Abstract
Understanding the mechanisms of cross-species virus transmission is critical to anticipating emerging infectious diseases. Canine parvovirus type 2 (CPV-2) emerged as a variant of a feline parvovirus when it acquired mutations that allowed binding to the canine transferrin receptor type 1 (TfR). However, CPV-2 was soon replaced by a variant virus (CPV-2a) that differed in antigenicity and receptor binding. Here we show that the emergence of CPV involved an additional host range variant virus that has circulated undetected in raccoons for at least 24 years, with transfers to and from dogs. Raccoon virus capsids showed little binding to the canine TfR, showed little infection of canine cells, and had altered antigenic structures. Remarkably, in capsid protein (VP2) phylogenies, most raccoon viruses fell as evolutionary intermediates between the CPV-2 and CPV-2a strains, suggesting that passage through raccoons assisted in the evolution of CPV-2a. This highlights the potential role of alternative hosts in viral emergence.
INTRODUCTION
The emergence of novel infectious diseases is often associated with pathogens jumping to new host species. However, the genetic and evolutionary mechanisms that determine how viruses cross species boundaries and adapt to new hosts are only partially understood (25). Well-documented examples of virus emergence in which both the donor and recipient species are known are rare but can reveal key aspects of this important evolutionary process.
Canine parvovirus (CPV) first emerged in the mid-1970s and then spread worldwide in 1978, causing a major disease pandemic in dogs (8). Feline panleukopenia virus (FPV) has long been known to infect many species within the order Carnivora, including large and small cats, mink, raccoons, and foxes (29, 33). CPV is clearly a variant of FPV that acquired the ability to infect canines through a small number of mutations in the capsid protein (VP2) gene that changed surface-exposed residues (1, 3, 19). These viruses use the transferrin receptor type 1 (TfR) as their primary receptor for attaching to and infecting cells. Canine cells resist infection by FPV because that virus cannot bind to the canine TfR, in particular because of a unique glycosylation site present in the canine TfR (3, 10, 16, 18). The first strain of CPV (named CPV type 2 [CPV-2]) was replaced globally during 1979 and 1980 by a genetic and antigenic variant named CPV type 2a (CPV-2a), which is the common ancestor of all the CPVs currently circulating in dogs worldwide (9, 24). The CPV-2a variant contained a number of important genetic changes, including replacements of four VP2 residues that alter capsid binding to the feline TfR and to antibodies and that likely increased the fitness of the virus in dogs and gave it the ability to infect cats (7, 16, 27, 30, 34). The CPV-2a strain has undergone additional evolution in dogs since 1979, generating genetic and antigenic variants known as CPV-2b and CPV-2c (9).
Here we further define the evolutionary basis of viral host range variation by refining our understanding of this classic example of a host-jumping virus—the emergence of CPV-2 and its variants in dogs—through the exploration of its evolution in alternative mammalian hosts.
MATERIALS AND METHODS
Cells and cell cultures.
CHO-derived TRVb cells, which lack an endogenous TfR (13), were grown in Ham's F-12 medium containing 10% fetal bovine serum. Crandell Rees feline kidney (CRFK) cells, Norden Laboratory feline kidney (NFLK) cells, and canine Cf2Th and A72 cells were grown in a 1:1 mixture of McCoy's 5A medium and Leibovitz L-15 medium with 5% fetal bovine serum.
Viruses.
The new viruses examined here are listed in Table 1 and are highlighted in Tables S1 and S2 in the supplemental material. For virus isolation, homogenized tissue or fecal samples were filtered and were used to infect CRFK or NLFK cells. Viral DNA was PCR amplified either directly from tissue extracts or after a single passage in cell culture. Amplification was performed using Phusion Hot Start high-fidelity DNA polymerase (Finnzymes, Espoo, Finland). Amplicons were sequenced either directly or after cloning using a PCR cloning kit (Qiagen, Valencia, CA) and covered the complete VP2 protein sequence or, in some cases, the entire nonstructural and structural coding regions. Control viruses were derived from infectious plasmid clones of FPV (FPV-b), CPV-2 (CPV-d), and CPV-2b (CPV-39) (10, 20) and were purified as described previously (1). Viral inocula were prepared by passing viruses in NLFK cells, homogenizing the supernatant and cell fractions using a freeze-thaw cycle, and clarifying the inoculum through a Steriflip filter (Millipore, Billerica, MA).
Table 1.
Parvovirus isolates recovered between 2007 and 2010a
| Isolate designation | Outbreak data |
GenBank accession no. | ||||
|---|---|---|---|---|---|---|
| Countyb and state | Date | No. of animals affected | Mortality (no. of deaths) | Ages of animals affected | ||
| CPV/Raccoon/VA/118-A/07 | Clarke Co., VA | Sept. 2007 | 11 | 8 | 4-5 mo | JN867610 |
| CPV/Raccoon/GA/287/08, -289/08, and -349/08 | Glynn Co., GA | Oct.-Nov. 2008 | Unknown | 3 | 2-7 mo | JN867615, JN867616, JN867617 |
| CPV/Raccoon/TN/351/09 | Knox Co., TN | May 2009 | 2 litters | Multiple | Newborn to 1.5 mo | JN867612 |
| CPV/Raccoon/FL/381/09 | Lee Co., FL | July 2009 | Unknown | 1 | <3 mo | JN867613 |
| CPV/Raccoon/VA/278-A/09 | Fairfax Co., VA | Aug. 2009 | 4 | 4 | <6 mo | JN867614 |
| CPV/Raccoon/KY/358-B/09, -39552/09, and -33817/09 | Scott Co., KY | July-Nov. 2009 | Unknown | 23 | Primarily juvenile animals | JN867611, JN867599, JN867600 |
| CPV/Raccoon/WI/37/10 | Green Co., WI | Jan. 2010 | 1 | 1 | <1 yr | JN867618 |
| CPV/Bobcat/KS/44/10 | Lyon Co., KS | Feb. 2010 | 1 | 1 | Adult | JN867598 |
| CPV/Raccoon/NY/94742/10 | Oswego Co., NY | Aug. 2010 | Unknown | 6 | <6 mo | JN867597 |
| CPV/Raccoon/NJ/76836/10 | Atlantic Co., NJ | Aug. 2010 | 8 | 7 | 3.5-4 mo | JN867601 |
| FPV/Raccoon/CA/208-A/10 | Sonoma Co., CA | Aug. 2010 | 2 | 2 | <1 yr | JN867593 |
Isolates obtained from the same outbreak/location are grouped. The clinical features of each outbreak, where available, are noted.
Co., county.
Evolutionary analysis.
A total of 433 VP2 sequences of different carnivore parvoviruses, 1 of which (GenBank accession number M24005) was sampled from a raccoon in 1978, were compiled from GenBank. These sequences were combined with 27 new sequences and with those determined here from other carnivore species, resulting in a final data set of 460 sequences of 1,755 bp (see Table S1 in the supplemental material). All VP2 sequences included in this analysis covered at least two-thirds of the VP2 gene (although similar phylogenetic results were obtained using 125 full-length VP2 sequences [data not shown]). An equivalent phylogenetic study was undertaken using 39 NS1 sequences collected from GenBank combined with 9 raccoon sequences determined here. This produced a total data set of 48 sequences of 2,007 bp (see Table S2 in the supplemental material). All sequence alignments were created using MUSCLE, version 3.7 (5).
Phylogenetic trees were inferred using the maximum-likelihood (ML) method implemented in PAUP*, version 4.0 (32), utilizing the general time-reversible model of nucleotide substitution incorporating invariant sites and a gamma distribution of among-site rate variation with four rate categories (model GTR+I+Γ4); this was the best-fit nucleotide substitution model determined by Modeltest, version 3.7 (28). Tree bisection-reconnection (TBR) branch swapping was employed in all cases. To assess the robustness of the phylogenetic groupings observed, a bootstrap resampling analysis was performed using 1,000 replicate neighbor-joining trees estimated under the ML substitution model described above, again utilizing PAUP*, version 4.0.
To assess the strength of temporal structure (i.e., clock-like behavior) in these sequence data, we performed a regression of root-to-tip genetic distances against the date (year) of sampling of each sequence by using the Path-O-Gen program (http://tree.bio.ed.ac.uk/software/pathogen/). This analysis was performed using the 48 NS1 and VP2 sequences that were available from the same viruses. Input genetic distances were obtained from the ML trees described above.
The time to the most recent common ancestor (TMRCA) of the raccoon sequences was estimated using the Bayesian Markov chain Monte Carlo (MCMC) method available in the BEAST package (4). This analysis incorporated the model of nucleotide substitution described above, in this case partitioned by codon positions, as well as a relaxed (uncorrelated lognormal) molecular clock and the conservative Bayesian skyline coalescent prior. Statistical support was depicted as values of the 95% highest probability density (HPD), and the MCMC analysis was run until convergence was achieved in all parameters. The BEAST analysis also allowed us to infer the maximum clade credibility (MCC) tree for these data, such that TMRCA values (i.e., divergence times) could be estimated for specific nodes. The MCC tree was topologically very similar to the tree produced in the ML analysis.
Finally, to determine whether recombination had occurred between the VP2 and NS1 genes, we employed the BOOTSCAN, GENECOV, and RDP methods available in the RDP3 package (12) using the default settings.
rTfR.
The raccoon transferrin receptor (rTfR) was independently sequenced from a raccoon cell line (Pl 1Ut cells; ATCC, Manassas, VA) and also from heart tissue from a wild raccoon. For subsequent experiments involving binding to the rTfR (see below), mRNA from the wild raccoon was extracted using an RNeasy kit (Qiagen), amplified using the SuperScript III one-step reverse transcription-PCR (RT-PCR) system (Invitrogen, Carlsbad, CA), cloned into the pcDNA3.1(−) plasmid (Invitrogen), and sequenced as described previously (18) (GenBank accession number JN600499).
Antigenic analysis.
Twenty-eight mouse or rat monoclonal antibodies (MAbs) against CPV-2, FPV, or CPV-2b have been described previously (21, 23, 27). For hemagglutination inhibition (HI) assays, antibodies were tested using standard methods, and titers were compared to the titer of the virus against which the MAb was raised (30).
Cell infection and binding assays.
Infection of NLFK, A72, or Cf2Th cells was assessed using standard methods for 50% tissue culture infective dose (TCID50) assays (34). Cell binding was analyzed using purified virus capsids at 10 μg/ml incubated in a solution for 60 min at 37°C with NLFK or Cf2Th cells, or with TRVb cells transfected with a plasmid expressing the feline, canine, or raccoon TfR (6). Virus capsids were detected with MAb 2 and an Alexa 488-conjugated goat anti-mouse antibody (Sigma, St. Louis, MO), and binding was quantified using a Guava EasyCyte Plus flow cytometer (Millipore).
Nucleotide sequence accession numbers.
All the virus sequences obtained in this study have been deposited in GenBank under accession numbers JN867593 to JN867618.
RESULTS
Raccoons (Procyon lotor) are native to North America but have become established in parts of Europe and Asia since the early 20th century (35). We sampled parvoviruses collected from raccoons that showed clinical disease during outbreaks in the United States between 2007 and 2010 (Table 1), and we compared these viruses to raccoon isolates collected between 1978 and 1990, as well as to the many FPV and CPV sequences from cats, dogs, and related hosts in GenBank (see Table S1 in the supplemental material). The infected raccoons exhibited disease symptoms characteristic of parvoviral enteritis, including lethargy, fever, diarrhea, and vomiting. Parvovirus infection was demonstrated by histopathology and immunohistochemistry of the intestine and lymphoid tissues (not shown).
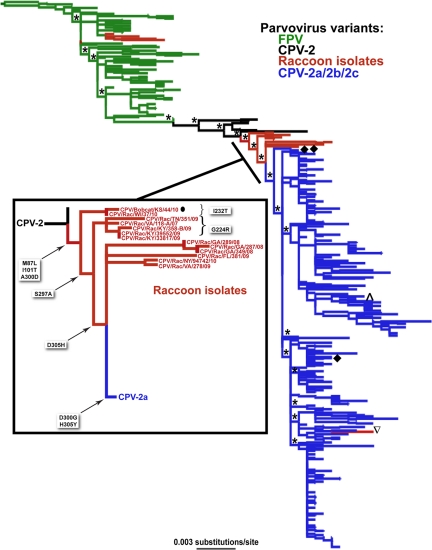
Phylogenetic analysis of VP2 sequences showed that the raccoon virus isolates fell into several unrelated clusters across the phylogeny, indicating that there have been multiple cross-species transmission events involving raccoons (Fig. 1). Remarkably, 12 of the 17 raccoon virus sequences and the sequence of a virus isolated from a wild bobcat (Lynx rufus) fell at intermediate positions between the CPV-2 and CPV-2a sequences (Fig. 1). Due to the recovery of only a single intermediate virus from a bobcat, the significance of this species either as a long-term host for maintaining these viruses or simply as an incidental host due to raccoon predation is currently unknown. The raccoon VP2 sequences appear to be evolutionary intermediates in that they did not form a separate monophyletic group but instead occupied interleaved positions between the CPV-2 and CPV-2a sequences. The isolates in this intermediate group were collected between 2007 and 2010 from several areas of the United States, including states located between Florida and New York, as well as Wisconsin, while the bobcat isolate was collected in Kansas in 2010.
Fig 1.
ML phylogeny of carnivore parvoviruses from various hosts based on 460 partial sequences of the VP2 gene. Red indicates that the sequence is from a raccoon, except for the marked (●) bobcat isolate (inset) that was sequenced during this study. Other sequences are colored by virus strain, regardless of host, with FPV-like sequences in green, CPV-2 sequences in black, and CPV-2a, -b, and -c sequences in blue. The phylogenetic positions of three leopard cat sequences from Southeast Asia (♦) and of one canine sequence from South Korea (∧) are also marked. The tree is rooted using the oldest sequence in the data set, FPV/Cat/US/FPV-d/64 (GenBank accession number U22189). Bootstrap values of >0.90 are marked by asterisks. Branch lengths are drawn to a scale of nucleotide substitutions per site. (Inset) Expansion of the phylogeny section containing the main group of virus sequences from raccoons, highlighting the amino acid substitutions that have occurred on each branch. All raccoon and bobcat viruses, including CPV/Raccoon/NJ/76836/10 (▿), have the 300Asp mutation.
A molecular-clock analysis of the VP2 data set revealed that the 12 intermediate raccoon isolates and the bobcat isolate diverged from CPV between 24 and 31 years ago (95% HPD values) and hence have been circulating undetected for this period. The remaining five raccoon isolates fell outside the main cluster and were located either among the FPV (FPV/Raccoon/CA/208-A/10, FPV/Raccoon/NJ/RPV-6/90, FPV/Raccoon/TX/Rac1.2/78, FPV/Raccoon/TX/Rac2.2/78) or among the CPV-2a-derived (CPV/Raccoon/NJ/76836/10) sequences. These viruses may represent short-term spillover of FPV and CPV-2a into raccoon populations without sustained transmission, although addressing this question will require additional sampling. Molecular-clock estimates place the origin of these other raccoon viruses at various times during the past 45 years (full results are available on request).
Contrasting evolutionary patterns in the capsid and nonstructural genes.
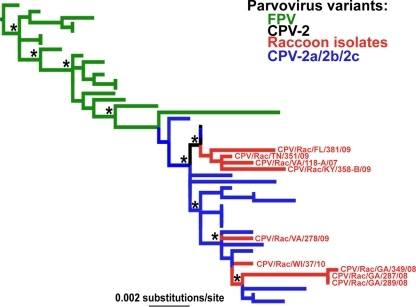
More-complex, and strikingly different, evolutionary patterns were observed in a phylogenetic analysis of 48 NS1 (nonstructural) gene sequences (Fig. 2). In this analysis, the raccoon viruses fell at three distinct phylogenetic positions, indicating multiple cross-species transmission events from canine viruses and giving no evidence that raccoons are intermediate hosts (Fig. 2). We used a Shimodaira-Hasegawa (SH) test to determine whether the NS1 and VP2 genes (the latter subsampled such that the same 48 viruses were used) gave different representations of the evolutionary history of carnivore parvoviruses. This confirmed that the NS1 and VP2 phylogenies were significantly different (P < 0.001) such that they have strongly contrasting evolutionary histories (which also means that they cannot be meaningfully combined into a complete genome phylogeny). Importantly, these dramatically contrasting phylogenetic patterns could not be explained by recombination, since we found no evidence for this process in our data (P > 0.1). Hence, the incongruent NS1 and VP2 trees are most likely the result of very different selection pressures acting on these genes.
Fig 2.
ML tree of 48 NS1 gene sequences. Branches are color coded as explained in the legend to Fig. 1. The tree is rooted using the oldest sequence in the data set, FPV/Cat/US/FPV-3/67 (GenBank accession number EU659111). Bootstrap values of >0.90 are marked by asterisks. Branch lengths are drawn to a scale of nucleotide substitutions per site, and sample names are given for the raccoon isolates.
There are good reasons to believe that the VP2 tree is the more accurate representation of evolutionary history. First, if the NS1 tree is correct, then both CPV-2 and CPV-2a, as well as the raccoon viruses, have evolved multiple times in parallel and do not form monophyletic groups, signifying a substantial amount of convergent evolution. Second, according to the NS1 tree, parvoviruses jumped from cats to dogs initially as the CPV-2a genotype and then evolved to CPV-2. Not only is this counterintuitive with respect to the known history of mutations in these viruses, but it runs counter to the time scale of parvovirus evolution. In particular, it is well documented that CPV-2 appeared in dogs and spread worldwide during 1978 and 1979, before CPV-2a emerged, as shown in the VP2 tree, and indeed CPV-2 isolates have earlier sampling times (1977 to 1980) than CPV-2a strains (late 1979 onward). Moreover, the correlation coefficients of root-to-tip genetic distance against sampling year were significant for the VP2 (r = 0.70; P < 1 × 10−5) but not the NS1 (r = 0.55; P = 0.171) data set. Because a strong temporal structure is present only in the VP2 phylogeny, we suggest that this is the correct representation of evolutionary history, although the sequencing of additional NS1 gene sequences will help resolve this issue.
Differences in capsid protein residues.
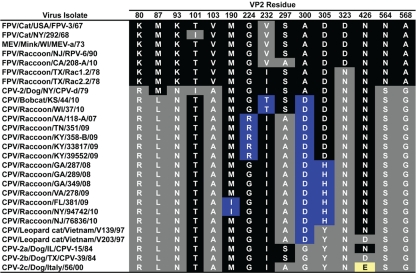
After it emerged in 1979, the CPV-2a lineage replaced the CPV-2 strain worldwide within 2 years, indicating increased fitness in dogs. Importantly, the CPV-derived raccoon viruses also accumulated changes in several of the capsid codons that later helped define the CPV-2a lineage (VP2 residues 87, 101, 297, 300, and 305) (Fig. 1 and 3). Residue 300 appears to be a particularly important component of the host adaptive process. This residue is an Ala in FPV, CPV-2, and FPV-like raccoon viruses but is a Gly in all CPV-2a-derived viruses from dogs. However, 300Asp was seen in all raccoon isolates derived from CPV, irrespective of their positions on the phylogeny (Fig. 1 and 3), and hence must have arisen more than once in the raccoon viruses derived from CPV. The importance of the 300Asp change for CPV-derived viruses in hosts other than dogs is confirmed by the finding of this mutation in some viruses from leopard cats in Southeast Asia and in a CPV-2 isolate that had been passaged in cultured feline cells (11, 14, 22). The variants at residues 297 and 305 also have complex evolutionary patterns. Residue 297 was a Ser in CPV-2 and some older CPV-2a and CPV-2b sequences but was mutated to Ala in many of the raccoon viruses, reverted to Ser in some early canine isolates of the CPV-2a variant, and became fixed in dog viruses as an Ala throughout most of the world in the early 1990s (2, 9, 15). Residue 305 was an Asp in FPV and CPV-2, mutated to His in some of the raccoon viruses, and became a Tyr when the virus transferred to dogs during the emergence of CPV-2a.
Fig 3.
Capsid amino acid residues associated with viruses isolated from raccoons in different parts of the United States between 1978 and 2010, as well as viruses isolated from leopard cats in Vietnam in 1997, a mink isolate from Wisconsin recovered in 1973, and a bobcat virus from Kansas recovered in 2010. Also included are the sequences for the FPV, CPV-2, and CPV-2a-related prototypes. FPV-like residues are highlighted in black. CPV-like residues are highlighted in gray or yellow. Mutations unique to the CPV-derived isolates from raccoons are highlighted in blue. The 426E in CPV-2c strains is highlighted in yellow.
Raccoon virus properties and host-specific receptor analysis.
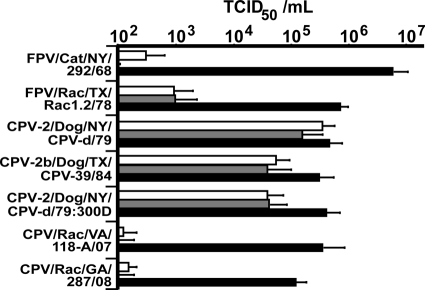
We confirmed that these mutations altered host range, TfR binding, and antigenic structure in the raccoon viruses. Tissue culture infectivity for dog cells reflects the canine host range of these viruses (34), and while the raccoon viruses tested infected feline cells similarly to the FPV, CPV-2, and CPV-2a prototypes, they did not infect canine cells (Fig. 4). Since a CPV-2 mutant with only the 300Asp change exhibited only 10-fold-lower infectivity for canine cells than for feline cells (Fig. 4), combinations of other residues in the raccoon viruses likely acted along with 300Asp to limit canine cell infection (Fig. 3 and 4).
Fig 4.
Relative infectivities of the different viruses for feline NLFK cells (filled bars), canine A72 cells (shaded bars), or Cf2Th cells (open bars). Three different raccoon isolates (Rac) were tested, along with prototype FPV, CPV-2, and CPV-2b sequences (FPV/Cat/NY/292/68, CPV-2/Dog/NY/CPV-d/79, and CPV-2b/Dog/TX/CPV-39/84) and the 300Asp mutant of CPV-2 (CPV-2/Dog/NY/CPV-d/79:300D). Virus titers were determined by TCID50 assays in each cell line. Error bars indicate standard deviations based on three independent experiments.
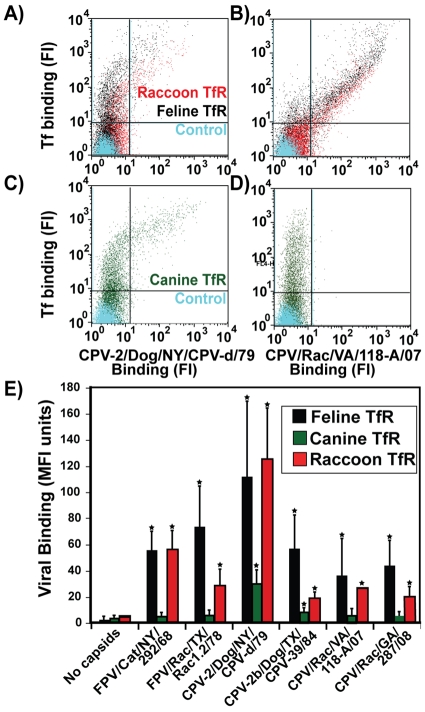
The TfR is a key determinant of feline and canine cell susceptibility, and host-specific virus binding to TfR is controlled by capsid surface residues (10, 16). We therefore cloned the cDNA from a raccoon tissue sample and found that it exhibited 88% and 89% amino acid identity to the dog and cat TfRs, respectively, and that it lacked the glycosylation site that blocks the binding of FPV capsids to the canine TfR (16, 17) (see Fig. S1 in the supplemental material). When the raccoon, feline, and canine TfRs were expressed on hamster cells under identical conditions, the raccoon virus capsids bound the raccoon and feline TfRs to similar levels but did not bind to the canine TfR (Fig. 5).
Fig 5.
Binding of capsids and canine transferrin (Tf) to the domestic cat, raccoon, and canine TfRs expressed on TRVb cells, as measured by flow cytometry. Cells were incubated with Cy5-conjugated Tf (to detect TfR-expressing cells) and with virus capsids. The cell-associated capsids were detected using Alexa 488-conjugated MAb 2. Binding is expressed as fluorescence intensity (FI) (A through D) or mean fluorescence intensity (MFI) (E). (A through D) Binding of Tf and CPV-2 (CPV-2/Dog/NY/CPV-d/79) (A and C) and of Tf and CPV/Rac/VA/118-A/07 (B and D) to feline and raccoon TfRs (A and B) or to canine TfR (C and D). (D) Tf binds the canine TfR, but CPV/Rac/VA/118-A/07 does not. (E) Comparison of the binding of viruses to all three TfRs. Error bars, standard deviations of the mean. Three independent experiments were performed. Asterisks indicate binding significantly higher than background levels (P < 0.05).
The CPV-derived raccoon viruses were also antigenically distinct when tested with a large panel of MAbs prepared against FPV, CPV-2, or CPV-2b capsids (Fig. 6). All CPV-like raccoon virus isolates lost reactivity with several antibodies that recognized antigenic site “B,” primarily due to the 300Asp mutation. The CPV/Raccoon/VA/118-A/07 isolate, which also contained the 224Arg mutation in the “A” site, lost reactivity with 26 of 28 MAbs tested (7, 30).
Fig 6.

Antigenic analysis of viral variants using the hemagglutination inhibition (HI) assay and MAbs prepared against capsids of FPV (red), CPV-2 (blue), or CPV-2b (black). The results are expressed as comparisons of HI titers to those with the original capsid. Four different viruses from raccoons were tested, along with the FPV, CPV-2, and CPV-2b prototypes (FPV/Cat/NY/292/68, CPV-2/Dog/NY/CPV-d/79, CPV-2b/Dog/TX/CPV-39/84) and the 300Asp mutant of CPV-2 (CPV-2/Dog/NY/CPV-d/79:300D). Filled rectangles indicate binding within ±2-fold of the original titer; shaded rectangles, 4- to 16-fold reductions in titer; open rectangles, >16-fold reductions in titer.
DISCUSSION
The most striking observation of our study was that a group of raccoon parvoviruses constituted a previously unidentified host range variant of CPV that has been circulating for at least 24 years and causing an epidemic of disease. In addition, the VP2 phylogeny revealed that most of these raccoon-associated viruses were evolutionary intermediates between CPV-2 and CPV-2a strains. Although virus surveillance has been conducted extensively in the domestic dog and cat populations since the emergence of CPV-2 in 1978, viruses intermediate between CPV-2 and CPV-2a have not been identified previously. In dogs, the CPV-2a strain rapidly replaced CPV-2 worldwide; this, coupled with the intermediate phylogenetic position occupied by most of the raccoon viruses, suggests that raccoons played a key role in the emergence of this fitter variant in dogs. The selective advantage of CPV-2a over CPV-2 may have involved both better adaptation to the canine host and partial evasion of the anti-CPV-2 antibodies that were widespread in the dog population by 1979 and early 1980 (24).
An unusual aspect of this adaptive process was that the evolutionary intermediates of CPV in raccoons could not themselves bind to the canine TfR or infect canine cells (Fig. 4 and 5). This system therefore illustrates the complexities of the evolutionary pathways and fitness landscapes that may occur during adaptation to new host species. While several raccoon-associated mutations (VP2 residues 87 and 101) were also found in the canine-adapted virus CPV-2a, the 300Asp mutation likely prevented binding of the capsids to the canine TfR in the background of the raccoon virus isolates (Fig. 5). The transfer of the raccoon virus back to dogs clearly required the crossing of a relatively low evolutionary barrier and involved transition mutations that converted VP2 codon 300 from Asp to Gly and VP2 codon 305 from His/Asp to Tyr and that resulted in the CPV-2a genotype, which infected and spread efficiently within dogs. However, it is possible that other carnivore hosts in addition to raccoons have played a role in the emergence of CPV-2a in dogs, such that more-comprehensive sampling of additional carnivore species would likely aid in determining those which are important in the evolution of these viruses.
Despite the clear phylogenetic links between the raccoon and canine viruses, the NS1 and VP2 phylogenies have strikingly different topologies, suggesting that NS1 and VP2 have been subject to very different selection pressures (especially given the apparent absence of recombination). Because our collection of canine isolates goes back to 1978, we can directly validate the inferred phylogenetic history through comparison with the known sampling dates of the viruses. In this case, the phylogenetic patterns seen for the VP2 gene fit well with the known history of the canine viruses. The raccoon virus sampling is less well distributed in time, but it is reasonable to expect that the VP2 gene analysis can similarly be used to more accurately infer the evolutionary history of these viruses. Future studies will be directed toward determining the selection pressures acting on NS1 and VP2 and how these might have shaped phylogenetic history.
At a more fundamental level, our study shows that the selection of small numbers of key host-adaptive mutations is readily accomplished in these single-stranded DNA viruses and can occur on multiple occasions; in this case, the transfers between a cat-like host, dogs, and raccoons were each associated with groups of such mutations. As a consequence, successful emergence is not always dependent on mutational availability (31). Similar observations have been made with respect to the adaptation of influenza viruses among mammalian species (25, 26) and further demonstrate that the strength of host phylogenetic relationships can act as a predictor of successful viral emergence (31).
Finally, and perhaps of most importance, this study highlights the potential role of alternative hosts (in this case raccoons) in providing indirect viral emergence pathways, even in cases where a fitter viral variant, such as the CPV-2a strain, appears to arise within the same host species as the original strain (CPV-2). While alternative and intermediate hosts are often proposed to provide a bridge for the transfer of viruses between widely separated host species, their role in facilitating increased adaptation to a single host has not been described previously. We therefore suggest that future attempts to survey virus biodiversity and to create theoretical models for predicting and forestalling viral emergence should pay close attention to the possible roles played by related hosts in providing favorable adaptive pathways.
Supplementary Material
ACKNOWLEDGMENTS
A.B.A., J.D.B., M.G.R., and M.K.K. were supported by funds obtained through sponsorship by the fish and wildlife agencies of the member states of the Southeastern Cooperative Wildlife Disease Study (SCWDS). A.B.A. thanks Jeremiah Saliki at the University of Georgia for graciously allowing the use of his laboratory space. I.P. was supported by Marie Curie fellowship PIOF-GA-2009-236470. This work was also supported by NIH grants GM080533-05 (to E.C.H. and C.R.P.), AI028385, and AI092571 (to C.R.P.).
Footnotes
Published ahead of print 9 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Agbandje M, McKenna R, Rossmann MG, Strassheim ML, Parrish CR. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155–171 [DOI] [PubMed] [Google Scholar]
- 2. Barros de Freitas R, et al. 2007. The “pressure pan” evolution of human erythrovirus B19 in the Amazon, Brazil. Virology 369:281–287 [DOI] [PubMed] [Google Scholar]
- 3. Chang SF, Sgro JY, Parrish CR. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 66:6858–6867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goodman LB, et al. 2010. Binding site on the transferrin receptor for the parvovirus capsid and effects of altered affinity on cell uptake and infection. J. Virol. 84:4969–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hafenstein S, et al. 2009. Structural comparison of different antibodies interacting with parvovirus capsids. J. Virol. 83:5556–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoelzer K, Parrish CR. 2010. The emergence of parvoviruses of carnivores. Vet. Res. 41:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoelzer K, Shackelton LA, Parrish CR, Holmes EC. 2008. Phylogenetic analysis reveals the emergence, evolution and dispersal of carnivore parvoviruses. J. Gen. Virol. 89:2280–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hueffer K, et al. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77:1718–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ikeda Y, et al. 2000. Predominance of canine parvovirus (CPV) in unvaccinated cat populations and emergence of new antigenic types of CPVs in cats. Virology 278:13–19 [DOI] [PubMed] [Google Scholar]
- 12. Martin DP, et al. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McGraw TE, Greenfield L, Maxfield FR. 1987. Functional expression of the human transferrin receptor cDNA in Chinese hamster ovary cells deficient in endogenous transferrin receptor. J. Cell Biol. 105:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miyazawa T, et al. 1999. Isolation of feline parvovirus from peripheral blood mononuclear cells of cats in northern Vietnam. Microbiol. Immunol. 43:609–612 [DOI] [PubMed] [Google Scholar]
- 15. Mohan RJ, Mukhopadhyay HK, Thanislass J, Antony PX, Pillai RM. 2010. Isolation, molecular characterization and phylogenetic analysis of canine parvovirus. Infect. Genet. Evol. 10:1237–1241 [DOI] [PubMed] [Google Scholar]
- 16. Palermo LM, Hafenstein SL, Parrish CR. 2006. Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J. Virol. 80:8482–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palermo LM, Hueffer K, Parrish CR. 2003. Residues in the apical domain of the feline and canine transferrin receptors control host-specific binding and cell infection of canine and feline parvoviruses. J. Virol. 77:8915–8923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parker JS, Murphy WJ, Wang D, O'Brien SJ, Parrish CR. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75:3896–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parker JS, Parrish CR. 1997. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 71:9214–9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parrish CR. 1991. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology 183:195–205 [DOI] [PubMed] [Google Scholar]
- 21. Parrish CR, Carmichael LE. 1983. Antigenic structure and variation of canine parvovirus type-2, feline panleukopenia virus, and mink enteritis virus. Virology 129:401–414 [DOI] [PubMed] [Google Scholar]
- 22. Parrish CR, Carmichael LE. 1986. Characterization and recombination mapping of an antigenic and host range mutation of canine parvovirus. Virology 148:121–132 [DOI] [PubMed] [Google Scholar]
- 23. Parrish CR, Carmichael LE, Antczak DF. 1982. Antigenic relationships between canine parvovirus type-2, feline panleukopenia virus and mink enteritis virus using conventional antisera and monoclonal antibodies. Arch. Virol. 72:267–278 [DOI] [PubMed] [Google Scholar]
- 24. Parrish CR, et al. 1988. The global spread and replacement of canine parvovirus strains. J. Gen. Virol. 69:1111–1116 [DOI] [PubMed] [Google Scholar]
- 25. Parrish CR, et al. 2008. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72:457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parrish CR, Kawaoka Y. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 59:553–586 [DOI] [PubMed] [Google Scholar]
- 27. Parrish CR, O'Connell PH, Evermann JF, Carmichael LE. 1985. Natural variation of canine parvovirus. Science 230:1046–1048 [DOI] [PubMed] [Google Scholar]
- 28. Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 29. Steinel A, Parrish CR, Bloom ME, Truyen U. 2001. Parvovirus infections in wild carnivores. J. Wildl. Dis. 37:594–607 [DOI] [PubMed] [Google Scholar]
- 30. Strassheim LS, Gruenberg A, Veijalainen P, Sgro J-Y, Parrish CR. 1994. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198:175–184 [DOI] [PubMed] [Google Scholar]
- 31. Streicker DG, et al. 2010. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 329:676–679 [DOI] [PubMed] [Google Scholar]
- 32. Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.Sinauer Associates, Sunderland, Mass [Google Scholar]
- 33. Truyen U, et al. 1995. Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J. Virol. 69:4702–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Truyen U, Parrish CR. 1992. Canine and feline host ranges of canine parvovirus and feline panleukopenia virus: distinct host cell tropisms of each virus in vitro and in vivo. J. Virol. 66:5399–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zeveloff SI. 2002. Raccoons: a natural history. Smithsonian Books, Washington, DC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.