the amino acid sequences of P9-1 and P9-2. P9-2 is not detected in RBSDV-infected plants and insects, while P9-1 accumulates in intracellular viroplasms (Vps), functioning as a probable component of Vps and therefore playing an important role in the formation of Vps and in viral morphogenesis (17). Molecules of P9-1 have the intrinsic ability to interact with one another in vitro and in vivo. Furthermore, P9-1 forms large, discrete, Vp-like structures in the absence of viral infection or of other RBSDV proteins when expressed transiently in Arabidopsis protoplasts (51).
At early times after infection with RBSDV, distinctive structures appear throughout the cytoplasm of infected cells. These cytoplasmic structures are known as viral factories (VFs) in the case of mammalian orthoreoviruses (MRVs) and avian orthoreoviruses (ARVs) (34, 47), as viral inclusion bodies (VIBs) in the case of orbivirus (11), and as Vps in the case of phytoreoviruses and rotaviruses (26, 49). In each case, these structures contain many viral proteins, particles, and dsRNAs, and they appear to be the sites of replication and packaging into progeny particles of viral RNA.
One or two of the nonstructural proteins of each virus are required for the formation of these cytoplasmic structures. In the case of MRVs and ARVs, the nonstructural protein μNS expressed in the absence of other viral proteins formed structures that resembled the VFs observed in infected cells (5, 28, 47). Likewise, orbiviruses such as Bluetongue virus (BTV) and Epizootic hemorrhagic disease virus encode a single nonstructural protein, NS2, that forms VIB-like structures when expressed in the absence of other viral proteins (6, 45, 46). Similarly, the nonstructural protein Pns12 of Rice dwarf virus (RDV) forms Vp-like structures (VLSs) when expressed alone (49). In contrast, rotaviruses encode two nonstructural proteins, NSP2 and NSP5, which, under most circumstances, must be coexpressed for formation of VLSs (12, 14). In the case of animal viruses in the family Reoviridae, reductions in levels of expression of genes for viral components by small interfering RNAs (siRNAs) directed against the respective viral genes interfere with the viral replication cycle and the production of infectious virions (21, 24, 36). Transgenic rice plants designed to thwart the expression of Pns12, a major component of the viroplasm matrix, are strongly resistant to infection by RDV (35).
In the case of rotaviruses, NSP2 forms Vps in the presence of NSP5 (12, 14), and NSP2 contributes to the binding of the innermost capsid protein and the RNA-dependent RNA polymerase (20, 33); to binding of single-stranded RNA (ssRNA); and to helix-destabilizing (40, 42), nucleotide triphosphatase (NTPase), and 5′-RNA triphosphatase (RTPase) activities (48). Full-length NSP2 of rotavirus has been analyzed by X-ray crystallography at 2.6-Å resolution (18). NSP2 consists of two domains, and eight monomers assemble to form an octamer. In addition, a recent study demonstrated the hydrolytic mechanism for nucleotide binding by NSP2 on the basis of structural information at the atomic level (22). Moreover, the regulation of the interaction of NSP2 with RNA by NSP5 was clarified by single-particle analysis of the octamer of NSP2 by electron microscopy (19). The crystallographic structure of the amino-terminal domain of NS2 of BTV indicated that this protein forms a large oligomer (7).
Thus, structural analyses of a variety of proteins have provided functional information about the macromolecules involved in viral replication. However, only a few components of Vps have been studied in detail at the molecular level. We present here the atomic structure of the Vp matrix protein P9-1 of RBSDV at 3.0-Å resolution, as determined by X-ray crystallography. This is the first report, to our knowledge, of the crystal structure of a protein that is important for Vp formation of a plant reovirus. The protein forms channel-like octamers, and the carboxy-terminal region of each monomer appears to play a significant role in oligomerization.
MATERIALS AND METHODS
Construction of P9-1 and P9-1ΔC expression plasmids.
DNA fragments encoding full-length P9-1 and a carboxy-terminal (amino acids 346 through 368) deletion mutant (P9-1ΔC) were amplified by PCR with KOD-Plus polymerase (Toyobo, Osaka, Japan), with viral cDNA as the template, the forward primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAATGGCAGACCAAGAGCGGA-3′ (the initiation codon is double underlined), and the reverse primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCAAACGTCCAATTTCAAGGAA-3′ (the termination codon is double underlined) for the wild-type sequence or 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTAGAATTTACGATATCTCTTGATGT-3′ (the termination codon is double underlined) for the P9-1ΔC sequence. The gene from RBSDV was supplemented with the site-specific recombination sequence (underlined) of bacteriophage lambda. The purified PCR products were cloned by use of the Gateway system (Invitrogen, La Jolla, CA) as described elsewhere (16). PCR products were subcloned into attP sites of pDONR-201 (Invitrogen), yielding entry clones pENTR/P9-1 and pENTR/P9-1ΔC. The DNAs of the entry clones were used to transfer sequences into the destination vector pDEST-17 (Invitrogen), yielding pDEST-17/P9-1 and pDEST-17/P9-1ΔC, respectively. The pDEST-17 vector introduces an amino-terminal hexahistidine tag (His tag) that is suitable for Ni-nitrilotriacetic acid (Ni-NTA) affinity purification. The resulting constructs contained the entire coding sequence of P9-1 or the deletion sequence with an amino-terminal His tag sequence and some inserted amino acids of vector origin (1MSYYHHHHHHLGSTSLYKKAG21). Recombinant plasmids were introduced into Escherichia coli DH5α for cloning and sequencing of DNA. Nucleotide sequences were confirmed by direct DNA sequence analysis.
Expression and purification of P9-1.
The native protein was expressed in E. coli BL21-AI (Invitrogen). BL21-AI cells were transformed with the expression plasmid and grown at 37°C overnight on Luria-Bertani (LB) agar plates with 50 μg/ml ampicillin and 15 μg/ml tetracycline. A single colony was inoculated and grown overnight in LB broth with 50 μg/ml ampicillin and 15 μg/ml tetracycline. The overnight culture was inoculated into LB broth and shaken at 37°C until the optical density at 600 nm (OD600) reached 0.6 to 0.9. Expression of P9-1 was induced by 0.1% l-arabinose and incubation for 4 h at 20°C. Cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C and then stored at −20°C. Selenomethionine-substituted proteins were expressed in E. coli B834(DE3) (Novagen, Madison, WI). B834 cells were transformed with an expression plasmid and grown at 37°C overnight on LB agar plates with 50 μg/ml ampicillin. A single colony was inoculated and cultured overnight in LB broth with 50 μg/ml ampicillin. The cells from the overnight culture were washed with minimal medium, inoculated into minimal medium that contained ampicillin and selenomethionine (2.5 mg/ml [final concentration]), and shaken at 37°C to an OD600 of 0.3. Expression was induced by 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubation for 12 h at 20°C. Cells were harvested by centrifugation at 8,000 × g for 10 min at 4°C and then stored at −20°C. Frozen cell pellets from a 1-liter culture were thawed on ice and then resuspended in 50 ml of lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 20 mM imidazole, pH 7.4). The suspension was sonicated, and the resulting lysate was centrifuged at 15,000 × g for 15 min. The final supernatant was added to 2 ml of Ni-NTA resin (Qiagen, Hilden, Germany) that had been preequilibrated with lysis buffer. After incubation for 1 h, the resin was loaded onto an empty column and washed with 200 ml of lysis buffer. P9-1 that had bound to the resin was eluted with elution buffer (50 mM Tris-HCl, 150 mM NaCl, 500 mM imidazole, pH 7.4). Fractions containing P9-1 were identified by SDS-PAGE, pooled, dialyzed overnight at 4°C against crystallization buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM dithiothreitol [DTT], pH 7.4), and concentrated to a final concentration of 10 mg/ml with Vivaspin 10,000-cutoff filters (Sartorius, Goettingen, Germany) and a Microcon YM-10 system (Millipore, Billerica, MA).
SEC.
Size exclusion chromatography (SEC) was performed with a Superdex 200 pg column (GE Healthcare, Buckinghamshire, England). The column was equilibrated with 10 mM Tris-HCl (pH 7.4) plus 150 mM NaCl. SEC was performed at a flow rate of 0.5 ml/min. Elution was monitored in terms of absorbance at 280 nm. The following standards were used to calibrate the column: blue dextran (to define the excluded volume), ferritin, catalase, conalbumin, ovalbumin, and carbonic anhydrase (GE Healthcare). Distribution coefficients (Kd) were calculated from the equation Kd = (Ve − V0)/Vi, where Ve is the elution volume for the protein of interest, V0 is the void volume, and Vi is the included volume.
Crystallization of P9-1.
Initial crystallization trials were performed by the hanging drop method with commercially available kits, namely, Crystal Screen, Crystal Screen II, SaltRX, Natrix, and PEG/Ion Screen kits (Hampton Research, Aliso Viejo, CA), at 22°C. The crystallization drops contained 1 μl of a solution of P9-1 at various concentrations mixed with 1 μl of screening solution and were set up for vapor diffusion against 500 μl of screening solution in 24-well VDX plates (Hampton Research). Crystals appeared after several hours under condition 11 of the Crystal Screen kit, which contained 100 mM trisodium citrate (pH 5.6) and 1 M ammonium dihydrogen phosphate monohydrate. Crystals also appeared after 24 h under condition 47 of the Crystal Screen kit and condition 73 of the PEG/Ion Screen kit, namely, in the presence of 100 mM sodium acetate (pH 4.6) plus 2 M ammonium sulfate and 100 mM sodium acetate (pH 4.6) plus 1 M magnesium sulfate, respectively. Refinement of conditions yielded the optimized composition of the reservoir solution, namely, 100 mM sodium acetate (pH 4.6) plus 0.8 M magnesium sulfate. The crystals reached maximum dimensions of 0.2 mm by 0.2 mm by 0.1 mm in 2 to 3 days. For collection of data, crystals were flash cooled to −173°C after soaking in a cryoprotectant solution, which consisted of the reservoir solution supplemented with 35% (vol/vol) glycerol.
Data collection and determination of structure.
All data sets were collected on beamline BL44XU at SPring-8 (Hyogo, Japan) and beamline AR-NW12A at the Photon Factory (KEK, Tsukuba, Japan). Measurements were performed at −173°C, with λ values of 1.00000 Å (native), 0.97907 Å (Se peak), 0.97931 Å (Se edge), and 0.96408 Å (high-remote). Complete data sets were collected through contiguous rotation ranges at a given wavelength before proceeding to the next wavelength. The data sets were integrated, scaled, and merged with the programs DENZO and SCALEPACK, as implemented in the HKL2000 program package (31). All crystals belonged to space group P4212, with two molecules per asymmetric unit. The structure of P9-1 was solved by the multiwavelength anomalous diffraction (MAD) method, with selenomethionyl P9-1 crystals. A total of six expected selenium sites were located with the program SOLVE (44), and an initial phasing density modification and initial model building were performed with the program RESOLVE (43). Further density modification was performed with noncrystallographic symmetry averaging with the program DM (8). The remainder of the model was built manually with the program Coot (13). The model was refined using the programs REFMAC 5 (30) and PHENIX (1, 2), with 5% of the data set aside as a free set. The final refined model, with an R factor of 24.26% and an Rfree value of 29.28%, included residues 25 through 368 of P9-1, with some missing internal fragments. The quality of the final model was checked with the program Molprobity (9). Data collection and refinement statistics are given in Table 1. All diagrams were prepared with the programs UCSF Chimera (32) and PyMOL (10). Electrostatic surface charges were calculated by APBS (4).
Table 1.
Data collection and refinement statistics
| Statistic | Value or description for crystal |
|||
|---|---|---|---|---|
| Native | Se-Met derivative |
|||
| Peak | Edge | Remote | ||
| Data collection statistics | ||||
| Beamline | SPring-8 BL44XU | PF AR-NW12A | ||
| Wavelength (Å) | 0.9000 | 0.9790 | 0.9793 | 0.9640 |
| Resolution limits (Å)a | 44.6-3.00 (3.05-3.00) | 150.0-3.55 (3.68-3.55) | ||
| Space group | P4212 | P4212 | ||
| Unit cell dimensions (Å) | a = b = 127.3, c = 143.2 | a = b = 126.6, c = 143.5 | ||
| No. of reflections | 341,495 | 208,603 | 104,483 | 105,545 |
| No. of unique reflectionsa | 24,043 (1,179) | 14,666 (1,456) | 14,717 (1,429) | 14,844 (1,441) |
| Redundancya | 14.2 (14.3) | 14.2 (14.2) | 7.1 (7.2) | 7.1 (7.2) |
| Completeness (%)a | 99.3 (100.0) | 99.9 (100.0) | 99.8 (100.0) | 99.8 (100.0) |
| <I>/<s(I)>a | 58.5 (5.20) | 49.4 (6.2) | 34.0 (3.8) | 32.6 (3.3) |
| Rmerge (%)a,b | 4.5 (41.2) | 4.8 (32.5) | 4.1 (34.8) | 4.1 (39.8) |
| No. of molecules per asymmetric unit | 2 | 2 | ||
| Refinement statistics | ||||
| Resolution limits (Å) | 42.18-3.00 | |||
| Rfactor (%)c | 24.26 | |||
| Rfree (%)d | 29.28 | |||
| No. of atoms | 4,475 | |||
| Average B factor | 92.80 | |||
| RMSD bond length | 0.016 | |||
| RMSD bond angle | 1.636 | |||
| Ramachandran statistics | ||||
| Favored (%) | 92.4 | |||
| Allowed (%) | 6.5 | |||
| Outlier (%) | 0.2 | |||
Numbers in parentheses refer to the appropriate outer shell.
Rmerge = 100 × [Shkl Si|I(hkl;i) − <I(hkl)> |/Shkl SiI(hkl;i)], where I(hkl;i) is the intensity of an individual measurement of a reflection and <I(hkl)> is the average intensity of that reflection.
Rfactor = 100 × (Shkl||Fobs| − |Fcalc||/Shkl |Fobs|), where |Fobs| and |Fcalc| are the observed and calculated structure factor amplitudes.
Rfree equals the R factor of the test set (5% of the data removed prior to refinement).
Baculoviral expression of P9-1 and the carboxy-terminal deletion mutant.
We used a baculovirus system for expression of P9-1 and P9-1ΔC, as described previously (29, 50). The DNAs of entry clones were used to transfer sequences that encoded P9-1 and P9-1ΔC into the destination vector pDEST-8 (Invitrogen), to yield pDEST-8/P9-1 and pDEST-8/P9-1ΔC, respectively. Recombinant pDEST-8/P9-1 and pDEST-8/P9-1ΔC were then introduced into E. coli DH10Bac (Invitrogen) for transposition into the bacmid. The recombinant bacmid was isolated and used to transfect Spodoptera frugiperda (Sf9) cells in the presence of CellFECTIN (Invitrogen), used according to the manufacturer's instructions. Then, 72 h after transfection, Sf9 cells were collected and the expression of proteins was examined by immunoblotting with antibodies specific for P9-1. For cytological observations, Sf9 cells were seeded, the day before transfection, at a density of 1.5 × 104 cells cm−2 on glass coverslips (15 mm in diameter). The culture medium was removed, and cells were infected with the recombinant baculovirus. Cells were then incubated for 12 to 72 h at 37°C before being processed for immunofluorescence microscopy.
Immunofluorescence staining.
Rabbit polyclonal antiserum against P9-1 was prepared as described previously (38, 39, 54). IgG was isolated from the polyclonal antiserum on a protein A Sepharose affinity column. Eluted IgG was dialyzed exhaustively against phosphate-buffered saline (PBS). IgG was then conjugated directly to fluorescein isothiocyanate (FITC; Invitrogen) according to the manufacturer's instructions. At various times after inoculation, Sf9 cells grown on glass coverslips were washed with PBS and fixed for 30 min at room temperature in 2% paraformaldehyde. Cells were washed with PBS and permeabilized in PBS that contained 1% bovine serum albumin (BSA) and 0.1% Triton X-100. After fixation, the cells were washed with PBS. Cells were then incubated with a 100-fold-diluted solution of the conjugated IgG for 1.0 to 1.5 h at 37°C. Coverslips were washed with PBS and then mounted on glass slides with ProLong antifade solution (Invitrogen). Cells were visualized under a confocal laser scanning microscope (LSM 510; Zeiss, Jena, Germany). Data were acquired with an oil immersion lens (663×). The green (FITC) fluorochrome was detected with narrow-band filter sets and argon laser lines (488 nm). Data were collected with the single-tracking feature on the microscope to avoid cross talk. All measurements, including cell depth calculations and scale bar lengths, were calculated with LSM 510 software (Zeiss).
Protein structure accession number.
The coordinates and structure factors of P9-1 have been deposited in the Protein Data Bank under accession code 3VJJ.
RESULTS
Purification, crystallization, and determination of the structure of P9-1.
We expressed the nonstructural protein P9-1 of RBSDV in E. coli as an amino-terminally His-tagged fusion protein. We purified P9-1 with 368 amino acid residues by Ni-NTA affinity chromatography, as confirmed by a single band of the protein after SDS-PAGE (Fig. 1). Purified P9-1 was crystallized with tetragonal space group P4212 and unit cell dimensions a = b = 127.3 Å and c = 143.2 Å (Table 1). The structure of P9-1 was solved by the MAD method, with native and selenomethionyl derivative data sets. The structure was refined at 3.0-Å resolution, with final R and Rfree factors of 24.26 and 29.28, respectively (Table 1). The crystallographic asymmetric unit contained two P9-1 monomers. Two molecules in the asymmetric unit were related by a noncrystallographic 2-fold axis that was almost perpendicular to the crystallographic 4-fold axis or the c axis of the crystal. The final, refined atomic model contained 278 of a total of 368 amino acid residues in two polypeptide chains of P9-1. The missing regions were the amino terminus (residues 1 through 24), including the His tag; a flexible loop (residues 129 through 164); and a β-turn (residues 42 through 62 and 314 through 324).
Fig 1.
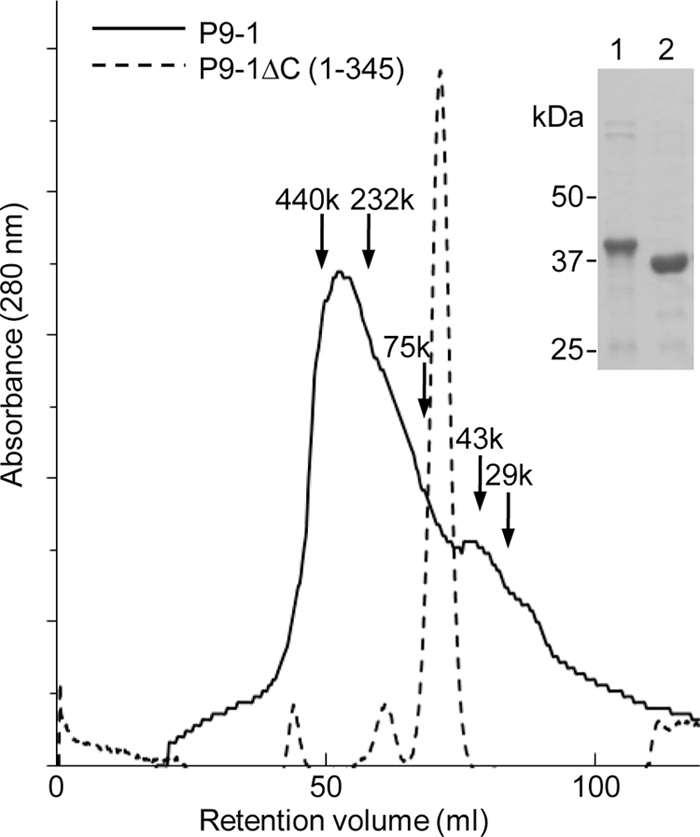
SEC and analysis by SDS-PAGE of P9-1 and P9-1ΔC. P9-1 and P9-1ΔC were purified by Ni-NTA affinity chromatography and then analyzed by SDS-PAGE and staining with Coomassie blue. Lane 1, purified P9-1; lane 2, purified P9-1ΔC. SEC was performed as described in the text. Solid line, purified P9-1; dotted line, purified P9-1ΔC. Each number with an arrow indicates the position and molecular mass of the respective marker protein (in kDa).
Atomic structure of P9-1.
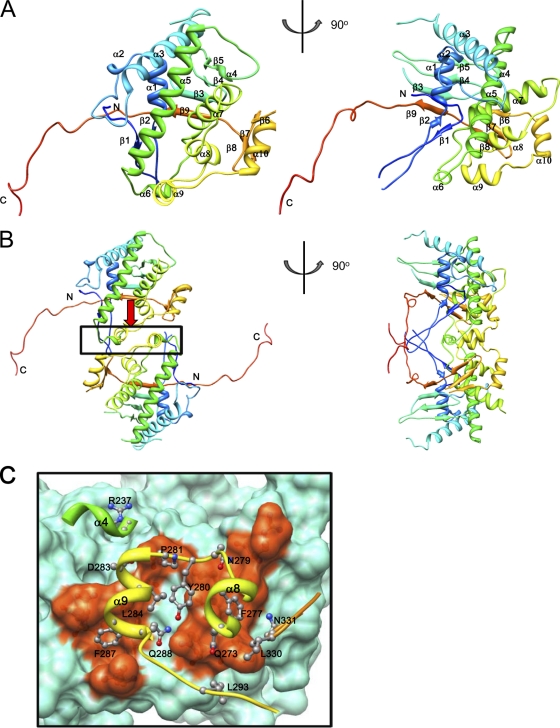
The overall structure of the P9-1 monomer consisted of 10 α-helices and 9 β-strands (Fig. 2A and 3). The amino-terminal loop (residues 34 through 68) was composed of a two-stranded antiparallel β-sheet (β1 and β2). This sheet interacted with β1 and β2 in a neighboring molecule to form a four-stranded antiparallel β-sheet. The α-helices α1 through α9 were gathered at the center of the molecule around a long α5 helix (residues 204 through 232) of 40 Å, forming a helical bundle. This helical bundle was in a strongly hydrophobic environment. Hydrophobic residues participated in leucine zipper-like interactions with the helical bundle. In particular, leucine residues were gathered in helices α5 through α8, forming a hydrophobic cluster. Thus, two molecules of P9-1 formed a noncrystallographic homodimer. As shown in Fig. 2B, three helices (α4 [residues 192 through 194], α8 [residues 269 through 276], and α9 [residues 281 through 288]) and the loop region (residues 329 through 332) between β7 and β8 contributed to the formation of the dimer. The area of surface contact in this region was 760 Å2 and was relatively small. Many hydrophobic residues were concentrated at the dimer interface, as shown in Fig. 2C, suggesting that the dimer was formed mainly via hydrophobic interactions. Together, β3, β4, β5, and β9 formed an antiparallel β-sheet. This region was covered by the helical bundle to form the internal structure of the P9-1 octamer, as described below. Similarly, β6, β7, and β8 formed an antiparallel β-sheet. The carboxyl terminus (C arm; residues 345 through 368) extended outwards toward the neighboring monomer. The length of the C arms was approximately 50 Å, and they extended from the main body of the dimer to each side, as shown in Fig. 2B. These extended arms interacted with neighboring dimers and appeared to be important for the formation of octamers (see below).
Fig 2.
Overall structure of P9-1. The ribbon diagrams show the P9-1 asymmetric monomer (A) and dimer (B). The diagrams in the right column were generated after rotation of the left images by 90°. Ribbon representations of P9-1 show polypeptide chains in rainbow colors from blue (amino-terminal region) to red (carboxy-terminal region). The amino and carboxyl termini are indicated. Secondary structural elements are labeled sequentially. (C) Dimer interface of P9-1. The upper molecule is shown by a ribbon, and intermolecular interacting side chains are shown as ball-and-stick models. The lower molecule is shown as a molecular surface model, and regions of intermolecular interaction are colored orange. Numbers of some residues and secondary structural elements are labeled to facilitate identification.
Fig 3.
Alignment of the amino acid sequences of P9-1 of RBSDV, NSP2 of rotavirus, and NS2 of BTV. Secondary structural elements are indicated schematically under the sequences, generated by ESPript (15), and are indicated as spirals (α-helices) or arrows (β-sheets). Red characters indicate residues that are similar or identical in the three proteins. Interruptions in sequences of amino acids are indicated with dotted lines. Green characters indicate residues of the amino-terminal His tag sequence and some inserted amino acids of vector origin (1MSYYHHHHHHLGSTSLYKKAG21).
The noncrystallographic symmetry (NCS) of two related subunits (A and B) had similar structures, with a root mean square deviation (RMSD) between the corresponding atoms of 0.862 Å. The RMSD with exclusion of the C-terminal arm was 0.330 Å. However, there were some structural differences between α4 and α5 (residues 196 through 202) and α7 and α8 (residues 262 through 265), in the loop regions of the respective subunits. These regions were disordered in the B subunit, while in the A subunit they were ordered. Moreover, the RMSD of the C arm, which interacted with neighboring dimers, was >3 Å, a large part of which might be due to the flexibility of the C arm, allowing formation of different types of oligomers.
Oligomer formation and the role of the C arm.
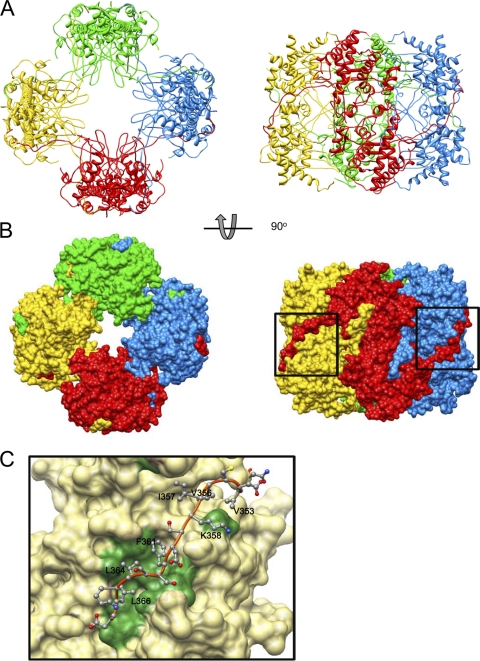
The arrangement of eight molecules of P9-1 in each crystallographic asymmetric unit suggests that they form an octamer (Fig. 4). Asymmetric dimers interact with neighbor dimers that are related by a 4-fold axis. Each octamer resembles a cylinder, with the dimensions 100 Å by 100 Å by 85 Å. To examine whether P9-1 is able to polymerize in solution, we performed gel filtration chromatography. As shown in Fig. 1, P9-1 eluted as a broad peak, with a main peak that corresponded to a molecular mass of 343 kDa, the mass of an octamer of P9-1. However, the shape of the peak indicates the presence of multiple species of oligomer. The long tail on the low-mass side of the peak is consistent with an equilibrium among various oligomers. However, the extensive interactions between dimers in the crystallographic octamer suggest that the exchange of subunits out of the octamer might be very slow.
Fig 4.
Structure of the P9-1 octamer. (A) Ribbon diagram of the P9-1 octamer. (B) Molecular surface model. The diagrams in the right column were generated after rotation of the left images by 90°. Each dimer is shown in one of four colors. (C) Dimer-dimer interface. The C arm of P9-1 is shown by a ribbon, and side chains involved in intermolecular interaction are shown as ball-and-stick models. The neighboring dimer is shown as a molecular surface model, and regions of intermolecular interaction are colored green. Numbers of some residues are labeled to facilitate identification.
As mentioned above, the loop at the amino terminus contributed to dimer formation with the C arm and seemed to contribute to binding to neighboring dimers and the eventual formation of octamers. Indeed, the surface of each octamer suggests that each C arm interacts with the adjoining dimer, with each holding the other at the “shoulder.” When we examined the contacts between the C arm and the adjoining dimer in detail, we found, as shown in Fig. 4C, that hydrophobic residues were “lined up” along the C arm and gathered on the surface of the contact region of the adjacent dimer. The area of surface contact between dimers was 2,204 Å2. The area occupied by the C arm (residues 345 through 367) was 1,875 Å2, accounting for 85% of the total area of contact between neighboring dimers.
The results described above suggest that the C arm might play an important role in the formation of the octamer. To confirm this hypothesis, we prepared a C arm deletion mutant (P9-1ΔC) and determined its mass by SEC. As shown in Fig. 1, P9-1ΔC eluted as a sharp single peak that corresponded to a molecular mass of 73.5 kDa, the expected mass of a dimer of this truncated protein. These results confirmed that the C arm plays an important role in the formation of higher-order oligomers, including octamers.
P9-1 can form Vp-like inclusions in vivo, but the C arm deletion mutant cannot.
To determine whether the C arm is also required for the formation of the matrix of viral inclusions in vivo, we inoculated Sf9 cells with a recombinant baculovirus that encoded either P9-1 or P9-1ΔC. Analysis by SDS-PAGE indicated that P9-1 and P9-1ΔC were first detectable at 24 h postinfection (hpi), with increasing levels that reached a maximum at 72 hpi. Immunofluorescence staining of P9-1 in Sf9 cells revealed the formation of Vp-like inclusions within cells (Fig. 5). In contrast, the expression of P9-1ΔC resulted in a diffuse pattern of immunofluorescence and an absence of detectable inclusions, demonstrating that the C arm is necessary for the formation of large inclusions that resemble Vps.
Fig 5.
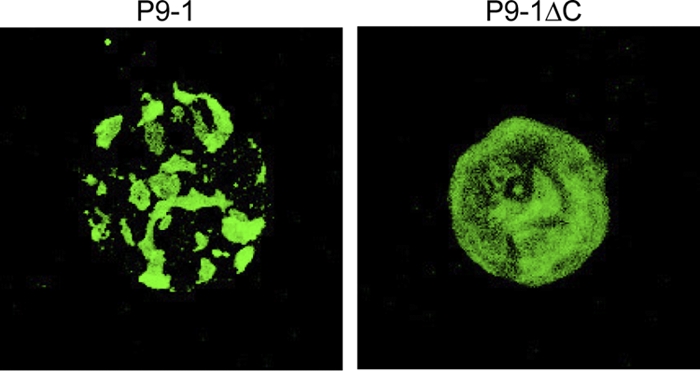
The C arm contributes to formation of the Vp. P9-1 and P9-1ΔC were expressed in Sf9 cells via the baculovirus system. Cells were fixed, immunostained with FITC-conjugated P9-1-specific IgG, and visualized by confocal microscopy.
DISCUSSION
Mechanism of assembly and formation of octamers of P9-1.
Crystallographic data suggested that the assembly of P9-1 oligomers occurred as follows. P9-1 appeared first to form dimers, four of which then united, via their “arms” and “shoulders,” to form a cylindrical octamer. Detailed analysis revealed high concentrations of hydrophobic residues at the various interfaces, suggesting that each dimer formed as a result of hydrophobic interactions. Each dimer extended an arm (C arm) to each side, binding to the neighboring dimer on each side. Hydrophobic forces contributed to this interaction as well as to the formation of dimers. Thus, hydrophobic interactions were effective in the formation of both dimers and octamers. P9-1 contains many hydrophobic residues. A total of 41 residues are leucine (11%), and these residues participate in leucine zipper-like interactions within molecules, such that the helical bundle is stabilized by these interactions. Our various results demonstrate that dimers of P9-1 are the building blocks of octamers and that the C arm plays an important role in the formation of octamers.
Our size exclusion chromatography data, confirming our crystallographic analysis, supported the above-mentioned hypothesis that the C arm plays a key role in the formation of octamers. We observed a shift from polymers, whose major component was octamers, to dimers when carboxy-terminal residues had been deleted from the protein (Fig. 1). Moreover, full-sized P9-1 formed Vp-like inclusions in Sf9 cells, while the C-arm-deleted protein did not (Fig. 5), confirming that the C arm of P9-1 is essential for the formation of large inclusions in vivo.
Construction of Vps.
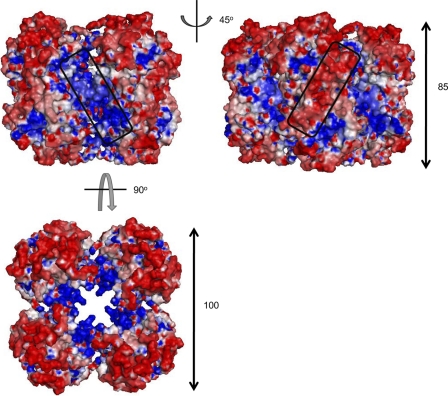
The fact that the expression of full-sized P9-1 in Sf9 cells resulted in the formation of Vp-like inclusions in the absence of any other viral proteins (Fig. 5) suggested that the P9-1 octamer has the intrinsic ability to form larger aggregates. When we analyzed the P9-1 octamer in this regard, we found that the surface charge on the side surface of each octamer consisted of clusters of positive and negative charges rotated by 45°, with negative charges being clearly abundant on the surface and positively charged amino acids being more abundant in the cleft between dimers (Fig. 6). Furthermore, positive charges were concentrated inside the octamer, with negative charges distributed on the outer sides when the octamer was observed from its “upper” surface. These clusters might allow binding to other macromolecules via electrostatic complementarity and might be involved in the formation of larger structures, such as the Vp.
Fig 6.
Surface electrostatic potential on the P9-1 octamer. The side surface of the octamer is shown as a molecular surface model. The diagram in the right column was generated after rotation of the left image by 45°. Positively and negatively charged regions are colored blue and red, respectively. Clusters of positive and negative charge are indicated by boxes. The third diagram (below the first two) was generated after rotation of the left image by 90°.
In preliminary experiments with purified P9-1, we found that the protein has nonspecific RNA-binding ability (data not shown), as reported for the P9-1 protein of Mal de Río Cuarto virus (MRCV) (25), a counterpart of the P9-1 protein of RBSDV. MRCV is in the same genus (Fijivirus), in the family Reoviridae, as RBSDV, and its P9-1 protein is 63% identical to P9-1 of RBSDV at the amino acid level. The interior of the pore of the octamer of P9-1 of RBSDV, in which positively charged clusters are located, might be a nucleic acid-binding site and/or a channel through which nucleic acids can pass. For biological reasons, the interaction of P9-1 with nucleic acid should not be too strong when the nucleic acid is stored inside viral particles. The presence of nucleic acid might neutralize and stabilize the pore of the P9-1 octamer.
Comparison of overall structures of P9-1 and other Vp proteins.
The biochemical properties of NSP2, namely, its RNA-binding, helix-destabilizing, NTPase, RTPase, nucleoside diphosphate (NDP) kinase, and core protein-binding activities, have been studied in detail, and ways in which this protein might function during viral assembly have been proposed. In addition, the structural features of the active sites of the protein have been discussed (41). As denoted above, the P9-1 proteins of RBSDV and MRCV have RNA-binding ability. In addition, P9-1 of MRCV has ATPase activity and is able to form Vp-like structures in Sf9 cells, provided that the carboxy-terminal half of the protein is intact (25). The parallels among NSP2 and the P9-1 proteins of MRCV and RBSDV suggest that the three proteins might have similar functions and might play similar roles during the viral replication cycle.
The NSP2 protein of rotavirus is the only reoviral nonstructural protein for which the X-ray crystallographic structure of the total protein is available (18). This protein, composed of two domains, has NTPase activity, RTPase activity, and NDP kinase-like activity (22, 48). Structural analysis of mutant derivatives and complexes with analogs revealed that two conserved histidine residues (H221 and H225) and three surrounding basic residues (K188, K223, and R227) contribute to its activities (22). These activities appear to occur inside a cleft between the two domains of NSP2 and structurally similar to the clefts in ubiquitous cellular HIT (histidine triad) proteins. The two histidine residues (H256 and H269) and some basic residues (R261, K265, and R300) in the primary structure and their location in the region between helices α7 and α10 in the tertiary structure of P9-1 suggest that P9-1 might have similar properties. However, there are slight differences in their respective locations: the histidine and basic residues mentioned above are located in an α-helix in P9-1, while the HIT-like fold is located in the β-strand in NSP2.
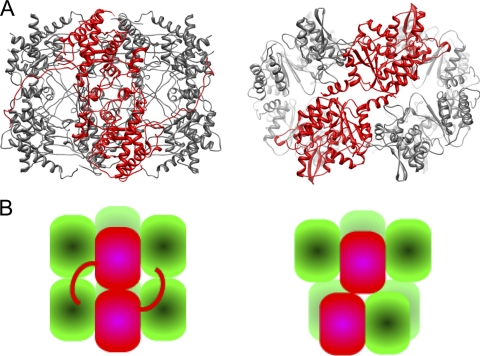
When we examined the structure of P9-1 by using the DALI server, we failed to find a folding motif in P9-1, while NSP2 includes a structure similar to the HIT, as described above. Furthermore, no similarities were apparent when we superimposed the two structures (data not shown). In spite of the differences between the monomeric structures of P9-1 and NSP2, the octameric structures formed by each protein resemble one another (Fig. 7A and B), even though there are no obvious similarities between the respective primary structures (13% identity) and secondary structures. Similarity in terms of octamer formation without significant similarity in terms of primary structure suggests that the structure of each octamer and its interior pore is crucial for the functioning of the respective proteins in the viroplasm, for example, in viral morphogenesis. Other differences between the two proteins are as follows: both total size and pore diameter are greater for NSP2 than for P9-1, namely, the dimensions are 100 Å by 100 Å by 85 Å in P9-1 versus 120 Å by 120 Å by 100 Å in NSP2 in terms of total size and 20 Å in P9-1 versus 35 Å in NSP2 in terms of pore diameter; the octamer of P9-1 has a vertically elongated cylindrical shape, while that of NSP2 is flatter and more doughnut-like; in the P9-1 octamer, the upper tetramer interacts vertically with the lower one, while the upper and lower tetramers are twisted and related via 4-2-2 symmetry in the NSP2 octamer; the amino-terminal region in P9-1 is disordered, whereas the amino-terminal region is part of the structural domain in NSP2; and, finally, side-by-side interactions, such as those involving the C arm of P9-1, are not apparent in the NSP2 octamer. It remains to be determined whether the His tag attached at the amino terminus of P9-1 alters the native structure.
Fig 7.
Comparison of structures of Vp proteins of members of the Reoviridae. (A) Ribbon diagrams of the octamers of P9-1 (left) and NSP2 (right). The Protein Data Bank entry number of rotavirus NSP2 is 1L9V. A dimer is shown in red. In each case in P9-1, the upper subunit of the dimer interacts vertically with the lower one. In contrast, the upper and lower subunits are twisted in the NSP2 dimer. (B) Schematic views of octamers of P9-1 (left) and NSP2 (right). Dimers are shown in red.
ACKNOWLEDGMENTS
We thank the staff of the Structural Biology Research Center of the Photon Factory (Tsukuba, Japan) and the staff of SPring-8 beamline BL44XU (Hyogo, Japan) for the collection of X-ray data.
This study was supported by a Grant-in-Aid for Scientific Research on Priority Areas (Structures of Biological Macromolecular Assemblies) from the Japanese Ministry of Education, Culture, Sports, Science and Technology and by the Program for Promotion of Basic Research Activities for Innovative Biosciences of the Bio-Oriented Technology Research Advancement Institution (BRAIN) (T.O.).
Footnotes
Published ahead of print 9 November 2011
REFERENCES
- 1. Adams P, et al. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 2. Afonine PV, et al. 2010. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 66:1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Azuhata F, Uyeda I, Kimura I, Shikata E. 1993. Close similarity between genome structures of rice black-streaked dwarf and maize rough dwarf viruses. J. Gen. Virol. 74:1227–1232 [DOI] [PubMed] [Google Scholar]
- 4. Baker N, Sept D, Joseph S, Holst M, McCammon J. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Broering T, Parker J, Joyce P, Kim J, Nibert M. 2002. Mammalian reovirus nonstructural protein microNS forms large inclusions and colocalizes with reovirus microtubule-associated protein micro2 in transfected cells. J. Virol. 76:8285–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brookes SM, Hyatt AD, Eaton BT. 1993. Characterization of virus inclusion bodies in bluetongue virus-infected cells. J. Gen. Virol. 74:525–530 [DOI] [PubMed] [Google Scholar]
- 7. Butan C, Van Der Zandt H, Tucker P. 2004. Structure and assembly of the RNA binding domain of bluetongue virus non-structural protein 2. J. Biol. Chem. 279:37613–37621 [DOI] [PubMed] [Google Scholar]
- 8. Collaborative Computational Project N 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 9. Davis IW, Murray LW, Richardson JS, Richardson DC. 2004. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32:W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeLano WL. 2008. The PyMOL molecular graphics system. DeLano Scientific LLC, San Carlos, CA: http://www.pmol.org [Google Scholar]
- 11. Eaton B, Hyatt A, White J. 1987. Association of bluetongue virus with the cytoskeleton. Virology 157:107–116 [DOI] [PubMed] [Google Scholar]
- 12. Eichwald C, Rodriguez JF, Burrone OR. 2004. Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J. Gen. Virol. 85:625–634 [DOI] [PubMed] [Google Scholar]
- 13. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 14. Fabbretti E, Afrikanova I, Vascotto F, Burrone OR. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333–339 [DOI] [PubMed] [Google Scholar]
- 15. Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hartley J. 2003. Use of the gateway system for protein expression in multiple hosts. Curr. Protoc. Protein Sci. Chapter 5:Unit 5.17 [DOI] [PubMed] [Google Scholar]
- 17. Isogai M, Uyeda I, Lee BC. 1998. Detection and assignment of proteins encoded by rice black streaked dwarf fijivirus S7, S8, S9 and S10. J. Gen. Virol. 79:1487–1494 [DOI] [PubMed] [Google Scholar]
- 18. Jayaram H, Taraporewala Z, Patton JT, Prasad BVV. 2002. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature 417:311–315 [DOI] [PubMed] [Google Scholar]
- 19. Jiang XF, et al. 2006. Cryoelectron microscopy structures of rotavirus NSP2-NSP5 and NSP2-RNA complexes: implications for genome replication. J. Virol. 80:10829–10835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kattoura MD, Chen X, Patton JT. 1994. The rotavirus RNA-binding protein NS35 (NSP2) forms 10S multimers and interacts with the viral RNA polymerase. Virology 202:803–813 [DOI] [PubMed] [Google Scholar]
- 21. Kobayashi T, Chappell JD, Danthi P, Dermody TS. 2006. Gene-specific inhibition of reovirus replication by RNA interference. J. Virol. 80:9053–9063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar M, et al. 2007. Crystallographic and biochemical analysis of rotavirus NSP2 with nucleotides reveals a nucleoside diphosphate kinase-like activity. J. Virol. 81:12272–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu HJ, Wei CH, Zhong YW, Li Y. 2007. Rice black-streaked dwarf virus outer capsid protein P10 has self-interactions and forms oligomeric complexes in solution. Virus Res. 127:34–42 [DOI] [PubMed] [Google Scholar]
- 24. López T, Rojas M, Ayala-Breton C, López S, Arias CF. 2005. Reduced expression of the rotavirus NSP5 gene has a pleiotropic effect on virus replication. J. Gen. Virol. 86:1609–1617 [DOI] [PubMed] [Google Scholar]
- 25. Maroniche GA, et al. 2010. Functional and biochemical properties of Mal de Río Cuarto virus (Fijivirus, Reoviridae) P9-1 viroplasm protein show further similarities to animal reovirus counterparts. Virus Res. 152:96–103 [DOI] [PubMed] [Google Scholar]
- 26. McNulty M, Curran W, McFerran J. 1976. The morphogenesis of a cytopathic bovine rotavirus in Madin-Darby bovine kidney cells. J. Gen. Virol. 33:503–508 [DOI] [PubMed] [Google Scholar]
- 27. Mertens PPC, Attoui H, Duncan R, Dermody TS. 2005. Reoviridae, p 447–454 In Fauquest CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom [Google Scholar]
- 28. Miller C, Arnold M, Broering T, Hastings C, Nibert M. 2010. Localization of mammalian orthoreovirus proteins to cytoplasmic factory-like structures via nonoverlapping regions of microNS. J. Virol. 84:867–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyazaki N, et al. 2005. Transcapsidation and the conserved interactions of two major structural proteins of a pair of phytoreoviruses confirm the mechanism of assembly of the outer capsid layer. J. Mol. Biol. 345:229–237 [DOI] [PubMed] [Google Scholar]
- 30. Murshudov G, Vagin A, Dodson E. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 31. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 32. Pettersen E, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 33. Prasad BVV, et al. 1996. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382:471–473 [DOI] [PubMed] [Google Scholar]
- 34. Sharpe A, Chen L, Fields B. 1982. The interaction of mammalian reoviruses with the cytoskeleton of monkey kidney CV-1 cells. Virology 120:399–411 [DOI] [PubMed] [Google Scholar]
- 35. Shimizu T, Yoshii M, Wei T, Hirochika H, Omura T. 2009. Silencing by RNAi of the gene for Pns12, a viroplasm matrix protein of rice dwarf virus, results in strong resistance of transgenic rice plants to the virus. Plant Biotechnol. J. 7:24–32 [DOI] [PubMed] [Google Scholar]
- 36. Silvestri LS, Taraporewala ZF, Patton JT. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 78:7763–7774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Supyani S, Hillman B, Suzuki N. 2007. Baculovirus expression of the 11 mycoreovirus-1 genome segments and identification of the guanylyltransferase-encoding segment. J. Gen. Virol. 88:342–350 [DOI] [PubMed] [Google Scholar]
- 38. Suzuki N, Hosokawa D, Matsuura Y, Kikuchi A, Omura T. 1999. In vivo and in vitro phosphorylation of rice dwarf phytoreovirus Pns12 cytoplasmic nonstructural protein. Arch. Virol. 144:1371–1380 [DOI] [PubMed] [Google Scholar]
- 39. Suzuki N, Sugawara M, Kusano T, Mori H, Matsuura Y. 1994. Immunodetection of rice dwarf phytoreoviral proteins in both insect and plant hosts. Virology 202:41–48 [DOI] [PubMed] [Google Scholar]
- 40. Taraporewala ZF, Chen D, Patton JT. 1999. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 73:9934–9943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taraporewala ZF, Kumar M, Prasad BVV, Patton JT. 2008. Structure and function of the rotavirus NSP2 octamer, an essential component of the viroplasm, p 273–289 In Patton JT. (ed), Segmented double-stranded RNA viruses: structure and molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 42. Taraporewala ZF, Patton JT. 2001. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 75:4519–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Terwilliger T. 2003. Improving macromolecular atomic models at moderate resolution by automated iterative model building, statistical density modification and refinement. Acta Crystallogr. D Biol. Crystallogr. 59:1174–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Terwilliger T, Berendzen J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55:849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Theron J, Huismans H, Nel LH. 1996. Site-specific mutations in the NS2 protein of epizootic haemorrhagic disease virus markedly affect the formation of cytoplasmic inclusion bodies. Arch. Virol. 141:1143–1151 [DOI] [PubMed] [Google Scholar]
- 46. Thomas CP, Booth TF, Roy P. 1990. Synthesis of bluetongue virus-encoded phosphoprotein and formation of inclusion bodies by recombinant baculovirus in insect cells: it binds the single-stranded RNA species. J. Gen. Virol. 71:2073–2083 [DOI] [PubMed] [Google Scholar]
- 47. Touris-Otero F, Martínez-Costas J, Vakharia V, Benavente J. 2004. Avian reovirus nonstructural protein microNS forms viroplasm-like inclusions and recruits protein sigmaNS to these structures. Virology 319:94–106 [DOI] [PubMed] [Google Scholar]
- 48. Vasquez-Del Carpio R, Gonzalez-Nilo FD, Riadi G, Taraporewala ZF, Patton JT. 2006. Histidine triad-like motif of the rotavirus NSP2 octamer mediates both RTPase and NTPase activities. J. Mol. Biol. 362:539–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wei T, et al. 2006. Pns12 protein of rice dwarf virus is essential for formation of viroplasms and nucleation of viral-assembly complexes. J. Gen. Virol. 87:429–438 [DOI] [PubMed] [Google Scholar]
- 50. Wei T, Shimizu T, Omura T. 2008. Endomembranes and myosin mediate assembly into tubules of Pns10 of rice dwarf virus and intercellular spreading of the virus in cultured insect vector cells. Virology 372:349–356 [DOI] [PubMed] [Google Scholar]
- 51. Zhang C, et al. 2008. Rice black streaked dwarf virus P9-1, an alpha-helical protein, self-interacts and forms viroplasms in vivo. J. Gen. Virol. 89:1770–1776 [DOI] [PubMed] [Google Scholar]
- 52. Zhang H, Chen J, Adams M. 2001. Molecular characterisation of segments 1 to 6 of rice black-streaked dwarf virus from China provides the complete genome. Arch. Virol. 146:2331–2339 [DOI] [PubMed] [Google Scholar]
- 53. Zhang LD, et al. 2005. Two virus-encoded RNA silencing suppressors, P14 of beet necrotic yellow vein virus and S6 of rice black streak dwarf virus. Chin. Sci. Bull. 50:305–310 [Google Scholar]
- 54. Zhong B, et al. 2003. A minor outer capsid protein, P9, of rice dwarf virus. Arch. Virol. 148:2275–2280 [DOI] [PubMed] [Google Scholar]