Abstract
Natural host sooty mangabeys (SM) infected with simian immunodeficiency virus SIVsmm do not develop AIDS despite high viremia. SM and other natural hosts express very low levels of CCR5 on CD4+ T cells, and we recently showed that SIVsmm infection and robust replication occur in vivo in SM genetically lacking CCR5, indicating the use of additional entry pathways. SIVsmm uses several alternative coreceptors of human origin in vitro, but which molecules of SM origin support entry is unknown. We cloned a panel of putative coreceptors from SM and tested their ability to mediate infection, in conjunction with smCD4, by pseudotypes carrying Envs from multiple SIVsmm subtypes. smCXCR6 supported efficient infection by all SIVsmm isolates with entry levels comparable to those for smCCR5, and smGPR15 enabled entry by all isolates at modest levels. smGPR1 and smAPJ supported low and variable entry, whereas smCCR2b, smCCR3, smCCR4, smCCR8, and smCXCR4 were not used by most isolates. In contrast, SIVsmm from rare infected SM with profound CD4+ T cell loss, previously reported to have expanded use of human coreceptors, including CXCR4, used smCXCR4, smCXCR6, and smCCR5 efficiently and also exhibited robust entry through smCCR3, smCCR8, smGPR1, smGPR15, and smAPJ. Entry was similar with both known alleles of smCD4. These alternative coreceptors, particularly smCXCR6 and smGPR15, may support virus replication in SM that have restricted CCR5 expression as well as SM genetically lacking CCR5. Defining expression of these molecules on SM CD4+ subsets may delineate distinct natural host target cell populations capable of supporting SIVsmm replication without CD4+ T cell loss.
INTRODUCTION
HIV-1 infection of humans and simian immunodeficiency virus SIVmac infection of non-natural host rhesus macaques (RM) result in high viral loads, peripheral CD4+ T cell loss, and progression to AIDS. In contrast, SIVsmm infection of natural host sooty mangabeys (SM) rarely leads to peripheral CD4+ T cell loss or disease despite viral loads comparable to those measured in non-natural hosts. Identifying the mechanisms responsible for the different outcomes of infection in natural hosts compared with non-natural simian or recent human hosts has become a central focus of efforts to understand AIDS pathogenesis. Although many models of natural host infection exist, SIVsmm is unique because cross-species transmission of SIVsmm from SM to humans and RM gave rise to pathogenic HIV-2 and SIVmac, respectively (2, 28). Similarly, experimental transmission of SIVsmm from infected SM hosts results in AIDS in non-natural host RM (40). While the factors regulating pathogenic versus nonpathogenic outcomes of infection are complex and not fully understood (9), comparison of infected SM with RM recently revealed differential targeting of CD4+ T cell subsets (45). Thus, an understanding of SIVsmm cellular tropism may identify cells in natural host SM that maintain viral replication without leading to CD4+ T cell depletion or AIDS.
Classically, SIVsmm was thought to exclusively use the entry coreceptor CCR5 in vivo. Highly restricted expression of CCR5 on CD4+ T cells is characteristic of SIV natural hosts, including SM, compared with non-natural host species (46, 47), and may contribute to distinct cellular targeting in vivo (45). However, SIVsmm use of non-CCR5 entry in vivo was revealed recently by the identification of common CCR5 deletion alleles among SM (CCR5Δ2 and CCR5Δ24) that abrogate CCR5 cell surface expression and entry coreceptor function (50). A substantial minority of captive SM in U.S. primate centers possess homozygous null (CCR5Δ/Δ) genotypes, but the absence of functional CCR5 expression does not restrict SIVsmm replication in vivo, as CCR5Δ/Δ animals are susceptible to natural as well as experimental infection and exhibit high viral loads (50). These findings indicate that SIVsmm infection, replication, and cell targeting occur in vivo through alternative pathways in addition to CCR5.
A number of 7-transmbrane receptors (7TMRs) of human origin have been shown to function as alternative coreceptors of HIV and SIV in vitro. SIVsmm can enter target cells through human CXCR6, GPR1, and GPR15 in addition to CCR5 (11, 44, 50). Red-capped mangabeys, the majority of which have a CCR5Δ/Δ genotype, are naturally infected with SIVrcm that enters cells through human CCR2b but not CCR5 (4, 12, 62), and experimental transmission of SIVrcm to pigtailed macaques resulted in expanded SIVrcm tropism in vitro for human CCR4 (24). Additional human 7TMR molecules that support entry by subsets of HIV-1 or SIV include APJ, CCR3, and CCR8 (13, 14, 18, 44, 52, 54). Although CXCR4 is a common coreceptor in late stages of HIV-1 infection, SIVsmm, like other SIV strains, does not use CXCR4 except in rare cases (42).
Notably, the majority of alternative coreceptor studies have employed 7TMR molecules of human origin and rarely utilized molecules derived from homologous species, even though species-specific amino acid differences in 7TMRs may markedly affect coreceptor function (49). Therefore, in order to define the sooty mangabey molecules that mediate SIVsmm entry and that might be involved in CCR5-independent cell targeting and replication in vivo, we cloned smAPJ, smCCR2b, smCCR3, smCCR4, smCCR8, smCXCR4, smCXCR6, smGPR1, and smGPR15. Because SM have two distinct alleles of CD4 that differ in their putative gp120 binding regions (10, 23), we utilized both smCD4 alleles in conjunction with SM 7TMRs for functional coreceptor analysis. Finally, we compared coreceptor use by a variety of SIVsmm isolates. While infected SM do not progress to AIDS, SIVsmm exists in several viral genetic lineages (2), and SM at the Yerkes National Primate Research Center (YNPRC) infected with subtype 5 SIVsmm generally have lower peripheral CD4+ T cell counts than uninfected SM or those infected with subtype 1, 2, or 3 SIVsmm (1). In addition, a small number of infected SM have developed rapid profound CD4+ T cell depletion and were found to harbor SIVsmm variants with expanded use of human coreceptors in fusion assays, including CXCR4 (41, 42), but their use of SM coreceptors has not been described. Thus, we assessed SM-derived coreceptor use by a variety of SIVsmm strains in pseudotype infection.
MATERIALS AND METHODS
Cloning of SM receptor molecules.
Full-length 7TMRs were cloned from SM genomic DNA (APJ, CCR3, CCR8, CXCR6, GPR1, and GPR15) or SM cDNA (CCR2b, CCR4, and CXCR4) from animals housed at the Yerkes National Primate Research Center (YNPRC). Primers were designed to anneal outside the 7TMR open reading frame 5′ start codons and 3′ stop codons (see Table S1 in the supplemental material) based on sequences previously used to clone receptors from rhesus macaques (39) or sequences matched to human and rhesus macaque mRNA published in GenBank. Genomic DNA was extracted from unfractionated SM peripheral blood mononuclear cells (PBMC). cDNA was obtained from SM CD4+ T cells stimulated for 120 h with concanavalin A and interleukin-2 (IL-2) and subjected to mRNA purification (RNeasy Minikit; Qiagen, Valencia, CA) and reverse transcription (SuperScript first-strand synthesis system for reverse transcription-PCR [RT-PCR]; Invitrogen, Carlsbad, CA). The 50-μl PCR mixtures contained 200 ng genomic DNA or 0.1 μl cDNA, 1 unit of high-fidelity Phusion polymerase in HF buffer (Finnzyme; Thermo Scientific, Waltham, MA), primers (0.5 mM), and deoxynucleoside triphosphates (dNTPs) (0.2 mM). Thermocycling conditions were as follows: initial denaturation at 98°C for 45 s, followed by 30 cycles of 98°C for 10 s, annealing for 30 s, and 72°C for 1.5 min, and a final extension at 72°C for 10 min. Annealing temperatures were 53°C (CCR2b), 60°C (GPR1), 62°C (CCR4 and CXCR4), 64°C (APJ and CCR3), and 72°C (CCR8, CXCR6, and GPR15). PCR products were column purified (QIAquick PCR purification; Qiagen). CCR8, CXCR6, and GPR15 were then cloned into the expression plasmid pcDNA3.1+ (Invitrogen) using dual restriction digests with HindIII, BamHI, and XhoI (see Table S1 in the supplemental material). APJ, CCR2b, CCR4, CXCR4, and GPR1 were TA cloned using a pcDNA3.1 directional TOPO expression kit (Invitrogen). Clones were screened by restriction analysis and nucleotide sequencing. Cloning of wild-type smCCR5 from SM genomic DNA has been previously described (50). SM CD4 alleles (smCD4 2.1 and smCD4 4.3) were PCR amplified from cDNA (CD4-F2, 5′-TGT TTG AGA AGC AGC GGG C-3′; CD4-R1, 5′-GGG GAG GCT GCA AGT GGG-3′) and TOPO cloned into pcDNA3.1 (Invitrogen). Individual clones were screened and verified by sequence analysis. Because our smCXCR4 clone differed from a previously published smCXCR4 sequence (11) at two predicted amino acids (residues 32 and 171), the relevant region was PCR amplified and sequenced from genomic DNAs of 10 additional SM using primers within the CXCR4 gene (forward, 5′-AGT GAA TGT CCA TTC CTT TG-3′; reverse, 5′-GAC AAT ACC AGG CAG GAT AA-3′). Nucleotide and amino acid alignments were performed using the ClustalW algorithm in MacVector 12.0 (Cary, NC). 7TMR membrane topology predictions were made using TMpred (http://www.ch.embnet.org/software/TMPRED_form.html).
SIVsmm envelope clones and pseudotyped virus.
SIVsmm env genes were cloned from virions isolated from plasma of infected SM by employing single-genome amplification (SGA) methods previously described (34, 35). Briefly, viral RNA was purified from plasma using the QIAamp viral RNA purification kit (Qiagen), and cDNA was prepared using Superscript III (Invitrogen) and the primer SM-ER1 (5′-CTA TCA CTG TAA TAA ATC CCT TCC AGT CCC-3′). First-round PCR amplification was performed on endpoint-diluted cDNA using primers H2SM-EF1 (5′-CCC TTG AAG GMG CMR GAG AGC TCA TTA-3′) and SM-ER1 (5′-ATA AAA TGA GAC ATG TCT ATT GCC AAT TTG-3′), followed by a second round of amplification using primers H2SM-EF2 (5′-CAC CTA AAA ART GYT GCT AYC ATT GCC AG-3′) and SM-ER2 (5′-ATA AAA TGA GAC ATG TCT ATT GCC AAT TTG-3′). PCR amplicons were cloned into the pcDNA3.1V5HisTOPO-TA expression vector (Invitrogen), and transformed colonies that contained an insert were identified by PCR screening.
SIVsmm Env-mediated entry was assessed using luciferase-expressing viral pseudotypes as previously described (50). Pseudotyped virions were generated in 293T cells by cotransfecting pNL-Luc-E−R+ plasmid encoding an NL4-3-based env-deleted luciferase-expressing HIV-1 viral backbone (15) along with plasmids carrying SIVsmm env genes. Pseudotypes lacking Env (pNL-Luc-E−R+ cotransfected with empty pcDNA3.1+) served as a negative control, and virions carrying vesicular stomatitis G protein (VSV-G) served as a positive control. Cells were transfected overnight using Fugene (Promega, Madison, WI) and washed the following day to remove DNA and transfection reagent. Supernatants were collected at 3 days posttransfection and stored at −80°C in 5% sucrose to enhance stability. Prior to use in infection assays, pseudotype stocks were treated with DNase (50 units/ml for 15 min). Pseudotype virion stocks were quantified by HIV-1 Gag p24 antigen content as measured by enzyme-linked immunosorbent assay (ELISA) (Perkin-Elmer, Waltham, MA) and by infectivity based on luciferase output (relative light units [RLU]) measured in U87 cells stably expressing human CD4 and CCR5 (5).
Virus infection and coreceptor function analyses.
Coreceptor function was evaluated by infection of 293T cells transfected with CD4 and 7TMR molecules of interest as described previously (50). Target cells were cotransfected with plasmids encoding CD4 and 7TMR plasmids using Fugene, washed the next day to remove transfection reagent, and replated at 2 × 104 cells/well in 96-well plates. The next day, cells were infected with pseudotype virus using equal amounts of each virus based on luciferase activity (RLU) measured in U87/CD4/CCR5 cells or based on p24 Gag antigen content. Infections were carried out by spin inoculation (1,200 × g for 2 h at 25°C). Three days later, cells were lysed (0.5% Triton X-100 in phosphate-buffered saline [PBS]), and luciferase was quantified by adding an equal volume of luciferase substrate (luciferase assay system; Promega) and measuring activity on either an MLX microplate luminometer (Dynex Technologies, Chantilly, VA) or a Luminoskan Ascent microplate luminometer (Thermo Scientific). The two luminometers have similar dynamic ranges, but absolute RLU values are 100-fold higher when read on the Luminoskan Ascent unit, so values read on the MLX microplate luminometer were scaled up by a factor of 102 to normalize data between machines. For infections in which blocking of endogenous human CXCR4 expressed by 293T cells was necessary, cells were treated for 1 h preinfection with 20 μM AMD3100 (Sigma-Aldrich, St. Louis, MO), which was also maintained over the course of infection.
Nucleotide sequence accession numbers.
The sequences of the SM CCR2b, CCR3, CCR4, CCR8, CXCR4, CXCR6, GPR1, GPR15, and APJ clones have been submitted to GenBank under the following accession numbers: CCR2b, JN701018; CCR3, JN701013; CCR4, JN701019; CCR8, JN701014; CXCR4, JN701020; CXCR6, JN701015; GPR1, JN701016; GPR15, JN701017; and APJ, JN701012.
RESULTS
Cloning and sequence analysis of sooty mangabey 7-transmembrane receptors.
We amplified the full open reading frames of smAPJ, smCCR2b, smCCR3, smCCR4, smCCR8, smCXCR4, smCXCR6, smGPR1, and smGPR15 using primers upstream and downstream of the coding regions matching both human and rhesus macaque genomic or mRNA sequences in order to maximize likely homology to SM sequences (see Table S1 in the supplemental material). SM genomic DNA was used for cloning of smAPJ, smCCR3, smCCR8, smCXCR6, smGPR1, and smGPR15, which lack introns in the open reading frames, and cDNA from SM CD4+ PBMC was used for cloning of smCCR2b, smCCR4, and smCXCR4, which contain introns within the open reading frames. Amplified receptors were cloned into expression plasmids for sequencing and functional studies.
We aligned the predicted amino acid sequences of these SM 7TMR molecules to sequences of human and rhesus macaque molecules. SM molecules showed greater than 91% identity to human sequences and greater than 98% identity to RM sequences (Table 1). While the domains of these 7TMR molecules that might interact with Env to facilitate entry have not been mapped, studies with human CCR5 and CXCR4 show that all extracellular regions may contribute but that the N-terminal domain (NTD) and extracellular loop 2 (ECL2) are likely to be most critical for entry (19). Thus, we aligned the predicted extracellular regions between human and SM 7TMR molecules (Fig. 1). Some 7TMR molecules are highly conserved between SM, RM, and humans within extracellular domains, such as CCR4 and GPR1, which differ by only two extracellular residues between SM and humans, whereas CXCR6, CCR3, and CCR8 are particularly divergent between the species in extracellular domains (Table 1 and Fig. 1).
Table 1.
Relatedness of sooty mangabey and human 7TMRs
| SM 7TMR | % Overall amino acid identity to 7TMR froma: |
No. of residues differing from human receptorb: |
||
|---|---|---|---|---|
| Human | Rhesus macaque | NTD | ECL2 | |
| APJ | 98.7 | 99.7 | 0 | 1 |
| CCR2b | 96.7 | 99.2 | 1 | 2 |
| CCR3 | 91.3 | 98.6 | 6 | 4 |
| CCR4 | 98.6 | 100.0 | 0 | 1 |
| CCR8 | 94.4 | 99.2 | 8 | 1 |
| CXCR4 | 98.3 | 99.4 | 3 | 2 |
| CXCR6 | 94.5 | 98.3 | 8 | 2 |
| GPR1 | 97.7 | 99.7 | 1 | 1 |
| GPR15 | 97.2 | 99.4 | 1 | 1 |
Alignment between predicted amino acid sequences of SM molecules cloned here and homologous human and rhesus macaque sequences from GenBank.
Number of predicted amino acids differences between SM and human molecules in the N-terminal domain (NTD) and extracellular loop 2 (ECL2), two regions most relevant to coreceptor function based on mapping studies with CCR5 and CXCR4 (19).
Fig 1.
Human and sooty mangabey putative alternative coreceptors differ in extracellular regions. Seven-transmembrane receptors APJ, CCR2b, CCR3, CCR4, CCR8, CXCR4, CXCR6, GPR1, and GPR15 were cloned and sequenced from sooty mangabey, and predicted protein sequences were aligned to human sequences. Alignments between human and sooty mangabey extracellular domains, including the N-terminal domain (NTD) and extracellular loops (ECL), are shown.
Sequences have not been described previously for smAPJ, smCCR2b, smCCR3, smCCR4, smGPR1, and smGPR15, but smCXCR6 and smCXCR4 sequences have been reported (11, 49). Our clones matched the published smCXCR6 sequence but differed at two predicted amino acids from the previously published smCXCR4 sequence (E32 instead of K in the NTD and D171 instead of E in transmembrane domain 4). To confirm that our smCXCR4 clone was not an artifact of PCR amplification or a unique sequence limited to one animal, we sequenced CXCR4 genes from 10 additional SM. Genomic sequencing revealed E32/D171 in all 10 SM, indicating that the allele of smCXCR4 that we cloned is neither an artifact nor rare among SM in the YNPRC colony.
Both sooty mangabey alleles of CD4 mediate SIVsmm entry through smCCR5 but not smCXCR4.
Studies of SIV Env entry have traditionally been performed using human CD4 and coreceptors. Sooty mangabeys are known to carry two distinct alleles of CD4 (smCD4 2.1 and smCD4 4.3) (23), whose putative gp120 interaction domains differ from both huCD4 and from each other (23, 32). Thus, we determined whether both alleles of smCD4 support SIVsmm entry through smCCR5 and whether they differ from huCD4 in this respect. The two smCD4 alleles were cloned from cDNA and found to match the sequences previously reported (23).
We then tested function of the two smCD4 alleles in conjunction with smCCR5. Virions were generated using an HIV-1 viral backbone lacking Env and carrying luciferase in place of Nef, pseudotyped with subtype 1 SIVsmm Envs cloned by single-genome amplification (SGA) from infected SM with CCR5-wild-type (CCR5wt/wt) or CCR5-deficient (CCR5Δ/Δ) genotypes (FFv and FNp, respectively) as previously described (50). Target 293T cells were transfected with huCD4, smCD4 2.1, and smCD4 4.3, with or without smCCR5, and then infected with pseudotype virions. Luciferase output was measured as an indication of entry and infection (Fig. 2). Flow cytometry confirmed that huCD4, smCD4 2.1, and smCD4 4.3 were expressed at similar levels on the cell surface (data not shown). Virions lacking Env served as a negative control, and VSV-G pseudotypes served as a positive control (Fig. 2 and data not shown).
Fig 2.
Two alleles of sooty mangabey CD4 support SIVsmm Env-mediated entry via CCR5 in vitro. Target 293T cells were transfected with human CD4 (huCD4) or each of two sooty mangabey alleles of CD4 (smCD4 2.1 and smCD4 4.3), with or without sooty mangabey CCR5 (smCCR5). Cells were then infected with luciferase-expressing pseudotype virions carrying Env glycoproteins from two subtype 1 SIVsmm-infected SM, using equal amounts of each virus based on infectivity in U87/CD4/CCR5 cells (106 RLU of each virus). FFv Envs were derived from an infected animal with a CCR5wt/wt genotype, and FNp Envs were derived from an infected animal with a CCR5Δ/Δ genotype. Virions lacking envelope glycoprotein served as negative controls. Infection was measured by relative light units (RLU) in cell lysates at 3 days postinfection (n = 3; values are means ± standard deviations [SD]).
Both smCD4 2.1 and smCD4 4.3 enabled SIVsmm infection when transfected along with smCCR5 (Fig. 2). While modest quantitative differences were seen within individual experiments, there was no consistent difference in efficiency between smCD4 2.1, smCD4 4.3, and huCD4 over repeated experiments. Target cells expressing smCCR5 in the absence of CD4 did not permit SIVsmm entry, confirming that SIVsmm Env-mediated infection does not occur through smCCR5 independent of CD4. Thus, despite sequence differences, both smCD4 molecules support efficient SIVsmm entry in conjunction with smCCR5.
These results also showed that 293T cells expressing CD4 alone were unable to support entry by these isolates. Because 293T cells express endogenous human CXCR4 (16), the lack of entry following transfection of huCD4, smCD4 2.1, or smCD4 4.3 alone shows that human CXCR4 is not a coreceptor for these SIVsmm isolates, even in conjunction with smCD4. Furthermore, given the breadth of human alternative coreceptors used by many SIV strains, lack of entry into CD4-transfected cells establishes 293T as a suitable cell line for our further coreceptor assays of SIVsmm.
Multiple studies show that most naturally occurring SIV isolates, including SIVsmm, do not use human CXCR4, and one prior study reported that SIVsmm did not enter cells through a previously described but distinct allele of smCXCR4 (11). Therefore, we asked whether SIVsmm could utilize the smCXCR4 we cloned, which we found is common among YNPRC animals, in conjunction with species-matched CD4 (Fig. 3). Neither the SIVsmm Envs derived from infected SM with wild-type CCR5 nor those from CCR5-deficient SM were capable of infecting cells through smCXCR4, regardless of which smCD4 allele was used. Efficient entry through smCXCR4 was seen with virions carrying the HIV-1 89.6 dual-tropic R5X4 Env (data not shown), confirming smCXCR4 expression and functionality. A similar lack of entry through smCXCR4/smCD4 was seen using an extended panel of additional SIVsmm isolates (discussed further below). These results confirm that smCXCR4 is not generally used by SIVsmm, in contrast to the case with HIV-1 infection, where acquisition of tropism for human CXCR4 is common.
Fig 3.
Sooty mangabey CXCR4 does not support entry by SIVsmm Envs that do not use human CXCR4. Target 293T cells were transfected with sooty mangabey CXCR4 (smCXCR4) in conjunction with smCD4 2.1 (A) or smCD4 4.3 (B), and then infected with luciferase-expressing SIVsmm Env pseudotype virions as described in the legend to Fig. 2 (using 5 × 105 RLU of each virus). Infection was measured by relative light units (RLU) in cell lysates at 3 days postinfection (n = 3; values are means ± SD).
Alternative coreceptors from sooty mangabey support subtype 1 SIVsmm infection.
Small differences in alternative coreceptor sequences between species can markedly alter coreceptor function (49), suggesting that prior studies using human 7TMR molecules may not necessarily reflect SIVsmm coreceptor use in natural host SM infection. Therefore, we determined the capacities of eight additional 7TMR molecules cloned from SM to mediate SIVsmm infection in conjunction with both smCD4 alleles. These coreceptors were as follows: smCXCR6, smGPR15, and smGPR1, since we previously showed that SIVsmm frequently uses human CXCR6, GPR15, and GPR1 (50); smCCR2b and smCCR4, since the human homologs of these receptors support entry by SIVrcm derived from natural host red-capped mangabeys or following passage into pigtail macaques (4, 12, 24, 62); and smAPJ, smCCR3, and smCCR8, since the human molecules are used by limited subsets of HIV or SIV (13, 14, 18, 44, 52, 54).
As shown in Fig. 4, smCXCR6 acted as a robust entry pathway for SIVsmm in conjunction with either smCD4 4.3 or smCD4 2.1, yielding infection levels generally comparable to those with smCCR5. smGPR15 also supported entry, although to a lesser extent than smCXCR6 or smCCR5. These results are similar to those seen previously using transfected human CXCR6 and GPR15 (50). We also observed low levels of infection mediated by smGPR1 and smAPJ, where luciferase output was above the background in mock-transfected cells for most Envs but well below output levels in cells expressing smCCR5, smCXCR6, or smGPR15. Surprisingly, neither smCCR2b nor smCCR4 supported infection by SIVsmm Env pseudotypes, even though the human 7TMR homologs support SIVrcm infection. smCCR3 and smCCR8 also failed to mediate infection by these SIVsmm isolates. smCD4 2.1 and smCD4 4.3 supported similar levels of infection (Fig. 4A and B), whereas none of the alternative coreceptors enabled infection in the absence of CD4 (data not shown). Similar coreceptor patterns were seen for the Envs from CCR5-deficient SM and SM with wild-type CCR5, consistent with the notion that SIVsmm alternative coreceptor use is not a feature exclusive to virus from CCR5-deficient SM (50).
Fig 4.
Alternative coreceptors of sooty mangabey origin support SIVsmm Env-mediated entry in vitro. 293T cells transfected with smCD4 along with smAPJ, smCCR2b, smCCR3, smCCR4, smCCR5, smCCR8, smCXCR4, smCXCR6, smGPR1, and smGPR15 were infected with pseudotypes carrying subtype 1 SIVsmm Envs from SM with CCR5wt/wt (FFv) and CCR5Δ/Δ genotypes (5 × 105 RLU of each virus). Infection was assessed in conjunction with smCD4 2.1 (A) and smCD4 4.3 (B) and measured by relative light units in cell lysates at 3 days postinfection (n = 3; values are means ± SD).
SIVsmm isolates of different subtypes show similar alternative coreceptor use in vitro.
SIVsmm isolates circulating in U.S. primate centers cluster in several genetic subtypes. Infected SM generally do not display CD4+ T cell loss, but collectively, animals at YNPRC infected with subtype 5 SIVsmm have lower peripheral CD4+ T cell counts than SM infected with other subtypes (1). Therefore, we asked whether coreceptor usage patterns differ among SIVsmm subtypes and, in particular, whether subtype 5 SIVsmm isolates exhibit a distinct pattern that might lead to expanded tropism and consequent CD4+ T cell loss. Pseudotyped virions were generated with SGA-derived Envs from SM infected with SIVsmm subtype 5, as well as subtypes 2 and 3, and tested for infection of 293T cells transfected with SM-derived coreceptors and smCD4. Since we previously observed no difference in entry between the two smCD4 alleles, we utilized smCD4 4.3 to test this extended panel of SIVsmm Envs.
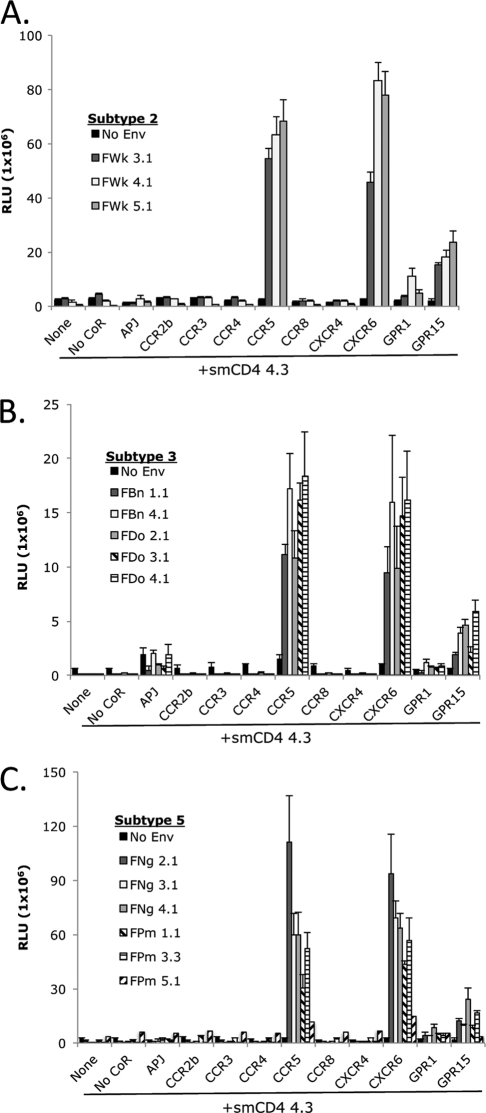
As shown in Fig. 5, SIVsmm subtypes 2, 3, and 5 exhibited alternative coreceptor use patterns comparable to those previously observed for subtype 1 SIVsmm (Fig. 4), with robust entry through smCXCR6 and smCCR5. These Envs displayed modest use of smGPR15 and levels of entry through smGPR1 and smAPJ that were variable and generally marginal. Subtype 2, 3, and 5 Envs did not enter cells expressing smCXCR4 in conjunction with smCD4. Notably, the subtype 5 SIVsmm did not have expanded alternative coreceptor use relative to other subtypes of SIVsmm examined, suggesting that factors other than distinct coreceptor use patterns underlie the relative CD4+ T cell loss associated with this SIVsmm lineage.
Fig 5.
Alternative coreceptor use by an extended panel of SIVsmm isolates of distinct subtypes. Target 293T cells expressing smCD4 4.3 in conjunction with SM-derived coreceptors were infected with pseudotype virions (3.5 × 105 RLU of each virus) carrying Envs from SIVsmm subtype 2 (FWk) (A), subtype 3 (FBn and FDo) (B), or subtype 5 (FNg and FPm) (C). Infection was measured by relative light units in cell lysates at 3 days postinfection (n = 3; values are means ± SD).
Rare SIVsmm isolates that cause rapid CD4+ T cell depletion in sooty mangabeys exhibit expanded SM coreceptor use.
In contrast to the case for the majority of infected animals, rare SIVsmm isolates that give rise to profound rapid CD4+ T cell depletion in infected SM hosts have been described (42). Previous studies employing cell-cell fusion assays and coreceptors of human origin showed that these SIVsmm isolates have expanded tropism for several coreceptors, including CXCR4 (41, 42). Therefore, we determined whether the expanded use of human coreceptors previously described also reflects expanded use of SM-derived 7TMR molecules and whether these coreceptors can support infection as well as cell-cell fusion. We tested pseudotyped virions carrying SGA-derived Envs isolated from three of these very-low-CD4 SM (FJv, FPy, and FTv) for infection of transfected 293T target cells, along with Envs from one animal with typical nonprogressive infection (FFv) for comparison (Fig. 6).
Fig 6.
SIVsmm Env from CD4-depleted sooty mangabeys uses sooty mangabey CXCR4 and a broad range of additional coreceptors for entry in vitro. (A) Target 293T cells were transfected with or without smCD4 4.3, smCXCR4, and smCCR5, treated with or without the CXCR4 antagonist AMD3100 (20 μM), and then infected with pseudotype virions. (B) 293T cells transfected with smCD4 4.3 in conjunction with SM 7TMR molecules (smAPJ, smCCR2b, smCCR3, smCCR4, smCCR5, smCCR8, smCXCR4, smCXCR6, smGPR1, an smGPR15) were treated with AMD3100 (20 μM) to block endogenous CXCR4 and then infected with pseudotype virions. Pseudotypes carried SIVsmm Envs from CD4-low sooty mangabeys FJv (red), FPy (blue), and FTv (orange) or from one animal with a typical course of infection, FFv (black). Inocula were equalized on the basis of p24 Gag antigen content (2 ng p24). Infection was measured by relative light units in cell lysates at 3 days postinfection (n = 3; values are means ± SD).
Because 293T cells express low levels of endogenous huCXCR4 (16), we first determined if transfection of smCD4 alone enabled infection. As shown in Fig. 6A, smCD4 alone enabled detectable levels of entry by pseudotypes carrying Envs from the low-CD4 animals FJv, FPy, and FTv. This pattern differed from that seen with previously tested SIVsmm isolates (Fig. 3 to 5 and FFv in Fig. 6). Pretreatment with the CXCR4 inhibitor AMD3100 completely abrogated entry of the SIVsmm isolates from FJv, FPy, and FTv into cells transfected with smCD4 alone, confirming that infection is mediated by endogenous human CXCR4 on 293T cells. Additionally, since AMD3100 completely blocked background entry by these isolates, 293T cells treated with AMD3100 can be used to examine other coreceptors.
Transfection of 293T cells with smCD4 and smCXCR4 together resulted in markedly increased levels of infection compared to those in cells transfected with smCD4 alone (Fig. 6A). Thus, SM-derived smCXCR4 supports robust infection by these low-CD4 SIVsmm isolates. AMD3100 pretreatment blocked the majority of infection in smCXCR4-transfected cells, suggesting that AMD3100 has substantial activity against smCXCR4 in addition to human CXCR4. Alignment of the human and SM CXCR4 sequences showed that amino acid residues previously identified as critical to the interaction between AMD3100 and human CXCR4 (D171, D262, and E288) were conserved between the two species (see Fig. S1 in the supplemental material) (27, 51). The SIVsmm isolates from FJv, FPy, and FTv retained the ability to infect cells via smCCR5. Based on this transfection system, entry by these isolates through smCXCR4 appeared to be at least as efficient as entry through smCCR5.
We then tested the extended panel of SM-derived 7TMR molecules with low-CD4-derived SIVsmm Envs. Target cells were transfected with 7TMR molecules and smCD4, pretreated with AMD3100 to block entry through endogenous 293T CXCR4, and infected with pseudotype viruses. As shown in Fig. 6B, the SIVsmm isolates from FJv, FPy, and FTv displayed broad and efficient use of multiple alternative coreceptors. Like those of all other SIVsmm isolates studied, these Envs used smCXCR6 very efficiently and smGPR15 slightly less so. In addition, while none of the typical SIVsmm Envs used smCCR3 or smCCR8 (Fig. 4 and 5 and FFv in Fig. 6B), both of these pathways supported infection by the SIVsmm isolates from FJv, FPy, and FTv. Finally, while most SIVsmm Envs used smAPJ and smGPR1 to a very limited extent, these 7TMRs supported substantially more robust infection by SIVsmm isolates from these low-CD4 SM. Notably, neither smCCR2b nor smCCR4 supported infection (Fig. 6B), nor did entry occur through any coreceptors in the absence of smCD4 (data not shown). Thus, in contrast to typical SIVsmm Envs, the SIVsmm Envs derived from rare CD4-depleted SM exhibited expanded use of SM alternative coreceptors smCXCR4, smCCR3, and smCCR8 and efficient entry though smAPJ, smCXCR6, smGPR1, and smGPR15, while retaining the ability to enter cells through smCCR5. This finding is consistent with earlier results using human CXCR4, CCR8, and CXCR6 in fusion assays and extends the breadth of coreceptor use to include SM-derived APJ, CCR3, GPR1, and GPR15.
DISCUSSION
Natural SIVsmm infection of SM is typically nonpathogenic despite robust virus replication. Key findings associated with the benign nature of this infection include the absence of chronic immune activation and a pattern of infected cells in vivo that shows relative sparing of CD4+ central memory T cells (Tcm) (45, 56). While the relative sparing of SM CD4+ Tcm from infection is associated with low CCR5 expression, previous studies from our group have shown that a substantial minority of SIVsmm-infected SM possess CCR5-null genotypes (50), indicating that alternative entry coreceptors are used in vivo and will need to be defined in order to understand mechanisms underlying viral tropism and targeting in vivo. In this study, we cloned multiple potential alternative coreceptors of SM origin and found that smCXCR6 was used consistently and very efficiently, with entry levels comparable to those for smCCR5, while smGPR15 use was also broad although somewhat less efficient. In contrast, entry through smGPR1 and smAPJ was variable and typically low level, and smCXCR4, smCCR3, and smCCR8 were used only by SIVsmm isolates from rare SM with profound CD4+ T cell loss that were previously demonstrated to have expanded human coreceptor use.
Robust use of the principal SM alternative coreceptors CXCR6 and GPR15, along with CCR5, was a feature of all SIVsmm isolates studied, including isolates from SM with CCR5 wild-type as well as CCR5-null genotypes. While CCR5-null SM have high-level viremia indicative of efficient alternative coreceptor function in vivo (mean, >104 copies/ml), a modest but significantly higher level of viremia (∼0.5-log-unit difference) was seen in SM with CCR5 homozygous wild-type genotypes, and intermediate viral loads were seen in heterozygous animals (50). Those findings suggest that alternative pathways are exclusively responsible for SIVsmm replication in animals lacking CCR5, while both CCR5 and alternative coreceptors may be used in hosts where both CCR5 and other pathways are available.
The SM model of SIV infection is of keen interest given the ability of natural hosts to avoid disease despite robust viral replication. Although multiple mechanisms likely contribute to this resistance, including distinct immune activation patterns and CD4-negative T helper cell function (3, 9, 41, 56, 61), entry coreceptor use by immunodeficiency viruses defines viral target cell tropism and can be an important determinant of CD4+ T cell loss. Acquisition of CXCR4 use by HIV-1 parallels rapid CD4+ T cell decline as HIV-1 gains the ability to infect CXCR4-expressing naïve T cells (6, 7, 31). In SIVmac-infected rhesus macaques, depletion of CD4+ Tcm correlates with CD4+ T cell loss and disease progression, suggesting a critical role for Tcm in maintaining T cell homeostasis (33, 43, 48). Natural hosts of SIV, including SM, have low CCR5 expression on CD4+ T cells relative to that in non-natural hosts (46, 47, 55, 59, 60). CCR5 is especially restricted on CD4+ Tcm of SM compared with rhesus macaques ex vivo and following in vitro stimulation (45). Tcm of SM are also relatively resistant to SIV infection in vitro compared with those of rhesus macaques, and they harbor lower levels of viral DNA in vivo (45). Thus, restricted coreceptor expression may protect critical cells from infection and preserve memory CD4+ T cell homeostasis in natural hosts. However, overall levels of viremia in SM are equivalent to or exceed those in rhesus macaques (56), indicating no lack of susceptible target cells in SM despite limited CCR5 expression. Therefore, combined with protection of critical target cells mediated by restricted CCR5 expression, alternative coreceptors may define populations of more dispensable target cells capable of supporting SIV replication in natural hosts without disruption of T cell homeostasis.
In human and mouse cells, CXCR6 is found on subsets of CD4+ TH1, TH17, and TREG memory/effector cells, as well as NKT cells, but is generally not coexpressed with naïve or central memory markers such as CD45RA, CCR7, or L-selectin (25, 30, 37, 53, 58). If patterns are similar in SIV natural hosts, the exclusion of CXCR6 from less differentiated and self-renewing naïve cells or Tcm cells would be consistent with shifting of viral targets away from subsets critical for T cell homeostasis into potentially more expendable cell targets. While GPR15 is expressed in human blood CD4+ T cells and in lymphoid tissues (17, 21, 22, 29, 36), its distribution on specific subsets has not been defined. CXCR6 and GPR15 are also both expressed in human intestinal tissue (17, 36), which is an important site for viral replication. Antibodies to define surface expression of both SM molecules are not presently available, but we have found smCXCR6 and smGPR15 RNA transcripts in SM CD4+ T cells (data not shown). Thus, it will be important to define the patterns of expression for these receptors by SM CD4+ cell subsets and their coreceptor function under physiological expression conditions in primary cells responsible for replication in vivo.
Coreceptors other than CCR5 and CXCR4 have not generally been thought to be important in human infection with HIV-1. However, a recent report described a transmitter/founder virus from an acutely infected person that was impaired in its ability to use CCR5 and CXCR4 but entered efficiently through GPR15 (29). Allelic variants of CXCR6 have also been associated with disease progression in HIV-1-infected subjects (20, 38), although a direct virological mechanism is unclear since most HIV-1 isolates do not utilize CXCR6 for entry. Thus, it remains to be determined if alternative coreceptors, including CXCR6 or GPR15, may also play a role in HIV-1 pathogenesis in rare situations or more broadly in as-yet-unrecognized ways.
While the majority of infected SM show neither CD4 loss nor disease, a small number of previously reported animals had extensive CD4+ T cell depletion, which was associated with expanded SIVsmm use of human coreceptors, including huCXCR4, huCXCR6, and huCCR8 (42). We now confirm expanded tropism by Envs from these animals for SM-derived coreceptor molecules and identify smCCR3, smGPR1, smGPR15, and smAPJ utilization as well. Acquisition of CXCR4-mediated tropism for naïve CD4+ T cells is strongly linked to accelerated CD4+ T cell loss in HIV-infected humans, so it is uncertain whether the additional coreceptors used by these CXCR4-tropic SIVsmm Envs contribute further to disrupted CD4+ T cell homeostasis. In contrast, we did not see distinct alternative coreceptor use or smCXCR4-mediated entry by isolates derived from SM naturally infected with subtype 5 SIVsmm isolates, which display more moderate levels of CD4+ T cell loss (1). Thus, CD4+ T cell loss in subtype 5 SIVsmm-infected SM is likely due to factors other than expanded tropism by the virus, at least for the coreceptors tested here.
Multiple complex and nonexclusive mechanisms are likely involved in protecting SM and other natural hosts both from CD4+ T cell loss in most individuals and from disease in the few animals where CD4+ T cell depletion does occur (3, 8, 9, 41, 45, 56, 57, 61), Thus, both the target cells spared from infection and those supporting replication are critical to understanding the nature of infection and pathogenesis (26). Future studies are needed to define CXCR6 and GPR15 coreceptor expression patterns on SM CD4+ cell subsets, the relationship between alternative coreceptor and CCR5 expression, and how alternative coreceptors are regulated. Finally, comparing coreceptor expression patterns between natural and non-natural host species may identify more expendable populations of CD4+ cells in the natural host that are capable of supporting viral replication through use of alternative entry pathways without leading to disease progression.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. Shaheen and S. Bryan for performing p24 ELISAs and members of the Collman and Silvestri labs for valuable advice and discussions.
This work was supported by NIH grants R01AI035502 and R56AI091516 (R.G.C.), R37AI66998 (G.S.), R01AI58706 (C.A.D.), and R01AI065325 (C.A.). We acknowledge valuable support from the Penn Center for AIDS Research (P30AI45008). S.T.C.E., N.E.R., and N.F. were supported in part by grant T32AI007632.
Footnotes
Published ahead of print 16 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Apetrei C, et al. 2007. Virus subtype-specific features of natural simian immunodeficiency virus SIVsmm infection in sooty mangabeys. J. Virol. 81:7913–7923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Apetrei C, et al. 2005. Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J. Virol. 79:8991–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beaumier CM, et al. 2009. CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat. Med. 15:879–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beer BE, et al. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75:12014–12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bjorndal A, et al. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blaak H, et al. 2000. In vivo HIV-1 infection of CD45RA(+)CD4(+) T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4(+) T cell decline. Proc. Natl. Acad. Sci. U. S. A. 97:1269–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 94:1925–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brenchley JM, et al. 2008. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112:2826–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brenchley JM, Silvestri G, Douek DC. 2010. Nonprogressive and progressive primate immunodeficiency lentivirus infections. Immunity 32:737–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chakrabarti LA. 2004. The paradox of simian immunodeficiency virus infection in sooty mangabeys: active viral replication without disease progression. Front. Biosci. 9:521–539 [DOI] [PubMed] [Google Scholar]
- 11. Chen Z, Gettie A, Ho DD, Marx PA. 1998. Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in west Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac. Virology. 246:113–124 [DOI] [PubMed] [Google Scholar]
- 12. Chen Z, et al. 1998. Natural infection of a homozygous delta24 CCR5 red-capped mangabey with an R2b-tropic simian immunodeficiency virus. J. Exp. Med. 188:2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choe H, et al. 1998. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J. Virol. 72:6113–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choe H, et al. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148 [DOI] [PubMed] [Google Scholar]
- 15. Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944 [DOI] [PubMed] [Google Scholar]
- 16. Del Real G, et al. 2002. Blocking of HIV-1 infection by targeting CD4 to nonraft membrane domains. J. Exp. Med. 196:293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng HK, Unutmaz D, KewalRamani VN, Littman DR. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296–300 [DOI] [PubMed] [Google Scholar]
- 18. Doranz BJ, et al. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149–1158 [DOI] [PubMed] [Google Scholar]
- 19. Dragic T. 2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol. 82:1807–1814 [DOI] [PubMed] [Google Scholar]
- 20. Duggal P, et al. 2003. Genetic influence of CXCR6 chemokine receptor alleles on PCP-mediated AIDS progression among African Americans. Genes Immun. 4:245–250 [DOI] [PubMed] [Google Scholar]
- 21. Elbim C, et al. 2008. Early divergence in neutrophil apoptosis between pathogenic and nonpathogenic simian immunodeficiency virus infections of nonhuman primates. J. Immunol. 181:8613–8623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farzan M, et al. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 186:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fomsgaard A, Hirsch VM, Johnson PR. 1992. Cloning and sequences of primate CD4 molecules: diversity of the cellular receptor for simian immunodeficiency virus/human immunodeficiency virus. Eur. J. Immunol. 22:2973–2981 [DOI] [PubMed] [Google Scholar]
- 24. Gautam R, et al. 2009. Simian immunodeficiency virus SIVrcm, a unique CCR2-tropic virus, selectively depletes memory CD4+ T cells in pigtailed macaques through expanded coreceptor usage in vivo. J. Virol. 83:7894–7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Germanov E, et al. 2008. Critical role for the chemokine receptor CXCR6 in homeostasis and activation of CD1d-restricted NKT cells. J. Immunol. 181:81–91 [DOI] [PubMed] [Google Scholar]
- 26. Grossman Z, Meier-Schellersheim M, Paul WE, Picker LJ. 2006. Pathogenesis of HIV infection: what the virus spares is as important as what it destroys. Nat. Med. 12:289–295 [DOI] [PubMed] [Google Scholar]
- 27. Hatse S, et al. 2001. Mutation of Asp(171) and Asp(262) of the chemokine receptor CXCR4 impairs its coreceptor function for human immunodeficiency virus-1 entry and abrogates the antagonistic activity of AMD3100. Mol. Pharmacol. 60:164–173 [DOI] [PubMed] [Google Scholar]
- 28. Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389–392 [DOI] [PubMed] [Google Scholar]
- 29. Jiang C, et al. 2011. Primary infection by a human immunodeficiency virus with atypical coreceptor tropism. J. Virol. 85:10669–10681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim CH, et al. 2001. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J. Clin. Invest. 107:595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koot M, et al. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681–688 [DOI] [PubMed] [Google Scholar]
- 32. Kwong PD, et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Letvin NL, et al. 2006. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science 312:1530–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li B, et al. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 80:5211–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li B, et al. 2010. Nonpathogenic simian immunodeficiency virus infection of sooty mangabeys is not associated with high levels of autologous neutralizing antibodies. J. Virol. 84:6248–6253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Q, et al. 2008. Simian immunodeficiency virus-induced intestinal cell apoptosis is the underlying mechanism of the regenerative enteropathy of early infection. J. Infect. Dis. 197:420–429 [DOI] [PubMed] [Google Scholar]
- 37. Lim HW, Lee J, Hillsamer P, Kim CH. 2008. Human Th17 cells share major trafficking receptors with both polarized effector T cells and FOXP3+ regulatory T cells. J. Immunol. 180:122–129 [DOI] [PubMed] [Google Scholar]
- 38. Limou S, et al. 2010. Multiple-cohort genetic association study reveals CXCR6 as a new chemokine receptor involved in long-term nonprogression to AIDS. J. Infect. Dis. 202:908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Margulies BJ, Hauer DA, Clements JE. 2001. Identification and comparison of eleven rhesus macaque chemokine receptors. AIDS Res. Hum. Retroviruses 17:981–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McClure HM, et al. 1989. Spectrum of disease in macaque monkeys chronically infected with SIV/SMM. Vet. Immunol. Immunopathol. 21:13–24 [DOI] [PubMed] [Google Scholar]
- 41. Milush JM, et al. 2011. Lack of clinical AIDS in SIV-infected sooty mangabeys with significant CD4+ T cell loss is associated with double-negative T cells. J. Clin. Invest. 121:1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Milush JM, et al. 2007. Virally induced CD4+ T cell depletion is not sufficient to induce AIDS in a natural host. J. Immunol. 179:3047–3056 [DOI] [PubMed] [Google Scholar]
- 43. Okoye A, et al. 2007. Progressive CD4+ central memory T cell decline results in CD4+ effector memory insufficiency and overt disease in chronic SIV infection. J. Exp. Med. 204:2171–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Owen SM, et al. 2000. Simian immunodeficiency viruses of diverse origin can use CXCR4 as a coreceptor for entry into human cells. J. Virol. 74:5702–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paiardini M, et al. 2011. Low levels of SIV infection in sooty mangabey central memory CD4(+) T cells are associated with limited CCR5 expression. Nat. Med. 17:830–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pandrea I, et al. 2007. Paucity of CD4+CCR5+ T cells is a typical feature of natural SIV hosts. Blood 109:1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pandrea I, et al. 2008. Paucity of CD4+ CCR5+ T cells may prevent transmission of simian immunodeficiency virus in natural nonhuman primate hosts by breast-feeding. J. Virol. 82:5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Picker LJ, et al. 2004. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J. Exp. Med. 200:1299–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pohlmann S, et al. 2000. Simian immunodeficiency virus utilizes human and sooty mangabey but not rhesus macaque STRL33 for efficient entry. J. Virol. 74:5075–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Riddick NE, et al. 2010. A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo. PLoS Pathog. 6:e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rosenkilde MM, et al. 2004. Molecular mechanism of AMD3100 antagonism in the CXCR4 receptor: transfer of binding site to the CXCR3 receptor. J. Biol. Chem. 279:3033–3041 [DOI] [PubMed] [Google Scholar]
- 52. Rucker J, et al. 1997. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J. Virol. 71:8999–9007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sharron M, et al. 2000. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood 96:41–49 [PubMed] [Google Scholar]
- 54. Shimizu N, et al. 2009. Broad usage spectrum of G protein-coupled receptors as coreceptors by primary isolates of HIV. AIDS 23:761–769 [DOI] [PubMed] [Google Scholar]
- 55. Silvestri G, Paiardini M, Pandrea I, Lederman MM, Sodora DL. 2007. Understanding the benign nature of SIV infection in natural hosts. J. Clin. Invest. 117:3148–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Silvestri G, et al. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441–452 [DOI] [PubMed] [Google Scholar]
- 57. Sodora DL, et al. 2009. Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat. Med. 15:861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Unutmaz D, et al. 2000. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern is conserved between human and mouse. J. Immunol. 165:3284–3292 [DOI] [PubMed] [Google Scholar]
- 59. Veazey R, et al. 2003. Decreased CCR5 expression on CD4+ T cells of SIV-infected sooty mangabeys. AIDS Res. Hum. Retroviruses 19:227–233 [DOI] [PubMed] [Google Scholar]
- 60. Villinger F, Brice GT, Mayne A, Bostik P, Ansari AA. 1999. Control mechanisms of virus replication in naturally SIVsmm infected mangabeys and experimentally infected macaques. Immunol. Lett. 66:37–46 [DOI] [PubMed] [Google Scholar]
- 61. Vinton C, et al. 2011. CD4-like immunological function by CD4− T cells in multiple natural hosts of simian immunodeficiency virus. J. Virol. 85:8702–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Y, et al. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 74:6893–6910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.