Abstract
Massive infection of memory CD4 T cells is a hallmark of early simian immunodeficiency virus (SIV) infection, with viral infection peaking at day 10 postinfection (p.i.), when a majority of memory CD4 T cells in mucosal and peripheral tissues are infected. It is not clear if mononuclear cells from the monocyte and macrophage lineages are similarly infected during this early phase of explosive HIV and SIV infections. Here we show that, at day 10 p.i., Lin− HLA-DR+ CD11c/123− CD13+ CD14− macrophages in the jejunal mucosa were infected, albeit at lower levels than CD4 memory T cells. Interestingly, Lin− HLA-DR+ CD11c/123− CD13+ CD14− macrophages in peripheral blood, like their mucosal counterparts, were preferentially infected compared to Lin− HLA-DR+ CD11c/123− CD13+ CD14+ monocytes, suggesting that differentiated macrophages were selectively infected by SIV. CD13+ CD14− macrophages expressed low levels of CD4 compared to CD4 T cells but expressed similar levels of CCR5 as lymphocytes. Interestingly, CD13+ CD14− macrophages expressed Apobec3G at lower levels than CD13+ CD14+ monocytes, suggesting that intracellular restriction may contribute to the differential infection of mononuclear subsets. Taken together, our results suggest that CD13+ CD14− macrophages in mucosal and peripheral tissues are preferentially infected very early during the course of SIV infection.
INTRODUCTION
Mucosal tissues play a central role in human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) pathogenesis (4, 14, 18, 21, 31, 35, 37, 53, 58). Acute infection is characterized by massive infection of memory CD4 T cells in the mucosa that peaks as early as day 10 postinfection (p.i.) and is followed by a nearly total loss of these cells (31, 35). Interestingly, memory CD4 T cells in peripheral tissues are simultaneously infected and destroyed at the same rate (35). Though memory CD4 T cells serve as the primary targets for early viral infection, a number of other mononuclear cells have been shown to be potential targets for HIV infection.
Mononuclear cells such as monocytes and macrophages have been thought to constitute an important and long-lasting viral reservoir in the infected host (1, 6, 20, 24, 36, 40, 60). Changes in these cells have been shown to correlate with disease progression (23, 27, 28, 34, 57). Others have shown that the level of monocyte turnover predicted disease progression in SIV-infected rhesus macaques (5, 15). Igarashi et al. (16) showed that macrophages were a principal reservoir in rhesus macaques after the depletion of CD4 T cells during SHIV infection, whereas macrophage-tropic SHIV-SF162 has been shown to infect rhesus macaques efficiently (33). On the other hand, massive covert infection of macrophages by HIV has been shown to occur during the incubation period of AIDS (12). Macrophages in other mucosal tissues, such as the vaginal mucosa, have been shown to be targets for SIV infection (38, 39).
Mucosal macrophages have been shown to be productively infected in vivo and in vitro (46, 54, 55). However, studies have shown that mucosal macrophages were less permissive to HIV infection than CD4 T cells, likely due to their terminally differentiated phenotype (41, 51, 55). Human mucosal tissue macrophages are predominantly CD13+ CD14− CD16− CD64− CD89− CD32−, which is characteristic of a macrophage-like phenotype, whereas peripheral blood mononuclear cells (PBMC) of monocytic lineage had a predominantly CD14+ phenotype (55). Clayton et al. (7) demonstrated that mononuclear macrophages in the rectal mucosa were one of the most highly infected target cells during HIV infection. Though the role of mononuclear cells has been extensively studied during HIV and SIV infections, little is known about the in vivo kinetics of infection in CD13+ CD14+ and CD14− mononuclear cells very early during the course of infection.
The primary goal of this study was to determine if CD13+ CD14+ and CD14− mononuclear cells were infected at levels similar to CD4 T cells at the peak of infection and to examine if the level of infection in these cells differs from that seen in peripheral blood mononuclear cells. To address these questions, we evaluated the changes in the proportions of Lin− HLA-DR+ CD11c/123− CD13+ CD14+ and CD14− mononuclear cells in peripheral blood and jejunal mucosa and determined the level of SIV infection in these subsets at day 10 p.i. and in chronic stages of infection. Additionally, we evaluated the expression of Apobec3G to determine if intracellular restriction was associated with differential infection of CD14+ and CD14− mononuclear cell subsets. Our results show CD13+ CD14− mononuclear cells in both peripheral blood and mucosal tissues are preferentially infected very early during the course of viral infection.
MATERIALS AND METHODS
Animals, infection, and samples.
Rhesus macaques (Macaca mulatta) of Indian origin were used in this study. Animals were housed in accordance with American Association for Accreditation of Laboratory Animal Care guidelines and were seronegative for SIV, simian retrovirus, and simian T-cell leukemia virus type 1. All animal care and procedures were reviewed and approved by the Institutional Animal Care and Use Committee. Peripheral blood and jejunal mucosa were collected at necropsy from SIV-infected animals during acute (day 10 postinfection [p.i.]; n = 8) and chronic (>1 year p.i.; n = 7) stages of infection. Animals were infected with 1,000 50% tissue culture infective doses (TCID50) of uncloned pathogenic SIVmac251 intravenously or intrarectally. Additionally, blood (n = 8) and jejunal (n = 3) samples were obtained from healthy animals as uninfected controls. PBMC were isolated by density gradient centrifugation and used for subsequent experiments. Jejunal mucosa was processed using enzymatic digestion followed by Percoll gradient centrifugation to enrich for mononuclear cells. We were unable to obtain cumulative blood counts from all of the animals; hence, we were unable to determine the absolute numbers of the various subsets.
Antibodies and flow cytometry.
All antibodies used in this study, except for HLA-DR-ECD (Beckman Coulter), CD13-phycoerythrin (CD13-PE; e-Biosciences, San Diego, CA), and CD4-Pacific blue (CD4-PB) and CD20-PB (Biolegend, San Diego, CA), were obtained from BD Biosciences (San Diego, CA) and titrated using rhesus macaque PBMC. For phenotypic analysis and sorting of mononuclear subsets, isolated cells were labeled with the following antibodies: anti-CD3/CD8/CD20–PB; anti-HLA-DR–ECD; anti-CD11c/CD123–allophycocyanin (anti-CD11c/CD123–APC); anti-CD16-Cy7–APC, -CD13–PE, -CD64-PE, -CCR5–PE, or -CD4–Alexa Fluor 700 and anti-CD14–fluorescein isothiocyanate (anti-CD14–FITC). Additionally, memory CD4 T cells were sorted based on the expression of CD28 and CD95 using the following combination of antibodies: anti-CD3-Cy7–APC, anti-CD4–PB, anti-CD8–Alexa Fluor 700, anti-CD28-Cy5–PE, and anti-CD95–FITC. Negative controls were stained with isotype control antibodies.
Labeled cells were fixed in 0.5% paraformaldehyde and analyzed using a Becton Dickinson (BD) LSR II. Fixed cells were sorted for quantification of viral DNA using a BD Aria sorter. Live CD4 T cells, total Lin− HLA-DR+ CD11c/123− cells, and Lin− HLA-DR+ CD11c/123− CD14+ and CD14− mononuclear cells were sorted and used for evaluating the expression levels of Apobec3G and CD4 mRNA. Due to the small number of cells recovered, CD14− and CD14+ subsets from four animals were individually pooled to determine the expression of mRNA.
Viral loads.
Plasma viral loads were determined by real-time reverse transcription-PCR (RT-PCR) assay (32). CD4 T-cell-associated viral DNA was measured by a quantitative PCR (qPCR) assay for SIV gag using SIV gag primers and a probe (32, 35). Albumin primers were used simultaneously to quantify the number of cells. The assay was calibrated using a cell line that carried a single copy of proviral SIV DNA as described previously (35). The SIV gag copy number is reported as number of SIV gag copies/100,000 cells.
Relative qPCR for Apobec3G and CD4.
RNA was isolated from purified populations of CD4 T cells and mononuclear cells using the RNeasy kit (Qiagen Sciences, Gaithersburg, MD). Isolated RNA was treated with Ambion Turbo DNase (Applied Biosystems, Austin, TX) to remove contaminating DNA. Purified RNA was reverse transcribed with Superscript III first strand synthesis kit (Invitrogen, Carlsbad, CA) to make cDNA that was used to determine the expression of Apobec3G and CD4 using the ABI 7500 instrument (Applied Biosystems). TaqMan qPCR was performed using high-fidelity Platinum Taq polymerase (Invitrogen) with Macaca mulatta macaque-specific Apobec3G (GenBank accession number AY331716) primers Apobec3G-F (CTGTGCTTCCTGGACCTGAT) and Apobec3G-R (CCAGGAGGTGAAGCAGGTAA) and probe Apobec3G-P (6-carboxyfluorescein [FAM]-TGGAAGCTGGATGGCCAGCA-black hole quencher 1 [BHQ1]) and CD4 (GenBank accession number NM_001042662) primers CD4-F (TATGCTGGCTCTGGAAACCT) and CD4-R (TCCTGGAACTGAGTGGCTCT) and probe CD4-P (ACGCTGGCCCTTGAAGCGAA); qPCR was normalized to the Macaca mulatta β2-M (GenBank accession number AY349163) housekeeping gene using the following β2-M-specific primers and probes: β2-M-F (GCTGGCGCTACTCTCTCTTTCT), β2-M-R (GGATGGCGTGAGTAAACCTGAA), and β2-M-P (FAM-CCTGGAGGCTATCCAGCGTACTCCAAAG-BHQ1). Collected data were analyzed using the 2−ΔΔCT (ddCT) method with ABI 7500 software, and fold differences were calculated as described previously (49).
Data analysis.
Flow cytometric data were analyzed using FlowJo version 8.6 (Tree Star, Inc., Ashland, OR). Statistical analysis was performed using the Mann-Whitney U test with GraphPad Prism version 4.0 software (GraphPad Prism Software, Inc., San Diego, CA). A P value of <0.05 was considered significant.
RESULTS
Chronic infection is associated with loss of peripheral and mucosal CD4 T cells.
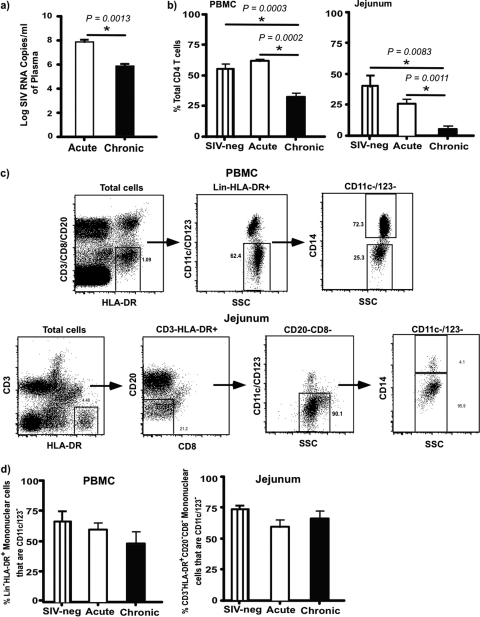
We first evaluated the plasma viral loads and proportions of CD4 T cells in peripheral blood and jejunal mucosa of SIV-infected animals and compared them to controls. Plasma viral loads were ∼8 logs/ml of plasma at day 10 p.i., whereas there were ∼6 logs/ml of plasma in chronically infected animals (Fig. 1a). There was a significant decrease in the proportion of total CD4 T cells in peripheral blood during chronic infection compared to those for uninfected and acutely infected animals (Fig. 1b). Likewise, as previously reported (4, 14, 18, 21, 31, 35, 37, 53, 58), chronic SIV infection was associated with a nearly total loss of CD4 T cells in the mucosa.
Fig 1.
Plasma viral loads and Lin− HLA-DR+ CD11c/123− mononuclear cell dynamics. (a) Plasma viral loads during acute SIV and chronic infection. Limit of detection is 30 copies/ml of plasma. (b) Percentage of CD4 T cells in peripheral blood mononuclear cells (PBMC) and jejunal mucosa in uninfected and acute and chronically SIV-infected animals. (c) Gating strategy used to identify Lin− HLA-DR+ CD11c/123− mononuclear cells in PBMC and jejunal mucosa. (d) Dynamics of Lin− HLA-DR+ CD11c/123− mononuclear cells in PBMC and jejunal mucosal of uninfected and acute and chronically SIV-infected animals.
Few Lin− HLA-DR+ CD11c/123− mononuclear cells are infected at day 10 p.i. compared to memory CD4 T cells.
Next we evaluated the effects of acute and chronic SIV infection on the dynamics of Lin− HLA-DR+ CD11c/123− mononuclear populations and infection in these cells from peripheral blood and compared them to those in the jejunal mucosa.
In line with previously published reports (15), the overall proportions of total Lin− HLA-DR+ CD11c/123− mononuclear cells identified based on the gating strategy (Fig. 1c and d) did not significantly change during acute and chronic infection compared to those for uninfected animals. As CD11c/CD123 are predominantly expressed on dendritic cells, we excluded all of the cells that expressed CD11c/123 on their surfaces to better delineate the effect of infection on mononuclear phagocyte populations.
Previous studies (35) have shown that the majority of memory CD4 T cells are infected and carry viral DNA at day 10 p.i. However, little is known about the level of infection in Lin− HLA-DR+ CD11c/123− mononuclear cells during the acute stages of infection. To determine the extent of SIV infection in these cells, we evaluated the levels of SIV gag DNA in total Lin− HLA-DR+ CD11c/123− mononuclear cells from peripheral blood and jejunal mucosa during acute and chronic infection and compared them to levels in CD4 memory T cells (Fig. 2).
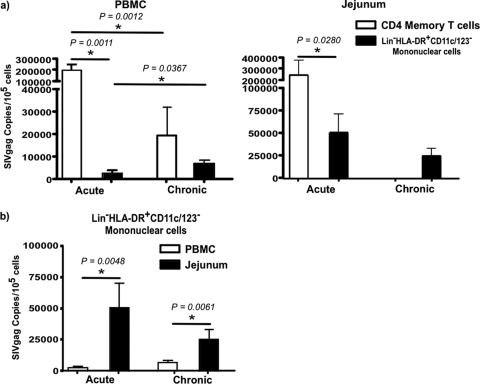
Fig 2.
Kinetics of cell-associated viral loads during SIV infection. (a) Level of SIV infection in memory CD4 T cells (CD95+ CD28+ and CD95+ 28−) and total Lin− HLA-DR+ CD11c/123− mononuclear cells in PBMC and jejunal mucosa during acute and chronic SIV infection. (b) Comparison of SIV infection of Lin− HLA-DR+ CD11c/123− mononuclear cells between PBMC and jejunal mucosa during acute and chronic infection.
In line with previous reports (35), at day 10 p.i., memory CD4 T cells in both peripheral blood and jejunal mucosa harbored ∼2 × 105 to 3 × 105 copies of SIV gag/105 cells, which significantly decreased to ∼2 × 104 copies of SIV gag/105 cells in peripheral blood memory CD4 T cells during chronic stages of disease (Fig. 2a). Due to the nearly total loss of CD4 T cells, few CD4 T cells could be recovered from jejunal mucosa of chronically infected animals for quantification of infection.
In contrast to memory CD4 T cells, few Lin− HLA-DR+ CD11c/123− mononuclear cells in peripheral blood carried viral DNA; at day 10 p.i., Lin− HLA-DR+ CD11c/123− mononuclear cells contained ∼2 × 103 to 5 × 103 copies of SIV gag/105 cells. The level of infection significantly increased to ∼1 × 104 to 2 × 104 SIV gag copies/105 cells during chronic infection. Interestingly, this level of infection did not significantly differ from that seen in memory CD4 T cells (Fig. 2a and b).
Compared to peripheral blood Lin− HLA-DR+ CD11c/123− mononuclear cells, jejunal Lin− HLA-DR+ CD11c/123− mononuclear cells were found to contain significantly higher levels of infection; at day 10 p.i., jejunal Lin− HLA-DR+ CD11c/123− mononuclear cells harbored ∼5 to 10 times more SIV gag DNA than peripheral blood. The level of infection in jejunal Lin− HLA-DR+ CD11c/123− mononuclear cells remained high during chronic infection and was significantly higher than that found in peripheral blood Lin− HLA-DR+ CD11c/123− mononuclear cells of chronically infected animals (Fig. 2a and b).
Mucosal and peripheral blood Lin− HLA-DR+ CD11c/123− mononuclear cells are CD13+ but differentially express CD14.
Next we sought to determine if Lin− HLA-DR+ CD11c/123− mononuclear cells found in peripheral blood were phenotypically different from those found in the jejunal mucosa. To address this question, we evaluated the expression of CD14, CD16, and CD64 on peripheral blood Lin− HLA-DR+ CD11c/123− mononuclear cells and compared it to that on jejunal Lin− HLA-DR+ CD11c/123− mononuclear cells (Fig. 3a and b).
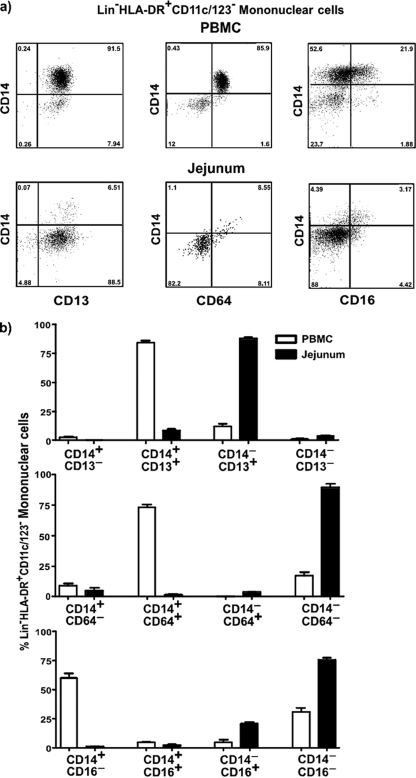
Fig 3.
Lin− HLA-DR+ CD11c/123− CD14− subsets express a predominantly CD13+ CD16− CD64− phenotype. (a) Representative dot plots showing the expression of CD13, CD64, and CD16 on Lin− HLA-DR+ CD11c/123− CD14+ and CD14− subsets in PBMC and jejunal mucosa of SIV-negative animals. (b) Proportions of Lin− HLA-DR+ CD11c/123− CD14+ and CD14− subsets in PBMC and jeunual mucosa expressing CD13, CD64, and CD16 in SIV-negative animals.
Our results showed that ∼75% of peripheral blood Lin− HLA-DR+ CD11c/123− mononuclear cells were CD14+, with the rest of them being CD14−. In contrast to peripheral blood, ∼95% of jejunal Lin− HLA-DR+ CD11c/123− mononuclear cells expressed a CD14− phenotype.
To confirm if CD14− mononuclear cells represent myeloid cells, we evaluated the expression of CD13 on CD14− mononuclear cells and compared it to expression on CD14+ cells (Fig. 3a and b). Nearly all of the CD14+ and CD14− mononuclear cells in peripheral blood and jejunum expressed CD13, suggesting that CD13+ CD14− mononuclear cells likely represent differentiated macrophages as previous studies in humans have shown (55).
Next we evaluated levels of expression of CD64 on Lin− HLA-DR+ CD11c/123− CD14+ and CD14− mononuclear cells in peripheral blood and compared them to the level in the jejunum from SIV-negative animals. Our results showed that Lin− HLA-DR+ CD11c/123− CD14− mononuclear cells in both peripheral blood and jejunal mucosa were predominantly CD64− whereas most of the Lin− HLA-DR+ CD11c/123− CD14+ mononuclear cells expressed CD64. Previous studies (55) have shown that human small intestinal macrophages were predominantly CD64−.
Earlier studies (55) have shown that CD14+ and CD14− mononuclear cells harbor a heterogeneous mix of subsets that can be discriminated based on the expression of CD16. A majority of the Lin− HLA-DR+ CD11c/123− CD14+ and CD14− subsets in both peripheral blood and jejunum were CD16−.
Lin− HLA-DR+ CD11c/123− CD14− CD16− mononuclear cells are significantly decreased in peripheral blood during chronic infection.
Next we sought to determine if SIV infection was accompanied by alterations in the proportions of Lin− HLA-DR+ CD11c/123− mononuclear cells in peripheral blood and jejunal mucosa. To address this question, we first evaluated the proportions of CD14+ and CD14− mononuclear cells in peripheral blood and jejunal mucosa of SIV-negative animals and compared them to those for SIV-infected animals (Fig. 4a).
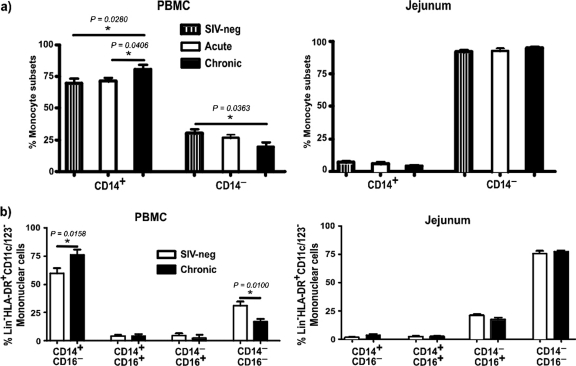
Fig 4.
Dynamics of CD14+ and CD14− subsets in PBMC and jejunal mucosa. (a) Proportions of Lin− HLA-DR+ CD11c/123− CD14+ and CD14− subsets of mononuclear cells in PBMC and jejunal mucosa of SIV-negative and acute and chronically infected animals. (b) Expression of CD16 on CD14+ and CD14− subsets in uninfected and chronically infected animals.
Chronic SIV infection was associated with a significant increase in the proportion of CD14+ mononuclear cells in peripheral blood and a coincident decrease in the proportion of CD14− mononuclear cells. No major changes were observed during acute infection.
To determine if the specific subsets of CD14+ and CD14− mononuclear cells in peripheral blood were altered during chronic infection, we evaluated the dynamics of CD16 expression on CD14+ and CD14− Lin− HLA-DR+ CD11c/123− mononuclear cells during chronic infection and compared them to those for uninfected animals (Fig. 4b).
No major changes in the proportions of CD14+ CD16+ and CD14− CD16+ subsets were observed during chronic infection compared to those for uninfected animals. However, chronic SIV infection was associated with a significant increase in the proportions of CD14+ CD16− mononuclear cells, whereas the proportions of CD14− CD16− mononuclear cells significantly declined compared to those for uninfected animals.
Lin− HLA-DR+ CD11c/123− CD14− mononuclear cells harbor higher levels of infection than CD14+ mononuclear cells.
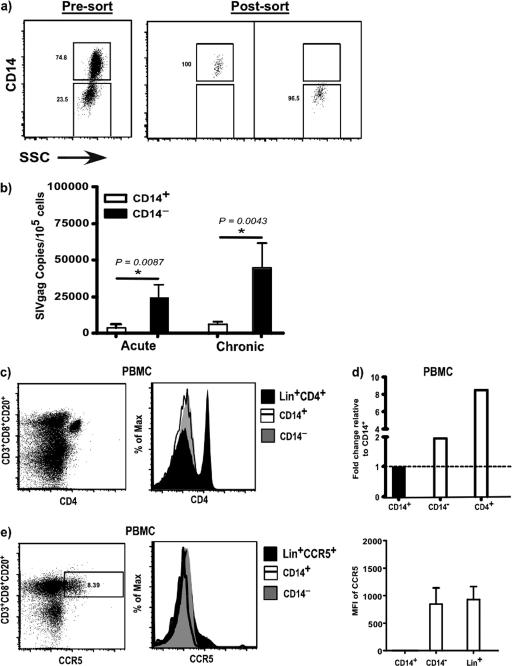
The predominance of Lin− HLA-DR+ CD11c/123− CD14− mononuclear cells in the jejunal mucosa accompanied by higher levels of infection in these subsets raised the possibility that CD14− subsets in peripheral blood may be preferentially infected compared to CD14+ subsets. To address this possibility, we sorted Lin− HLA-DR+ CD11c/123− CD14+ and CD14− subsets from peripheral blood (Fig. 5a) and quantified the level of infection (Fig. 5b) in these subsets. Since a majority of the jejunal mononuclear cells were Lin− HLA-DR+ CD11c/123− CD14− and few CD14+ cells could be recovered from the jejunal mucosa, we restricted our analysis to peripheral blood. Our results showed CD14− subsets in peripheral blood, like their mucosal counterparts, harbored significantly higher levels of SIV gag DNA than the CD14+ subsets. The level of infection in CD14− subsets did not significantly change during chronic infection compared to acute infection.
Fig 5.
Lin− HLA-DR+ CD11c/123− CD14− mononuclear cells are preferentially infected compared to CD14+ subsets. (a) Sort purity of Lin− HLA-DR+ CD11c/123− CD14+ and CD14− subsets. (b) Level of SIV infection in CD14+ and CD14− subsets of Lin− HLA-DR+ CD11c/123− mononuclear cells in PBMC of acute and chronically infected animals. (c) Representative dot plot showing CD4 expression on Lin+ cells and representative histograms showing the expression of CD4 on CD14+ and CD14− subsets of Lin− HLA-DR+ CD11c/123− mononuclear cells and Lin+ CD4+ T cells in PBMC from uninfected animals. (d) Expression of CD4 mRNA in CD14− subsets and CD4+ T cells relative to CD14+ subsets from PBMC of uninfected animals. The line represents baseline. (e) Representative dot plot showing CCR5 expression on Lin+ cells and representative histograms showing the expression of CCR5 on CD14+ and CD14− subsets of Lin− HLA-DR+ CD11c/123− mononuclear cells and Lin+ cells in PBMC from uninfected animals. The MFIs of CCR5 expression on CD14+ and CD14− subsets and Lin+ cells are shown.
Lin− HLA-DR+ CD11c/123− CD14− mononuclear cells express low levels of CD4.
Next we sought to determine if the preferential infection of CD14− subsets was associated with differences in the expression of CD4 and CCR5, the primary receptor and coreceptors for SIVmac251, on these subsets. To address this question, we evaluated the expression of CD4 on CD14+ and CD14− subsets in peripheral blood by flow cytometry and compared them to CD4 expression on Lin+ (CD3+ CD20+ CD8+) CD4+ T cells (Fig. 5c).
Neither the CD14+ nor the CD14− subsets expressed detectable CD4 on their surfaces compared to CD4 T cells. The expression of CD4 on CD14+ and CD14− subsets did not change following SIV infection. Given the CD4-tropic nature of SIVmac251, we hypothesized that CD14− subsets likely express sufficient CD4 for SIV to infect these cells but at levels that were not easily detectable by flow cytometric analysis. To address this question, we evaluated the level of CD4 mRNA in CD14− subsets of healthy animals and compared it to levels in CD14+ subsets and CD4+ T cells (Fig. 5d). Our results showed that CD14− subsets express twice the level of CD4 mRNA as CD14+ subsets but at levels that were 6-fold less than those expressed by CD4+ T cells.
Next we assessed if the differential infection was associated with differences in the expression of CCR5 on CD14+ and CD14− subsets in healthy animals using flow cytometry and compared the expression levels to levels in lymphocytes (Fig. 5e). No CCR5 was detectable on CD14+ subsets, whereas CD14− subsets were found to express CCR5 at levels similar to that of Lin+ lymphocytes. The gates were set based on the expression of CCR5 on Lin+ lymphocytes, and mean fluorescence intensity (MFI) values were determined based on isotype controls.
Lin− HLA-DR+ CD11c/123− mononuclear cells express high levels of Apobec3G compared to CD4 T cells.
Previous studies (11, 54, 55) have suggested that the difference in infection between CD4 T cells and mononuclear phagocytes was likely due to the low permissiveness of these cells to infection. We hypothesized that this low permissiveness may be due to higher levels of intracellular restriction factors such as Apobec3G.
To determine if Apobec3G levels play a role in the differential infection of Lin− HLA-DR+ CD11c/123− mononuclear cells, we first evaluated the expression of Apobec3G in total Lin− HLA-DR+ CD11c/123− mononuclear cells from peripheral blood and compared it to expression in CD4 T cells (Fig. 6a). Due to the lack of sufficient mucosal samples, we restricted our analysis to peripheral blood.
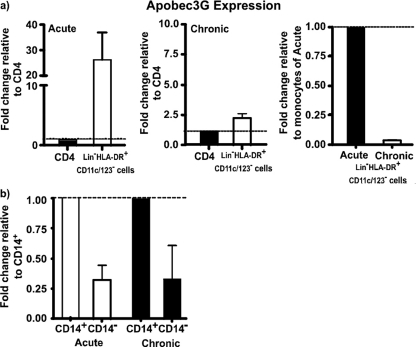
Fig 6.
Lin− HLA-DR+ CD11c/123− CD14− mononuclear cells express lower levels of Apobec3G than CD14+ subsets. Shown is expression of Apobec3G mRNA in total Lin− HLA-DR+ CD11c/123− mononuclear cells relative to CD4 T cells of acute and chronically infected animals (a) and in Lin− HLA-DR+ CD11c/123− CD14− subsets relative to CD14+ subsets of acute and chronically infected animals (b). Data from three repeat experiments are shown. The lines represent baseline.
At day 10 p.i., total Lin− HLA-DR+ CD11c/123− mononuclear cells were found to have ∼30 times more Apobec3G mRNA than memory CD4 T cells; Apobec3G levels declined significantly in chronically infected animals. This suggests that Apobec3G levels likely contribute to the differential permissiveness of Lin− HLA-DR+ CD11c/123− mononuclear cells to SIV infection.
Next we evaluated if the significantly higher levels of infection in peripheral blood Lin− HLA-DR+ CD11c/123− CD14− macrophages than in CD14+ monocytes were associated with differential expression of Apobec3G. Lin− HLA-DR+ CD11c/123− CD14+ macrophages and CD14− monocytes from four animals were sorted and used for analysis. Since the number of cells recovered from the Lin− HLA-DR+ CD11c/123− CD14− mononuclear subsets was low, approximately similar numbers of cells from each subset from the 4 animals were pooled prior to RNA isolation and pooled RNA was used for subsequent qPCR analysis. As shown in Fig. 6b, both during acute and chronic infections, Lin− HLA-DR+ CD11c/123− CD14− mononuclear cells had lower levels of Apobec3G than CD14+ mononuclear cells.
DISCUSSION
Mucosal tissues are extensively involved in HIV pathogenesis and constitute a major site for latent infection. Most of the primary target cells in the mucosa, namely, the CD4 T cells, are depleted within weeks after infection. Our data show that, like the CD4 T cells, Lin− HLA-DR+ CD11c/123− CD14− macrophages in the mucosa are infected very early during the course of infection, albeit at a level lower than memory CD4 T cells. Interestingly, like their mucosal counterparts, Lin− HLA-DR+ CD11c/123− CD14− macrophages in peripheral blood were found to be preferentially infected, suggesting that CD14− mononuclear cells serve as early targets for SIV infection. Our results support previous reports showing that the CD14− CD16+ subset of monocytes was the major infected population in peripheral blood during HIV and SIV infections (11, 13, 17, 19, 29, 42, 46, 62).
Interestingly, chronic infection was associated with a significant mobilization of CD14+ mononuclear cells in peripheral blood. CD14+ mononuclear cells develop in the bone marrow and migrate into peripheral blood, with recent migrants expressing a predominantly CD14+ CD16− phenotype. The significant increase in the proportions of CD14+ CD16− mononuclear cells during chronic infection may be due to the high level of turnover during chronic infection as previous studies have reported (15). Kim et al. (22) demonstrated that CD14hi subsets of mononuclear cells were significantly elevated during acute and chronic SIV infection.
The significantly lower level of infection in Lin− HLA-DR+ CD11c/123− CD14− macrophages than in memory CD4 T cells suggests that there are likely inherent differences in permissiveness between these two cellular compartments to SIV infection. In fact, CD14− mononuclear cells, compared to CD4 T cells, expressed much lower levels of CD4 on their surfaces whereas the levels of expression of CCR5 were similar between the two compartments. The highly CD4- and CCR5-tropic nature of SIV infection (3, 9) suggests that CD4 expression likely plays a key role in limiting infection in CD14− mononuclear cells compared to CD4 T cells. Numerous studies have shown that CD4 expression plays a critical role in HIV and SIV infection of macrophages (30, 44, 45). Kozak et al. (26) showed that HIV binds weakly to cells expressing low levels of CD4, whereas low levels of CD4 expression on macrophages played a role in restricting the entry of T-tropic SIV strains such as SIVmac239 (2), which could not be overcome by the overexpression of CCR5. Likewise Kozak et al. (25) found that CD4 rather than CCR5 or CXCR4 expression determines the kinetics and pathways for gp120 binding, endocytosis, and proteolysis on cells that contain sufficient coreceptors for efficient infection. On the other hand, Platt et al. (45) showed that cells with high expression of CD4 and low levels of CCR5 were sufficient for maximum susceptibility to HIV-1 strains, whereas Pesenti et al. (44) demonstrated that susceptibility of macrophages to HIV-1 was highly increased when they expressed higher levels of CD4 than macrophages that expressed low levels of CD4, with no significant difference in susceptibility based on CCR5 expression. Recent studies (52) have shown that low levels of CD4/CCR5 likely contribute to the low permissiveness of intestinal macrophages to HIV infection. Human small intestinal macrophages have been shown to express low levels of CD4 (54, 55), and minimal levels of CCR5 were sufficient for efficient infection of monocytes and macrophages (44, 45) and T cells (10, 18, 35). Though CD4 and CCR5 expression may explain the differential expression in macrophage and monocyte subsets, our data do not rule out other mechanisms that SIV may use to infect these cells.
The low permissiveness of Lin− HLA-DR+ CD11c/123− CD14− mononuclear cells to SIV infection compared to that of memory CD4 T cells may also be due to differences in intracellular restriction factors such as Apobec3G. Numerous studies (8, 50, 59, 61) have shown that Apobec3G could restrict SIV infection, whereas Sui et al. (56) demonstrated that plasma viral loads inversely correlated with Apobec3G levels. Saez-Cirion et al. (48) showed that intracellular restriction contributes to the intrinsic cell resistance of monocytes to HIV-1 infection, whereas Peng et al. (43) showed that monocytes contain higher levels of Apobec3G than macrophages. In line with this argument, we found low levels of Apobec3G in CD4 T cells compared to those in Lin− HLA-DR+ CD11c/123− mononuclear cells. Likewise, CD14− macrophages expressed lower levels of Apobec3G than CD14+ monocytes. Though the relative differences in Apobec3G mRNA are not directly indicative of protein level differences, our data seem to suggest that lower levels of Apobec3G may contribute to the lower level of infection in CD14− subsets. Additional studies are, however, needed to address this question in more detail.
A number of other mechanisms have also been shown to limit infection in macrophages during SIV infections. Sacha et al. (47) showed that SIV gag and nef-specific CD4 T cells displayed direct a effector function in eliminating macrophages. Though we did not address this specific question in our study, the results reported here suggest that multiple mechanisms likely play a role in limiting the extent of infection in macrophages.
Taken together, our data provide new insights into effect of SIV infection on the dynamics of infection in Lin− HLA-DR+ CD11c/123− CD14− macrophages in peripheral blood and mucosal tissues and demonstrate that a significant fraction of differentiated CD14− macrophages are infected very early during the course of infection. The low level of CD4 expression along with differential intracellular restriction likely plays a role in limiting infection in CD14− mononuclear subsets compared to CD4 T cells. The relative abundance of differentiated CD14− mononuclear subsets in the mucosa, however, suggests that these cells likely play a major role disease pathogenesis and progression.
ACKNOWLEDGMENTS
We thank Jeffy George, Olusegun Onabajo, and Sean Maynard at the Uniformed Services University for assistance with processing some of the samples and Karen Wolcott and Kateryna Lund at the Biomedical Instrumentation Center. We thank Matt Collins at Bioqual, Inc., Rockville, MD, for expert assistance with the animals, Jeffrey Lifson and Michael Piatak at NCI for their help in determining plasma viral loads, and Bernard Lafont at NIAID for valuable suggestions.
A.C.M. and S.L.B. performed the experiments, analyzed the data, and assisted in writing the paper; D.V. and M.L. provided the samples and assisted in writing the paper; J.J.M. designed and supervised the study and wrote the paper.
We declare no financial conflict of interest.
The project was supported by AI07812, DE018339, and DE019397 awarded to J.J.M. Studies were supported in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract NO1-CO-124000.
The content is solely the responsibility of the authors and does not necessarily represent the official views of NIAID, NIDCR, NCI, or the National Institutes of Health.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Alexaki A, Liu Y, Wigdahl B. 2008. Cellular reservoirs of HIV-1 and their role in viral persistence. Curr. HIV Res. 6:388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bannert N, Schenten D, Craig S, Sodroski J. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J. Virol. 74:10984–10993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berger EA. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl. A):S3–S16 [PubMed] [Google Scholar]
- 4. Brenchley JM, et al. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200:749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burdo TH, et al. 2010. Increased monocyte turnover from bone marrow correlates with severity of SIV encephalitis and CD163 levels in plasma. PLoS Pathog. 6:e1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chun TW, et al. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188 [DOI] [PubMed] [Google Scholar]
- 7. Clayton F, et al. 1992. Rectal mucosal pathology varies with human immunodeficiency virus antigen content and disease stage. Gastroenterology 103:919–933 [DOI] [PubMed] [Google Scholar]
- 8. Depboylu C, Eiden LE, Weihe E. 2007. Increased APOBEC3G expression is associated with extensive G-to-A hypermutation in viral DNA in rhesus macaque brain during lentiviral infection. J. Neuropathol. Exp. Neurol. 66:901–912 [DOI] [PubMed] [Google Scholar]
- 9. Doms RW, Peiper SC. 1997. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology 235:179–190 [DOI] [PubMed] [Google Scholar]
- 10. Eberly MD, et al. 2009. Increased IL-15 production is associated with higher susceptibility of memory CD4 T cells to simian immunodeficiency virus during acute infection. J. Immunol. 182:1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ellery PJ, et al. 2007. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J. Immunol. 178:6581–6589 [DOI] [PubMed] [Google Scholar]
- 12. Embretson J, et al. 1993. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature 362:359–362 [DOI] [PubMed] [Google Scholar]
- 13. Fischer-Smith T, et al. 2001. CNS invasion by CD14+/CD16+ peripheral blood-derived monocytes in HIV dementia: perivascular accumulation and reservoir of HIV infection. J. Neurovirol. 7:528–541 [DOI] [PubMed] [Google Scholar]
- 14. Guadalupe M, et al. 2003. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J. Virol. 77:11708–11717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hasegawa A, et al. 2009. The level of monocyte turnover predicts disease progression in the macaque model of AIDS. Blood 114:2917–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Igarashi T, et al. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc. Natl. Acad. Sci. U. S. A. 98:658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jaworowski A, et al. 2007. CD16+ monocyte subset preferentially harbors HIV-1 and is expanded in pregnant Malawian women with Plasmodium falciparum malaria and HIV-1 infection. J. Infect. Dis. 196:38–42 [DOI] [PubMed] [Google Scholar]
- 18. Kader M, et al. 2009. a4+b7hi CD4+ memory T cells harbor most Th-17 cells and are preferentially infected during acute SIV infection. Mucosal Immunol. 2:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kedzierska K, Crowe SM. 2002. The role of monocytes and macrophages in the pathogenesis of HIV-1 infection. Curr. Med. Chem. 9:1893–1903 [DOI] [PubMed] [Google Scholar]
- 20. Kelly J, et al. 2008. Human macrophages support persistent transcription from unintegrated HIV-1 DNA. Virology 372:300–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kewenig S, et al. 1999. Rapid mucosal CD4(+) T-cell depletion and enteropathy in simian immunodeficiency virus-infected rhesus macaques. Gastroenterology 116:1115–1123 [DOI] [PubMed] [Google Scholar]
- 22. Kim WK, et al. 2010. Monocyte heterogeneity underlying phenotypic changes in monocytes according to SIV disease stage. J. Leukoc. Biol. 87:557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitagawa M, Lackner AA, Martfeld DJ, Gardner MB, Dandekar S. 1991. Simian immunodeficiency virus infection of macaque bone marrow macrophages correlates with disease progression in vivo. Am. J. Pathol. 138:921–930 [PMC free article] [PubMed] [Google Scholar]
- 24. Koenig S, et al. 1986. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science 233:1089–1093 [DOI] [PubMed] [Google Scholar]
- 25. Kozak SL, Kuhmann SE, Platt EJ, Kabat D. 1999. Roles of CD4 and coreceptors in binding, endocytosis, and proteolysis of gp120 envelope glycoproteins derived from human immunodeficiency virus type 1. J. Biol. Chem. 274:23499–23507 [DOI] [PubMed] [Google Scholar]
- 26. Kozak SL, et al. 1997. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J. Virol. 71:873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuroda MJ. 2010. Macrophages: do they impact AIDS progression more than CD4 T cells? J. Leukoc. Biol. 87:569–573 [DOI] [PubMed] [Google Scholar]
- 28. Kuwata T, et al. 2007. Contribution of monocytes to viral replication in macaques during acute infection with simian immunodeficiency virus. AIDS Res. Hum. Retroviruses 23:372–380 [DOI] [PubMed] [Google Scholar]
- 29. Lewin SR, et al. 1998. HIV-1 DNA and mRNA concentrations are similar in peripheral blood monocytes and alveolar macrophages in HIV-1-infected individuals. AIDS 12:719–727 [DOI] [PubMed] [Google Scholar]
- 30. Lewin SR, et al. 1996. Surface CD4 is critical to in vitro HIV infection of human alveolar macrophages. AIDS Res. Hum. Retroviruses 12:877–883 [DOI] [PubMed] [Google Scholar]
- 31. Li Q, et al. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434:1148–1152 [DOI] [PubMed] [Google Scholar]
- 32. Lifson JD, et al. 2001. Role of CD8+ lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 75:10187–10199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luciw PA, Pratt-Lowe E, Shaw KE, Levy JA, Cheng-Mayer C. 1995. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. U. S. A. 92:7490–7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mandell CP, Jain NC, Miller CJ, Dandekar S. 1995. Bone marrow monocyte/macrophages are an early cellular target of pathogenic and nonpathogenic isolates of simian immunodeficiency virus (SIVmac) in rhesus macaques. Lab. Invest. 72:323–333 [PubMed] [Google Scholar]
- 35. Mattapallil JJ, et al. 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 434:1093–1097 [DOI] [PubMed] [Google Scholar]
- 36. McElrath MJ, Pruett JE, Cohn ZA. 1989. Mononuclear phagocytes of blood and bone marrow: comparative roles as viral reservoirs in human immunodeficiency virus type 1 infections. Proc. Natl. Acad. Sci. U. S. A. 86:675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mehandru S, et al. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200:761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller CJ. 1998. Localization of simian immunodeficiency virus-infected cells in the genital tract of male and female rhesus macaques. J. Reprod. Immunol. 41:331–339 [DOI] [PubMed] [Google Scholar]
- 39. Miller CJ, Shattock RJ. 2003. Target cells in vaginal HIV transmission. Microbes Infect. 5:59–67 [DOI] [PubMed] [Google Scholar]
- 40. Montaner LJ, et al. 2006. Advances in macrophage and dendritic cell biology in HIV-1 infection stress key understudied areas in infection, pathogenesis, and analysis of viral reservoirs. J. Leukoc. Biol. 80:961–964 [DOI] [PubMed] [Google Scholar]
- 41. Montaner LJ, et al. 1993. Interleukin 13 inhibits human immunodeficiency virus type 1 production in primary blood-derived human macrophages in vitro. J. Exp. Med. 178:743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Orenstein JM, Fox C, Wahl SM. 1997. Macrophages as a source of HIV during opportunistic infections. Science 276:1857–1861 [DOI] [PubMed] [Google Scholar]
- 43. Peng G, et al. 2007. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood 110:393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pesenti E, et al. 1999. Role of CD4 and CCR5 levels in the susceptibility of primary macrophages to infection by CCR5-dependent HIV type 1 isolates. AIDS Res. Hum. Retroviruses 15:983–987 [DOI] [PubMed] [Google Scholar]
- 45. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rich EA, Chen IS, Zack JA, Leonard ML, O'Brien WA. 1992. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1 (HIV-1). J. Clin. Invest. 89:176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sacha JB, et al. 2009. Gag- and Nef-specific CD4+ T cells recognize and inhibit SIV replication in infected macrophages early after infection. Proc. Natl. Acad. Sci. U. S. A. 106:9791–9796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saez-Cirion A, et al. 2011. Restriction of HIV-1 replication in macrophages and CD4+ T cells from HIV controllers. Blood 118:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108 [DOI] [PubMed] [Google Scholar]
- 50. Schrofelbauer B, Senger T, Manning G, Landau NR. 2006. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 80:5984–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schuitemaker H, et al. 1992. Proliferation-dependent HIV-1 infection of monocytes occurs during differentiation into macrophages. J. Clin. Invest. 89:1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shen R, et al. 2011. Stromal down-regulation of macrophage CD4/CCR5 expression and NF-kappaB activation mediates HIV-1 non-permissiveness in intestinal macrophages. PLoS Pathog. 7:e1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smit-McBride Z, Mattapallil JJ, McChesney M, Ferrick D, Dandekar S. 1998. Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4+ T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J. Virol. 72:6646–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smith PD, Fox CH, Masur H, Winter HS, Alling DW. 1994. Quantitative analysis of mononuclear cells expressing human immunodeficiency virus type 1 RNA in esophageal mucosa. J. Exp. Med. 180:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith PD, Meng G, Shaw GM, Li L. 1997. Infection of gastrointestinal tract macrophages by HIV-1. J. Leukoc. Biol. 62:72–77 [DOI] [PubMed] [Google Scholar]
- 56. Sui Y, et al. 2010. Innate and adaptive immune correlates of vaccine and adjuvant-induced control of mucosal transmission of SIV in macaques. Proc. Natl. Acad. Sci. U. S. A. 107:9843–9848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tajima Y, et al. 2009. Use of a modified alpha-N-acetylgalactosaminidase in the development of enzyme replacement therapy for Fabry disease. Am. J. Hum. Genet. 85:569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Veazey RS, et al. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280:427–431 [DOI] [PubMed] [Google Scholar]
- 59. Williams KC, Burdo TH. 2009. HIV and SIV infection: the role of cellular restriction and immune responses in viral replication and pathogenesis. Apmis 117:400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Williams KC, et al. 2001. Perivascular macrophages are the primary cell type productively infected by simian immunodeficiency virus in the brains of macaques: implications for the neuropathogenesis of AIDS. J. Exp. Med. 193:905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu Q, et al. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379–53386 [DOI] [PubMed] [Google Scholar]
- 62. Zhu T. 2002. HIV-1 in peripheral blood monocytes: an underrated viral source. J. Antimicrob. Chemother. 50:309–311 [DOI] [PubMed] [Google Scholar]