Abstract
Highly pathogenic avian influenza (HPAI) H5N1 viruses continue to cause sporadic human infections with a high fatality rate. Respiratory failure due to acute respiratory distress syndrome (ARDS) is a complication among hospitalized patients. Since progressive pulmonary endothelial damage is the hallmark of ARDS, we investigated host responses following HPAI virus infection of human pulmonary microvascular endothelial cells. Evaluation of these cells for the presence of receptors preferred by influenza virus demonstrated that avian-like (α2-3-linked) receptors were more abundant than human-like (α2-6-linked) receptors. To test the permissiveness of pulmonary endothelial cells to virus infection, we compared the replication of selected seasonal, pandemic (2009 H1N1 and 1918), and potentially pandemic (H5N1) influenza virus strains. We observed that these cells support productive replication only of HPAI H5N1 viruses, which preferentially enter through and are released from the apical surface of polarized human endothelial monolayers. Furthermore, A/Thailand/16/2004 and A/Vietnam/1203/2004 (VN/1203) H5N1 viruses, which exhibit heightened virulence in mammalian models, replicated to higher titers than less virulent H5N1 strains. VN/1203 infection caused a significant decrease in endothelial cell proliferation compared to other subtype viruses. VN/1203 virus was also found to be a potent inducer of cytokines and adhesion molecules known to regulate inflammation during acute lung injury. Deletion of the H5 hemagglutinin (HA) multibasic cleavage site did not affect virus infectivity but resulted in decreased virus replication in endothelial cells. Our results highlight remarkable tropism and infectivity of the H5N1 viruses for human pulmonary endothelial cells, resulting in the potent induction of host inflammatory responses.
INTRODUCTION
Since 2003, highly pathogenic avian influenza (HPAI) A H5N1 viruses have spread to Asia, the Middle East, Africa, and Europe, and they present a continuing threat to global public health. As of October 2011, more than 565 laboratory-confirmed human cases of H5N1 virus infection have been reported, with a high fatality rate of approximately 60% (http://www.who.int/en/). In humans, common symptoms of H5N1 infection are fever, cough, and pneumonia with impairment of gas exchange (1, 18, 53). The disease can progress to acute respiratory distress syndrome (ARDS), multiorgan failure including combined hepatic/renal dysfunction, and death (24, 33, 53, 56). High viral load and exacerbated cytokine production in the lower respiratory tracts of patients have been shown to be associated with fatal cases (16, 24, 37, 43). Although pulmonary endothelial injury is expected to contribute to the complication of H5N1-induced ARDS, H5N1 infection of human pulmonary endothelial cells has not been well studied.
Receptor specificity is an important determinant of host range restriction among influenza viruses (41). The influenza virus hemagglutinin (HA) protein is responsible for binding to sialic acid (SA)-containing cell surface receptors for virus entry. In general, human influenza viruses express HA that preferentially bind to α-2,6-linked SA receptors, which are the predominant linkage expressed in the upper respiratory tracts (URT) of humans, whereas avian influenza viruses, such as contemporary avian H5N1 viruses, preferentially bind to α-2,3-linked SA receptors (21, 39, 52). Higher proportions of α-2,3-linked SA receptors in the human lower respiratory tract compared with the URT may partially explain the severity of H5N1 viral pneumonia in humans resulting from H5N1 viral attachment deep in the lungs (21, 39, 52). Detailed studies of virus attachment to human respiratory tissue have shown that H5N1 viruses bind to type II pneumocytes, alveolar macrophages, and nonciliated epithelial cells in the terminal bronchioles of the lower respiratory tract (48, 49). Therefore, the composition of a particular SA species present on cells can influence influenza virus tropism and pathogenesis.
The cleavage properties of HA0 and the distribution of functional proteases in the host are main factors for tissue tropism and systemic virus spread. Cleavage of the HA precursor molecule HA0 is required for the release of the fusion peptide and the conformational changes necessary for viral infectivity. Human influenza viruses contain a single basic amino acid at the cleavage site and can be cleaved by extracellular trypsin-like proteases present in certain respiratory cell types, restricting virus spread beyond the respiratory tract (8, 10, 23, 28). More recently, type II transmembrane serine proteases (TTSPs), such as TMPRSS2 (transmembrane protease, serine 2), have been identified in the human lung and may also support the spread of human influenza viruses by intracellular cleavage activation of HA (10). HA cleavability is a critical determinant of HPAI H5N1 virus pathogenicity in poultry and mammals (14, 20, 32, 40). The HA of HPAI H5N1 viruses invariably contain multiple basic amino acid residues (-RRRKK-) at the cleavage loop and can be cleaved intracellularly by ubiquitously expressed furin-like proteases, which are expressed in most organs of mammals, enabling the virus to infect many cell types and causing systemic infections (14, 20, 32, 40). Systemic spread of virus to multiple extrapulmonary organs is a characteristic of HPAI H5N1 virus infection in mammals (29, 30). In limited autopsy cases, viral genomic sequences or antigens have been detected not only in lung pneumocytes but also in tissue samples from spleen, brain, liver, and placenta (19, 24, 42, 56). Viral RNA has also been detected in blood samples from several fatal H5N1 cases (12, 15, 16, 42).
The nature of the interactions between H5N1 influenza virus and the host pulmonary vasculature is largely unknown. Pulmonary endothelium, the intimal lining of blood vessels, is the barrier between the blood and interstitium and has important regulatory functions (reviewed in reference 2). The alveolar capillaries are terminal networks in the pulmonary circulation, with microvascular endothelial cells present in the alveolar gas exchange units. The gross pulmonary pathology of H5N1 virus-infected lung shows that the diffuse alveolar damage and extensive consolidation with various degrees of hemorrhage is usually caused by alveolar capillary damage (24, 33). Moreover, the interaction of leukocytes and mediators, such as cytokines, oxygen radicals, and complement, is believed be responsible for endothelial damage and increased permeability to fluid and proteins (13). Recently published papers reported replication of HPAI H5N1 virus in lung microvascular endothelial cells (11, 35). However, the interaction of HPAI H5N1 viruses and those of other subtypes with pulmonary endothelial cells, which comprise one-third of the lung cell population, warrants further investigation.
In this study, we characterized the influenza virus receptors on human pulmonary microvascular endothelial cells and used an in vitro model to assess the infection and replication of selected seasonal, pandemic (2009 H1N1 and 1918), and potentially pandemic H5N1 influenza virus strains. The data demonstrated that pulmonary endothelial cells support productive replication only of HPAI H5N1 viruses and that the polybasic amino acids at the cleavage site play an important role in H5N1 virus replication. Furthermore, in comparison to infection with H1N1 subtype virus, HPAI H5N1 virus infection results in substantial induction of cytokines and adhesion molecules. These differences shed light on understanding the pathogenic mechanisms of pulmonary endothelial injury associated with H5N1 virus infections.
MATERIALS AND METHODS
Viruses.
The viruses used in this study are listed in Table 1. HPAI H5N1 and H7 subtype viruses were grown in the allantoic cavities of 10-day-old embryonated hen's eggs for 24 to 40 h at 37°C. Seasonal human H1N1 and H3N2 viruses were grown in eggs for 48 h at 33.5°C. Allantoic fluid was clarified by centrifugation, aliquoted, and stored at −70°C. The 2009 H1N1 virus, recombinant 1918 virus, and Tx/91 virus were propagated in MDCK cells for 48 h at 37°C. The supernatants were clarified by centrifugation, aliquoted, and stored at −70°C. Virus titers were determined by standard plaque assay (54). The identity of virus genes was confirmed by sequence analysis to verify that no inadvertent mutations were present during the generation of virus stocks. All research with HPAI H5 and H7 subtype viruses was conducted under biosafety level 3 containment, including enhancements required by the U.S. Department of Agriculture and the Select Agent Program (11a).
Table 1.
Viruses used in this study
| Virus | Abbreviation | Subtype | Cleavage sitea |
|---|---|---|---|
| A/Vietnam/1203/04 | VN/1203 | H5N1 | *PQRERRRKKR/GLF |
| A/Thailand/16/2004 | Thai/16 | H5N1 | *PQRERRRKKR/GLF |
| A/Thailand/SP/83/2004 | SP/83 | H5N1 | *PQRERRRKKR/GLF |
| A/Chicken/Korea/ES/2003 | CK/Korea | H5N1 | *PQRE-KRKKR/GLF |
| A/Netherlands/219/2003 | NL/219 | H7N7 | *PEIP-KRRRR/GLF |
| A/Netherlands/230/2003 | NL/230 | H7N7 | *PEIP-KRRRR/GLF |
| A/Canada/504/2004 | Can/504 | H7N3 | PENPKQAYQKRMTR/GLF |
| A/Canada/444/2004 | Can/444 | H7N3 | PENPKQAYQKQMTR/GLF |
| A/New York/107/2003 | NY/107 | H7N2 | PEKPKPR/GLF |
| A/Duck/New York/15024/96 | Dk/NY | H1N1 | PSIQSR/GLF |
| A/South Carolina/1/1918 | SC/18 | H1N1 | PEKQTR/GIF |
| A/Brisbane/59/2007 | Brisbane/59 | H1N1 | PEKQTR/GIF |
| A/Texas/36/1991 | Tx/91 | H1N1 | PEKQTR/GIF |
| A/Mexico/4482/2009 | Mexico/4482 | H1N1 | PSIQSR/GLF |
| A/Panama/2007/1999 | Pan/99 | H3N2 | PSIQSR/GLF |
| A/Wisconsin/67/2005 | Wis/05 | H3N2 | PSIQSR/GLF |
*, polybasic cleavage site on HA protein.
Generation of recombinant virus.
The HAdelVN/1203 mutant (virus lacking the polybasic cleavage site), wild-type HAVN/1203 virus, and the HA and neuraminidase (NA) of VN/1203 in the 6-gene backbone of A/Puerto Rico/8/34 (PR/8) virus were generated using reverse genetics plasmids (kindly provided by Adolfo Garcia-Sastre, Mount Sinai School of Medicine, New York, NY) (36). The construction of plasmids and the removal of nucleotides that encode the polybasic amino acids at the cleavage site of VN/1203 HA gene were as described by Park et al. (36). For virus rescue, plasmids were transfected into 293T cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Forty-eight hours later, the supernatants were transferred to MDCK cells, and virus was harvested upon observation of cytopathic effect (CPE). The rescued viruses were propagated in MDCK cells, and the identity of virus genes was confirmed by sequence analysis.
Cells and viral infection.
Primary human lung blood microvascular endothelial cells (HMVEC-LBI) (Lonza, Walkersville, MD) and an immortalized human lung microvascular endothelial cell (HULEC) line were grown in endothelial cell basal medium 2 (EBM-2) (500 ml) containing the following growth supplements: human epidermal growth factor (hEGF), 0.5 ml; hydrocortisone, 0.2 ml; GA-1000, 0.5 ml; fetal bovine serum (FBS), 25 ml; vascular endothelial growth factor (VEGF), 0.5 ml; human fibroblast growth factor B (hFGF-B), 2 ml; rat insulin-like growth factor 1 (R-IGF-1), 0.5 ml; and ascorbic acid, 0.5 ml (Lonza). The HULEC line possesses many features of primary pulmonary microvascular endothelial cells (9, 31). Cells were seeded onto six-well (5 × 105/well) or 12-well (3 × 105/well) plates and cultured for 24 h. Monolayers were then washed with EBM-2 supplemented with 0.3% bovine serum albumin (EBM-2–BSA) and infected with virus at a multiplicity of infection (MOI) ranging from 0.01 to 5 for 1 h. After washing, EBM-2–BSA was added to each well and left for the duration of the experiment. Human bronchial epithelium cells (Calu-3) were grown and inoculated as previously described (54).
Rat pulmonary microvascular endothelial cells (PMVEC) and rat pulmonary arterial endothelial cells (PAEC) (22) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS and penicillin-streptomycin. The cells were seeded onto six-well plates at 5 × 105/well and infected in a manner similar to that described above using DMEM supplemented with 0.3% BSA.
In experiments that required supplementation of exogenous protease for cleavage of the HA protein, either 1 μg/ml of N-p-tosyl-l-phenylalanine chloromethyl ketone (TPCK)-treated trypsin (Sigma-Aldrich, St. Louis, MO) or 10% chicken egg allantoic fluid was used.
Immunofluorescence staining and microscopy.
For surface receptor analysis, endothelial cells were seeded onto collagen-coated 8-well chamber slides. After 30 min of fixation with 3.7% formaldehyde in phosphate-buffered saline (PBS), cell monolayers were blocked with 3% BSA in PBS for 30 min and then sequentially incubated with either biotinylated Maackia amurensis lectins I and II (MAA I and II) (20 μg/ml) or biotinylated Sambucus nigra lectin (SNA) (20 μg/ml) (Vector Laboratories, Burlingame, CA) for 1 h, followed by the addition of fluorescein-conjugated avidin D (Vector Laboratories). To detect influenza A virus nucleoprotein (NP) antigen, cells seeded on chamber slides were infected with influenza virus at an MOI of 1. At 10 and 24 h postinoculation (p.i.), cells were fixed, permeabilized with 0.5% Triton X-100 in PBS for 20 min, and incubated with mouse anti-NP monoclonal antibody A-3 (50) followed by rhodamine-conjugated secondary antibody (Becton Dickinson [BD] Biosciences, San Diego, CA). Immunostained cells were mounted with 4′,6-diamidino-2-phenyindole (DAPI) (Sigma-Aldrich) and examined under a Zeiss Axioskop 2 fluorescence microscope.
Detection of α-2,6- and α-2,3-linked sialic acid residues by flow cytometry.
Endothelial cells grown in tissue culture flasks were trypsinized, and the cell suspension was passed through a cell strainer (BD Biosciences) to obtain single cells. After counting, cells were separated to 1.5-ml tubes containing 1 × 106 cells each, centrifuged at 1,000 × g for 10 min, and washed twice with EBM-2 medium and once with fluorescence-activated cell sorter (FACS) washing buffer (PBS with 2% FBS). A 200-μl aliquot of SNA-fluorescein isothiocyanate (FITC) or MAA-FITC (1 mg/ml; EY Laboratory Inc., San Mateo, CA) in different dilutions was added to the cells and incubated for 1 h at 4°C. Cells were washed and resuspended in FACS washing buffer. Data acquisition was performed on a BD LRS II flow cytometer and analyzed using BD FACSDiva software (BD Biosciences).
Virus binding assay.
The procedure for labeling of virus was modified and adapted from previously published methods (6, 48). Concentrated virus was labeled using the FluoroTag FITC conjugation kit (Sigma-Aldrich) according to the manufacturer's protocols. FITC-labeled virus was diluted and passed through a Millex-HV 0.45-μm filter, followed by determination of hemagglutination units (HAU). The confluent HMVEC-LBI grown on collagen-coated chamber slides were fixed with 2% paraformaldehyde for 30 min and blocked with 3% BSA. FITC-labeled virus at a comparable concentration was added to the cell monolayer and incubated overnight at 4°C in humidified chamber. After a PBS wash, cell monolayers were incubated with 3% H2O2 for 10 min and anti-FITC-horseradish peroxidase (HRP) antibody for 1 h, followed by 3-amino-9-ethylcarbazole (AEC) solution for 10 min in dark. The slides were mounted and evaluated under a Zeiss Axioskop 2 microscope.
Real-time quantitative PCR and Human Endothelial Cell Biology PCR array.
Total RNA from virus-infected or uninfected cells was extracted using the RNeasy Mini Kit (Qiagen; Carlsbad, CA) with DNase digestion, and 1.0 μg of total RNA was reverse transcribed with the QuantiTect reverse transcription kit (Qiagen). The cDNA products were subjected to real-time PCR assay using the QuantiTect SYBR green PCR kit (Qiagen) and analyzed using previously published methods (54). Additionally, cDNAs were analyzed using Human Endothelial Cell Biology RT2 Profiler PCR array (Qiagen), containing 84 genes involved in endothelial cell permissibility and vessel tone, angiogenesis, and endothelial cell activation and injury, according to the manufacturer's instructions. The expression data were analyzed through the vendor's web module, and fold changes in differential expression with significant P values were calculated and are presented.
Cytokine quantification.
Endothelial cells were infected with virus at an MOI of 1. Supernatants were collected at 24 h p.i., and cytokine levels were examined using the Bio-Plex Pro assay according to the manufacturer's instructions (Bio-Rad, Hercules, CA). The cytokines included in the assay were interleukin-6 (IL-6), IL-7, IL-8, granulocyte-macrophage colony-stimulating factor (GM-CSF), gamma interferon (IFN-γ), IP-10, monocyte chemoattractant protein 1 (MCP-1), RANTES, tumor necrosis factor alpha (TNF-α), and VEGF.
Cell proliferation assay.
Endothelial cells were seeded onto a 96-well plate at 5 × 104 per well 24 h before cells were inoculated with virus at different MOIs. Uninfected and infected cells (44 h p.i.) were incubated with 10 μl of cell proliferation reagent WST-1 (Roche, Indianapolis, IN) for 4 h. The stable tetrazolium salt WST-1 is cleaved to a soluble formazan through a complex cellular mechanism as a marker for cellular metabolic activity. The absorbance (450 nm) of each sample was then measured against a background control using a microplate reader according to the manufacturer's instructions. The measured absorbance directly correlates to the number of viable cells.
RESULTS
α-2,3-Sialic acid (SA) is more abundant than α-2,6-SA on the surface of pulmonary endothelial cells.
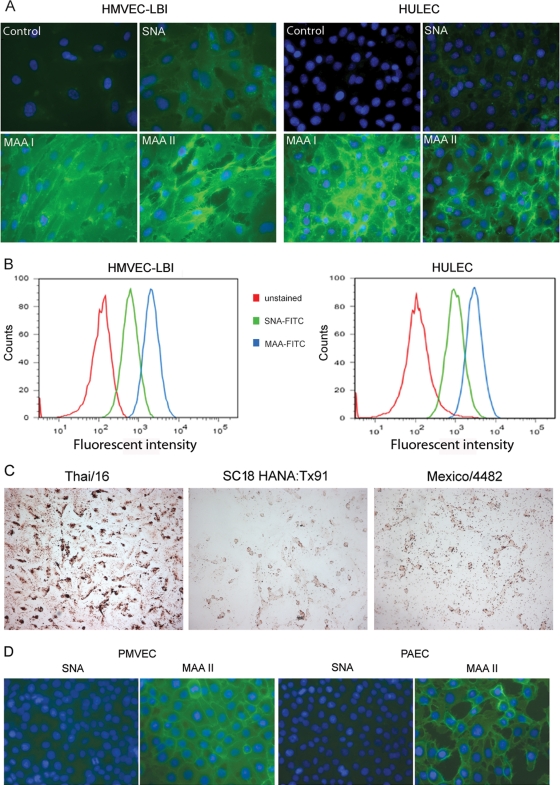
Influenza virus infection is initiated due to interactions between the HA protein and sialic acid-containing glycans on the surface of host cells. To determine the susceptibility of lung endothelial cells to influenza virus infection, we first examined the distribution of SA receptors on the cell surface using lectin staining. Sambucus nigra lectin (SNA) binds predominantly to the α-2,6-SA configuration, the receptor preferred by human influenza viruses. Maackia amurensis lectin I (MAA I) and II (MAA II) are two isoforms that identify the α-2,3-SA configuration, the preferred receptor of avian influenza viruses. MAA I is specific for SAα2-3Galβ1-4GalNAc, whereas MAA II is specific for SAα2-3Galβ1-3GalNAc (34).
Both α-2,6- and α-2,3-SA receptors were detected on the surfaces of HMVEC-LBI (primary cells) and HULEC (cell line) by lectin staining; however, the intensity of MAA (I and II) staining was visually stronger than that of SNA staining (Fig. 1A). To perform a relative quantification of lectin binding on pulmonary endothelial cell surfaces, human endothelial cells were removed from the culture flasks by a brief trypsinization, stained with FITC-conjugated lectin at different concentrations, and analyzed using flow cytometry. At a concentration of 20 μg/ml, almost 100% of human endothelial cells were stained positive for both α-2,6-SA (FITC-SNA) and α-2,3-SA (FITC-MAA) receptors; however, the mean fluorescence intensity (MFI) of MAA was three times higher than that of SNA. Using a lower concentration of 5 μg/ml labeled lectin, 53.8% and 99.5% of HMVEC-LBI stained positive for FITC-SNA and FITC-MAA, with MFIs of 422 and 1,275, respectively. At the same concentration, 57.8% and 93% of HULEC stained positive for FITC-SNA and FITC-MAA, with MFIs of 389 and 1,944, respectively (Fig. 1B). To further investigate the binding preferences of different influenza viruses, we assessed the attachment of FITC-labeled virus to HMVEC-LBI. As shown in Fig. 1C, the HPAI H5N1 virus (Thai/16) demonstrated extensive attachment to the cell surface, whereas attachment was much less abundant with human influenza viruses 1918 HA/NA:Tx/91 (SC/18 HA/NA and internal genes of A/Texas/36/91 virus) (7, 45, 54, 55) or 2009 H1N1 (Mexico/4482) virus.
Fig 1.
Detection and quantification of α-2,3- and α-2,6-linked sialic acid (SA) receptors on the cell surface of pulmonary microvascular endothelial cells. (A) Fluorescence lectin staining. Primary human lung blood microvascular endothelial cells (HMVEC-LBI) and human lung microvascular endothelial cells (HULEC) were grown on collagen-coated chamber slides overnight. The cells were incubated with no lectin (control), biotinylated SNA, MAA I, or MAA II, followed by FITC-conjugated avidin D (green) and DAPI (blue) for nuclei. (B) Trypsinized endothelial cells were incubated with FITC-conjugated SNA or MAA (5 μg/ml) and analyzed using flow cytometry. Fluorescent intensities for the α-2,6-SA configuration (SNA, green) and the α-2,3-SA configuration (MAA, blue) were compared to that for unstained cells (red). (C) Virus binding assay. HMVEC-LBI cells grown on collagen-coated chamber slides were incubated with FITC-labeled influenza virus at comparable HAU titers, followed by anti-FITC–horseradish peroxidase (HRP) antibody and AEC detection. The images were taken using the same exposure. (D) Fluorescent lectin staining. Primary rat pulmonary microvascular endothelial cells (PMVEC) and primary rat pulmonary arterial endothelial cells (PAEC) were stained green.
To confirm our observations made with human endothelial cells, these initial studies were repeated using well-defined endothelial cells derived from nonhuman mammalian species (22). Primary rat pulmonary microvascular endothelial cells (PMVEC) and primary rat pulmonary arterial endothelial cells (PAEC) seeded on chamber slides were stained with the lectins as described above. Similar to the binding pattern of human pulmonary endothelial cells, much greater amounts of α-2,3-linked SA than of α-2,6-linked SA were observed on the cell surfaces of both PAEC and PMVEC (Fig. 1D). Taken together, these data demonstrate that the α-2,3-linked SA configuration is much more abundant than the α-2,6-linked SA configuration on the surface of pulmonary endothelial cells.
HPAI H5N1 viruses replicate productively in human pulmonary microvascular endothelial cells.
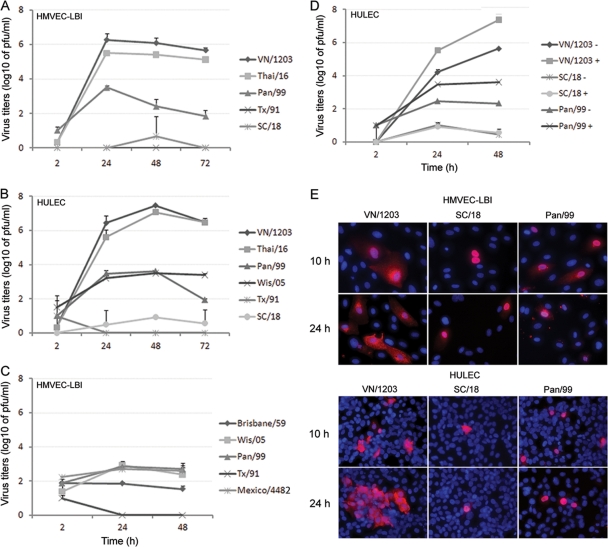
Targeted influenza virus infection of pulmonary vascular endothelial cells may acutely impair normal lung function and contribute to the pathogenesis of severe disease. Here, we compared the replication kinetics of multiple subtypes of influenza A viruses (listed in Table 1) in HMVEC-LBI and HULEC grown on six-well plates. As shown in Fig. 2A and B, both types of human endothelial cells tested supported productive replication of the HPAI H5N1 viruses, VN/1203 and Thai/16. The H5N1 viruses reached high titers of 106 to 107 PFU/ml at 24 and 48 h p.i. In contrast, despite the addition of a protease supplement to the culture medium to promote HA cleavage, the peak infectious titers of human H3N2 viruses (Pan/99 and Wis/05) reached only 103.6 PFU/ml (Fig. 2A and B). Comparatively poor replication was also observed among human H1N1 influenza viruses, including the pandemic 1918 (SC/18) virus. Similar replication kinetics were obtained in rat pulmonary endothelial cells, PMVEC and PAEC, following virus inoculation at an MOI of 0.01. Rat PMVEC and PAEC cultures were found to support productive replication of VN/1203 H5N1 virus (peak titers of 106 to 107 PFU/ml), whereas comparatively low virus titers (<102 PFU/ml) were observed following inoculation with seasonal influenza H1N1 and H3N2 virus subtypes (data not shown).
Fig 2.
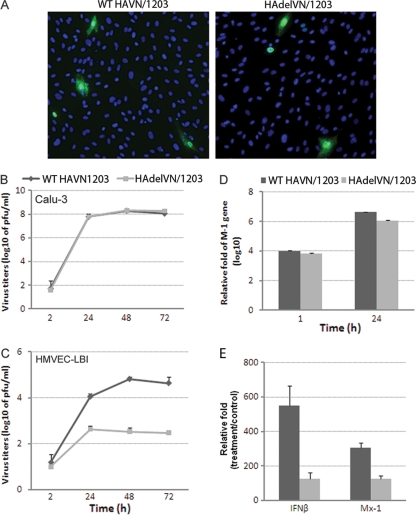
Replication of influenza viruses in human pulmonary microvascular endothelial cells. Endothelial cells grown on six-well plates were infected with influenza virus at an MOI of 0.01. Culture supernatants were collected, and viral titers were determined by a standard plaque assay. Values represent means from three independent experiments plus standard deviations. (A and C) Virus replication in HMVEC-LBI. Ten percent normal chicken egg allantoic fluid was added to the medium during virus infection. (B) Virus replication in HULEC. TPCK-trypsin (0.3 μg/ml) was added to the medium during infection of H1 and H3 subtype viruses. (D) Virus replication in HULEC in the presence or absence of TPCK-trypsin. (E) Immunofluorescence detection of nucleoprotein (NP) antigen in infected HMVEC-LBI and HULEC. Cells seeded on chamber slides were infected with virus at an MOI of 1 and fixed for NP detection at 10 and 24 h p.i.
We next determined the replication kinetics of a 2009 H1N1 pandemic virus that began circulating in the spring of 2009. For this experiment, primary HMVEC-LBI medium was supplemented with 10% normal allantoic fluid to promote extracellular HA cleavage. Only subtle differences were observed among the influenza viruses tested, and in comparison to the first data point (2 h) of the growth curve, cultures did not demonstrate substantial virus titer increases up to 48 h p.i. (Fig. 2C). Human seasonal H3N2 viruses (Pan/99 and Wis/05) and 2009 pandemic H1N1 virus (Mexico/4482) titers reached approximately 103 PFU/ml at 24 h p.i., with no further increases measured at 48 h p.i. Seasonal H1N1 virus (Tx/91 and Brisbane/59) cultures showed no increase in viral titers at any time point.
Because the growth of influenza viruses in many cell lines requires the addition of trypsin to the culture medium to ensure HA cleavage and multicycle infection, replication kinetics of selected viruses were measured in the presence or absence of exogenous protease (TPCK-treated trypsin). In the absence of TPCK-trypsin, only H5N1 VN/1203 virus replicated to substantial titers (>105 PFU/ml) (Fig. 2D). Conversely, even in the presence of TPCK-trypsin, the 1918 (SC/18) virus failed to replicate above the input titer, and Pan/99 virus failed to reach viral titers above 103.5 PFU/ml.
To further test the permissiveness of pulmonary endothelial cells for influenza virus infection, cells grown on chamber slides were inoculated with VN/1203, SC/18 (1918), or Pan/99 virus at an MOI of 1 and intracellular expression of influenza virus NP was evaluated at 10 h and 24 h p.i. Overall, influenza virus infection of pulmonary endothelial cells was less efficient than infection of airway epithelial cells, which possess a reported infection rate of 30% to 40% within 24 h p.i. (7, 45, 54, 55). In this study, greater numbers of NP-positive cells (8 to 24% infection rate) were observed at 24 h p.i. in endothelial cells infected with H5N1 VN/1203 virus than in SC/18 or Pan/99 virus-infected cells (3 to 7% infection rate); SC/18 virus showed the lowest number (0.5 to 1.1%) of NP-positive cells (Fig. 2E). HULEC, which can reach a higher cell density, allowed the visualization of more NP-positive cells, some of which were observed in clusters in VN/1203 virus-infected cultures at 24 h p.i. (Fig. 2E, bottom left panel).
In a detailed side-by-side comparison of avian influenza subtype viruses possessing α-2,3-SA binding preference, we next compared the replication kinetics of avian H1N1, H5N1, and H7 subtype viruses in HULEC and HMVEC-LBI cultures. H7 viruses were included for comparison purposes because they represent avian influenza viruses of another subtype and they largely maintain the classic avian binding preference for α-2,3-linked SA. However, the North American lineage H7N2 (NY/107) virus was found to possess increased affinity toward α-2,6-linked SA and reduced binding to α-2,3-linked SA (5). Although possessing an HA with a polybasic cleavage site (Table 1), both SP/83 and Ck/Korea HPAI H5N1 viruses exhibit a low-pathogenicity phenotype in ferrets and mice (30). As shown in Fig. 3A, SP/83 and Ck/Korea HPAI H5N1 viruses replicated significantly less efficiently than the highly virulent H5N1 viruses, VN/1203 and Thai/16 (P < 0.05). The LPAI avian H1N1 (Dk/NY) virus exhibited minimal growth in HULEC. In HMVEC-LBI, Thai/16 virus showed a clear distinction in replication efficiency compared to H7 subtype viruses, for which productive replication was generally not observed (Fig. 3B). Only the highly pathogenic H7N7 (NL/219) virus, isolated from a fatal case, replicated to a moderate level of 102 to 103 PFU/ml at 24 and 48 h p.i. Taken together, the data suggest that endothelial cells support entry and efficient replication of HPAI H5N1 viruses, whereas human and other avian viruses, including the HPAI H7 subtype viruses tested, showed poor infectivity in these cells.
Fig 3.
Replication of avian influenza viruses in human pulmonary microvascular endothelial cells. Endothelial cells grown on six-well plates were infected with influenza virus at an MOI of 0.01. Culture supernatants were collected, and viral titers were determined by a standard plaque assay. Values represent the means from three independent experiments plus standard deviations. (A) Virus replication in HULEC. TPCK-trypsin (0.3 μg/ml) was added to the medium only for cells infected with Dk/NY virus. (B) Virus replication in HMVEC-LBI. Ten percent normal chicken egg allantoic fluid was added to the medium during virus infection. *, H5N1 virus replicated to a significantly higher titer than H7 subtype viruses (P < 0.05) at 48 h and 72 h p.i.
HPAI H5N1 virus preferentially enters through and is released from the apical surface of human pulmonary endothelial cells.
To assess the polarity of HPAI H5N1 virus infection and release of progeny virus particles, we compared the replication kinetics of VN/1203 virus following infection of the apical or basolateral side of the cell monolayer. HULEC were seeded onto 12-well transwell membrane inserts with pore size of 0.3 μm and cultured for 1 week for polarization of the pulmonary endothelial cells. Unlike human bronchial epithelial cells, which exhibit high transepithelial resistance (54), HULEC displayed very low transepithelial resistance. Cells were inoculated with virus from either the apical surface of the monolayers or the basolateral surface at an MOI of 0.01. Supernatants from both compartments were collected for virus titer determination. When VN/1203 virus was inoculated via the apical surface of endothelial cells, the H5N1 strain replicated to a much higher titer than after basolateral inoculation (Fig. 4). Moreover, following apical surface inoculation, virus was released from the apical side at titers significantly higher (P < 0.05) than those released from the basolateral side at 24 h p.i. These results demonstrate that avian H5N1 viruses enter and release primarily from the apical surface of polarized human pulmonary microvascular endothelial cells.
Fig 4.
HPAI H5N1 viruses preferentially enter from the apical side of polarized human pulmonary microvascular endothelial cells. HULEC monolayers grown on transwells for 1 week were infected with VN/1203 virus apically or basolaterally at an MOI of 0.01. The supernatant from both apical and basolateral compartments was collected for virus titration using standard plaque assay. Values represent the means from three independent experiments plus standard deviation. aa, apical infection and apical release; ab, apical infection and basolateral release; ba, basolateral infection and apical release; bb, basolateral infection and basolateral release.
Removal of the HA polybasic cleavage site diminishes HPAI H5N1 virus replication in human pulmonary microvascular endothelial cells.
We next evaluated the contribution of the multibasic HA cleavage site on H5N1 virus replication in pulmonary endothelial cells. Using NP staining of HMVEC-LBI, a similar rate of infection was observed for the mutant H5N1 virus (HAdelVN/1203) with deletion of the HA polybasic cleavage site and the wild-type rescued VN/1203 virus (Fig. 5A). The replication kinetics of this pair of viruses was further evaluated in HMVEC-LBI and an established human bronchial epithelial (Calu-3) cell line which possesses host proteases capable of cleaving monobasic HA cleavage sites (54). Calu-3 cell cultures were found to support productive replication of H5N1 virus containing either the wild-type or mutant HA (peak titers of 108 PFU/ml) (Fig. 5B); however, in HMVEC-LBI cultures, the HAdelVN/1203 virus displayed significantly (P < 0.001) lower viral titers than wild-type VN/1203 virus at 24, 48, and 72 h p.i. (Fig. 5C). Although HAdelVN/1203 virus displayed reduced replication capacity in pulmonary endothelial cells, a relatively similar infection rate was observed for the HA mutant virus based on M1 gene expression levels (Fig. 5D). At 1 and 24 h p.i., M1 gene levels were comparable for both viruses in HMVEC-LBI cultures, suggesting that H5N1 viruses could successfully infect pulmonary endothelial cells independent of the cleavage site motif, but efficient H5N1 replication and production of infectious progeny virus in these cells requires a multibasic cleavage site.
Fig 5.
Contribution of the HA multiple basic amino acid cleavage site to virus replication in human endothelial cells. Cells were infected with virus at an MOI of 0.01, and supernatants were collected for virus titration by plaque assay. (A) Immunofluorescence detection of nucleoprotein (NP) antigen in infected HMVEC-LBI. Cells seeded on chamber slides were infected with virus at an MOI of 1 and fixed for NP detection at 8 h p.i. (B) Virus replication in polarized bronchial epithelial cells (Calu-3). (C) Virus replication in HMVEC-LBI. Ten percent normal chicken egg allantoic fluid was added to the medium. For real-time PCR analysis, cells were infected with each virus at an MOI of 1 and RNA was collected at 24 h p.i. (D) Relative M1 gene levels in infected cells determined by real-time PCR. (E) Relative fold change of gene expression of type I interferon genes in infected cells.
Because the expression of the beta interferon gene (IFN-β) and the Mx1 gene (an interferon-stimulated antiviral gene) is a well-characterized component of the innate immune response directed against viral infections, we also compared the expression levels of these IFN signature genes in pulmonary endothelial cell cultures collected at 24 h p.i. The 24-hour time point was selected because previous studies have demonstrated low expression of IFN-β at earlier time points (such as 8 h) following H5N1 virus infection of cultured cells (54). Consistent with replication data, endothelial cells elicited a significantly higher induction of IFN-β and Mx-1 genes in response to wild-type VN/1203 virus compared to the mutant HAdelVN/1203 virus (Fig. 5E). Taken together, these data support an important role for the HA polybasic cleavage site in both HPAI H5N1 virus replication and host innate antiviral defense in HMVEC-LBI. However, other viral genes, or combination of genes, most likely contribute to the replication efficiency of HPAI H5N1 virus in human pulmonary endothelial cells.
HPAI H5N1 virus elicits a higher level of gene expression and production of proinflammatory cytokine/chemokines and adhesion molecules in human pulmonary microvascular endothelial cells.
We further examined host responses to infection with HPAI H5N1 viruses in human pulmonary endothelial cells and compared the results with those responses induced by the 1918 H1N1 virus. As indicated above, dramatic differences in replication were observed between H5N1 (VN/1203) and 1918 (SC/18) viruses in pulmonary endothelial cells (Fig. 2). In this experiment, HULEC cultures grown on six-well plates were infected with virus (MOI = 1) and at 24 h p.i., total RNA from uninfected (mock) or virus-infected cells was extracted and analyzed for host gene expression using a Human Endothelial Cell Biology RT2 Profiler PCR array. Detailed gene expression data for genes with greater than 3-fold induction or reduction over mock-infected cells are presented in Table 2. With few exceptions, VN/1203 virus infection induced the greatest number of host genes with the highest level of expression; VN/1203 virus infection induced 24 host genes, whereas SC/18 virus infection resulted in the induction of 5 genes with greater than 3-fold induction over mock-infected HULEC cultures (Table 2). In particular, VN/1203 virus induced expression of key cytokines/chemokines, including IFN-β, IL-7, TNF, CCL2, and CCL, that were at least 10-fold higher than the host responses observed following SC/18 virus infection. Significant increases in gene expression of several adhesion molecules, including members of the selectin family (E-selectin, L-selectin, and P-selectin ligand), ICAM1, and VCAM1, were observed following VN/1203, but not SC/18, virus infection. Genes related to endothelial cell function regulation were also upregulated, including genes involved in angiogenesis (TYMP and FGF1), coagulation (PF4), homeostasis (NPPB), cell survival (SPHK1), and thrombosis (SERPINE1). During VN/1203 virus infection, the transcriptional level of endothelial NO synthase (eNOS) did not change significantly; however, inducible nitric oxide synthase 2A was highly induced. Among genes related to apoptosis, TNFAIP3, FASLG, and CASP1 were upregulated; CASP6 and TNFRSF10C were downregulated 3-fold.
Table 2.
Genes induced in HULEC infected with influenza viruses
| Category and gene product | Gene symbol | Fold changea |
|
|---|---|---|---|
| VN/1203 | SC/18 | ||
| Cytokines/chemokines | |||
| Beta 1 interferon, fibroblast | IFNB1 | 746.2 | 5.6 |
| Interleukin-7 | IL-7 | 268.1 | 2.8 |
| Tumor necrosis factor | TNF | 53.3 | 4.8 |
| Interleukin-6 (beta 2 interferon) | IL-6 | 35.3 | 5.3 |
| Chemokine (C-C motif) ligand 2 | CCL2 | 28.6 | 1.4 |
| Chemokine (C-C motif) ligand 5 | CCL5 | 23.0 | 1.4 |
| Tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | 11.7 | 1.5 |
| Chemokine (C-X3-C motif) ligand 1 | CX3CL1 | 9.0 | 1.5 |
| Colony-stimulating factor 2 | CSF2 | 8.9 | 1.9 |
| Adhesion molecules | |||
| Selectin P ligand | SELPLG | 138.1 | 4.6 |
| Selectin L | SELL | 8.2 | 1.7 |
| Selectin E | SELE | 7.7 | 0.9 |
| Intercellular adhesion molecule 1 | ICAM1 | 6.9 | 1.4 |
| Vascular cell adhesion molecule 1 | VCAM1 | 3.0 | 1.1 |
| Endothelial cell function regulation | |||
| Nitric oxide synthase 2, inducible | NOS2 | 63.1 | 1.3 |
| Thymidine phosphorylase | TYMP | 13.2 | 1.9 |
| Natriuretic peptide precursor B | NPPB | 5.9 | 1.4 |
| Serpin peptidase inhibitor, clade E, member 1 | SERPINE1 | 4.1 | 1.2 |
| Fibroblast growth factor 1 (acidic) | FGF1 | 3.9 | 1.4 |
| Sphingosine kinase 1 | SPHK1 | 3.7 | 1.3 |
| Platelet factor 4 | PF4 | 3.6 | 1.2 |
| Placental growth factor | PGF | 0.3 | 1.2 |
| Apoptosis | |||
| Tumor necrosis factor alpha-induced protein 3 | TNFAIP3 | 12.5 | 1.5 |
| Fas ligand (TNF superfamily, member 6) | FASLG | 9.6 | 1.2 |
| Caspase 1, apoptosis-related cysteine peptidase | CASP1 | 3.7 | 1.3 |
| Caspase 6, apoptosis-related cysteine peptidase | CASP6 | 0.3 | 1.1 |
| Tumor necrosis factor receptor superfamily, member 10c | TNFRSF10C | 0.3 | 1.2 |
Fold changes in host gene expression between infected and uninfected cultures are represented. The subset of 29 genes were induced (bold) or suppressed (underlined) by VN/1203 virus infection at 24 h p.i., using a 3-fold difference over the value for mock infection with a P value of <0.01 as the cutoff.
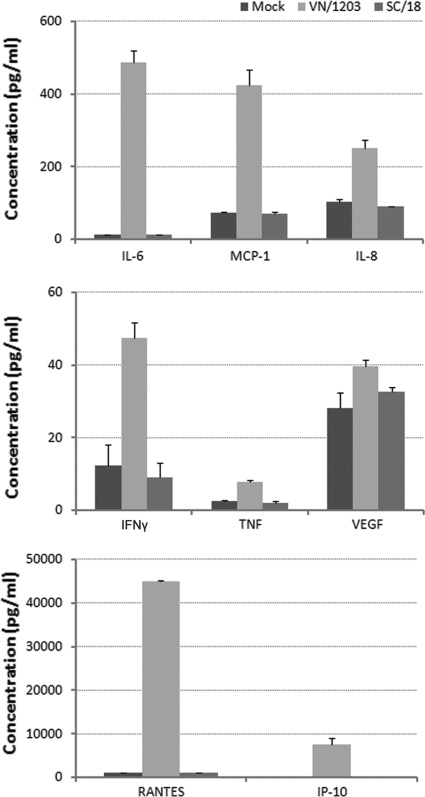
We further investigated differences in host responses to VN/1203 virus compared with 1918 H1N1 infection by measuring cytokine protein levels released into pulmonary endothelial cell cultures. HULEC were infected with virus at an MOI of 1, and supernatants were collected at 24 h p.i. for Bio-Plex assay (Fig. 6). The results showed that VN/1203 virus-infected cultures released substantial amounts of cytokines/chemokines (RANTES, IP-10, IL-6, IL-8, MCP-1, VEGF, TNF, and IFN-γ) that were significantly (>4-fold) higher than those induced by the pandemic SC/18 virus (P < 0.05). These data further demonstrate that in comparison to H1N1 virus, HPAI H5N1 virus infection resulted in a substantial induction of the innate immune response in human pulmonary endothelial cells.
Fig 6.
Evaluation of cytokine production in virus-infected endothelial cells. HULEC were infected with virus at an MOI of 1, and supernatants were collected at 24 h p.i. for Bio-Plex assay. Values represent the means from three independent experiments plus standard deviations.
Proliferation and viability of human pulmonary microvascular endothelial cells are affected by HPAI H5N1 virus infection.
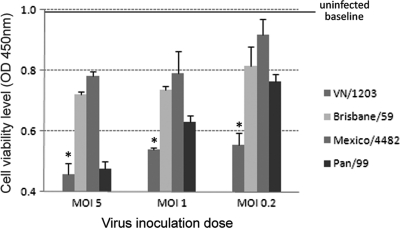
Overall, influenza virus infection of pulmonary endothelial cells (HULEC and HMVEC-LBI) resulted in less CPE and cell death observed microscopically than those previously observed in infected airway epithelial cells (7, 45, 54, 55). In fact, following VN/1203 virus infection, HMVEC-LBI monolayers remained generally intact for up to 72 h p.i. (data not shown). To investigate the impact of influenza virus infection on the growth and survival of pulmonary endothelial cells, infected HMVEC-LBI (MOIs of 5.0, 1.0, and 0.2) were subjected to the WST-1 proliferation/viability assay at 44 h p.i. (Fig. 7). In comparison to uninfected controls, which showed an absorbance reading of 0.997, influenza virus infection of HMVEC-LBI cultures resulted in a decrease in cell proliferation/viability, with the greatest decrease observed at the highest MOI. VN/1203 virus infection caused a significant decrease in cell proliferation/viability at all inoculation doses in comparison to pandemic 2009 H1N1 and seasonal influenza viruses. The seasonal H3N2 (Pan/99) virus induced an intermediate decrease, whereas virus infection with both H1N1 (Brisbane/59 and Mexico/4482) viruses resulted in a minimal decrease in cell proliferation/viability following virus infection of pulmonary endothelial cells. These results demonstrate that one outcome of HPAI H5N1 viral infection of pulmonary endothelial cells is an accelerated loss of cell viability compared with that caused by other influenza virus strains of lesser virulence.
Fig 7.
Examination of endothelial cell proliferation/viability during influenza virus infection. HMVEC-LBI were seeded onto 96-well plates, inoculated with virus at an MOI of 5, 1, or 0.2, and examined at 44 h p.i. using WST-1 reagent. *, infection with VN/1203 virus resulted in significantly lower levels of cell variability compared to other tested viruses at all virus dilutions (P < 0.05). Values represent the means from three independent experiments plus standard deviations.
DISCUSSION
In contrast to the case for seasonal influenza virus, human infection with HPAI H5N1 viruses causes severe disease of the lower respiratory tract, and many patients progress rapidly to ARDS and multiorgan failure (16, 24, 37, 43). Although endothelial cell injury is a general characteristic feature of ARDS (51), the role of pulmonary endothelial cells in H5N1 pathogenesis is still largely unknown. Here, we provide data obtained using a pulmonary endothelial cell line and primary cells isolated from human lung tissue and compare host cell permissiveness to infection with influenza viruses of multiple subtypes. The human pulmonary endothelial cell model was also utilized to evaluate the cell receptors for influenza virus and innate immune responses induced by HPAI H5N1 virus compared with other influenza virus subtypes. We found that unlike human bronchial epithelial cells, which are permissive to productive replication of both avian and human influenza viruses and express avian-like α-2,3-linked and human-like α-2,6-linked SA receptors equally well (7, 45, 54, 55), pulmonary endothelial cells possess α-2,3-linked SA configuration as the dominant receptor type and support efficient replication only of HPAI H5N1 virus. The polybasic HA cleavage site was found to be necessary but not sufficient for the high replication efficiency of HPAI H5N1 virus in pulmonary endothelial cells. The H5N1 virus replication correlated with high-level expression of proinflammatory cytokines/chemokines and adhesion molecules and also resulted in a marked decrease in cell viability and proliferation, which could be attributable to virus-mediated cell death and/or a cytokine-mediated process.
The clinical course of H5N1 virus infection in humans often results in the progression of severe lower respiratory tract disease, which includes dyspnea, chest pain, and pulmonary infiltrates (1, 18, 53). Infiltration of neutrophils, alveolar edema, endothelial hypertrophy, and necrosis among H5N1-infected patients are thought to contribute to the severe lung pathology of fatal human infection. Although in situ hybridization failed to show positive viral RNA or influenza NP antigen staining in lung endothelial cells from postmortem examination of patients who succumbed to H5N1 infection (19), the detection of IFN-β-positive endothelial cells in lungs of H5N1-infected patients suggests that these cells are responding to viral infection (17). However, in situ evidence for H5N1 virus infection of pulmonary endothelial cells in humans is limited due to the paucity of postmortem tissues for study. In the macaque model, endothelial cells appeared to be productively infected and displayed necrosis in H5N1-challenged animals (4, 25). In standard cultures, we observed that both human and rat pulmonary microvascular endothelial cells could be productively infected by VN/1203 (H5N1) virus, which was highly lethal in ferrets and mice (29, 30). Utilizing a polarized endothelial cell model, in which cells were seeded on transwells creating two distinct surfaces, i.e., apical and basolateral domains, we found preferential viral entry and release from the apical surface. HPAI H5N1 virus particles bud from the apical (luminal) side of endothelial cells, in contact with blood, so that released virus could enter the general circulation and cause viremia. Unlike for seasonal influenza viruses, which are confined mainly to the upper respiratory tracts of mammals, viral RNA has been detected in the blood of H5N1-infected patients with extrapulmonary complications, including multiorgan failure, which are common in fatal cases (reviewed in reference 46). It is reasonable to speculate that efficient replication of H5N1 virus in pulmonary endothelial cells may promote systemic spread of virus and contribute to the pathogenicity of HPAI H5N1 viruses in the mammalian host. Since transport of H5N1 virus from the basolateral to the apical chamber was minimal in the polarized endothelial cell model, it is not entirely clear how initial apical infection occurs in these cells.
In this study, we have addressed the possible contribution of the HA glycoprotein to the replication efficiency of H5N1 virus in pulmonary endothelial cells. The two main functions of the HA are to facilitate (i) virus binding to target host cells, via sialic acid-containing receptors, and (ii) virus entry into the cell, which is dependent on HA cleavage. Because the availability of suitable receptors on the host cell surface can determine efficiency of influenza virus infection and replication (41), the abundance of surface-associated SA on pulmonary endothelial cells was analyzed through careful flow cytometric analysis coupled with lectin staining in a dose-dependent manner. It has been reported that α-2,3-SA and α-2,6-SA receptors were homogenously expressed on microvascular endothelial cells (52). However, we found that the α-2,3-SA configuration was much more abundant than α-2,6-SA receptors, which corresponded to a lower rate of infectivity for human influenza viruses possessing the α-2,6-SA receptor binding preference. Moreover, avian-origin viruses with preferential α-2,3-SA binding attached to and infected pulmonary endothelial cells at a higher frequency than human influenza viruses. Taken together, the data suggest that the receptor specificity most likely contributes to the efficient H5N1 virus replication in these cells; however, additional data are needed to definitively assess the role of this molecular determinant. The generation of mutant influenza viruses that carry substitutions in the H5 HA receptor binding site conferring a change in binding preference from the avian- to the human-type receptor is required.
The cleavage properties of HA0 and the distribution of functional proteases in the host contribute to tissue tropism (14, 20, 32, 40). Unlike airway epithelial cells (54), pulmonary endothelial cells do not appear to produce sufficient endogenous proteases capable of cleaving HA at a monobasic cleavage site. We found that these cells support multiple-cycle growth only of H5N1 viruses containing a polybasic cleavage site. Removal of the polybasic cleavage site in VN/1203 virus did not affect the initial viral infectivity in pulmonary endothelial cells; however, it greatly attenuated viral growth kinetics relative to that of wild-type virus. The results lend support to the concept that in vivo, the polybasic HA cleavage site is a molecular determinant not only for extrapulmonary virus replication but also for enhancing H5N1 virus replication in pulmonary endothelial cells. H5N1 infection of endothelial cells may contribute to the high viral load in lung tissue and potential virus spread beyond the respiratory tract.
Although HA appears to play an important role in efficient H5N1 virus replication through its receptor binding specificity and polybasic cleavage site, additional virulence determinants of the virus most certainly exist. We observed that a reassortant virus containing six internal genes from A/PR/8/34 (H1N1) and the HA and NA genes from VN/1203 virus exhibited attenuated replication in pulmonary endothelial cells compared with wild-type VN/1203 virus despite comparable efficiencies of initial infectivity (based on NP staining) (data not shown). Moreover, H7 subtype viruses and the less virulent SP/83 and Ck/Korea H5N1 viruses, which possess an “avian-like” α-2,3-SA binding preference and a polybasic cleavage site, could not productively replicate as efficiently as HPAI H5N1 viruses in human pulmonary endothelial cells, further suggesting that additional molecular factors are associated with HPAI H5N1 virus virulence.
ARDS is characterized by diffuse alveolar damage usually following an intense inflammatory response to infection (51). The suggestion that the fatal outcome in patients with H5N1 virus infection may be attributed to elevated levels of proinflammatory cytokines (4, 16, 30, 45) prompted us to characterize the mediators of inflammation produced by human pulmonary endothelial cells. We found that H5N1 virus infection resulted in elevated production of multiple cytokines, including TNF, IP-10, IL-6, IL-8, MCP-1, IFN-γ, VEGF, and RANTES. TNF and IL-8 are of particular interest because of their association with ARDS and can instigate a cascade of physiological changes, including recruitment of neutrophils leading to alveolar capillary damage (51). Moreover, it has been demonstrated that H5N1-infected, but not H1N1-infected, TNFR-1-deficient knockout mice exhibit a substantial reduction in lung inflammation and delay in mortality compared to wild-type mice, suggesting that TNF contributes to H5N1-induced inflammation of lung tissue (38). Blocking of TNF has been shown to decrease vascular permeability through the destabilization of the endothelial cell cytoskeleton (47) and to affect subsequent immune cell transmigration across the endothelium (26). When exposed to TNF, a normally “quiescent” endothelium becomes activated and expresses additional proinflammatory factors, including chemokines and adhesion molecules (51). In our analysis we also observed that human pulmonary endothelial cell cultures generated high levels of adhesion molecules, including selectin P ligand, selectins L and E, ICAM1, and VCAM1, in response to H5N1 but not H1N1 virus infection. Production of chemokines and adhesion molecules by endothelial cells could contribute to the tissue damage by causing vascular injury and destruction of the parenchymal cells through the accumulation of inflammatory cells (3, 27). The resulting loss of functional alveolar surface area could result in inadequate gas exchange, lower respiration, and ultimately death.
The pulmonary endothelium is strategically located within the lung, and its functional and structural integrity are essential for adequate pulmonary function. The majority of patients with confirmed H5N1 virus infection have an aggressive clinical course and often present with respiratory failure frequently complicated with ARDS (44). ARDS induced by H5N1 viral infection is most likely to be linked to patient death, and pulmonary endothelial injury is expected to contribute to the abnormalities seen in H5N1-induced ARDS. Our results demonstrate a unique virulence trait of HPAI H5N1 following virus infection of pulmonary endothelial cells which leads to a high virus load, an overwhelming immune reaction, and a marked decrease in cell viability. Future investigation of the interaction between H5N1-induced inflammation and the pulmonary endothelium, especially in in vivo models, is warranted.
ACKNOWLEDGMENTS
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
This research was funded in part by NIH grants HL-60024 and HL-66299 to T.S.
Footnotes
Published ahead of print 9 November 2011
REFERENCES
- 1. Abdel-Ghafar AN, et al. 2008. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 358:261–273 [DOI] [PubMed] [Google Scholar]
- 2. Alom-Ruiz SP, Anilkumar N, Shah AM. 2008. Reactive oxygen species and endothelial activation. Antioxid. Redox Signal. 10:1089–1100 [DOI] [PubMed] [Google Scholar]
- 3. Azoulay E, et al. 2002. Deterioration of previous acute lung injury during neutropenia recovery. Crit. Care Med. 30:781–786 [DOI] [PubMed] [Google Scholar]
- 4. Baskin CR, et al. 2009. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc. Natl. Acad. Sci. U. S. A. 106:3455–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belser JA, et al. 2008. Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc. Natl. Acad. Sci. U. S. A. 105:7558–7563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belser JA, Wadford DA, Xu J, Katz JM, Tumpey TM. 2009. Ocular infection of mice with influenza A (H7) viruses: a site of primary replication and spread to the respiratory tract. J. Virol. 83:7075–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belser JA, Zeng H, Katz JM, Tumpey TM. 2011. Infection with highly pathogenic H7 influenza viruses results in an attenuated proinflammatory cytokine and chemokine response early after infection. J. Infect. Dis. 203:40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bertram S, Glowacka I, Steffen I, Kuhl A, Pohlmann S. 2010. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev. Med. Virol. 20:298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Birkness KA, et al. 1999. An in vitro tissue culture bilayer model to examine early events in Mycobacterium tuberculosis infection. Infect. Immun. 67:653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bottcher-Friebertshauser E, et al. 2010. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J. Virol. 84:5605–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan MC, et al. 2009. Influenza H5N1 virus infection of polarized human alveolar epithelial cells and lung microvascular endothelial cells. Respir. Res. 10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a. Chosewood LC, Wilson DE. 2009. Biosafety in microbiological and medical laboratories. U. S. Department of Health and Human Services, Washington, DC: http://www.cdc.gov/biosafety/publications/bmbl5/BMBL.pdf. [Google Scholar]
- 12. Chutinimitkul S, et al. 2006. H5N1 influenza A virus and infected human plasma. Emerg. Infect. Dis. 12:1041–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cotran RS, Kumar V, Collins T. (ed). 1999. Robbins pathologic basis of disease, 6th ed W.B. Saunders Company, Philadelphia, PA [Google Scholar]
- 14. Decha P, et al. 2008. Source of high pathogenicity of an avian influenza virus H5N1: why H5 is better cleaved by furin. Biophys. J. 95:128–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Jong MD, et al. 2005. Fatal avian influenza A (H5N1) in a child presenting with diarrhea followed by coma. N. Engl. J. Med. 352:686–691 [DOI] [PubMed] [Google Scholar]
- 16. de Jong MD, et al. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng R, et al. 2008. Distinctly different expression of cytokines and chemokines in the lungs of two H5N1 avian influenza patients. J. Pathol. 216:328–336 [DOI] [PubMed] [Google Scholar]
- 18. Gambotto A, Barratt-Boyes SM, de Jong MD, Neumann G, Kawaoka Y. 2008. Human infection with highly pathogenic H5N1 influenza virus. Lancet 371:1464–1475 [DOI] [PubMed] [Google Scholar]
- 19. Gu J, et al. 2007. H5N1 infection of the respiratory tract and beyond: a molecular pathology study. Lancet 370:1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo XL, Li L, Wei DQ, Zhu YS, Chou KC. 2008. Cleavage mechanism of the H5N1 hemagglutinin by trypsin and furin. Amino Acids 35:375–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ibricevic A, et al. 2006. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 80:7469–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. King J, et al. 2004. Structural and functional characteristics of lung macro- and microvascular endothelial cell phenotypes. Microvasc. Res. 67:139–151 [DOI] [PubMed] [Google Scholar]
- 23. Klenk HD, Garten W. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39–43 [DOI] [PubMed] [Google Scholar]
- 24. Korteweg C, Gu J. 2008. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am. J. Pathol. 172:1155–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuiken T, van den Brand J, van Riel D, Pantin-Jackwood M, Swayne DE. 2010. Comparative pathology of select agent influenza a virus infections. Vet. Pathol. 47:893–914 [DOI] [PubMed] [Google Scholar]
- 26. Kusters S, et al. 1997. In vivo evidence for a functional role of both tumor necrosis factor (TNF) receptors and transmembrane TNF in experimental hepatitis. Eur. J. Immunol. 27:2870–2875 [DOI] [PubMed] [Google Scholar]
- 27. La Gruta NL, Kedzierska K, Stambas J, Doherty PC. 2007. A question of self-preservation: immunopathology in influenza virus infection. Immunol. Cell Biol. 85:85–92 [DOI] [PubMed] [Google Scholar]
- 28. Lamb RA, Krug RM. (ed). 1996. Fields virology, 3rd ed Lippincott-Raven, Philadelphia, PA [Google Scholar]
- 29. Lu X, et al. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 73:5903–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maines TR, et al. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 79:11788–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mehta PK, Karls RK, White EH, Ades EW, Quinn FD. 2006. Entry and intracellular replication of Mycobacterium tuberculosis in cultured human microvascular endothelial cells. Microb. Pathog. 41:119–124 [DOI] [PubMed] [Google Scholar]
- 32. Nakayama K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327:625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ng WF, To KF, Lam WW, Ng TK, Lee KC. 2006. The comparative pathology of severe acute respiratory syndrome and avian influenza A subtype H5N1—a review. Hum. Pathol. 37:381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nicholls JM, et al. 2007. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat. Med. 13:147–149 [DOI] [PubMed] [Google Scholar]
- 35. Ocana-Macchi M, et al. 2009. Hemagglutinin-dependent tropism of H5N1 avian influenza virus for human endothelial cells. J. Virol. 83:12947–12955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park MS, Steel J, Garcia-Sastre A, Swayne D, Palese P. 2006. Engineered viral vaccine constructs with dual specificity: avian influenza and Newcastle disease. Proc. Natl. Acad. Sci. U. S. A. 103:8203–8208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peiris JS, et al. 2004. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363:617–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perrone LA, Szretter KJ, Katz JM, Mizgerd JP, Tumpey TM. 2010. Mice lacking both TNF and IL-1 receptors exhibit reduced lung inflammation and delay in onset of death following infection with a highly virulent H5N1 virus. J. Infect. Dis. 202:1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shinya K, et al. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436 [DOI] [PubMed] [Google Scholar]
- 40. Steinhauer DA. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1–20 [DOI] [PubMed] [Google Scholar]
- 41. Suzuki Y, et al. 2000. Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 74:11825–11831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thanh TT, van Doorn HR, de Jong MD. 2008. Human H5N1 influenza: current insight into pathogenesis. Int. J. Biochem. Cell Biol. 40:2671–2674 [DOI] [PubMed] [Google Scholar]
- 43. To KF, et al. 2001. Pathology of fatal human infection associated with avian influenza A H5N1 virus. J. Med. Virol. 63:242–246 [DOI] [PubMed] [Google Scholar]
- 44. Tran TH, et al. 2004. Avian influenza A (H5N1) in 10 patients in Vietnam. N. Engl. J. Med. 350:1179–1188 [DOI] [PubMed] [Google Scholar]
- 45. Tumpey TM, et al. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77–80 [DOI] [PubMed] [Google Scholar]
- 46. Uyeki TM. 2009. Human infection with highly pathogenic avian influenza A (H5N1) virus: review of clinical issues. Clin. Infect. Dis. 49:279–290 [DOI] [PubMed] [Google Scholar]
- 47. Vandenbroucke E, Mehta D, Minshall R, Malik AB. 2008. Regulation of endothelial junctional permeability. Ann. N. Y. Acad. Sci. 1123:134–145 [DOI] [PubMed] [Google Scholar]
- 48. van Riel D, et al. 2006. H5N1 virus attachment to lower respiratory tract. Science 312:399. [DOI] [PubMed] [Google Scholar]
- 49. van Riel D, et al. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 171:1215–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Walls HH, Harmon MW, Slagle JJ, Stocksdale C, Kendal AP. 1986. Characterization and evaluation of monoclonal antibodies developed for typing influenza A and influenza B viruses. J. Clin. Microbiol. 23:240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ware LB. 2006. Pathophysiology of acute lung injury and the acute respiratory distress syndrome. Semin. Respir. Crit. Care Med. 27:337–349 [DOI] [PubMed] [Google Scholar]
- 52. Yao L, Korteweg C, Hsueh W, Gu J. 2008. Avian influenza receptor expression in H5N1-infected and noninfected human tissues. FASEB J. 22:733–740 [DOI] [PubMed] [Google Scholar]
- 53. Yu H, et al. 2008. Clinical characteristics of 26 human cases of highly pathogenic avian influenza A (H5N1) virus infection in China. PLoS One 3:e2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zeng H, et al. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type i interferon response in polarized human bronchial epithelial cells. J. Virol. 81:12439–12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zeng H, Pappas C, Katz JM, Tumpey TM. 2011. The 2009 pandemic H1N1 and triple-reassortant swine H1N1 influenza viruses replicate efficiently but elicit an attenuated inflammatory response in polarized human bronchial epithelial cells. J. Virol. 85:686–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang Z, et al. 2009. Systemic infection of avian influenza A virus H5N1 subtype in humans. Hum. Pathol. 40:735–739 [DOI] [PMC free article] [PubMed] [Google Scholar]