Abstract
Coronavirus replication and transcription are processes mediated by a protein complex, with the RNA-dependent RNA polymerase (RdRp) as a main component. Proteomic analysis of highly purified transmissible gastroenteritis virus showed the RdRp to be a component of the viral particles. This finding was confirmed by Western blotting, immunofluorescence, and immunoelectron microscopy analyses. Interestingly, the replicase nonstructural proteins 2, 3, and 8 colocalized with the RdRp in the viral factories and were also incorporated into the virions.
TEXT
Transmissible gastroenteritis virus (TGEV) is an enveloped virus with a 28.6-kb positive-stranded RNA genome that belongs to the genus α of the Coronaviridae family within the Nidovirales order (4, 6) (see http://ictvonline.org/virusTaxonomy.asp for official coronavirus taxonomy). Coronavirus (CoV) replication and transcription are complex processes that take place at cytoplasmic double-membrane vesicles (DMVs) and involve coordinated processes of both continuous and discontinuous RNA synthesis (8, 11, 13, 16, 24). Both processes are mediated by a protein complex encoded by the replicase gene together with the participation of cellular factors (7, 25, 26). The replicase gene, which occupies the 5′ two-thirds of the genome, is translated at the beginning of the infection into two co-amino-terminal polyproteins. Both proteins are autoproteolytically cleaved into 16 nonstructural proteins (nsp1 to nsp16), including two proteinases (nsp3 and nsp5), the RNA-dependent RNA polymerase (RdRp, or nsp12), the RNA helicase (nsp13), and a primase (nsp8), which are believed to be part of the replication-transcription complex (10, 26, 27). In addition to the replicase proteins, the viral nucleoprotein (N) has also been described to play an important role in CoV RNA synthesis (1, 23, 28). In contrast to negative-strand RNA viruses, it is currently accepted that in positive-strand RNA viruses, the replicase proteins are not incorporated into viral particles. However, a recent study based on a proteomic analysis of purified severe acute respiratory syndrome CoV (SARS-CoV) showed the presence of nsps 2, 3, and 5 in the virions (17).
In the present study, we report the presence of RdRp, nsp2, nsp3, and nsp8 in highly purified TGEV particles by Western blotting, immunofluorescence, and immunoelectron microscopy assays.
Analysis of the RdRp presence in TGEV viral particles.
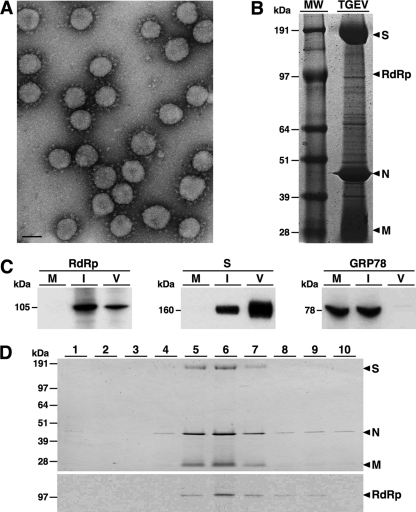
To analyze the presence of RdRp in viral particles, highly purified TGEV (PUR46-MAD strain) was prepared as described previously (5). Briefly, the virus from clarified infected cell culture supernatants was concentrated by centrifugation through a 31% sucrose cushion and then purified over a continuous 30% to 42% sucrose density gradient. The purity and integrity of the purified virus were confirmed by conventional electron microscopy. More than 99% of the viral particles preserved their structural properties, and no vesicles or other contaminants were detected (Fig. 1A). The protein profile of the purified virus was then analyzed by SDS-PAGE, and in addition to the structural proteins S, N, and M, a large number of proteins with different molecular masses were also detected (Fig. 1B). These proteins were excised from the gel, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry in an ABI 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystem), as described previously (9). Most of the identified proteins were cellular proteins, including membrane alanine aminopeptidase, eukaryotic translation elongation factor 2, the heat shock 90-kDa AB1 protein, calnexin, the poly(A) binding protein, the chaperonin containing TCP1 (subunit 7), F1 ATP synthase, glyceraldehyde-3-phosphate dehydrogenase, tubulin (β5), and β-actin. All of these proteins were identified with significant Mascot scores (P < 0.05) and sequence coverage between 45% and 87%. With the exception of the membrane alanine aminopeptidase, these proteins have been previously identified either in purified SARS-CoV (17) or infectious bronchitis virus (14). Interestingly, a minor polypeptide with apparent mass of 105 kDa was identified as the viral RdRp (Fig. 1B), suggesting that the RdRp was incorporated into the viral particles. However, other replicase proteins were not detected in this analysis.
Fig 1.
Identification of RdRp in purified TGEV. (A) Electron microscopy of purified TGEV. Purified virus was adsorbed to glow-discharged carbon-coated copper grids, processed for negative staining with 2% uranyl acetate as described previously (2), and examined in a JEOL JEM-1020 transmission electron microscope. Bar, 100 nm. (B) Proteomic analysis of purified TGEV. Proteins from 30 μg of purified TGEV were separated by SDS-PAGE (4 to 12%), stained with Coomassie blue, excised from the gel, and analyzed by mass spectrometry. Arrowheads indicate the identified viral proteins. Molecular mass markers are shown in kDa. (C) Western blot of purified TGEV virions. Proteins from 10 μg of purified TGEV (V) were separated by SDS-PAGE (4 to 12%) together with extract from ST cells mock infected (M) or infected with TGEV (I) and analyzed by immunoblotting with specific antibodies recognizing RdRp, S, and GRP78 (Abcam). Protein molecular masses in kDa are shown to the left. (D) Analysis of RdRp during the virus purification process. Fractions from the sucrose density gradient were collected from the bottom to the top, resolved by SDS-PAGE (4 to 12%), and analyzed by Coomassie blue staining (upper panel) or immunoblotting using the RdRp MAb 4D10 (lower panel). The viral proteins S, N, M, and RdRp are indicated by arrowheads. Molecular mass markers are shown in kDa.
To confirm the RdRp encapsidation, its presence in purified virions was initially analyzed by Western blotting. To this end, 10 μg of purified TGEV and extracts from porcine swine testis (ST) cells either mock infected or infected with TGEV at a multiplicity of infection (MOI) of 5 for 12 h were resolved by SDS-PAGE and analyzed by immunoblotting using the RdRp monoclonal antibody (MAb) 4D10 (diluted 1:100), as described previously (18). The blots were also probed with the MAb 5BH1 (diluted 1:4,000) (12) specific for TGEV S protein as a positive control and a rabbit polyclonal antibody (pAb) against the endoplasmic reticulum marker GRP78 (Abcam) (diluted 1:4,000) as a negative control. Bound antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit IgG (Sigma) diluted 1:30,000 and the Immobilon Western chemiluminescent substrate (Millipore), according to the manufacturer's recommendations. Similarly to the S protein, the RdRp was detected in infected cells and purified virus, while the GRP78 was detected in mock-infected and infected cells but not in purified virus (Fig. 1C). To discard that the RdRp was a general contaminant of the virus purification process, its presence in the different fractions of the sucrose gradient was analyzed by Western blotting. The RdRp was only detected in the fractions containing the virus (Fig. 1D), indicating that the RdRp copurified with the viral particles.
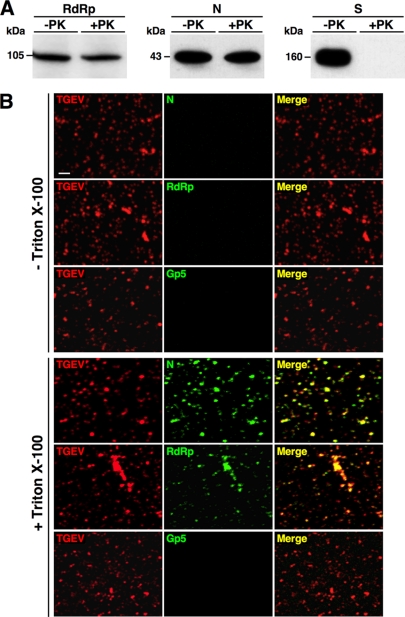
To further investigate whether the RdRp was in fact incorporated into the viral particles, the purified virus was treated with proteinase K and analyzed by Western blotting (Fig. 2A). Protease treatment would degrade proteins outside the virus particle or exposed on the virus surface, such as S protein, while proteins inside the virions, such as N protein, must be protected by the lipid envelope. Purified virus was either not treated or treated with 0.01 μg of proteinase K (Roche) per μg of virus at 25°C for 30 min and analyzed by immunoblotting using the RdRp MAb 4D10 (diluted 1:100) (18), the S MAb 5BH1 (diluted 1:4,000) (12), and the N MAb 3DC10 (diluted 1:3,000) (15) as described above. Similarly to the N protein, more than 75% of the RdRp present in the purified virus was detected after proteinase K treatment, while the S protein was completely degraded (Fig. 2A), indicating that the RdRp was inside the virions.
Fig 2.
Identification of RdRp in TGEV particles. (A) Analysis of RdRp in viral particles treated with proteinase K. Ten micrograms of highly purified TGEV was not treated (−PK) or treated (+PK) with 0.1 μg of proteinase K, and the presence of the viral proteins RdRp, N, and S was analyzed by Western blotting using specific MAbs. Protein molecular masses in kDa are shown to the left. (B) Identification of RdRp in purified virions by immunofluorescence analysis. Virions on glass coverslips were fixed, not permeabilized or permeabilized with Triton X-100, and analyzed by immunofluorescence using specific antibodies against TGEV (red), RdRp (green), N (green), and PRRSV GP5 (green). Colocalization is indicated by yellow pixels in the merge panels. Bar, 2 μm.
To provide additional evidence of the RdRp encapsidation, purified TGEV was analyzed by immunofluorescence before and after permeabilization with Triton X-100 (Fig. 2B). Purified TGEV virions were placed on glass coverslips previously treated with 0.1 mg/ml poly-l-lysine (Sigma), fixed by incubation with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) buffer for 45 min at room temperature, and either not permeabilized or permeabilized with 0.5% Triton X-100 for 20 min at room temperature. After incubating for 1 h in blocking solution (10% fetal bovine serum [FBS] in PBS), the viral particles were incubated with RdRp MAb 2C11 (diluted 1:20 in blocking solution) for 90 min at room temperature. Among the previously described RdRp MAbs, this antibody was chosen because it provides the highest signal in immunofluorescence (18). Alternatively, virions were probed with a rabbit pAb against TGEV (20) (diluted 1:1,200) as a control for virus identity, the N MAb 3DC10 (15) (diluted 1:100) as a control for an internal protein, and an irrelevant MAb recognizing the porcine reproductive and respiratory syndrome virus (PRRSV) GP5 envelope protein (22) (diluted 1:100) as a negative control. After washing with PBS, the samples were incubated for 1 h at room temperature with secondary antibodies conjugated to Alexa Fluor 488 or 549 (Invitrogen) diluted 1:500 in blocking solution, mounted on glass slides with Prolong Gold antifade reagent (Invitrogen), and analyzed with a Leica SP5 laser scanning microscope. Intact virions were labeled with only the TGEV pAb, which recognized the N, S, and M proteins (Fig. 2B, left panels). In contrast, when TGEV particles were permeabilized, a clear labeling was observed both with the N or RdRp MAbs, which colocalized with the viral particles detected with the TGEV pAb (Fig. 2B, right panels). As expected, no labeling was observed with the PRRSV GP5 antibody in both permeabilized and nonpermeabilized samples (Fig. 2B). These data indicated that similarly to the N protein, RdRp was incorporated into the viral particles.
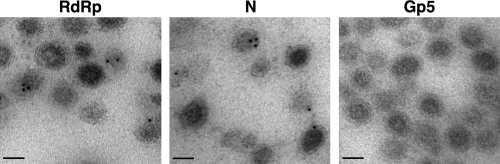
The presence of the RdRp in the virions was further analyzed by immunoelectron microscopy. Purified TGEV virions were fixed with a mixture of 4% PFA and 0.1% glutaraldehyde in PBS for 30 min at 4°C, cryoprotected with 30% glycerol, and quickly frozen in liquid ethane at −180°C. Then, vitrified samples were transferred to a Leica AFS2 EM FSP freeze-substitution unit and embedded in Lowicryl HM20 (TAAB) as described previously (19). Ultrathin sections (90 nm) on gold grids were immunogold labeled with purified RdRp MAb 2C11 (2 mg/ml) diluted 1:50 and a 5-nm-diameter colloidal gold-conjugated rabbit anti-mouse IgG (BBI) diluted 1:40, as described previously (21). Immunogold labeling was also performed with the PRRSV GP5 and TGEV N 3DC10 MAbs diluted 1:5 (15, 22) as negative and positive controls, respectively. Finally, the samples were negative stained with 2% uranyl acetate for 20 min at room temperature and examined in a JEOL JEM-1020 transmission electron microscope. Virions with gold particles were detected in the samples probed with the RdRp and N MAbs but not in that probed with the PRRSV GP5 MAb (Fig. 3). Interestingly, the proportion of virions labeled with the RdRp MAb (328 labeled viruses out of 3,645 counted viruses [9%]) was very similar to that observed with the N MAb (369 labeled viruses out of 3,720 counted viruses [10%]), and it was consistent with the expected proportion of virions that are accessible to the antibodies in one section of 90 nm (the viral particle size). In addition, a careful examination of the gold particle distribution in the virions labeled either with the RdRp or N MAbs indicated that the majority (about 65%) were located inside the viral particles (Table 1). All together, these data confirmed that the RdRp was incorporated into the viral particles.
Fig 3.
Analysis of RdRp in purified TGEV virions by immunoelectron microscopy. Ultrathin sections of purified virions were immunogold labeled with specific antibodies recognizing TGEV RdRp and N proteins or an irrelevant antibody against PRRSV GP5, followed by 5-nm-diameter gold-conjugated rabbit anti-mouse IgG. Bars, 50 nm.
Table 1.
Gold particle distribution in purified TGEV virions
| Gold labeling location | No. (%) of gold particles in: |
||||
|---|---|---|---|---|---|
| −Ca | RdRp | Nsp2 | Nsp3 | N | |
| Inside | 0 | 264 (65.2) | 252 (63.6) | 232 (59.5) | 260 (61.3) |
| Edge | 0 | 54 (13.3) | 48 (12.2) | 68 (17.4) | 48 (11.3) |
| Outside | 32 (100) | 87 (21.5) | 96 (24.2) | 90 (23.1) | 116 (27.4) |
| Total | 32 (100) | 405 (100) | 396 (100) | 390 (100) | 424 (100) |
−C, negative control (irrelevant antibody).
Analysis of the presence of nsps 2, 3, and 8 in TGEV viral particles.
Taken into consideration that RdRp is a main component of the replication-transcription complex, the encapsidation of other replicase proteins potentially involved in CoV RNA synthesis was also analyzed. Based on the availability of pAbs recognizing the TGEV nsps 2, 3, and 8, these replicase proteins were selected for this study. The nsp3 antibody was generated in rabbit using the His-tagged N-terminal 345-amino-acid (aa) fragment expressed in Escherichia coli as immunogen (data not shown). This fragment contains the acidic and most of the PLP1 protease domains. The pAbs recognizing TGEV nsp2 and nsp8 were kindly provided by John Ziebuhr (Justus Liebig University, Giessen, Germany).
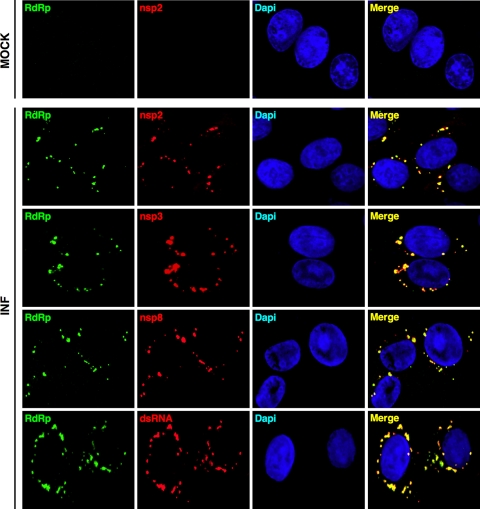
Before studying the incorporation of nsps 2, 3, and 8 into the viral particles, their presence in TGEV replication-transcription factories was analyzed by confocal immunomicroscopy. To this end, subconfluent ST cells grown on glass coverslips were mock infected or infected with TGEV at an MOI of 2. At 9 h postinfection, cells were fixed and permeabilized with methanol for 10 min at −20°C and analyzed by confocal immunomicroscopy using the RdRp MAb 2C11 (diluted 1:20) and the pAbs against nsps 2, 3, and 8 (diluted 1:300), as described above. In infected cells, a clear colocalization of RdRp with nsps 2, 3, and 8 was observed in cytoplasmic perinuclear structures that might correspond to the DMVs (3, 11, 13) (Fig. 4). In addition, when the subcellular distribution of RdRp and double-stranded RNA (dsRNA; a marker of active viral RNA synthesis) was analyzed using a MAb specific for dsRNA (English & Scientific Consulting) and a pAb against the TGEV RdRp (generated in rabbit using the RdRp expressed in baculovirus), a clear colocalization was also found (Fig. 4). In contrast, in mock-infected cells, no labeling was detected with any antibody for the nsps and dsRNA, as expected (Fig. 4, upper panels). These results indicated that nsps 2, 3, and 8 together with RdRp were accumulated in the replication-transcription complexes responsible for viral RNA synthesis.
Fig 4.
Colocalization analysis of RdRp with nsp2, nsp3, nsp8, and dsRNA. Confocal immunomicroscopy analysis was performed on ST cells mock infected (MOCK) or infected (INF) with TGEV (MOI, 2) for 9 h. RdRp (green) was detected with either MAb 2C11 or a rabbit pAb (bottom left panel). Nsps 2, 3, and 8 (red) were visualized with specific rabbit pAbs and dsRNA (red) with a commercial MAb (English & Scientific Consulting). Cell nuclei (blue) were stained with DAPI (4′,6-diamidino-2-phenylindole). Colocalization is indicated by yellow pixels in the merge panels. In mock-infected cells, the staining with the RdRp and nsp2 antibodies is shown as a representative example of the labeling with the other antibodies.
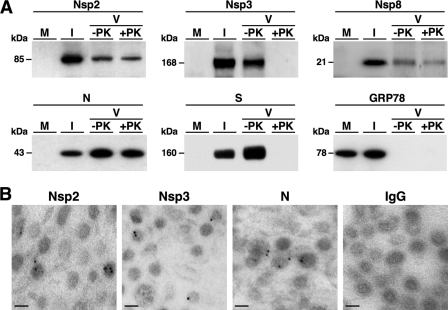
The presence of nsps 2, 3, and 8 in the viral particles was first analyzed by Western blotting as described above. Extracts from ST cells either mock infected or infected with TGEV (MOI of 5 for 12 h) and 10 μg of purified TGEV either treated or not treated with proteinase K were resolved by SDS-PAGE and analyzed by immunoblotting with the pAbs specific for nsps 2 (diluted 1:4,000), 3 (diluted 1:8,000), and 8 (diluted 1:2,000). As controls, the blots were also probed with the S MAb 5BH1 (diluted 1:4,000) (12), the N MAb 3DC10 (diluted 1:3,000) (15), and the GRP78 pAb (diluted 1:4,000) (Abcam). Similarly to N and S proteins, and in contrast to GRP78, nsps 2, 3, and 8 were detected in purified virus (Fig. 5A). After treatment with proteinase K, the levels of nsp2, nsp8 and N were not significantly reduced, while nsp3 and S were not detected at all (Fig. 5A). These data suggested that nsps 2 and 8 were incorporated into the viral particles and nsp3 was either a contaminant of the purification process or an envelope protein such as the S protein. To provide additional evidence of the encapsidation of these nsps, their presence in purified virions was analyzed by immunoelectron microscopy using the specific pAbs diluted 1:25, following the same procedure described above for the RdRp. The same experiment was performed with irrelevant antibodies and the N MAb 3DC10 (15) diluted 1:5 as negative and positive controls, respectively. Unfortunately the nsp8 pAb did not work in this technique (data not shown). However, clear immunogold labeling was detected in the virions probed with the nsp2 and nsp3 pAbs (Fig. 5B). In addition, the gold particle distribution and the proportion of labeled virions with the nsp2 (285 labeled viruses out of 3,452 counted viruses [8.3%]) and nsp3 (291 labeled viruses out of 3,427 counted viruses [8.5%]) pAbs were very similar to those observed for the RdRp and N proteins (Table 1). Overall, these data indicated that nsp2 and most likely nsp8 were incorporated into the viral particles and that nsp3 was probably present in the viral envelope exposing the N-terminal region (recognized by the pAb) to outside the virion.
Fig 5.
Identification of nsps 2, 3, and 8 in TGEV viral particles. (A) Western blot analysis. Proteins from purified TGEV (V) treated (+PK) or not treated (−PK) with proteinase K were separated by SDS-PAGE (4 to 12%) together with extracts from ST cells mock infected (M) or infected (I) with TGEV and analyzed by immunoblotting with specific antibodies recognizing nsp2, nsp3, nsp8, S, N, and GRP78. Protein molecular masses in kDa are shown to the left. (B) Immunoelectron microscopy analysis. Ultrathin sections of purified virions were immunogold labeled with specific antibodies recognizing the indicated proteins or with mouse and rabbit irrelevant antibodies (IgG), followed by 5-nm-diameter gold-conjugated secondary antibodies. Bars, 50 nm.
Although the presence of nsp2 and nsp3 in purified SARS-CoV has been previously reported using a proteomic approach (17), this is the first complete study indicating that RdRp, nsp2, and most likely nsp3 and nsp8 are incorporated into the CoV virions. Studies to investigate whether other viral and cellular components of the replication-transcription complex are also encapsidated and their role in the CoV life cycle are in progress. It could be postulated that the encapsidated RdRp together with other replicase proteins involved in CoV RNA synthesis could act as a starting replication machinery, allowing a first round of genome amplification before genome translation to improve the efficiency of virus infection.
ACKNOWLEDGMENTS
We thank John Ziebuhr for providing us with nsp2 and nsp8 pAbs and S. Zúñiga and I. Sola for critically reading the manuscript.
This work was supported by grants from the Ministry of Science and Innovation of Spain (MCINN) (BIO2010-16705), the Spanish National Research Council (CSIC) (project 200920I024), the Community of Madrid (S-SAL-0185-2006), the European Community's Seventh Framework Programme (FP7/2007-2013) under the projects “PLAPROVA” (KBBE-227056) and “PoRRSCon” (EC grant agreement number 245141), and Pfizer Animal Health. S.M.-J. received a predoctoral fellowship from National Institute of Health (ISCIII) of Spain. A.N. and C.G. received a contract from the Spanish National Research Council (CSIC).
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Almazán F, Galán C, Enjuanes L. 2004. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 78:12683–12688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bremer A, Häner M, Aebi U. 1998. Negative staining, p 277–284 In Celis JE. (ed), Cell biology, a laboratory handbook, 2nd ed, vol 3 Academic Press, San Diego, CA [Google Scholar]
- 3. Brockway SM, Clay CT, Lu XT, Denison MR. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515–10527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carstens EB. 2010. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009). Arch. Virol. 155:133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Correa I, Jiménez G, Suñé C, Bullido MJ, Enjuanes L. 1988. Antigenic structure of the E2 glycoprotein from transmissible gastroenteritis coronavirus. Virus Res. 10:77–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Groot RJ, et al. Coronaviridae, p 774–796 In Fauquet CM, Mayo MA, Maniloff J, Desselberg U, King A. (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses Academic Press, San Diego, CA [Google Scholar]
- 7. Enjuanes L, Almazán F, Sola I, Zuñiga S. 2006. Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol. 60:211–230 [DOI] [PubMed] [Google Scholar]
- 8. Enjuanes L, Sola I, Zuñiga S, Almazán F. 2008. Coronavirus replication and interaction with host, p 149–202 In Mettenleiter TC, Sobrino F. (ed), Animal viruses. Molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 9. Galán C, et al. 2009. Host cell proteins interacting with the 3′ end of TGEV coronavirus genome influence virus replication. Virology 391:304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 17:4847–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiménez G, Correa I, Melgosa MP, Bullido MJ, Enjuanes L. 1986. Critical epitopes in transmissible gastroenteritis virus neutralization. J. Virol. 60:131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Knoops K, et al. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:1957–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kong Q, et al. 2010. Proteomic analysis of purified coronavirus infectious bronchitis virus particles. Proteome Sci. 8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martín Alonso JM, et al. 1992. Antigenic structure of transmissible gastroenteritis virus nucleoprotein. Virology 188:168–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Masters PS. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Neuman BW, et al. 2008. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 82:5279–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nogales A, et al. 2011. Immunogenic characterization and epitope mapping of transmissible gastroenteritis virus RNA dependent RNA polymerase. J. Virol. Methods 175:7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ortego J, Ceriani JE, Patiño C, Plana J, Enjuanes L. 2007. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology 368:296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Risco C, Antón IM, Enjuanes L, Carrascosa JL. 1996. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 70:4773–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Risco C, et al. 1995. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J. Virol. 69:5269–5277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodriguez MJ, et al. 2001. Identification of an immunodominant epitope in the C terminus of glycoprotein 5 of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 82:995–999 [DOI] [PubMed] [Google Scholar]
- 23. Schelle B, Karl N, Ludewig B, Siddell SG, Thiel V. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 79:6620–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Snijder EJ, et al. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sola I, Mateos-Gomez PA, Almazán F, Zuñiga S, Enjuanes L. 2011. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 8:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ziebuhr J. 2005. The coronavirus replicase, p 57–94 In Enjuanes L. (ed), Coronavirus replication and reverse genetics, vol 287 Springer-Verlag, Berlin, Germany [Google Scholar]
- 27. Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853–879 [DOI] [PubMed] [Google Scholar]
- 28. Zúñiga S, et al. 2010. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 84:2169–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]