Abstract
The transmembrane domains (TMDs) of integral membrane proteins do not merely function as membrane anchors but play active roles in many important biological processes. The downregulation of the CD4 coreceptor by the Vpu protein of HIV-1 is a prime example of a process that is dependent on specific properties of TMDs. Here we report the identification of Trp22 in the Vpu TMD and Gly415 in the CD4 TMD as critical determinants of Vpu-induced targeting of CD4 to endoplasmic reticulum (ER)-associated degradation (ERAD). The two residues participate in different aspects of ERAD targeting. Vpu Trp22 is required to prevent assembly of Vpu into an inactive, oligomeric form and to promote CD4 polyubiquitination and subsequent recruitment of the VCP-UFD1L-NPL4 dislocase complex. In the presence of a Vpu Trp22 mutant, CD4 remains integrally associated with the ER membrane, suggesting that dislocation from the ER into the cytosol is impaired. CD4 Gly415, on the other hand, contributes to CD4-Vpu interactions. We also identify two residues, Val20 and Ser23, in the Vpu TMD that mediate retention of Vpu and, by extension, CD4 in the ER. These findings highlight the exploitation of several TMD-mediated mechanisms by HIV-1 Vpu in order to downregulate CD4 and thus promote viral pathogenesis.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) targets helper T cells and macrophages/monocytes through interaction of the viral envelope glycoprotein (Env) with a combination of the CD4 and CCR5/CXCR4 cell surface receptors (59). Once inside the cell, the genetic material of the virus directs rapid and sustained downregulation of CD4 (26, 62), a phenomenon that promotes viral spread by preventing superinfection, enabling the release of progeny virions, and interfering with the host immune response (3, 6, 34, 53). The ability of HIV-1 to downregulate CD4 depends on two accessory proteins encoded in the viral genome, Nef and Vpu (32, 38, 45, 61). Nef is a myristoylated protein that attaches to the cytosolic leaflet of the plasma membrane, where it functions to link the cytosolic tail of CD4 to the clathrin-associated adaptor protein 2 (AP-2) complex (1, 12, 16, 17, 19, 24, 39). These interactions lead to rapid internalization of cell surface CD4 by a clathrin-dependent pathway (14, 16, 30, 60). Nef exerts a second function in endosomes, namely, the targeting of internalized CD4 to the multivesicular body pathway for eventual degradation in lysosomes (20).
Vpu is a type III integral membrane protein that, in contrast to Nef, acts on newly synthesized CD4, causing its retention in the endoplasmic reticulum (ER) (42) and subsequent delivery to the ER-associated degradation (ERAD) pathway (7, 42, 63, 79). The mechanism of CD4 downregulation by Vpu involves a physical interaction of the cytosolic domains of both proteins (11, 47). A phosphoserine (pS)-based motif, DpSGxxpS, in the cytosolic domain of Vpu then acts as a binding site for the β-TrCP1/2 component of the SCFβ-TrCP1/2 E3 ubiquitin (Ub)-ligase complex (15, 25, 48), which in turn mediates lysine- and serine/threonine-dependent polyubiquitination of the CD4 cytosolic tail (42). Polyubiquitination partly arrests CD4 in the ER while enabling recruitment of the VCP-UFD1L-NPL4 dislocase complex, which extracts CD4 from the ER membrane into the cytosol (7, 42). Dislocated CD4 is subsequently delivered to the proteasome for degradation (7, 42, 63).
Although most studies on Vpu-induced CD4 downregulation have focused on reactions involving the cytosolic domains of the proteins, some studies have also implicated the corresponding transmembrane domains (TMDs) in this process (13, 42, 57, 73). Indeed, replacement of the vesicular stomatitis virus G glycoprotein (VSV-G) TMD for the CD4 TMD abolished Vpu-induced degradation of the resulting chimeric protein (13). Moreover, we recently showed that substitution of the VSV-G TMD for the Vpu TMD abrogated both CD4 retention in the ER and ERAD targeting induced by Vpu (42). These findings indicate that the TMDs of both CD4 and Vpu play important roles in the process of Vpu-induced CD4 downregulation.
In this study, we have examined the specific features of the Vpu and CD4 TMDs that are required for CD4 downregulation. We report the presence of a cluster of amino acids on the Vpu TMD, centered on Trp22, which is critical for Vpu-mediated CD4 degradation. Mutation of Trp22 does not prevent interaction of Vpu with CD4 but enhances Vpu oligomerization and also reduces CD4 polyubiquitination and recruitment of the VCP-UFD1L-NPL4 dislocase complex. In the presence of a Vpu Trp22 mutant, CD4 remains integrated into the ER membrane, suggesting that dislocation from the ER to the cytosol is impaired. We also define an additional determinant comprising Gly415 in the CD4 TMD that is required for both Vpu-CD4 interaction and induction of CD4 degradation by Vpu. Finally, we identify Val20 and Ser23 in the Vpu TMD as components of a separate determinant of Vpu and CD4 retention in the ER. These findings highlight the crucial requirement of specific TMD residues at several steps in the mechanisms of CD4 downregulation by Vpu.
MATERIALS AND METHODS
Recombinant DNA constructs.
pFLAG-CMV2-human β-TrCP1 was a gift from Y. Ben-Neriah (Hadassah Medical School, Hebrew University). pcDNA3.1-FLAG-Ub was already described (42). Site-directed mutagenesis (QuikChange II kit; Stratagene, Cedar Creek, TX) was performed on pcDNA3.1-codon-optimized HIV-1 NL4-3 Vpu (54) and pCMV-human CD4 (11). A nucleotide sequence encoding a hemagglutinin (HA) epitope was also added to the 3′ terminus of the codon-optimized Vpu cDNA by site-directed mutagenesis. Alternatively, the codon-optimized Vpu cDNA (without a stop codon) was amplified by PCR from the pcDNA3.1-codon-optimized Vpu construct and cloned as an EcoRI/XhoI fragment into the pcDNA3.1/myc-His A vector (Invitrogen, Carlsbad, CA). All mutagenesis and cloning products were verified by DNA sequencing.
Antibodies.
The following mouse monoclonal antibodies were used: anti-human CD4, clone 4B12 (Leica Microsystems, Bannockburn, IL) and clone OKT4 (eBiosciences, San Diego, CA); anti-actin (clone Ab-5) and anti-VCP (clone 18) (BD Biosciences, San Jose, CA); anti-human transferrin receptor (TfR), clone H68.4 (Zymed, San Francisco, CA); unconjugated/horseradish peroxidase (HRP)-conjugated anti-FLAG (clone M2) and anti-myc (clone 9E10) (Sigma-Aldrich, Saint Louis, MO); anti-HA, clone 16B12 (Covance, Berkeley, CA); and anti-RGS-His (Qiagen, Valencia, CA). Rabbit polyclonal antibodies to the human CD4 (residues 420 to 447) and Vpu (residues 32 to 81) were previously described (44, 63). Goat polyclonal antibody to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and HRP-conjugated donkey anti-goat IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). HRP-conjugated donkey anti-mouse IgG and donkey anti-rabbit IgG were from GE Healthcare Biosciences (Piscataway, NJ).
Cell culture and transfections.
Media and reagents for cell culture were purchased from Mediatech, Inc. (Manassas, VA). HeLa cells (American Type Culture Collection, Manassas, VA) were grown in Dulbecco's modified Eagle's medium, supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM l-glutamine at 37°C in a humidified atmosphere of 5% carbon dioxide in air. Cells were transiently transfected using Lipofectamine 2000 (Invitrogen). Plasmids encoding human CD4, codon-optimized Vpu, and FLAG-Ub were transfected at a 1:1:0.5 ratio (42). Cells were analyzed at 8 to 16 h after transfection.
Pulse-chase analysis, immunoprecipitation, electrophoresis, and immunoblotting.
Cells were pulse-labeled with [35S]methionine-cysteine and chased in complete medium, and cell lysates were subjected to immunoprecipitation as described previously (9). Immunoprecipitated proteins were analyzed by SDS-PAGE (42) and visualized by fluorography on a Typhoon 9200 PhosphorImager (Amersham Biosciences). Data analysis and quantification were performed using the ImageQuant software (GE Healthcare). Immunoblotting studies were carried out as reported previously (42) and analyzed using the Image J software (http://rsbweb.nih.gov/ij).
In vivo ubiquitination.
Cells expressing FLAG-tagged Ub were extracted in denaturing lysis buffer as described previously (42). Ubiquitinated CD4 was immunoprecipitated using a conformation-independent antibody to CD4 and analyzed by immunoblotting using an HRP-conjugated antibody to the FLAG epitope (42).
Fractionation of permeabilized cells.
Cells grown on 10-cm plates were detached with trypsin, washed twice with ice-cold phosphate-buffered saline (PBS) supplemented with 0.9 mM CaCl2, and permeabilized in ice-cold permeabilization buffer (PB) (25 mM HEPES [pH 7.3], 115 mM potassium acetate, 5 mM sodium acetate, 2.5 mM MgCl2, 0.5 mM EGTA) containing 10 mM α-iodoacetamide, 5 mM N-ethylmaleimide, 0.028% digitonin, and the complete Mini protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) for 5 min on ice. Cytosol was extracted from permeabilized cells by centrifugation at 20,000 × g for 10 min at 4°C. The pellet was resuspended in ice-cold PB, and all fractions were adjusted to 1% SDS and 10 mM dithiothreitol (DTT). Samples were heated for 10 min at 100°C and processed for in vivo ubiquitination as previously described (42).
Isolation of microsomes.
Cells (5 × 106) were scraped off the plates and incubated with ice-cold 20 mM HEPES (pH 7.2), 10 mM α-iodoacetamide, 5 mM N-ethylmaleimide, and the complete Mini protease inhibitor cocktail for 10 min before passage through a 1-ml syringe fitted with a 21-gauge needle (∼20 strokes). Cell lysates were supplemented with 5 mM MgCl2 and 100 mM potassium acetate and then sequentially centrifuged at 600 × g for 2 min and 15,000 × g for 10 min at 4°C. The resulting supernatants were ultracentrifuged at 200,000 × g for 1 h at 4°C, and pelleted microsomes were resuspended in membrane buffer (50 mM HEPES [pH 7.4], 150 mM potassium acetate, 10 mM magnesium acetate, 250 mM sucrose, 10 mM α-iodoacetamide, 5 mM N-ethylmaleimide) supplemented (or not) with 100 mM Na2CO3, pH 11.3. The samples were fractionated into soluble and microsomal membrane fractions by ultracentrifugation at 200,000 × g for 10 min at 4°C. All fractions were adjusted to 1% SDS and 10 mM dithiothreitol (DTT), heated for 10 min at 100°C, and processed for in vivo ubiquitination as described previously (42).
Cell surface biotinylation.
Cells grown on 6-well plates were incubated with 2 mM sulfo-NHS-LC biotin (Thermo Scientific, Rockford, IL) in ice-cold PBS supplemented with 0.1 mM CaCl2 and 1 mM MgCl2 for 30 min at 4°C. After extensive washing, cells were treated with 50 mM Tris-HCl (pH 7.5) for 10 min at 4°C and then extracted with ice-cold lysis buffer (0.5% Triton X-100, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA) supplemented with the complete Mini protease inhibitor cocktail. Equivalent amounts of cell lysates were incubated with a NeutrAvidin-coupled resin (Thermo Scientific) for 2 h at 4°C. Pulled-down biotinylated proteins or whole-cell lysates (10 μg of protein) were subjected to SDS-PAGE and immunoblotting with antibodies to CD4, Vpu, and TfR (loading control) as previously reported (42).
Endo H and PNGase F digestion.
Cell lysates made under denaturing conditions were processed for endoglycosidase H (endo H) and peptide:N-glycosidase F (PNGase F) digestion as described previously (42).
RESULTS
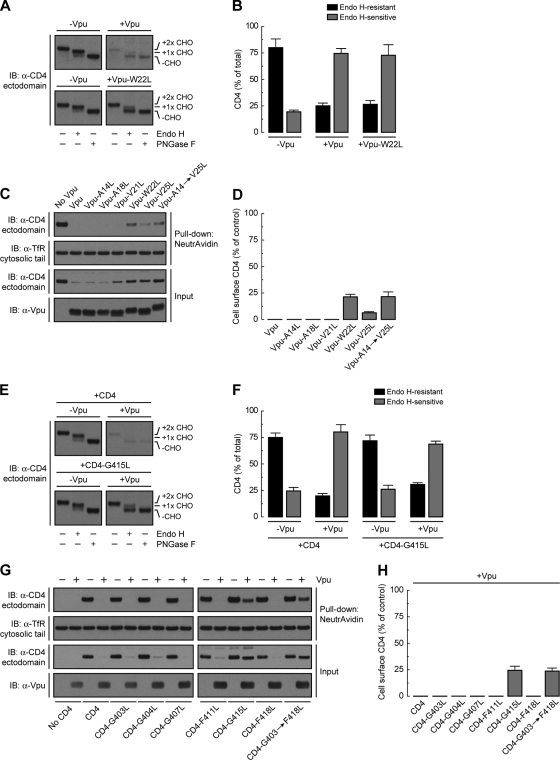
Mutational analysis of the Vpu TMD identifies Trp22 as a critical determinant for induction of CD4 degradation.
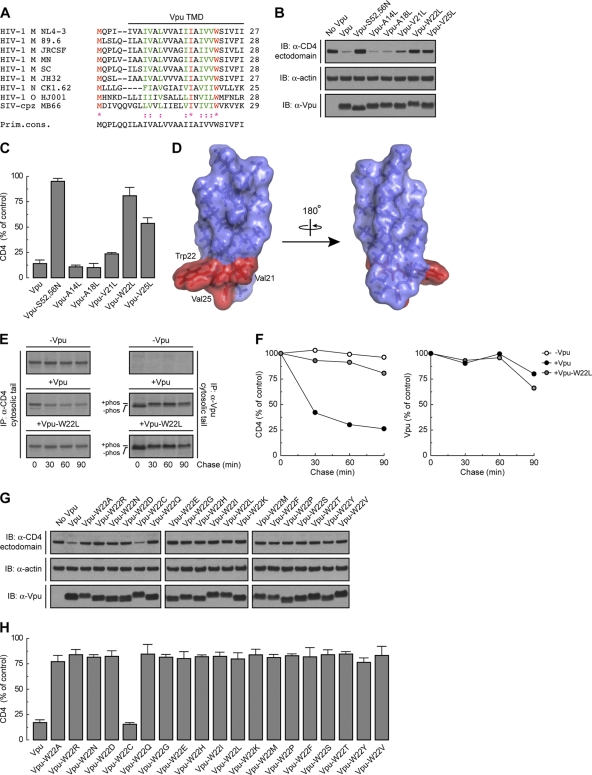
The amino acid sequence of the Vpu TMD is highly conserved among strains of the pandemic HIV-1 group M, which account for more than 90% of all HIV-1 infections (4, 33) (Fig. 1A). To determine the requirement for specific amino acid residues within this TMD for induction of CD4 downregulation, we mutated each residue other than Leu/Ile to Leu in the context of codon-optimized Vpu from the HIV-1 group M strain NL4-3 (54) (Fig. 1A). These mutants, along with wild-type Vpu and an inactive Vpu mutant (Vpu-S52,56N) bearing substitutions of the Ser52 and Ser56 residues that mediate interaction with β-TrCP1/2 (64) (positive and negative controls, respectively), were coexpressed with human CD4 by transient transfection of HeLa cells. The ability of the different Vpu constructs to induce CD4 degradation was determined by immunoblot analysis of total CD4 levels (42) (Fig. 1B). As expected, wild-type Vpu induced CD4 degradation whereas Vpu-S52,56N did not (Fig. 1B and C). Most mutations in the Vpu TMD had no effect on Vpu-mediated CD4 degradation (Fig. 1B and C and unpublished observations). However, mutation of Trp22 almost completely abolished CD4 degradation by Vpu, whereas mutation of Val21 or Val25 had intermediate effects (Fig. 1B and C). These three residues form a patch on the Vpu TMD α-helix (56), at a location proximal to the membrane-cytosol interface (Fig. 1D). Notably, the Trp residue is conserved in virtually all HIV-1 Vpu variants, including those of HIV-1 groups M, N, and O, as well as in some simian immunodeficiency virus (SIV) variants (77) (Fig. 1A). Val21 and Val25 are also highly conserved in HIV-1 group M but are replaced by other hydrophobic residues in most other variants (77) (Fig. 1A). A number of HIV-1 group M subtype C variants have Ala in place of Val at position 25. Replacement of Val25 by Ala had no effect on the ability of NL4-3 Vpu to induce CD4 degradation (see Fig. S1A and B in the supplemental material), indicating that this is a conservative substitution.
Fig 1.
Trp22 within the Vpu TMD is a key determinant of Vpu-induced CD4 degradation. (A) ClustalW alignment of the TMD sequences of HIV-1 groups M, N, and O and SIV-cpz MB66 Vpu variants, highlighting conservation of the residues (71). The TMD sequence shown on top corresponds to the NL4-3 strain used in this study. Notice that NL4-3 Vpu Trp22 is highly conserved among all HIV-1 and some SIV-cpz strains. (B) HeLa cells were transfected with plasmids encoding human CD4, with or without wild-type Vpu, Vpu-S52,56N, or Vpu mutants where all TMD non-Leu/Ile residues were replaced by Leu. At 12 h after transfection, cell lysates were prepared and analyzed by SDS-PAGE and immunoblotting (IB) with antibodies to the CD4 ectodomain, actin (loading control), and Vpu. For conciseness, a representative blot showing the effect of the Vpu-A14L, -A18L, -V21L, -W22L, and -V25L mutants on CD4 stability is shown. (C) CD4 levels in the presence of Vpu constructs were quantified by densitometry and expressed as the percentage of the total amount of CD4 in the absence of Vpu (100% control). Values represent the means ± standard errors of the means (SEM) from three independent experiments. (D) PyMOL images of the solid-state NMR structure of the NL4-3 Vpu TMD (56) (RSCB protein database entry 1PJE). Val21, Trp22, and Val25 (red) form a patch on the same face of the Vpu TMD α-helix. (E) HeLa cells expressing human CD4, with or without wild-type Vpu or Vpu-W22L, were labeled with [35S]methionine-cysteine for 2 min and then chased for the indicated times at 37°C. Cells extracts were subjected to immunoprecipitation using antibodies to the CD4 or Vpu cytosolic tails. Immunoprecipitated proteins were analyzed by SDS-PAGE and fluorography. (F) Percentage of CD4 or Vpu at each chase time relative to CD4 at time zero (100% control). (G) HeLa cells were transfected with plasmids encoding human CD4 and no Vpu (empty vector), wild-type Vpu, or an array of Vpu mutants where Trp22 was replaced by all amino acids. Cell lysates were treated as for panel B. (H) CD4 levels in the presence of Vpu were quantified as for panel C.
To confirm that the effect of the Vpu Trp22 mutation was on the ability of Vpu to induce degradation of newly synthesized CD4, we performed pulse-chase analysis (Fig. 1E). HeLa cells expressing human CD4, with or without wild-type Vpu or Vpu-W22L, were labeled for 2 min with [35S]methionine-cysteine and chased for up to 90 min in complete medium. Cells were extracted with Triton X-100, and CD4 was isolated by immunoprecipitation (42). Using this methodology, we observed that Vpu caused rapid degradation of CD4, with only ∼25% remaining after 90 min of chase (42) (Fig. 1E and F). In contrast, Vpu-W22L failed to promote CD4 degradation (Fig. 1E and F). This failure was not due to destabilization of Vpu by the mutation, since both wild-type and Vpu-W22L were equally stable over the time course of the experiment (Fig. 1E and F). These findings thus demonstrated a crucial requirement of Vpu Trp22 for CD4 targeting to degradation.
Trp has a bulky, aromatic side chain—the most hydrophobic side chain among all amino acids, according to some scales (52, 66). However, the Trp side chain also has an indole nitrogen atom that can act as a hydrogen bond donor (43). This dual character endows Trp with a particular affinity for membrane-water interfaces (80, 84), where Trp22 in the Vpu TMD is likely located (Fig. 1A and D). To determine whether these unique features of Trp are essential for Vpu-induced CD4 degradation, we mutated Vpu Trp22 to each of the other amino acids and tested for the ability of the resulting mutants to decrease total CD4 levels. We observed that with the exception of Cys, none of the other amino acids could substitute for Trp in this assay (Fig. 1G and H). These findings demonstrated the critical requirement for Trp at this position, likely explaining the strict conservation of this residue in all Vpu variants (Fig. 1A).
Mutation of Vpu Trp22 does not prevent Vpu-CD4 interaction but enhances Vpu oligomerization.
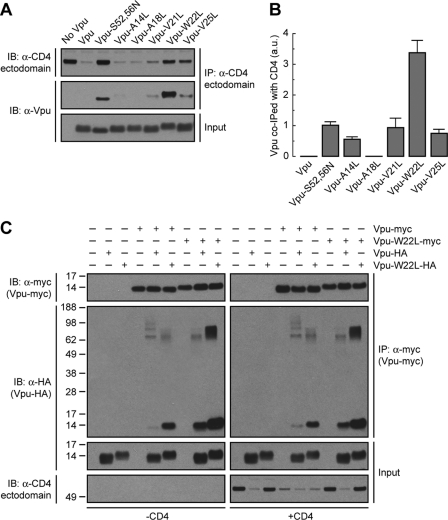
The cytosolic domains of Vpu and CD4 are known to interact with one another and to harbor critical determinants of Vpu-induced CD4 downregulation (11, 36, 47, 78). The Vpu and CD4 TMDs are also essential for Vpu-induced CD4 degradation as well as ER retention of CD4 (13, 42, 57, 73), but their roles in mediating protein interactions have not been addressed. To investigate whether Trp22 and other residues located on the same face of the Vpu TMD are involved in interaction with CD4, HeLa cells were transfected with plasmids encoding human CD4, with or without wild-type or mutant Vpu constructs. CD4 was isolated by immunoprecipitation under nondenaturing conditions and associated Vpu detected by immunoblotting (Fig. 2A). Because wild-type Vpu promotes CD4 degradation, coprecipitation of these two proteins could be visualized only after prolonged exposure of the films (unpublished observations). The inability of Vpu-S52,56N to induce CD4 degradation, on the other hand, allowed visualization of its coprecipitation with CD4 upon short exposure times (11) (Fig. 2A and B). Surprisingly, replacement of Leu for Trp22 in the Vpu TMD greatly increased the amount of Vpu that coprecipitated with CD4 (Fig. 2A and B). All other mutations in the Vpu TMD had little or no effect on Vpu-CD4 interactions (Fig. 2A and B).
Fig 2.
Mutation of Vpu Trp22 does not prevent Vpu-CD4 or Vpu-Vpu interactions but enhances Vpu oligomerization. (A) HeLa cells were transfected with plasmids encoding human CD4 and no Vpu (empty vector), wild-type Vpu, Vpu-S52,56N, or the Vpu-A14L, -A18L, -V21L, -W22L, or -V25L mutant. At 12 h after transfection, cell lysates were prepared under nondenaturing conditions and subjected to immunoprecipitation using an antibody to the CD4 ectodomain. Coprecipitation of Vpu with CD4 was detected by SDS-PAGE and immunoblotting with an antibody to Vpu. (B) The amount of Vpu coprecipitated (co-IPed) with CD4 was quantified by densitometry, normalized to the amount of immunoprecipitated CD4 in each sample, and expressed as arbitrary units (a.u.). Values represent the means ± SEM from three independent experiments. (C) HeLa cells were transfected with plasmids encoding no Vpu (empty vector), wild-type Vpu-myc or Vpu-W22L-myc and no Vpu (empty vector), wild-type Vpu-HA, or Vpu-W22L-HA, with or without CD4. Cell extracts were prepared as for panel A and subjected to immunoprecipitation with an antibody to the myc epitope. The resultant immunoprecipitates were resuspended in nonreducing sample buffer without boiling before being analyzed by SDS-PAGE and immunoblotting with an antibody to the HA peptide. Numbers on the left indicate molecular masses (in kDa). Notice the increased amount of Vpu-HA oligomers that coprecipitate with Vpu-myc upon mutation of Vpu Trp22.
The increased coprecipitation of Vpu-W22L with CD4 could be due to an enhanced affinity between the two proteins or to a change in the stoichiometry of the interaction. The latter interpretation is warranted by nuclear magnetic resonance (NMR) studies indicating that the Vpu TMD can occur in both monomeric and oligomeric forms (22, 55, 68). In this context, mutations in the TMD could alter the Vpu monomer/oligomer ratio and thus change the amount of Vpu coprecipitated with CD4. To determine the effect of the Trp22-to-Leu mutation on Vpu-Vpu interactions, HeLa cells were transfected with combinations of plasmids encoding wild-type Vpu and Vpu-W22L tagged at the C termini with the myc or HA epitopes, with or without CD4. Triton X-100 extracts of these cells were subjected to immunoprecipitation with an antibody to the myc epitope, followed by SDS-PAGE without a reducing agent and without boiling and immunoblotting with an antibody to the HA peptide (Fig. 2C). Under these conditions, CD4 was efficiently degraded when at least one of the Vpu constructs corresponded to the wild-type version, but it failed to be degraded when both constructs had the W22L mutation (Fig. 2C). Coexpression of wild-type Vpu-HA and Vpu-myc resulted in only a small amount of coprecipitation (Fig. 2C), suggesting that Vpu monomers predominate or that Vpu oligomers are unstable under these conditions. Interestingly, coexpression of wild-type Vpu with Vpu-W22L-myc or Vpu-W22L-HA resulted in intermediate levels of coprecipitation (Fig. 2C). Finally, coexpression of Vpu-W22L-myc with Vpu-W22L-HA resulted in much greater coprecipitation as evidenced by the detection of coprecipitating species that migrate as both monomers and stable oligomers on SDS-PAGE (Fig. 2C). Similar results were obtained in the absence or presence of CD4 (Fig. 2C), indicating that these changes are intrinsic to Vpu. Substitution of Ala or Gly for Trp22 also increased coprecipitation of epitope-tagged Vpu pairs (see Fig. S2A and B in the supplemental material), indicating that this effect is due to the removal of Trp rather than the addition of another specific residue at position 22. From these experiments we concluded that Vpu Trp22 hinders Vpu-Vpu interactions and that its mutation stabilizes Vpu oligomers. The above results thus indicated that the Vpu Trp22 residue is not required for either Vpu-CD4 or Vpu-Vpu interactions. Rather, mutation of this amino acid increases coprecipitation of Vpu with CD4 due to enhanced Vpu oligomerization.
Decreased CD4 polyubiquitination caused by replacement of Vpu TMD residues.
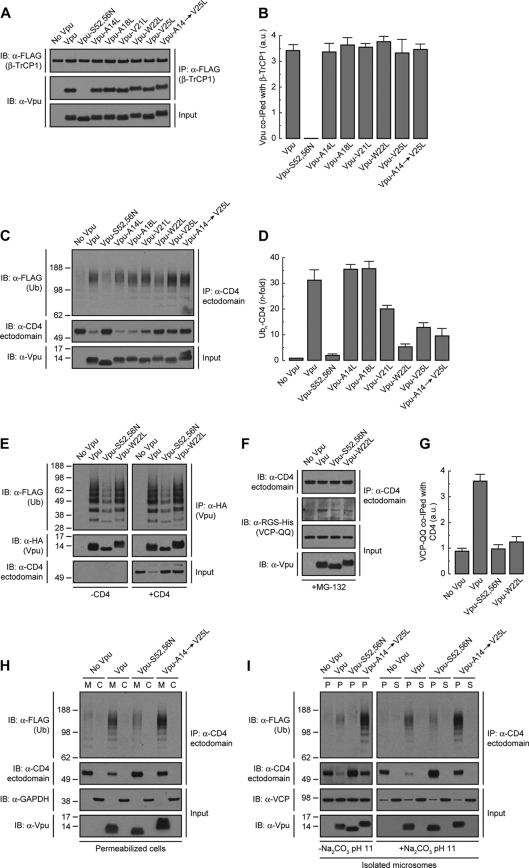
If mutation of Vpu Trp22 does not prevent Vpu-CD4 interactions, how does it inhibit CD4 degradation? To address this question, we initially examined the ability of Vpu TMD mutants to bind the β-TrCP1 component of the SCFβ-TrCP1/2 E3 Ub-ligase complex (48). HeLa cells were transfected with plasmids encoding FLAG-tagged human β-TrCP1 and Vpu, Vpu-S52,56N (negative control), or Vpu TMD mutants. β-TrCP1 was isolated by immunoprecipitation with an antibody to the FLAG epitope and coprecipitated Vpu species detected with an antibody to Vpu (Fig. 3A). As expected (48), wild-type Vpu coprecipitated with β-TrCP1 whereas Vpu-S52,56N did not (Fig. 3A and B). Vpu mutants having TMD substitutions of Leu for Ala14, Ala18, Val21, Trp22, Val25, or all residues together (Vpu-A14→V25L mutant) coprecipitated with β-TrCP1 to the same extent as wild-type Vpu (Fig. 3A and B). These observations indicated that the functional inactivation of Vpu by mutation of TMD residues, particularly Trp22, was not due to decreased recruitment of β-TrCP1 and, by extension, of the SCFβ-TrCP1/2 E3 Ub-ligase complex.
Fig 3.
The Vpu TMD promotes polyubiquitination and dislocation of CD4 from the ER membrane. (A) HeLa cells expressing FLAG-tagged human β-TrCP1, with or without wild-type Vpu, Vpu-S52,56N, or the Vpu-A14L, -A18L, -V21L, -W22L, -V25L, or -A14,A18,V21,W22,V25L (Vpu-A14→V25L) mutant, were lysed under nondenaturing conditions and extracts subjected to immunoprecipitation using an antibody to the FLAG peptide. Coprecipitation of Vpu with FLAG-β-TrCP1 was detected by SDS-PAGE and immunoblotting with an antibody to Vpu. Notice that only phosphorylatable wild-type Vpu and Vpu TMD mutants coprecipitate with FLAG-β-TrCP1. (B) The amount of coprecipitated Vpu with FLAG-β-TrCP1 was quantified by densitometry, normalized to the amount of immunoprecipitated FLAG-β-TrCP1 in each sample, and expressed as arbitrary units (a.u.). Values represent the means ± SEM from three independent experiments. (C) HeLa cells were transfected with plasmids encoding FLAG-tagged Ub and human CD4, with or without wild-type Vpu, Vpu-S52,56N, or the Vpu-A14L, -A18L, -V21L, -W22L, -V25L, or -A14→V25L mutant. At 12 h after transfection, equivalent amounts of cell extracts made under denaturing conditions were subjected to immunoprecipitation with a conformation-independent antibody to the CD4 ectodomain. Ubiquitination of CD4 was detected by immunoblotting with an HRP-conjugated antibody to the FLAG epitope. (D) Ubn-CD4 levels in the presence of Vpu were quantified by densitometry and expressed as the percentage of the total amount of Ubn-CD4 in the absence of Vpu (100% control). These values were normalized to the remaining nonubiquitinated CD4 (i.e., 1 for Ubn-CD4 in the absence of Vpu). Values are the means ± SEM from three independent experiments. (E) Denatured cell lysates from HeLa cells expressing FLAG-Ub and no Vpu or HA-tagged versions of wild-type Vpu, Vpu-S52,56N, or Vpu-W22L, with or without human CD4, were subjected to immunoprecipitation using an antibody to the HA peptide. Ubiquitination of Vpu was detected by immunoblotting with an HRP-conjugated antibody to the FLAG epitope. (F) HeLa cells were transfected with plasmids encoding human CD4 and RGS-His-tagged VCP-QQ plus no Vpu (empty vector), wild-type Vpu, Vpu-S52,56N, or Vpu-W22L. At 12 h after transfection, cells were treated with a 20 μM concentration of the proteasome inhibitor MG-132 in complete medium for 8 h at 37°C, lysed, and then subjected to immunoprecipitation with an antibody to the CD4 ectodomain. Coprecipitation of RGS-His-tagged VCP-QQ with CD4 in the presence of wild-type Vpu was detected by immunoblotting with an antibody to the RGS-His epitope. (G) The amount of coprecipitated RGS-His-VCP-QQ with CD4 was quantified by densitometry, normalized to the amount of immunoprecipitated CD4 in each sample, and expressed as arbitrary units (a.u.). Values represent the means ± SEM from three independent experiments. (H) HeLa cells expressing FLAG-Ub and human CD4, with or without wild-type Vpu, Vpu-S52,56N, or Vpu-A14→V25L, were permeabilized with 0.028% digitonin before being fractionated into membrane (M) and cytosol (C) by centrifugation. Samples were adjusted to 1% SDS and 10 mM DTT and then processed for in vivo ubiquitination as described for panel C. Transmembrane Vpu and cytosolic GAPDH were used as fractionation controls. (I) Microsomes from HeLa cells described for panel H were resuspended in membrane buffer supplemented (or not) with 100 mM Na2CO3, pH 11.3. After ultracentrifugation of the samples, membrane (P) and supernatant (S) fractions were collected and adjusted to 1% SDS and 10 mM DTT before being processed for in vivo ubiquitination as described for panel C. Transmembrane Vpu and the peripherally associated membrane protein VCP were used as fractionation controls. Numbers on the left in panels C, H, and I indicate molecular masses (in kDa). Notice that both nonubiquitinated and polyubiquitinated CD4 species are membrane anchored in the presence of the Vpu-A14→V25L mutant even after the high-pH treatment.
Next, we examined the effect of Vpu TMD mutations on the ability of Vpu to induce CD4 polyubiquitination. To this end, HeLa cells were cotransfected with plasmids encoding FLAG-tagged Ub, human CD4, and the Vpu TMD mutants mentioned above. Cells were solubilized under denaturing conditions and CD4 species subsequently immunoprecipitated with a conformation-independent antibody to the CD4 ectodomain. Polyubiquitinated CD4 in the immunoprecipitates was detected by immunoblotting using an antibody to the FLAG epitope (42) (Fig. 3C). The amount of polyubiquitinated CD4 was normalized to the amount of nonubiquitinated CD4 in each sample (42) (Fig. 3C and D). Using this procedure, we observed that wild-type Vpu increased CD4 polyubiquitination by 32-fold (Fig. 3C and D), consistent with previous results (42). The Vpu-S52,56N mutant elicited only background CD4 polyubiquitination (Fig. 3C and D), as expected from the inability of this mutant to recruit the SCFβ-TrCP1/2 E3 Ub-ligase complex (42, 48) (Fig. 3A and B). Single mutation of Val21, Trp22, or Val25 or combined mutation of Ala14, Ala18, Val21, Trp22, and Val25 (Vpu-A14→V25L), reduced the extent of Vpu-induced CD4 polyubiquitination to 5- to 20-fold, whereas single mutation of Ala14 and Ala18 had no effect (Fig. 3C and D). We also tested for polyubiquitination of HA-tagged Vpu under identical experimental conditions. We observed that Vpu was polyubiquitinated in a SCFβ-TrCP1/2-dependent fashion, as previously described (5) (Fig. 3E), and independently of CD4 coexpression (Fig. 3E). Importantly, polyubiquitination of Vpu was not affected by replacement of Trp22 by Leu (Fig. 3E). These experiments demonstrated that mutation of Val21, Trp22, and/or Val25 decreased CD4 polyubiquitination without affecting SCFβ-TrCP1/2 recruitment to Vpu as well as Vpu polyubiquitination.
Accumulation of CD4 in the ER membrane in the presence of a Vpu TMD mutant.
Vpu-mediated CD4 polyubiquitination allows recruitment of the VCP-UFD1L-NPL4 dislocase complex, promoting CD4 dislocation from the ER membrane for eventual targeting to the cytosolic proteasome (7, 42, 87). We observed that mutation of Vpu Trp22 to Leu reduced the amount of the substrate-trapping VCP-QQ mutant (42, 85) that coprecipitated with CD4 (Fig. 3F and G), in line with the reduction of CD4 polyubiquitination caused by this mutation (Fig. 3C and D). Decreased recruitment of the VCP-UFD1L-NPL4 complex to CD4 in the presence of the Vpu-W22L mutant would be expected to result in accumulation of CD4 in the ER membrane. To test this prediction, we developed a permeabilized cell system in which HeLa cells expressing FLAG-tagged Ub and human CD4, with or without Vpu, Vpu-S52,56N, or Vpu-A14→V25L were treated with a low concentration of digitonin and soluble, cytosolic proteins were extruded from the cells by centrifugation. The resulting pellet and supernatant fractions were fully denatured prior to immunoprecipitation with a conformation-independent antibody to CD4 and immunoblotting with an antibody to the FLAG epitope (42) (Fig. 3H). This procedure was efficient at separating membrane and cytosolic proteins as demonstrated by detection of transmembrane Vpu and cytosolic GAPDH in the corresponding fractions (Fig. 3H). As expected, CD4 was completely associated with membranes in the absence of Vpu (Fig. 3H). The small amount of polyubiquitinated and nonubiquitinated CD4 that remained in the presence of wild-type Vpu was also found to be membrane associated, as was the case for nonubiquitinated CD4 that accumulated in the presence of the Vpu-S52,56N mutant (Fig. 3H). Importantly, we observed that both polyubiquitinated and nonubiquitinated CD4 remained membrane associated upon expression of the Vpu-A14→V25L construct having mutations in the Trp22-centered patch of residues (Fig. 3H). Moreover, neither CD4 species could be removed from isolated microsomal membranes by treatment with 100 mM Na2CO3, pH 11.3 (23, 42), indicating that they behaved as integral membrane proteins (Fig. 3I). Therefore, both polyubiquitinated and nonubiquitinated CD4 that cannot be targeted to degradation by the Vpu TMD mutant (Fig. 3C and H) accumulate as integral membrane proteins rather than being released into the cytosol. These findings suggest that Vpu TMD residues contribute to dislocation of CD4 from the ER membrane.
Gly415 within the CD4 TMD is required for both Vpu-induced CD4 degradation and Vpu-CD4 interaction.
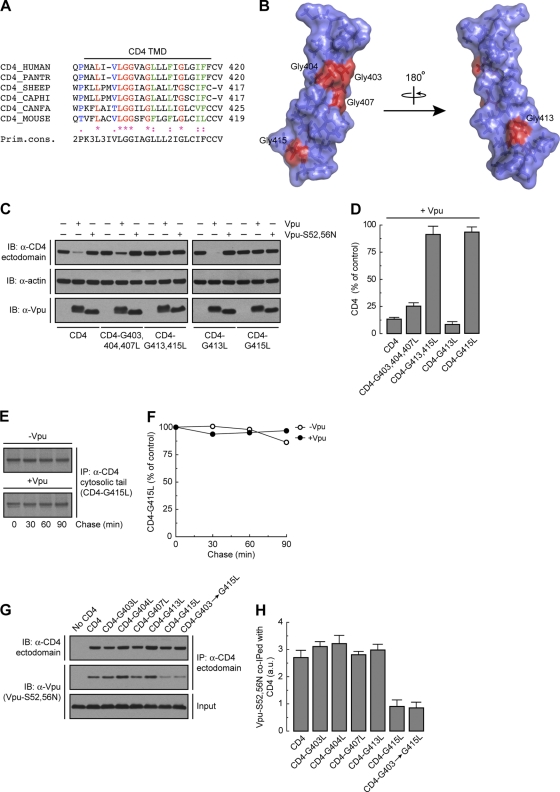
Since the TMD of CD4 is also important for Vpu-induced CD4 downregulation (13, 57), we next examined the requirement for specific residues within this domain. A notable feature of the CD4 TMD is the occurrence of several Gly residues at regular intervals within its sequence (Fig. 4A). Of these, Gly403, Gly404, Gly407, and Gly413 are highly conserved in all mammals (Fig. 4A), whereas Gly415 is present only in humans and monkeys (Fig. 4A), the natural hosts for HIV and SIV, respectively. All of these Gly residues except Gly413 are located on the same face of the CD4 TMD α-helix (81) (Fig. 4B). This arrangement of Gly residues has been previously shown to promote interhelical interactions that are stabilized by van der Waals forces (18, 31, 41) and Cα—H · · · O hydrogen bonds (65). To analyze the requirement of these Gly residues, we mutated each of them to Leu and examined the susceptibility of the mutant proteins to Vpu-induced degradation (Fig. 4C). We observed that mutation of Gly415 completely abrogated the decrease in CD4 levels elicited by Vpu, whereas mutation of the other Gly residues, either singly or in combinations, had little or no effect (Fig. 4C and D). Pulse-chase analysis confirmed the stability of newly synthesized CD4-G415L in the presence of Vpu (Fig. 4E and F). Mutation of each of the other non-Gly residues also had no effect on the susceptibility of CD4 to Vpu-mediated degradation (unpublished observations). From these experiments, we concluded that CD4 Gly415 is a critical determinant for Vpu-induced CD4 targeting to degradation.
Fig 4.
CD4 Gly415 is a key determinant for both Vpu-CD4 interaction and Vpu-induced CD4 degradation. (A) ClustalW alignment of CD4 TMD sequences, highlighting conservation of the residues. Notice that human CD4 Gly403, Gly404, Gly407, and Gly413 are highly conserved among all mammals, whereas Gly415 is present only in humans (CD4_HUMAN) and monkeys (CD4_PANTR; chimpanzee). (B) PyMOL images of the solid-state NMR structure of the human CD4 TMD (81) (RSCB protein database entry 2KLU). Gly residues within the CD4 TMD are shown in red. (C) HeLa cells were transfected with plasmids encoding human CD4 or the CD4 TMD mutant CD4-G403,404,407L, -G413,415L, -G413L, or -G415L, with or without wild-type Vpu or Vpu-S52,56N. At 12 h after transfection, cell lysates were processed as described in the legend to Fig. 1B. (D) CD4 levels were quantified as described in the legend to Fig. 1C. Values are the means ± SEM from three independent experiments. (E) HeLa cells expressing human CD4-G415L, with or without wild-type Vpu, were labeled with [35S]methionine-cysteine for 2 min and chased for the indicated times at 37°C. Radiolabeled CD4 was detected as described in the legend to Fig. 1E. (F) Percentage of CD4 at each chase time relative to CD4 at time zero (100% control). (G) HeLa cells were transfected with plasmids encoding no CD4 (empty vector), human CD4, or CD4 TMD mutants bearing mutations on Gly 403, 404, 407, 413, or 415 or all glycines together (CD4-G403→G415L), plus the Vpu-S52,56N mutant. Cell lysates were processed as described in the legend to Fig. 2A. (H) The amount of coprecipitated Vpu-S52,56N with CD4 was quantified as described in the legend to Fig. 2B. Values are the means ± SEM from three independent experiments. Notice that Vpu-S52,56N is poorly coprecipitated with CD4 in the absence of CD4 Gly415 compared to wild-type CD4 or the other CD4 TMD mutants.
To determine whether CD4 Gly415 and other TMD Gly residues are required for interaction with Vpu, we transfected HeLa cells with plasmids encoding wild-type CD4 or CD4 TMD Gly mutants plus Vpu-S52,56N, which binds to CD4 but does not induce its degradation (11). Interactions were analyzed by coprecipitation under nondenaturing conditions as described above (Fig. 2A). We observed that wild-type CD4 as well as CD4 TMD constructs bearing mutations of Gly403, Gly404, Gly407, and Gly413 equally coprecipitated with Vpu-S52,56N (Fig. 4G and H). However, coprecipitation of Vpu-S52,56N with CD4 was greatly impaired when CD4 Gly415 was replaced by Leu (Fig. 4G and H). The interaction between Vpu and the CD4 Gly415 mutants was not completely abrogated, presumably due to the contribution of the cytoplasmic domains of the proteins to their mutual binding (11, 47). These experiments indicated that the requirement of CD4 Gly415 for Vpu-CD4 degradation is at least in part due to its involvement in Vpu-CD4 TMD interaction.
Vpu Trp22 and CD4 Gly415 are not involved in Vpu-mediated inhibition of CD4 export to the Golgi complex.
We recently uncovered a new function of Vpu in preventing transport of CD4 from the ER to the Golgi complex, which is distinct from its ability to target CD4 to the ERAD pathway (42). Because this new function is also dependent on the Vpu TMD (42), we tested the effect of mutating several Vpu TMD residues on its ability to cause CD4 retention in the ER. HeLa cells were transfected with plasmids encoding CD4, with or without various Vpu TMD mutants, and the intracellular transport of CD4 was analyzed by endoglycosidase H (endo H) treatment (Fig. 5A) and cell surface biotinylation (Fig. 5C). Newly synthesized CD4 contains two N-linked oligosaccharides within its ectodomain, one of which acquires endo H resistance when CD4 is transported through the Golgi complex (72) (Fig. 5A and B). In cells expressing wild-type Vpu, the small amount of remaining CD4 was ∼75% sensitive to endo H, consistent with its predominant localization to the ER (42) (Fig. 5A and B). In cells expressing the Vpu-W22L mutant, CD4 was not degraded, but it remained mostly sensitive to endo H (Fig. 5A and B), indicating that Vpu Trp22 is required for CD4 targeting to ERAD but not for CD4 retention in the ER.
Fig 5.
Vpu Trp22 and CD4 Gly415 are not required for Vpu-mediated ER retention of CD4. (A) HeLa cells were transfected with plasmids encoding human CD4 and no Vpu (empty vector), wild-type Vpu, or Vpu-W22L. Total cell lysates were prepared and then digested with endo H or PNGase F or left untreated before immunoblotting with an antibody to the CD4 ectodomain. CHO, N-linked carbohydrate chain. (B) Endo H-resistant and -sensitive CD4 were quantified by densitometry and expressed as percentage of the total amount of CD4 in each sample (100%). Values are the means ± SEM from three independent experiments such as that for panel A. (C) The surface of HeLa cells expressing human CD4, with or without wild-type Vpu or the Vpu-A14L, -A18L, -V21L, -W22L, -V25L, or -A14→V25L mutant was biotinylated by incubation with ice-cold PBS supplemented with sulfo-NHS-LC biotin before lysis under nondenaturing conditions. Biotinylated proteins were pulled-down using a NeutrAvidin-coupled resin, and surface CD4 was detected by SDS-PAGE and immunoblotting with an antibody to the CD4 ectodomain. Cell surface TfR was used as both a biotinylation and loading control. (D) Bar graphs showing percentages of surface CD4 levels (normalized to total CD4) in cells expressing Vpu relative to normalized surface CD4 levels in the absence of Vpu (100% control). Values are the means ± SEM from three independent experiments. Notice that CD4 is not expressed at the cell surface even in the absence of those Vpu TMD residues that are required for ERAD targeting of CD4. (E) HeLa cells were transfected with plasmids encoding human CD4 or CD4-G415L, with or without wild-type Vpu. Detergent extracts were prepared and processed as for panel A. (F) Endo H-resistant and -sensitive CD4 were quantified as for panel B. Values are the means ± SEM from three independent experiments such as that for panel E. (G) The surface of HeLa cells expressing no CD4 (empty vector), human CD4, or the CD4-G403L, -G404L, -G407L, -F411L, -G415L, -F418L, or -G403,G404,G407,F411,G415,F418L (CD4-G403→F418L) mutant, with or without wild-type Vpu, was biotinylated as described for panel C before lysis under nondenaturing conditions. Surface biotinylated CD4 was detected as for panel C. (H) Quantification of surface CD4 levels was performed as for panel D. Values are the means ± SEM from three independent experiments. Notice that CD4 Gly415 is not required for Vpu-mediated ER retention of CD4, even though this residue is a critical determinant for CD4 ERAD targeting by Vpu (Fig. 4).
The effects of Vpu on CD4 at the ER were reflected in changes of CD4 levels at the cell surface as observed in surface biotinylation experiments (Fig. 5C). HeLa cells were transfected with plasmids encoding human CD4, with or without Vpu or the Vpu-A14L, -A18L, -V21L, -W22L, -V25L, and -A14→V25L mutants. Cell surface proteins were biotinylated at 4°C, followed by cell lysis and pulldown with a NeutrAvidin-coupled resin. CD4, Vpu, and TfR were then visualized by immunoblotting using specific antibodies (Fig. 5C). Since TfR stability and maturation are not affected by Vpu (42), this protein was used as a loading control (Fig. 5C). For quantification purposes, the amounts of CD4 present at the cell surface were normalized to the amounts of CD4 in each sample (100% for the control without Vpu) (Fig. 5D). No CD4 could be detected at the cell surface in the presence of Vpu (Fig. 5C and D), as expected from the ER retention and ERAD targeting of CD4 under these conditions. Despite the inhibition of ERAD targeting caused by mutation of Vpu Trp22, however, only a small fraction (up to ∼25% of the control) of CD4 reached the cell surface in the presence of the Vpu-W22L or Vpu-A14→V25L mutant (Fig. 5C and D). This result is consistent with Vpu Trp22 not being involved in CD4 retention in the ER.
We next investigated the contribution of CD4 Gly415 to Vpu-mediated ER retention of CD4 by performing endo H sensitivity (Fig. 5E) and cell surface biotinylation (Fig. 5G) assays, as described above. In the absence of Vpu, both CD4 and CD4-G415L efficiently exited the ER en route to the plasma membrane (Fig. 5E to H). In contrast, in the presence of Vpu, most of the stable CD4-G415L mutant remained in the ER (Fig. 5E and F) and did not reach the plasma membrane (∼25% of the control) (Fig. 5G and H). The same result was obtained when not only Gly415 but also all non-Leu residues on the same face of the CD4 TMD α-helix were replaced by Leu (CD4-G403→F418L mutant) (Fig. 5G and H). Altogether, these results demonstrated that Vpu conserved the ability to retain CD4 in the ER when a key CD4 TMD residue required for its degradation, Gly415, was mutated.
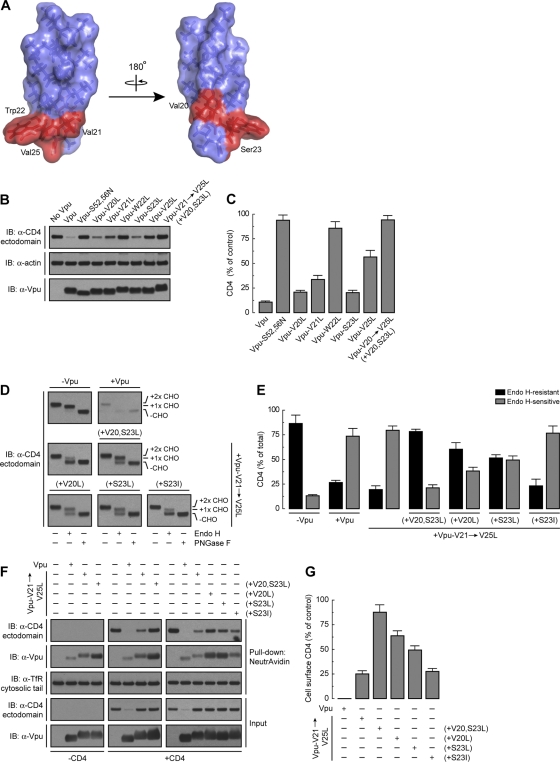
Val20 and Ser23 within the Vpu TMD are critical for Vpu-induced CD4 retention in the ER.
Since the Vpu TMD contributes to CD4 retention in the ER (42) and Vpu Trp22 (and to a lesser extent Val21 and Val25) is required for CD4 ERAD targeting but not ER retention (this study), we hypothesized that other residues in the Vpu TMD must be involved in ER retention of CD4. We focused our analysis on Vpu Val20 and Ser23, two residues that are contiguous on a face opposite to that displaying Val21, Trp22, and Val25 (Fig. 6A). Vpu Val20 is conserved in most HIV-1 group M variants (Fig. 1A), whereas Ser23 is highly conserved only in HIV-1 group M subtype B Vpu sequences (Fig. 1A). Mutational analysis showed that replacement of Val20 or Ser23 by Leu had little or no effect on the ability of Vpu to cause CD4 degradation (Fig. 6B and C). Substitution of Leu for Val21, Trp22, and Val25 (Vpu-V21→V25L mutant) in addition to Val20 and Ser23 [Vpu-V21→V25L (+V20,S23L) mutant] had the expected effect of preventing CD4 degradation (Fig. 6B and C). We next examined the effect of replacing Val20 and Ser23 on the ability of Vpu to induce CD4 retention in the ER, as determined by endo H digestion analysis (Fig. 6D). Interestingly, CD4 became mostly endo H resistant (∼80%) in the presence of Vpu-V21→V25L (+V20,S23L) (Fig. 6D and E). This was in contrast to the endo H sensitivity of CD4 in the presence of the Vpu-V21→V25L mutant (Fig. 6D and E). Individual mutation of Val20 or Ser23 to Leu [Vpu-V21→V25L (+V20L) and Vpu-V21→V25L (+S23L) mutants, respectively] had intermediate effects (∼50 to 60% of CD4 remained resistant to endo H) (Fig. 6D and E). Interestingly, most CD4 remained endo H sensitive when Vpu Ser23 was replaced by Ile [Vpu-V21→V25L (+S23I) mutant] instead of Leu (Fig. 6D and E). The latter finding is consistent with a significant percentage of HIV-1 group M Vpu sequences carrying Ile at position 23. These differential effects of the Vpu TMD mutants on the ER retention of CD4 were reflected in the expression of CD4 at the plasma membrane, as measured by the surface biotinylation assay (Fig. 6F and G). We observed undetectable levels of surface CD4 in cells expressing wild-type Vpu, low levels in cells expressing Vpu-V21→V25L or Vpu-V21→V25L (+S23I) (∼25% of control), intermediate levels in cells expressing Vpu-V21→V25L (+V20L) or Vpu-V21→V25L (+S23L) (∼50 to 60% of control), and high levels in cells expressing Vpu-V21→V25L (+V20,S23L) (∼90% of control) (Fig. 6F and G), even though none of these Vpu TMD mutants induced CD4 degradation (Fig. 6F). Remarkably, replacement of Leu for Vpu Val20 and Ser23 together or in isolation also enhanced Vpu trafficking from the ER to the cell surface in a CD4-independent fashion (Fig. 6F). From these experiments we concluded that Vpu Val20 and Ser23 are both components of a TMD determinant for retention of not only CD4 but also Vpu in the ER, which is distinct from the ERAD-targeting TMD determinant conformed by Val21, Trp22, and Val25.
Fig 6.
ER retention of CD4 by Vpu requires Val20 and Ser23 within the Vpu TMD. (A) PyMOL images of the solid-state NMR structure of the NL4-3 Vpu TMD. Notice that Val20 and Ser23 are positioned opposite to the patch made up of Val21, Trp22, and Val25 on the TMD α-helix. (B) HeLa cells were transfected with plasmids encoding human CD4, with or without wild-type Vpu, Vpu-S52,56N, or the Vpu-V20L, -V21L, -W22L, -S23L, -V25L, or -V20,V21,W22,S23,V25L [Vpu-V21→V25L (+V20,S23L)] mutant. At 12 h after transfection, cell lysates were prepared and analyzed as described in the legend to Fig. 1B. (C) CD4 levels in the presence of Vpu were quantified as described in the legend to Fig. 1C. Values are the means ± SEM from three independent experiments. (D) Lysates from HeLa cells expressing human CD4 and no Vpu, wild-type Vpu, Vpu-V21,W22,V25L (Vpu-V21→V25L mutant), Vpu-V21→V25L (+V20,S23L), Vpu-V20,V21,W22,V25L [Vpu-V21→V25L (+V20L) mutant], Vpu-V21,W22,S23,V25L [Vpu-V21→V25L (+S23L) mutant], or Vpu-V21L,W22L,S23I,V25L [Vpu-V21→V25L (+S23I) mutant] were prepared and processed as described in the legend to Fig. 5A. (E) Endo H-resistant and -sensitive CD4 were quantified as described in the legend to Fig. 5B. Values are the means ± SEM from three independent experiments such as that for panel D. (F) Surface biotinylated CD4 from HeLa cells transfected as for panel D was detected as described in the legend to Fig. 5C. The same experiment was also done with cells expressing no Vpu, wild-type Vpu, Vpu-V21→V25L, or Vpu-V21→V25L (+V20,S23L), without CD4 for comparison. Surface biotinylated Vpu was also detected by immunoblotting using an antibody to Vpu. (G) Quantification of surface CD4 levels was performed as described in the legend to Fig. 5D. Values are the means ± SEM from three independent experiments. Notice that the Vpu-V21→V25L and Vpu-V21→V25L (+V20,S23L) mutants exhibit different abilities to retain CD4 in the ER even though neither mutant is able to induce CD4 degradation.
DISCUSSION
It is now well established that TMDs do not merely act as membrane anchors or connectors between the luminal/extracellular domains (i.e., ectodomains) and cytosolic domains (i.e., endodomains) of integral membrane proteins. Rather, they play active roles in processes such as transport of molecules across membranes (58), intramembrane proteolysis (82), assembly of multiprotein complexes (46), protein localization to intracellular organelles (67), and targeting to the ERAD pathway (8, 10, 76). For this reason, many TMDs are not just random sequences of hydrophobic amino acid residues but possess specific characteristics, including conserved residues at defined positions within the transmembrane α-helices. The results presented in this study show that Vpu exploits these properties of TMDs in order to downregulate CD4. Indeed, we demonstrate that the TMDs of both Vpu and CD4 contain specific residues that are required for Vpu-induced CD4 targeting to ERAD and retention in the ER, as well as for assembly of Vpu-CD4 and Vpu-Vpu complexes.
A cluster of residues centered on Trp22 in the Vpu TMD is required for CD4 targeting to the ERAD pathway.
Although initially overlooked, the role of TMDs in Vpu-induced CD4 degradation has now been demonstrated by several studies (13, 42, 57, 73). The specific features of the Vpu and CD4 TMDs that are required for this process, however, have remained poorly understood. Here we show that a cluster of residues centered on Trp22 (i.e., Val21, Trp22, and Val25) on the Vpu TMD is required for CD4 targeting to ERAD. Vpu Trp22 is the most critical residue on this cluster, since it cannot be replaced by any residue other than Cys without complete loss of activity (Fig. 1G and H). The fact that Cys can substitute for Trp at position 22 is puzzling, since these two amino acids have very different chemical properties. Trp22 is at a position near the membrane-cytosol interface, the most frequent location for Trp within TMDs (80, 84). The affinity of TMD Trp residues for membrane-cytosol interfaces stems from the dual bulky-hydrophobic/hydrogen-bonding nature of the indole ring (43, 52, 66). Vpu Trp22 could participate in CD4 targeting to the ERAD pathway by mediating protein-protein interactions or by fixing the position of the Vpu TMD relative to the lipid bilayer. These properties could be conferred on CD4 through assembly with Vpu.
An earlier study showed that scrambling of some residues in the Vpu TMD did not affect the ability of Vpu to downregulate CD4 (VpuRD mutant) (64), an observation that seems at odds with the conclusion from this and previous studies (42, 73). However, the scrambled VpuRD mutant has Val22, Trp24, and Val27 residues at positions that are roughly equivalent to those of Val21, Trp22, and Val25 in the wild-type Vpu TMD. The location of Trp24 near the membrane-cytosol interface in VpuRD could explain why this mutant, like wild-type Vpu, remains active for CD4 downregulation.
Effects of the Vpu Trp22 mutation on CD4 polyubiquitination and recruitment of the VCP-UFD1L-NPL4 dislocase complex.
How might Trp22 participate in the mechanism by which Vpu induces targeting of CD4 to ERAD? We initially hypothesized that Vpu Trp22 could be required for Vpu-CD4 interaction at the level of the TMDs. However, we found that this was not the case, as mutation of Trp22 to Leu did not abrogate, but instead enhanced, coprecipitation of Vpu with CD4 (Fig. 2A and B). This enhancement was not likely due to increased affinity of the Vpu-CD4 TMD interaction but rather to increased oligomerization of Vpu and coprecipitation of Vpu oligomers with CD4 (Fig. 2). Indeed, Vpu had previously been shown to occur in equilibrium between monomeric and oligomeric forms (29, 40, 44, 56). We found that mutation of Vpu Trp22 to Leu, Ala, or Gly shifts the equilibrium toward oligomeric forms (Fig. 2C; see Fig. S2A and B in the supplemental material), implying that the Vpu TMD Trp residue hinders Vpu-Vpu interactions. In agreement with these findings, the Vpu TMD containing a Trp-to-Leu substitution remains open as an ion channel for much longer than the wild type, behaving more like a pore (50). A corollary of these findings is that monomeric, and not oligomeric, Vpu is the active form for CD4 targeting to the ERAD pathway. Therefore, at a minimum, Vpu Trp22 functions to maintain Vpu in its active, monomeric form.
To determine whether Vpu Trp22 has additional functions, we examined the possible involvement of this residue in ERAD-targeting steps downstream of the Vpu-CD4 interaction. We observed that mutation of Vpu Trp22 did not affect the recruitment of the cytosolic SCFβ-TrCP1/2 E3 Ub-ligase complex to the Vpu cytosolic domain (Fig. 3A and B). However, we found that Vpu-induced polyubiquitination of CD4 was reduced from 32-fold to 5- to 20-fold by single or combined mutation of several Vpu TMD residues, including Trp22 (Fig. 3C and D). A possible explanation of these findings is that Vpu TMD mutations, though not decreasing CD4-Vpu interactions, alter the orientation of the CD4 cytosolic tail relative to the SCFβ-TrCP1/2 complex such that CD4 polyubiquitination is impaired. Vpu TMD mutations could also prevent the function of another transmembrane protein that contributes to CD4 polyubiquitination such as, for example, an E2 Ub-conjugating enzyme (86). Finally, decreased CD4 polyubiquitination could be due to partial deubiquitination of the accumulated CD4 that cannot be targeted to degradation by the Vpu TMD mutants (74).
Binding of VCP (and, by extension, of the VCP-UFD1L-NPL4 complex) to CD4 was also impaired by mutation of Vpu Trp22 (Fig. 3F and G), likely as a consequence of reduced CD4 polyubiquitination. Since the VCP-UFD1L-NPL4 complex functions to extract CD4 from the ER membrane in the presence of Vpu (7, 42), we expected that CD4 would remain associated with membranes upon expression of Vpu TMD mutants. Indeed, subcellular fractionation experiments showed that both nonubiquitinated and polyubiquitinated CD4 species accumulated as integral membrane proteins in the presence of Vpu having mutations of several TMD residues, including Trp22 (Vpu-A14→V25L mutant) (Fig. 3H and I). This result should be interpreted with caution, however, as CD4 that was protected from Vpu-induced degradation by incubation with the proteasomal inhibitor MG-132 also remained integrally associated with membranes (unpublished observations). The latter observation indicates that proteasomal activity is a prerequisite for membrane dislocation of CD4 by Vpu, as previously shown for some ERAD substrates (35, 49, 83) but not others (27, 28). Nevertheless, our observations are most consistent with a role for the Vpu TMD, and in particular Trp22, in allowing extraction of CD4 from the ER membrane.
Gly415 in the CD4 TMD is critical for assembly with Vpu and for Vpu-induced CD4 degradation.
The TMD of CD4 is also required for Vpu-mediated CD4 degradation (13, 57). In this study, we found that mutation of a single CD4 TMD residue, Gly415, completely abrogated CD4 degradation induced by Vpu (Fig. 4C and D). Unlike mutation of Vpu Trp22, however, mutation of CD4 Gly415 impaired the Vpu-CD4 interaction (Fig. 4G and H), thus establishing a clear basis for the inhibition of Vpu-induced CD4 degradation caused by this mutation. This finding represents the first demonstration that mutation of a TMD residue decreases Vpu-CD4 interactions. Thus, both the TMD (this study) and the cytosolic domains (11, 47) contribute to the physical interaction of Vpu with CD4. Based on the solid-state NMR structure of the human CD4 TMD α-helix (81) (Fig. 4B), Gly415 is predicted to be part of a “Gly strip,” a structural motif that mediates TMD interactions (18, 31) (Fig. 4A and B). With the exception of CD4 Gly415, single substitution of each Gly residue on the strip (Fig. 4) or of other, non-Gly residues in the CD4 TMD (unpublished observations), however, had no effect on assembly with Vpu (Fig. 4G and H) or Vpu-induced CD4 degradation (Fig. 4C and D). These results do not rule out that other CD4 TMD residues are involved, as inhibition of those functions could require combined mutation of two or more residues.
An ER retention determinant comprising Val20 and Ser23 in the Vpu TMD.
In addition to promoting targeting of CD4 to the ERAD pathway, the Vpu TMD contributes to retention of CD4 in the ER (42). Mutation of Vpu TMD Trp22 prevents Vpu-induced CD4 degradation but not CD4 ER retention (Fig. 5), an observation that is in agreement with the ability of this mutant to decrease expression of CD4 at the cell surface as reported by one group (69) but not another (77). Additional mutagenesis revealed two other Vpu TMD residues, Val20 and Ser23, which are critical for CD4 ER retention (Fig. 6). Val20 and Ser23 are located on a face of the TMD α-helix opposite to that of the cluster comprising Val21, Trp22, and Val25 (Fig. 6A). Moreover, mutation of Val20 and Ser23 alone did not inhibit Vpu-induced CD4 degradation (Fig. 6B and C). These observations indicate that the determinants for ERAD targeting and ER retention of CD4 in the Vpu TMD are spatially and mechanistically separable. How does Vpu retain CD4 in the ER? Vpu itself lacks a known ER retention motif, but replacement of Val20 and Ser23 on its TMD increased Vpu transport out of the ER en route to the plasma membrane independently of CD4 (Fig. 6F). This indicates that Vpu might be at least partially retained in the ER by establishment of Val20- and Ser23-mediated TMD interactions with a putative ER resident protein. In this regard, calnexin has been shown to modulate ER localization and stability of a number of ERAD substrates by recognition of specific features within their TMDs (37, 51, 70). It is also possible that Val20 and Ser23 confer ER retention through interactions with the ER lipid bilayer. In any case, CD4 appears to be retained in the ER in trans, through assembly with Vpu. It is worth noting that Vpu has been localized not just to the ER (42) but also to the trans-Golgi network and endosomal compartments (21, 75, 77). In fact, the post-ER population of Vpu is likely the form that functions in BST-2 antagonism (2, 69).
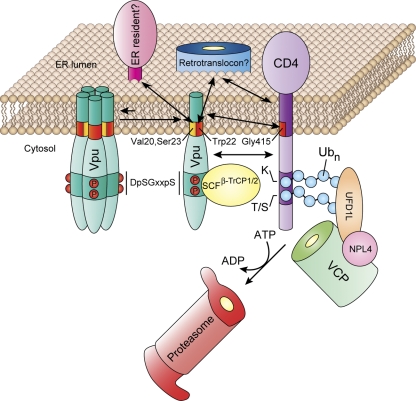
A working model for the role of TMDs in Vpu-induced CD4 downregulation.
Our findings suggest the working model shown in Fig. 7. A Vpu TMD determinant comprising Val20 and Ser23 retains a substantial fraction of the protein in the ER through interaction with an ER resident protein or the ER lipid bilayer. This population of Vpu exists in equilibrium between monomeric and oligomeric forms, which are active and inactive, respectively, for CD4 targeting to the ERAD pathway. Vpu Trp22 helps maintains Vpu in its monomeric, active form. CD4 binds to Vpu via the cytoplasmic domains as well as the TMDs of both proteins. TMD binding involves an interaction between the CD4 TMD Gly415 residue and an unidentified site within the Vpu TMD. Despite the ability of the Vpu TMDs to oligomerize, we do not think that Vpu itself functions as a retrotranslocon because (i) monomeric Vpu seems to be the active form, (ii) the Vpu TMD does not have enough hydrophilic or charged residues that could line a protein translocation channel, and (iii) the dimensions of the Vpu TMD oligomers and the putative channel are too small to support protein translocation. Rather, Vpu is likely to deliver CD4 to a host cell retrotranslocon, in a process that might be dependent on Vpu Trp22. This residue also promotes CD4 polyubiquitination, possibly by controlling the spatial positioning of the SCFβ-TrCP1/2 E3 Ub-ligase complex relative to the CD4 cytosolic tail. Finally, this residue could be required to promote CD4 dislocation itself by forcing the CD4 TMD into a position from which it could be more easily extracted from the membrane. Concomitant with these TMD-mediated processes, the cytosolic domain of Vpu enables SCFβ-TrCP1/2-dependent polyubiquitination of the CD4 cytosolic tail, followed by VCP-UFD1L-NPL4-mediated dislocation and delivery to the proteasome.
Fig 7.
A working model for the role of TMDs in the mechanism of CD4 downregulation by Vpu. See Discussion for details.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NIH Intramural AIDS Targeted Antiviral Program (IATAP) and the Intramural Program of the NICHD. J.G.M. is the recipient of a fellowship from The Pew Charitable Trusts.
We thank X. Zhu and N. Tsai for expert technical assistance and Y. Ben-Neriah, K. Strebel, and Y. Ye for generous gifts of reagents.
Footnotes
Published ahead of print 16 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76:853–864 [DOI] [PubMed] [Google Scholar]
- 2. Andrew AJ, Miyagi E, Strebel K. 2011. Differential effects of human immunodeficiency virus type 1 Vpu on the stability of BST-2/tetherin. J. Virol. 85:2611–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arganaraz ER, Schindler M, Kirchhoff F, Cortes MJ, Lama J. 2003. Enhanced CD4 down-modulation by late stage HIV-1 nef alleles is associated with increased Env incorporation and viral replication. J. Biol. Chem. 278:33912–33919 [DOI] [PubMed] [Google Scholar]
- 4. Arien KK, et al. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 79:8979–8990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belaidouni N, Marchal C, Benarous R, Besnard-Guerin C. 2007. Involvement of the betaTrCP in the ubiquitination and stability of the HIV-1 Vpu protein. Biochem. Biophys. Res. Commun. 357:688–693 [DOI] [PubMed] [Google Scholar]
- 6. Benson RE, Sanfridson A, Ottinger JS, Doyle C, Cullen BR. 1993. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J. Exp. Med. 177:1561–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Binette J, et al. 2007. Requirements for the selective degradation of CD4 receptor molecules by the human immunodeficiency virus type 1 Vpu protein in the endoplasmic reticulum. Retrovirology 4:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonifacino JS, Cosson P, Klausner RD. 1990. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell 63:503–513 [DOI] [PubMed] [Google Scholar]
- 9. Bonifacino JS, Dell'Angelica EC. 2001. Immunoprecipitation. Curr. Protoc. Cell Biol. 7:Unit 7.2 [DOI] [PubMed] [Google Scholar]
- 10. Bonifacino JS, Suzuki CK, Klausner RD. 1990. A peptide sequence confers retention and rapid degradation in the endoplasmic reticulum. Science 247:79–82 [DOI] [PubMed] [Google Scholar]
- 11. Bour S, Schubert U, Strebel K. 1995. The human immunodeficiency virus type 1 Vpu protein specifically binds to the cytoplasmic domain of CD4: implications for the mechanism of degradation. J. Virol. 69:1510–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bresnahan PA, Yonemoto W, Greene WC. 1999. SIV Nef protein utilizes both leucine- and tyrosine-based protein sorting pathways for down-regulation of CD4. J. Immunol. 163:2977–2981 [PubMed] [Google Scholar]
- 13. Buonocore L, Turi TG, Crise B, Rose JK. 1994. Stimulation of heterologous protein degradation by the Vpu protein of HIV-1 requires the transmembrane and cytoplasmic domains of CD4. Virology 204:482–486 [DOI] [PubMed] [Google Scholar]
- 14. Burtey A, et al. 2007. Dynamic interaction of HIV-1 Nef with the clathrin-mediated endocytic pathway at the plasma membrane. Traffic 8:61–76 [DOI] [PubMed] [Google Scholar]
- 15. Butticaz C, Michielin O, Wyniger J, Telenti A, Rothenberger S. 2007. Silencing of both beta-TrCP1 and HOS (beta-TrCP2) is required to suppress human immunodeficiency virus type 1 Vpu-mediated CD4 down-modulation. J. Virol. 81:1502–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. 2007. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J. Virol. 81:3877–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaudhuri R, Mattera R, Lindwasser OW, Robinson MS, Bonifacino JS. 2009. A basic patch on alpha-adaptin is required for binding of human immunodeficiency virus type 1 Nef and cooperative assembly of a CD4-Nef-AP-2 complex. J. Virol. 83:2518–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cosson P, Bonifacino JS. 1992. Role of transmembrane domain interactions in the assembly of class II MHC molecules. Science 258:659–662 [DOI] [PubMed] [Google Scholar]
- 19. Craig HM, Reddy TR, Riggs NL, Dao PP, Guatelli JC. 2000. Interactions of HIV-1 nef with the mu subunits of adaptor protein complexes 1, 2, and 3: role of the dileucine-based sorting motif. Virology 271:9–17 [DOI] [PubMed] [Google Scholar]
- 20. daSilva LL, et al. 2009. Human immunodeficiency virus type 1 Nef protein targets CD4 to the multivesicular body pathway. J. Virol. 83:6578–6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dube M, et al. 2009. Suppression of tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J. Virol. 83:4574–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ewart GD, Sutherland T, Gage PW, Cox GB. 1996. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J. Virol. 70:7108–7115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Greenberg ME, et al. 1997. Co-localization of HIV-1 Nef with the AP-2 adaptor protein complex correlates with Nef-induced CD4 down-regulation. EMBO J. 16:6964–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guatelli JC. 2009. Interactions of viral protein U (Vpu) with cellular factors. Curr. Top. Microbiol. Immunol. 339:27–45 [DOI] [PubMed] [Google Scholar]
- 26. Hoxie JA, et al. 1986. Alterations in T4 (CD4) protein and mRNA synthesis in cells infected with HIV. Science 234:1123–1127 [DOI] [PubMed] [Google Scholar]
- 27. Hughes EA, Hammond C, Cresswell P. 1997. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc. Natl. Acad. Sci. U. S. A. 94:1896–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huppa JB, Ploegh HL. 1997. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity 7:113–122 [DOI] [PubMed] [Google Scholar]
- 29. Hussain A, Das SR, Tanwar C, Jameel S. 2007. Oligomerization of the human immunodeficiency virus type 1 (HIV-1) Vpu protein—a genetic, biochemical and biophysical analysis. Virol. J. 4:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin YJ, et al. 2005. HIV Nef-mediated CD4 down-regulation is adaptor protein complex 2 dependent. J. Immunol. 175:3157–3164 [DOI] [PubMed] [Google Scholar]
- 31. Kim S, et al. 2005. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 102:14278–14283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kirchhoff F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8:55–67 [DOI] [PubMed] [Google Scholar]
- 33. Korber B, et al. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789–1796 [DOI] [PubMed] [Google Scholar]
- 34. Lanzavecchia A, Roosnek E, Gregory T, Berman P, Abrignani S. 1988. T cells can present antigens such as HIV gp120 targeted to their own surface molecules. Nature 334:530–532 [DOI] [PubMed] [Google Scholar]
- 35. Lee RJ, et al. 2004. Uncoupling retro-translocation and degradation in the ER-associated degradation of a soluble protein. EMBO J. 23:2206–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lenburg ME, Landau NR. 1993. Vpu-induced degradation of CD4: requirement for specific amino acid residues in the cytoplasmic domain of CD4. J. Virol. 67:7238–7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Q, et al. 2010. Transmembrane segments prevent surface expression of sodium channel Nav1.8 and promote calnexin-dependent channel degradation. J. Biol. Chem. 285:32977–32987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lindwasser OW, Chaudhuri R, Bonifacino JS. 2007. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr. Mol. Med. 7:171–184 [DOI] [PubMed] [Google Scholar]
- 39. Lindwasser OW, et al. 2008. A diacidic motif in human immunodeficiency virus type 1 Nef is a novel determinant of binding to AP-2. J. Virol. 82:1166–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu JX, Sharpe S, Ghirlando R, Yau WM, Tycko R. 2010. Oligomerization state and supramolecular structure of the HIV-1 Vpu protein transmembrane segment in phospholipid bilayers. Protein Sci. 19:1877–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. MacKenzie KR, Prestegard JH, Engelman DM. 1997. A transmembrane helix dimer: structure and implications. Science 276:131–133 [DOI] [PubMed] [Google Scholar]
- 42. Magadan JG, et al. 2010. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 6:e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mahalakshmi R, Sengupta A, Raghothama S, Shamala N, Balaram P. 2005. Tryptophan-containing peptide helices: interactions involving the indole side chain. J. Pept. Res. 66:277–296 [DOI] [PubMed] [Google Scholar]
- 44. Maldarelli F, Chen MY, Willey RL, Strebel K. 1993. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type I integral membrane protein. J. Virol. 67:5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malim MH, Emerman M. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398 [DOI] [PubMed] [Google Scholar]
- 46. Manolios N, Bonifacino JS, Klausner RD. 1990. Transmembrane helical interactions and the assembly of the T cell receptor complex. Science 249:274–277 [DOI] [PubMed] [Google Scholar]
- 47. Margottin F, et al. 1996. Interaction between the cytoplasmic domains of HIV-1 Vpu and CD4: role of Vpu residues involved in CD4 interaction and in vitro CD4 degradation. Virology 223:381–386 [DOI] [PubMed] [Google Scholar]
- 48. Margottin F, et al. 1998. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1:565–574 [DOI] [PubMed] [Google Scholar]
- 49. Mayer TU, Braun T, Jentsch S. 1998. Role of the proteasome in membrane extraction of a short-lived ER-transmembrane protein. EMBO J. 17:3251–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mehnert T, et al. 2008. Biophysical characterization of Vpu from HIV-1 suggests a channel-pore dualism. Proteins 70:1488–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Molinari M, Galli C, Piccaluga V, Pieren M, Paganetti P. 2002. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J. Cell Biol. 158:247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Monera OD, Sereda TJ, Zhou NE, Kay CM, Hodges RS. 1995. Relationship of sidechain hydrophobicity and alpha-helical propensity on the stability of the single-stranded amphipathic alpha-helix. J. Pept. Sci. 1:319–329 [DOI] [PubMed] [Google Scholar]
- 53. Nethe M, Berkhout B, van der Kuyl AC. 2005. Retroviral superinfection resistance. Retrovirology 2:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nguyen KL, et al. 2004. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology 319:163–175 [DOI] [PubMed] [Google Scholar]
- 55. Park SH, De Angelis AA, Nevzorov AA, Wu CH, Opella SJ. 2006. Three-dimensional structure of the transmembrane domain of Vpu from HIV-1 in aligned phospholipid bicelles. Biophys. J. 91:3032–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Park SH, et al. 2003. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J. Mol. Biol. 333:409–424 [DOI] [PubMed] [Google Scholar]
- 57. Raja NU, Vincent MJ, abdul Jabbar M. 1994. Vpu-mediated proteolysis of gp160/CD4 chimeric envelope glycoproteins in the endoplasmic reticulum: requirement of both the anchor and cytoplasmic domains of CD4. Virology 204:357–366 [DOI] [PubMed] [Google Scholar]
- 58. Rapoport TA. 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450:663–669 [DOI] [PubMed] [Google Scholar]
- 59. Ray N, Doms RW. 2006. HIV-1 coreceptors and their inhibitors. Curr. Top. Microbiol. Immunol. 303:97–120 [DOI] [PubMed] [Google Scholar]
- 60. Rhee SS, Marsh JW. 1994. Human immunodeficiency virus type 1 Nef-induced down-modulation of CD4 is due to rapid internalization and degradation of surface CD4. J. Virol. 68:5156–5163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ruiz A, Guatelli JC, Stephens EB. 2010. The Vpu protein: new concepts in virus release and CD4 down-modulation. Curr. HIV Res. 8:240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Salmon P, et al. 1988. Loss of CD4 membrane expression and CD4 mRNA during acute human immunodeficiency virus replication. J. Exp. Med. 168:1953–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schubert U, et al. 1998. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 72:2280–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schubert U, et al. 1996. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 70:809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Senes A, Ubarretxena-Belandia I, Engelman DM. 2001. The Calpha—H … O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc. Natl. Acad. Sci. U. S. A. 98:9056–9061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sereda TJ, Mant CT, Sonnichsen FD, Hodges RS. 1994. Reversed-phase chromatography of synthetic amphipathic alpha-helical peptides as a model for ligand/receptor interactions. Effect of changing hydrophobic environment on the relative hydrophilicity/hydrophobicity of amino acid side-chains. J. Chromatogr. A 676:139–153 [DOI] [PubMed] [Google Scholar]
- 67. Sharpe HJ, Stevens TJ, Munro S. 2010. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142:158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sharpe S, Yau WM, Tycko R. 2006. Structure and dynamics of the HIV-1 Vpu transmembrane domain revealed by solid-state NMR with magic-angle spinning. Biochemistry 45:918–933 [DOI] [PubMed] [Google Scholar]
- 69. Skasko M, et al. 2011. BST-2 is rapidly down-regulated from the cell surface by the HIV-1 protein Vpu: evidence for a post-ER mechanism of Vpu-action. Virology 411:65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Swanton E, High S, Woodman P. 2003. Role of calnexin in the glycan-independent quality control of proteolipid protein. EMBO J. 22:2948–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tifft CJ, Proia RL, Camerini-Otero RD. 1992. The folding and cell surface expression of CD4 requires glycosylation. J. Biol. Chem. 267:3268–3273 [PubMed] [Google Scholar]
- 73. Tiganos E, et al. 1998. Structural and functional analysis of the membrane-spanning domain of the human immunodeficiency virus type 1 Vpu protein. Virology 251:96–107 [DOI] [PubMed] [Google Scholar]
- 74. Tsai YC, Weissman AM. 2011. Ubiquitylation in ERAD: reversing to go forward? PLoS Biol. 9:e1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Varthakavi V, et al. 2006. The pericentriolar recycling endosome plays a key role in Vpu-mediated enhancement of HIV-1 particle release. Traffic 7:298–307 [DOI] [PubMed] [Google Scholar]
- 76. Vembar SS, Brodsky JL. 2008. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9:944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vigan R, Neil SJ. 2010. Determinants of tetherin antagonism in the transmembrane domain of the human immunodeficiency virus type 1 Vpu protein. J. Virol. 84:12958–12970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vincent MJ, Raja NU, Jabbar MA. 1993. Human immunodeficiency virus type 1 Vpu protein induces degradation of chimeric envelope glycoproteins bearing the cytoplasmic and anchor domains of CD4: role of the cytoplasmic domain in Vpu-induced degradation in the endoplasmic reticulum. J. Virol. 67:5538–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Willey RL, Maldarelli F, Martin MA, Strebel K. 1992. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 66:7193–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wimley WC, White SH. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3:842–848 [DOI] [PubMed] [Google Scholar]
- 81. Wittlich M, Thiagarajan P, Koenig BW, Hartmann R, Willbold D. 2010. NMR structure of the transmembrane and cytoplasmic domains of human CD4 in micelles. Biochim. Biophys. Acta 1798:122–127 [DOI] [PubMed] [Google Scholar]
- 82. Wolfe MS, Kopan R. 2004. Intramembrane proteolysis: theme and variations. Science 305:1119–1123 [DOI] [PubMed] [Google Scholar]
- 83. Yang M, Omura S, Bonifacino JS, Weissman AM. 1998. Novel aspects of degradation of T cell receptor subunits from the endoplasmic reticulum (ER) in T cells: importance of oligosaccharide processing, ubiquitination, and proteasome-dependent removal from ER membranes. J. Exp. Med. 187:835–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yau WM, Wimley WC, Gawrisch K, White SH. 1998. The preference of tryptophan for membrane interfaces. Biochemistry 37:14713–14718 [DOI] [PubMed] [Google Scholar]
- 85. Ye Y, Meyer HH, Rapoport TA. 2003. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162:71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ye Y, Rape M. 2009. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10:755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429:841–847 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.