Abstract
Tankyrase 1 is a poly(ADP-ribose) polymerase (PARP) which localizes to multiple subcellular sites, including telomeres and mitotic centrosomes. Poly(ADP-ribosyl)ation of the nuclear mitotic apparatus (NuMA) protein by tankyrase 1 during mitosis is essential for sister telomere resolution and mitotic spindle pole formation. In interphase cells, tankyrase 1 resides in the cytoplasm, and its role therein is not well understood. In this study, we found that herpes simplex virus (HSV) infection induced extensive modification of tankyrase 1 but not tankyrase 2. This modification was dependent on extracellular signal-regulated kinase (ERK) activity triggered by HSV infection. Following HSV-1 infection, tankyrase 1 was recruited to the nucleus. In the early phase of infection, tankyrase 1 colocalized with ICP0 and thereafter localized within the HSV replication compartment, which was blocked in cells infected with the HSV-1 ICP0-null mutant R7910. In the absence of infection, ICP0 interacted with tankyrase 1 and efficiently promoted its nuclear localization. HSV did not replicate efficiently in cells depleted of both tankyrases 1 and 2. Moreover, XAV939, an inhibitor of tankyrase PARP activity, decreased viral titers to 2 to 5% of control values. We concluded that HSV targets tankyrase 1 in an ICP0- and ERK-dependent manner to facilitate its replication.
INTRODUCTION
Herpes simplex virus types 1 and 2 (HSV-1 and HSV-2), members of the Herpesviridae family (17), possess large DNA genomes, share virion structures and replication mechanisms, and establish lifelong latency in host cells. The HSV genome comprises a 152-kb double-stranded DNA molecule that encodes approximately 80 gene products expressed in a temporally regulated cascade (6, 68). HSV genes are classified into three groups: immediate-early, early, and late genes. Immediate-early genes are expressed first upon infection and encode several transactivators, which in turn initiate transcription of the other early and some late genes; the latter are called leaky late or γ1 genes (12, 20, 48, 73). Early gene products include viral DNA replication factors that initiate viral DNA synthesis, which in turn stimulate expression of γ1 and true late (γ2) genes, encoding mainly virion structural proteins.
The immediate-early viral proteins ICP4, ICP27, ICP0, and ICP22 allow the virus to create an environment conducive to infection and counteract the intrinsic ability of cells to inhibit viral infection (29, 34, 55, 59). ICP4 and ICP27 play essential roles in stimulating robust viral gene expression (34). The immediate-early protein ICP0 activates viral and cellular gene expression and functions as an E3 ubiquitin ligase that degrades several cellular proteins (29). ICP0 targets the promyelocytic leukemia protein (PML), a major component of nuclear foci called ND10 bodies that repress viral gene expression. ICP0 interferes with several intrinsic host defense mechanisms, including the host interferon responses (29), thereby playing a major role in establishing permissive conditions for viral infection. ICP22 is required for efficient growth and expression of viral late genes in some, but not all, cultured cells (59). It also plays a role in posttranslational modification of cellular RNA polymerase II (see reference 27 and references therein).
HSV replication dramatically reorganizes the nuclear structure in the host cell (1). Incoming HSV genomes interact with ICP4 and ICP27 (24) and nucleate the formation of ND10/PML-like bodies (23), which are subsequently disrupted by ICP0 (21). The viral genomes then associate with several other viral and cellular proteins, forming complexes that ultimately develop into large structures called viral replication compartments (37, 39). HSVs are believed to synthesize DNA (51) and RNA (49) and to perform capsid assembly (36) in the replication compartments, defined by the presence of the HSV single-stranded DNA-binding protein ICP8 (40, 43). Other viral proteins known to accumulate within replication compartments include the origin-binding protein (UL9), heterotrimeric helicase-primase complex (UL5/UL8/UL52) polymerase and its accessory factor (UL30/UL42), and immediate-early ICP27, ICP4 viral transactivator, and ICP5 capsid proteins (8, 13, 24, 42, 52, 66). Cellular proteins involved in DNA replication, such as proliferating cell nuclear antigen, DNA ligase, and replication protein A (RPA), as well as those involved in DNA damage, such as ataxia-telangiectasia mutated and Rad3-related-interacting protein, are also translocated to the replication compartments (45, 53, 69, 70). The specific roles of these proteins in HSV infection and replication have not been well characterized.
Poly(ADP-ribose) polymerases (PARPs) mediate posttranslational protein modification by poly(ADP-ribosyl)ation and catalyze the transfer and polymerization of ADP ribose units from NAD+ to form branched ADP-ribose polymers covalently linked to heterologous acceptor proteins or PARPs themselves (44). Tankyrases 1 and 2 (TRF1-interacting, ankyrin [ANK]-related ADP-ribose polymerases) belong to the PARP family. The central domain of tankyrase 1 contains 24 ANK repeats and a 33-amino-acid motif that mediates protein-protein interactions, and its COOH-terminal region has homology to the catalytic domain of PARP, a highly conserved nuclear enzyme found in most eukaryotes. The catalytic domain of tankyrase 1 resides within the C-terminal region, which flanks a sterile alpha module (SAM) (63). Tankyrase 1 interacts with multiple proteins, including telomere repeat binding factor 1 (TRF1) (63), insulin-responsive aminopeptidase (IRAP) (14), 182-kDa tankyrase-binding protein (TAB182) (56, 60), and nuclear mitotic apparatus (NuMA) protein (10, 56). Tankyrase 2, a highly related paralogue, interacts with TRF1 (33) and several additional cellular proteins, including the aminopeptidase IRAP4, a vesicular protein that undergoes regulated translocation to the cell surface (14). Tankyrases 1 and 2 have approximately 85% amino acid sequence identity (35, 63) and interact via the SAM domains (18, 57). The role of tankyrase 1 in telomere maintenance has been assessed extensively (15), and it has been shown to be essential for sister telomere resolution and mitotic spindle pole formation by poly(ADP-ribosyl)ating (PARsylating) the NuMA protein during mitosis (9). Despite its known effect on telomeres, its role in interphase cells is not clearly understood. Only a fraction of tankyrase colocalizes with telomeres (61), while most tankyrase 1 is found in the cytoplasm and the vicinity of centrosomes (60), but its roles in the cytoplasm are also unclear.
In this study, we show by various techniques that tankyrases, especially tankyrase 1, are targeted by HSV to promote its replication. Tankyrase 1 but not tankyrase 2 was phosphorylated extensively upon HSV infection. HSV-1 infection induced tankyrase 1 redistribution involving its nuclear recruitment and the formation of complexes resembling replication compartments. Tankyrase 2 remained in the cytoplasm, similar to the case in mock infection. Inhibition of viral DNA synthesis by phosphonoacetic acid (PAA) prevented tankyrase 1 phosphorylation but not its redistribution, suggesting that viral DNA synthesis and/or viral γ2 protein synthesis is necessary. Tankyrase 1 underwent extracellular signal-regulated kinase (ERK)-dependent phosphorylation, which could be blocked by U0126. Its nuclear translocation was independent of ERK activity. In the early phase of infection, tankyrase 1 colocalized with nuclear ICP0 foci. We found that in the absence of infection, both HSV-1 and HSV-2 ICP0 proteins interact with tankyrase 1. The HSV-1 ICP0-null mutant R7091 failed to induce redistribution and phosphorylation of tankyrase 1. Thus, HSV infection induces ERK-dependent phosphorylation and ICP0-dependent nuclear accumulation of tankyrase 1. Depletion of tankyrases 1 and 2 by RNA interference (RNAi) indicated that they are required but functionally redundant for viral growth. Finally, inhibition of tankyrase PARP activity by XAV939 suppressed viral protein expression and decreased viral growth. We concluded that PARP activity of tankyrase is required for efficient HSV replication.
MATERIALS AND METHODS
Cells and viruses.
Vero (African green monkey kidney) and HEp-2 (human laryngeal carcinoma) cell lines were obtained from the RIKEN BioResource Center (Ibaraki, Japan). The U2OS (human osteosarcoma) cell line was a gift from Bernard Roizman (University of Chicago, IL). Vero cells were maintained in Eagle's minimum essential medium (MEM) supplemented with 8% calf serum (CS) (58), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine at 37°C in 5% CO2. HEp-2 and U2OS cells were maintained in Dulbecco's modified Eagle's medium and McCoy's 5A medium (Invitrogen, Carlsbad, CA), respectively, supplemented with 10% fetal CS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine, at 37°C in 5% CO2. The HSV-1 wild-type virus F and ICP0-null virus R7910 were kindly provided by Bernard Roizman (University of Chicago, IL), and HSV-2 wild-type virus 186 was also used. Virus stocks were propagated and titrated on Vero cell monolayers. UV-irradiated HSV-1 (UV-F) was prepared by exposure to a germicidal lamp (254 nm) for 3 min at a distance of 10 cm.
Plaque reduction assay.
We determined the effect of XAV939 on HSV-1 replication by using Vero cells in a plaque reduction assay. The assay was performed according to a previously described method, with some modifications (65). In brief, Vero cells were grown as a monolayer in a 6-well plate. Approximately 100 PFU of HSV-1 was added to the cells. The plate was incubated at 37°C in 5% CO2 for 1 h, with intermittent rocking at 15-min intervals, and then various amounts of XAV939 in normal medium were added to the monolayers. After incubation for 1 day at 37°C, the infected cells were fixed with 10% formalin, and the plaque number and size were evaluated.
Preparation of mitotic cells.
Exponentially growing HEp-2 cells were supplemented with 250 μg/ml nocodazole (Sigma) and cultured for 15 h to arrest cells at the mitotic phase. The cells undergoing mitosis were released into the culture medium by extensive agitation of the culture medium and vigorous shaking of the flasks (mitotic shake-off) (72). The detached cells were harvested by centrifugation, washed, and designated the mitotic fraction.
Phosphatase assays.
HEp-2 cells infected for 16 h with HSV at a multiplicity of infection (MOI) of 3 PFU/cell were washed in cold, sterile phosphate-buffered saline (PBS), harvested in Nonidet P-40 (NP-40) lysis buffer (10 mM Tris-HCl [pH 7.8], 150 mM NaCl, 1% NP-40, 1 mM EDTA, and protease inhibitor cocktail [Sigma]), and kept on ice. After 10 min, the lysates were centrifuged at 14,000 × g for 10 min at 4°C, and the supernatants were transferred to a fresh tube. Next, phosphatase buffer and MnCl2 were added to the lysates and dephosphorylated with 2,000 U of protein phosphatase (Lambda PP; New England BioLabs) for 30 min at 30°C. The reaction was terminated by the addition of 4× sodium dodecyl sulfate (SDS) buffer (1× final concentration), and the samples were boiled and analyzed by 7% SDS-polyacrylamide gel electrophoresis (11) and Western blotting with an anti-tankyrase (TNKS1) polyclonal antibody.
Extraction of cell lysates and Western blot analysis.
For subcellular fractionation, infected HEp-2 cells were washed once in sterile PBS, harvested, and pelleted. Nuclear and cytoplasmic extracts were prepared with NE-PER nuclear and cytoplasmic extraction reagents (Thermo) according to the manufacturer's instructions. For blotting, cells were lysed directly with SDS sample buffer containing 50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 6% 2-mercaptoethanol, and 0.0025% bromophenol blue. Cell lysates were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon-P membranes; Millipore). Membranes were blocked with blocking buffer (5% skim milk, 0.1% Tween 20 in PBS) for 1 h at room temperature. After incubation with appropriate primary antibodies for 1 h at room temperature, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies, detected using Westone (iNtRON Biotechnology). One membrane was sequentially incubated with different primary antibodies in Restore Plus Western blot stripping buffer (Thermo), blocked, and probed with secondary antibodies.
Immunofluorescence microscopy.
Cells grown on coverslips were washed in PBS three times and fixed for 10 min in 4% paraformaldehyde in PBS at room temperature. For indirect immunofluorescence microscopy, fixed cells were permeabilized in 1% Triton X-100 in PBS for 5 min at room temperature. The coverslips were inverted and touched to droplets (20 μl) of blocking buffer (4% goat serum, 1% bovine serum albumin in PBS-Tween [0.05%]) on a clean Parafilm sheet for 45 min at room temperature. Primary and Alexa Fluor-conjugated secondary antibodies (Molecular Probes) were diluted in blocking buffer and incubated for 60 min at room temperature. Coverslips were additionally incubated with DRAQ5 (Biostatus, Leicestershire, United Kingdom) for 20 min at room temperature to stain nuclear DNA. Samples were examined under a Zeiss LSM 510 confocal immunofluorescence microscope.
Plasmids and transfection.
The expression plasmid pCMV-Myc HSV-1 ICP0 was constructed for expression of the HSV-1 ICP0 gene in cultured cells. To construct pCMV-Myc HSV-1 ICP0, the HSV-1 ICP0 coding sequence was amplified by PCR with the primers HSV1 ICP0 EcoRI U (CGGAATTCCCATGGAGCCCCGCCCCGGAGCG) and HSV1 ICP0 BglII L (GCTCTAGATCTTATTGTTTTCCCTCGTCCCG; reverse primer). EcoRI and BglII sites were incorporated into the forward and reverse primers, respectively, to facilitate cloning. The PCR product was digested with EcoRI and BglII and cloned into pCMV-Myc (Clontech). The Myc-HSV-2 ICP0 expression plasmid was kindly provided by Yoko Ushijima (Nagoya University, Nagoya, Japan). In transfection experiments, cells were plated in 35-mm dishes and incubated for 24 h before transfection. Cells were transfected with 1 μg of each plasmid by use of Lipofectamine 2000 (Invitrogen) following the manufacturer's recommendations. The medium was replaced with growth medium (without antibiotics) 5 h after transfection. Cells were subjected to Western blotting and immunofluorescence analyses at the indicated times.
Coimmunoprecipitation assay.
In assays with transfected cells, HEp-2 cells in 60-mm dishes were transfected with plasmids encoding the HSV-1 or HSV-2 ICP0 protein, harvested at 18 h posttransfection, and washed twice in PBS. Harvested cells were lysed with 1 ml of NP-40 cell lysis buffer (Invitrogen) with protease inhibitor cocktail (Sigma) and clarified by centrifugation at 15,000 rpm for 15 min at 4°C. Immunoprecipitation was performed using Dynabeads protein G (Invitrogen) according to the manufacturer's instructions. Lysed protein was immunoprecipitated with Dynabeads protein G for 1 h at 4°C after coating with anti-c-Myc (Santa Cruz Biotechnology) antibody or normal rabbit IgG. Protein complexes were eluted with SDS sample buffer and subjected to Western blot analysis.
TNKS1 and TNKS2 siRNA transfection.
Stealth short interfering RNAs (siRNAs) for silencing the expression of the tankyrase 1 (siTNKS1) and 2 (siTNKS2) target genes and a nontarget gene (siCON) (as a control) were purchased from Invitrogen. HEp-2 cells were seeded at 1 × 105 cells per dish into 35-mm culture dishes and transfected with 4 μl siRNA and 5 μl Lipofectamine 2000 (Invitrogen) in 500 μl Opti-MEM (Gibco) the next day. The medium was replaced with growth medium (without antibiotics) 5 h after transfection. The cells were analyzed at 3 days posttransfection. For normalization of the viral yield per cell, infected cells were trypsinized and counted. Each viral PFU result was normalized to 106 cells.
Antibodies.
Anti-tankyrase 1 (N-20) and anti-tankyrase 1/2 (H-350) polyclonal antibodies were purchased from Santa Cruz Biotechnology. Anti-NuMA polyclonal antibody was purchased from Novus, and anti-p42 mitogen-activated protein kinase (MAPK; ERK2) and anti-phospho-p44/42 MAPK (Erk1/2) polyclonal antibodies were purchased from Cell Signaling Technology. Monoclonal antibodies and their sources were as follows: anti-ICP0, Virusys; anti-VP5, anti-ICP8, anti-PAR, and anti-β-actin, Abcam; and anti-c-Myc, Santa Cruz Biotechnology. Normal rabbit serum was obtained from Dako.
Chemicals.
PAA, nocodazole (Sigma), XAV939 (Sigma), and U0126 (Calbiochem) were all solubilized in dimethyl sulfoxide (DMSO; Wako, Osaka, Japan) and filter sterilized before use.
RESULTS
HSV infection induces phosphorylation of tankyrase 1.
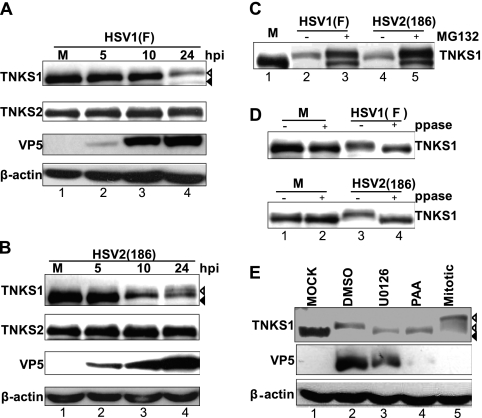
Tankyrase 1 is modified extensively by glycogen synthase kinase 3 (GSK3) and ERK signaling in mitotic cells (72). To determine whether tankyrase 1 is modified in HSV-infected cells, HEp-2 cells were infected with HSV-1 or HSV-2 at an MOI of 3 PFU/cell. Cells were harvested at various intervals, and the expression and phosphorylation state of tankyrases 1 and 2 were determined by Western blotting using anti-tankyrase 1/2 (H-350) antibody. The electrophoretic mobility of tankyrase 1 (Fig. 1A and B, top panels), but not that of tankyrase 2 (Fig. 1A and B, second panels), gradually decreased as infection proceeded. This effect on tankyrase 1 was observed earlier and to a greater extent in HSV-2-infected cells (Fig. 1B, top panel, lanes 1 to 4) than in HSV-1-infected cells (Fig. 1A, top panel, lanes 1 to 4). Furthermore, the amount of tankyrase 1 reduced greatly as the infection proceeded (Fig. 1A and B, lanes 1 and 4), suggesting that infection led to tankyrase 1 instability. When cells were infected with HSV in the presence of the proteasome inhibitor MG132, tankyrase 1 was observed as double bands, and its stability increased dramatically (Fig. 1C, lanes 2 to 5). Thus, tankyrase 1 was degraded in a proteasome-dependent manner during HSV infection.
Fig 1.
ERK-dependent phosphorylation of tankyrase 1 during HSV infection is modified by inhibition of viral DNA synthesis. (A and B) Extensive modification of tankyrase 1 (TNKS1) during HSV infection. HEp-2 cells were mock infected (M) or infected with HSV-1 F (A) or HSV-2 186 (B) at an MOI of 3 PFU/cell. Cell lysates were collected at 5, 10, or 24 hpi and analyzed by Western blotting. Mock-infected tankyrase 1 is indicated by black arrowheads, and the faster-migrating band observed in lysates prepared from HSV-infected cells is indicated by white arrowheads. (C) Proteasome-dependent degradation involved in reduction of tankyrase 1. HEp-2 cells were mock infected (M) or infected with HSV-1 F or HSV-2 186 in the absence or presence of MG132 (50 μM). Cell lysates were collected at 24 hpi and analyzed by Western blotting. (D) Phosphorylation is involved in tankyrase 1 modification. Uninfected HEp-2 cells (M) or HEp-2 cells infected for 14 h with HSV-1 at an MOI of 3 PFU/cell were harvested and dephosphorylated with Lambda PP (ppase) (+) or incubated without it (−). (E) Tankyrase 1 phosphorylation is ERK dependent and affected by the inhibition of viral DNA synthesis. HEp-2 cells were mock infected or infected with HSV-1 strain F at an MOI of 3 PFU/cell in the absence or presence of PAA (300 μg/ml). An equivalent amount of DMSO was used as a control. For U0126 treatment, cells were pretreated with 30 μM U0126 4 h before infection, and cells were infected with HSV-1 strain F at an MOI of 3 PFU/cell with U0126. Cell lysates were collected 24 h after infection and analyzed by Western blotting. The black arrowhead indicates the tankyrase 1 band detected in lysates prepared from mock-infected cells. HSV infection induced a faster-migrating band, and mitotic tankyrase 1 is indicated by white arrowheads.
To examine if phosphorylation was responsible for the slower-migrating population of tankyrase 1, lysates prepared from uninfected and infected cells were treated with Lambda protein phosphatase. Tankyrase 1 migrated faster in phosphatase-digested extracts prepared from HSV-infected cells than in nondigested samples (Fig. 1D, lanes 3 and 4), indicating that tankyrase 1 was phosphorylated during infection.
Tankyrase 1 phosphorylation is ERK dependent and affected by viral DNA synthesis inhibition.
Tankyrase is a signaling target of MAPK and is stoichiometrically phosphorylated upon insulin stimulation (14). To investigate the role of the ERK pathway in tankyrase 1 phosphorylation in HSV-infected cells, we used the MAPK kinase inhibitor U0126. In lysates prepared from U0126-treated cells, tankyrase 1 migrated faster than in control cells (Fig. 1E, lanes 2 and 3). Interestingly, ERK-dependent phosphorylation did not enhance degradation of tankyrase 1 (Fig. 1E, lanes 1 and 3). Tankyrase 1 from HSV-infected cells migrated faster than that from a mitotic cell extract (Fig. 1E, lanes 2 and 5). U0126 treatment did not reduce the phosphorylation of tankyrase 1 in mitotic extracts (data not shown), indicating that ERK activity was dispensable for tankyrase 1 phosphorylation during mitosis (72).
PAA is a potent inhibitor of HSV polymerase. Inhibiting HSV DNA synthesis reduces the expression of viral late γ1 proteins and blocks γ2 proteins completely. PAA treatment of infected cells resulted in faster migration of tankyrase 1 than that in untreated infected cells (Fig. 1E, lanes 2 and 4), suggesting that viral DNA synthesis and/or viral γ2 protein synthesis promoted the phosphorylation of tankyrase 1. At 24 h postinfection (hpi), tankyrase 1 was degraded (Fig. 1E, lanes 1 and 4). Collectively, these data indicate that tankyrase 1 phosphorylation did not occur efficiently when viral DNA synthesis was inhibited.
Tankyrase 1 but not tankyrase 2 is redistributed in response to HSV-1 infection.
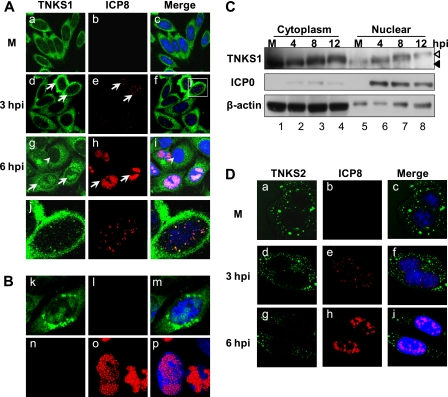
We examined the intracellular localization of tankyrase 1 in HSV-1-infected cells. HEp-2 cells were infected with HSV-1 at an MOI of 1 PFU/cell, fixed at various intervals, and then prepared for indirect immunofluorescence microscopy using polyclonal anti-TNKS1 and monoclonal anti-ICP8 antibodies. ICP8 is the major DNA-binding protein of HSV and acts as a marker for replication compartments. Tankyrases 1 and 2 localized predominantly to the cytoplasm of uninfected interphase cells (Fig. 2A, panel a, and D, panel a). HSV infection induced recruitment of tankyrase 1 to the nucleus (Fig. 2A, panels d, g, and j), whereas tankyrase 2 remained unaffected (Fig. 2D, panels d and g). During the early phase of infection, punctate staining of tankyrase 1 that partially associated with ICP8 was observed in the nucleus (Fig. 2A, panel j). These puncta were later localized within the replication compartment (Fig. 2A, panel g). We stained only for tankyrase 1, and a similar signal consistent with recruitment into replication compartments was detected (Fig. 2B, panel k). In addition, the results of single staining of ICP8 indicated no cross-reactivity between the anti-tankyrase 1 and anti-ICP8 antibodies (Fig. 2B, panels k and n). However, the accumulation of tankyrase 1 in the perinuclear region is likely a nonspecific signal, as it is known that viral glycoprotein Fc receptors also accumulate in this compartment (26).
Fig 2.
Nuclear translocation of tankyrase 1 in HSV-1-infected cells. (A) Tankyrase 1 is recruited into the nucleus and localized in the replication compartment in HSV-1-infected cells. The intracellular localization of tankyrase 1 in HSV-1-infected cells (MOI of 1) was analyzed by indirect immunofluorescence using antibodies against tankyrase 1 (N-20) (green) and a marker of the HSV replication compartment (ICP8) (red). Arrows indicate infected cells, and arrowheads indicate uninfected cells. (B) Nuclear localization of tankyrase 1 was not due to channel overlap. HEp-2 cells were infected with the F strain for 6 h and subjected to indirect immunofluorescence analysis using anti-tankyrase 1 (N-20) (green) or anti-ICP8 (red) antibody. (C) Subcellular localization of tankyrase 1 in infected cells (MOI of 3). Nuclear and cytoplasmic extracts of infected cells were analyzed by Western blotting using the indicated antibodies. (D) Effect of infection on tankyrase 2 localization. Intracellular localization of tankyrase 2 in HSV-1-infected cells (MOI of 1) was analyzed by indirect immunofluorescence using antibodies against tankyrase 2 (green) and ICP8 (red). Imaging was performed using a Zeiss LSM 510 Meta confocal microscope with a 64× objective lens. Images were acquired at a 2× or 4× digital zoom.
Next, we took a biochemical approach to analyze the subcellular localization of tankyrase 1 in infected cells (Fig. 2C). A small fraction of tankyrase 1 localized to the nucleus in uninfected cells (Fig. 2C, lane 5), consistent with the indirect immunofluorescence microscopy data (Fig. 2A, panel a). The nuclear fraction of phosphorylated tankyrase 1 increased at 4 hpi (Fig. 2C, lane 6) and 8 hpi (lane 7). Therefore, HSV-1 infection induces tankyrase 1 phosphorylation and its translocation to the nucleus.
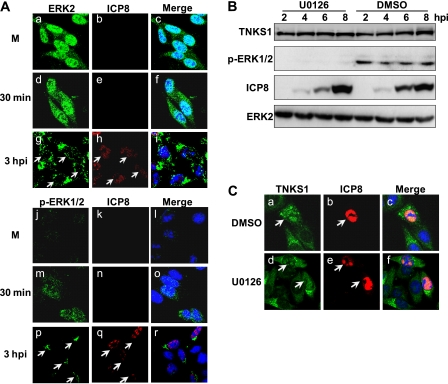
Nuclear localization of tankyrase 1 is independent of its phosphorylation.
Cell fractionation showed that the level of phosphorylated tankyrase 1 in the nucleus increased during the early phase of HSV-1 infection. Was phosphorylation required for nuclear translocation of tankyrase 1 in HSV-1-infected cells? We first examined the localization of ERK2 and p-ERK1/2 (Fig. 3A). In mock-infected cells, ERK2 showed nuclear staining with a weak cytoplasmic signal (Fig. 3A, panel a). Thirty minutes after exposure of cells to HSV, staining of ERK2 remained unchanged (Fig. 3A, panel d). At 3 hpi, however, ERK2 was excluded from the nuclei of many infected cells (Fig. 3A, panel g). In contrast, in mock-infected cells, active ERK (p-ERK1/2) was observed faintly in the nucleus (Fig. 3A, panel j). As early as 30 min after infection, active ERK was detected in both the cytoplasm and the nucleus (Fig. 3A, panel m). At 3 hpi, it was observed juxtaposed to the nuclei of most cells (Fig. 3A, panel p). These data are consistent with previous reports that the US2 protein present in the incoming virion is capable of sequestering activated ERK in the cytoplasm (40a). We next used U0126 to inhibit ERK1/2 and downstream tankyrase 1 phosphorylation (Fig. 3B, top panel) and performed Western blotting to determine ERK1/2 activity (Fig. 3B, second panel). ICP8 was used as a marker of infection (Fig. 3B, third panel), and ERK2 was used as a loading control (Fig. 3B, bottom panel). HEp-2 cells were infected with HSV-1 in the presence or absence of U0126 and fixed at 8 hpi. The drug did not block nuclear translocation of tankyrase 1, indicating that ERK-dependent phosphorylation was not required (Fig. 3C, panel d).
Fig 3.
Tankyrase 1 phosphorylation is not essential for nuclear translocation in HSV-1-infected cells. (A) Subcellular localization of ERK2 and active ERK (p-ERK) in infected cells. HEp-2 cells were mock infected (M) or infected with HSV-1 strain F at an MOI of 1 PFU/cell, and the intracellular localization of ERK2 and p-ERK was analyzed by indirect immunofluorescence using antibodies against ICP8 (red) and ERK2 or p-ERK (green) at the indicated times. (B) ERK activity and tankyrase 1 phosphorylation are inhibited by U0126. HEp-2 cells were treated with 30 μM U0126 4 h before infection, and cells were then infected with HSV-1 strain F at an MOI of 3 PFU/cell with U0126. Cell lysates were collected at the indicated times after infection and were analyzed by Western blotting. (C) Phosphorylation is not required for tankyrase 1 nuclear translocation during HSV-1 infection. HEp-2 cells were treated with 30 μM U0126 4 h before infection, and cells were then infected with HSV-1 strain F at an MOI of 1 PFU/cell with U0126 for 8 h. Cells were then fixed, and localization of tankyrase 1 was determined by indirect immunofluorescence using anti-tankyrase 1 (N-20) (green) and anti-ICP8 (red) antibodies. The arrow indicates infected cells, and imaging was performed using a Zeiss LSM 510 Meta confocal microscope with a 64× objective lens. Images were collected at a 2× digital zoom.
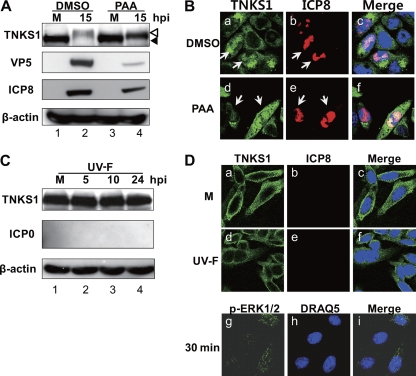
We next examined the effect of viral DNA synthesis on tankyrase 1 localization (Fig. 4A and B). At 15 hpi, PAA blocked the expression of the major capsid protein VP5 (Fig. 4A, second panel, lanes 2 and 4), but tankyrase 1 phosphorylation was not significantly decreased (Fig. 4A, top panel, lanes 2 and 4). Nuclear translocation of tankyrase 1 was not blocked (Fig. 4B, panel d), suggesting that viral DNA synthesis and/or viral γ2 protein synthesis was not required for tankyrase 1 redistribution. In addition, tankyrase 1 seemed significantly more stable with PAA treatment than with DMSO treatment (Fig. 4A, lanes 2 and 4), whereas it was degraded at 24 hpi (Fig. 1E, lanes 3 and 4).
Fig 4.
Localization of tankyrase 1 in cells treated with PAA or infected with the UV-F strain. (A and B) Viral DNA synthesis is not essential for tankyrase 1 translocation in HSV-infected cells. (A) HEp-2 cells were infected with HSV-1 strain F at an MOI of 3 PFU/cell in the absence or presence of PAA (300 μg/ml). An equivalent amount of DMSO was used as a control. Cell lysates were collected 15 h after infection, and inhibition of viral DNA synthesis was monitored by Western blotting. (B) HEp-2 cells were mock infected (M) or infected with the F strain at an MOI of 1 PFU/cell in the presence of PAA (300 μg/ml) or DMSO. Cells were fixed 8 h after infection and stained for ICP8 (red) and tankyrase 1 (green). (C and D) Tankyrase 1 shows no modification or nuclear redistribution in UV-F-infected cells. (C) HEp-2 cells were mock infected (M) or infected with the UV-F strain at an MOI of 10 PFU/cell, and cell lysates were analyzed by Western blotting at the indicated times. (D) HEp-2 cells were mock infected (M) or infected with the UV-F strain at an MOI of 10 PFU/cell and fixed for indirect immunofluorescence using tankyrase 1 (green) and ICP8 antibodies at 12 hpi. Active ERK1/2 was stained at 30 min postinfection, using p-ERK1/2 antibody. Imaging was performed using a Zeiss LSM 510 Meta confocal microscope with a 64× objective lens. Images were collected at a 2× digital zoom.
To determine the effect of viral attachment to the cell surface or viral internalization on tankyrase 1, we prepared a UV-irradiated HSV-1 F strain (UV-F). In the UV-F-infected cells, ICP0 expression was blocked, as was tankyrase phosphorylation (Fig. 4C, top panel), while ERK activity was observed at 30 min postinfection (Fig. 4D, panel g). Tankyrase 1 remained in the cytoplasm of UV-F-infected cells (Fig. 4D, panel d). These data indicate that de novo viral protein expression is required for nuclear translocation of tankyrase 1 and for its phosphorylation by ERK.
Tankyrase 1 colocalizes with ICP0 during the early phase of HSV-1 infection.
The HSV genome encodes 5 immediate-early proteins: ICP0, ICP4, ICP22, ICP27, and ICP47. ICP4 and ICP27 are essential gene products that activate expression of early and late viral gene products (54). ICP0 is a multifunctional protein that enhances replication, especially at low MOIs, partly by its ability to block chromatin silencing of viral lytic genes (16, 28). ICP0 possesses E3 ubiquitin ligase activity (29) and has been shown to induce in vivo degradation of a number of cellular proteins, including the major ND10 proteins PML and Sp100 and the catalytic subunit of DNA protein kinase, in a proteasome-dependent manner (5, 22, 67), thereby playing a major role in establishing permissive conditions for viral infection.
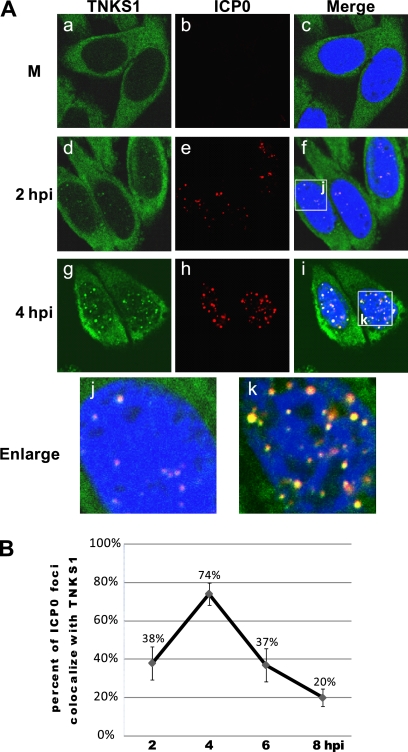
We next determined the intracellular localization of ICP0 and tankyrase 1 in HSV-1-infected cells. ICP0 was not detected in mock-infected cells (Fig. 5A, panel b), while tankyrase 1 was detected mostly in the cytoplasm (Fig. 5A, panel a). In the early phase of HSV-1 infection, punctate staining of tankyrase 1 colocalized with ICP0 in the nucleus (Fig. 5A, panels d and g). Thereafter, tankyrase 1 localized within the replication compartment, while ICP0 was observed partially in the cytoplasm. The levels of colocalization of ICP0 with tankyrase 1 were 38% (2 hpi), 74% (4 hpi), 37% (6 hpi), and 20% (8 hpi) (Fig. 5B). In addition, we observed at early times of infection that approximately 15% of tankyrase 1 foci associated with ICP8, while 90% colocalized with ICP0 (Fig. 2A and 5A; data not shown). These data suggest that incoming tankyrase 1 is associated more closely with ICP0 than with ICP8 protein during the early phase of infection.
Fig 5.
Tankyrase 1 colocalizes with ICP0 during the early phase of HSV-1 infection. (A) HEp-2 cells were mock infected (M) or infected with HSV-1 strain F at an MOI of 1 PFU/cell. Intracellular localization of tankyrase 1 and ICP0 in HSV-1-infected cells was analyzed using indirect immunofluorescence with tankyrase 1 (green) and ICP0 (red) antibodies. Imaging was performed using a Zeiss LSM 510 Meta confocal microscope with a 64× objective lens. Images were collected at a 3× digital zoom. (B) Percentages of ICP0 foci colocalized with tankyrase 1 in HSV-1-infected cells. HEp-2 cells were infected with strain F and fixed for immunofluorescence at various time points between 2 and 8 h postinfection. Infected cells were stained with anti-ICP0 monoclonal antibody and anti-tankyrase 1 polyclonal antibody. Over 50 cells for each time point were counted, and results are shown as means ± standard deviations.
HSV ICP0 interacts with tankyrase 1 and mediates its nuclear translocation.
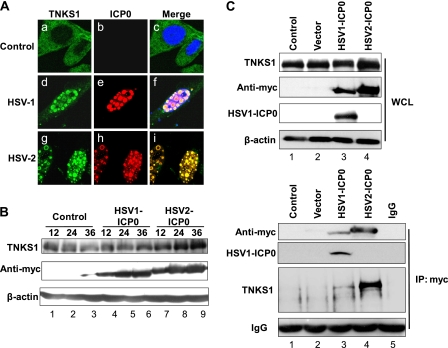
We next examined whether ICP0 expression regulated tankyrase 1 distribution in the absence of infection. HEp-2 cells were transfected with a Myc-tagged HSV-1 or HSV-2 ICP0 expression plasmid, and the localization of endogenous tankyrase 1 was determined by indirect immunofluorescence confocal microscopy 18 h after transfection (Fig. 6A). Tankyrase 1 was detected mainly in the cytoplasm in nontransfected cells (Fig. 6A, panel a). In cells expressing either HSV-1 or HSV-2 ICP0-Myc, tankyrase 1 translocated completely to the nucleus and colocalized with ICP0 (Fig. 6A, panels d to f and g to i). Thus, ICP0 was sufficient to recruit tankyrase 1 to the nucleus.
Fig 6.
ICP0 interacts with tankyrase 1 and is sufficient to redistribute tankyrase 1 in ICP0-transfected cells. (A) HEp-2 cells were mock transfected (control) or transfected with plasmids carrying the ICP0 gene (HSV-1 or HSV-2). Cells were obtained using tankyrase 1 (green) and anti-Myc (red) antibodies. Merged images of the transfected cells are shown in the right panels. Imaging was performed using a Zeiss LSM 510 Meta confocal microscope with a 64× objective lens. Images were collected at a 3× digital zoom. (B) Western blotting. Transfected cells were collected at 12, 24, and 36 h posttransfection and analyzed by Western blotting using the indicated antibodies. (C) Coimmunoprecipitation assay. Transfected cells were collected at 24 h posttransfection, and whole-cell lysates (WCL) were immunoprecipitated (IP) with anti-Myc antibodies or subjected directly to Western blotting with the indicated antibodies. Normal rabbit IgG was used as a negative control for immunoprecipitation. Immunoprecipitated ICP0 was Western blotted using anti-Myc and HSV-1 ICP0 antibodies.
When we performed Western blotting with HSV ICP0-transfected cell lysates (Fig. 6B), there was no significant alteration in the electrophoretic mobility of tankyrase 1. However, tankyrase 1 levels seemed to increase gradually in HSV-2 ICP0-transfected cells but not in HSV-1 ICP0-transfected cells (Fig. 6B, top panel, lanes 4 to 6 and lanes 7 to 9, respectively). ICP0 was sufficient to relocalize tankyrase 1 to the nucleus and increase its stability in the absence of infection, whereas the phosphorylation of tankyrase 1 apparently required additional viral proteins.
These observations prompted us to examine whether tankyrase 1 interacted with ICP0. We carried out coimmunoprecipitation assays with lysates obtained from HEp-2 cells overexpressing Myc-tagged HSV-1 or HSV-2 ICP0. Lysates were immunoprecipitated using an anti-Myc antibody or normal rabbit IgG as a control. The anti-Myc antibody specifically precipitated ICP0 and endogenous tankyrase 1 (Fig. 6C, lanes 3 and 4). The control rabbit IgG precipitated neither (Fig. 6C, lane 5). Therefore, tankyrase 1 interacted with both HSV-1 and HSV-2 ICP0 proteins.
ICP0 is essential for tankyrase 1 redistribution in HSV-1-infected cells.
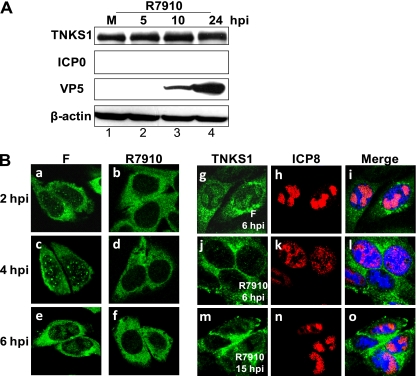
To determine the role of ICP0 in the redistribution of tankyrase 1 in the context of HSV infection, we used HSV-1 R7910, a strain that lacks both copies of ICP0 (38). Tankyrase 1 was not phosphorylated, even at 24 hpi, and degradation of tankyrase 1 was not observed as the infection proceeded (Fig. 7A, top panel). Tankyrase 1 was detected in the nucleus and replication compartments in wild-type-infected cells (Fig. 7B, panels a, c, e, and g). In R7910-infected cells, tankyrase 1 remained mostly in the cytoplasm, similar to that in mock-infected cells (Fig. 7B, panels b, d, f, and j). Even as late as 15 hpi, tankyrase 1 was predominantly cytoplasmic (Fig. 7B, panel m), and we therefore concluded that tankyrase 1 requires ICP0 for nuclear accumulation. However, a trace of tankyrase 1 was detectable in the R7910-infected cells (Fig. 7B, panel m), so we do not exclude the possibility that other viral proteins also promote the nuclear translocation process. Our observations also suggest that in the context of infection, tankyrase 1 is apparently degraded only after it is translocated into the nucleus (Fig. 1E).
Fig 7.
ICP0 is essential for tankyrase 1 redistribution and phosphorylation in HSV-1-infected cells. (A) HEp-2 cells were mock infected (M) or infected with R7910 at an MOI of 3 PFU/cell. Cell lysates were collected 5, 10, or 24 h after infection and analyzed by Western blotting using the indicated antibodies. (B) HEp-2 cells were infected with HSV-1 strain F (left) or the mutant virus R7910 (right) at an MOI of 3 or 1 PFU/cell, respectively. Cells were fixed at the indicated times, and tankyrase 1 localization was analyzed using indirect immunofluorescence with tankyrase 1 (green) and ICP8 (red) antibodies. Imaging was performed using a 64× objective lens and a Zeiss LSM 510 Meta confocal microscope. Images were acquired at a 3× digital zoom.
Tankyrases 1 and 2 are essential but redundant for efficient HSV replication.
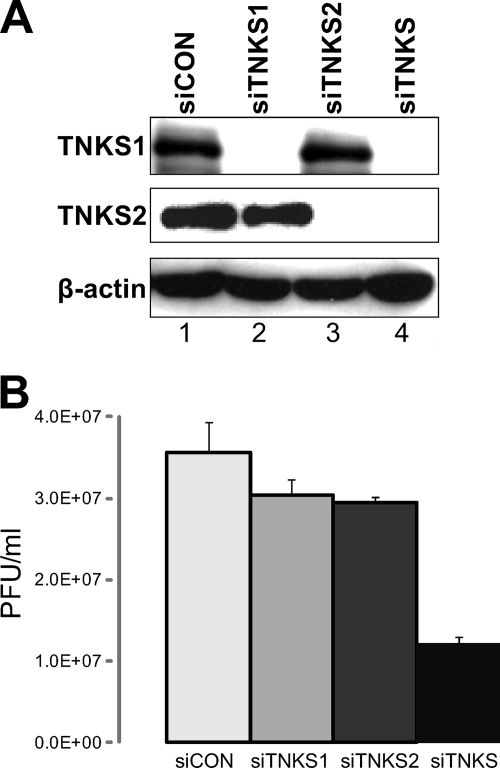
To investigate whether tankyrase 1 was required for HSV-1 replication, we examined the effect of tankyrase 1 depletion on viral growth. HEp-2 cells were transfected with siRNA targeting tankyrase 1 (siTNKS1) or tankyrase 2 (siTNKS2) or with a control siRNA (siCON). The depletion of target proteins proved to be highly efficient 3 days after transfection (Fig. 8A). No cell toxicity was observed (data not shown). Using the plaque assay, we analyzed the viral titers of HSV-1 in the tankyrase-depleted cells at 24 hpi (Fig. 8B). Depletion of either tankyrase 1 or tankyrase 2 decreased the viral titers moderately, to 82% and 80% of control levels, respectively. However, simultaneous depletion of both tankyrases 1 and 2 resulted in stronger impairment of viral replication. The viral titer decreased to 34% (P = 0.02) compared to that of control cells. This observation is consistent with a previous study showing that there is substantial redundancy between tankyrases 1 and 2 (15). This indicates that efficient HSV replication requires tankyrase function, but tankyrases 1 and 2 are apparently redundant.
Fig 8.
siRNA-mediated tankyrase knockdown inhibits HSV growth. (A) siCON-, siTNKS1-, siTNKS2-, or siTNKS1-plus-siTNKS2 (siTNKS)-transfected HEp-2 cells were analyzed at 3 days posttransfection for protein expression. Tankyrase levels were greatly decreased in siRNA-treated cells, whereas actin levels were constant. (B) Analysis of viral growth in siRNA-treated cells. HEp-2 cells transfected with siCON, siTNKS1, siTNKS2, and siTNKS were infected with F at an MOI of 3, cells were collected at 24 hpi and lysed by a freeze-thaw cycle, and progeny viruses were titrated on Vero cells. Results for three independent experiments are shown as means with standard deviations.
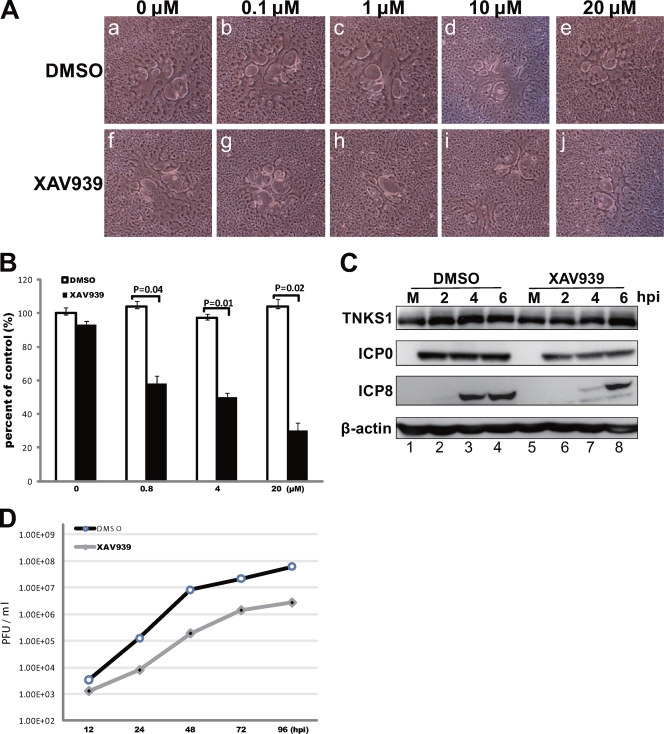
Efficient HSV-1 replication requires PARP activity.
To investigate whether the PARP activity of tankyrases is required for HSV infection, we used the small-molecule inhibitor XAV939, which specifically inhibits tankyrase PARP activity by binding the conserved catalytic domains of both tankyrases (31). We first analyzed the cell-to-cell spread of HSV-1 in the presence of XAV939 (Fig. 9A and B). The plaques were significantly smaller in XAV939-treated cells than in DMSO-treated control cells. XAV939 reduced cell-to-cell spread even at a concentration of 100 nM (Fig. 9A, panels b and g). The plaque numbers decreased to 58% (0.8 μM XAV939), 50% (4 μM), and 30% (20 μM) compared to that of control cells (Fig. 9B). When HSV-1 was used to infect cells at an MOI of 3 PFU/cell in the presence of 1 μM XAV939 (Fig. 9C), ICP8 expression was significantly delayed (Fig. 9C, third panel, lanes 3, 4, 7, and 8), but ICP0 expression was not (Fig. 9C, second panel). PARP activity of tankyrases was apparently required for the temporal expression of viral proteins. Finally, we analyzed the effect of the drug on HSV-1 replication. XAV939 (1 μM)-treated HEp-2 cells were infected with HSV-1 at an MOI of 0.001 PFU/cell (Fig. 9D). The reduction in overall virus yield was approximately 20- to 50-fold. XAV939 also suppressed one-step virus replication in cells infected at an MOI of 3 PFU/cell. The average total viral yields at 24 hpi were (1.7 ± 0.4) × 108 and (7.9 ± 1.3) × 106 PFU/ml for DMSO- and XAV939-treated cells, respectively. These data indicate that PARP activity of tankyrase is essential for efficient HSV replication.
Fig 9.
Inhibition of tankyrase PARP activity significantly impairs viral replication. (A) Plaque size depends on XAV939 concentration. Vero cell monolayers were infected with approximately 100 PFU of HSV-1 in the presence of the indicated concentration of XAV939 or DMSO (control). (B) The cells were fixed at 24 hpi, and plaque size and number were determined to assess viral spread. Results for three independent experiments are shown as means ± standard deviations, and P values were determined by the two-tailed t test. (C) Inhibition of tankyrase 1 PARP activity results in defective viral protein expression. HEp-2 cells were infected with HSV-1 strain F at an MOI of 3 PFU/cell in the absence (DMSO) or presence of XAV939 (1 μM), and then lysates were analyzed by Western blotting. (D) Multistep growth curve of HSV-1 in XAV939-treated HEp-2 cells. The results shown are representative of 3 independent experiments. After similar treatment with XAV939 (1 μM) or DMSO, HEp-2 cells were infected with HSV-1 at an MOI of 0.001 PFU/cell. The virus replication kinetics was significantly diminished when PARP activity was inhibited.
DISCUSSION
HSV often manipulates infected host cell proteins to promote their replication. For example, ND10 proteins are disrupted upon HSV-1 infection and translocate to sites associated with viral genome complexes (23, 25). US3 induces protein kinase A (PKA) activity in HSV-1-infected cells by upregulating PKA expression (2). Multiple cellular proteins, including p53, are recruited to the HSV-1 DNA replication compartment in response to infection (4, 45).
We showed previously that HSV infection specifically targets NuMA, a nuclear matrix apparatus protein which is hyperphosphorylated upon infection and becomes solubilized, a phenomenon normally observed in noninfected cells only during mitosis (71). In the present study, we discovered tankyrase 1, a telomere length regulator, as a new target of HSV. There were multiple effects of HSV-1 on tankyrase 1, such as phosphorylation, nuclear translocation, and degradation. Efficient HSV-1 replication required tankyrase PARP activity. ICP0 interacted with tankyrase 1 and apparently plays a key role in its nuclear import, although the biological significance of the interaction remains unclear. Does ICP0 regulate tankyrase 1 function? This remains unknown, but ICP0 could, for example, prime the infected cell for efficient replication by recruiting tankyrase 1 to viral replication compartments.
HSV-1 replication was impaired when both tankyrases 1 and 2 were codepleted using siRNAs or deprived of their enzymatic activity with the inhibitor XAV939. Therefore, HSV-1 replication was promoted by the PARP activity of tankyrases. ICP8 expression was significantly delayed (Fig. 9B). Although it has been shown that HSV ICP4 is PARsylated (3, 50), its significance to the viral life cycle is not known. We also observed that in HSV-1-infected HEp-2 cells, TRF1 localization was affected (not shown). TRF1 is a substrate of tankyrase and is important for telomere structure and stabilization (46, 47). It is possible that PARsylation redistributes cellular proteins to favor viral gene transcription and/or DNA synthesis. However, we cannot exclude the possibility that cytoplasmic tankyrase also promotes viral replication.
It is known that the PARP activity of tankyrase is enhanced by ERK-dependent phosphorylation (14). Mitotic phosphorylation of tankyrase does not seem to alter its PARP activity, based on in vitro auto-PARsylation assays (10). We have shown that HSV infection induces hyperphosphorylation and solubilization of NuMA, which is a substrate of tankyrase (71). Tankyrase 2 shares 83% sequence identity with tankyrase 1 but lacks the N-terminal HPS (homopolymeric runs of His, Pro, and Ser) domain (41). It is unclear why tankyrase 2 is not modified during HSV infection, despite the fact that it is required for efficient replication. The lack of tankyrase 2 phosphorylation could be due to this discrepancy. Our results suggest that cytoplasmic activity of tankyrase 2 is required for efficient HSV replication.
Tankyrase 1 was degraded during the late phase of HSV infection, in a proteasome-dependent manner. Infection is required to trigger the degradation process. The results from experiments using PAA and U0126 indicated that there was no clear correlation between phosphorylation of tankyrase 1 and its degradation (Fig. 1E). Neither PARsylation of ICP0 nor ubiquitination of tankyrase 1 was detected in ICP0-transfected cells (not shown). It is known that overexpression of tankyrase 1 releases TRF1 from telomeres, and long-term overexpression of tankyrase 1 in telomerase-positive human cells results in a gradual and progressive elongation of telomeres (62). This suggests that accumulation of tankyrase 1 in the nucleus is not favorable for the host cell. Infection with the ICP0-null mutant R7910 showed that tankyrase 1 is degraded only following a nuclear translocation event. Viral DNA synthesis and late gene expression were not required for degradation. The stability of tankyrase 1 also increased when HSV-2 ICP0 was overexpressed. Therefore, ICP0 may stabilize tankyrase 1 in the nucleus at the onset of infection, which is later followed by proteasome-dependent degradation.
Early in infection, ICP0 performs two key functions: it enhances viral gene expression and replication, which are essential at low MOIs (64), and it inhibits the silencing of viral DNA by cellular proteins and the interferon-stimulated host response (32). During the course of infection, ICP0 localizes to the nucleus and disrupts ND10 bodies, followed by inhibition of host chromatin silencing mechanisms that in turn block the transition from immediate-early to early gene expression. At high MOIs, viral replication appears to circumvent the requirements of ICP0, because under these conditions, replication is not dependent on ICP0 (67). A previous study reported that HSV-1 ICP0 interacts specifically with transforming growth factor-activated protein kinase 1 and stimulates c-Jun N-terminal kinase activity but has no effect on ERK activity (19). Our results are consistent with this report; tankyrase 1 was not phosphorylated in ICP0-overexpressing cells. ICP0 does not contain tankyrase 1 binding motifs (30), and the interaction between ICP0 and tankyrase 1 could be indirect. It has been shown that Hsc70 is also redistributed to punctate nuclear dots upon ICP0 overexpression (7), similar to what we observed for tankyrase 1.
In summary, we found that HSV manipulates tankyrase 1 by hyperphosphorylation, by its nuclear transport and retention via ICP0, and, finally, by inducing its degradation in a proteasome-dependent manner during the late phase of infection. Most importantly, efficient viral replication, especially at low MOIs, requires the PARP activity of tankyrases. Given the importance of tankyrases in the viral life cycle, studies of other tankyrase substrates are under way.
ACKNOWLEDGMENTS
We are grateful to all of the members of the Nishiyama lab for helpful discussions and constructive advice. We are especially thankful to Bernard Roizman for the gift of R7910 virus, U2OS cells, and useful technical information. We also thank Yoko Ushijima for the gift of the Myc-HSV-2 ICP0 plasmid and for excellent technical assistance.
Footnotes
Published ahead of print 19 October 2011
REFERENCES
- 1. Bastian TW, Livingston CM, Weller SK, Rice SA. 2010. Herpes simplex virus type 1 immediate-early protein ICP22 is required for VICE domain formation during productive viral infection. J. Virol. 84:2384–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benetti L, Roizman B. 2004. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc. Natl. Acad. Sci. U. S. A. 101:9411–9416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blaho JA, et al. 1992. Differences in the poly(ADP-ribosyl)ation patterns of ICP4, the herpes simplex virus major regulatory protein, in infected cells and in isolated nuclei. J. Virol. 66:6398–6407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boutell C, Everett RD. 2003. The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with and ubiquitinates p53. J. Biol. Chem. 278:36596–36602 [DOI] [PubMed] [Google Scholar]
- 5. Boutell C, Orr A, Everett RD. 2003. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J. Virol. 77:8686–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brandt CR. 2005. The role of viral and host genes in corneal infection with herpes simplex virus type 1. Exp. Eye Res. 80:607–621 [DOI] [PubMed] [Google Scholar]
- 7. Burch AD, Weller SK. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J. Virol. 78:7175–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carrington-Lawrence SD, Weller SK. 2003. Recruitment of polymerase to herpes simplex virus type 1 replication foci in cells expressing mutant primase (UL52) proteins. J. Virol. 77:4237–4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang P, Coughlin M, Mitchison TJ. 2009. Interaction between poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol. Biol. Cell 20:4575–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang W, Dynek JN, Smith S. 2005. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem. J. 391:177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen C, et al. 2006. Activating KIR genes are associated with CMV reactivation and survival after non-T-cell depleted HLA-identical sibling bone marrow transplantation for malignant disorders. Bone Marrow Transplant. 38:437–444 [DOI] [PubMed] [Google Scholar]
- 12. Chen X, et al. 2000. Herpes simplex virus type 1 ICP0 protein does not accumulate in the nucleus of primary neurons in culture. J. Virol. 74:10132–10141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y, Livingston CM, Carrington-Lawrence SD, Bai P, Weller SK. 2007. A mutation in the human herpes simplex virus type 1 UL52 zinc finger motif results in defective primase activity but can recruit viral polymerase and support viral replication efficiently. J. Virol. 81:8742–8751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chi NW, Lodish HF. 2000. Tankyrase is a Golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 275:38437–38444 [DOI] [PubMed] [Google Scholar]
- 15. Chiang YJ, et al. 2008. Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PLoS One 3:e2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cliffe AR, Knipe DM. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82:12030–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davison AJ, et al. 2009. The order Herpesvirales. Arch. Virol. 154:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Rycker M, Venkatesan RN, Wei C, Price CM. 2003. Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem. J. 372:87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Diao L, et al. 2005. Activation of c-Jun N-terminal kinase (JNK) pathway by HSV-1 immediate early protein ICP0. Exp. Cell Res. 308:196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Everett RD, Cross A, Orr A. 1993. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell type dependent manner. Virology 197:751–756 [DOI] [PubMed] [Google Scholar]
- 21. Everett RD, Maul GG. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Everett RD, Maul GG, Orr A, Elliott M. 1995. The cellular RING finger protein PML is not a functional counterpart of the herpes simplex virus type 1 RING finger protein Vmw110. J. Gen. Virol. 76:791–798 [DOI] [PubMed] [Google Scholar]
- 23. Everett RD, Murray J. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Everett RD, Sourvinos G, Orr A. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Everett RD, Zafiropoulos A. 2004. Visualization by live-cell microscopy of disruption of ND10 during herpes simplex virus type 1 infection. J. Virol. 78:11411–11415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frank I, Friedman HM. 1989. A novel function of the herpes simplex virus type 1 Fc receptor: participation in bipolar bridging of antiviral immunoglobulin G. J. Virol. 63:4479–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fraser KA, Rice SA. 2007. Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J. Virol. 81:5091–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gu H, Roizman B. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc. Natl. Acad. Sci. U. S. A. 104:17134–17139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hagglund R, Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsiao SJ, Smith S. 2008. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 90:83–92 [DOI] [PubMed] [Google Scholar]
- 31. Huang SM, et al. 2009. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461:614–620 [DOI] [PubMed] [Google Scholar]
- 32. Kalamvoki M, Roizman B. 2008. Nuclear retention of ICP0 in cells exposed to HDAC inhibitor or transfected with DNA before infection with herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 105:20488–20493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaminker PG, et al. 2001. TANK2, a new TRF1-associated poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J. Biol. Chem. 276:35891–35899 [DOI] [PubMed] [Google Scholar]
- 34. Kuddus RH, DeLuca NA. 2007. DNA-dependent oligomerization of herpes simplex virus type 1 regulatory protein ICP4. J. Virol. 81:9230–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuimov AN, et al. 2001. Cloning and characterization of TNKL, a member of tankyrase gene family. Genes Immun. 2:52–55 [DOI] [PubMed] [Google Scholar]
- 36. Lamberti C, Weller SK. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liptak LM, Uprichard SL, Knipe DM. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lopez P, Jacob RJ, Roizman B. 2002. Overexpression of promyelocytic leukemia protein precludes the dispersal of ND10 structures and has no effect on accumulation of infectious herpes simplex virus 1 or its proteins. J. Virol. 76:9355–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lukonis CJ, Burkham J, Weller SK. 1997. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J. Virol. 71:4771–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lukonis CJ, Weller SK. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71:2390–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a. Lyman MG, Randall JA, Calton CM, Banfield BW. 2006. Localization of ERK/MAP kinase is regulated by the alphavirus tegument protein. Us2. J. Virol. 80:7159–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lyons RJ, et al. 2001. Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J. Biol. Chem. 276:17172–17180 [DOI] [PubMed] [Google Scholar]
- 42. Malik AK, Shao L, Shanley JD, Weller SK. 1996. Intracellular localization of the herpes simplex virus type-1 origin binding protein, UL9. Virology 224:380–389 [DOI] [PubMed] [Google Scholar]
- 43. Martin JR, Jenkins FJ, Henken DB. 1991. Targets of herpes simplex virus type 1 infection in a mouse corneal model. Acta Neuropathol. 82:353–363 [DOI] [PubMed] [Google Scholar]
- 44. Miwa M, Masutani M. 2007. PolyADP-ribosylation and cancer. Cancer Sci. 98:1528–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mohni KN, Livingston CM, Cortez D, Weller SK. 2010. ATR and ATRIP are recruited to herpes simplex virus type 1 replication compartments even though ATR signaling is disabled. J. Virol. 84:12152–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Okamoto K, Iwano T, Tachibana M, Shinkai Y. 2008. Distinct roles of TRF1 in the regulation of telomere structure and lengthening. J. Biol. Chem. 283:23981–23988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Okamoto K, Shinkai Y. 2009. TRFH domain is critical for TRF1-mediated telomere stabilization. Cell Struct. Funct. 34:71–76 [DOI] [PubMed] [Google Scholar]
- 48. Phelan A, Carmo-Fonseca M, McLaughlan J, Lamond AI, Clements JB. 1993. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc. Natl. Acad. Sci. U. S. A. 90:9056–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Phelan A, Dunlop J, Patel AH, Stow ND, Clements JB. 1997. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J. Virol. 71:1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Preston CM, Notarianni EL. 1983. Poly(ADP-ribosyl)ation of a herpes simplex virus immediate early polypeptide. Virology 131:492–501 [DOI] [PubMed] [Google Scholar]
- 51. Randall RE, Dinwoodie N. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 67:2163–2177 [DOI] [PubMed] [Google Scholar]
- 52. Rojas S, Corbin-Lickfett KA, Escudero-Paunetto L, Sandri-Goldin RM. 2010. ICP27 phosphorylation site mutants are defective in herpes simplex virus 1 replication and gene expression. J. Virol. 84:2200–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rojek JM, et al. 2010. Targeting the proteolytic processing of the viral glycoprotein precursor is a promising novel antiviral strategy against arenaviruses. J. Virol. 84:573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Samaniego LA, Webb AL, DeLuca NA. 1995. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J. Virol. 69:5705–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sandri-Goldin RM. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front. Biosci. 13:5241–5256 [DOI] [PubMed] [Google Scholar]
- 56. Sbodio JI, Chi NW. 2002. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 277:31887–31892 [DOI] [PubMed] [Google Scholar]
- 57. Sbodio JI, Lodish HF, Chi NW. 2002. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase). Biochem. J. 361:451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Scherag A, et al. 2010. Two new loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and German study groups. PLoS Genet. 6:e1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sears AE, Halliburton IW, Meignier B, Silver S, Roizman B. 1985. Herpes simplex virus 1 mutant deleted in the alpha 22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J. Virol. 55:338–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Seimiya H, Smith S. 2002. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J. Biol. Chem. 277:14116–14126 [DOI] [PubMed] [Google Scholar]
- 61. Smith S, de Lange T. 1999. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112:3649–3656 [DOI] [PubMed] [Google Scholar]
- 62. Smith S, de Lange T. 2000. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10:1299–1302 [DOI] [PubMed] [Google Scholar]
- 63. Smith S, Giriat I, Schmitt A, de Lange T. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282:1484–1487 [DOI] [PubMed] [Google Scholar]
- 64. Stow ND, Davison AJ. 1986. Identification of a varicella-zoster virus origin of DNA replication and its activation by herpes simplex virus type 1 gene products. J. Gen. Virol. 67:1613–1623 [DOI] [PubMed] [Google Scholar]
- 65. Tanaka M, Kodaira H, Nishiyama Y, Sata T, Kawaguchi Y. 2004. Construction of recombinant herpes simplex virus type I expressing green fluorescent protein without loss of any viral genes. Microbes Infect. 6:485–493 [DOI] [PubMed] [Google Scholar]
- 66. Taus NS, Salmon B, Baines JD. 1998. The herpes simplex virus 1 UL 17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology 252:115–125 [DOI] [PubMed] [Google Scholar]
- 67. van Lint AL, et al. 2010. Herpes simplex virus immediate-early ICP0 protein inhibits Toll-like receptor 2-dependent inflammatory responses and NF-kappaB signaling. J. Virol. 84:10802–10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Weber PC, Levine M, Glorioso JC. 1987. Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science 236:576–579 [DOI] [PubMed] [Google Scholar]
- 69. Wilcock D, Lane DP. 1991. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349:429–431 [DOI] [PubMed] [Google Scholar]
- 70. Wilkinson DE, Weller SK. 2006. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J. Cell Sci. 119:2695–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yamauchi Y, Kiriyama K, Kimura H, Nishiyama Y. 2008. Herpes simplex virus induces extensive modification and dynamic relocalisation of the nuclear mitotic apparatus (NuMA) protein in interphase cells. J. Cell Sci. 121:2087–2096 [DOI] [PubMed] [Google Scholar]
- 72. Yeh TY, Sbodio JI, Chi NW. 2006. Mitotic phosphorylation of tankyrase, a PARP that promotes spindle assembly, by GSK3. Biochem. Biophys. Res. Commun. 350:574–579 [DOI] [PubMed] [Google Scholar]
- 73. Zhao Y, Holden VR, Smith RH, O'Callaghan DJ. 1995. Regulatory function of the equine herpesvirus 1 ICP27 gene product. J. Virol. 69:2786–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]