Abstract
Sequence analysis of duck hepatitis virus type 1 (DHV-1) led to its classification as the only member of a new genus, Avihepatovirus, of the family Picornaviridae, and so was renamed duck hepatitis A virus (DHAV). The 5′ untranslated region (5′ UTR) plays an important role in translation initiation and RNA synthesis of the picornavirus. Here, we provide evidence that the 651-nucleotide (nt)-long 5′ UTR of DHAV genome contains an internal ribosome entry site (IRES) element that functions efficiently in vitro and within BHK cells. Comparative sequence analysis showed that the 3′ part of the DHAV 5′ UTR is similar to the porcine teschovirus 1 (PTV-1) IRES in sequence and predicted secondary structure. Further mutational analyses of the predicted domain IIId, domain IIIe, and pseudoknot structure at the 3′ end of the DHAV IRES support our predicted secondary structure. However, unlike the case for the PTV-1 IRES element, analysis of various deletion mutants demonstrated that the optimally functional DHAV IRES element with a size of approximately 420 nt is larger than that of PTV-1 and contains other peripheral domains (Id and Ie) that do not exist within the type IV IRES elements. The domain Ie, however, could be removed without significant loss of activity. Surprisingly, like the hepatitis A virus (HAV) IRES element, the activity of DHAV IRES could be eliminated by expression of enterovirus 2A protease. These findings indicate that the DHAV IRES shares common features with type IV picornavirus IRES elements, whereas it exhibits significant differences from type IV IRESs. Therefore, we propose that DHAV possesses a distinct type IV IRES element of picornavirus.
INTRODUCTION
The internal ribosome entry site (IRES) elements have the function to direct cap-independent internal initiation of protein synthesis, which is mechanistically quite different from the canonical cap-dependent mechanism for the translation initiation of the majority of cellular mRNAs (3). Earlier studies have indicated that all cytoplasmic cellular mRNAs possess a 5′ terminal cap structure (m7GpppG…) that is recognized by the translation initiation factor complex—eukaryotic initiation factor 4 (eIF4F), which contains three proteins including eIF4E (cap-binding protein); eIF4A (ATP-dependent RNA helicase); and eIF4G, which acts as a protein scaffold. This complex of proteins acts as a bridge between the mRNA and the 40S ribosomal subunit via its interaction with eIF3 by multiple protein-protein interactions (23). In contrast to the cellular mRNAs, no cap structure exists at the 5′ terminus of some viral mRNAs, such as those of picornaviruses and relevant viruses like hepatitis C virus (HCV). Initiation of protein synthesis for these viruses which rely on cap structure is termed internal initiation; in this case, the IRES element located within the 5′ untranslated region (5′ UTR) of the viral genome can exert the function to direct translation initiation (4, 19). Previous studies have showed that these IRES elements contain diverse secondary structures with sizes ranging from 280 to 450 nucleotides (nt) and are able to interact with various cellular proteins (3). The majority of these elements can function without the requirement of eIF4E and remain able to work even though the synthesis of cap-dependent protein is inhibited.
For picornavirus, their IRES elements are currently divided into four groups based on distinct secondary structures and biological properties (38). One group (type I) contains the IRES elements from entero-/rhinoviruses, with poliovirus (PV) as the prototype. The type II IRES element contains the cardio-/aphthovirus IRES elements, with encephalomyocarditis virus (EMCV) and foot-and-mouth disease virus (FMDV) as prototypes. The secondary structure of the type I IRES element appears completely different from that of the type II IRES element. Previous experiments showed that the type I IRES element functions poorly in the rabbit reticulocyte lysate (RRL) in vitro translation system unless it is supplemented with extracts from HeLa cells, while the type II IRES element can function very efficiently in this system (18). The type III IRES element has been identified only in hepatitis A virus (HAV), showing a distinct secondary structure and low activity compared to the IRES of EMCV both in vitro and within cells (11). When the entero- and rhinovirus 2A or aphthovirus L protease cleaved eIF4G to release the N terminus of eIF4G, including its eIF4E binding site, the type I and II picornavirus IRES elements can function efficiently (9, 34, 43). In some cell lines (e.g., BHK cells), the activity of the type I IRES element could be stimulated by the expression of the 2A protease (9, 43). The HAV IRES is unique among the IRESs of picornavirus, however, in that it is inactive under these conditions (8). It has been revealed that the activity of HAV IRES is sensitive to recombinant eIF4E-binding protein, which prevents the association of the cap-binding protein eIF4E to eIF4G and to cap analogue, suggesting that the unique feature of the HAV IRES is its requirement for the intact eIF4F complex (including eIF4E, eIF4G, and eIF4A) (1, 10). Several studies have confirmed that type I and II IRES elements also require the initiation factor eIF4A, because dominant negative mutants and inhibitors of eIF4A can block their activities (6, 14, 47). Recently, the type IV IRES element has been characterized in the 5′ UTR of porcine teschovirus 1 (PTV-1) (26), simian virus 2 (SV2), porcine enterovirus 8 (PEV-8) (13), simian picornavirus type 9 (SPV9) (15), avian encephalomyelitis virus (AEV) (2), and Seneca Valley virus (SVV) (52). These elements are highly similar to those of classical swine fever virus (CSFV) and hepatitis C virus, members of the Flaviviridae, and are predicted to share a very similar secondary structure that is critical for function, including the highly conserved domain IIIe and a pseudoknot near the 3′ end of the IRES (2, 14, 41). These IRES elements are generally shorter than other previously characterized picornavirus elements. The type I, II, and III IRES elements are about 450 nt in length, while the PTV-1 IRES is shorter than 280 nt (41). Unlike the type I and II IRES elements, the type IV IRES element not only functions without intact eIF4G (3, 38) but also is resistant to dominant negative mutants of eIF4A (13, 38) and hippuristanol, a small-molecule inhibitor of eIF4A (6). Similar to the IRES element of HCV, that of PTV-1 does not require any of the eIF4 initiation factors for assembly of 48S initiation complexes on the RNA (14, 26, 41).
Duck hepatitis virus type 1 (DHV-1) can cause an acute, contagious, rapidly spreading, and highly lethal infection in young ducklings of less than 4 weeks of age. This DHV was recently classified as a member of Picornaviridae and renamed duck hepatitis A virus (DHAV) (28). Complete genome sequence determination showed that DHAVs form a genetically distinct group within the picornaviruses. The different types of DHAV constitute a novel genus, Avihepatovirus, which was proposed by the International Committee on Taxonomy of Viruses (ICTV) in August 2009, within the Picornaviridae family (http://www.picornastudygroup.com). The DHAVs are genetically divided into three different types: the original type (DHAV-1) (17, 30, 50), a type recently isolated in Taiwan (DHAV-2) (51), and the recently described type isolated in South Korea and China (DHAV-3) (21, 29). The 5′ untranslated regions (5′ UTRs) are 625, 655, and 651 nt in types 1 to 3, respectively (17, 29, 51). Previous studies proposed that the 5′ UTR of DHAV-1 may possess a type II IRES (30, 50) similar to Ljungan virus (LV) and human parechovirus (HPeV). Subsequently, it was suggested that DHAV-1 possesses a type IV IRES (17, 24), similar to PTV-1 and HCV. The two proposals were based on only partial secondary structure predictions, without comparative sequence analyses of genetically diverse viruses for covariant nucleotide substitutions that predict conserved helical structures, because DHAV-2 and DHAV-3 were not characterized at that time. Moreover, comparative sequence analysis of available sequences of the 5′ UTR of DHAV has not been performed, possibly due to the highly conserved nucleotide sequences of the 5′ UTR of DHAV strains within the same type. Furthermore, the IRES of DHAV has not been characterized.
In the present study, we identified a number of covariant nucleotide substitutions by comparing the 5′ UTR sequences of three DHAV strains and predicted the secondary structure of the DHAV 5′ UTR based on different approaches. We analyzed for the first time the functional and structural properties of the DHAV IRES element by testing a series of dicistronic constructs in vitro and in vivo in order to determine the characteristics and type of the IRES in DHAV.
MATERIALS AND METHODS
Virus.
DHAV C-GY, a strain of DHAV-3, was used in this study (GenBank accession number: EU352805) (21). The virus was propagated in the allantoic cavities of 10-day-old duck embryos and harvested.
Sequences.
DHAV strains used for 5′ UTR nucleotide sequence analyses include the following: DHAV-1 strains C80 (17) (GenBank accession number: DQ864514), R85952 (30) (DQ226541), and DRL-62 (30) (DQ219396); DHAV-2 strains 04G (51) (EF067923) and 90D (51) (EF067924); and DHAV-3 strains AP-03337 (29) (DQ256132) and AP-04114 (29) (DQ812093). The PTV-1 Talfan IRES is contained within nt 125 to 405 of the 5′ UTR (14, 26, 41) (AB038528). The partial IRES elements of the AEV Calnek strain (36) (AJ225173) and HCV (46) (AB016785) were used for comparison.
Sequence analysis and secondary structure prediction.
Nucleotide sequences of the PTV-1 IRES element and the 5′ UTR of DHAV were aligned with CLUSTAL-W (http://www.ebi.ac.uk/clustalw/index.html) (49) using the default parameters. To illustrate the conservation of selected alignments and potential helices, the results of sequence alignments were manually edited using the GeneDoc program (37). Secondary structure elements in the 5′ UTRs of DHAV were modeled using Pfold (http://www.daimi.au.dk/∼compbio/rnafold/) (32). Pfold analysis was performed using aligned sequences of the DHAV types 1 to 3 isolates listed above. Sequences were submitted for analysis in FASTA format. Secondary structure elements within the 5′ UTR of DHAV C-GY sequence and the partial IRES elements of HCV, PTV-1, and AEV sequences (other than the pseudoknot) were generated by Mfold (54) using the default parameters and drawn with the RnaViz 2.0 program (16).
Plasmid constructions.
DNA preparations and manipulations were performed by standard methods as described previously (45) or according to the manufacturers' instructions.
The dicistronic luciferase plasmid was constructed. Briefly, the pRL-CMV vector (Promega, Madison, WI) was used as a template for PCR using primers RlucF and RlucR (Table 1) to generate a fragment with KpnI, XhoI, and NcoI sites containing the Renilla luciferase (Rluc) gene linked to the T7 promoter. The amplified products were then digested with KpnI and NcoI and inserted into the corresponding sites of the pGL3-basic vector (Promega) to yield a dicistronic luciferase plasmid termed pRF-NO that express a dicistronic mRNA encoding Rluc and firefly luciferase (Fluc) from a T7 promoter (see Fig. 2A). Each cistron of the plasmid has its own initiation and termination codons. Rluc and Fluc cistrons are separated by a short sequence containing XhoI and NcoI sites used to insert DHAV sequences.
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| RlucF | CGGGGTACCCCGTACTTAATACGACTCACTATAGGCTAG |
| RlucR | CTACAGCCATGGCTCAACGACTACCGCTCGAGCTAGAATTATTGTTCATTTTTGAGAA |
| EMCVF | CCGCTCGAGTAACGTTACTGGCCGAAGCC |
| EMCVR | CTACAGCCATGGATTATCATCGTGTTTTTCAAAGGAA |
| GYF1 | CCGCTCGAGTTGAAAGCGGCTGTGGTGTAGAC |
| GYF125 | CCGCTCGAGCCCCTATCCATTTGTTTTACCTTTA |
| GYF230 | CCGCTCGAGTAGTGGTTAGCCTACCACCCCTTG |
| GYF248 | CCGCTCGAGCCCTTGGCCACTAATTCTTGGC |
| GYF372 | CCGCTCGAGTTGGACAAGGAAAGGCTAGTGTTTG |
| GYF390 | CCGCTCGAGGTGTTTGGTCTGGGTACAAACCC |
| GYF401 | CCGCTCGAGGGGTACAAACCCTTGTTGTGAAACG |
| GYF411 | CCGCTCGAGCCTTGTTGTGAAACGGATTACC |
| GYF421 | CCGCTCGAGAAACGGATTACCGGTAGTAGCATC |
| GYR651 | CTACAGCCATGGTGCATGAAAGTCAACTGGTTTTATAGTG |
| GYR641 | CTACAGCCATGGTCAACTGGTTTTATAGTGTATGGGACTC |
| GYR630 | CTACAGCCATGGTATAGTGTATGGGACTCGACCAGC |
| GYR600 | CTACAGCCATGGCCTATCAGGCAGTGTGGATCAAAG |
| GYR681 | CTACAGCCATGGATCTTCAATGTTTTTAGTTAGAGTGTCC |
| GYFas | CTACAGCCATGGTTGAAAGCGGCTGTGGTGTAGAC |
| GYRas | CCGCTCGAGTGCATGAAAGTCAACTGGTTTTATAGTG |
| fuGYR126 | GGGTGGTAGGCTAACCACTAGGGTCCTATATCGCTAGGACTA |
| fuGYF230 | TAGCGATATAGGACCCTAGTGGTTAGCCTACCACCCCTT |
| fuGYR231 | AGAATTAGTGGCCAAGGGTACAAAATTCATAGAGACCTAGACGTG |
| fuGYF248 | GGTCTCTATGAATTTTGTACCCTTGGCCACTAATTCTTGG |
| fuGYR252 | CTAGCCTTTCCTTGTCCAAAAGGGGTGGTAGGCTAACCACTACAA |
| fuGYF372 | TAGTGGTTAGCCTACCACCCCTTTTGGACAAGGAAAGGCTAGTG |
| fuGYR389 | CACTCTAGACCATACACTCTAGCCTTTCCTTGTCCAAAAC |
| fuGYF467 | GGACAAGGAAAGGCTAGAGTGTATGGTCTAGAGTGGACATAG |
| Fm-Id-L1 | CTATGAATTTTGTAGTGGTTCGCCTACCACCCCTTGGCCA |
| Rm-Id-L1 | TGGCCAAGGGGTGGTAGGCGAACCACTACAAAATTCATAG |
| Fm-Id-L2 | CTATGAATTTTGTAGTGGTTACCCTACCACCCCTTGGCCA |
| Rm-Id-L2 | TGGCCAAGGGGTGGTAGGGTAACCACTACAAAATTCATAG |
| Fm-Id-L3 | CTATGAATTTTGTAGTGGTTAGACTACCACCCCTTGGCCA |
| Rm-Id-L3 | TGGCCAAGGGGTGGTAGTCTAACCACTACAAAATTCATAG |
| Fm-Id-L4 | CTATGAATTTTGTAGTGGTTAGCATACCACCCCTTGGCCA |
| Rm-Id-L4 | TGGCCAAGGGGTGGTATGCTAACCACTACAAAATTCATAG |
| Fm-Id-L5 | CTATGAATTTTGTAGTGGTTCCCCTACCACCCCTTGGCCA |
| Rm-Id-L5 | TGGCCAAGGGGTGGTAGGGGAACCACTACAAAATTCATAG |
| Fm-Id-L6 | CTATGAATTTTGTAGTGGTTAAACTACCACCCCTTGGCCA |
| Rm-Id-L6 | TGGCCAAGGGGTGGTAGTTTAACCACTACAAAATTCATAG |
| Fm-Id-L7 | CTATGAATTTTGTAGTGGTTAGAATACCACCCCTTGGCCA |
| Rm-Id-L7 | TGGCCAAGGGGTGGTATTCTAACCACTACAAAATTCATAG |
| Fm-Id-L8 | CTATGAATTTTGTAGTGGTTCCAATACCACCCCTTGGCCA |
| Rm-Id-L8 | TGGCCAAGGGGTGGTATTGGAACCACTACAAAATTCATAG |
| Fm-Id-S1 | CTAGGTCTCTATGAATTTTGTACCGGTTAGCCTACCACCCCTTG |
| Rm-Id-S1 | CAAGGGGTGGTAGGCTAACCGGTACAAAATTCATAGAGACCTAG |
| Fm-Id-S2 | CTAGGTCTCTATGAATTTTGTAGACCTTAGCCTACCACCCCTTGG |
| Rm-Id-S2 | CCAAGGGGTGGTAGGCTAAGGTCTACAAAATTCATAGAGACCTAG |
| Fm-Id-S1comp | TTTTGTACCGGTTAGCCTACCGGCCCTTGGCCACTAATTCTTG-3 |
| Rm-Id-S1comp | CAAGAATTAGTGGCCAAGGGCCGGTAGGCTAACCGGTACAAAA |
| Fm-Id-S2comp | GAATTTTGTAGACCTTAGCCTAGGTCCCCTTGGCCACTAATTCTTG |
| Rm-Id-S2comp | CAAGAATTAGTGGCCAAGGGGACCTAGGCTAAGGTCTACAAAATTC |
| Fm-Ie-L1 | ATATCTTGGAGGTGGTGCCACAATATTGCAAGCCACATG |
| Rm-Ie-L1 | CATGTGGCTTGCAATATTGTGGCACCACCTCCAAGATAT |
| Fm-Ie-L2 | ATATCTTGGAGGTGGTGCTGAGGCATTGCAAGCCACATG |
| Rm-Ie-L2 | CATGTGGCTTGCAATGCCTCAGCACCACCTCCAAGATAT |
| Fm-Ie-L3 | GGAGGTGGTGCTGAAATCAAGCAAGCCACATGGTATC |
| Rm-Ie-L3 | GATACCATGTGGCTTGCTTGATTTCAGCACCACCTCC |
| Fm-Ie-S1 | ACCATATCTTGGAGGTGGACCTGAAATATTGCAAGCCAC |
| Rm-Ie-S1 | GTGGCTTGCAATATTTCAGGTCCACCTCCAAGATATGGT |
| Fm-Ie-S2 | GATCCACCATATCTTGGAGCGGGTGCTGAAATATTGCAAG |
| Rm-Ie-S2 | CTTGCAATATTTCAGCACCCGCTCCAAGATATGGTGGATC |
| Fm-Ie-S1comp | GAGGTGGACCTGAAATATTGGTAGCCACATGGTATCTGTGTG |
| Rm-Ie-S1comp | CACACAGATACCATGTGGCTACCAATATTTCAGGTCCACCTC |
| Fm-Ie-S2comp | GGTGCTGAAATATTGCAACGCACATGGTATCTGTGTGTTT |
| Rm-Ie-S2comp | AAACACACAGATACCATGTGCGTTGCAATATTTCAGCACC |
| Fm-IIId | CAAGTTATGAGGGGCTATAGCCAAACCCCTTTGATCCACAC |
| Rm-IIId | GTGTGGATCAAAGGGGTTTGGCTATAGCCCCTCATAACTTG |
| Fm-IIIe | TTGATCCACACTGCCTAAAAGGGTCGCGGCTGGTC |
| Rm-IIIe | GACCAGCCGCGACCCTTTTAGGCAGTGTGGATCAA |
| Fm-mutS1a | CAGTCCATAACATGAGTGTCCCGTCTAGAGTGGACATAGCTTG |
| Rm-mutS1a | CAAGCTATGTCCACTCTAGACGGGACACTCATGTTATGGACTG |
| Fm-mutS1b | GCGGCTGGTCGAGTCCGGGACACTATAAAACCAGTTG |
| Rm-mutS1b | CAACTGGTTTTATAGTGTCCCGGACTCGACCAGCCGC |
| Fm-mutS2a | CCTGATAGGGTCGCGGCTCATCGAGTCCCATACACTATAAA |
| Rm-mutS2a | TTTATAGTGTATGGGACTCGATGAGCCGCGACCCTATCAGG |
| Fm-mutS2b | GAGTCCCATACACTATAAAATGAGTTGACTTTCATGCAGAATTC |
| Rm-mutS2b | GAATTCTGCATGAAAGTCAACTCATTTTATAGTGTATGGGACTC |
| Fm-mutS3a | CACTGCCTGATAGGGTGCCGGCTGGTCGAGTCC |
| Rm-mutS3a | GGACTCGACCAGCCGGCACCCTATCAGGCAGTG |
| Fm-mutS3b | GATAGGGTCGCGGCTGGTGCAGTCCCATACACTATAAA |
| Rm-mutS3b | TTTATAGTGTATGGGACTGCACCAGCCGCGACCCTATC |
| Fm-mutS3comp | CACACTGCCTGATAGGGTGCCGGCTGGTGCAGTCCCATAC |
| Rm-mutS3comp | GTATGGGACTGCACCAGCCGGCACCCTATCAGGCAGTGTG |
| F1SVDV | GTGGCTAGCCACCATGGGCGCCTTCGGGCAGCAGTCTGGTGCCGTGTACGTTGGCAAC |
| R1SVDV | CCGCGCGCGTTGCGAGATGTCTATTCACCACTCTATAGTTGCCAACGTACACGGCACC |
| F2SVDV | GACATCTCGCAACGCGCGCGGACTGGCAAAACTGTGTGTGGGAAGACTACGACAGAGAC |
| R2SVDV | GTCGCAGCCATGTGCAGTGGTGGTGCTCACTAGAAGGTCTCTGTCGTAGTCTTCCCAC |
| F3SVDV | CCACCACTGCACATGGCTGCGACACCATTGCCAGGTGCGATTGCACAGCAGGAGTGTAC |
| R3SVDV | TGTGACTGGATAGTGCTTATTTCTGGAGGCGCAGAAGTACACTCCTGCTGTGCAATCG |
| F4SVDV | AGAAATAAGCACTATCCAGTCACATTTGAGGGGCCCGGTCTTGTGGAGGTTCAAGAGAG |
| R4SVDV | GCAGTACATGGGATTGGTGCTTTTTCGGGTAATACTCACTCTCTTGAACCTCCACAAG |
| F5SVDV | GCACCAATCCCATGTACTGCTCGCAGCTGGATTTGCAGAGCCGGGTGATTGTGGAGGG |
| R5SVDV | ACGGTAACTATGCCAATCACCCCATGTTGGCATCTGAGAATCCCTCCACAATCACCCGG |
| F6SVDV | GTGATTGGCATAGTTACCGTGGGGGGAGAAGGTGTTGTTGGTTTTGCCGATGTAAGAGA |
| R6SVDV | CCGACTCTAGAATTATTGCTCCATGGCATCGTCCTCCAGCCACAACAAGTCTCTTACATCGGCAAAACC |
Underlined sequences indicate the restriction sites included in the sequence. Nucleotides in bold italics are those that modified the WT sequence.
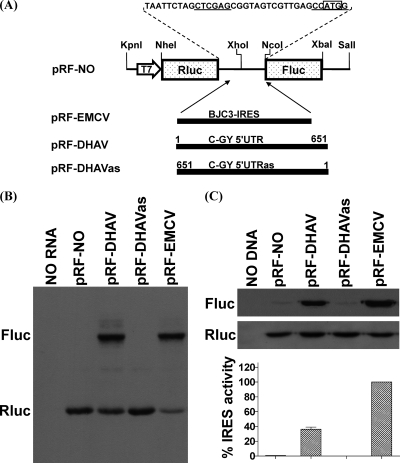
Fig 2.
Identification of an IRES element within the DHAV 5′ UTR in vitro and in vivo. (A) The structures of dicistronic luciferase plasmids used in this study are shown. The fragments of the 5′ UTR of the DHAV C-GY genome were inserted between the Rluc and Fluc genes at XhoI and NcoI sites of plasmid pRL-NO in sense or antisense orientation as described in Materials and Methods. Nucleotide numbers corresponding to the fragments are shown. The EMCV IRES was cloned similarly to yield the construct pRF-EMCV and was used as a positive control. (B) In vitro translation reactions containing RRL and biotinylated lysine-tRNA complex were programmed with RNA transcripts derived from the dicistronic plasmids containing the indicated virus sequences. Reaction products were analyzed by Transcend nonradioactive translation detection systems (Promega). The positions of the Rluc and Fluc proteins are indicated. (C) Transient-expression assay in BHK cells. The dicistronic plasmids containing the indicated virus sequences were transfected into vTF7-3-infected BHK cells. After 20 h, cell lysates were prepared and analyzed for Rluc and Fluc expression by Western blotting. The Rluc and Fluc activities were measured. IRES activity was calculated as the mean of three independent experiments, and the results were standardized to the values for the Fluc/Rluc ratio from the pRL-EMCV, which was set at 100%. The mean values (plus standard errors of the means [error bars]) are shown.
By using the cDNA of DHAV C-GY as a template, the known 5′ UTR was amplified by PCR with the primers GYF1 and GYR651 (Table 1) to generate a fragment that was then digested with XhoI and NcoI. The purified fragment was ligated into similarly digested plasmid pRF-NO between the two open reading frames (ORFs) to yield plasmid pRF-DHAV (see Fig. 2A). A construct containing the fragment in the antisense orientation called pRF-DHAVas (see Fig. 2A) was created in a similar manner using the primers GYFas and GYRas (Table 1). To create pRF-EMCV, the preferred region of EMCV IRES (5) was amplified by PCR from the cDNA of EMCV BJC3 strain (53) (GenBank accession number: DQ464062) with the primers (Table 1) and was digested and ligated into pRF-NO as described above. The orientation and veracity of these inserts in pRF-NO were confirmed by DNA sequencing.
Dicistronic luciferase plasmids with deletions in the 5′ UTR of DHAV C-GY were constructed by deleting specific regions within the pRF-DHAV. A set of cDNA fragments with 5′-terminal deletions was generated by PCR using the plasmid pRF-DHAV (see Fig. 2A) as a template with one of a series of forward primers plus the reverse primer GYR651 (Table 1). A set of cDNA fragments with 3′-terminal deletions was generated by PCR using the plasmid pRF-DHAV (see Fig. 2A) as a template with the forward primer GYF1 plus one of a series of reverse primers (Table 1). A set of cDNA fragments with internal deletions was generated by overlap PCR. The plasmid pRF-DHAV was used as a template to amplify two fragments corresponding to nt 1 to 126 and 230 to 651 of the 5′ UTR of DHAV C-GY by PCR using the primers GYF1 with fuGYR126 and fuGYF230 with GYR651, respectively (Table 1). The two purified products were mixed and used in a further PCR with primers GYF1 and GYR651 to create the single fragment DHAVΔ127-229 (inclusive of nt 1 to 126 and 230 to 651 of the 5′ UTR of DHAV C-GY). Three other derivatives of cDNA fragments with internal deletions (DHAVΔ232-247, DHAVΔ253-371, and DHAVΔ390-466) were generated in a similar way (all primers are given in Table 1). The fragment DHAVΔ1-229/253-371 (containing nt 230 to 252 and 372 to 651 of the 5′ UTR of DHAV C-GY) was amplified by PCR with primers GYF230 and GYR651 using the DHAVΔ253-371 as a template. All of the fragments obtained were purified and digested with XhoI and NcoI, and the products were ligated into pRF-NO as described above to generate a series of plasmids with truncated forms of 5′ UTR cDNA of DHAV C-GY (see Fig. 3). The deleted nucleotides were confirmed by DNA sequencing.
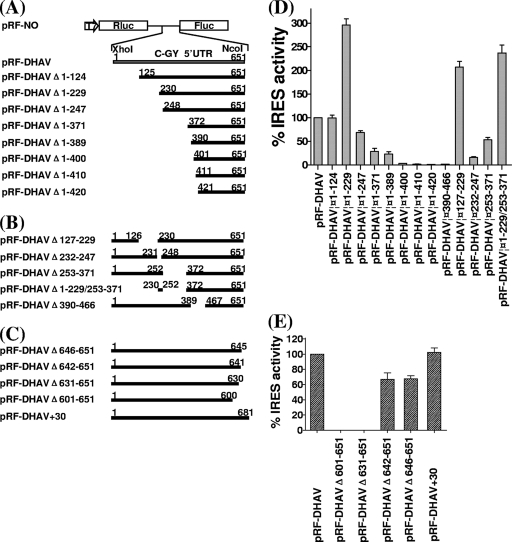
Fig 3.
Delimitation of the DHAV 5′ UTR sequences required for IRES activity in BHK cells. The cDNA fragments indicated were generated by using the PCR as described in Materials and Methods and inserted into the XhoI and NcoI-digested vector pRL-NO. Dicistronic luciferase plasmids with 5′-terminal (A), internal (B), and 3′-terminal (C) deletions made in the DHAV 5′ UTR and inclusion of 30 nt of coding sequence downstream of the DHAV 5′ UTR were used to transfect vTF7-3-infected BHK cells. After 20 h, cell extracts were prepared and analyzed for the expression of Rluc and Fluc. (C and D) The Rluc and Fluc activities were measured and IRES activity was calculated as the mean of three independent experiments. The results were standardized to the values for the Fluc/Rluc ratio directed by the plasmid pRL-DHAV, which was set at 100%. The mean values (plus standard errors of the means [error bars]) are shown.
To construct a monocistronic plasmid expressing Fluc from a T7 promoter, the upstream Rluc-coding region was removed from pRF-NO by digestion with NheI and NcoI. To construct the monocistronic plasmid pDHAV, which contains the sequence of 5′ UTR of DHAV C-GY between a T7 promoter and Fluc, the upstream Rluc-coding region was removed from pRF-DHAV by digestion with NheI and XhoI. Monocistronic plasmids with a series of 5′-terminal deletions in the 5′ UTR of DHAV C-GY were constructed in a similar way from the corresponding dicistronic luciferase plasmids with 5′-terminal deletions in the 5′ UTR of DHAV. For example, the monocistronic construct pDHAVΔ1-124 (inclusive of nt 125 to 651 of the 5′ UTR of DHAV C-GY) was made from the dicistronic luciferase plasmid pRF-DHAVΔ1-124. All of the digested plasmid cDNA were blunt ended by T4 DNA polymerase prior to religation (see Fig. 4A). Nucleotide deletions were confirmed by DNA sequencing.
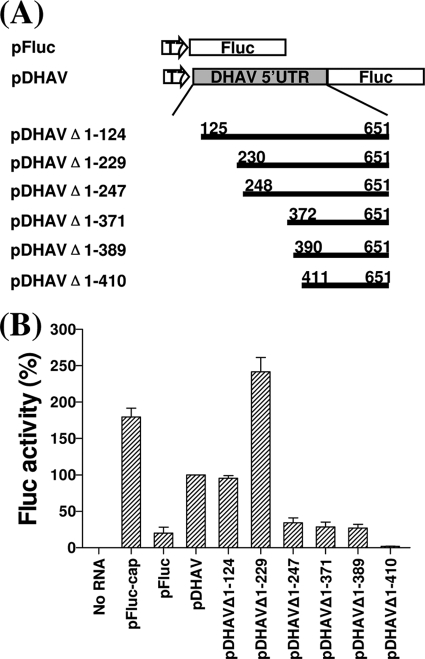
Fig 4.
Schematic representation of transcriptional units present in monocistronic plasmid constructs used as templates for synthesis of RNAs transiently expressed in BHK cells. (A) Monocistronic constructs pFluc and pDHAV and 5′-terminal deletion mutants made from pDHAV. (B) Uncapped or capped mRNA transcribed from the indicated monocistronic plasmids were transfected into BHK cells. After 20 h, cell extracts were prepared and analyzed for Fluc expression. The Fluc expression was measured using the Promega assay kit and a luminometer. Results are the mean of three independent transfections. The results were standardized to the values for Fluc activity in pDHAV, which was set at 100%. The mean values (plus standard errors of the means [error bars]) are shown. pFluc-cap is capped and the rest are uncapped.
Plasmid pT7-2A was constructed to express the 2A protease from an enterovirus, the swine vesicular disease virus (SVDV), under the control of a T7 promoter as described below. The segment containing 2A protease cDNA of SVDV J1′ 73 strain (44) with its own initiation and termination codon was synthesized by an approach described previously (12). Briefly, we synthesized this segment flanked with NheI and XbaI by assembling 12 gel-purified oligonucleotides in an asymmetric PCR assay (40). The lengths of the oligonucleotides varied between 58 and 69 nt (Table 1) and the overlapping regions between two oligonucleotides were on average 20 nt long. The PCR products were purified and ligated directly into a pCR-Blunt II-TOPO vector (Invitrogen, Carlsbad, CA). Ten clones were sequenced to identify error-free DNA segments. The correct DNA fragment was excised from the pCR-Blunt II-TOPO vector by NheI and XbaI, purified, and ligated into the similarly digested pRL-NO to yield plasmid pT7-2A. The sequence of the plasmid was verified by DNA sequencing.
Mutagenesis of the cDNA of DHAV.
Mutagenesis and analysis were performed using a QuikChange site-directed mutagenesis kit (Agilent Technologies, La Jolla, CA) according to the manufacturer's instructions.
Mutations were introduced into the AGCC loop (nt 238 to 241) of the predicted domain Id to generate the sequence CGCC (to create pRF-Id-L1), ACCC (pRF-Id-L2), AGAC (pRF-Id-L3), AGCA (pRF-Id-L4), CCCC (pRF-Id-L5), AAAC (pRF-Id-L6), or AGAA (pRF-Id-L7) by using the pRF-DHAV plasmid as the template and the appropriate primers (Table 1). In order to disrupt base-pairing interactions, mutations within the GUGGU stem sequence (nt 232 to 236) of the predicted domain Id were also created with the sequence CCGGU (pRF-Id-S1) or GACCU (pRF-Id-S2) by using the pRF-DHAV plasmid as the template and the appropriate primers (Table 1), Compensatory mutations were produced (termed pRF-Id-S1comp and pRF-Id-S2comp) in the same way with the pRF-Id-S1 and pRF-Id-S2 plasmids as templates, respectively. The mutagenic primers specified the compensatory mutations (Table 1). All of the expected initial and compensatory mutations were confirmed by sequencing.
Mutations within the apical portion of the predicted domain Ie were created with the pRF-DHAV plasmid as the template and the appropriate primers to change the UGAAAUAUU loop (nt 308 to 316) to CACAAUAUU (pRF-Ie-L1), UGAGGCAUU (pRF-Ie-L2), or UGAAAUCGG (pRF-Ie-L3), the stem UG (nt 305 to 306) to AC (pRF-Ie-S1), and GU (nt 301 to 302) to CG (pRF-Ie-S2). Compensatory mutations were produced (termed pRF-Ie-S1comp and pRF-Ie-S2comp) in the same way with the pRF-Ie-S1 and pRF-Ie-S2 plasmids as templates, respectively. The mutagenic primers specified the compensatory mutations (Table 1). All of the expected initial and compensatory mutations were confirmed by sequencing.
Mutations were also created in the loop sequences of the predicted domain IIId and domain IIIe regions in order to change the sequence in the loop region from GGG to AGC (nt 567 to 569) and GAUA to AAAA (nt 595 to 598), respectively, by using the pRF-DHAV plasmid as the template and the primer pairs Fm-IIId with Rm-IIId and Fm-IIIe with Rm-IIIe (Table 1). The resultant plasmids were named pRF-mutIIId and pRF-mutIIIe and were sequenced to verify the presence of the expected mutations.
Mutations within the stem sequences of the predicted pseudoknot were also introduced (termed pRF-mutS1a, pRF-mutS1b, pRF-mutS2a, pRF-mutS2b, pRF-mutS3a, and pRF-mutS3b) with the pRF-DHAV plasmid as the template and the appropriate primers (Table 1) in order to disrupt base-pairing interactions. For the pRF-mutS1a plasmid, AUG (nt 473 to 475) was changed to CCC to create the plasmid pRF-mutS1a. For the plasmid pRF-mutS1b, CAU (nt 620 to 622) was changed to GGG. For the pRF-mutS2a plasmid, GG (nt 610 to 611) was changed to CA. Similarly, for the pRF-mutS2b plasmid, UG (nt 634 to 635) was changed to GGG; for the pRF-mutS3a plasmid, CG (nt 603 to 604) was changed to GC, and for the pRF-mutS3b plasmid, CG (nt 613 to 614) was changed to GC. Compensatory mutations (termed pRF-mutS1comp, pRF-mutS2comp, and pRF-mutS3comp) were produced by the same approach using the pRF-mutS1a, pRF-mutS2a, or pRF-mutS3b plasmids as templates, respectively. The mutagenic primers specified the compensatory mutations (Table 1). All of the expected initial and compensatory mutations in the pRF-DHAV plasmids were confirmed by sequencing.
In vitro translation reactions.
The dicistronic luciferase plasmids (1 μg) were assayed in TNT T7 quick coupled transcription/translation system (Promega) using biotinylated lysine-tRNA complex (1 μl) according to the manufacturer's instructions. A 2-μl aliquot of the biotin-containing translation products was resolved directly on sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gels (10%) and transferred to 0.2 μm polyvinylidene difluoride (PVDF) membranes (Millpore, Temecula, CA), and the biotinylated proteins were visualized by binding streptavidin-horseradish peroxidase (streptavidin-HRP, 1:10,000), followed by chemiluminescent detection onto X-ray film according to the manufacturer's instructions for the Transcend nonradioactive translation detection systems (Promega). Alternatively, the dicistronic luciferase plasmids were linearized by the restriction enzyme SalI. The uncapped mRNA was transcribed in vitro with T7 RNA polymerase using MegaScript kit (Ambion, Austin, TX) according to the manufacturer's instructions. Each mRNA was translated in the rabbit reticulocyte lysate system (Promega) with biotinylated lysine-tRNA complex, and the products were also detected by the same method as described above. The capped mRNA transcripts were produced from pFluc with T7 RNA polymerase by using a Message high-yield capped RNA transcription kit (Ambion, Austin, TX) following linearization of the plasmid DNAs with SalI. These capped RNA transcripts are designated by the suffix “-cap.”
Transient-expression assays.
The plasmids were transfected into cells previously infected with the recombinant vaccinia virus vTF7-3 expressing the T7 RNA polymerase (22). The dicistronic luciferase plasmids (4 μg per well) described above were transfected into cells alone or with the plasmid pT7-2A (1 μg per well) by using Lipofectamine 2000 (Invitrogen) and Opti-MEM I medium (Invitrogen) according to the manufacturer's instructions. The RNAs were transfected into BHK cells by using DMRIE-C reagent (Invitrogen) according to the manufacturer's instructions. Cell lysates were prepared 20 h after transfection by passive lysis buffer (Promega) and were analyzed by immunoblotting from 10% SDS-PAGE minigels and probed with anti-Rluc monoclonal antibody (1:2,000; Millipore) or anti-Fluc polyclonal antibody (1:1,000; Promega) followed by HRP-labeled anti-mouse IgG (1:10,000; Sigma-Aldrich, St. Louis, MO) or anti-goat IgG (Sigma-Aldrich) antibodies, respectively. Detection was conducted using chemiluminescence reagents (Promega). For detection of eIF4G cleavage products, samples were analyzed on 7% SDS-PAGE minigels, blotted, and detected with a goat anti-eIF4G polyclonal antibody at a dilution of 1:300 (raised against the C-terminal fragment) (Santa Cruz, CA).
Measurement of bioluminescence activities.
The Rluc and Fluc bioluminescent activities were measured using the Dual Glo luciferase kit (Promega) and GloMax 96 microplate luminometer (Promega). Posttransfection cell lysate (20 μl) was incubated sequentially with firefly and then Renilla luciferase-specific substrates according to the protocol supplied by the manufacturer. The Fluc/Rluc signal ratio controls the balancing of the IRES activity with transfection efficacy, and the IRES activity was calculated as the mean of three independent experiments. Fluc expression from the monocistronic plasmid was quantified using a firefly luciferase assay kit (Promega) with a luminometer.
RESULTS
Secondary structure prediction for the 5′ UTR of DHAV.
The 5′ UTR sequences of eight strains of DHAV prototypes (representatives for types 1 to 3) were aligned together with the sequence of PTV-1 IRES. To generate a model of the secondary structure of the whole 5′ UTR of DHAV, we conducted a manually revised alignment which was helpful for the identification of covariant substitutions predictive of conserved helical structures within the 5′ UTR. The alignment analyses indicate that the conserved potential base pairs exist as folding constraints for the prediction of the 5′ UTR secondary structure as described previously (25). Previous studies showed that the 5′ UTR sequences of DHAV shared higher nucleotide identity with PTV-1 IRES than with type I-to-III picornavirus IRES (17, 24). As shown in Fig. 1A, the 5′ UTR sequences of DHAV types 1 to 3 also exhibit higher nucleotide identities (49 to 51%) with the sequence (nt 122 to 414) of PTV-1 IRES; out of the 12 nt in domain IIIe within the PTV-1 IRES, there are 11 nt identical to a sequence within the 5′ UTR of DHAV. The 5′ UTR sequences of DHAVs are highly conserved, with 96 to 99% identity within the same serotype, but are clearly distinguished from those of the heterologous serotype (69 to 72% identity). These results are consistent with those reported previously (21).
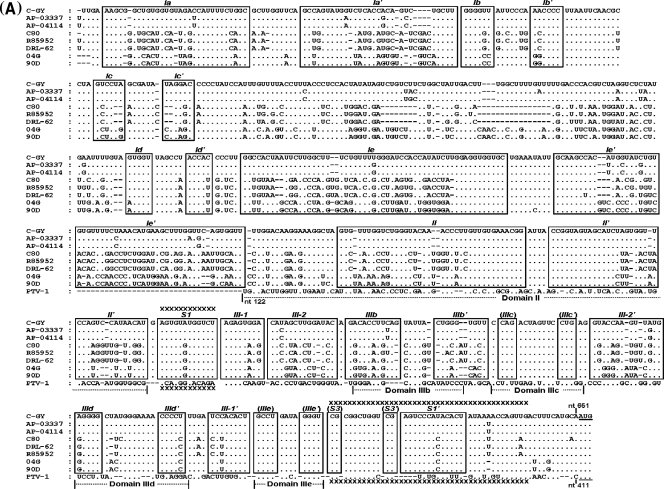
Fig 1.
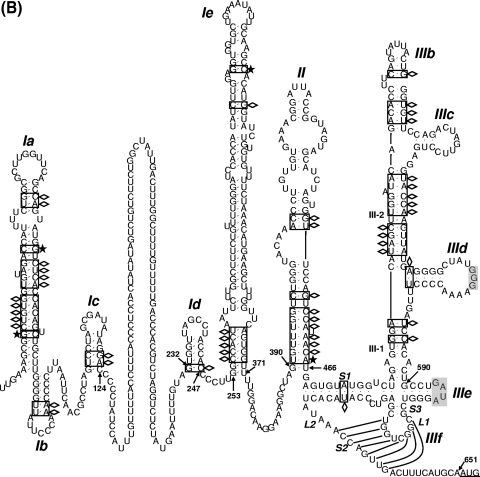
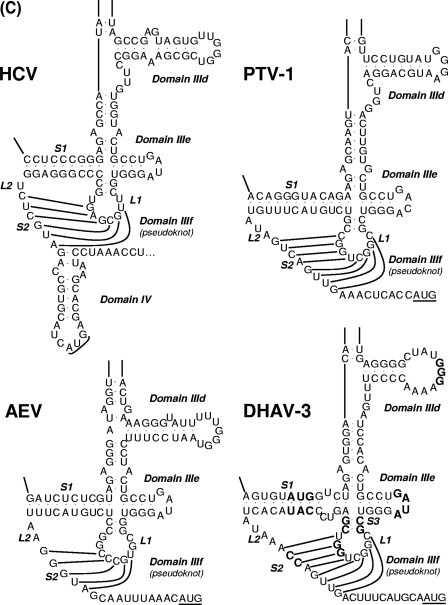
Sequence and predicted secondary structure of the 5′ UTR and IRES element of DHAV. (A) Alignment of the DHAV 5′ UTR and PTV-1 IRES sequences. Sequences were aligned with ClustalW and manually edited in light of the predicted secondary structure models. Dots indicate conserved bases (relative to the C-GY sequence); dashes indicate deletions. The initiation codon (AUG) is underlined. Boxes indicate approximate locations of the proposed helical structures. From the 5′ terminus, complementary sequences involved in potential helices are labeled sequentially by major domain (Ia, Ia′; Ib, Ib′, etc.). Potential helices that are not supported by the presence of covariant base substitutions have labels enclosed by parentheses, but their sequences are completely conserved among different strains. The regions of the PTV-1 IRES corresponding to domain II and domain III are indicated. The sequences involved in base pairing to form the pseudoknot (domain IIIf) structure within the PTV-1 IRES and DHAV IRES elements are marked with xxxx. (B) Predicted secondary structure of the DHAV C-GY 5′ UTR RNA. The structure was predicted by comparative sequence analysis and by using Pfold and Mfold, as described in the text. Domain II and domain III are labeled according to corresponding domains in the PTV-1 IRES. The two stems (S1 and S2) and loop regions (L1 and L2) that form the pseudoknot are shown. The stem of L1 is labeled S3; individual helical segments in domain III are labeled III-1 and III-2. The initiation codon (AUG) is underlined. Lightly shaded rectangles indicate base pairs that are maintained despite sequence variation between types, strains, and isolates of designated viruses; single-site substitutions that do not disrupt base pairing are indicated by asterisks, and base pairs maintained by paired covariant substitutions are indicated by lozenges. The conserved apical loops of domain IIIe and the apical GGG element of the domain IIId loop conserved in the HCV IRES are indicated by gray shading. (C) Comparison of predicted secondary structures for the domains IIId, IIIe, and IIIf among HCV, PTV-1, AEV, and DHAV IRES elements. The two stems (S1 and S2) and loop regions (L1 and L2) that form the pseudoknot are indicated. The stem of L1 within DHAV IRES is labeled S3. Initiation codons (AUG) are underlined. Note that for HCV, this codon is within a stem-loop structure (domain IV) but no such structure is predicted within the PTV-1, AEV, or DHAV sequences. Note the complete identity of domain IIIe in DHAV, HCV, and AEV, and the conserved GGG motif within domain IIId. Within the DHAV sequence, the nucleotides indicated in bold type are those that were modified in the experiments shown in Fig. 6.
To model the structure of 5′ UTR, we applied Pfold, which predicts secondary structure based on an explicit evolutionary model (32), and Mfold, which generates complementary structural predictions on the basis of energy minimization (54). The resulting model of the 5′ UTR structure of DHAV C-GY is shown in Fig. 1B, representing the combined results of a search for nucleotide covariant substitutions predictive of (or at least permissive for) conserved helices, thermodynamic predictions, and an evolutionary model of comparative RNA sequence analysis. Our analyses suggest that the 5′ UTR contains two highly folded regions (nt 1 to 124 and nt 232 to 651) separated by a single-stranded and pyrimidine-rich region that is highly variable among strains of different serotypes (Fig. 1B). Compared to the sequence of DHAV-3, DHAV-1 shares a 23-nt deletion in this region, while DHAV-2 has no deletions.
The final model structure contains three major secondary structural domains, labeled I through III and beginning at the 5′ terminus of the 5′ UTR (Fig. 1B). Domains Ia to Ic (nt 1 to 124) located upstream of the pyrimidine-rich region has a relatively conserved primary structure with the predicted covariant mutations. Domains Ia to Ic are likely to form a large hairpin structure at the extreme 5′ end of the genome (20), analogous to that found in other picornaviruses (33). The 3′ end of the 5′ UTR of DHAV shares high sequence identity with the PTV-1 IRES, implying that it is relatively easy to map certain features of the PTV-1 IRES onto the 3′ end of the 5′ UTR sequence of DHAV. As mentioned above, PTV-1 domain IIIe differs at only one position from the DHAV sequence; in this case, the difference is within the 4-nt loop (Fig. 1C). Notably, the domain IIIe of HCV and AEV is identical to a region of 12 nt within the DHAV sequence (Fig. 1C). The PTV-1 domain IIIf is a pseudoknot (Fig. 1C) composed of two base-paired stem regions (S1 and S2) connected by two loops (L1 and L2). The sequences forming this structure are distinct between PTV-1 and DHAV, but structural conservation is striking. Like the HCV IRES, stem 1 of the PTV-1 IRES is composed of uninterrupted base-paired nucleotides, whereas DHAV stem 1 is bipartite. Like the PTV-1 IRES, stem 2 within the DHAV-2 and DHAV-3 IRES is up to 8 nt long, but it is unknown that whether each of these base pairs is able to form. In addition, the DHAV-1 IRES stem 2 is up to 6 nt long (17).
Like the PTV-1 IRES, the 3′ end of DHAV 5′ UTR (nt 390 to 651, C-GY) is composed of two major domains, and it is convenient to refer to these elements with the same nomenclature as used for the PTV-1 IRES. Thus, domain II is now predicted to comprise nt 390 to 466 of the DHAV (C-GY) sequence. Between the pyrimidine-rich region and domain II, two stem-loop structures comprise domains Id and Ie (Fig. 1B). Domain Id has a highly conserved primary structure. The structure of domain Ie contains a long stem with sequence variability among different DHAV serotypes and a highly conserved apical nonanucleotide loop.
Identification of the IRES element within the 5′ UTR of DHAV.
To confirm the existence of an IRES within the 5′ UTR of DHAV, a dicistronic luciferase plasmid was constructed in which cDNA corresponding to the 5′ UTR of DHAV (nt 1 to 651; C-GY) was inserted between two reporter gene sequences for Rluc and Fluc (Fig. 2A). Plasmid pRF-DHAVas, containing the DHAV sequence in the antisense orientation, and plasmid pRF-NO, lacking any IRES sequence, were both used as negative controls. A positive control (pRF-EMCV) containing the EMCV IRES was also constructed.
Dicistronic RNA transcripts were analyzed in translation assays in RRLs. The level of Rluc expression represents the translation of the first ORF, and the expression of Fluc represents the translation directed by functional IRES (Fig. 2A). In the assays, each of the RNAs efficiently expressed Rluc as expected (Fig. 2B). The dicistronic RNAs containing the EMCV IRES and the DHAV 5′ UTR sequence in the sense orientation also expressed Fluc efficiently (Fig. 2B). Only a background level of Fluc expression could be detected from the construct lacking any IRES element or from the DHAVas construct. The cap-dependent translation of the first cistron (Rluc) and the internally initiated translation of the second cistron (Fluc) involved competition for translation factors present in limiting quantities in RRLs. The expression of Rluc from pRF-EMCV IRES was much lower than that of pRF-IRES (Fig. 2B), indicating that the 5′ UTR of DHAV is less active in this system than the EMCV IRES. Similar results were obtained in the TNT RRL system (data not shown).
In order to confirm and extend results from these in vitro assays, the same dicistronic plasmids were tested using the transient-expression assay system within vTF7-3-infected BHK cells. Consistent with the in vitro assays, each of the plasmids efficiently expressed Rluc, and as expected, the plasmids pRF-DHAV and pRF-EMCV expressed Fluc efficiently (Fig. 2C). Dual luciferase assays showed that the 5′ UTR of DHAV was more than 3-fold less active than the EMCV IRES in producing translation product. These results indicate that the 5′ UTR of DHAV contains an IRES element that is functional both in the RRL system and within the cells.
Delimitation of the IRES element of DHAV C-GY.
To define the 5′ border and 3′ boundary of the DHAV IRES precisely, a series of progressive 5′ deletions, 3′ deletions, and internal deletions in the 5′ UTR of DHAV were made (Fig. 3), and the residual sequences were inserted into the pRF-NO vector. All of the plasmids were analyzed in TNT RRL assays and transient-expression assays within BHK cells as described above.
Deletion of 124 nt from the 5′ end of the 5′ UTR (pRF-DHAVΔ1-124) (Fig. 3A), leading to the removal of predicted domain Ia, domain Ib, and domain Ic from the complete 5′ UTR, had almost no effect on IRES activity in cells (Fig. 3D). Surprisingly, the loss of 229 nt from the 5′ end of the 5′ UTR (pRF-DHAVΔ1-229) (Fig. 3A), leading to the removal of a pyrimidine-rich track from DHAVΔ1-124, resulted in IRES activity approximately 3-fold greater than that detected from the complete 5′ UTR (Fig. 3D). A construct with an internal deletion of the pyrimidine-rich region from the complete 5′ UTR, pRF-DHAVΔ127-229 (Fig. 3B), also resulted in IRES activity approximately 2-fold greater than that detected from complete 5′ UTR (Fig. 3D). The results of these experiments suggest that these deletions in the 5′ UTR of DHAV may increase translational efficiency. The IRES activity from pRF-DHAVΔ1-247 (Fig. 3A), with a removed predicted domain Id from DHAVΔ1-229, was markedly decreased to approximately 25% of that from pRF-DHAVΔ1-229 (Fig. 3D). Internal deletion of the predicted domain Id from the complete 5′ UTR (pRF-DHAVΔ232-247) (Fig. 3B) markedly inhibited IRES activity to approximately 16% of the activity of the complete 5′ UTR (Fig. 3D). These results indicate that predicted domain Id is critical for IRES activity. Similar results were obtained in the TNT RRL system (data not shown).
The DHAV IRES derived from pRF-DHAVΔ1-371, which was obtained from DHAVΔ1-247 by removing the predicted domain Ie (Fig. 3A), showed activity lower than that from pRF-DHAVΔ1-247 (Fig. 3D). The plasmid pRF-DHAVΔ253-371, with an internal deletion of domain Ie from complete 5′ UTR (Fig. 3B), reduced the activity to 52% of that from the complete 5′ UTR. Surprisingly, the same internal deletion of predicted domain Ie from DHAVΔ1-229 (pRF-DHAVΔ1-229/253-371) (Fig. 3B) reduced the activity to about 80% of that from pRF-DHAVΔ1-229, but its activity was still more than 2-fold greater than that observed from the complete 5′ UTR (Fig. 3D). These findings suggest that the loss of 230 nt from the 5′ end of the 5′ UTR still allows for internal initiation of translation, although the deletion of predicted domain Ie from 5′ UTR complete has some effect on IRES activity. The IRES activity assays within the TNT RRLs (data not shown) were entirely consistent with the transient-expression assays within BHK cells.
Although the region containing nt 390 to 651 of the 5′ UTR of DHAV was predicted to form a structurally conserved type IV-like IRES element (Fig. 1B), deletion of nt 1 to 389 (pRF-DHAVΔ1-389) (Fig. 3A) markedly reduced the IRES activity to approximately 23% of that from complete 5′ UTR and to 8% of the activity of DHAVΔ1-229 (Fig. 3D). Further deletions (pRF-DHAVΔ1-400, pRF-DHAVΔ1-410, pRF-DHAVΔ1-420) (Fig. 3A) completely abolished IRES activity (Fig. 3D). Internal deletion of domain II from the 5′ UTR (pRF-DHAVΔ390-466) (Fig. 3B) markedly inhibited IRES activity (Fig. 3D). These data indicate that the region containing nt 390 to 651 of DHAV 5′ UTR, a predicted type IV-like IRES element, is not able to direct efficient internal initiation of translation independently, but it is critical for IRES activity, suggesting that this region is an important part of the DHAV IRES. Similar results were also obtained using TNT RRL assays in vitro (data not shown).
Deletion of 12 nt or 6 nt from the 3′ end of the 5′ UTR (pRF-DHAVΔ642-651 or pRF-DHAVΔ646-651) (Fig. 3C), leading to the removal of the sequence between the initiation codon and the predicted pseudoknot, resulted in a small decrease in DHAV IRES activity (Fig. 3E). The other two deletions (pRF-DHAVΔ601-651 and pRF-DHAVΔ631-651) (Fig. 3C) completely inhibited IRES activity (Fig. 3E). These data indicate that the 3′ sequences of 5′ UTR are critical for DHAV IRES activity. pRF-DHAV+30, containing the viral coding sequence downstream of the 5′ UTR of DHAV (Fig. 3C), did not enhance the IRES activity within cells (Fig. 3E), suggesting that this sequence is not essential for the function of DHAV IRES. Similar results were also obtained using TNT RRL system in vitro (data not shown).
In order to rule out the possibility that sequences adjacent to the 5′-terminal region of the DHAV sequences or within the dicstronic mRNAs can affect its ability to form the correct structure, this region of DHAV needed to be assayed within a monocistronic construct. Therefore, we made progressive 5′-terminal deletions in the 5′ UTR of DHAV within a series of monocistronic plasmids (Fig. 4A). The transcript from each plasmid was analyzed by transient-expression assays within BHK cells as described above. As with dicistronic deletion mutants, deletion of nt 1 to 124 from the DHAV 5′ UTR in monocistronic plasmid pDHAVΔ1-124 (Fig. 4A) barely effected translation in cells (Fig. 4B). Removal of 229 nt from the 5′ end of the 5′ UTR (pDHAVΔ1-229) (Fig. 4A) resulted in IRES activity approximately two and half times higher than that from complete 5′ UTR (Fig. 4B). Thus, the loss of 230 nt from the 5′ end of the 5′ UTR still significantly stimulated internal initiation of translation in the monocistronic construct. The IRES activity from pDHAVΔ1-247 (Fig. 4A), which had the predicted domain Id removed from pDHAVΔ1-229, was markedly decreased, to nearly 15% of that from pDHAVΔ1-229 (Fig. 4B). Removal of domain Id in the monocistronic construct reduced IRES activity more significantly than that in the dicistronic construct. The DHAV IRES derived from pDHAVΔ1-371 and pDHAVΔ1-389 (Fig. 4A) both showed almost the same (lower) activity as that from pDHAVΔ1-247 (Fig. 4B), providing a contrast to the dicistronic constructs (pRF-DHAVΔ1-247 and pRF-DHAVΔ1-371), because the removal of predicted domain Ie from pDHAVΔ1-247 did not appear to decrease translation. Deletion of nt 1 to 371 and nt 1 to 389 still resulted in low levels of IRES activity in the monocistronic construct, although these sequences were able to direct expression of Fluc more efficiently than pDHAVΔ1-410 (Fig. 4A), which completely abolished IRES activity (Fig. 4B).
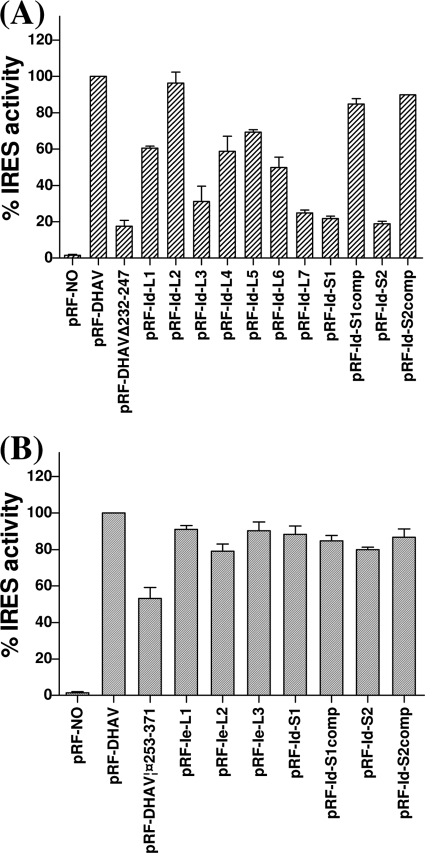
The loop of domain Id contains an AGCC motif that is highly conserved in three genotypes of DHAV (Fig. 1A). The GUGGU stem sequence (nt 232 to 236) of the predicted domain Id was also relatively conserved and predicted a conserved stem maintained by paired covariant substitution within nt 232 (Fig. 1A). To investigate the effect of the stem-loop of domain Id on DHAV IRES activity, mutational analysis of the stem-loop was conducted in transient-expression assays. Eight different mutant forms were produced with single or double substitutions within this loop motif; the sequences generated were CGCC (pRF-Id-L1), ACCC (pRF-Id-L2), AGAC (pRF-Id-L3), AGCA (pRF-Id-L4), CCCC (pRF-Id-L5), AAAC (pRF-Id-L6), and AGAA (pRF-Id-L7) (mutations are in boldface and underlined). Two mutations within the stem sequence were made to disrupt the predicted base pairs, and two other compensatory mutations were introduced to restore the interaction. The wild-type (WT) and mutant plasmids were transfected separately into vTF7-3-infected BHK cells and analyzed by dual luciferase assays. Removal of domain Id (pRF-DHAVΔ232-247) greatly reduced the IRES activity (17% of WT efficiency). The IRES elements with the double mutation in the loop (AGAA) displayed only about 25% of the WT activity. The IRES with the single mutation (AGAC) was also severely impaired in activity, with translation directed by this element reduced to approximately 30% of WT levels. The elements with the ACCC mutation, however, did not affect translation (96% of WT efficiency). Additionally, the other four mutations in loop maintained a reasonable capacity to direct translation (50 to 70% of WT activity) (Fig. 5A). These results suggested that this motif is important for the activity of DHAV IRES. In the stem of domain Id, nt 232 to 236 (GUGGU) are predicted to base pair with ACCAC (nt 243 to 247). To disrupt this interaction, nt 232 to 233 (GU) were changed to CC (to create pRF-Id-S1) and nt 233 to 235 (UGG) were changed to ACC (to create pRF-Id-S2). These changes are expected to inhibit the activity of the IRES; indeed, the two mutations significantly inhibited the activity of IRES (approximately 20% of WT activity) (Fig. 6A). To make compensatory changes in the stem, nt 246 to 247 (AC) was changed to GG on the pRF-Id-S1 background (to create pRF-Id-S1comp) and nt 244 to 246 (CCA) was changed to GGU on the pRF-Id-S2 background (to create pRF-Id-S2comp) to restore the predicted base pairing. Introduction of these compensatory mutations greatly increased the IRES activity (to about 80 to 90% of WT activity) (Fig. 5A), strongly suggesting that the base-pairing interactions within the stem were restored and further demonstrating that the sequence-function relations were predicted correctly. These results indicate that the domain Id is important for IRES activity and that the structure and primary sequence of the loop may be critical for IRES function.
Fig 5.
Mutational analysis of domain Id and domain Ie. Dicistronic plasmids containing the indicated mutations within the predicted stem-loops regions of domain Id (A) and Ie (B) were transfected into vTF7-3-infected BHK cells and analyzed for the expression of Rluc and Fluc by using the Dual Glo luciferase kit (Promega). The IRES activities were standardized to the values for Fluc/Rluc ratio directed by the plasmid pRL-DHAV, which was set at 100%. The results are the mean values for Fluc/Rluc ratios from three experiments.
Fig 6.
Analysis of DHAV IRES activity by mutation of the loop of domain IIId and IIIe or pseudoknot structure. Dicistronic plasmids containing the indicated mutations within the predicted pseudoknot region and the loop IIId and IIIe were transfected into vTF7-3-infected BHK cells and analyzed for the expression of Rluc and Fluc as described above. The IRES activities were standardized to the values for Fluc/Rluc ratio directed by the plasmid pRL-DHAV, which was set at 100%. The results are the mean values for Fluc/Rluc ratios from three experiments.
The predicted domain Ie was about 120 nt in length. On the basis of our alignment results, the stem sequence of domain Ie exhibits relatively low nucleotide identities among the different DHAV serotypes compared to the other domains. This long stem is composed of interrupted base-paired nucleotides with a relatively conserved structure and many predicted paired covariant mutations. In addition, the structure of domain Ie also contains a highly conserved apical nonanucleotide loop, and the terminal part of the adjacent stem has very high nucleotide identity among serotypes. To further explore the effects of domain Ie on DHAV IRES activity, mutational analysis of the terminal stem-loop was conducted in transient-expression assays. Three different mutations were produced with triple substitutions within this loop: CACAAUAUU (pRF-Ie-L1), UGAGGCAUU (pRF-Ie-L2), and UGAAAUCGG (pRF-Ie-L3). Two mutations within the stem sequence were made to disrupt the predicted base pairs, and two compensatory mutations were introduced to restore the interaction. Removal of domain Ie (pRF-DHAVΔ253-371) decreased the IRES activity (to 60% of WT efficiency) (Fig. 5B). All the IRES elements with mutations in loop and mutations in stem displayed nearly 80 to 90% of the WT activity (Fig. 5B), as did the IRES element with the compensatory mutations (Fig. 5B). These results indicated that disruption of this conserved loop and stem in domain Ie did not significantly inhibit the activity of DHAV IRES. Combined with the results of deletion mutations within the dicistronic construct (pRF-DHAVΔ1-229/253-371) and within the monocistronic construct (pDHAVΔ1-371), it appears that domain Ie is not essential for the activity of DHAV IRES.
Above all, we conclude that the optimally functional DHAV IRES element lies within nt 230 to 651 of the DHAV (C-GY) RNA, while nt 253-371 can be deleted without significant loss of activity.
Mutational analysis of the putative domain IIId region within the DHAV IRES.
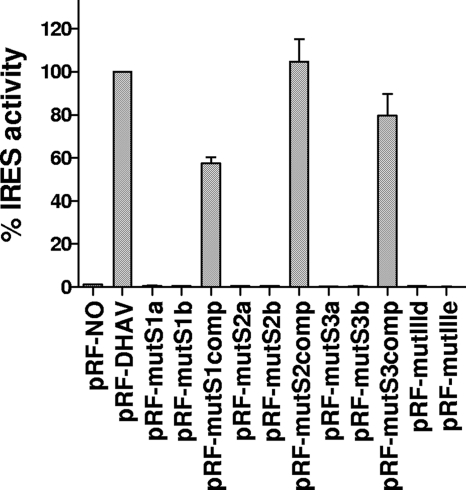
The domain IIId within IRES is an important region for the interaction of the HCV IRES with the 40S subunit (27). The terminal loop of domain IIId contains a GGG motif that is highly conserved in all HCV-like IRESs (24). In the HCV sequence, domain IIId has a stem structure with an apical hexanucleotide loop (31, 46). The domain IIId of PTV-1 is predicted to be composed of a simple stem with an apical tetraloop (GGGA) (14), while the domain IIId of DHAV is predicted to be a simple stem with a terminal hendecaloop that is larger than any other terminal loop of the domain IIId within HCV-like IRESs (Fig. 1C). To investigate the importance of the predicted GGG motif within the DHAV IRES, which corresponds to the domain IIId tetraloop of PTV-1, we mutated this motif to AGC (pRF-mutIIId). The IRES activity of the mutant was completely abolished in RRLs (data not shown) and within BHK cells (Fig. 6), suggesting that this motif is necessary for the activity of DHAV IRES.
Mutational analysis of the putative domain IIIe region within the DHAV IRES.
The domain IIId within IRES is an important region for the interaction of the HCV IRES with the 40S subunit The domain IIIe, a simple stem-loop structure of 12 nt, is part of the most conserved region in HCV-like IRESs (24). Together with the domain IIId, it plays an essential role in binding the 40S ribosomal subunit. Previous studies have shown that each of the nucleotides within the highly conserved GAUA tetraloop of domain IIIe is crucial for HCV IRES activity (27, 39, 48). The GAUA tetraloop is also highly conserved in the IRES of CSFV, GBV-B, AEV, and DHAV (24). The only difference between the HCV IIIe element and the PTV-1 sequence is within the loop, GAUA in HCV and GACA in PTV-1 (14). To assess the importance of the highly conserved tetraloop within the DHAV IRES element, which corresponds to the domain IIIe loop of the HCV-like IRESs, we mutated this motif from GAUA to AAAA (pRF-mutIIIe). Consistent with the results from similar mutation within the AEV IRES elements (2), these changes abolished DHAV IRES activity in vitro (data not shown) and in vivo (Fig. 6).
Mutational analysis of the predicted pseudoknot region within the DHAV IRES.
Mutagenesis studies demonstrated that the pseudoknot structures within the type IV IRES elements were critical for the IRES activity (2, 3, 14, 24, 38). In order to test the structure of the predicted pseudoknot of the DHAV IRES, we made mutations to disrupt the predicted base pairs and compensatory mutations introduced to restore the interactions.
Three sets of mutant forms were produced within the base-paired stem regions of the predicted pseudoknot (Fig. 1C). These mutations were analyzed in TNT assays (data not shown) and by transient-expression assays within BHK cells (Fig. 6).
In stem 1 of the DHAV IRES, AUG (nt 473 to 475) is predicted to base pair with CAU (nt 620 to 622). In order to disrupt the pseudoknot structure, AUG (nt 473 to 475) was changed to CCC (pRF-mutS1a), and CAU (nt 620 to 622) was changed to GGG (pRF-mutS1b). As expected, these mutations in stem 1 completely abrogated IRES activity (Fig. 6).
To confirm that it was the disruption of the stem structure rather than the change in primary sequence alone that was responsible for the inhibition of DHAV IRES activity, a compensatory mutation that changed nt 620 to 622 (CAU) within pRF-mutS1a to GGG was created to restore the predicted base pairing (pRF-mutS1comp); these compensatory mutations restored IRES activity to about 60% of WT IRES activity (Fig. 6).
Thus, the predicted structure of stem 1 appears to be correct. The fact that full restoration of IRES activity was not achieved was consistent with preciously described pseudoknot mutations in PTV-1 and AEV. The incomplete rescue of function suggests that these nucleotides not only form the pseudoknot structure but also are involved in other interactions (such as RNA-protein interactions) (2, 14).
In stem 2 of the DHAV IRES, nt 610 to 611 (GG) was changed to CA (pRF-mutS2a), and nt 634 to 635 (CC) was changed to UG (pRF-mutS2b). These changes were predicted to disrupt the base pair interaction with nt 634 to 635 (CC) and nt 610 to 611 (GG), respectively. These mutations completely abolished IRES activity as expected (Fig. 6). To make compensatory changes in stem 2, nt 634 to 635 (CC) within pRF-mutS2a was changed to UG (to create pRF-mutS2comp) to restore the predicted base pairing. Introduction of these compensatory mutations resulted in fully restoration of WT IRES activity (Fig. 6), strongly suggesting that the base-pairing interactions within stem 2 were restored and that the critical interactions were predicted correctly.
In stem 3 of the DHAV IRES, nt 603 to 604 (CG) are predicted to base pair with nt 613 to 614 (CG). Hence, to disrupt this interaction, nt 603 to 604 (CG) was changed to GC (to create pRF-mutS3a) and CG (nt 613 to 614) to GC (to create pRF-mutS3b). As expected (Fig. 6), these mutations severely inhibited IRES activity. Furthermore, compensatory mutations (pRF-mutS3comp) that changed nt 603 to 604 (CG) to GC within pRF-mutS3a to restore the base pair interactions greatly increased IRES activity, to about 80% of WT IRES activity (Fig. 6). These results also suggest that the predicted secondary structure is correct.
Enterovirus 2A protease-mediated inhibition of DHAV IRES activity in BHK cells.
Cap-dependent translation is strongly inhibited by the expression of an enteroviruses 2A protease or aphthovirus L protease that cleaves eIF4G, but these proteases have different effects on the various picornavirus IRES elements (3). Type II and type IV IRES elements function very efficiently both in the presence and absence of the 2A and L proteases, for example, in the EMCV and PTV IRESs (26). Type I IRES elements, like those from PV and other enteroviruses, are stimulated by these proteases within certain cell types, including BHK cells (43). Due to the requirement of an intact eIF4F complex, HAV IRES (the only representative of type III IRES) is the only one whose efficiency is severely inhibited in the presence of the enteroviruses 2A proteinase (8, 10).
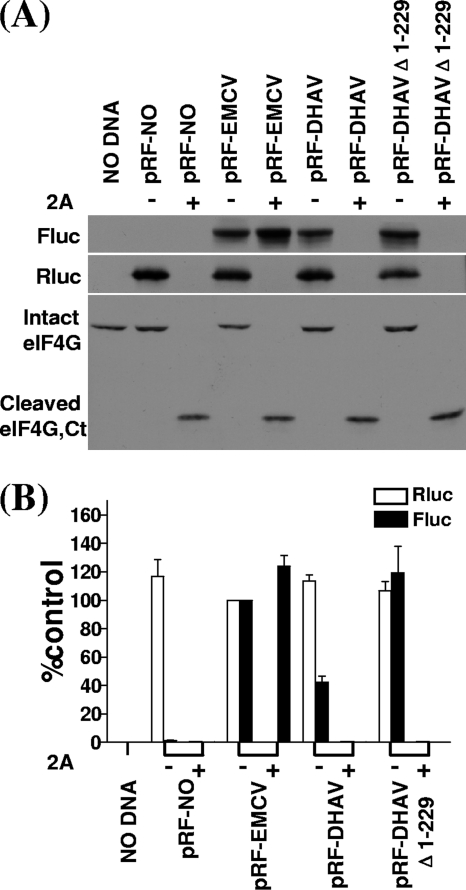
To analyze the effect of the enteroviruses 2A protease on the DHAV IRES activity in cells, the dicistronic luciferase plasmids were transfected into BHK cells either alone or with the pT7-2A plasmid that expresses the SVDV 2A protease. After 20 h, cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting to detect Rluc and Fluc expression. As expected, each plasmid expressed Rluc efficiently when assayed alone, but expression was severely inhibited in the presence of SVDV 2A protease (Fig. 7). The DHAV and EMCV IRES elements directed efficient Fluc expression in cells without 2A protease (Fig. 7). EMCV IRES-driven translation was minimally stimulated by 2A protease (∼0.5-fold stimulation) (7). However, the expression of Fluc directed by DHAV IRES was severely inhibited in the presence of 2A protease.
Fig 7.
Inhibition of the DHAV IRES activity in the presence of cleaved eIF4G. (A) Dicistronic plasmids (4 μg) containing the indicated IRES sequences were transfected into BHK cells in the absence (−) or presence (+) of plasmid pT7-2A encoding SVDV 2A protease (1 μg). After 20 h, cell extracts were prepared and analyzed for Rluc and Fluc expression as described in the legend for Fig. 2. Samples were also analyzed by immunoblotting to analyze the status of eIF4G. The position of the C-terminal cleavage product of eIF4G is indicated (Ct). (B) Dual luciferase assays were performed on cell extracts from three separate transfections. The relative luciferase activity of Rluc (open bars) and Fluc (solid bars) from each DNA construct was normalized to that from the plasmid pRL-EMCV in the absence of pT7-2A, with the value set at 100%. Error bars indicate standard errors of the means.
The state of eIF4G in cells expressing the SVDV 2A protease was examined by immunoblotting analysis for eIF4G (Fig. 7A); the C-terminal cleavage product was observed only in the presence of 2A protease. Inhibition of the activity of DHAV IRES correlated very closely with eIF4G cleavage. These data indicate that the intact eIF4G is required for DHAV internal initiation of translation and that the HAV IRES element is no longer the only one abolished by cleavage of eIF4G. These results also demonstrate that the DHAV IRES is different from the type I, II, and IV IRES elements.
DISCUSSION
In this study, we have demonstrated that the 5′ UTR of the DHAV contains an IRES element that functions efficiently in RRLs and in mammalian cells. Recently, the IRES elements of picornaviruses have been classified into four groups (3). The type IV picornavirus IRES elements, also called HCV-like IRES elements, have recently been described; this group includes the elements of PTV-1, PEV-8, SV2, SPV9, AEV, and SVN (2, 13–15, 52). DHAV has been predicted to contain HCV-like IRES elements (17, 24) and to possess a type II IRES similar to that of LV and HPeV (30, 50). Here, we describe the functional and structural analysis of the IRES of DHAV and provide evidence that this IRES element shares similar structural characteristics with those of type IV IRES elements; meanwhile, it has significant differences from other type IV IRESs. Thus, we propose that the DHAV IRES element is a distinct type IV IRES element of picornavirus.
Our mutagenesis data demonstrated that the optimal functional DHAV IRES locates within nt 232 to 651 of the 5′ UTR. This region was assayed within both monocistronic constructs and dicistronic constructs. Like SV2 (approximately 440 nt) (13), the functional element within the 5′ UTR of DHAV (approximately 420 nt) is somewhat larger than those of PEV-8, HCV, and PTV-1 IRES elements. However, additional sequences are within the predicted DHAV domains Id and Ie, and these two extra domains are the unique feature of DHAV IRES among type IV IRESs. The domain Id containing a small stem-loop structure is a highly conserved region among different types of DHAV. Mutation of the AGCC motif in the loop disrupted predicted base-pairs in the stem and resulted in the inhibition of DHAV IRES activity. Compensatory mutations designed to restore the base pairing in this structure efficiently rescued activity (Fig. 5). Thus, domain Id is important for DHAV IRES activity. In contrast, domain Ie appears not to be an essential region for DHAV IRES, although domain Ie contains a highly conserved apical nonanucleotide and a long stem composed of interrupted base-paired nucleotides with a relatively conserved structure and many predicted paired covariant mutations. Although mutational analysis predicates that the terminal stem-loop structure of domain Ie does not apparently inhibit the IRES activity, removal of domain Ie from the region of DHAVΔ1-229 within the dicistronic construct (pRF-DHAVΔ1-229/253-371) and within the monocistronic construct (pDHAVΔ1-371) both resulted in very small declines in activity.
Like PTV-1 IRES (14), DHAV IRES is predicted to lack a structure analogous to domain IIIa within the HCV IRES. The PEV-8 and SV2 IRES elements, in contrast, were predicted to contain a domain IIIa but lack a domain IIIc (13).
In contrast to picornavirus IRES types I to III, the DHAV IRES lacks a typical polypyrimidine tract located about 20 nt upstream of an AUG codon at the 3′ end of the element. The DHAV IRES can function well without the need for any viral coding sequence, and the secondary structure predictions indicate that there is no region equivalent to the HCV IRES domain IV (46). In this respect, there are some differences between the picornavirus “HCV-like” IRES (type IV IRES) elements in this respect, such as the SV2 IRES is predicted to contain a domain IV region (13), and the addition of the coding sequence resulted in more-efficient IRES activity from the SVN sequence (52), whereas the PTV-1 and AEV do not contain a domain IV, and they do not require any coding sequence for optimal activity (2, 14).
The PTV-1 and DHAV IRES elements have higher similarity in the nucleotide sequences (about 49 to 51% overall identity). The most conserved region between the PTV-1 and DHAV IRES elements includes domain IIIe, which is composed of a stem with an apical loop consisting of GACA in PTV-1 and GAUA in DHAV (Fig. 1C). Indeed, the corresponding region of the DHAV IRES exhibits 100% sequence identity with that in domain IIIe of HCV and AEV (Fig. 1C). Mutation of this highly conserved GAUA motif within the DHAV IRES sequence severely disrupted DHAV IRES activity. The activities of HCV and PTV-1 IRES were also inhibited by the mutation of this loop region (14, 35, 42). It is known that domain IIId and domain IIIe of the HCV IRES interact with the 40S ribosomal subunit (27, 39, 48). Mutation of the conserved GGG motif within the apical loop of domain IIId abolished the DHAV IRES activity. However, the stem of domain IIId of DHAV IRES is shorter than that of HCV-like IRES elements, and the loop is larger than that of any other HCV-like IRES elements. A critical feature of the secondary structure models for the HCV-like IRES elements is the presence of a pseudoknot structure (domain IIIf). The DHAV sequence is also proposed to form this structure (Fig. 1B). Through mutational analysis of the sequences predicted to form the pseudoknot structure, we have fully proved the formation of this structure in the DHAV IRES. The pseudoknot consists of two stem structures linked by two loops. Mutations within the S1 or S2 regions (stems of the pseudoknot) that were expected to disrupt the predicted base-pairs in the stems of pseudoknot structure greatly inhibited IRES activity (Fig. 6), and mutations within the S3 region (stem of loop 2) also abolished the DHAV IRES activity (Fig. 6). Compensatory mutations designed to restore the base pairing in this structure, albeit with altered original sequences, efficiently rescued activity (Fig. 6). Therefore, these results strongly support the predicted structure.
Although the DHAV IRES possesses core structure domains of type IV IRES elements, it is inhibited significantly by the coexpression of an enterovirus 2A protease that induces cleavage of eIF4G. This feature completely distinguishes the DHAV IRES from the type I, II, and IV IRES elements, but it is very similar to that of the type III IRES elements. Henceforth, the HAV IRES is no longer unique among all picornavirus IRESs in that it was inhibited if translation initiation factor eIF4G was cleaved. In contrast, type I, II, and IV IRES elements can function when eIF4G is cleaved, and the activity of type I IRES elements was actually stimulated quite dramatically within certain cell types (like BHK cells) (3).
A key difference between the properties of HCV-like IRES elements and most picornavirus IRES elements is the role of the translation initiation complex eIF4F. For example, neither the cleavage of eIF4G nor dominant negative mutant forms of eIF4A can affect PTV-1 IRES (14, 26). Thus, the PTV-1 IRES functions independently of eIF4F within RRL. This result is consistent with the lack of a requirement for any component of the eIF4F protein for the formation of 48S preinitiation complexes in vitro from purified components (41). However, it is interesting that the AEV IRES showed some requirement for components of eIF4F (2). A new study has found that the SVV IRES was also shown to be inhibited by about 50% in the presence of cleaved eIF4G (52). When eIF4G was cleaved, their IRES activities were reduced, demonstrating an unusual type IV picornavirus IRES element that partially depends on an intact eIF4G. These findings provide strong support for the main findings in this study. If the DHAV IRES element is considered a type IV picornavirus IRES element, it must be a very special one, because IRES activity is almost completely inhibited by the coexpression of an enterovirus 2A protease that induces eIF4G cleavage. Although AEV IRES displayed significant resistance to hippuristanol (an inhibitor of eIF4A) (6), the activity was still moderately inhibited by this reagent. Indeed, it was also shown that a dominant negative mutant of eIF4A, which does not block HCV and PTV-1 IRES activity, did have some inhibitory effect on the SPV9 IRES (15). We suggest that not all type IV picornavirus IRESs function independently of eIF4F, although the role of eIF4F in the activity of the IRES of DHAV, AEV, and SPV9 is still not clear. Further studies are required to determine whether eIF4A is essential for the function of the DHAV IRES element.
DHAV was recently classified as the only member of a new genus Avihepatovirus of the family Picornaviridae, and complete genome sequence determination showed that it exhibits several unique features among picornaviruses. For example, the most unique feature is the association of an aphthovirus-like 2A1 and parechovirus-like 2A2 and the unprecedented presence of an AIG1 domain in the N terminus of 2A2 (17, 30). It is possible that recombination has occurred. Another unique feature is its IRES element. Despite the presence of structure domains similar to type IV picornavirus IRES, the DHAV IRES element is clearly distinct from type IV IRES in that it is severely inhibited in the presence of cleaved eIF4G, is as much as 140 nt larger than that of PTV-1, and possesses two extra domains, Id and Ie, not found in any other type IV IRES (although domain Ie does not appear to be essential for DHAV IRES activity). Due to these significant similarities and differences between DHAV IRES and type IV IRES, we suggest that IRES elements have been exchanged between unrelated virus families by recombination and support the hypothesis that RNA viruses consist of modular coding and noncoding elements that can exchange and evolve independently (3). Further studies on DHAV IRES element are necessary to more fully understand the origin and evolutionary potential of DHAV, including picornavirus.
ACKNOWLEDGMENT
This work was supported by the earmarked fund for Modern Agro-industry Technology Research System (CARS-43) and Innovative Research Team in University of China from the Ministry of Education (no. IRT0866).
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Ali IK, McKendrick L, Morley SJ, Jackson RJ. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 75:7854–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bakhshesh M, et al. 2008. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J. Virol. 82:1993–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belsham GJ. 2009. Divergent picornavirus IRES elements. Virus Res. 139:183–192 [DOI] [PubMed] [Google Scholar]
- 4. Belsham GJ, Jackson RJ. 2000. Translation initiation on picornavirus RNA, p 869–900 In Sonenberg N, Hershey JWB, Mathews MB. (ed), Translational control of gene expression. Cold Spring Harbor monograph 39. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 5. Bochkov YA, Palmenberg AC. 2006. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. Biotechniques 41:283–284, 286, 288 passim [DOI] [PubMed] [Google Scholar]
- 6. Bordeleau ME, et al. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2:213–220 [DOI] [PubMed] [Google Scholar]
- 7. Borman AM, Bailly JL, Girard M, Kean KM. 1995. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 23:3656–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borman AM, Kean KM. 1997. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology 237:129–136 [DOI] [PubMed] [Google Scholar]
- 9. Borman AM, Le Mercier P, Girard M, Kean KM. 1997. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 25:925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Borman AM, Michel YM, Kean KM. 2001. Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J. Virol. 75:7864–7871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown EA, Zajac AJ, Lemon SM. 1994. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 68:1066–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cello J, Paul AV, Wimmer E. 2002. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297:1016–1018 [DOI] [PubMed] [Google Scholar]
- 13. Chard LS, Bordeleau ME, Pelletier J, Tanaka J, Belsham GJ. 2006. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J. Gen. Virol. 87:927–936 [DOI] [PubMed] [Google Scholar]
- 14. Chard LS, Kaku Y, Jones B, Nayak A, Belsham GJ. 2006. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J. Virol. 80:1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Breyne S, Yu Y, Pestova TV, Hellen CU. 2008. Factor requirements for translation initiation on the simian picornavirus internal ribosomal entry site. RNA 14:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Rijk P, Wuyts J, De Wachter R. 2003. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics 19:299–300 [DOI] [PubMed] [Google Scholar]
- 17. Ding C, Zhang D. 2007. Molecular analysis of duck hepatitis virus type 1. Virology 361:9–17 [DOI] [PubMed] [Google Scholar]
- 18. Dorner AJ, et al. 1984. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J. Virol. 50:507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doudna J, Sarnow P. 2007. Translation initiation by viral internal ribosome entry sites, p 129–153 In Mathews MB, Sonenberg N, Hershey JWB. (ed), Translational control in biology and medicine. Cold Spring Harbor monograph 48. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20. Ekstrom JO, et al. 2007. Replication of Ljungan virus in cell culture: the genomic 5′-end, infectious cDNA clones and host cell response to viral infections. Virus Res. 130:129–139 [DOI] [PubMed] [Google Scholar]
- 21. Fu Y, et al. 2008. Molecular detection and typing of duck hepatitis A virus directly from clinical specimens. Vet. Microbiol. 131:247–257 [DOI] [PubMed] [Google Scholar]
- 22. Fuerst TR, Niles EG, Studier FW, Moss B. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83:8122–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gingras AC, Raught B, Sonenberg N. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913–963 [DOI] [PubMed] [Google Scholar]
- 24. Hellen CU, de Breyne S. 2007. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 81:5850–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. James BD, Olsen GJ, Pace NR. 1989. Phylogenetic comparative analysis of RNA secondary structure. Methods Enzymol. 180:227–239 [DOI] [PubMed] [Google Scholar]
- 26. Kaku Y, Chard LS, Inoue T, Belsham GJ. 2002. Unique characteristics of a picornavirus internal ribosome entry site from the porcine teschovirus-1 talfan. J. Virol. 76:11721–11728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kieft JS, Zhou K, Jubin R, Doudna JA. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7:194–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim MC, et al. 2009. Development of duck hepatitis A virus type 3 vaccine and its use to protect ducklings against infections. Vaccine 27:6688–6694 [DOI] [PubMed] [Google Scholar]
- 29. Kim MC, et al. 2007. Recent Korean isolates of duck hepatitis virus reveal the presence of a new geno- and serotype when compared to duck hepatitis virus type 1 type strains. Arch. Virol. 152:2059–2072 [DOI] [PubMed] [Google Scholar]
- 30. Kim MC, et al. 2006. Molecular analysis of duck hepatitis virus type 1 reveals a novel lineage close to the genus Parechovirus in the family Picornaviridae. J. Gen. Virol. 87:3307–3316 [DOI] [PubMed] [Google Scholar]
- 31. Klinck R, et al. 2000. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA 6:1423–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Knudsen B, Hein J. 2003. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 31:3423–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larsen GR, Semler BL, Wimmer E. 1981. Stable hairpin structure within the 5′-terminal 85 nucleotides of poliovirus RNA. J. Virol. 37:328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lloyd RE. 2006. Translational control by viral proteinases. Virus Res. 119:76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. 2000. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 7:1105–1110 [DOI] [PubMed] [Google Scholar]
- 36. Marvil P, et al. 1999. Avian encephalomyelitis virus is a picornavirus and is most closely related to hepatitis A virus. J. Gen. Virol. 80:653–662 [DOI] [PubMed] [Google Scholar]
- 37. Nicholas KB, Nicholas HB, Jr, Deerfield DW. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14 [Google Scholar]
- 38. Niepmann M. 2009. Internal translation initiation of picornaviruses and hepatitis C virus. Biochim. Biophys. Acta 1789:529–541 [DOI] [PubMed] [Google Scholar]
- 39. Otto GA, Puglisi JD. 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119:369–380 [DOI] [PubMed] [Google Scholar]
- 40. Pan W, et al. 1999. Vaccine candidate MSP-1 from Plasmodium falciparum: a redesigned 4917 bp polynucleotide enables synthesis and isolation of full-length protein from Escherichia coli and mammalian cells. Nucleic Acids Res. 27:1094–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pisarev AV, et al. 2004. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 78:4487–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Psaridi L, Georgopoulou U, Varaklioti A, Mavromara P. 1999. Mutational analysis of a conserved tetraloop in the 5′ untranslated region of hepatitis C virus identifies a novel RNA element essential for the internal ribosome entry site function. FEBS Lett. 453:49–53 [DOI] [PubMed] [Google Scholar]
- 43. Roberts LO, Seamons RA, Belsham GJ. 1998. Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA 4:520–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sakoda Y, Ross-Smith N, Inoue T, Belsham GJ. 2001. An attenuating mutation in the 2A protease of swine vesicular disease virus, a picornavirus, regulates cap- and internal ribosome entry site-dependent protein synthesis. J. Virol. 75:10643–10650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sambrook J, Russell DW. 2006. The condensed protocols from Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46. Sarnow P. 2003. Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis. J. Virol. 77:2801–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Svitkin YV, et al. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7:382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tallet-Lopez B, et al. 2003. Antisense oligonucleotides targeted to the domain IIId of the hepatitis C virus IRES compete with 40S ribosomal subunit binding and prevent in vitro translation. Nucleic Acids Res. 31:734–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tseng CH, Knowles NJ, Tsai HJ. 2007. Molecular analysis of duck hepatitis virus type 1 indicates that it should be assigned to a new genus. Virus Res. 123:190–203 [DOI] [PubMed] [Google Scholar]
- 51. Tseng CH, Tsai HJ. 2007. Molecular characterization of a new serotype of duck hepatitis virus. Virus Res. 126:19–31 [DOI] [PubMed] [Google Scholar]
- 52. Willcocks MM, et al. 2011. Structural features of the Seneca Valley virus internal ribosome entry site (IRES) element: a picornavirus with a pestivirus-like IRES. J. Virol. 85:4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang GQ, Ge XN, Guo X, Yang HC. 2007. Genomic analysis of two porcine encephalomyocarditis virus strains isolated in China. Arch. Virol. 152:1209–1213 [DOI] [PubMed] [Google Scholar]
- 54. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]