Abstract
To prove that the peptidic HIV-1 fusion inhibitors containing the pocket-binding domain (PBD) mainly target the hydrophobic pocket in the gp41 N-terminal heptad repeat (NHR), we constructed pseudoviruses by replacement of Q64 in the gp41 pocket region with Ala (Q64A) or Leu (Q64L). These viruses were highly resistant to C34 and CP32M containing the PBD, while they were susceptible to T20 (enfuvirtide) lacking the PBD but containing the GIV-motif-binding domain (GBD) and lipid-binding domain (LBD). They were also sensitive to C52L, which contains the PBD, GBD, and LBD. Those mutations may disrupt the hydrophilic interaction between Q64 in the NHR and N113 in the peptides containing the PBD. This report provides insights into the mechanisms of drug resistance, with implications for the design of novel HIV fusion and entry inhibitors.
TEXT
Entry of human immunodeficiency virus type 1 (HIV-1) into the target cell is initiated by binding of the viral envelope (Env) glycoprotein surface subunit gp120 to the primary receptor CD4 and a coreceptor, CXCR4 or CCR5, followed by a cascade of conformational arrangements in the gp41 transmembrane subunit (5, 9, 30). Insertion of the fusion peptide (FP) into the target cell membrane results in exposure of the trimeric coiled coil formed by three molecules of the N-terminal heptad repeat (NHR), representing the pre-hairpin fusion intermediate (PFI). Three cognate C-terminal heptad repeats (CHR) bind along the NHR trimer to form a thermostable six-helix bundle (6-HB) core structure (4, 25, 40) that brings the viral and host cell membranes into proximity, leading to virus-cell fusion (5). A highly conserved hydrophobic pocket is formed in each groove of the NHR trimer (Fig. 1A), and it plays an important role in 6-HB stabilization as well as viral fusion (3, 8, 17). Therefore, the gp41 NHR trimer in the PFI state, especially in the hydrophobic pocket region, serves as an attractive target for the development of HIV fusion and entry inhibitors (2, 32).
Fig 1.
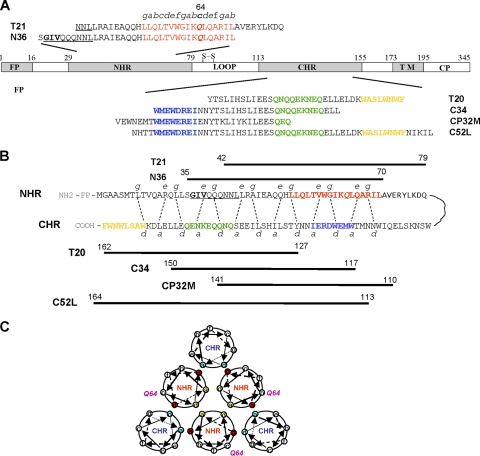
Schematic representation of the HIV-1 gp41 molecule and its functional domains. (A) Functional domains of the HIV-1 gp41 and the corresponding N- and C-peptide sequences. FP, fusion peptide; NHR, N-terminal heptad repeat; CHR, C-terminal heptad repeat; TM, transmembrane domain; CP, cytoplasmic domain. The pocket-forming sequence in the NHR domain is marked in red. The pocket-binding domain (PBD), GIV-motif-binding domain (GBD), and lipid-binding domain (LBD) in the CHR domain are highlighted in blue, green, and orange, respectively. The GIV motif in the N-terminal region of the NHR domain is underlined. The letters a to g indicate the positions of the corresponding residues in the helical wheel of the pocket-forming sequence. The residue at position 64 is indicated in italics and boldface. (B) Interactions between NHR and CHR of gp41 and between N and C peptides. The dashed lines between NHR and CHR indicate interactions between the residues located at the e and g positions in the NHR and at the a and d positions in the CHR. The numbers of residues of peptides corresponding to T21, N36, T20, C34, and CP32M are shown. (C) Schematic view of the HIV-1 gp41 6-HB core. Residue Q64 is located at the c site in the helical wheel of the gp41 NHR domain, which seems not to be involved in direct interactions with the key residues in NHR for the formation of N trimers or those in CHR for the formation of 6-HB. The internal N trimer is formed via the interactions of residues located at the a and d positions (in blue) among the NHR helices. The 6-HB is formed through the interaction of residues located at the e and g positions (in red) in the N helices and those at a and d positions (in green) in the C helices. Residue Q64, located at the c site, is also shown.
Several peptides derived from the gp41 CHR domain, including SJ-2176 (18), DP-178 (also known as T20) (43), and C34 (25, 26), were previously shown to inhibit HIV-1 fusion by interacting with the viral gp41 NHR trimer (22). T20 (brand name, Fuzeon; generic name, enfuvirtide) is the first peptide HIV fusion inhibitor to be licensed by the U.S. FDA for clinical use. However, one of the major disadvantages of T20 as an antiviral drug is its quick induction of drug-resistant mutations in the GIV motif (residues 36 to 44 GIVQQQNNL) in the N-terminal region of the gp41 NHR domain (10, 29, 35–37, 39), suggesting that T20 mainly interacts with this region in gp41. Based on these findings, scientists then shifted their interest to the development of peptides that could interact with the entire gp41 NHR domain, including the pocket region, such as C34, sifuvirtide (16), T1249, and C52L (6, 7, 11, 20, 28). Although T20-resistant HIV-1 strains are more susceptible to the peptides containing the pocket-binding domain (PBD) than to T20, they still exhibited some resistance to C34 (1), T1249 (6, 10), and sifuvirtide (24) since those peptides also contain the GIV motif-binding domain (GBD). To avoid the problem posed by this resistance, CP32 and CP32M, two peptides that contain the PBD but lack the GBD (Fig. 1B), were designed and were shown to be exceptionally effective against T20-resistant viruses (12, 13). However, none of the peptide HIV fusion inhibitors containing the PBD have been reported to induce drug-resistant mutations in the gp41 pocket region, leading us to ask whether the gp41 pocket is indeed a good target for drug development.
To address this question, we applied a site-directed mutagenesis approach to identify which residues in the gp41 pocket region are critically associated with the binding of HIV fusion and entry inhibitors. Previous studies have shown that the residues at the a and d sites in the NHR helical wheel are important for formation of the internal trimer by NHR domains (Fig. 1C), while the residues at the e and g sites in the NHR helical wheel are involved in interactions between the NHR and CHR domains that result in the formation of 6-HB (Fig. 1C) (3–5). Since these highly conserved residues play critical roles in maintaining the fusogenic conformation of gp41 (3, 8, 17), mutations of these residues, such as Q66R/E, would render the viruses incapable of fusing with the target cells (42). However, some residues, such as G36, that are not at the a, d, e, or g position but are at the c position in NHR are critical for the viral resistance to T20 (27). Therefore, we selected Q64, a residue at the c site in the NHR helical wheel, for mutation analysis in this study.
Using a QuikChange site-directed mutagenesis kit (Stratagene), we constructed a plasmid encoding HIV-1 HXB2-Env, replacing Gln, a polar residue, at position 64 in the pocket region with Ala, a smaller amino acid with no polarity (Q64A), or with Leu, a hydrophobic reside (Q64L). A pseudovirus was produced by cotransfecting HEK293T cells with the Env-coding plasmid and a plasmid carrying a Env-defective, luciferase-expressing HIV-1 genome (pNL4-3.Luc.RE) by the use of FuGENE 6 reagents as previously described (14, 15). A single-cycle infection assay was performed to determine the infectivity of the pseudovirus (using equal amounts of 250 ng of p24 as the input) and the inhibitory activity of the peptide HIV fusion and entry inhibitors by the use of a luciferase kit (Promega, Madison, WI) following the manufacturer's instructions.
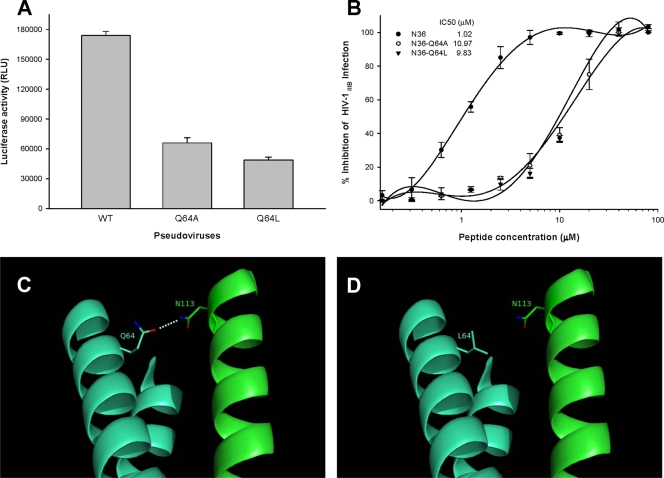
In virus with either a Q64A or Q64L mutation in gp41, infectivity was reduced by about 64% or 70%, respectively (Fig. 2A); the mutations did not significantly affect Env expression and proteolytic processing (data not shown). However, since the mutant strains still retained substantial infectivity, we tested their susceptibility to the selected peptide HIV fusion inhibitors, including T20, C34, CP32M, and C52L. We found that the viruses with Q64 mutations in the gp41 pocket region showed resistance to C34 and CP32M but were sensitive to T20 and C52L. In contrast, the virus with V38E/N42S mutation in the N-terminal region of gp41 NHR domain exhibited high resistance to T20 and low resistance to C34 but was sensitive to CP32M and C52L (Table 1). These results suggest that the peptide HIV fusion inhibitors T20, C34, CP32M, and C52L must target different sites in the gp41 NHR domain, employing different mechanisms of action. Specifically, T20 contains the GBD and lipid-binding domain (LBD) but lacks the PBD, mainly interacting with the N-terminal region of the gp41 NHR domain. As a consequence, viruses with mutations in the GIV motif are highly resistant to T20 whereas those with mutations in the pocket region are still sensitive to T20. CP32M contains the PBD, but it lacks the GBD and LBD, mainly binding to the pocket region. According to test results, viruses with mutations in the pocket region are highly resistant to CP32M, while those with mutations in the GIV motif are susceptible to CP32M. Peptide C34 contains both the GBD and PBD. Consequently, the HIV-1 variants with mutations in either the GIV motif or the pocket region are resistant to C34. Interestingly, C52L, which contains all functional domains in the CHR, including PBD, GBD, and LBD, was effective in inhibiting infection by the pseudovirus mutants that were resistant to both T20 and CP32M.
Fig 2.
Effect of Q64 mutations on viral infectivity, N-peptide-mediated anti-HIV-1 activity, and interactions between NHR and CHR. (A) Infectivity of HIV-1 pseudoviruses bearing a wild-type (WT) or mutant sequence of gp41. Single-cycle infection of the viruses in MT-2 cells was measured by a luciferase-based assay. (B) Anti-HIV-1 activity of wild-type and mutant N36 peptides. The infectivity of HIV-1IIIB in MT-2 cells in the presence or absence of the peptides was measured by a p24-based assay. The IC50 of each peptide is shown in the figure. The experiment was performed in triplicate, and the data are presented as means ± standard deviations (SD). (C and D) Predicted interactions between N113 in the CHR domain and Q64 (C) or L64 (D) in the NHR domain. Molecular modeling analysis of the complex formed by CP32 (amino acids 110 to 141) and T21 (14) was performed using PyMol software (38). Two NHR domains are shown in cyan blue, while the CHR peptide is shown in green. A hydrophilic interaction between Q64 in NHR and N113 in CHR is indicated. Mutation of Q64 with the hydrophobic residue leucine resulted in the loss of the polar contact.
Table 1.
Susceptibility of HIV-1 NL4-3 variants with mutations in the GIV motif and in the pocket-forming region of the HIV-1 gp41 NHR domain to peptide HIV fusion inhibitorsa
| Virus | T20 |
C34 |
CP32 M |
C52L |
||||
|---|---|---|---|---|---|---|---|---|
| IC50 (nM) | Resistance (n-fold) | IC50 (nM) | Resistance (n-fold) | IC50 (nM) | Resistance (n-fold) | IC50 (nM) | Resistance (n-fold) | |
| WT | 13.6 ± 0.39 | 1.0 | 2.2 ± 0.23 | 1.0 | 4.5 ± 0.37 | 1.0 | 8.2 ± 1.3 | 1.0 |
| V38E/N42S | 1,165.9 ± 80.5 | 85.7 | 9.24 ± 1.1 | 4.2 | 6.2 ± 0.72 | 1.3 | 13.0 ± 2.1 | 1.5 |
| Q64A | 22.6 ± 4.21 | 1.6 | 123.6 ± 10.6 | 56.2 | 35.6 ± 3.50 | 8.0 | 15.3 ± 3.4 | 1.8 |
| Q64L | 30.2 ± 0.52 | 2.2 | 220.6 ± 21.4 | 100.3 | 278.5 ± 5.61 | 62.1 | 20.1 ± 4.2 | 2.4 |
IC50 data represent means ± standard deviations.
To further investigate the mechanism of drug resistance to C34 and CP32M, we synthesized the peptides N36, N36-Q64A, N36-Q64L, T21, T21-Q64A, and T21-Q64L by a standard solid-phase FMOC (9-fluorenylmethoxy carbonyl) method as previously described (34). A previous study had shown that N36 is also a potent HIV fusion inhibitor that acts by binding the gp41 CHR domain (15). We therefore compared the inhibitory activity of N36 with respect to HIV-1IIIB infection with that of N36-Q64A and N36-Q64L in MT-2 cells as described previously (19). As shown in Fig. 2B, wild-type N36 could inhibit viral infectivity with a 50% inhibitory concentration (IC50) of 1.02 μM, while the mutant peptides N36-Q64A and N36-Q64L exhibited significantly reduced anti-HIV-1 activity, with IC50 values of 10.97 and 9.83 μM, respectively.
Using a Jasco circular dichroism (CD) spectropolarimeter (model J-715; Jacobin, Japan) and a protocol described before (23), we compared the secondary structures of the N peptides N36 and T21 and their mutants or the complexes formed by the N peptides with those of the corresponding C peptides, C34 and CP32M. As shown in Table 2, N36, N36-Q64A, and N36-Q64L displayed low (29.9%, 22.2%, and 25.9%, respectively) α-helicity as expected. A mixture of N36 and C34 at an equimolar concentration formed a 6-HB conformation with high (88%) α-helicity. N36-Q64A and N36-Q64L also interacted with C34 to form 6-HB, but the N36-Q64L/C34 6-HB had greatly reduced (56%) α-helical content. A thermal denaturation analysis was then performed to determine the stability of the 6-HB formed by N and C peptides by monitoring the CD signal at 222 nm over a range of temperatures. The thermal unfolding transition (thermal denaturation [Tm]) of the 6-HB formed by wild-type N36 and C34 occurred at 66°C, while the thermal unfolding transition of those formed by C34 and the mutant N36-Q64A and N36-Q64L peptides occurred at lower temperatures of 57°C and 56°C, respectively. Similarly, the α-helicity content and Tm values seen with the 6-HB formed by CP32M and mutant peptide T21-Q64A or T21-Q64L were significantly lower than the α-helicity content and Tm values seen with the 6-HB formed by CP32M and WT T21.
Table 2.
The α-helical content of N peptide alone and in mixtures with C34 or CP32 Ma
| N peptide | [θ] at 222 nm (×103) | % Helicity | Tm (°C) |
|---|---|---|---|
| N36 | −9.9 | 29.9 | NAb |
| N36-Q64A | −7.3 | 22.2 | NA |
| N36-Q64L | −8.5 | 25.9 | NA |
| N36 + C34 | −29.1 | 88.0 | 66 |
| N36-Q64A + C34 | −27.8 | 84.0 | 57 |
| N36-Q64L + C34 | −18.5 | 56.0 | 56 |
| T21 | −2.1 | 6.3 | NA |
| T21-Q64A | −3.0 | 9.1 | NA |
| T21-Q64L | −3.3 | 10.0 | NA |
| T21 + CP32 M | −28.6 | 86.7 | 88.2 |
| T21-Q64A + CP32 M | −23.3 | 70.6 | 82.0 |
| T21-Q64L + CP32 M | −11.7 | 35.5 | 71.2 |
All of the experiments were performed with 10 μM peptide solutions in phosphate-buffered saline (pH 7.0). The midpoint of thermal denaturation (Tm) was calculated from the thermal dependence of the CD signal at 222 nm.
NA, not applicable.
These results suggest that Q64, although it is located at the c position in the helical wheel, is the pivotal binding site for the HIV fusion inhibitors targeting the gp41 pocket. Molecular modeling analysis suggested that N113 in the CHR may bind to Q64 in the NHR through the polar contact, with a donor-acceptor distance in the range of 3.01 to 3.33 Å as calculated using the I-TASSER server (38) (Fig. 2C). Replacements of Q64 with hydrophobic residues, such as alanine or leucine, may disrupt this hydrophilic interaction (Fig. 2D), resulting in a weakened binding affinity between the NHR and CHR and the inhibitors bearing PBD and reduced α-helicity content and stability of the 6-HB. Nevertheless, we could not exclude the possibility that there are still other essential interactions that may be affected by the Q64 mutations.
We previously reported that A12, a small-molecule HIV-1 fusion inhibitor, could interact with Q64 and Q66 in the gp41 hydrophobic pocket via H bonds (21). Kay and colleagues have demonstrated that the polar contact of Q64 in the pocket with the hydroxyl of dTyr7 in PIE7, a pocket-binding D peptide, is critical for the potent anti-HIV-1 activity of PIE7 (41). Notably, our previous study showed that viruses with mutations in the GIV motif in NHR and an N113 substitution in CHR are much more resistant to sifuvirtide, which contains the PBD, than those with mutations in the GIV motif only (24). Mutation of G36, which is located at the c position in the α-helical wheel, resulted in decreased binding of T20 to the gp41 NHR, leading to high resistance to T20 (27). These findings support our hypothesis that the viral resistance to CP32M caused by a Q64A or Q64L mutation may be ascribed to the mutation-associated disruption of the polar contact between Q64 in NHR and N113 in CHR or to other hydrophilic interactions.
Based on our results, the combinational application of HIV-1 fusion inhibitors targeting different sites in NHR, such as CP32M and T20, which mainly target the C-terminal pocket region and the N-terminal GIV motif in the gp41 NHR domain, respectively, is expected to have a synergistic anti-HIV-1 effect, especially against the drug-resistant viral strains, since our previous studies demonstrated this effect when the first- and-next generation HIV fusion inhibitors were combined (31, 33). Similarly, a longer peptide containing all the functional domains in CHR may be effective against viruses with mutations resistant to both CP32M and T20. Indeed, we found that C52L, which contains the PBD in CP32M as well as the GBD and LBD in T20, is highly effective against viruses that bear substitutions of Q64 or mutations in the GIV motif and are resistant to CP32M or T20, respectively, suggesting a new strategy to combat the drug resistance. Therefore, this report provides a better understanding of the mechanisms underlying drug resistance and, consequently, sheds light on the rational design of novel HIV-1 fusion inhibitors or new combinational therapeutic regimens.
ACKNOWLEDGMENTS
This work was supported by a grant (2007CB914402) from the Ministry of Science and Technology of China to Y.-H.C., by a grant (11CG03) from the “Chen Guang” Project of Shanghai Municipal Education Commission and Shanghai Education Development Foundation to L.L., and by a grant (AI46221) from the U.S. NIH to S.J.
Footnotes
Published ahead of print 19 October 2011
REFERENCES
- 1. Armand-Ugón M, Gutierrez A, Clotet B, Este JA. 2003. HIV-1 resistance to the gp41-dependent fusion inhibitor C-34. Antiviral Res. 59:137–142 [DOI] [PubMed] [Google Scholar]
- 2. Cai L, Jiang S. 2010. Development of peptide and small-molecule HIV-1 fusion inhibitors that target gp41. ChemMedChem 5:1813–1824 [DOI] [PubMed] [Google Scholar]
- 3. Chan DC, Chutkowski CT, Kim PS. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. U. S. A. 95:15613–15617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273 [DOI] [PubMed] [Google Scholar]
- 5. Chan DC, Kim PS. 1998. HIV entry and its inhibition. Cell 93:681–684 [DOI] [PubMed] [Google Scholar]
- 6. Chinnadurai R, Rajan D, Munch J, Kirchhoff F. 2007. Human immunodeficiency virus type 1 variants resistant to first- and second-version fusion inhibitors and cytopathic in ex vivo human lymphoid tissue. J. Virol. 81:6563–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng Y, Zheng Q, Ketas TJ, Moore JP, Lu M. 2007. Protein design of a bacterially expressed HIV-1 gp41 fusion inhibitor. Biochemistry 46:4360–4369 [DOI] [PubMed] [Google Scholar]
- 8. Dwyer JJ, et al. 2003. The hydrophobic pocket contributes to the structural stability of the N-terminal coiled coil of HIV gp41 but is not required for six-helix bundle formation. Biochemistry 42:4945–4953 [DOI] [PubMed] [Google Scholar]
- 9. Eckert DM, Kim PS. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777–810 [DOI] [PubMed] [Google Scholar]
- 10. Eggink D, et al. 2008. Selection of T1249-resistant human immunodeficiency virus type 1 variants. J. Virol. 82:6678–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eron JJ, et al. 2004. Short-term safety and antiretroviral activity of T-1249, a second-generation fusion inhibitor of HIV. J. Infect. Dis. 189:1075–1083 [DOI] [PubMed] [Google Scholar]
- 12. He Y, et al. 2008. Identification of a critical motif for the human immunodeficiency virus type 1 (HIV-1) gp41 core structure: implications for designing novel anti-HIV fusion inhibitors. J. Virol. 82:6349–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He Y, et al. 2008. Potent HIV fusion inhibitors against enfuvirtide-resistant HIV-1 strains. Proc. Natl. Acad. Sci. U. S. A. 105:16332–16337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He Y, et al. 2008. Conserved salt bridge between the N- and C-terminal heptad repeat regions of the human immunodeficiency virus type 1 gp41 core structure is critical for virus entry and inhibition. J. Virol. 82:11129–11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He Y, et al. 2007. Conserved residue Lys574 in the cavity of HIV-1 gp41 coiled-coil domain is critical for six-helix bundle stability and virus entry. J. Biol. Chem. 282:25631–25639 [DOI] [PubMed] [Google Scholar]
- 16. He Y, et al. 2008. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 283:11126–11134 [DOI] [PubMed] [Google Scholar]
- 17. Ji H, Shu W, Burling T, Jiang S, Lu M. 1999. Inhibition of human immunodeficiency virus type 1 infectivity by the gp41 core: role of a conserved hydrophobic cavity in membrane fusion. J. Virol. 73:8578–8586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang S, Lin K, Strick N, Neurath AR. 1993. HIV-1 inhibition by a peptide. Nature 365:113. [DOI] [PubMed] [Google Scholar]
- 19. Jiang S, et al. 2004. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrob. Agents Chemother. 48:4349–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lalezari JP, et al. 2005. T-1249 retains potent antiretroviral activity in patients who had experienced virological failure while on an enfuvirtide-containing treatment regimen. J. Infect. Dis. 191:1155–1163 [DOI] [PubMed] [Google Scholar]
- 21. Liu K, et al. 2008. Design, synthesis, and biological evaluation of N-carboxyphenylpyrrole derivatives as potent HIV fusion inhibitors targeting gp41. J. Med. Chem. 51:7843–7854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu S, et al. 2007. HIV gp41 C-terminal heptad repeat contains multifunctional domains: relation to mechanisms of action of anti-HIV peptides. J. Biol. Chem. 282:9612–9620 [DOI] [PubMed] [Google Scholar]
- 23. Liu S, Lu H, Xu Y, Wu S, Jiang S. 2005. Different from the HIV fusion inhibitor C34, the anti-HIV drug Fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J. Biol. Chem. 280:11259–11273 [DOI] [PubMed] [Google Scholar]
- 24. Liu Z, et al. 2011. In vitro selection and characterization of HIV-1 variants with increased resistance to sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 286:3277–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075–1082 [DOI] [PubMed] [Google Scholar]
- 26. Lu M, Kim PS. 1997. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J. Biomol. Struct. Dyn. 15:465–471 [DOI] [PubMed] [Google Scholar]
- 27. McGillick BE, Balius TE, Mukherjee S, Rizzo RC. 2010. Origins of resistance to the HIVgp41 viral entry inhibitor T20. Biochemistry 49:3575–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Menzo S, et al. 2004. Genotype and phenotype patterns of human immunodeficiency virus type 1 resistance to enfuvirtide during long-term treatment. Antimicrob. Agents Chemother. 48:3253–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mink M, et al. 2005. Impact of human immunodeficiency virus type 1 gp41 amino acid substitutions selected during enfuvirtide treatment on gp41 binding and antiviral potency of enfuvirtide in vitro. J. Virol. 79:12447–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moore JP, Doms RW. 2003. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. U. S. A. 100:10598–10602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pan C, Cai L, Lu H, Qi Z, Jiang S. 2009. Combinations of the first and next generations of human immunodeficiency virus (HIV) fusion inhibitors exhibit a highly potent synergistic effect against enfuvirtide-sensitive and -resistant HIV type 1 strains. J. Virol. 83:7862–7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pan C, Liu S, Jiang S. 2010. HIV-1 gp41 fusion intermediate: a target for HIV therapeutics. J. Formos. Med. Assoc. 109:94–105 [DOI] [PubMed] [Google Scholar]
- 33. Pan C, Lu H, Qi Z, Jiang S. 2009. Synergistic efficacy of combination of enfuvirtide and sifuvirtide, the first- and next-generation HIV-fusion inhibitors. AIDS 23:639–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qi Z, et al. 2008. Rationally designed anti-HIV peptides containing multifunctional domains as molecule probes for studying the mechanisms of action of the first and second generation HIV fusion inhibitors. J. Biol. Chem. 283:30376–30384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ray N, et al. 2007. Clinical resistance to enfuvirtide does not affect susceptibility of human immunodeficiency virus type 1 to other classes of entry inhibitors. J. Virol. 81:3240–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ray N, Blackburn LA, Doms RW. 2009. HR-2 mutations in human immunodeficiency virus type 1 gp41 restore fusion kinetics delayed by HR-1 mutations that cause clinical resistance to enfuvirtide. J. Virol. 83:2989–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rimsky LT, Shugars DC, Matthews TJ. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei X, et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–428 [DOI] [PubMed] [Google Scholar]
- 41. Welch BD, VanDemark AP, Heroux A, Hill CP, Kay MS. 2007. Potent D-peptide inhibitors of HIV-1 entry. Proc. Natl. Acad. Sci. U. S. A. 104:16828–16833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weng Y, Weiss CD. 1998. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 72:9676–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U. S. A. 91:9770–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]