Abstract
In addition to its central role as a template for replication and translation, the viral plus-strand RNA genome also has nontemplate functions, such as recruitment to the site of replication and assembly of the viral replicase, activities that are mediated by cis-acting RNA elements within viral genomes. Two noncontiguous RNA elements, RII(+)-SL (located internally in the tombusvirus genome) and RIV (located at the 3′-terminus), are involved in template recruitment into replication and replicase assembly; however, the importance of each of these RNA elements for these two distinct functions is not fully elucidated. We used an in vitro replicase assembly assay based on yeast cell extract and purified recombinant tombusvirus replication proteins to show that RII(+)-SL, in addition to its known requirement for recruitment of the plus-strand RNA into replication, is also necessary for assembly of an active viral replicase complex. Additional studies using a novel two-component RNA system revealed that the recruitment function of RII(+)-SL can be provided in trans by a separate RNA and that the replication silencer element, located within RIV, defines the template that is used for initiation of minus-strand synthesis. Collectively, this work has revealed new functions for tombusvirus cis-acting RNA elements and provided insights into the pioneering round of minus-strand synthesis.
INTRODUCTION
Plus-strand RNA viruses exhibit many similarities during genome replication, including the formation of membrane-bound viral replicase complexes, the production of minus-strand and abundant plus-strand RNAs, and the use of co-opted host factors (1, 15, 16, 18). One of the best-studied plus-strand RNA viruses is Tomato bushy stunt virus (TBSV), which has a single 4,800-nucleotide (nt) RNA genome (43). The virus-encoded replication proteins p33 RNA chaperone and p92pol RNA-dependent RNA polymerase (RdRp) participate in the formation of the membrane-bound viral replicase complex, which also contains several host proteins (3, 12, 16, 38, 40).
Replication of TBSV and other plus-strand RNA viruses involve several sequential steps, including selection of the viral plus-strand RNA template for replication, recruitment of the viral plus-strand RNA and the viral replication proteins from the cytosol to the subcellular membrane surfaces where replication takes place, assembly and activation of the viral replicase, minus-strand and then plus-strand RNA synthesis, and the release of progeny plus-strand RNAs from the replicase complex (5, 15, 17). This complex process helps to ensure that authentic viral templates are replicated and that the replication process is rapid and efficient.
The TBSV plus-strand RNA plays multiple roles during viral replication. In addition to its main function as a store of genetic information, the viral plus-strand RNA also regulates its own intracellular localization and recruitment to the site of RNA replication (20, 26, 32, 33, 44). Moreover, the TBSV plus-strand RNA serves as an assembly platform for the viral replicase, consisting of viral replication proteins, co-opted host proteins, and host lipids and/or membranes (11, 31, 32, 38). These replication-related functions are guided by various cis-acting elements within the TBSV plus-strand RNA, most notably by RII(+)-SL, which is located internally, and RIV, which is positioned 3′-terminally (14, 26, 33, 44). These two RNA segments (RII and RIV), along with the additional cis-acting elements RI and RIII, are retained in TBSV defective interfering (DI) RNAs, which are small virus-derived replicons used to study sequence functions (Fig. 1A) (28).
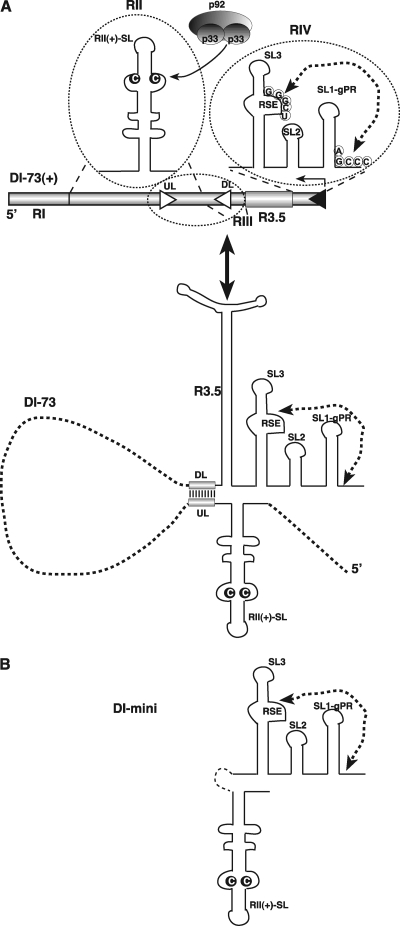
Fig 1.
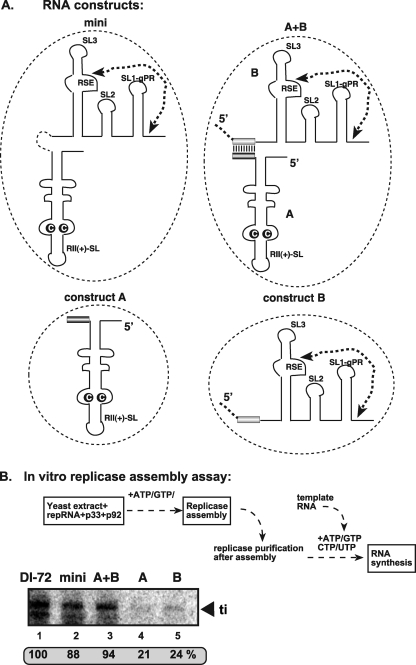
Schematic representation of TBSV DI-73 plus-strand repRNA and its derivatives carrying the three known cis-acting replication elements. (A) (Top) The three cis-acting sequences are circled. The characteristic C·C mismatch, which is critical for binding of p33/p92 replication proteins, within the RII(+)-SL is highlighted. The complementary nucleotides in the replication silencer element (RSE) and the genomic promoter (gPR) that form a 5-bp region are indicated with an arrow. Note that R3.5 serves as a translation enhancer, which is missing from DI-72 repRNA. (Bottom) A long-range interaction between UL-DL elements brings RII(+)-SL and RSE/gPR into proximal positions. (B) Predicted secondary structure of mini-RNA used as a model template for testing the assembly of the TBSV replicase complex in vitro in yeast CFE. Note that the UL-DL interaction and other portions of DI-73 repRNA are replaced by a short sequence from RIV(+).
Other plant viruses also contain specific sequences in their viral plus-strand RNAs that affect RNA recruitment and the assembly of their cognate replicase complexes. For example, short stem-loops within the 3′ UTR of Tobacco mosaic virus (TMV) plus-strand RNA can bind specifically to the TMV 126K replication protein in vitro (6, 19). The Brome mosaic virus (BMV) 1a protein participates in template selection and recruitment via interaction with the 1a-responsive element present in BMV RNAs (41, 42). A Y-shaped RNA element in the 3′ UTR in RNA2 of Red clover necrotic mosaic virus is specifically recognized by its cognate viral replication proteins, aiding recruitment of this viral RNA into replication (2, 7).
Dissection of the actual function(s) of cis-acting RNA elements such as those mentioned above can be hindered by the sequential and cycling nature of the plus-strand RNA replication, where a particular replication step depends on the previous step(s) and portions of the process are repeated in many cycles. Consequently, mutations introduced into a viral RNA could directly or indirectly affect multiple steps in replication, making assignment of functions of specific cis-acting RNA sequences and structures challenging. To circumvent some of the above problems, we previously developed an in vitro replicase assembly assay based on yeast cell-free extract (CFE) and purified recombinant tombusvirus replication proteins (32). In our CFE assay, the viral plus-strand RNA has to be recruited to the membrane (derived from the organelles of Saccharomyces cerevisiae), followed by the assembly of the viral replicase complex and a single cycle of replication producing minus-strand and abundant plus-strand RNA progeny, which is released into the solution (31, 32).
In this study, we employed the CFE assay to dissect the functions of various known cis-acting elements in the TBSV plus-strand RNA. We show that an essential internal stem-loop structure, RII(+)-SL, has a dual function, serving both as a plus-strand RNA recruitment element and as a mediator of viral replicase assembly, with the former activity being able to function in trans. Using a novel two-component RNA system in the replication assay, we also showed that another cis-acting element, located in RIV and termed the replication silencer element (RSE), has to be present in the viral plus-strand RNA in order for it to serve as a functional template. These findings have provided important insights into the detailed steps of the replication process in tombusviruses.
MATERIALS AND METHODS
Yeast and bacterial strains.
Saccharomyces cerevisiae strain BY4741α (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was obtained from Open Biosystems (Huntsville, AL). Escherichia coli Top10 (Invitrogen, Carlsbad CA) and Epicurion BL21-codon-plus (DE3)-RIL cells (Stratagene, La Jolla, CA) were used to propagate plasmids and for expression of recombinant proteins, respectively. E. coli strain Stbl2 (Invitrogen, Carlsbad, CA) was used for maximizing the stability of the plasmids containing direct repeats [such as (MS2)233]. Also, we changed the typical growing temperature from 37°C to 30°C when using Stbl2.
E. coli expression plasmids.
pMAL-33 and pMAL92 were described earlier (34). pET-His-MBP-p33, expressing p33 with dual 6×His and maltose-binding protein (MBP) tags, was also obtained earlier (32). pMAL-MS233, containing TBSV p33 fused in-frame with bacteriophage MS2 coat protein (MS2-CP), was obtained by PCR amplification of the MS2-CP open reading frame (ORF) from pGBK-MS2-CFP (20) using primers 1576 (5′-GGAGTCTAGAGCTTCTAACTTTACTCAG) and 3269 (5′-CCGCCATGGGTAGATGCCGGAGTTTGC) containing XbaI and NcoI restriction sites. The TBSV p33 ORF was amplified from pMAL92 using primers 3313 (5′-CGGACCATGGGAGACCATCAAGAGAATG) and 2744 (5′-CGGCTGCAGCTATTTGACACCCAGGGAC) containing NcoI and PstI restriction sites, respectively. To get the desired clone, after gel isolation of the restriction enzyme-digested PCR products, we ligated the PCR products into pMAL-c2X digested with XbaI and PstI.
To obtain pMAL-MS292, the TBSV p92 ORF was fused in-frame to the MS2-CP ORF, which was PCR amplified from pGBK-MS2-CFP (20) using primers 1576 and 3269 containing XbaI and NcoI restriction sites. The TBSV p92 ORF was PCR amplified from pMAL92 using primers 3313 and 3529 (5′-CCAGCTGCAGTCAAGCTACGGCGGAGTCGAGG) containing NcoI and PstI restriction sites, respectively. After gel isolation of the restriction enzyme-digested PCR products, we ligated the PCR products into pMAL-c2X digested with XbaI and PstI.
Yeast expression plasmids.
The plasmids used to construct TBSV repRNAs are shown in Table 1. pGBK-His33 and pGAD-His92, expressing only 6×His-tagged p33 and p92, respectively, from the ADH1 promoter and pYC-DI72, were described previously (23). pGBK-Cup-(MS2)2-33 was obtained by fusing the CNV p33 ORF in frame with two copies of bacteriophage MS2-CP [(MS2)2, representing direct repeats of the MS2-CP ORF linked with a short linker (GAPGIHPGM) and also containing an internal poly-His tag]. The sequence of (MS2)2 was amplified from p(MS2)2PCBP2 (39) using primers 4194 (5′-CGGACCATGGCGGATATCGAAGGTCCCACC) and 4196 (5′ CCAGCCATGGGTCGTTTGGGTGATGGTGATGGTGGTGGCTGCCGCGTGG) containing an NcoI restriction site and cloned into NcoI-digested and dephosphorylated vector pGBK-His33/Cup1 (8). Similarly, pGAD-Cup-(MS2)2-92 was obtained by fusing the CNV p92 ORF in frame with (MS2)2 using primers 4194 and 4196 containing NcoI restriction site and cloned into NcoI-digested and dephosphorylated vector pGAD-His92-Cup1 (10).
Table 1.
Construction of TBSV repRNAs used in this work
| RNA | Plasmid used for PCR (reference) | Primer no. | Forward primer | Reverse primer |
|---|---|---|---|---|
| Mini | pYC-R2(SL)-4(ΔS4) (26) | 1300/1190 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGGCTGCATTTCTGCAATG |
| Mini(G·C) | pYC-R2(SL)-4(ΔS4)#57 (26) | 1300/1190 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGGCTGCATTTCTGCAATG |
| Mini-ΔGPR | pYC-R2(SL)-4(ΔS4)#24 (26) | 1300/1192 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGGCTTGTTCCGGTTGTCCGGT |
| Mini-ΔSL2 | pYC-R2(SL)-4(ΔS4)#23 (26) | 1300/1191 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGGCTGATTTCTGCAATGGTAGTGCTTCCAGCGAT |
| Mini-ΔSL3 | pYC-R2(SL)-4(ΔS4)#22 (26) | 1300/1190 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGGCTGCATTTCTGCAATG |
| Mini-52 (ΔSL2) | pYC-R2(SL)-4(ΔS4)#52 (26) | 1300/1190 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGGCTGCATTTCTGCAATG |
| Mini-SL3(C·G) | pYC-R2(SL)-4(ΔS4)#59 (26) | 1300/1190 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGGCTGCATTTCTGCAATG |
| Mini-MS2 (ΔSL2) | pYC-R2(SL)-4(ΔS4) (26) | 1300/4629 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGGCTGCATTTCTGCAATGTTACATGGGTGATCCTCATGTTAGTGCTTCCAGCGATCTCT |
| RI/III(−) | pYC-DI-ΔRIIΔRIV (26) | 20/23 | GGAAATTCTCCAGGATTTCTC | GTAATACGACTCACTATAGGGACCCAACAAGAGTAACCTG |
| DI72(+) | pYC-DI72(+) | 359/1190 | GTAATACGACTCACTATAGGAAATTCTCCAGGATTTC | GGGCTGCATTTCTGCAATG |
| WM | pYC-DI-6XMS2(+) | 359/1190 | GTAATACGACTCACTATAGGAAATTCTCCAGGATTTC | GGGCTGCATTTCTGCAATG |
| RII-G/C-M | pYC-DI(C·G)6XMS2(+) | 359/1190 | GTAATACGACTCACTATAGGAAATTCTCCAGGATTTC | GGGCTGCATTTCTGCAATG |
| WRM | pYC-DI-6XMS2(−) | 359/1190 | GTAATACGACTCACTATAGGAAATTCTCCAGGATTTC | GGGCTGCATTTCTGCAATG |
| RII-G/C-RM | pYC-DI(C·G)-6XMS2(−) | 359/1190 | GTAATACGACTCACTATAGGAAATTCTCCAGGATTTC | GGGCTGCATTTCTGCAATG |
| A(23) | pYCDI72(+) | 1300/693 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | CTCCACAAACTCAGACTG |
| B | PDI70(+)BR (37) | 268/1190 | GTAATACGACTCACTATAGGAGATTTACACTCATCTC | GGGCTGCATTTCTGCAATG |
| A(12) | pYCDI72(+) | 1300/1581 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGAGCTGCAGTCAGACTGAAGAGTCTGTC |
| A(2) | pYCDI72(+) | 1300/1782 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | CGAGGTCGACGTGATATGCAGACTCTCCACGGCTC |
| A(5′-29) | DNA A | 2964/1782 | GTAATACGACTCACTATAGGGAGTTTGTGGAGATGAGTGTAAACAAAGGAGCCTTTGGAC | CGAGGTCGACGTGATATGCAGACTCTCCACGGCTC |
| A/GPR | DNA A | 1300/3199 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGGCTGCATTTCTGCAATGTTCTCCACAAACTCAGACT |
| A/GPR/SL2 | DNA A | 1300/3202 | GTAATACGACTCACTATAGAGGTTTGTGAGAAGGTTGG | GGGCTGCATTTCTGCAATGTTCCGGTTGTCCGGTAGTGCTTCCCTCCACAAACTCAGACT |
pYC-DI(C·G)6XMS2(+) was generated by PCR amplifying regions I and II of DI-72(+) from pYC-DI/(C99·G) (33) using primers 542 and 1565 (21). The PCR product was digested with HindIII and BamHI and used to replace the corresponding region in pYC-DI72(+)/MS2 (20) treated with the same pair of enzymes. To create pYC-DI(C·G)-6XMS2−, a similar strategy based on the vector pYC-DI72−/MS2 was used (20).
RNA template production and annealing.
Single-stranded RNA (ssRNA) templates were obtained by in vitro transcription with T7 RNA polymerase using PCR-amplified DNA templates (21, 25). The reaction mixture was incubated at 37°C for 2 h; 2 μl of DNase I (10 U/ml from Roche) was added, and mixtures were incubated for ∼20 min at 37°C. After phenol-chloroform extraction, RNA was precipitated twice with isopropanol-ammonium acetate (10:1). To synthesize radioactively labeled RNA, a similar T7 reaction was performed, except that 5 μl of 10 mM rATP, rGTP, and rCTP and 1 mM rUTP supplemented with 0.1 μl of [32P]UTP was used. RNA annealing was done with RNA samples mixed in an equimolar ratio (20 pmol each) in 20 μl buffer containing 50 mM NaCl, 10 mM Tris-Cl, and 1 mM EDTA. The samples were incubated in a thermocycler at 95°C for 5 min and then cooled to 25°C by reducing the annealing temperature by 1°C per minute.
In vitro replication assay based on yeast CFE.
The cell extract was prepared from yeast strain BY4741, untransformed or transformed with pairwise combinations of pGBK-His33/Cup1, pGAD-His92-Cup1, pGBK-Cup1-(MS2)2-33, and pGAD-Cup1-(MS2)2-92 (see Fig. 9) as described previously (31, 32). Briefly, the CFE (1 μl) was preincubated on ice for 10 min in 10 μl cell-free replication buffer containing 50 mM HEPES-KOH (pH 7.4), 150 mM potassium acetate, 5 mM magnesium acetate, 0.2 M sorbitol, and 0.4 μl actinomycin D (5 mg/ml). Then, the reaction volume was adjusted to 20 μl with 1× cell-free replication buffer also containing 2 μl of 150 mM creatine phosphate; 2 μl of 10 mM ATP, CTP, and GTP; 0.25 mM UTP; 0.3 μl of [32P]UTP; 0.2 μl of 10-mg/ml creatine kinase; 0.2 μl of RNase inhibitor; 0.2 μl of 1 M dithiothreitol (DTT); and 0.5 μg RNA transcript. The reaction mixture also contained 4 pmol MBP-tagged TBSV p33 and 1 pmol MBP-p92 purified from E. coli cells. This reaction mixture was incubated at 25°C for 3 h. The reaction was terminated by adding 110 μl stop buffer (1% sodium dodecyl sulfate [SDS] and 0.05 M EDTA, pH 8.0), followed by phenol-chloroform extraction, isopropanol-ammonium acetate precipitation, and a washing step with 70% ethanol as described earlie(26). The RNA samples were electrophoresed under denaturing conditions (5% polyacrylamide gels containing 8 M urea) and analyzed by phospho-imaging using a Typhoon (GE) instrument as described previously (26).
Fig 9.
RNA templates containing the MS2-CP hairpins are efficiently recruited, but they are replication incompatible in vitro. (A) Denaturing-PAGE analysis of the CFE-based replication assay. The in vitro replication assay contained CFE prepared from yeast coexpressing p33 and p92 or (MS2)233 fusion protein (containing the dimeric CP of MS2) and p92, and equal amounts of the indicated TBSV repRNA templates. The full-length products are indicated with arrowheads on the right. Note that the repRNAs serve as both assembly factors and templates in this assay. (B) Western blot analysis of p33, p92, and (MS2)233 fusion proteins in yeast CFE. (C) Denaturing-PAGE analysis of the in vitro RNA recruitment assay. The assay contained CFE (as in panel A) and equal amounts of P32-labeled TBSV repRNA templates. After the recruitment assay, the membrane-associated P32-labeled repRNAs were quantified.
Purification of the recombinant tombusvirus replicase from yeast.
Yeast cells transformed with pGBK-His33, pGAD-His92, and pYC-DI72 were pregrown in SC-ULH− medium containing 2% glucose for 15 h at 23°C with shaking at 250 rpm. The affinity purification of the solubilized tombusvirus replicase was performed using ProBond resin (Invitrogen) as described previously (27). The obtained template-dependent replicase was then used in a standard replicase reaction using either DI-72− or RI/RIII− exogenous templates and [32P]UTP (26, 27).
In vitro assembly and purification of the TBSV replicase.
The cell-free replication assay was conducted at 20°C for 1 h as described above, except that the reaction volume was increased to 200 μl, while the final concentration of DTT was reduced from 10 mM to 2.5 mM. In addition, only rATP and rGTP were used, while [32P]UTP was omitted. The recombinant p33 was dually tagged with both MBP and 6×His. After incubation, the assay mixture was diluted with 800 μl chilled solubilization buffer, and affinity purification was done exactly as described previously (26, 27).
Protein purification from E. coli.
The MBP-tagged p33, p92, MS233, MS292, and Turnip crinkle virus p88C were purified from E. coli as described previously (34, 35). Briefly, expression of the MBP-tagged proteins was induced by isopropyl-d-thiogalactopyranoside (IPTG) in Epicurion BL21-codon-plus (DE3)-RIL cells (Stratagene). Cells were suspended in the column buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 25 mM NaCl, 10 mM β-mercaptoethanol) and were broken by sonication, and then the cell lysate was passed through the equilibrated amylose columns to bind MBP-tagged proteins. After passing the cell lysate, the columns were washed three times with ice-cold column buffer, eluted with column buffer containing 10 mM maltose, and stored at −80°C until further use.
In vitro RNA recruitment assay.
The recruitment assay was performed as described previously (12). Briefly, the recruitment assay based on yeast CFE and recombinant p33/p92 is similar to the replication assay, except for the following changes. 32P-labeled RNAs (2 pmol) were added to the CFE. In addition, only rATP and rGTP were used, while [32P]UTP was omitted. The assay was performed at room temperature for 1 h. Then the mixture was suspended in 980 μl of prechilled buffer A and centrifuged at 35,000 × g for 30 min at 4°C. Supernatant was discarded, and the pellet was washed twice more. After a final washing, the pellet was dissolved in 120 μl of stop buffer (1% sodium dodecyl sulfate [SDS] and 0.05 M EDTA, pH 8.0). Afterwards, standard RNA extraction and purification were performed, followed by autoradiography of the electrophoresed RNA samples as described previously (26).
In vitro RdRp reaction.
We used affinity-purified recombinant TCV p88C or affinity-purified CNV replicase from yeast in an RdRp assay as described previously (27, 35). Briefly, the RdRp reaction was performed in a 100-μl volume containing RdRp buffer (40 mM Tris [pH 8.0], 10 mM MgCl2, 10 mM DTT, 0.2 μl RNase inhibitor, 1 mM ATP, CTP, and GTP, 0.1 μl radioactive [32P]UTP, and 50 μl RdRp fraction). As an external template, 300 ng of DI-72− RNA or RI/III− RNA was added. Samples were incubated at 25°C for 2 h. The reaction was terminated by adding 70 μl SDS-EDTA (1% SDS, 50 mM EDTA [pH 8.0]) and 100 μl phenol-chloroform (1:1). The RdRp products were analyzed as described previously (27, 35).
S1 nuclease digestion of the RdRp products.
The RNA products from the in vitro RdRp assays were purified and divided into two aliquots of 8 μl each. S1 digestion was performed with one aliquot in a 20-μl reaction mixture containing 0.1 μl S1 nuclease (400 U/μl; Boehringer), 1× S1 nuclease buffer, and 2.5 μl 3 M NaCl. The other aliquot was treated similarly, except without S1 nuclease. After incubation at 37°C for 30 min, standard RNA analysis was performed (27, 35).
RESULTS
Defining minimal RNA sequences required for the assembly of the tombusviral replicase complex in vitro.
Previous work with TBSV identified the extended stem-loop RII(+)-SL in RII and the two subelements RSE and SL1-gPR in RIV as distinct cis-acting RNA elements, required for the assembly of the TBSV replicase in vivo, either in plant cells or in yeast, a surrogate host (26, 44) (Fig. 1A). The RSE and SL1-gPR interact via a 5-bp interaction, and this feature is important for replicase assembly (Fig. 1A) (26, 30). In the context of the TBSV genome, RII and RIV are separated by ∼3 kb, but they can be brought into close proximity by an RNA base-pairing bridge that forms between the UL (upstream linker) sequence just 3′ of RII(+)-SL and its complementary DL (downstream linker) sequence, positioned near RIV (Fig. 1A, bottom). Formation of this UL-DL bridge mediates efficient replicase assembly in vivo in both yeast and plant cells (44).
To further define and dissect the functions of the above-mentioned cis-acting RNA sequences during replication, we measured their effects on the assembly of the tombusvirus replicase complex in vitro (i.e., separate from their effects on template amplification) using our recently developed in vitro replicase assembly assay based on yeast CFE (32). In this assay, the recombinant viral proteins are affinity purified from E. coli, the various plus-stranded TBSV DI-RNA-based templates, termed replicon RNAs (repRNAs), are made via T7 transcription, while the CFE is prepared from yeast BY4741 (free of any TBSV components). The assembly assay contains ATP and GTP but lacks CTP and UTP, thus preventing cRNA synthesis or replication (Fig. 2A). After these components are mixed, assembly is allowed to occur, and then any assembled replicase complex is solubilized and affinity purified, a process that leads to the loss of the original repRNA template. Subsequently, a minus-strand RNA template, DI-72(-), is added to the purified replicase preparations to measure the copying activity of the replicase in vitro, which provides a measure of the efficiency of replicase assembly (Fig. 2A). In this replicase assembly assay, the originally added repRNA functions only in template recruitment and replicase assembly and does not act as a template for complementary-strand synthesis or replication.
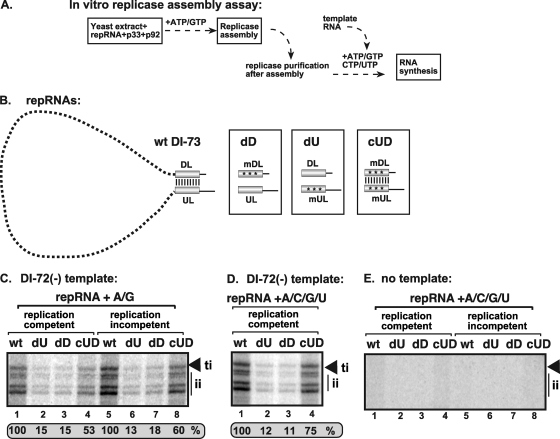
Fig 2.
UL-DL cis-acting element functions as an enhancer element for the replicase assembly (EERA). (A) Scheme of the in vitro TBSV replicase assembly assay performed with yeast CFE. Note that the recombinant p33 and p92pol are purified from E. coli, while the CFE was prepared from BY4741. After a 1-hour reconstitution, the membrane-bound replicase was solubilized with Triton X-100/SB3-10 detergent, followed by purification on an Ni column of the 6×His/MBP-tagged p33, which is an integral part of the replicase complex. The activity of the affinity-purified TBSV replicase was tested on DI-72(-) RNA added to each sample using the same amount of RNA. (B) Mutations within UL-DL, which interfere with base pairing or reform base pairing due to complementary mutagenesis. (C and D) Representative denaturing gels of 32P-labeled RNA products synthesized by affinity-purified TBSV replicase preparations obtained in TBSV replicase assembly assays in vitro with yeast CFE in the presence of all four or two ribonucleotide triphosphates. The replicase assembly assay contained the wt DI-73 plus-strand repRNA or versions with mutations in the UL-DL region, as shown in panel B. We used DI-73 plus-strand repRNA in the assembly assay because it contains the UL-DL elements, while DI-72 plus-strand repRNA lacks the corresponding RNA sequences. The replication-competent RNA was DI-73 based, while the replication-incompetent DI-73 carried a debilitating mutation. The template RNA was DI-72(-) repRNA, which produces both full-length (due to terminal initiation [ti]) and shorter (due to internal initiation [ii]) complementary products in the in vitro assay with the purified TBSV replicase. The level of full-length RNA synthesis was compared to that of the replicase activity obtained with DI-73 plus-strand repRNA (100%). (E) No RNA template was added to the in vitro assays with the purified TBSV replicase preparations, while the indicated repRNAs were used during the TBSV replicase assembly assays in vitro with yeast CFE prior to affinity purification of the TBSV replicase. Each experiment was repeated.
To determine if the UL-DL long-distance base-pairing interaction is required for the assembly of the tombusvirus replicase complex in vitro, we used wild-type (wt) DI-73 repRNA and mutants containing substitutions in UL, DL, or both (44), as shown in Fig. 2B. Mutations in either the UL or DL region (mutants dD and dU in Fig. 2B), which reduced base pairing between UL and DL, decreased the in vitro assembly of the replicase by ∼85% (Fig. 2C and D, lanes 2, 3, 6, and 7). Restoring the base pairing between UL and DL via complementary mutations in UL and DL (mutant cUD in Fig. 2B) resulted in ∼3- to 4-fold more efficient replicase assembly, versus the single mutants, for both replication-competent and incompetent repRNAs (Fig. 2C, lanes 4 and 8, respectively). Including all four ribonucleotides in the assembly assay led to slightly increased recovery levels for the compensatory mutant (Fig. 2D, lane 4), while omitting the template under the same conditions resulted in no products (Fig. 2E). Overall, as observed in vivo (44), the UL-DL interaction is also important for promoting replicase assembly in vitro. Since the UL-DL interaction was not essential for this process, but did stimulate the in vitro assembly of the replicase (albeit in a sequence-independent manner), we define it as an enhancer element for replicase assembly (EERA).
RII(+)-SL and RSE-SL2-gPR constitute the minimal RNA elements required for the assembly of the tombusviral replicase complex in vitro.
To test if RII(+)-SL and RSE-gPR elements (Fig. 1A) together were sufficient for the assembly of the replicase in vitro, we constructed a minimal RNA (mini-RNA) containing only these elements, as shown in Fig. 1B (26). Comparing the in vitro replicase assembly efficiency of the mini-RNA to the full-length DI-72 plus-strand repRNA revealed comparable levels of replicase assembly in the CFE-based replicase assembly assay (Fig. 3A, lanes 2 and 4 versus 1 and 3).
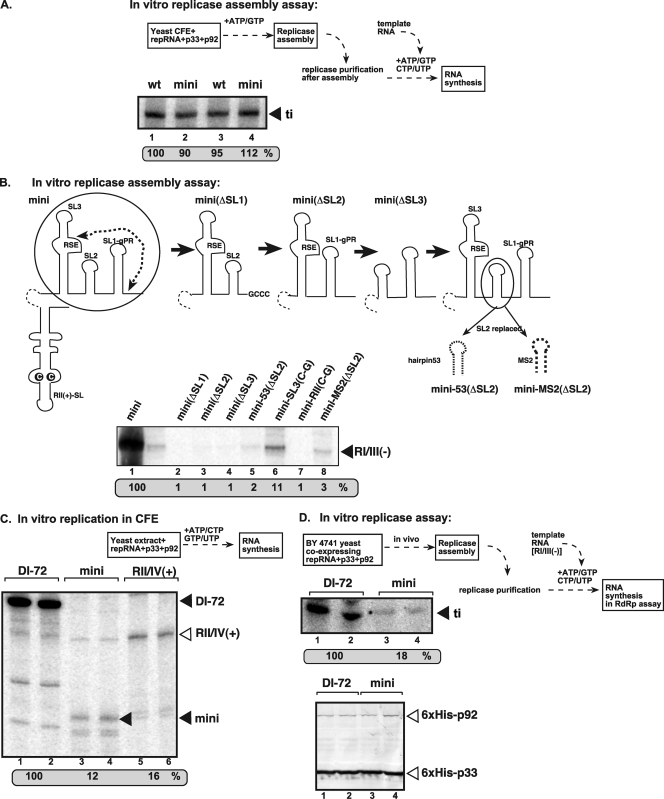
Fig 3.
A mini-RNA template with RII(+)-SL and RSE-gPR can efficiently support the in vitro assembly of the TBSV replicase. (A) Denaturing-PAGE analysis of the in vitro-reconstituted TBSV replicase in the presence of repRNAs. The replicase reconstitution assay contained CFE, affinity-purified recombinant TBSV p33 and p92pol, ATP/GTP, and equal amounts of TBSV repRNAs. After assembly and affinity purification, the activity of the replicase preparations was tested on DI-72(-) RNA template in vitro. See further details in Fig. 2. (B) Denaturing-PAGE analysis of the in vitro-reconstituted TBSV replicase in the presence of the indicated mini-repRNAs. Note that each construct had RII(+)-SL, and deletions and mutations were introduced only at the 3′ end (circled) of the DI mini-RNA construct. The only exception is mini-RII(C-G), which carried a single C-to-G mutation within the C·C mismatch in RII(+)-SL. The in vitro replicase assembly assay was performed as described for panel A, except that RI/III(-) was used as a template in the replicase activity assay. (C) Denaturing-PAGE analysis of the CFE-based replication assay. The in vitro replication assay contained yeast CFE, affinity-purified recombinant TBSV p33 and p92pol, and equal amounts of various TBSV repRNAs (as shown in Fig. 1). The full-length products are depicted with arrowheads on the right. Note that the repRNA serves as both an assembly factor and a template in this assay. (D) Denaturing-PAGE analysis of the in vitro replicase assay. The assay contained both the purified replicase from yeast coexpressing p33 and p92pol and the plus-strand repRNAs (as shown). The activity of the affinity-purified TBSV replicase was tested on RI/RIII(-) RNA, which was added to all samples in equal amounts. Note that the assembly of the replicase took place in yeast cells in the presence of coexpressed p33/p92 and actively replicating repRNA. The bottom image shows a Western blot demonstrating equivalent amounts of p33/p92 replication proteins in the purified tombusvirus replicase preparations used for the in vitro assay.
To test if each hairpin within DI mini-RNA is needed for supporting the assembly of the tombusvirus replicase in vitro, we made single deletions of each SL or mutations in DI mini-RNA, as shown schematically (Fig. 3B). Interestingly, deletion of any of the three SLs in the 3′ UTR led to complete abolishment of the assembly of the tombusvirus replicase in vitro (Fig. 3B, lanes 2 to 4). Also, as expected, a single point mutation within RII(+)-SL (Fig. 3B, lane 7) completely abolished while a point mutation within RSE [construct mini-SL3(C-G); Fig. 3B, lane 6] greatly reduced the assembly of the tombusvirus replicase in vitro. Since SL2 might serve as a “spacer” within SL1 and SL3, we replaced SL2 with either an artificial hairpin (hairpin 53) (26) or the MS2 hairpin (20), both of which are similar in size to SL2. Both hairpins greatly debilitated the assembly of the tombusvirus replicase in vitro (Fig. 3B, lanes 5 and 8), suggesting that neither of these heterologous hairpins could complement SL2 function during replicase assembly.
Thus, these data demonstrate that RII(+)-SL and RSE-SL2-gPR elements together are sufficient for the efficient assembly of the TBSV replicase in vitro.
To test the in vitro template function of the DI mini-RNA construct, all four ribonucleotides were added, along with the repRNA and purified p33 and p92, to the yeast CFE, allowing both assembly and RNA synthesis from the repRNA in the reaction (Fig. 3C). Comparing the replication efficiency of the mini-RNA template versus DI-72 repRNA in the CFE revealed that the mini-RNA template was highly deficient in this replication assay (Fig. 3C, lanes 3 and 4 versus 1 and 2). The larger RII/IV(+) template, carrying the additional sequences flanking RII(+)-SL and RSE-gPR, was also very inactive for RNA replication in vitro (Fig. 3C, lanes 5 to 6). Thus, while the DI mini-RNA is good at supporting replicase assembly, it is a poor template during replication (also see below).
The efficiency of in vivo assembly of replicase by DI-72 repRNA and DI mini-RNA template was also assessed using a modified in vitro replicase assay (Fig. 3D, top). The affinity-purified replicase from yeast cells coexpressing the mini-RNA template and His-tagged p33/p92 (Fig. 3D, bottom) showed a reduced ability to copy the exogenously provided RI/III(-) template compared with the replicase preparation from yeast coexpressing DI-72 and p33/p92 (Fig. 3D, middle, lanes 3 and 4 versus 1 and 2). The decreased isolated replicase activity from yeast cells for the mini-RNA is consistent with its being a poor template for replication in vitro (Fig. 3C); thus, there would be less of this template available for replicase assembly. Altogether, the in vivo and in vitro results show that the DI mini-repRNA is good at facilitating the assembly of the TBSV replicase but is a poor replicon. This is likely due, at least in part, to the absence of important cis-acting replication elements such as RI, which is present in DI-72 plus-strand repRNA (21, 22, 24, 36, 43).
Template competition reveals that the C·C mismatch in RII(+)-SL is important for competitiveness of an RNA template during in vitro replication.
Previous work indicated that RSE-gPR and possibly RII(+)-SL are important for forming an assembly platform for the replicase, while RII(+)-SL is proposed to aid in RNA recruitment to the cellular membranes (32). TBSV p33 is targeted to peroxisomal membranes (9, 13, 20, 29) and binds to RII(+)-SL with high affinity (33). Accordingly, it has been proposed that a key function of RII(+)-SL is to facilitate the recruitment of the TBSV repRNA to the site of viral replication on membranes via its interaction with p33 (14, 20, 33).
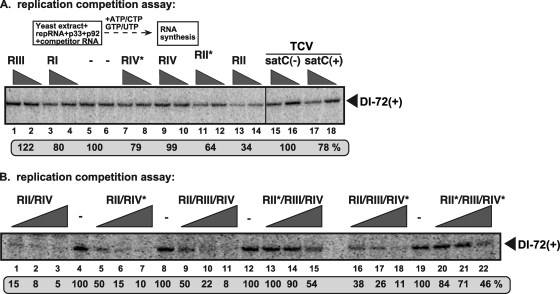
To determine the importance of RII(+)-SL and RSE-gPR for replication in the CFE, we performed template competition experiments in our CFE replication assay (Fig. 4, top). This involved adding defined viral segments containing different cis-acting elements as competitors to a replication assay for DI-72 plus-strand repRNA. Added plus-strand RI, RIII, or RIV, as well as heterologous TCV satC RNA, exhibited relatively poor competition against the accumulation of DI-72 plus-strand repRNA template in vitro (Fig. 4A, lanes 3 and 4, 1 and 2, 9 and 10, and 15 to 18, respectively), while RII was more competitive (Fig. 4A, lanes 13 and 14). The C·C mismatch in mutant RII*, which prevents the p33-RII(+)-SL interaction (33), reduced its competiveness in vitro compared to wt RII (Fig. 4A, compare lanes 11 and 12 with lanes 13 and 14). The mutant RIV*, in which the RSE-gPR interaction was disrupted (Fig. 1A) (30), was slightly more competitive than its wt RIV counterpart (Fig. 4A, compare lanes 7 and 8 with lanes 9 and 10). Since none of these short RNA templates are replication competent in the CFE assay, these RNAs likely inhibit replication of DI-72 plus-strand repRNA in vitro by competing for diffusible factors during RNA recruitment or replicase assembly. Results from competition assays with larger RNAs containing two or more of the above-described segments were also consistent with the single-segment results and indicated that wt RII has the greatest negative effect on DI-72(+) replication (Fig. 4B). This effect was partially related to RII's ability to bind to p33, as RNAs containing mutant RII* [which are unable to bind to p33 (33)] showed a marked reduction in competitiveness (Fig. 4B). Thus, in this factor-limited in vitro environment, RII, alone or with other RNA elements, likely sequesters p33 or p92 away from DI-72(+), leading to reduced replicase assembly and replication of DI-72(+).
Fig 4.
RII(+) RNA inhibits the in vitro assembly of the TBSV replicase. (A) Denaturing-PAGE analysis of the CFE-based replication assay. The in vitro replicase reconstitution assay contained yeast CFE, affinity-purified recombinant TBSV p33 and p92pol, and equal amounts of TBSV DI-72 plus-strand repRNA plus increasing amounts of competitor TBSV-derived RNA templates (RI, RII, RIII, and RIV [Fig. 1]) or the TCV-associated satC templates. The samples contained 0, 2, and 8 μg of competitor RNA, and each had 0.4 μg of DI-72(+) RNA template. The full-length DI-72-derived RNA products are depicted with arrowheads on the right. Note that the repRNA serves as both an assembly factor and a template in this assay, while the competitor RNA cannot assemble a functional replicase but can interfere with the replicase assembly process. The level of DI-72 repRNA replication in the no-competitor samples was set as 100%. (B) Another template competition in CFE-based replication assays. The competitor RNA contains the wt regions (RII, RIII, or RIV), a mutated RII (asterisk; C-to-G mutation in the C·C mismatch [Fig. 1A]), or a mutated RIV (asterisk; G-to-C mutation in gPR [Fig. 1A]). These mutations are known to interfere with the cis-acting replication functions of these regions. See further details in panel A. The samples contained 0, 3, and 9 μg of competitor RNA, and each had 0.4 μg of DI-72(+) RNA template.
A novel two-component RNA system supports TBSV replication in vitro.
To further dissect the roles of RII(+)-SL and RSE-gPR elements in TBSV RNA replication, we developed a novel two-component RNA replicase assembly assay based on CFE. One RNA, construct A (Fig. 5A), contained RII(+)-SL, and the other RNA, construct B (Fig. 5A), carried the RSE-gPR sequence. Constructs A and B also contained a 23-nt region of complementarity at their 3′ and 5′ ends, respectively, that would allow them to interact via base pairing (Fig. 5A). Testing the replicase activity of the affinity-purified TBSV replicase from CFE revealed that neither construct A nor construct B was able to efficiently support the assembly of the TBSV replicase when provided individually in the assembly assay (Fig. 5B, lanes 4 and 5, respectively). However, when both RNAs were present, the assembly of the TBSV replicase was as efficient as the single-component mini-RNA construct or DI-72 plus-strand repRNA (Fig. 5B, compare lane 3 with lanes 1 and 2). This result demonstrates that both RII(+)-SL and RSE-gPR RNA elements are required for replicase assembly, but they do not have to be present in the same RNA molecule.
Fig 5.
An efficient two-component RNA-based TBSV replicase assay. (A) Schematic representation of the RNA constructs used in the replicase assembly assay. Construct A contains functional RII(+)-SL, while construct B carries a RSE-gPR element. Note that constructs A and B can form a 23-bp heteroduplex that holds the two RNAs together as shown for the A+B construct. (B) Denaturing-PAGE analysis of the CFE-based replication assay. The in vitro reconstitution assay contained yeast CFE, affinity-purified recombinant TBSV p33 and p92pol, and equal amounts of the indicated TBSV repRNA templates. After in vitro reconstitution, the activity of the purified replicase preparations was tested using an RI/RIII(-) template. The full-length RNA product is depicted with an arrowhead on the right. Note that the repRNAs (A) serve only as assembly factors in this assay.
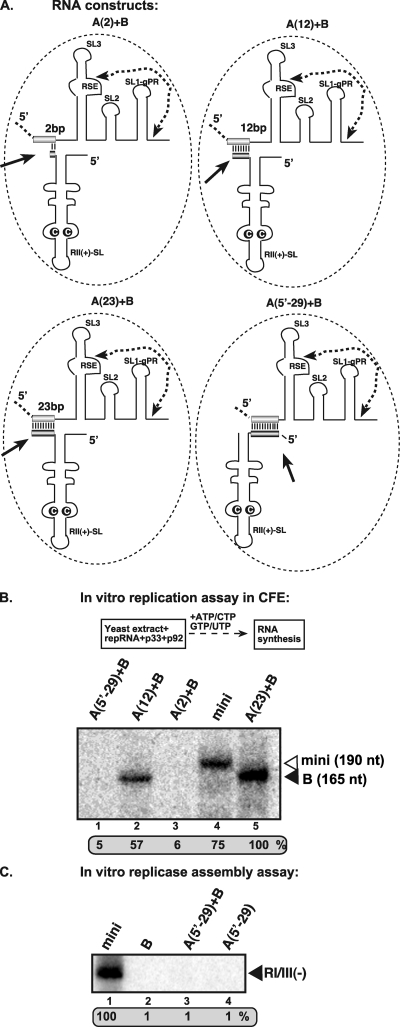
The complementarity between constructs A and B was important for in vitro RNA synthesis in the CFE replication assay, since constructs with 23 nt of complementarity supported RNA synthesis more efficiently than constructs with 12 nt of complementarity (Fig. 6B, lanes 5 versus 2) or that lacking extensive base pairing (Fig. 6B, lanes 5 versus 3). Interestingly, mixing construct A(5′-29) and construct B, which share a 29-bp tract of complementarity, did not support RNA synthesis in the CFE assay (Fig. 6B, lane 1). However, relative to construct A (23), the section of complementarity in construct A(5′-29) is at the opposite end of the RNA, which would result in a different and less proximal positioning of the RII(+)-SL and RSE-gPR [Fig. 6, compare A(23)+B with A(5′-29)]. Importantly, construct A(5′-29) alone or in combination with construct B did not support replicase assembly in the CFE-based assay (Fig. 6C), demonstrating that the replicase assembly step is defective, not the RNA synthesis step. This result suggests that not only do the two RNAs have to be physically close together in order to efficiently promote the assembly of the replicase and allow RNA synthesis, but also there are additional structural requirements with respect to their precise spatial orientation and proximity relative to each other. Another interesting finding from this analysis is that the functional replicase that assembled with the two-component system showed a preference for copying construct B, which contained RSE-gPR.
Fig 6.
RII(+)-SL and RSE-gPR sequences must be located in close proximity during the assembly of the TBSV replicase. (A) Schematic representation of the two-component RNA constructs used in the replicase assay. See further details in Fig. 5A. (B) Denaturing-PAGE analysis of the CFE-based replication assay. The in vitro reconstitution assay contained yeast CFE, affinity-purified recombinant TBSV p33 and p92pol, and equal amounts of the indicated TBSV repRNA templates. Note that the repRNAs (A) serve as both assembly factors and templates in this assay. The full-length RNA products are depicted with arrowheads on the right. (C) Denaturing-PAGE analysis of the in vitro reconstituted TBSV replicase in the presence of mini-repRNAs. The replicase assembly assay contained CFE, affinity-purified recombinant TBSV p33 and p92pol, ATP/GTP, and equal amounts of TBSV repRNAs. After assembly and affinity purification, the activity of the replicase preparations was tested on an RI/III(-) RNA template in vitro. See further details in Fig. 2.
RSE-gPR defines the template for RNA synthesis in the two-component RNA system in vitro.
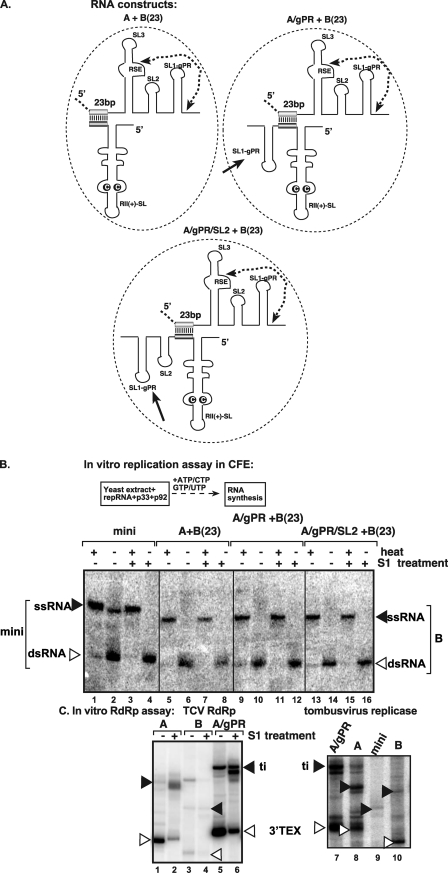
The results shown in Fig. 6 indicated that the TBSV replicase preferentially used construct B, carrying the RSE-gPR, as a template to make minus-strand RNA, and this notion was confirmed by polyacrylamide gel electrophoresis (PAGE) analysis of the RNA products synthesized (seen as a double-stranded RNA [dsRNA] in Fig. 7B, lane 6). As expected, when heat-denatured, the dsRNA product became single stranded (ssRNA in Fig. 7B, lane 5), and similar results were observed when the samples were treated with S1 nuclease prior to heating and gel analysis (Fig. 7B, lanes 7 and 8), supporting the double-stranded nature of the faster-moving product.
Fig 7.
cis replication of the template RNA carrying RSE-gPR in vitro. (A) Schematic representation of the two-component RNA constructs used in the replicase assay. See further details in Fig. 5A. (B) Nondenaturing-PAGE analysis of the CFE-based replication assay. The in vitro reconstitution assay contained yeast CFE, affinity-purified recombinant TBSV p33 and p92pol, and equal amounts of the indicated TBSV repRNA templates. Note that the repRNAs (A) serve as both assembly factors and templates in this assay. The full-length ssRNA and dsRNA products are depicted with arrowheads on the right. Note that only construct B, carrying RSE-gPR, can produce a complementary minus-strand RNA product in vitro, since ssRNA is visible only after denaturation of the dsRNA product. (C) Representative denaturing gel of 32P-labeled RNA products synthesized by TCV p88C RdRp (left) or the affinity-purified tombusvirus replicase preparation (right) in vitro in the presence of 1 μg of the indicated RNA transcripts. ti, de novo-initiated terminal products (depicted by black arrowheads). The samples were treated with S1 nuclease to show the 3′-terminal extension products (3′TEX), which change migration after treatment (35). Each experiment was repeated three times.
To test if construct A could be converted to an active template, we introduced SL1-gPR sequence (construct A/gPR; Fig. 7A) or SL1-gPR and SL2 sequences (construct A/gPR/SL2) into RII(+)-SL-containing construct A at a 3′ position. These new construct A derivatives were able to serve as promoters in an in vitro replicase assay based on an active TCV RdRp or the purified tombusvirus replicase (Fig. 7C, lanes 5 to 7). In contrast, constructs A/gPR and A/gPR/SL2 did not produce minus-strand RNA products when mixed with construct B in the CFE replication assay (Fig. 7B, lanes 9 to 16), and only construct B was copied in these two-component RNA systems. These data suggest that the RSE is an important determinant of template copying by the replicase and that, for in vitro replicase assembly in association with viral RNA, only an RNA carrying a complete RSE-gPR is used in cis by the replicase as a template for RNA synthesis.
The RII(+)-SL is needed for the assembly of the TBSV replicase in vitro.
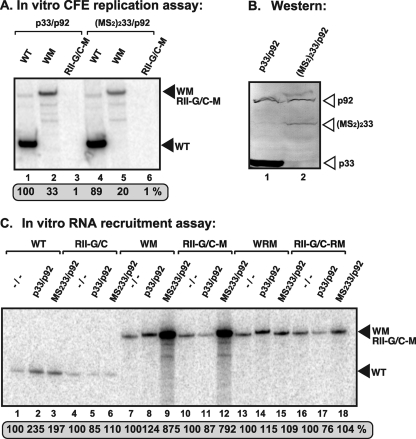
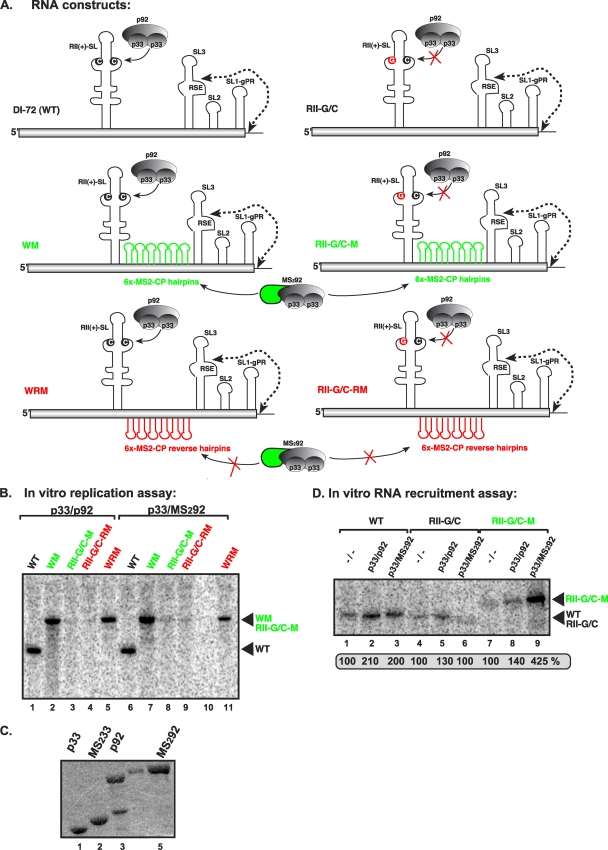
RII(+)-SL has been designated a template recruitment element (17, 33); however, it is not known if this RNA element is also required for the subsequent replicase assembly step. To test this, we inactivated the recruitment function of RII(+)-SL (via a C-to-G mutation in the critical C·C mismatch) (Fig. 1A) (33) but also introduced a heterologous “recruitment element,” namely, six RNA hairpins from bacteriophage MS2 that bind selectively to the coat protein (CP) of MS2 (4, 20), to obtain construct RII-G/C-M (Fig. 8A). We also tagged the TBSV p92 protein with a monomer of MS2-CP (39), creating MS292 (Fig. 8A) in order to promote binding of RII-G/C-M to the TBSV replicase via the heterologous MS2-CP domain (Fig. 8A). This arrangement was predicted to promote the viral RNA recruitment into replication by the binding of the MS2 hairpins in RII-G/C-M to the MS2-CP part of the p92 fusion protein, MS292.
Fig 8.
RII(+)-SL is required for in vitro assembly of the TBSV replicase. (A) Schematic representation of the RNA constructs used in the in vitro replication assay. Constructs WM and RII-G/C-M contain six copies of the MS2-CP hairpin (green), which can specifically bind to MS2-CP. p92 was fused to MS2-CP (green) as indicated. Constructs WRM and RII-G/C-RM contain six copies of the complementary MS2-CP hairpin sequence (red), which cannot bind to MS2-CP. The predicted status of binding of the RNA constructs to p33/p92 or the MS2-CP fusion proteins is shown with plain arrows (positive binding) or arrows crossed with red (no binding). (B) Denaturing-PAGE analysis of the CFE-based replication assay. The in vitro replication assay contained yeast CFE, affinity-purified recombinant TBSV p33 and p92pol or the MS2-CP fusion p92 protein, and equal amounts of the indicated TBSV repRNA templates. The full-length products are indicated with arrowheads on the right. Note that the repRNAs serve as both assembly factors and templates in this assay. (C) SDS-PAGE analysis of the affinity-purified recombinant proteins. (D) Denaturing-PAGE analysis of the in vitro RNA recruitment assay. The assay contained CFE, affinity-purified recombinant TBSV p33 and p92pol or the MS2-CP fusion p92 protein, and equal amounts of 32P-labeled TBSV repRNA templates. After the recruitment assay, the membrane-associated P32-labeled repRNAs were quantified.
In vitro assembly of the TBSV replicase with purified recombinant p33 and MS292 fusion protein (Fig. 8C) in the CFE assay revealed that construct RII-G/C-M did not support replication (Fig. 8B, lane 8). However, the in vitro RNA recruitment assay in CFE showed that wt p33 and the MS292 fusion protein did recruit RII-G/C-M RNA to the membrane ∼3-fold more efficiently than did wt p33 and p92 (Fig. 8D, compare lane 9 with 8). Thus, we conclude that the heterologous MS2-CP hairpins work with p33/MS292 fusion proteins in template recruitment. Importantly, the six MS2-CP hairpins did not hinder replication when inserted in wt DI-72 (construct WM in Fig. 8A; Fig. 8B, lane 2), and the hybrid MS292 was functional for replication (Fig. 8B, lane 6).
We also performed a second test with a dually MS2-CP-tagged p33 [(MS2)233] in which the viral replication proteins were expressed in yeast (31) via coexpression of (MS2)233 fusion protein and p92 only (in the absence of TBSV repRNA) (Fig. 9B). After preparation of the CFE from this yeast, various RNA templates were introduced and an in vitro replication assay was performed (Fig. 9A). Although (MS2)233 was expressed at a lower level than wt p33 in yeast (Fig. 9B), the CFE containing (MS2)233/p92 supported the replication of wt DI-72 plus-strand repRNA to levels similar to those obtained with p33/p92 (Fig. 9A, compare lanes 4 and 1). The same CFEs also allowed replication of WM repRNA, albeit at reduced levels (Fig. 9A, lanes 2 and 5) but did not support the replication of RII-G/C-M repRNA (Fig. 9A, lanes 3 and 6). The in vitro RNA recruitment experiments with CFE containing (MS2)233/p92 revealed efficient recruitment of RII-G/C-M RNA to the membrane (Fig. 9C, lane 12), suggesting that this step was performed more efficiently by (MS2)233/p92 with RII-G/C-M RNA than by p33/p92 with the wt DI-72 plus-strand repRNA (Fig. 9C, lane 2). From these data, we conclude that replacing the recruitment function of RII(+)-SL with the heterologous MS2-CP hairpins is sufficient for repRNA recruitment to the membrane but not sufficient to promote the functional assembly of the TBSV replicase. Thus, RII(+)-SL may have an additional essential function during the assembly of the TBSV replicase complex.
DISCUSSION
Role of UL-DL in replication assembly.
The two critical cis-acting elements, RII(+)-SL and RSE-gPR, are located ∼3 kb apart in the TBSV genomic RNA. However, a long-distance base-pairing interaction between UL-DL sequences brings RII(+)-SL and RSE-gPR into close proximity (Fig. 2B) (44). We found that UL-DL interaction is not necessary for the assembly of the TBSV replicase in vitro, but it boosts the assembly ∼10-fold (Fig. 2). The UL-DL interaction is not needed when RII(+)-SL and RSE-gPR are located at nearby positions (see the “mini” template lacking UL-DL in Fig. 3); therefore, it represents an unconventional type of enhancer element for replicase assembly (EERA) that does not directly interact with proteins.
The two-component system also confirmed the importance of the UL-DL interaction and revealed that RII(+)-SL and RSE-gPR could function when not covalently linked to each other. This suggests that there is no processive tracking of components along the RNA between RII(+)-SL and RSE-gPR during replicase assembly. Nonetheless, the requirement for base pairing between the two RNAs underscored the importance of proximity for these two elements. In addition to general proximity, their relative orientations and/or precise proximity also seems to be significant, as base pairing between sites that were not similar in location relative to the UL-DL interaction did not lead to functional replicase assembly [Fig. 6, A(5′-29)+B]. Thus, the UL-DL interaction acts indirectly to enhance the assembly process by optimally positioning RII(+)-SL and RSE-gPR relative to one another.
Dual role of RII(+)-SL during template recruitment and the assembly of the viral replicase.
Two of the most intriguing nontemplate roles of the viral plus-strand RNA are its essential function for template recruitment and assembly of the viral replicase complex (26, 32, 44). These functions are mediated by distinct cis-acting elements in the viral plus-strand RNA. In tombusviruses, one of these elements is the internally located RII(+)-SL (Fig. 1A), which binds to p33/p92 replication proteins via a C·C mismatch present in an internal loop (14, 33). Template competition experiments using our CFE replication assay revealed that RII(+)-SL, and particularly the C·C mismatch, is required for the template to inhibit the replication of the full-length DI-72 repRNA in vitro (Fig. 4). In contrast, other important regions of DI-72 plus-strand repRNA did not efficiently compete in the CFE-based replication assay, suggesting the lack of contribution by these sequences to viral RNA recruitment or direct binding to the viral replicase. These observations are consistent with the model that RII(+)-SL is an authentic RNA recruitment element that determines if a particular RNA is selected or recruited for replication by the tombusvirus replicase.
However, neither this result nor previous data (26, 33, 44) have precluded the possibility that, independent of template recruitment, RII(+)-SL also plays a role in the assembly of the replicase complex. These two activities are difficult to separate, since the assembly of the replicase depends on the prior RNA recruitment step. Consequently, in order to separate these two functions, we utilized a heterologous recruitment approach based on the specific MS2-CP–MS2 RNA hairpin interaction (4, 20, 39) using chimeric RNAs and fusion proteins. This alternative recruitment scheme was able to direct RNA templates to membranes; however, no replicase assembly was observed (Fig. 8 and 9). This suggests that the RII(+)-SL–p33/p92 interaction mediated by the C·C mismatch is also critical for replicase assembly. Indeed, it is possible that this interaction both tethers the template for protein-mediated transport to membranes and, at the same time, establishes the foundation from which a replicase complex can assemble. The requirement for this specific interaction may be related to allosteric effects on p33/p92 that are important for subsequent interactions and/or for precise positioning of these viral factors relative to host factors, which facilitates complex formation. Indeed, such a strategy of coupling the RNA-protein interaction with template recruitment and replicase assembly would streamline these consecutive processes and potentially provide a fitness benefit to the virus by minimizing unnecessary steps.
cis replication of the RNA template by the tombusvirus replicase depends on the presence of RSE.
The novel two-component RNA system was used to confirm that the RSE-gPR cis-acting sequence is also absolutely necessary for the assembly of the viral replicase in vitro (Fig. 5 and 6). Surprisingly, however, this system also revealed that only one of the two RNAs is used as a template by the newly assembled and activated membrane-bound replicase. Analysis of various modified template pairs indicated that selection of an RNA for copying required the presence of the RSE element in that template and that neither the promoter for minus-strand synthesis, gPR, nor a longer version of it, gPR+SL2, would suffice for this function (Fig. 7). This finding is in contrast with the data obtained with the solubilized and purified tombusvirus replicase or the related recombinant TCV RdRp, which efficiently used RNAs carrying gPR or gPR+SL2 sequences at 3′-terminal positions but lacking the RSE (Fig. 7). Indeed, this solubilized replicase is not associated with viral RNA and thus is able to encounter promoter sequences in the template RNAs added to the reaction. Conversely, our data indicate that in de novo replicase assembly, the nascent replicase may assemble with the viral RNA in a manner that positions its active site in proximity to the gPR, where minus-strand synthesis initiates. Our results also suggest that the RSE, in cooperation with covalently linked gPR, would be necessary for this to occur. Alternatively, it is possible that the proper positioning of RII(+)-SL and RIV might define the initiation site by the assembled replicase.
This type of tight coupling between replicase assembly/activation and template use could be limited to the pioneering round of minus-strand synthesis, as initial copying of the template would presumably dislodge any replicase or cofactor contacts involved in replicase assembly that were not involved in the RNA synthesis step. Nonetheless, this strategy would help to ensure that only templates capable of assembling functional replicase would be templates for replication. This and other cis-preferential replication strategies would be particularly beneficial at the early stage of infections initiated at a low multiplicity of infection.
Summarizing template recruitment and replicase assembly.
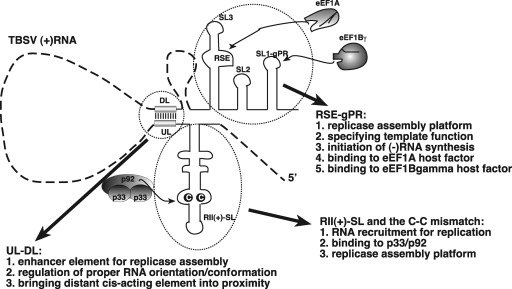
Replication of a viral RNA requires the preceding processes of template recruitment and replicase assembly. In TBSV, RII(+)-SL interacts with p33/p92, which targets the RNA for replication by shuttling it to membranes (Fig. 10). RII also likely directly contributes to replicase complex assembly in a manner that is, at least partially, linked to its p33/p92 binding activity. RSE-gPR forms part of the replicase assembly platform (Fig. 10) and binds to eEF1A, which is a component of the replicase complex (11, 12). RSE-gPR also specifies the template for minus-strand synthesis and harbors the core promoter for initiation, gPR. UL-DL plays an indirect role in replicase assembly by bringing RII(+)-SL and RSE-gPR into proximity in the proper orientation. Collectively, this diverse group of core RNA elements function jointly to mediate efficient replicase complex assembly.
Fig 10.
Known functions of the cis-acting replication elements in TBSV plus-strand RNA. Since RII(+)-SL could not be replaced by a heterologous RNA recruitment element, we propose that RII(+)-SL not only is needed for RNA recruitment into replication but also is required for assembly of the replicase complex. The RSE-gPR element is required for assembly of the replicase and also determination of the template for the replicase, since only RNAs which carry the RSE element are used as templates by the TBSV replicase. We propose that the role of UL-DL is not only to bring the RII(+)-SL and RSE-gPR elements into close proximity but also to ensure their proper orientation for replicase assembly.
ACKNOWLEDGMENTS
We thank Zhenghe Li for very helpful suggestions. p(MS2)2PCBP2 was a generous gift from J. B. Flanegan (University of Florida, Gainesville).
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (NIH-NIAID AI05767001A1) and the Kentucky Tobacco Research and Development Center at the University of Kentucky, awarded to P.D.N., and by a Philip Morris fellowship, awarded to K.P.
Footnotes
Published ahead of print 19 October 2011
REFERENCES
- 1. Ahlquist P, Noueiry AO, Lee W-M, Kushner DB, Dye BT. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. An M, et al. 2010. A Y-shaped RNA structure in the 3′ untranslated region together with the trans-activator and core promoter of Red clover necrotic mosaic virus RNA2 is required for its negative-strand RNA synthesis. Virology 405:100–109 [DOI] [PubMed] [Google Scholar]
- 3. Barajas D, Jiang Y, Nagy PD. 2009. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 5:e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertrand E, et al. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2:437–445 [DOI] [PubMed] [Google Scholar]
- 5. den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64:241–256 [DOI] [PubMed] [Google Scholar]
- 6. Goregaoker SP, Culver JN. 2003. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J. Virol. 77:3549–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iwakawa HO, et al. 2011. Template recognition mechanisms by replicase proteins differ between bipartite positive-strand genomic RNAs of a plant virus. J. Virol. 85:497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jaag HM, Stork J, Nagy PD. 2007. Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology 368:388–404 [DOI] [PubMed] [Google Scholar]
- 9. Jonczyk M, Pathak KB, Sharma M, Nagy PD. 2007. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362:320–330 [DOI] [PubMed] [Google Scholar]
- 10. Li Z, Barajas D, Panavas T, Herbst DA, Nagy PD. 2008. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 82:6911–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Z, et al. 2009. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology 385:245–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Z, et al. 2010. Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus-strand synthesis. PLoS Pathog. 6:e1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Monkewich S, et al. 2005. The p92 polymerase coding region contains an internal RNA element required at an early step in tombusvirus genome replication. J. Virol. 79:4848–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagy PD. 2008. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 46:217–242 [DOI] [PubMed] [Google Scholar]
- 16. Nagy PD, Pogany J. 2010. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against tomato bushy stunt virus. Adv. Virus Res. 76:123–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagy PD, Pogany J. 2006. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 344:211–220 [DOI] [PubMed] [Google Scholar]
- 18. Novoa RR, et al. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97:147–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Osman TA, Buck KW. 2003. Identification of a region of the tobacco mosaic virus 126- and 183-kilodalton replication proteins which binds specifically to the viral 3′-terminal tRNA-like structure. J. Virol. 77:8669–8675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Panavas T, Hawkins CM, Panaviene Z, Nagy PD. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology 338:81–95 [DOI] [PubMed] [Google Scholar]
- 21. Panavas T, Nagy PD. 2005. Mechanism of stimulation of plus-strand synthesis by an RNA replication enhancer in a tombusvirus. J. Virol. 79:9777–9785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panavas T, Nagy PD. 2003. The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis. J. Virol. 77:258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Panavas T, Nagy PD. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology 314:315–325 [DOI] [PubMed] [Google Scholar]
- 24. Panavas T, Pogany J, Nagy PD. 2002. Analysis of minimal promoter sequences for plus-strand synthesis by the Cucumber necrosis virus RNA-dependent RNA polymerase. Virology 296:263–274 [DOI] [PubMed] [Google Scholar]
- 25. Panavas T, Pogany J, Nagy PD. 2002. Internal initiation by the cucumber necrosis virus RNA-dependent RNA polymerase is facilitated by promoter-like sequences. Virology 296:275–287 [DOI] [PubMed] [Google Scholar]
- 26. Panaviene Z, Panavas T, Nagy PD. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 79:10608–10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Panaviene Z, Panavas T, Serva S, Nagy PD. 2004. Purification of the cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 78:8254–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pathak KB, Nagy PD. 2009. Defective interfering RNAs: foes of viruses and friends of virologists. Viruses Basel 1:895–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pathak KB, Sasvari Z, Nagy PD. 2008. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology 379:294–305 [DOI] [PubMed] [Google Scholar]
- 30. Pogany J, Fabian MR, White KA, Nagy PD. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 22:5602–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pogany J, Nagy PD. 2008. Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J. Virol. 82:5967–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pogany J, Stork J, Li Z, Nagy PD. 2008. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. U. S. A. 105:19956–19961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pogany J, White KA, Nagy PD. 2005. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 79:4859–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rajendran KS, Nagy PD. 2003. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J. Virol. 77:9244–9258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rajendran KS, Pogany J, Nagy PD. 2002. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 76:1707–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ray D, Na H, White KA. 2004. Structural properties of a multifunctional T-shaped RNA domain that mediate efficient tomato bushy stunt virus RNA replication. J. Virol. 78:10490–10500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ray D, White KA. 2003. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol. 77:245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Serva S, Nagy PD. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 80:2162–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spear A, Sharma N, Flanegan JB. 2008. Protein-RNA tethering: the role of poly(C) binding protein 2 in poliovirus RNA replication. Virology 374:280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stork J, Kovalev N, Sasvari Z, Nagy PD. 2011. RNA chaperone activity of the tombusviral p33 replication protein facilitates initiation of RNA synthesis by the viral RdRp in vitro. Virology 409:338–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sullivan ML, Ahlquist P. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang X, et al. 2005. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 79:13747–13758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. White KA, Nagy PD. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 78:187–226 [DOI] [PubMed] [Google Scholar]
- 44. Wu B, et al. 2009. A discontinuous RNA platform mediates RNA virus replication: building an integrated model for RNA-based regulation of viral processes. PLoS Pathog. 5:e1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]