Abstract
The four Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded interferon (IFN) regulatory factor homologues (vIRF1 to vIRF4) are used to counter innate immune defenses and suppress p53. The vIRF genes are arranged in tandem but differ in function and expression. In KSHV-infected effusion lymphoma lines, K10.5/vIRF3 and K11/vIRF2 mRNAs are readily detected during latency, whereas K9/vIRF1 and K10/vIRF4 mRNAs are upregulated during reactivation. Here we show that the K10/vIRF4 promoter responds to the lytic switch protein RTA in KSHV-infected cells but is essentially unresponsive in uninfected cells. Coexpression of RTA with vIRF4 is sufficient to restore regulation, a property not shared by other vIRFs. The K9/vIRF1 promoter behaves similarly, and production of infectious virus is enhanced by the presence of vIRF4. Synergy requires the DNA-binding domain (DBD) and C-terminal IRF homology regions of vIRF4. Mutations of arginine residues within the putative DNA recognition helix of vIRF4 or the invariant cysteines of the adjacent CxxC motif abolish cooperation with RTA, in the latter case by preventing self-association. The oligomerization and transactivation functions of RTA are also essential for synergy. The K10/vIRF4 promoter contains two transcription start sites (TSSs), and a 105-bp fragment containing the proximal promoter is responsive to vIRF4/RTA. Binding of a cellular factor(s) to this fragment is altered when both viral proteins are present, suggesting a possible mechanism for transcriptional synergy. Reliance on coregulators encoded by either the host or viral genome provides an elegant strategy for expanding the regulatory potential of a master regulator, such as RTA.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) is a gammaherpesvirus 2 and etiological agent of KS, primary effusion lymphoma (PEL), and variant multicentric Castleman's disease (16). The KSHV genome encodes at least 90 gene products that are expressed in the latent and lytic phases of the viral life cycle. While the majority of these genes have counterparts in other herpesviruses, a surprising number are unique to KSHV or related gammaherpesviruses 2 that infect other primates. The latter category includes the viral interferon (IFN) regulatory factors (vIRFs), which are inserted between open reading frames (ORFs) 57 and 58 but transcribed in the opposite orientation (reviewed in reference 36). In KSHV, there are four vIRF genes: vIRF1 (encoded by gene K9), vIRF2 (K11), vIRF3/LANA2 (K10.5), and vIRF4 (K10). The literature is complicated by the fact that vIRF2, vIRF3, and vIRF4 are each encoded by two exons that were assigned separate gene names in early annotations. In this study we consider K10 to be synonymous with K10 and K10.1. The genome of rhesus rhadinovirus (RRV) contains nine vIRFs arranged in an analogous tandem array inserted at the same genomic position. The predicted amino acid sequences have diverged significantly from the KSHV counterparts (1). Little is known about the expression or function of the RRV vIRFs, but the sheer number of genes suggests an unprecedented degree of specialization.
Incorporation of the vIRFs is one of many examples of “molecular piracy” evident in the KSHV genome (11). Functional studies have shown that vIRF1 to -4 can counteract innate antiviral defenses mediated by either IFN or the tumor suppressor protein p53 (23). The mechanisms are varied, involving interactions with cellular regulators of the IFN response, such as IRF1, IRF3, IRF5, and IRF7, or transcriptional cofactors, such as p300/CBP, which is required for the activation of IRF and p53 gene targets (5, 17, 25, 53, 57). The vIRF4 protein has been shown to antagonize p53-mediated apoptosis by stabilizing the human double minute 2 (HDM2) E3 ubiquitin ligase, resulting in the accelerated turnover of p53 by the proteosome (24). It is notable that vIRF1 also targets the p53 pathway but does so by preventing phosphorylation by ATM kinase and by limiting its transactivation potential (35, 45, 46). In addition, vIRF4 binds to the cellular poly(A)-binding protein C (PABP-C) and promotes its nuclear accumulation (21). This may help to curtail the antiviral response by limiting the export and translation of newly synthesized cellular mRNAs encoding antiviral effectors. Recently, vIRF4 was found to interact with CSL (RBP-Jκ), a cellular transcription factor that acts as a critical coregulator for RTA, the master regulator of the reactivation program (18). The biological consequences are still to be determined but may enable the virus to modulate Notch signaling or provide negative feedback by competing with RTA on a subset of lytic promoters.
Here we examined the regulation of the K10/vIRF4 gene by RTA. The isolated K10 promoter showed a robust response to RTA in several KSHV-infected cells but was essentially unresponsive in uninfected cells. Subsequent analysis showed that coexpression of RTA with vIRF4 resulted in strong transcriptional synergy and presumably accounts for the activity in the infected cells. The K9/vIRF1 promoter responds to RTA alone, but this activity is also significantly enhanced by coexpression of vIRF4. Synergy is not recapitulated with the other vIRFs. Depletion of vIRF4 during reactivation from latency results in a reduced yield of infectious KSHV virions, whereas overexpression enhances virus yield. Analysis of truncations and point mutants established a critical role for the N terminus of vIRF4, which shows imperfect homology to the DNA-binding domains (DBDs) of cellular IRFs. The vIRFs are distinguished from their cellular counterparts by a CxxC motif located immediately C-terminal to the putative DNA-binding domain. Mutation of either of the cysteines in vIRF4 abolishes self-association and synergy with RTA, providing the first known function for this signature motif. Lastly, biochemical studies identified a cellular factor(s) that binds to the K10/vIRF4 proximal promoter but is displaced or otherwise altered when vIRF4 and RTA are present. These findings establish vIRF4 as an important coregulator for RTA that contributes to the efficiency of KSHV reactivation.
MATERIALS AND METHODS
Plasmids.
Promoter regions and open reading frames were amplified from KSHV genomic DNA by using Expand high-fidelity PCR system (Roche) with custom primers (available on request). Amplification products were captured using pCR2.1-TOPO (Invitrogen), sequenced, and then excised using customized restriction endonucleases sites. Promoters were subcloned into the unique XhoI and HindIII sites of a modified pGL3-Basic luciferase reporter. ORFs were subcloned into the XbaI and BamHI sites of pCGFlag (30). The vIRF4 full-length ORF was subcloned using the following primers: 5′-CGGTCTAGACCTAAAGCCGGTGGCTCAGAATGG-3′ and 5′-CATGGATCCTCAATGTAGACTATCCCAAATGGAGCC-3′ (added restriction sites are underlined). Expression plasmids encoding full-length RTA, RTAΔSTAD, and HCF-1c and the luciferase reporters PAN-luc, LTc-luc (pLT7-luc), and LTi-luc have been described previously (33, 38, 54). Amino acid substitutions in vIRF4 and RTA and internal deletion mutant RTAΔLR were generated by QuikChange II XL site-directed mutagenesis (Stratagene). For in vitro translation, the coding regions of RTA and vIRF4 were subcloned into derivatives of the pCITE2a+ vector (Novagen). Expression plasmids encoding IRF7 (32), vIRF2 (2), and truncated vIRF4 (24) were kind gifts of Isabelle Marié (New York University School of Medicine), David Blackbourn (Cancer Research UK Cancer Centre, University of Birmingham), and Hye-Ra Lee and Jae Jung (University of Southern California), respectively.
Cell lines and transfections.
KSHV-positive lymphoma cells (BC3 and JSC-1) were cultured in RPMI 1640 medium (HyClone) supplemented with 20% fetal bovine serum (FBS), 2 mM l-glutamine, and antibiotics. For luciferase reporter assays, a total of 0.5 × 107 cells were electroporated with 12.5 μg luciferase reporter and 1.25 μg pCGFlag-RTA or empty vector by using a Gene Pulser II (Bio-Rad) set at 250 V, 975 μF. HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Cellgro) supplemented with 10% FBS, 2 mM l-glutamine, and antibiotics. SLK cells (KS tumor-derived human endothelial cells) were grown in RPMI 1640 with 10% Fetalplex, 2 mM l-glutamine, and antibiotics. Latently infected iSLK.219 cells (34), a kind gift of Jinjong Myoung and Don Ganem, were maintained in DMEM supplemented with 250 μg/ml G418, 400 μg/ml hygromycin, 10 μg/ml puromycin, 10% FBS, 2 mM l-glutamine, and antibiotics and were transfected using either Lipofectamine 2000 (Invitrogen) (for expression plasmids) or Lipofectamine RNAiMax (Invitrogen) (for shRNA).
Luciferase reporter assays.
For luciferase reporter assays, 2 × 104 HeLa or SLK cells were seeded 24 h before transfection in 24-well plates and then transfected using Polyfect (Qiagen). Luciferase activity was measured 24 h after transfection. In the majority of the luciferase assays shown, 250 ng of reporter plasmid was cotransfected into 1 well of a 24-well plate with 200 ng of vIRF4 expression plasmid and/or 25 ng of RTA expression plasmid. The relative activity of the reporter in the presence or absence of RTA was calculated for each cell line after normalization and plotted as the fold induction. Each cell line was assayed three or more times, and the replicates gave similar results. When comparing cell lines with different transfection efficiencies (see Fig. 1, below), we included an additional sample with the constitutively active LTc promoter, and all values were normalized to the relative activity of this construct.
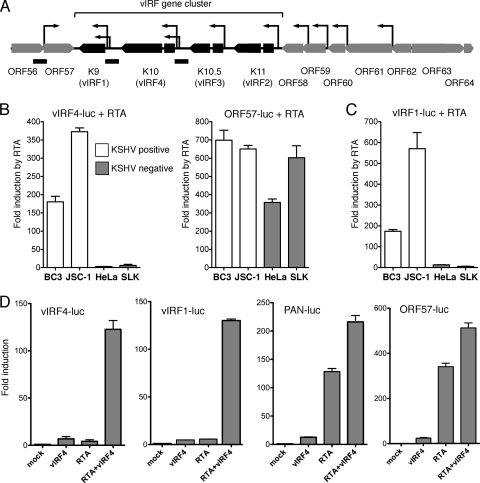
Fig 1.
vIRF4 and RTA cooperate to activate the K10/vIRF4 and K9/vIRF1 promoters in uninfected cells. (A) Schematic showing the vIRF gene cluster and surrounding open reading frames. The four vIRFs (K9/vIRF1, K10/vIRF4, K10.5/vIRF3, and K11/vIRF2) are arranged in tandem and are flanked by ORF57 (posttranscriptional regulator Mta) and ORF58 (EBV BMRF2 homologue). The TSSs (arrows) of the genes in this region have been mapped by 5′ rapid amplification of cDNA ends or primer extension (12, 31, 50, 51). Promoter fragments extending from immediately upstream of the initiation codon and incorporating the TSSs upstream of ORF57, K9/vIRF1, and K10/vIRF4 are shown as black bars below the map. (B) The K10/vIRF4 (vIRF4-luc) and Mta/ORF57 (ORF57-luc) reporters were transiently transfected into KSHV-positive BC3 and JSC-1 cells (open bars) or KSHV-negative HeLa and SLK cells (filled bars) together with an expression vector encoding RTA or an empty vector. The constitutively active LTc promoter was assayed in parallel in order to normalize transfection efficiency. The relative activity of the reporter in the presence or absence of RTA was calculated for each cell line after normalization and plotted as the fold induction, with standard deviations. (C) Response of the K9/vIRF1 reporter (vIRF1-luc) to RTA in KSHV-positive BC3 and JSC-1 cells (open bars) and KSHV-negative HeLa and SLK cells (filled bars). Values were calculated as described for panel B. (D) HeLa cells were transfected with the indicated reporters (250 ng/well of a 24-well plate) and expression plasmids encoding vIRF4 (200 ng) and/or RTA (25 ng). Fold induction was calculated relative to the reporter alone (mock). Values represent the means and standard errors of the means of three independent transfections.
Immunoblotting, coimmunoprecipitation, and indirect immunofluorescence microscopy.
Cells were lysed in SDS sample buffer and heat denatured. Lysates were separated by SDS–10% PAGE and then transferred to a Protran nitrocellulose membrane (Whatman) and blocked in Tris-borate-EDTA buffer containing 5% nonfat milk. Extracts for coimmunoprecipitation assays were prepared using medium salt extraction buffer (250 mM KCl, 20 mM Tris-HCl [pH 7.9], 10% glycerol, 0.25% NP-40, 0.5 mM phenylmethylsulfonyl fluoride) with protease inhibitors, and precipitates were washed in the buffer containing 200 mM KCl and 5% glycerol. Membranes were probed with anti-Flag (M2, diluted 1:1,000; Sigma), anti-T7 (diluted 1:1,000; Novagen), anti-Rho GDIα (A-20/sc-360, diluted 1:5,000; Santa Cruz Biotechnology), or anti-alpha-tubulin (diluted 1:5,000; Sigma) primary antibodies and detected using horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (diluted 1:5,000; Amersham). Chemiluminescence detection was performed with SuperSignal West Pico (Thermo Scientific). A rabbit polyclonal antibody against vIRF4 was generously provided by Hye-Ra Lee and Jae Jung (University of Southern California). For subcellular localization studies, HeLa cells were seeded onto sterile coverslips in a 24-well plate and transfected with 200 ng of each expression plasmid by using Polyfect reagent (Qiagen). Cells were fixed after 24 h and probed with a mix of anti-Flag and anti-T7 antibodies, washed, and then incubated with a mix of fluorescence-coupled secondary antibodies. After drying, coverslips were applied to slides by using fluorescence mounting medium (Dako Corporation) and visualized by laser scanning confocal microscopy with a Zeiss LSM 510 META microscope. Images were captured using the Zeiss AIM software, and .tiff images were exported into Adobe Photoshop 7.0 for cropping and minor adjustments.
Secondary structure prediction.
Secondary structure predictions were made using the PredictProtein server (41). The helical wheel projection was drawn using a shareware application written by Don Armstrong and Raphael Zidovetzki (University of California, Riverside).
Gel shift analysis.
Recombinant RTA and vIRF4 were synthesized by in vitro translation using the TNT quick-coupled transcription/translation system (Promega) in the presence of Easytag l-[35S]methionine (Perkin-Elmer). Radiolabeled double-stranded DNA probes were prepared by PCR amplification using 32P-labeled primers to amplify subcloned promoter fragments. Binding reaction mixtures containing labeled probe, 1 μg poly(dI-dC), 10 mM HEPES (pH 7.9), 75 mM KCl, 1 mM EDTA, 10 mM dithiothreitol and up to 4 μl reticulocyte lysate were incubated at room temperature for 20 min and loaded on 4% native polyacrylamide gels with 1× Tris-glycine-EDTA buffer. Electrophoresis was carried out at room temperature and run at a constant 170 V. Gels were fixed in methanol-acetic acid and dried, and the probe was visualized by autoradiography using an X-ray film to block the 35S signal.
Detection of infectious KSHV.
KSHV-infected iSLK.219 cells were induced to reactivate with 100 ng/ml doxycycline. After 72 h, the culture medium was collected, passed through a 0.45-μm filter, and used to infect 293-PAN-Luc cells (56). A total of 500 μl of the filtered medium was added to each well of 80% confluent reporter cells in a 24-well plate and centrifuged at 450 × g for 20 min to promote viral adherence. Infected cultures were incubated at 37°C for 2 h and then refed with fresh medium. After 48 h, the 293-PAN-Luc cells were harvested and assayed for luciferase activity.
Depletion of vIRF4 in KSHV-infected cells.
27/25-mer Dicer substrate RNAs (dsiRNAs) targeting the KSHV K10/vIRF4 transcript were custom synthesized (Integrated DNA Technologies) and evaluated by transfection into iSLK.219 cells by using Lipofectamine RNAiMax (Invitrogen) following the manufacturer's protocol. Predesigned dsiRNA against enhanced green fluorescent protein (EGFP) was used as a control. Cells were induced to reactivate with doxycycline, and total RNA was isolated after 72 h. The extent of knockdown was determined by quantitative reverse transcription-PCR (qRT-PCR). Three dsiRNA duplexes gave a reduction in K10/vIRF4 mRNA of 90% or greater and were used in subsequent analyses. The sequences were as follows: vIRF4 dsiRNA#1, 5′-CAAUGUAGACUAUCCCAAAUGGAGCCU-3′ and 5′-GCUCCAUUUGGGAUAGUCUACAUTG-3′; vIRF4 dsiRNA#2, 5′-ACAUAUAUCCUCUCCUUGAAGGUUCGA-3′ and 5′-GAACCUUCAAGGAGAGGAUAUAUGT-3′; vIRF4 dsiRNA#1, 5′-UACCAAACCACCAGCCUUCCACUUGAG-3′ and 5′-CAAGUGGAAGGCUGGUGGUUUGGTA-3′.
Quantitative RT-PCR analysis.
Total RNA was prepared using TRIzol (Invitrogen) and converted to cDNA with SuperScript III (Invitrogen) and random hexamer primers (Fermentas). qRT-PCR analysis was performed using FastStart Universal SYBR green Master-ROX (Roche) and a MyiQ single-color real-time thermal cycler (Bio-Rad). Samples were normalized using 18S rRNA primers (SA Biosciences), and the relative changes in transcript levels were calculated using the ΔΔCT method. Viral transcripts were measured with the following primer sets: K9/vIRF1, 5′-TCGCGGACCCCTGTTTTGAA-3′ and 5′-TCCACGGGCATGTTACCCCTTT-3′; K10/vIRF4, 5′-CCGGGTATGCGAAACCATCA-3′ and 5′-TCCAAAGCGGTGTGCGAACT-3′.
RESULTS
The K10 (vIRF4) promoter responds to RTA in KSHV-infected cells.
The four KSHV genes encoding the vIRFs (K9/vIRF1, K10/vIRF4, K10.5/vIRF3, and K11/vIRF2) are situated between ORF57 (Mta) and ORF58 (Epstein-Barr virus [EBV] BMRF2 homologue) (42). This section of the KSHV genome (shown schematically in Fig. 1A) is relatively well characterized, and the transcription start sites (TSSs), alternative splices, and polyadenylation sites of each vIRF mRNA have been described (8, 12, 28, 29, 31, 48, 50, 51). Each of the vIRF genes can be loosely classified as constitutive or inducible. Transcripts originating from K10/vIRF4 are barely detectable in latently infected PEL cells but accumulate with early kinetics upon reactivation (12, 20, 22). K9/vIRF1 is transcribed at low levels in latency but then is significantly induced at early times in reactivation, paralleling K10/vIRF4 (8, 48). In PEL-derived cell lines at least, K10.5/vIRF3 and K11/vIRF2 transcripts are readily detected during latency but accumulate further during reactivation (5, 12, 40).
Activation of the K10/vIRF4 promoter by RTA has not been reported. To address this, we generated a luciferase reporter construct using a 0.9-kbp fragment taken immediately upstream of the K10/vIRF4 initiator codon (nucleotides 88,911 to 89,815 of the prototype M-type genome; NCBI reference sequence NC_003409N). This genomic fragment includes the two previously mapped TSSs and is expected to include all of the critical proximal promoter elements (12). Note that this new construct is distinct from the K10/vIRF4 reporter described by Ellison and colleagues (nucleotides 88,202 to 88,879), which begins within the second exon and omits all of these sequences (14). The reporter (vIRF4-luc) was tested in PEL-derived BC3 and JSC-1 cells by cotransfection with an expression vector encoding RTA or the empty equivalent (Fig. 1B). Differences in transfection efficiencies were normalized using the constitutive promoter (LTc) that lies upstream of ORF73/LANA (33). Expression of RTA resulted in a substantial increase in luciferase activity (150- to 350-fold), consistent with the notion that the K10/vIRF4 promoter is a regulatory target of RTA. The same analysis was performed in two additional cell lines (HeLa and SLK) that do not contain the KSHV genome. Surprisingly, the vIRF4-luc reporter was induced only weakly (2.4- and 9.5-fold) in each of these cell contexts (Fig. 1B). This behavior was unexpected and contrasts markedly to more-typical RTA-responsive promoters, exemplified here by the ORF57 promoter (Fig. 1B), that can be induced to high levels in both uninfected HeLa and SLK cells (300- and 600-fold) and latently infected BC3 and JSC-1cells (>600-fold). The K9/vIRF1 promoter also showed a significantly greater response to RTA in BC3 and JSC-1 cells (175- and 550-fold) than HeLa or SLK cells (12-fold and 6-fold) (Fig. 1C). Thus, both the K10/vIRF4 and K9/vIRF1 promoters show a requirement for additional viral gene products, or at least some other aspect of the latently infected cell environment, in order to respond fully to RTA.
vIRF4 and RTA cooperate to activate the vIRF4 promoter.
To identify the viral factors responsible for activation of K10/vIRF4, we tested a panel of KSHV regulatory proteins for the ability to activate transcription in the presence of RTA (data not shown). Of these, vIRF4 showed a unique ability to cooperate with RTA to stimulate transcription from the vIRF4 and vIRF1 promoters (Fig. 1D). Expression of vIRF4 alone or RTA alone had little or no effect on the activity of any of the reporter constructs tested but conferred a substantial increase in the activity of vIRF4-luc and vIRF1-luc when coexpressed with RTA. Canonical RTA-responsive promoters, such as PAN and ORF57, were induced by more than 100-fold in the presence of RTA, and this was increased by less than 2-fold in the presence of vIRF4. Thus, synergy between RTA and vIRF4 appears to be relatively promoter specific.
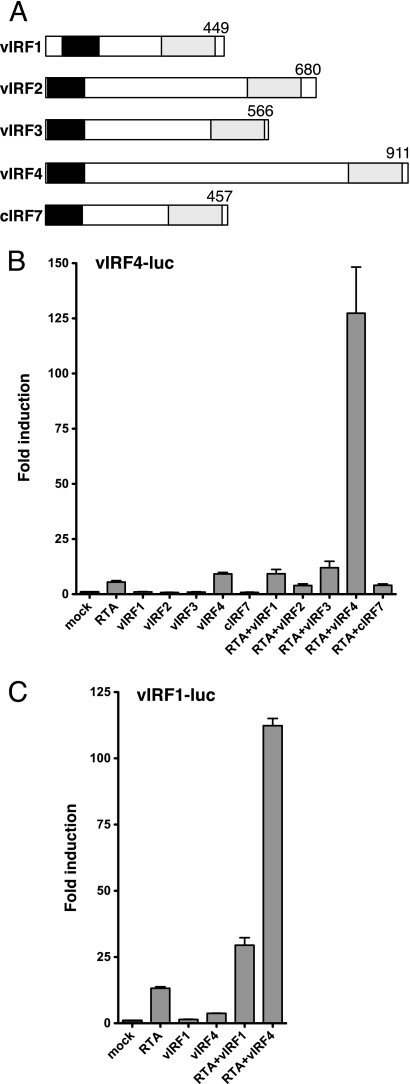
Next we asked if the ability to cooperate with RTA was shared by other vIRF proteins. The predicted products of the vIRF locus are shown schematically in Fig. 2A. The four vIRFs have diverged substantially but retain two regions of limited homology, an N-terminal region that corresponds to the DBD of cellular IRFs and a C-terminal region that forms part of the larger dimerization and transactivation domains (10). The central region of vIRF4 is large and lacks significant homology to other proteins. The vIRF4-luc reporter was transfected into HeLa cells together with a vector encoding each vIRF, either alone or with RTA (Fig. 2B). Only the combination of vIRF4 and RTA resulted in activation. Cellular IRF7 was included as a control and also failed to synergize with RTA. It is conceivable that the vIRF4-RTA synergy represents a positive (feed-forward) autoregulatory loop, and we therefore asked if vIRF1 was capable of stimulating its own promoter. As shown in Fig. 2C, expression of vIRF1 gave a 2-fold increase over RTA alone, perhaps consistent with a previous report of autoactivation by vIRF1 (52). However, this increase was modest compared to the increase achieved with vIRF4. Thus, vIRF4 appears to be unique in being able to cooperate with RTA to stimulate transcription from both the K10/vIRF4 and K9/vIRF1 promoters.
Fig 2.
Synergy with RTA is a unique property of vIRF4. (A) Schematic comparing the general structure of the four KSHV vIRFs with cellular IRF7. Two regions of sequence homology are indicated: the putative DNA-binding domain at the N terminus (black) and part of the C-terminal transactivation/dimerization domain (shaded). (B) HeLa cells were transfected with 250 ng vIRF4-luc and expression plasmids carrying RTA (25 ng) and/or vIRF1, vIRF2, vIRF3, vIRF4, and IRF7 (200 ng). Fold induction was calculated relative to the reporter alone (mock). Values represent the means and standard errors of the means of three independent transfections. (C) Assay using vIRF1-luc cotransfected with plasmids expressing RTA (25 ng) and/or vIRF1 and vIRF4 (200 ng).
vIRF4 contributes to efficient reactivation by KSHV.
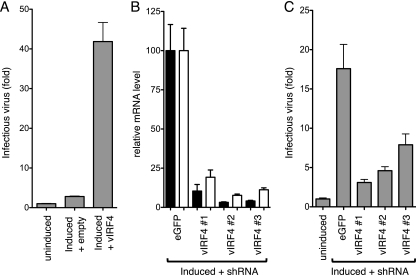
To determine the contribution of vIRF4 to lytic reactivation, we performed overexpression and depletion experiments in iSLK.219 cells (Fig. 3). Cultures were transfected with empty expression plasmid or one encoding full-length vIRF4 and then induced to reactivate by addition of doxycycline to the medium. Release of infectious virus into the medium was measured using a reporter cell line (293-PAN-Luc) that is stably transfected with a luciferase gene driven by the RTA-responsive PAN promoter (56). Induction of iSLK.219 cells that were transfected with the empty plasmid resulted in a small increase in reporter activity, reflecting a modest level of infectious virus. Note that this is lower than that observed using untransfected cells and reflects toxicity of the transfection reagents. However, introduction of the vIRF4 plasmid resulted in a 10-fold increase in reporter activity, which we interpreted as an increase in the yield of infectious virus.
Fig 3.
vIRF4 contributes to KSHV reactivation. (A) KSHV latently infected iSLK.219 cells were either mock treated or transfected with empty vector or vector expressing full-length vIRF4. The next day, the transfected cultures were induced with 100 ng/ml doxycycline (DOX) to drive expression of a DOX-regulated RTA cDNA stably integrated into the cell genome and maintained for 72 h. Culture medium was collected, filtered to remove debris, including cells, and used to infect 293-PAN-Luc reporter cells. After 48 h, lysates were prepared and assayed for luciferase activity. Values represent the means and standard errors of the means of three independent transfections. (B) iSLK.219 cells were transfected with dsiRNAs against EGFP or K10/vIRF4 mRNA sequences. Cultures were induced to reactivate by using doxycycline, and RNA was harvested 72 h later and analyzed by quantitative RT-PCR using primers to detect K9/vIRF1 (open bars) and K10/vIRF4 (filled bars). (C) Results of an experiment similar to that shown in panel B, except that the culture medium was collected from induced and uninduced iSLK.219 cells and assayed for infectious KSHV virions by using 293-PAN-Luc cells. Values are expressed relative to the averages of the uninduced samples.
Next, we used RNA interference to reduce the levels of vIRF4 encoded by the viral episome. For maximal incorporation into the RNA-induced silencing complex, we synthesized dsiRNAs. The degree of knockdown was assessed by qRT-PCR to measure K10/vIRF4 transcript levels (Fig. 3B). Three different dsiRNAs were identified that decreased transcript levels by 90% or more compared to cells transfected with a control dsiRNA against EGFP. iSLK.219 cells express EGFP from a constitutive promoter integrated into the viral episome, providing a convenient measure of transfection efficiency. By flow cytometry, green fluorescence was significantly reduced in at least 60% of the culture (data not shown). Transcripts from K9/vIRF1 were assayed in parallel and showed a similar reduction supporting the role of vIRF4 in activating the K9/vIRF1 promoter. Culture medium was collected from transfected cultures and tested on 293-PAN-Luc cells (Fig. 3C). Surrogate viral titers were reduced in cells transfected with each of all three vIRF4 dsiRNA complexes compared to the EGFP dsiRNA. These results indicated that vIRF4 contributes to reactivation of latent KSHV, presumably through cooperation with RTA to activate a subset of lytic promoters as well as through additional roles, such as antagonizing p53-mediated apoptosis (24).
Analysis of the K10/vIRF4 promoter.
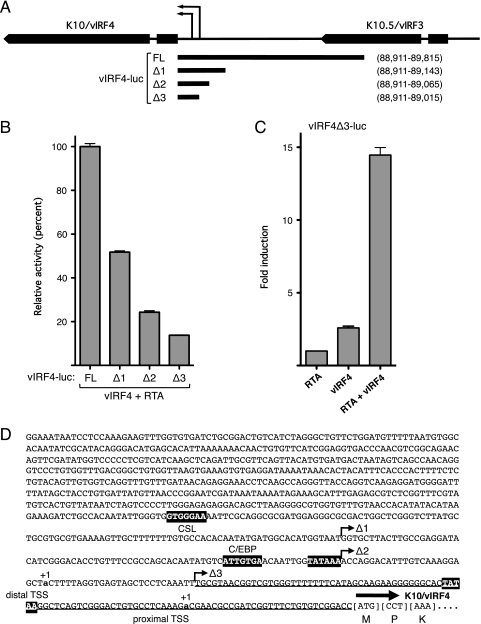
Previous studies found that tetradecanoyl phorbol acetate (TPA)-inducible K10/vIRF4 mRNAs initiate at two discrete TSSs within the intergenic region (12). The sites are separated by 102 bp and are positioned 30 or 31 nucleotides downstream of a consensus TATA box, indicative of two core promoters (Fig. 4A and D). To define the sequences necessary for the response to RTA/vIRF4, we generated three truncations of the original vIRF4-luc reporter and analyzed their activities in HeLa cells (Fig. 4A and B). Overall activity was reduced in stepwise fashion with each successive truncation (vIRF4-luc set to 100%), suggesting that multiple promoter elements contribute to the overall activity. The shortest fragment tested (vIRF4Δ3-luc) lacked the distal TSS but retained significant activity in the presence of vIRF4 and RTA (Fig. 4B). Importantly, this fragment was induced up to 15-fold in the presence of RTA and vIRF4 compared to RTA alone (Fig. 4C). Thus, sequences capable of responding to vIRF4 and RTA in a synergistic manner lie within the 105 bp immediately upstream of the initiation codon. This does not exclude the possibility of additional response elements elsewhere in the starting fragment that contribute to the overall level of activation.
Fig 4.
Analysis of the K10/vIRF4 promoter. (A) Schematic showing K10/vIRF4 and K10.5/vIRF3 and the intergenic region. Transcription of K10/vIRF4 initiates at two discrete sites (thin arrows). The 0.9-kb fragment used in the vIRF4-luc reporter is shown below, with the three additional truncations (Δ1, Δ2, and Δ3). Genomic coordinates of each promoter fragment are given on the right. (B) Activity assay results in HeLa cells transfected with full-length (FL) and truncated versions of vIRF4-luc (250 ng), vIRF4 (200 ng), and RTA (37.5 ng). Values are plotted relative to the mean activity of FL vIRF4-luc (set at 100%). Data are derived from three independent transfections. (C) Response of vIRF4Δ3-luc to RTA (37.5 ng) and/or vIRF4 (200 ng). Activity is plotted relative to the reporter in the presence of RTA only. (D) Nucleotide sequence of the K10/vIRF4 promoter region (vIRF4-luc) and the first codons of exon 1 (corresponding to nucleotides 88,902 to 89,815 of the BC-1 reference genome). Distal and proximal TPA-inducible transcription start sites (+1) (12) are shown in bold lowercase letters. Putative TATA boxes located 30 to 31 nucleotides upstream of each TSS are highlighted. Matches to the CSL and C/EBP binding site consensus sequences are also indicated. The minimal vIRF4/RTA-responsive fragment (vIRF4Δ3-luc) is underlined.
Transcriptional cooperation requires multiple regions of vIRF4.
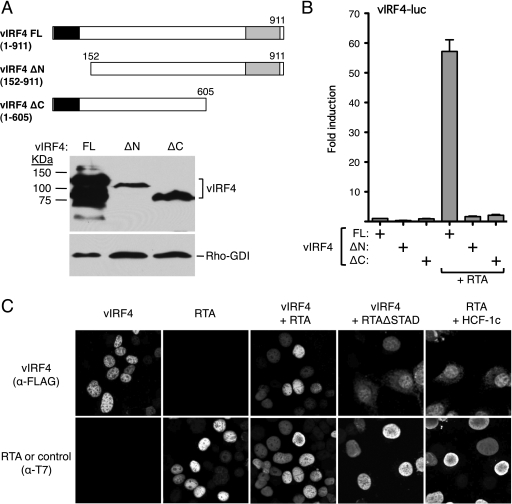
As a first step toward understanding how vIRF4 collaborates with RTA, we examined the contribution of the two IRF homology regions within vIRF4. Specifically, we compared the full-length vIRF4 (Fig. 5A, residues 1 to 911) to truncations that removed the N-terminal 151 residues (ΔN; residues 152 to 911) and C-terminal 307 residues (ΔC; residues 1 to 605). Previously, Lee and colleagues showed that vIRF4ΔN retains the ability to interact with and stabilize MDM2 but that this activity was lost with C-terminal truncation (24). Immunoblotting with a vIRF4 antibody confirmed that both truncations were expressed, albeit at lower levels than the full-length protein (Fig. 5A). When cotransfected into HeLa cells with the vIRF4-luc reporter, neither truncation supported significant levels of transcription in the presence of RTA, arguing that the putative DBD and the C-terminal transactivation/dimerization regions are important.
Fig 5.
N- and C-terminal domains of vIRF4 are required for RTA synergy. (A) Schematic showing limits of the N- and C-terminal vIRF4 truncations. Expression levels were assessed by immunoblotting using anti-vIRF4 antiserum (kind gift of Jae Jung and Hye-Ra Lee). (B) Activity assay in HeLa cells transfected with vIRF4-luc reporter (250 ng) and expression plasmids carrying full-length or truncated vIRF4 (200 ng) in the presence or absence of RTA (37.5 ng). Activity (fold induction) was calculated relative to that of the reporter with full-length vIRF4 only. Values represent the means and standard errors of the means of three independent transfections. (C) vIRF4 and RTA independently localize to the nucleus. Indirect immunofluorescence analysis of HeLa cells transfected with plasmids expressing T7-epitope-tagged RTA and Flag-epitope-tagged vIRF4 proteins (200 ng each). After fixation, coverslips were probed with a mix of anti-T7 (rabbit polyclonal) and anti-Flag (mouse monoclonal) antibodies followed by a mixture of anti-mouse and anti-rabbit fluorescence-coupled secondary antibodies.
The subcellular localizations of the major virus-inducible IRFs (IRF3, IRF5, and IRF7) are tightly controlled, with the latent forms residing in the cytoplasm until activation (47). With this in mind, we examined the localization of vIRF4 in the presence or absence of RTA by indirect immunofluorescence microscopy (Fig. 5C). When expressed on its own, vIRF4 was predominantly nuclear with a diffusing distribution throughout the nucleoplasm. RTA showed a similar distribution, and there was no obvious difference when the two proteins were expressed together. The cellular chromatin-associated cofactor HCF-1c served as a control for nucleoplasmic localization and had no discernible impact on the localization of vIRF4. Thus, synergy does not arise through an obvious change in the subcellular localization of either vIRF4 or RTA.
Mutations in the vIRF4 DNA recognition helix abrogate synergy.
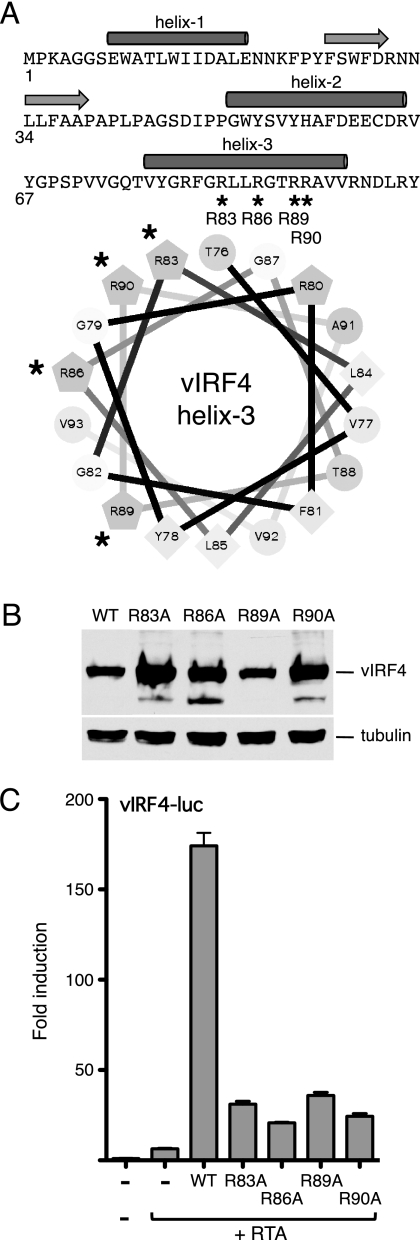
The N-terminal DBD of the cellular IRFs are characterized by five invariant tryptophan residues (tryptophan pentad repeat) and the ability to bind DNA sequences based on the consensus sequence 5′-AANNGAAA-3′ (15). Several structures have been determined and found to resemble a winged helix-turn-helix, with three α-helices, three long loops, and four stranded antiparallel β-sheets. The third helix slots into the major groove of the 5′-GAAA-3′ subsequence and is the major determinant of sequence-specific binding through contacts made by arginine residues on the hydrophilic face with the phosphate backbone. Residues within the loops and other helices provide additional contacts. In vIRF4, three of the hallmark tryptophans are missing, and the degree of sequence conservation is very limited; however, folding programs predict an equivalent trihelical arrangement (Fig. 6A). This suggests that the N terminus of vIRF4 might still be capable of binding DNA. This idea is strengthened by the fact that the putative DNA recognition helix (helix 3) retains its amphipathic nature, with hydrophobic residues clustered on one face and hydrophilic residues, including several arginines, on the other (Fig. 6A, helical wheel). To address the possibility that vIRF4 interacts with promoter DNA through its N-terminal domain, we generated a set of point mutations that changed four of the five arginines in helix 3 to alanine. The resulting mutants (R83A, R86A, R89A, and R90A) were expressed at equal or higher levels than the wild type (Fig. 6B). When assayed for synergy with RTA and the vIRF4 promoter, we found all four of these missense mutants had significantly reduced activity (Fig. 6C). Thus, in spite of the degenerate appearance of the N-terminal DBD region, point mutations that would be predicted to disrupt DNA binding abolished transcriptional activation in the presence of RTA.
Fig 6.
Mutations in helix 3 of vIRF4 disrupt synergy with RTA. (A) Primary sequence of the vIRF4 N terminus (residues 1 to 98), showing the locations of the predicted α-helices (filled cylinders) and β-strands (horizontal arrows). Helix 3 is also shown as a helical wheel projection with hydrophobic residues (F, L, V, and Y) clustered on one face (lower right). The four arginine residues (R83, R86, R89, and R90) that were changed in turn to alanine are indicated with an asterisk. (B) Immunoblot (with anti-Flag antibody) of lysates prepared from cells transfected with each vIRF4 plasmid. The species corresponding to full-length vIRF4 is indicated. Equal loading was demonstrated by blotting for α-tubulin. (C) Activity assay using HeLa cells transfected with vIRF-luc (250 ng) and each vIRF4 derivative (200 ng) in the presence of RTA (37.5 ng). Fold induction was calculated relative to the reporter alone. Values represent the means and standard errors of the means of three independent transfections.
The vIRF CxxC signature motif is necessary for synergy.
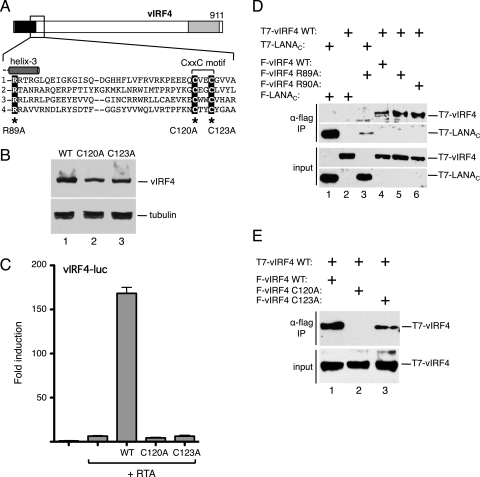
Adjacent to the DBD region is a double-cysteine motif (CxxC, where x is any residue) that is present in all four KSHV vIRFs (Fig. 7A). The CxxC motif is also found in all eight vIRFs encoded by rhesus rhadinoviruses (12). Remarkably, the motif is not found in any cellular IRFs and thus can be considered a distinctive signature of viral IRFs. The functional significance of the CxxC motif is unknown. We therefore generated two additional mutants in vIRF4, changing Cys-120 or Cys-123 to alanine. Again, both of the missense mutants were expressed at approximately the wild-type level (Fig. 7B) but were severely compromised for transactivation of the vIRF4 promoter with RTA (Fig. 7C). This is the first demonstration that the CxxC motif is functionally important for any vIRF protein.
Fig 7.
The signature cysteine motif in vIRF4 is essential for the transactivation function. (A) Schematic of vIRF4. The inset shows the sequence immediately downstream of helix 3, including the CxxC motif common to all KSHV vIRFs (and RRV vIRFs [data not shown]). Corresponding sequences of KSHV vIRF1, vIRF2, and vIRF3 are shown using the invariant arginine (R89 in vIRF4) to anchor the alignment. Cysteine-120 and cysteine-123 were individually changed to alanine. (B) Immunoblot (with anti-Flag antibody) of lysates prepared from cells transfected with each vIRF4 plasmid. The species corresponding to full-length vIRF4 is indicated. Equal loading was demonstrated by blotting for α-tubulin. (C) Activity assay using HeLa cells transfected with vIRF4-luc (250 ng) and each vIRF4 derivative (200 ng) in the presence of RTA (25 ng). Fold induction was calculated relative to the reporter alone. Values represent the means and standard errors of the means of three independent transfections. (D) vIRF4 is capable of self-association. Lysates were prepared from HeLa cells transfected with plasmids encoding combinations of T7 and Flag-tagged proteins and subjected to immunoprecipitation using Flag-coupled beads. Precipitates were resolved by SDS-PAGE and blotted with anti-T7 antibody. Expression was confirmed by blotting lysates directly. (E) Self-association is disrupted by mutation of the signature cysteine residues. Coimmunoprecipitation analysis was conducted as described for panel D, using wild-type (WT) and mutant (C120A and C123A) versions of vIRF4.
Mutations in the CxxC motif compromise vIRF4 self-association.
Cellular IRF proteins function as homo- or heterodimers, and it is conceivable that the CxxC motif contributes to self-association by vIRF4. To test this, we performed coimmunoprecipitation experiments with T7-tagged and Flag epitope-tagged proteins (Fig. 7D). As a positive control, we used the C terminus of LANA, which is known to form a stable homodimer (44) and, as expected, T7-LANAC was efficiently coimmunoprecipitated by Flag-LANAC (lane 1) but not with Flag-vIRF4R89A (lane 3). When versions of wild-type vIRF4 carrying both tags were coexpressed, blotting showed that T7-vIRF4 was readily recovered as shown with Flag-vIRF4 (lane 4). This was not seen if T7-vIRF4 was coexpressed with the control, Flag-LANAC (lane 2). We also tested two of the helix 3 mutants, R89A and R90A, and observed similar or greater levels of association with wild type. Thus, although both of these mutants are defective for synergy with RTA, they each retain the ability to self-associate. Similar coimmunoprecipitation experiments were performed using the CxxC motif mutants (Fig. 7E). Under these conditions, the C120A mutant (Flag-vIRF4C120A) failed to coprecipitate wild-type T7-vIRF4 (lane 2), and the C123A mutant showed a reduced ability (lane 3). We conclude from this result that the CxxC motif contributes to self-association and that this property is critical for synergy with RTA (Fig. 7C).
RTA contributes to DNA recognition by vIRF4/RTA.
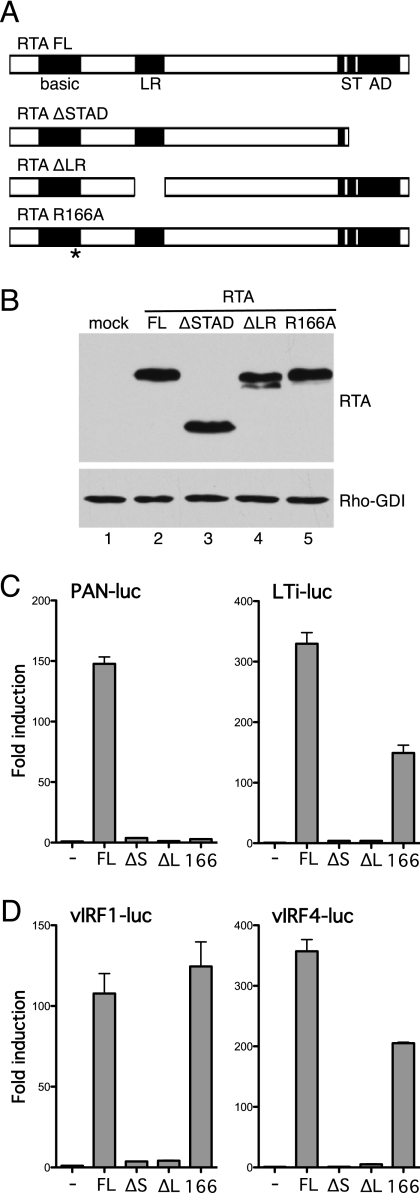
RTA comprises several functional domains and must assemble into a tetramer to function as a transactivator (3). To better understand the contribution of RTA, we tested two derivatives of RTA, which are illustrated in Fig. 8A. The first, RTAΔSTAD, lacks the C-terminal activation domain, whereas the second, RTAΔLR, contains an internal deletion that omits the leucine-rich region (residues 244 to 275) that is critical for oligomerization. Each version was FLAG-epitope tagged at the N terminus, and expression was demonstrated by immunoblotting (Fig. 8B). Removal of the activation domain or disruption of oligomerization abolished activation of RTA-responsive promoters, such as PAN and LTi (Fig. 8C), and vIRF4/RTA-responsive promoters K9/vIRF1 and K10/vIRF4 (Fig. 8D). As shown in Fig. 5C, the subcellular localization of RTAΔSTAD is indistinguishable from that of wild-type RTA, consistent with the idea that the lack of vIRF4-luc activation reflects the absence of the RTA transactivation domain.
Fig 8.
Transactivation and DNA-binding functions of RTA contribute to synergy. (A) Schematic showing known functional domains in RTA. DNA binding is mediated by the N terminus, which includes a region of positively charged residues (basic) and a leucine-rich region (LR) that is critical for oligomerization. The C terminus is required for transcriptional activation mediated by a serine/threonine-rich region (ST) and a separate activation domain (AD). Derivatives lacking the transactivation region (RTAΔSTAD) or leucine-rich region (RTAΔLR) are shown below. Mutation of R166 within the basic region disrupts DNA binding. (B) Immunoblot (with anti-Flag antibody) of lysates prepared from HeLa cells transfected with each RTA plasmid. Detection of Rho-GDI showed equal loading. (C) Activity assay using HeLa cells transfected with PAN-luc or LTi-luc (250 ng) and each RTA expression plasmid (25 ng). Fold induction was calculated relative to the reporter alone. Values represent the means and standard errors of the means of three independent transfections. (D) Activity assay results with HeLa cells transfected with vIRF1-luc or vIRF4-luc (250 ng) and each RTA expression plasmid (25 ng) in the presence of wild-type vIRF4 (200 ng).
The N terminus of RTA includes a basic residue-rich region (residues 131 to 170) that is critical for DNA binding (6, 7). Several arginines (R160, R161, and R166) within this region are critical for activation of promoters reliant on direct recognition of RTA response elements, such as PAN and K12, but have less impact on promoters that use cellular coregulators to ensure RTA binding and activity. To address the contribution of direct DNA binding to vIRF4-RTA synergy, we changed arginine 166 to alanine (R166A) and compared the response to wild-type RTA. As expected, R166A was unable to activate the PAN promoter (Fig. 8C) but retained significant activity on the LTi promoter, which includes two CSL binding sites that function as RTA response elements (27, 33). The R166A mutant also activated the K9/vIRF1 and K10/vIRF4 promoters (Fig. 8D), consistent with the idea that vIRF4 facilitates recruitment of RTA to vIRF4/RTA-responsive promoters.
vIRF4 and RTA cooperate to alter the association of a cellular factor with the K10/vIRF4 proximal promoter.
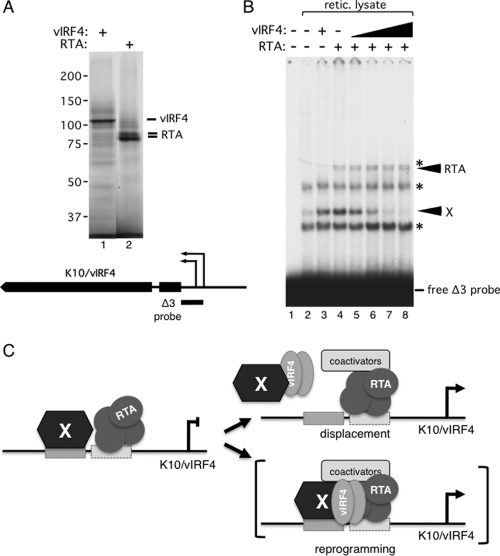
To determine whether vIRF4 and RTA can form stable complexes on the K10/vIRF4 promoter, we expressed both proteins in a coupled in vitro transcription/translation system using T7 RNA polymerase and rabbit reticulocyte lysate and then used the recombinant proteins in gel shift analyses with a 32P-labeled probe corresponding to the minimal responsive fragment (Fig. 4, Δ3) of the proximal K10/vIRF4 promoter. SDS-PAGE followed by autoradiography confirmed that each recombinant protein was expressed well and was predominantly full length (Fig. 9A). When the labeled probe was incubated with mock lysate (lane 2) and resolved on a 4% native polyacrylamide gel, four distinct complexes were resolved. Three complexes (indicated with an asterisk) were also detected with unrelated probes of similar size and are presumably nonspecific (data not shown). The fourth complex (labeled X) was unique to the Δ3 probe and reflects the binding of one or more cellular factors present in the reticulocyte lysate. Addition of RTA resulted in an additional slowly migrating complex (lane 4, labeled RTA), indicating that RTA can bind independently to the K10/vIRF4 promoter region. Inclusion of RTA also resulted in a small but reproducible increase in the abundance of complex X. Addition of vIRF4 alone did not result in any new complexes, but again there was a modest increase in the cellular complex (compare lane 3 with lane 2). Interestingly, when vIRF4 and RTA were included simultaneously in the binding reaction mixture (lanes 5 to 8), the abundance of complex X decreased markedly. Loss of complex X was proportional to the amount of input vIRF4 and, at the same time, was dependent on the presence of RTA (lanes 3 and 8 contain equal amounts of vIRF4). We conclude that one or more cellular factors can form a complex on the K10/vIRF4 proximal promoter that is sensitive to the presence of both RTA and vIRF4. Complex X may either be displaced by RTA/vIRF4 or assembled into a larger complex that is not resolved under the gel conditions used.
Fig 9.
vIRF4 and RTA antagonize binding of a cellular factor(s) to the K10/vIRF4 proximal promoter. (A) Reticulocyte lysates were programmed with T7 RNA polymerase-transcribed plasmids carrying full-length vIRF4 and RTA in the presence of [35S]methionine. In vitro translation products were resolved by 8% SDS-PAGE and visualized by autoradiography. Relative molecular masses (in kDa) were determined using prestained protein standards run concurrently. The position of the K10/vIRF4 promoter Δ3 probe fragment encompassing the proximal promoter is shown. (B) Gel mobility shift analysis results with a 32P-labeled K10/vIRF4 promoter Δ3 fragment probe incubated with rabbit reticulocyte lysates programmed with empty vector (lane 2) or plasmids encoding RTA or vIRF4 (lanes 3 to 8). Nonspecific complexes observed with multiple probes are marked with an asterisk. Arrows mark a low-mobility complex that incorporated RTA and an uncharacterized complex (X) that was sensitive to the presence of vIRF4 and RTA. (C) Schematic of a working model for activation of the K10/vIRF4 promoter by RTA and vIRF4. RTA and a cellular factor(s) (X) associate with the proximal promoter but do not activate transcription. Inclusion of vIRF4 leads to reprogramming of the complex, either by displacement of X or by incorporation of vIRF4 into a larger complex that is not resolved on the gel. Cellular coactivators recruited by RTA and/or other components of the complex are then able to activate transcription.
DISCUSSION
RTA is often introduced as the “master regulator” of the switch from latency to lytic reactivation in KSHV. This accolade seems well justified: transcription of the ORF50/RTA gene is the first detectable response of latent genomes to various reactivation stimuli, and unlike other herpesvirus lytic initiators that activate the immediate-early (IE) or early (E) genes only, RTA stimulates at least 40 viral promoters belonging to the IE, E, and even late (L) kinetic classes (9, 14, 39). How one regulator achieves such far-reaching control is not fully understood, but the ability of RTA to interface with a variety of cellular transcription factors (termed coregulators) may be a critical aspect. The mediator of cellular Notch signaling, CSL (RBP-Jκ, CBF-1), is a particularly important coregulator for KSHV RTA, and functionally important binding sites have been demonstrated in at least 20 RTA-responsive promoters (39). Other cellular transcription factors, such as C/EBP and Oct-1, perform similar roles, albeit for a more limited set of viral promoters (43, 49). Viral gene products, such as LANA and K8, also modulate RTA function (14, 26).
In the current study we showed that vIRF4, a viral homologue of a cellular transcription factor, serves as a positive coregulator for RTA, and we identified two lytic promoters that utilize vIRF4 to achieve a full response to RTA. It should be noted that a previous study found that the K9/vIRF1 promoter responded well to the expression of RTA in the absence of other viral gene products (8), while a third study reported that vIRF1 could stimulate transcription from its own promoter independent of RTA (52). The reasons for these discrepancies are unclear, but it is worth noting that in some experiments we saw a significant response of vIRF1-luc to RTA, although this was always lower than with the combination of RTA and vIRF4 (data not shown). The differences in the response to individual factors might relate to differences in the assays, including the amounts of reporter and effector plasmids used. We chose to focus on the K10/vIRF4 promoter for further analysis, because the response to either RTA or vIRF4 alone was consistently modest and was robust in the presence of the two factors together. However, it makes sense that the K10/vIRF4 promoter would not be entirely dependent on the two factors, because at early times only RTA is present and some level of transcription is needed to synthesize the first molecules of vIRF4. The subsequent feed-forward loop would lead to more significant accumulation of vIRF4 protein, perhaps enhancing other functions, such as antagonizing p53 (24) or directing the relocalization of cytoplasmic poly(A)-binding protein (21).
Many developmental systems rely on combinatorial control to achieve context-specific transcription. A dependence on vIRF4 and RTA for maximal expression may define a specific set of KSHV genes that are functionally related. We know already that both vIRF1 and vIRF4 help to block p53-mediated apoptosis, and it seems likely that other genes with related functions are scattered elsewhere in the genome. Additional degrees of control (fine-tuning) could be obtained by regulating vIRF4/RTA synergy in such a way that other aspects of the lytic reactivation program that are also driven by RTA can, if necessary, be uncoupled from these ancillary processes.
Cellular IRFs function as multisubunit complexes formed through a combination of dimerization with the same or other IRFs and interactions with unrelated transcription factors, such as STATs, NF-κB, and SMAD3 (19). The working model presented in Fig. 9 imagines an analogous complex composed of viral and possibly cellular components. Additional experimentation will be needed to determine whether RTA and vIRF4 interact directly or indirectly. Gel shift analysis showed that RTA can stably associate with the K10/vIRF4 proximal promoter, and this requires its DNA-binding function (data not shown). Whether vIRF4 is capable of binding to DNA independently remains speculative. The N terminus of vIRF4 is predicted to fold into a helical structure resembling the winged helix DBD of cellular IRFs, and even though it lacks some of the critical tyrosine residues, substitutions in the putative DNA recognition helix (helix 3) interfere with its transcription function. It is worth emphasizing that the equivalent region of vIRF1 has also diverged from the IRF pattern and yet binds to palindromic DNA sequences identified through an in vitro binding site selection assay (37). As might be expected, the consensus vIRF1-binding sequence (5′-GCGTCnnGACGC-3′, where n is any nucleotide) shares no obvious similarity to cellular IFN-stimulated response elements (ISREs) (5′-GAAAnnGAAACT-3′) (13), providing a strong hint that the vIRFs may have evolved to recognize their own unique response elements. These may be composite in nature, bringing a weak vIRF4-binding site into juxtaposition with binding sites for other factors, thereby ensuring robust activation of the target promoter only when there are sufficient levels of multiple factors: in other words, combinatorial control. The intriguing relationship between RTA and cellular or viral IRFs may extend even deeper; Zhang and colleagues found that RTA activated the promoters of several interferon-stimulated genes and was capable of binding to probes containing an ISRE (55). Moreover, they identified sequence similarities between the DNA-binding domains of RTA and cellular IRFs.
It will be interesting to discover exactly how many lytic promoters rely on the synthesis of additional viral products to achieve a full response to RTA. When protein synthesis is blocked with hygromycin, only 10% of lytic genes are activated in the presence of preexisting RTA, suggesting that in fact the majority of lytic genes rely on de novo protein synthesis for efficient transcription (4). The hygromycin experiment does not distinguish between viral and cellular proteins, but the findings presented in the current study argue that the absence of vIRF4 should account for some or many of the hygromycin-sensitive lytic genes. Profiling studies and the analysis of individual promoters will shed further light on this important question.
ACKNOWLEDGMENTS
We thank Melissa Victoria Fernandez, Christopher Rusnak, and Carlos Sanchez for their valuable contributions at various stages of this project. David Blackbourn, Don Ganem, Jae Jung, Hye-Ra Lee, Isabelle Marié, and Jinjong Myoung provided helpful suggestions or reagents.
This work was supported by grants from the NIH (GM61139 and S10 RR017970) and pilot project awards from the NYU Center for AIDS Research and NYU Cancer Institute for HIV-Associated Malignancy Research (NIH/NIAID P30 AI027742) to I.M. and A.C.W.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Alexander L, et al. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26 to 95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Areste C, Mutocheluh M, Blackbourn DJ. 2009. Identification of caspase-mediated decay of interferon regulatory factor-3, exploited by a Kaposi sarcoma-associated herpesvirus immunoregulatory protein. J. Biol. Chem. 284:23272–23285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bu W, Carroll KD, Palmeri D, Lukac DM. 2007. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50/Rta lytic switch protein functions as a tetramer. J. Virol. 81:5788–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bu W, et al. 2008. Identification of direct transcriptional targets of the Kaposi's sarcoma-associated herpesvirus Rta lytic switch protein by conditional nuclear localization. J. Virol. 82:10709–10723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burysek L, et al. 1999. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J. Virol. 73:7334–7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang PJ, Miller G. 2004. Autoregulation of DNA binding and protein stability of Kaposi's sarcoma-associated herpesvirus ORF50 protein. J. Virol. 78:10657–10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang PJ, Shedd D, Miller G. 2005. Two subclasses of Kaposi's sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J. Virol. 79:8750–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J, Ueda K, Sakakibara S, Okuno T, Yamanishi K. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623–8634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen J, Ye F, Xie J, Kuhne K, Gao SJ. 2009. Genome-wide identification of binding sites for Kaposi's sarcoma-associated herpesvirus lytic switch protein, RTA. Virology 386:290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen W, Royer WE., Jr 2010. Structural insights into interferon regulatory factor activation. Cell. Signal. 22:883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi J, Means RE, Damania B, Jung JU. 2001. Molecular piracy of Kaposi's sarcoma associated herpesvirus. Cytokine Growth Factor Rev. 12:245–257 [DOI] [PubMed] [Google Scholar]
- 12. Cunningham C, Barnard S, Blackbourn DJ, Davison AJ. 2003. Transcription mapping of human herpesvirus 8 genes encoding viral interferon regulatory factors. J. Gen. Virol. 84:1471–1483 [DOI] [PubMed] [Google Scholar]
- 13. Darnell JE, Jr, Kerr IM, Stark GR. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415–1421 [DOI] [PubMed] [Google Scholar]
- 14. Ellison TJ, Izumiya Y, Izumiya C, Luciw PA, Kung HJ. 2009. A comprehensive analysis of recruitment and transactivation potential of K-Rta and K-bZIP during reactivation of Kaposi's sarcoma-associated herpesvirus. Virology 387:76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujii Y, et al. 1999. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 18:5028–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ganem D. 2010. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J. Clin. Invest. 120:939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao SJ, et al. 1997. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene 15:1979–1985 [DOI] [PubMed] [Google Scholar]
- 18. Heinzelmann K, et al. 2010. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 (vIRF4/K10) is a novel interaction partner of CSL/CBF1, the major downstream effector of Notch signaling. J. Virol. 84:12255–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Honda K, Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644–658 [DOI] [PubMed] [Google Scholar]
- 20. Jenner RG, Alba MM, Boshoff C, Kellam P. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kanno T, Sato Y, Sata T, Katano H. 2006. Expression of Kaposi's sarcoma-associated herpesvirus-encoded K10/10.1 protein in tissues and its interaction with poly(A)-binding protein. Virology 352:100–109 [DOI] [PubMed] [Google Scholar]
- 22. Katano H, Sato Y, Kurata T, Mori S, Sata T. 2000. Expression and localization of human herpesvirus 8-encoded proteins in primary effusion lymphoma, Kaposi's sarcoma, and multicentric Castleman's disease. Virology. 269:335–344 [DOI] [PubMed] [Google Scholar]
- 23. Lee HR, Kim MH, Lee JS, Liang C, Jung JU. 2009. Viral interferon regulatory factors. J. Interferon Cytokine Res. 29:621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee HR, et al. 2009. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 targets MDM2 to deregulate the p53 tumor suppressor pathway. J. Virol. 83:6739–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li M, et al. 2000. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol. Cell. Biol. 20:8254–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Q, Zhou F, Ye F, Gao SJ. 2008. Genetic disruption of KSHV major latent nuclear antigen LANA enhances viral lytic transcriptional program. Virology 379:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang Y, Ganem D. 2004. RBP-J (CSL) is essential for activation of the K14/vGPCR promoter of Kaposi's sarcoma-associated herpesvirus by the lytic switch protein RTA. J. Virol. 78:6818–6826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y, et al. 2008. Kaposi's sarcoma-associated herpesvirus RTA activates the processivity factor ORF59 through interaction with RBP-Jκ and a cis-acting RTA responsive element. Virology 380:264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lukac DM, Garibyan L, Kirshner JR, Palmeri D, Ganem D. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mahajan SS, Little MM, Vazquez R, Wilson AC. 2002. Interaction of HCF-1 with a cellular nuclear export factor. J. Biol. Chem. 277:44292–44299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Majerciak V, Yamanegi K, Zheng ZM. 2006. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J. Virol. 80:11968–11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marie I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsumura S, Fujita Y, Gomez E, Tanese N, Wilson AC. 2005. Activation of the Kaposi's sarcoma-associated herpesvirus major latency locus by the lytic switch protein RTA (ORF50). J. Virol. 79:8493–8505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 174:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakamura H, Li M, Zarycki J, Jung JU. 2001. Inhibition of p53 tumor suppressor by viral interferon regulatory factor. J. Virol. 75:7572–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Offermann MK. 2007. Kaposi sarcoma herpesvirus-encoded interferon regulator factors. Curr. Top. Microbiol. Immunol. 312:185–209 [DOI] [PubMed] [Google Scholar]
- 37. Park J, et al. 2007. Identification of the DNA sequence interacting with Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 1. J. Virol. 81:12680–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pearce M, Matsumura S, Wilson AC. 2005. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J. Virol. 79:14457–14464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Persson LM, Wilson AC. 2010. Wide-scale use of Notch signaling factor CSL/RBP-Jκ in RTA-mediated activation of Kaposi's sarcoma-associated herpesvirus lytic genes. J. Virol. 84:1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rivas C, Thlick AE, Parravicini C, Moore PS, Chang Y. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75:429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rost B, Yachdav G, Liu J. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Russo JJ, et al. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sakakibara S, Ueda K, Chen J, Okuno T, Yamanishi K. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwam DR, Luciano RL, Mahajan SS, Wong L, Wilson AC. 2000. Carboxy terminus of human herpesvirus 8 Latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532–8540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Seo T, Park J, Lee D, Hwang SG, Choe J. 2001. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus binds to p53 and represses p53-dependent transcription and apoptosis. J. Virol. 75:6193–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shin YC, et al. 2006. Inhibition of the ATM/p53 signal transduction pathway by Kaposi's sarcoma-associated herpesvirus interferon regulatory factor 1. J. Virol. 80:2257–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tamura T, Yanai H, Savitsky D, Taniguchi T. 2008. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26:535–584 [DOI] [PubMed] [Google Scholar]
- 48. Ueda K, et al. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) replication and transcription factor activates the K9 (vIRF) gene through two distinct cis elements by a non-DNA-binding mechanism. J. Virol. 76:12044–12054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang SE, et al. 2003. Role of CCAAT/enhancer-binding protein alpha (C/EBPα) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang SE, Wu FY, Yu Y, Hayward GS. 2003. CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 77:9590–9612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang SS, et al. 2010. Transcriptional regulation of the ORF61 and ORF60 genes of Kaposi's sarcoma-associated herpesvirus. Virology 397:311–321 [DOI] [PubMed] [Google Scholar]
- 52. Wang XP, Gao SJ. 2003. Auto-activation of the transforming viral interferon regulatory factor encoded by Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Gen. Virol. 84:329–336 [DOI] [PubMed] [Google Scholar]
- 53. Wies E, et al. 2008. The viral interferon-regulatory factor-3 is required for the survival of KSHV-infected primary effusion lymphoma cells. Blood 111:320–327 [DOI] [PubMed] [Google Scholar]
- 54. Wilson AC, Boutros MJ, Johnson KM, Herr W. 2000. HCF amino- and carboxy-terminal subunit association through two separate sets of interaction modules: involvement of fibronectin type 3 repeats. Mol. Cell. Biol. 20:6721–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang J, Wang J, Wood C, Xu D, Zhang L. 2005. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 replication and transcription activator regulates viral and cellular genes via interferon-stimulated response elements. J. Virol. 79:5640–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao J, et al. 2007. K13 blocks KSHV lytic replication and deregulates vIL6 and hIL6 expression: a model of lytic replication induced clonal selection in viral oncogenesis. PLoS One 2:e1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zimring JC, Goodbourn S, Offermann MK. 1998. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J. Virol. 72:701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]