Abstract
Hepatitis C virus core protein forms the viral nucleocapsid and plays a critical role in the formation of infectious particles. In this study, we demonstrate that the highly conserved residue G33, located within domain 1 of the core protein, is important for the production of cell culture-infectious virus (HCVcc). Alanine substitution at this position in the JFH1 genome did not alter viral RNA replication but reduced infectivity by ∼2 logs. Virus production by this core mutant could be rescued by compensatory mutations located immediately upstream and downstream of the original G33A mutation. The examination of the helix-loop-helix motif observed in the core protein structure (residues 15 to 41; Protein Data Bank entry 1CWX) indicated that the residues G33 and F24 are in close contact with each other, and that the G33A mutation induces a steric clash with F24. Molecular simulations revealed that the compensatory mutations increase the helix-loop-helix flexibility, allowing rescue of the core active conformation required for efficient virus production. Taken together, these data highlight the plasticity of core domain 1 conformation and illustrate the relationship between its structural tolerance to mutations and virus infectivity.
INTRODUCTION
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma (10). It has been estimated that between 123 and 170 million people are infected with the virus worldwide (3, 25, 38). No vaccine is available, and the current antiviral therapies fail to cure approximately 50% of treated patients (41). HCV has been classified within the Flaviviridae family as a unique member in the Hepacivirus genus (34). The positive-sense, single-stranded RNA genome (∼9.6 kb long) encodes a polyprotein that is cleaved by cellular and viral proteases to yield the mature structural (core protein, E1, and E2) and nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (31). HCV is classified into six major genetic groups, each further divided into more closely related subtypes, and within an infected individual the virus exists as a constantly evolving quasispecies (40). The genetic variability of HCV is owed to a high viral replication rate and an error-prone RNA-dependent polymerase (NS5B) with an estimated mutation rate of 10−4 per nucleotide per generation (12).
The core protein is a highly conserved basic, RNA-binding protein that forms the viral nucleocapsid (28). The mature form of the protein that forms the viral nucleocapsid can be separated into two domains based on its hydropathic profile. Domain 1 and domain 2 are mainly α-helical, and the folding of the former depends on the presence of the latter (8). Domain 1, spanning residues 1 to 118, contains a high proportion of basic residues involved in RNA binding, which promotes nucleocapsid assembly (8, 35, 39). Domain 2 (residues 118 to approximately 169) consists of two amphipathic α-helices connected by a hydrophobic loop and mediates the association of core with lipid droplets (LDs) and endoplasmic reticulum membranes (4, 6, 20).
Although the major role of core protein is to encapsulate the virus genome, it is a multifunctional protein known to have a wide range of interactions with viral and cellular proteins (13, 28). Many of the host-protein interactions have been mapped to residues within domain 1 of the core protein (28). Using alanine scanning mutagenesis, we previously identified six residues (F24, G27, I30, G33, V34, and Y35) within domain 1 of the core protein that determined its interaction with the cellular RNA helicase DDX3 (2). These residues are located between core amino acids 24 and 35, which are highly conserved across all HCV genotypes (8, 9). Alanine substitution of these six residues revealed that the Y35A change resulted in the greatest knockdown of the core-DDX3 interaction. However, this mutation caused no alteration to the replication of the HCV JFH1 cell culture infectious virus (HCVcc) (44), implying that the core-DDX3 interaction is dispensable for HCV replication in vitro (2). We also showed that JFH1 genomes harboring the I30A, G33A, and V34A mutations could undergo efficient RNA replication; however, their ability to produce infectious virus was not tested.
In the present study, we show that the G33A mutation severely impairs infectious virus production. Furthermore, the passage of assembly-defective JFH1G33A genomes identified compensatory mutations in close proximity to the original G33A substitution, indicating that domain 1 of the core protein has important functions in virion morphogenesis. Molecular dynamic simulations performed on the core protein structure (residues 2 to 45, designated segment 2-45; Protein Data Bank [PDB]entry 1CWX) established correlations between the structural features of the various core mutants and their phenotypes in terms of infectious virus production.
MATERIALS AND METHODS
Cell culture and antibodies.
Human hepatoma Huh-7 and the Huh-7/J20 reporter cell lines were propagated as described previously (22). The anti-NS5A mouse monoclonal antibody (MAb) 9E10 (26) was a kind gift from Charles M. Rice. The anti-HCV E2 MAb 3/11 and the rabbit anti-core protein antisera CII575 (raised against HCV core protein amino acids [aa] 1 to 81), CII680 (core aa 1 to 116), and CII830 (core aa 1 to 166) were obtained from Jane McKeating and Helmut Jacobsen, respectively (16, 21). The rabbit anti-core antiserum R526 was raised against amino acids 1 to 59 of core protein (11). The anti-bovine serum albumin (BSA) antiserum was obtained from Millipore.
Plasmid constructs and mutagenesis.
The plasmid pUC-JFH1 carries the full-length cDNA of the genotype 2a HCV strain JFH-1 (44). The plasmids pUC-GND JFH1 and pUC-JFH1 ΔE1E2 are identical to pUC-JFH1, except they carry the GND mutation in the NS5B-encoding sequence or an in-frame deletion in the E1 and E2 sequences (44). Site-directed mutagenesis was carried out using the QuikChange II kit (Stratagene) to introduce substitutions at the target sites in the core protein-coding sequence. Briefly, various substitutions in the core protein-coding region were individually introduced into the plasmid pGEM-T (Promega) carrying nucleotides 1 to 2614 (corresponding to the 5′ untranslated region [UTR] and the coding sequence of core to E2 proteins) of HCV strain JFH1 using appropriate primers (the sequences of which are available upon request). The presence of the desired mutation in the resulting clones was confirmed by nucleotide sequencing. Sequences carrying the appropriate mutation were subcloned back into pUC-JFH1 to generate mutant viruses (see below). The generation and characterization of the monocistronic N17/JFH1 subgenomic replicon construct will be described elsewhere. This construct encodes the firefly luciferase reporter and the puromycin resistance marker (separated by the foot-and-mouth-disease virus [FMDV] 2a self-cleavage site) in the JFH1 ΔE1E2 background (Fig. 1Bi). The G33A or F130E mutation in core protein was introduced in the N17/JFH1 background by site-directed mutagenesis.
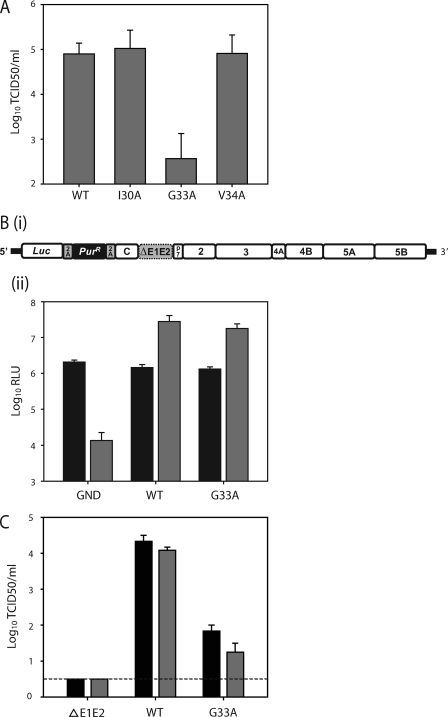
Fig 1.
Effect of G33A mutation of virus replication. (A) Core mutation G33A impairs infectious virus production. Huh-7 cells were electroporated with the viral genomes specified, and 72 h later the infectious virus present in the culture medium was determined by TCID50 assay. (Bi) A schematic representation of the N17/JFH1 genome. The N17/JFH1 viral genome contains a luciferase and puromycin resistance gene preceding HCV polyprotein carrying an in-frame deletion in E1E2 (dotted box). The FMDV 2A peptide has been inserted at either side of the puromycin (PurR) gene to liberate luciferase and PurR following translation. (Bii) Replication of N17/JFH1 containing the G33A mutation. Huh-7 cells were electroporated with N17/JFH1 viral genomes, and replication was assayed by luciferase activity at 4 (black bars) and 48 h (gray bars) postincubation. Means and standard errors of duplicate electroporations measured in triplicate are shown. (C) Huh-7 cells were electroporated with the indicated viral RNA, and 48 h later the cell-released (black bars) and cell-associated (gray bars) infectivity levels were determined by TCID50 assay. Mean values and standard errors from two independent experiments measured in triplicate are shown. The dotted line indicates assay sensitivity.
Determination of virus infectivity and RNA replication.
Infectious viruses were generated as previously described (44). Virus titers in the supernatants were determined following immunostaining for NS5A by a 50% tissue culture infective dose (TCID50) assay as described previously (26). The level of intracellular RNA replication was determined by measuring the secreted alkaline phosphatase (SEAP) activity in the culture medium as described previously (22). To monitor virus infectivity during serial passaging, 2 × 106 Huh-7 cells were electroporated with 10 μg of in vitro-transcribed viral RNA and seeded into a T80 flask in a total of 25 ml of medium. Cells were passaged at the time of subconfluency into a new flask containing 24 ml fresh medium. At each passage the cell culture supernatants were harvested, and the released virus infectivity was determined by TCID50 assay. To determine the replication of each mutant virus, 2 × 106 Huh-7/J20 cells were electroporated with 10 μg of viral RNA and resuspended in 4 ml of Dulbecco's modified essential medium (DMEM). Aliquots of 1 ml were then seeded into triplicate wells of a 12-well plate. Following incubation at 37°C for 48 h, the SEAP activity and infectious virus yields in cell culture supernatants were determined by SEAP and TCID50 assay, respectively. Cell-associated virus was obtained essentially as described previously (19, 37). Briefly, 3 × 106 Huh-7 cells were electroporated with 10 μg of viral RNA, resuspended in 15 ml of DMEM, and seeded into 90-mm culture dishes. Cells were harvested at 48 h postelectroporation, washed in medium, resuspended in 0.8 ml medium, and freeze-thawed three times. The samples were centrifuged to remove cell debris, and the supernatant was assayed by TCID50 assay to determine virus infectivity.
N17/JFH1 replicon RNA was generated by in vitro transcribing 1 μg of full-length plasmid using the T7 Megascript kit (ABI). To measure the intracellular RNA replication of N17/JFH1, 2 × 106 Huh-7 cells were electroporated with 10 μg of viral RNA and resuspended in 3 ml of culture medium. Aliquots of 250 μl then were seeded into triplicate wells of a 48-well plate. Following incubation at 37°C for 4 and 48 h, cells were lysed for luciferase assay.
Western blot analysis.
Western blot analysis was performed as described previously (11), with some modifications. To detect intracellular antigens, cultured cells were washed once in phosphate-buffered saline (PBS) and lysed directly in SDS-PAGE sample loading buffer (200 mM Tris-HCl, pH 6.7; 0.5% SDS; β-mercaptoethanol; 10% glycerol). Lysates were homogenized by passing through a 22-gaugle needle five times before use. To obtain extracellular lysates, the cells from four electroporations were seeded into a T175 flask in a total of 34 ml of medium. At 48 h postincubation, the culture medium was harvested, filtered, and overlaid onto 3 ml of a 20% (wt/vol) sucrose cushion made with PBS and centrifuged at 25,000 rpm for 4 h using a Sorvall Discovery 90SE ultracentrifuge. The pellets then were lysed directly in 50 μl of SDS-PAGE sample loading buffer and stored at −20°C until use. The proteins in 20 μl of sample then were resolved by 12.5% SDS-PAGE and transferred onto nitrocellulose membranes (Hybond-ECL; Amersham).
Sedimentation equilibrium gradient analysis.
Gradients were formed by layering equal volumes of 10 to 40% OptiPrep (iodixanol; Axis-Shield) solutions in PBS (in 5% increments) and incubated at 4°C for 16 h. Cell supernatants, concentrated beforehand by ultracentrifugation without the sucrose cushion, were layered onto the top of the gradient. Equilibrium was reached by ultracentrifugation at 36,000 rpm for 16 h at 4°C on an AH629 rotor (Sorvall). After centrifugation, l-ml gradient fractions were harvested from the top of the tube, and 100-μl portions were weighed to calculate fraction densities. The infectivity of each fraction was determined by TCID50 as described above. RNA was extracted from each fraction and quantified by reverse transcription-quantitative PCR (RT-qPCR) assay as described previously (45).
Indirect immunofluorescence.
Cells on coverslips were fixed in methanol, washed with PBS, blocked for 10 min with PBS containing 2% fetal calf serum (FCS), and incubated at room temperature for 1 h with primary antibody in the blocking buffer. Cells were washed with PBS, stained with secondary antibody conjugated with either fluorescein isothiocyanate (FITC) or tetramethyl rhodamine isothiocyanate (TRITC) in blocking buffer for 1 h, and washed with PBS, and then the coverslips were mounted on a glass slide and examined with a Zeiss laser-scanning LSM510 META inverted confocal microscope (Carl Zeiss Ltd., United Kingdom). The images were analyzed using LSM510 software.
Nucleotide sequencing.
Total RNA was prepared using the RNAEasy kit (Qiagen) from cells infected with virus collected from passage 6 cells (see Results). Approximately 100 ng of RNA was converted to first-strand DNA by using a Superscript III first-strand synthesis kit (Invitrogen) with primer 5′-TTGCGAGTGCCCCGGGA-3′. After digestion with 1 U of RNase H (Invitrogen) for 20 min at 37°C, one-quarter of the RT reaction mix was amplified with appropriate primers to yield four fragments of HCV cDNA (nucleotides [nt] 322 to 930, 538 to 3038, 2544 to 5542, and 5412 to 7890) covering the core to NS5A regions of the viral genome. The PCR products were gel purified by gel extraction (Qiagen) and used directly for nucleotide sequencing.
Molecular modeling.
The modeling of the different mutants was carried out with the program XPLOR-NIH (36). The modeling protocol started with the generation of a large number of extended conformers with good covalent geometry, followed by a molecular dynamic-simulated annealing protocol with torsion angles as internal degrees of freedom (33, 42). The nuclear Overhauser effect (NOE)-based distance restraints obtained from the nuclear magnetic resonance (NMR)-analyzed HCV core protein (segment 15-45) (PDB entry 1CWX) were used as input structural information. For each conformer generated, a high-temperature searching phase at 30,000 K (500 steps), followed by 1,000 cycles while cooling to 1,000 K and 1,000 cycles of molecular dynamics in Cartesian space. A final Powell minimization of 300 cycles was applied at the end of the protocol. One thousand conformers that satisfied acceptance criteria were selected (no NOE violations of >0.5 Å and no bond violation of >0.05 Å). Structures were superimposed, and root mean squared deviation (RMSD) values were computed using MolMol (24). We computed the interhelical angle distribution with MolMol using the least-squares method by generating two helical axes based on the Cα coordinates of α-helices 1 and 2.
RESULTS
Mutation G33A impairs infectious virus production.
Previously, we reported that the alanine substitutions I30A, G33A, and V34A in the HCV core protein had no noticeable effects on viral protein expression (2). To further analyze these mutants, we measured the level of infectious virus released from cells electroporated with the in vitro-transcribed viral RNAs. As shown in Fig. 1A, the infectivity of JFH1I30A and JFH1V34A was comparable to that of wild-type JFH1 (JFH1WT), whereas JFH1G33A showed a large (∼2 log) reduction in titer. Based on this result, we sought to further characterize the replication properties of JFH1G33A. To test if the reduced infectivity caused by G33A was due to an effect on viral RNA replication, we introduced this mutation into a noninfectious ΔE1E2/JFH1 monocistronic subgenomic replicon (N17/JFH1) (Fig. 1B). This replicon was constructed to contain sequences carrying the HCV 5′ UTR followed by the luciferase reporter gene, the puromycin selectable marker, the viral polyprotein lacking E1E2, and the 3′ UTR. The inclusion of the FMDV 2a protease sequence allows the self-cleavage of the puromycin resistance marker during translation. As the N17/JFH1 replicon is noninfectious, it allows the accurate measurement of HCV RNA replication in the absence of viral spread. Huh-7 cells were electroporated with the N17/JFH1G33A RNA, and luciferase readings were measured at 4 and 48 h postincubation. As shown in Fig. 1Bii, the N17/JFH1 replication was unaffected by the G33A substitution, indicating that this mutation impairs virus infectivity without affecting viral RNA replication. To determine more precisely the effect of the G33A mutation on infectious particle production, the intracellular and extracellular virus titers were assayed at 48 h postelectroporation. As shown in Fig. 1C, the cell-free and cell-associated infectivity of JFH1G33A RNA-electroporated cells shared reductions similar to those of JFH1WT, suggesting that this core protein mutant has a defect acting prior to infectious particle assembly.
Detection of G33A core protein.
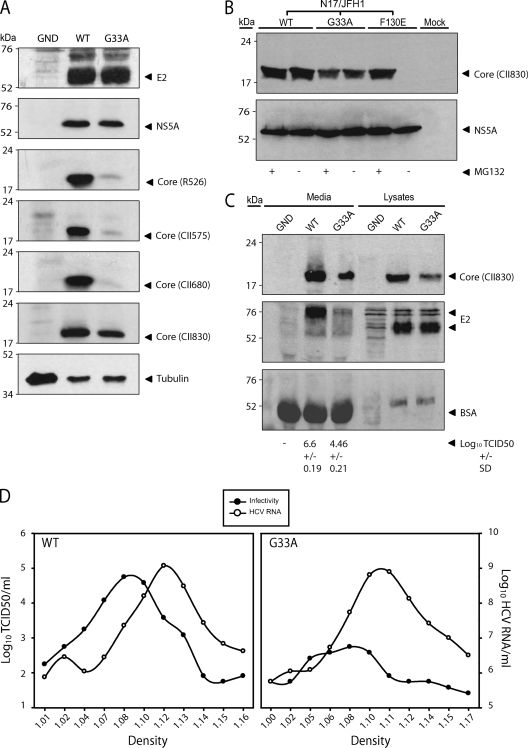
In our previous study, Western blot analysis found that JFH1G33A core protein was undetectable using the anti-core MAb C7-50 and barely detectable using the anti-core rabbit serum R308 (2). These results suggested that this mutation either lowered the stability of the protein or simply reduced the binding efficiency of these antibodies. To distinguish between these two possibilities, we used four additional anti-core rabbit sera (R526, CII575, CII830, and CII680 [11, 21]) raised against various N-terminal portions of core protein (see Materials and Methods). These sera were tested against the JFH1G33A core protein obtained from electroporated cells. As shown in Fig. 2A, the G33A core protein was barely or not at all detectable when immunoblotting with R526, CII575, and CII680. In contrast, the CII830 antiserum detected greater quantities of the mutant core protein. Nevertheless, the levels of G33A core protein were slightly lower than that of the WT compared to the equal quantities of NS5A and E2 detected from both lysates, indicating either that this mutant core protein is not fully reactive to this antiserum or that it is more sensitive to degradation. To test if G33A enhances core protein degradation, cells electroporated with N17/JFH1G33A were treated with the proteasome inhibitor MG132. Mutation F130E is known to render core protein susceptible to complete degradation (6), an effect that can be overcome by MG132 treatment. In line with these reports, the addition of MG132 to cells electroporated with N17/JFH1F130E rescued the core protein from degradation (Fig. 2B). In contrast, MG132 treatment had no impact on the quantities of G33A protein detected by immunoblotting, indicating that the lower level of core protein observed is not due to degradation. Amino acids 29 to 37 are reported to form an immunodominant epitope in core protein (29). Therefore, antibodies generated against the first domain of core protein, such as those used in this study, are likely to target this region of the protein. The mutation of G33 may inflict gross conformational changes to this immunodominant epitope and therefore prevent the proper binding of antibodies targeting this region. Experiments using antibodies that target different regions of core protein, such as domain 2, may provide key information to support this concept.
Fig 2.
Detection of intracellular and extracellular G33A core protein. (A) Western blot analysis of cell lysates harvested at 48 h postelectroporation with the indicated viral RNAs. Results for core protein using various anti-core antibodies (shown in parentheses), E2, NS5A, and tubulin are indicated. (B) Stability of G33A core protein. Huh-7 cells were electroporated with N17/JFH1 containing the indicated mutations. At 72 h postincubation, cells were treated with or without the proteasome inhibitor MG132 at a final concentration of 2.5 mg/ml for 12 h prior to the preparation of cell extracts for Western blot analysis. Cell lysates were probed with core antiserum CII830 and NS5A MAb 9E10. (C) Western blot for extracellular proteins. Culture medium of Huh-7 cells was collected and concentrated 48 h postelectroporation with the indicated viral RNAs and subjected to Western blotting to detect core protein, E2, and BSA using antiserum CII830, MAb 3/11, and anti-BSA MAb, respectively. The amount of infectious virus loaded into each well is indicated. (D) Concentrated culture medium from JFH1WT- and JFH1G33A-electroporated cells was fractionated using 10 to 40% iodixanol density gradient centrifugation. For each fraction, the amounts of infectivity and HCV RNA are plotted against the buoyant density.
Production of noninfectious particles by JFH1G33A.
Recent studies have shown that infectious hepatitis C virions have a low buoyant density, likely due to the association of virus particles with low-density and very-low-density lipoproteins (reviewed in reference 5). Interestingly, for every infectious virion secreted, there are approximately 1,000 accompanying noninfectious particles (19). Biochemical and electron microscopy analyses imply that these higher-density noninfectious particles are nonenveloped genome-containing capsids that are secreted via a pathway different from that of virions (18, 19, 30, 43). To determine what effect the G33A mutation had on the secretion of noninfectious particles, immunoblotting for core and E2 proteins was performed on the concentrated culture medium obtained at 48 h postelectroporation using the CII830 antiserum and the rat MAb 3/11, respectively. The titration of the culture medium confirmed that the JFH1G33A electroporated cells produced virus that was ∼2 logs less infectious than JFH1WT (Fig. 2C, bottom). In line with this, the levels of JFH1G33A extracellular E2 were barely detectable. It is important to note that the molecular weight of the extracellular E2 protein was noticeably higher than that of its intracellular form. This alteration in E2 migration profile is likely caused by the posttranslational modification of the secreted proteins and possibly high levels of fetal calf serum albumin present in the concentrated virus preparation (Fig. 2C) (43). Interestingly, JFH1G33A extracellular core protein was readily detectable, albeit at slightly lower levels than those of JFH1WT. However, these reduced levels may be accounted for by the reduced affinity of the core protein antiserum to the mutant protein (Fig. 2A). To further examine the noninfectious particle production of JFH1G33A, equilibrium buoyant density centrifugation in iodixanol gradients was performed on the concentrated culture medium obtained from electroporated cells. Similar JFH1WT and JFH1G33A HCV RNA titers were found at densities ranging between 1.1 and 1.14 g/ml (Fig. 2D). Despite the low level of infectious virus detected for JFH1G33A throughout the gradient, most of this infectivity was concentrated at a density similar to that of the WT virus. Collectively, these data indicate that the G33A mutation affects the assembly and secretion of noninfectious particles to a lesser extent than it does the infectious virions.
Intracellular localization of G33A core protein.
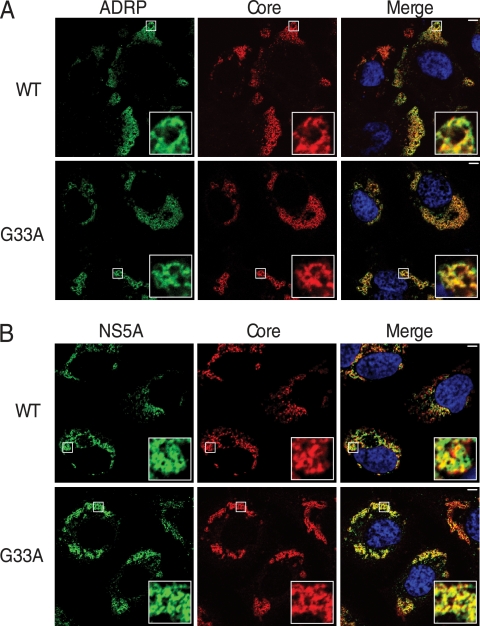
As previously described, the association of core protein with LDs is essential for infectious virus production (7, 30). We therefore analyzed the subcellular localization of the G33A mutant core protein to determine the effect of this mutation on its association with LDs. Huh-7 cells electroporated with the viral RNA were analyzed by indirect immunofluorescence after 48 h for HCV core and adipocyte differentiation-related protein (ADRP), a protein abundant on the surface of LDs (17). As shown in Fig. 3A, G33A core colocalized with ADRP in a manner similar to that of the WT, indicating that this mutation does not disturb the core-LD association. NS5A is another viral protein that plays a vital role in the virion assembly process through its interaction with core protein (27). To test whether the G33A mutation altered this interaction, we measured the level of colocalization between these two viral proteins at 72 h postelectroporation. As shown in Fig. 3B, the partial colocalization expected between core protein and NS5A was observed in cells replicating JFH1WT and JFH1G33A, indicating that the G33A mutation does not disrupt the subcellular localization between core protein and NS5A.
Fig 3.
Subcellular localization of G33A core protein. Huh-7 cells electroporated with the indicated viral RNAs were fixed and processed for immunofluorescence for core (CII830 antiserum) and ADRP (anti-ADRP) (A) and core (CII830 antiserum) and NS5A (9E10 MAb) (B). Nuclei were stained with DAPI (blue). Bar, 5 μm.
Identification of compensatory mutations within core protein.
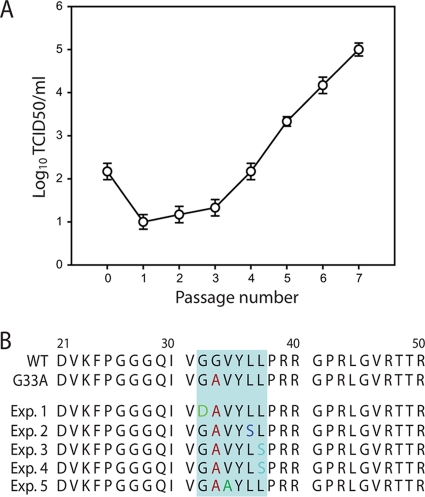
To determine if the infectivity of JFH1G33A could be rescued by compensatory mutations, cells electroporated with this mutant genome were serially passaged. The level of infectious virus released into the culture medium was monitored throughout the passaging experiment. As shown in Fig. 4A, there was a progressive increase in extracellular virus release from passage 1 onwards that eventually achieved peak titers similar to those expected for JFH1WT. To identify the mutation(s) responsible for this increased infectivity, total RNAs were prepared from cells infected with passage 6 virus, and the core to NS5A regions of the HCV genome were sequenced by RT-PCR. Interestingly, the sequencing revealed the presence of a single mutation in core protein (G32D) located adjacent to the original alanine substitution. Further independent passaging experiments on JFH1G33A observed similar increases in infectivity (data not shown), which coincided with the detection of second-site core mutations. Overall, we obtained a total of four different second-site mutations in core protein (namely G32D, V34A, L36S, or L37S), all of which were located in very close proximity to the original engineered substitution (Fig. 4B). Importantly, similar passaging experiments performed on the JFH1WT virus revealed no changes to the core protein sequence (data not shown).
Fig 4.
Second-site core protein mutations isolated from passaged JFH1G33A genomes. (A) Cells electroporated with JFH1G33A genomes were passaged 7 times. At each passage the infectivity of the cell culture supernatant was determined by TCID50 assay. Results of a single representative experiment of four independent experiments are shown. Error bars demonstrate the variability of the TCID50 assay. (B) Mutations identified in the JFH1G33A core protein sequence from viruses obtained at passage 6 in five independent experiments. The original G33A mutation is shown in red and the second-site mutations in green (G32D), blue (L36S), light blue (L37S), or dark green (V34A).
Effect of second-site mutations on virus replication.
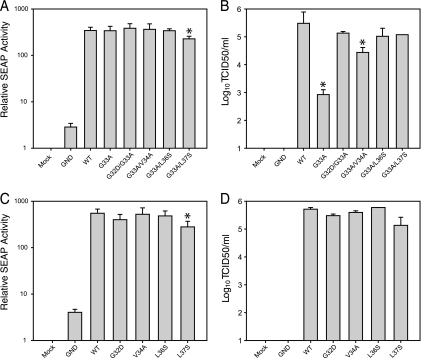
To determine if these additional core mutations were responsible for the enhanced infectivity of JFH1G33A seen during passaging, we individually engineered these second-site changes into the original JFH1G33A genome. The viral replication levels of these mutants were tested using the Huh-7/J20 reporter cell line (22). This cell line stably expresses the enhanced green fluorescent protein fused in frame to the secreted alkaline phosphatase (SEAP) via a recognition sequence of the viral NS3/4A serine protease. During HCV replication, the SEAP reporter is released from the fusion protein and secreted into the extracellular culture medium following cleavage by NS3/4A. The level of SEAP activity in the culture medium directly correlates with the level of intracellular viral RNA replication. The viral RNA from each core mutant was electroporated into Huh-7/J20 cells, and after 48 h the supernatant was harvested for TCID50 and SEAP assays. As shown in Fig. 5A, all of the mutants showed SEAP activities similar to that of JFH1WT, except JFH1G33A/L37S, which showed a slight yet significant decrease. SEAP activities approximately 4-fold higher than background levels were released from cells electroporated with the replication-deficient JFH1GND RNA. Such levels are likely caused by a low level of viral NS3/4A protein expression produced from single rounds of JFH1GND genome translation. The presence of each second-site core mutation greatly increased infectious virus production (Fig. 5B). In fact, all of the double core mutants displayed cell-free infectivity comparable to that of JFH1WT except JFH1G33A/V34A, whose titers were ∼10-fold less. These data indicate that these second-site core mutations function as compensatory mutations. It is possible that the second-site core mutations considerably increase the fitness of JFH1WT, and that this enhancement accounts for the suppression of the JFH1G33A defects. To test this hypothesis, we introduced the G32D, V34A, L36S, and L37S mutations into the JFH1WT genome and analyzed virus replication at 48 h postelectroporation. The SEAP assay revealed that each core mutant genome replicated efficiently; however, JFH1L37S displayed lower values (Fig. 5C). Similar decreases were observed for JFH1G33A/L37S (Fig. 5A), indicating that the L37S change slightly decreases RNA replication. None of the core reversion mutants differed significantly in their ability to produce infectious virus (Fig. 5D). The lack of enhancement of JFH1WT fitness in the presence of the G32D, V34A, L36S, and L37S changes suggests that these mutations specifically compensate for the defects brought about by the G33A mutation.
Fig 5.
Second-site core protein mutations rescue JFH1G33A infectivity. (A to D) Huh-7/J20 cells were electroporated with the viral RNA containing the second-site core protein mutations in the JFH1G33A (A and B) or the JFH1WT (C and D) genetic backbone. Infectious virus release (B and D) and replication (A and C) were determined at 48 h postelectroporation by TCID50 and SEAP assay, respectively. Means and error ranges of two independent electroporations performed in triplicate are shown (*, P < 0.05 by t test).
Effect of core mutations on protein plasticity using molecular dynamic modeling.
The N-terminal domain of core protein (aa 2 to 117) behaves as an intrinsically unstructured protein when isolated. However, this domain also includes several potential α-helices that fold upon interaction with its C-terminal membrane-binding domain (8) or in the presence of cosolvent (e.g., trifluoroethanol [TFE]) used to probe potent polypeptide conformation (14, 15, 23). Similarly to other intrinsically unstructured proteins, core domain 1 is expected to adopt different conformations depending on the absence or presence of specific biological partners. In nuclear magnetic resonance (NMR) structure analysis of the core segment 2-45 in water, TFE mixtures revealed the existence of a helix-loop-helix motif involving amino acids 15 to 41 (PDB entry 1CWX) (23). Besides constituting an immunodominant antigenic site (23, 29), this helix-loop-helix conformation should be relevant to one of the functional roles of the HCV core protein.
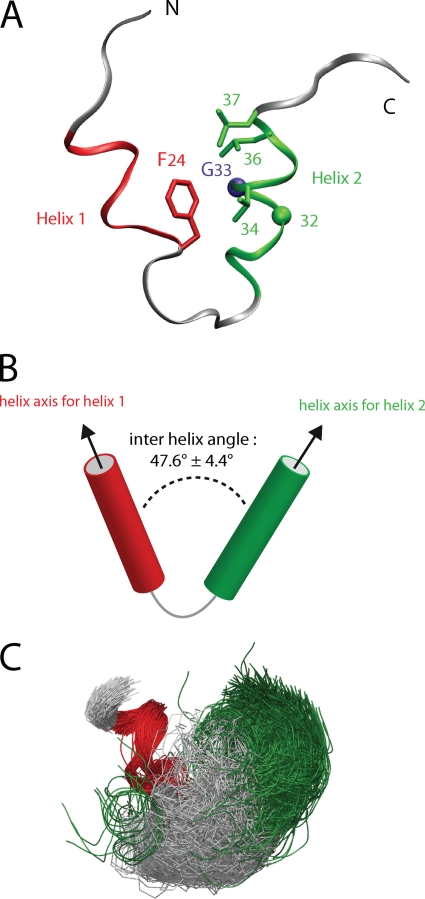
Using molecular dynamic modeling, we analyzed the conformational changes induced by the G33A compensatory mutations in the core protein three-dimensional (3D) structure. This structure consists of a helix-loop-helix motif (aa 15 to 41) (Fig. 6A) formed by α-helix 1 (P19-F24), the loop P25-Q29, and α-helix 2 (I30-L37) (Fig. 7). As illustrated in Fig. 6A, the F24 aromatic ring plays a crucial role in the interaction between helices 1 and 2 due to its contact with the hydrophobic residues of helix 2. Notably, the F24 aromatic ring is close to G33, and the mutation of the latter to alanine induces a steric clash. Furthermore, except for position 32, the G33A compensatory mutations at positions 34, 36, and 37 are facing F24 and are located on the same side of helix 2 (Fig. 6A).
Fig 6.
Structure of segment 15-41 of core protein. (A) Ribbon illustration of the NMR representative structure of core segment 15-41 (PDB entry 1CWX) showing the helix-loop-helix motif. The α-helices 1 (aa 19 to 24) and 2 (aa 30 to 37) are colored red and green, respectively. The side chains of the relevant residues are displayed as sticks (F24, L36, L37, and V34A), except glycine residues 32 and 33, which are represented by green and violet spheres, respectively. (B) Scheme showing the interhelical angle between the two α-helices 1 and 2. (C) Bundle of the 1,000 conformers calculated for the modeling studies (WT core). The conformers are superimposed on α-helix 1.
Fig 7.
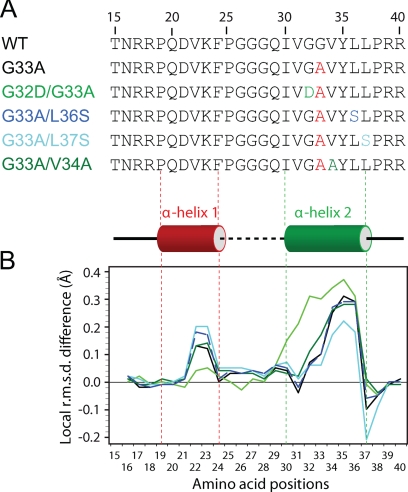
Local structure analogies between core protein segment 15-41 and mutants. (A) Amino acid sequence of WT and core mutants used for the molecular dynamic modeling studies. (B) Variation of the local structure along the sequence for the mutants compared to the WT core (represented as the baseline). The local RMSD of backbone heavy atoms (N, Cα, and CO) first were calculated for each residue with a tripeptide sliding window along the sequences. These values were averaged for each set of 1,000 calculated conformers of mutants and subtracted from the averaged local RMSD calculated for the WT conformers. The colors used for the lines in panel B correspond to the color coding of the mutants for panel A.
To investigate the potential plasticity of this helix-loop-helix motif as well as to evaluate the impact of mutations on its structural properties, sets of 1,000 structures were calculated using molecular dynamics for the WT and each of the reversion mutants (see Materials and Methods for details). The bundles of 1,000 conformers generated (Fig. 6C, aligned on α-helix 1) were compared on the basis of pairwise atomic RMSD for the backbone atom coordinates (N, Cα, and CO). The local structure similarities of each mutant to the WT core protein were analyzed by comparing the average local RMSD for each residue along the sequence to those of WT core protein (represented by the baseline in Fig. 7B). As expected, the local RMSD of mutants G33A, G32D/G33A, G33A/V34A, G33A/L36S, and G33A/L37S indicate important local variations in the α-helix 2 containing the mutated positions (Fig. 7A). Interestingly, these mutations also affect the local conformation of segment D21-F24, indicating that compensatory mutations indirectly induce conformational changes in α-helix 1. To evaluate the conformational changes between these two helices, the orientational preferences between interacting helices were characterized by measuring the interhelical angle formed by the two helical axes (Fig. 6B). While a broad range of interhelical angles was observed (16 to 155°) for the bundle of 1,000 conformers generated for WT core protein, most of the conformer angles fluctuated around 56° (Fig. 8, top left). This relatively narrow distribution of most interhelical angles among the 1,000 calculated structures indicate a limited conformational flexibility for the WT core protein structure. This dominant and favored conformation ensemble is expected to confer an essential functional role to the core protein.
Fig 8.
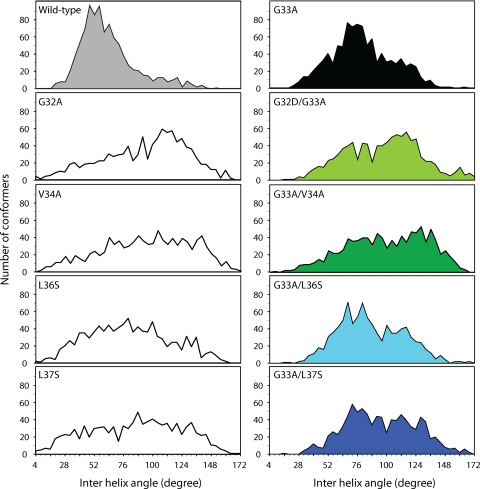
Comparison of interhelical angles for the calculated conformers of core segment 15-41 and mutants. For each mutant, the distributions of the interhelical angles between helices 1 and 2 are shown. For the double core mutants, the means and interquartile ranges for each distribution are shown in Table 1.
The distributions of interhelical angles for the core mutants display significant differences from that of WT (Fig. 8). For the core protein single mutations G32D, V34A, L36S, and L37S, which do not alter the production of infectious virus (Fig. 5D), the broad distributions of interhelical angles illustrate the functional plasticity of the helix-loop-helix motif. For the G33A mutant (Fig. 8, right), the mean interhelical angle (about 76°) was greater than that of the WT (about 60°) and should be related to the steric clash observed between side chains of A33 and F24. In addition, the relatively narrow interhelical angle distribution (interquartile range of about 28°) for the favored conformation indicates that the flexibility of the core structure of the G33A mutant is limited (Table 1). The fact that this mutation severely impairs infectious virus production may be related to this particular helix-loop-helix conformation being unfavorable for some specific function of core protein. In contrast, broader distributions of interhelical angles (interquartile ranges from 35° to 45°) were observed for the compensatory mutants G32D/G33A, G33A/L36S, G33A/L37S, and G33A/V34A, indicating a larger conformational plasticity of the helix-loop-helix motif (Table 1). An F test confirmed that the variance of the interhelical angles was significantly different (P < 10−4) between the compensatory mutants and G33A. The F test was not applicable between WT and G33A, as the intrahelical angles of the WT were not normally distributed. As these compensatory mutants displayed cell-free infectivity comparable to that of JFH1WT, one can suppose that this plasticity allows the helix-loop-helix motif to easily adopt the required active conformation.
Table 1.
Average interhelical angles for the WT and mutants and their interquartile rangesa
| Allele | Avg angle (°) | Interquartile range |
|---|---|---|
| WT | 59.96 | 23.25 |
| G33A | 76.2 | 28.33 |
| G32D/G33A | 98.18 | 44.45 |
| G33A/L36S | 95.54 | 40.95 |
| G33A/L37S | 105 | 40.43 |
| G33A/V34A | 98.12 | 52 |
The distributions are represented graphically in Fig. 8.
Characterization of F24 mutations in the JFH1WT and JFH1G33A virus.
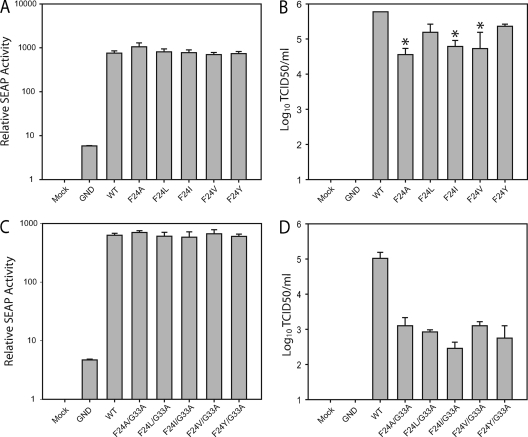
The steric clash predicted between G33A and F24 in our core protein model (Fig. 6) prompted us to test whether mutating F24 to other hydrophobic amino acids (A, V, I, L, and Y) altered infectious virus production in both the JFH1WT and JFH1G33A genetic background. These substitutions were chosen to minimize effects on core secondary structure folding and to evaluate the importance of the hydrophobic and/or cyclic character of residue 24. As shown in Fig. 9A and C, replacing F24 with A, V, I, L, or Y caused no changes to the intracellular RNA replication of JFH1WT or JFH1G33A. However, JFH1WT infectious virus production was significantly reduced by the alanine, isoleucine, and valine changes, while tyrosine and leucine changes had limited effect (Fig. 9B). This indicates that the bulky but flexible leucine residue, as well as the cyclic tyrosine residue, mimic the properties of phenylalanine at position 24. In contrast, small residues, such as alanine, as well as rigid bulky hydrophobic residues, including valine and isoleucine, are not well tolerated at this position. This mutational study indicates that a bulky but flexible or cyclic hydrophobic residue is required at position 24. In addition, JFH1G33A harboring each substitution at position 24 showed a decrease (∼100- to 200-fold) in infectious virus production compared to that of JFH1WT (Fig. 9D) that was similar to the decrease found for JFH1G33A alone (Fig. 5A). Therefore, the suppression of the predicted steric clash between F24 and A33 is not sufficient to restore the production of infectious particles. This steric clash in the G33A mutant is likely overcome due to the flexibility of the core conformation in this region.
Fig 9.
Effect of F24 residue on virus replication. (A and B) Huh-7/J20 cells were electroporated with viral RNA containing the F24A, F24L, F24I, F24V, and F24Y mutations in the JFH1WT (A and B) or the JFH1G33A (C and D) genetic backbone. Infectious virus release (B and D) and replication (A and C) were determined at 48 h postelectroporation by TCID50 and SEAP assay, respectively. Means and error ranges of two independent electroporations performed in triplicate are shown (*, P < 0.05 by t test).
DISCUSSION
The advent of the HCVcc system has permitted studies on the role of the viral core protein in virion morphogenesis. Using chimeric J6/JFH1, Murray and colleagues performed extensive mutagenesis across a large part (amino acids 57 to 191) of the core protein-coding region, where a total of 34 mutant viruses were constructed, each containing alanine changes in blocks of four amino acids (32). That study revealed numerous residues that were critical for infectious virus production but not for viral RNA replication. A more recent core mutagenesis study focused on the two highly conserved clusters of basic amino acids located between residues 6 to 23 and 39 to 62 (1). No specific residues or motifs affecting virus infectivity were identified within the first basic cluster. However, the alanine substitution of four residues (R50, K51, R59, and R62) within the second cluster was shown to completely abolish infectious virus assembly. Although the exact stage at which these mutations inhibited virus assembly was not ascertained, defects to core protein stability, colocalization with LDs and NS5A, RNA encapsidation, oligomerization, and envelopment by intracellular membranes were excluded experimentally.
In the present study, we show that the alanine substitution of residue G33 grossly reduces virus infectivity in the context of the HCVcc system. G33 and the compensatory mutations identified are all located within the helix-loop-helix motif we previously identified by NMR structural analysis in the N-terminal 2-45 segment of core protein (23). This motif is expected to be essential to one of the functional roles of the HCV core protein. This assumption is supported by the correlations we could establish in this study between the structural features of the various core mutants and their phenotypes in terms of infectious virus production. Mutation G33A, which might be expected to have a mild effect from a structural point of view, exhibits a dramatic effect on infectious virus assembly. As shown by our modeling analyses, this G33A mutation disturbs the amino acid pattern interaction as well as the optimal geometry between helices 1 and 2. However, the large range of interhelical angle distribution also revealed that the helix-loop-helix motif in mutant G33A is rather flexible, and that it could at least transiently adopt geometry close to that of the optimal WT conformation. This flexibility may explain why the G33A mutant nevertheless could replicate and produce low levels of infectious particles. Intriguingly, compared to its effect on infectious virus production, the G33A mutation appears to cause only minor reductions of noninfectious particle secretion. One explanation is that the G33A mutation allows the formation of capsids containing the genomic RNA, but their subsequent transition into mature virions is far less efficient. Although we can only speculate on the underlying reason for the phenotype of G33A, our data suggest that these effects are not a result of protein instability, the association of core protein with LDs and NS5A, capsid formation, or RNA encapsidation.
It is intriguing that none of the JFH1G33A passaging experiments saw the reversion of the engineered mutation back to its WT form, since alanine and glycine are just a single mutational step apart. Then again, an A33G change would require a transversion mutation of C to G (GCA to GGA). In contrast, the compensatory mutations G32D (GGC to GAC), V34A (GUA to GCA), L36S (UUG to UCG), and L37S (UUG to UCG) all are created by transition mutations, which generally occur at higher frequencies than transversions. This may partly explain the preference of JFH1G33A to select compensatory mutations rather than reverting to the WT sequence. In the study by Murray and colleagues, alanine substitutions between residues 69 and 72 reduced infectious virus production by ∼3 logs. However, the infectivity of this mutant could be rescued by compensatory mutations within p7 (F26L/S) or NS2 (A67P) that were selected during multiple cell passages (32). Such compensatory mutations indicate specific interactions between core protein and these nonstructural proteins. Although the compensatory mutations identified in our study all were confined to the originally mutated core protein, they also may act to increase interactions of core protein with any of the numerous viral and/or cellular factors involved in particle generation. The fact that the compensatory mutations at residues 34, 36, and 37 each reside on the same side of helix 2 reinforces the relevance of the helix-loop-helix motif from a functional point of view. Interestingly, the compensatory mutations at residues 36 and 37 both replace the bulky hydrophobic residue leucine with the smaller and more polar residue serine. This substitution yields higher flexibility to the helix-loop-helix motif, as illustrated by the wide distribution of interhelix angles in Fig. 8. This flexibility is thought to allow the reversion mutants to easily adopt the optimal WT conformation of helix-loop-helix required for the functionality of core protein. This is also true for the G32D/G33A and G33A/V34A reversion mutants. Interestingly, as the side chain of residue 32 is not involved in the cluster of hydrophobic residues that maintains the interactions between helix 1 and helix 2, this glycine-to-aspartic acid alteration is not expected to disturb the overall structure of the helix-loop-helix motif. Taken together, these findings indicate that both the proper folding of the helix-loop-helix motif as well as the flexibility of its conformation are essential features required for the functionality of core protein. This study highlights the functional plasticity of the N-terminal part of core protein, which is expected to ensure the numerous functions of this protein.
ACKNOWLEDGMENTS
This work was supported by the Medical Research Council, United Kingdom (to A.H.P.), the French Centre National de la Recherche Scientifique, grants from the French National Agency for Research on AIDS and Viral Hepatitis (ANRS), and the FINOVI foundation (to F.P.).
We thank T. Wakita, J. McLauchlan, P. Targett-Adams, C. Rice, M. Ryan, J. McKeating, and H. Jacobsen for the provision of reagents and N. Jeffery for expert technical assistance.
Footnotes
Published ahead of print 9 November 2011
REFERENCES
- 1. Alsaleh K, et al. 2010. Identification of basic amino acids at the N-terminal end of the core protein that are crucial for hepatitis C virus infectivity. J. Virol. 84:12515–12528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angus AG, et al. 2010. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J. Gen. Virol. 91:122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anonymous 2004. Global burden of disease (GBD) for hepatitis C. J. Clin. Pharmacol. 44:20–29 [DOI] [PubMed] [Google Scholar]
- 4. Barba G, et al. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. U. S. A. 94:1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartenschlager R, Penin F, Lohmann V, Andre P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 19:95–103 [DOI] [PubMed] [Google Scholar]
- 6. Boulant S, et al. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281:22236–22247 [DOI] [PubMed] [Google Scholar]
- 7. Boulant S, Targett-Adams P, McLauchlan J. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88:2204–2213 [DOI] [PubMed] [Google Scholar]
- 8. Boulant S, Vanbelle C, Ebel C, Penin F, Lavergne JP. 2005. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J. Virol. 79:11353–11365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bukh J, Purcell RH, Miller RH. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. U. S. A. 91:8239–8243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen SL, Morgan TR. 2006. The natural history of hepatitis C virus (HCV) infection. Int. J. Med. Sci. 3:47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clayton RF, et al. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672–7682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cuevas JM, Gonzalez-Candelas F, Moya A, Sanjuan R. 2009. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J. Virol. 83:5760–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Chassey B, et al. 2008. Hepatitis C virus infection protein network. Mol. Syst. Biol. 4:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Duvignaud JB, Leclerc D, Gagne SM. 2010. Structure and dynamics changes induced by 2,2,2-trifluoro-ethanol (TFE) on the N-terminal half of hepatitis C virus core protein. Biochem. Cell Biol. 88:315–323 [DOI] [PubMed] [Google Scholar]
- 15. Duvignaud JB, et al. 2009. Structure and dynamics of the N-terminal half of hepatitis C virus core protein: an intrinsically unstructured protein. Biochem. Biophys. Res. Commun. 378:27–31 [DOI] [PubMed] [Google Scholar]
- 16. Flint M, et al. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujimoto Y, et al. 2004. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta 1644:47–59 [DOI] [PubMed] [Google Scholar]
- 18. Gastaminza P, et al. 2010. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J. Virol. 84:10999–11009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gastaminza P, Kapadia SB, Chisari FV. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hope RG, McLauchlan J. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913–1925 [DOI] [PubMed] [Google Scholar]
- 21. Hüssy P, Langen H, Mous J, Jacobsen H. 1996. Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology 224:93–104 [DOI] [PubMed] [Google Scholar]
- 22. Iro M, et al. 2009. A reporter cell line for rapid and sensitive evaluation of hepatitis C virus infectivity and replication. Antiviral Res. 83:148–155 [DOI] [PubMed] [Google Scholar]
- 23. Jolivet M, Penin F, Dalbon P, Ladaviere L, Lacoux X. 1997. Antigenic structural peptide, antigenic and immunogenic compounds and uses for detecting, preventing and treating an HCV infection. Patent application EP 1015481, WO 98/39360
- 24. Koradi R, Billeter M, Wuthrich K. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 25. Lavanchy D. 2009. The global burden of hepatitis C. Liver Int. 29(Suppl. 1):74–81 [DOI] [PubMed] [Google Scholar]
- 26. Lindenbach BD, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 27. Masaki T, et al. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McLauchlan J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral. Hepat. 7:2–14 [DOI] [PubMed] [Google Scholar]
- 29. Ménez R, et al. 2003. Crystal structure of a hydrophobic immunodominant antigenic site on hepatitis C virus core protein complexed to monoclonal antibody 19D9D6. J. Immunol. 170:1917–1924 [DOI] [PubMed] [Google Scholar]
- 30. Miyanari Y, et al. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097 [DOI] [PubMed] [Google Scholar]
- 31. Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5:453–463 [DOI] [PubMed] [Google Scholar]
- 32. Murray CL, Jones CT, Tassello J, Rice CM. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rice LM, Brunger AT. 1994. Torsion angle dynamics: reduced variable conformational sampling enhances crystallographic structure refinement. Proteins 19:277–290 [DOI] [PubMed] [Google Scholar]
- 34. Robertson B, et al. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch. Virol 143:2493–2503 [DOI] [PubMed] [Google Scholar]
- 35. Santolini E, Migliaccio G, La Monica N. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. 2003. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160:65–73 [DOI] [PubMed] [Google Scholar]
- 37. Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158–37169 [DOI] [PubMed] [Google Scholar]
- 38. Shepard CW, Finelli L, Alter MJ. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558–567 [DOI] [PubMed] [Google Scholar]
- 39. Shimoike T, Mimori S, Tani H, Matsuura Y, Miyamura T. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73:9718–9725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simmonds P, et al. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973 [DOI] [PubMed] [Google Scholar]
- 41. Soriano V, Peters MG, Zeuzem S. 2009. New therapies for hepatitis C virus infection. Clin. Infect. Dis. 48:313–320 [DOI] [PubMed] [Google Scholar]
- 42. Stein EG, Rice LM, Brunger AT. 1997. Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J. Magn. Reson. 124:154–164 [DOI] [PubMed] [Google Scholar]
- 43. Vieyres G, et al. 2010. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J. Virol. 84:10159–10168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wakita T, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Witteveldt J, et al. 2009. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J. Gen. Virol. 90:48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]