Abstract
Epstein-Barr virus (EBV) infection is causatively associated with a variety of human cancers, including nasopharyngeal carcinoma (NPC). The only viral nuclear protein expressed in NPC is EBNA1, which can alter cellular properties in ways that may promote oncogenesis. Here, we used 2-dimensional difference gel electrophoresis (2-D DiGE) to profile changes in the nuclear proteome that occur after stable expression of EBNA1 in the EBV-negative NPC cell line CNE2. We found that EBNA1 consistently altered the levels of a small percentage of the nuclear proteins. The identification of 19 of these proteins by mass spectrometry revealed that EBNA1 upregulated three proteins affecting metastatic potential (stathmin 1, maspin, and Nm23-H1) and several proteins in the oxidative stress response pathway, including the antioxidants superoxide dismutase 1 (SOD1) and peroxiredoxin 1 (Prx1). Western blot analysis verified that EBNA1 expression upregulated and EBNA1 silencing downregulated these proteins. In addition, transcripts for stathmin 1 were induced by EBNA1, whereas EBNA1 only affected Prx1 and SOD1 at the protein level. Further investigation of the EBNA1 effects on the redox pathway showed that long-term EBNA1 expression in NPC resulted in increased reactive oxygen species (ROS) and increased levels of the NADPH oxidases NOX1 and NOX2, known to generate ROS. In addition, EBNA1 depletion in EBV-positive cells decreased NOX2 and ROS. The results show multiple roles for EBNA1 in the oxidative stress response pathway and suggest mechanisms by which EBNA1 may promote NPC metastases.
INTRODUCTION
Epstein-Barr virus (EBV) is a ubiquitous human gamma-herpesvirus that is harbored in at least 90% of the world population (40). Transmitted through saliva, EBV infects B lymphocytes as well as the epithelium of the orthopharynx. Upon primary infection, EBV immediately establishes latent infection in cells by maintaining multiple copies of its genome in the form of episomes. While most infected individuals control the virus efficiently and remain free of EBV-associated diseases, latent EBV infection is linked to the development of both lymphoid and epithelial tumors. Nasopharyngeal carcinoma (NPC) is one such epithelial tumor in which EBV is widely recognized as a causative agent, since most NPC tumors are monoclonal proliferations of latently EBV-infected cells (38). These tumor cells express only one EBV nuclear protein, EBNA1, in addition to the EBV latent membrane proteins LMP1 and LMP2 (38).
Epstein-Barr nuclear antigen 1 (EBNA1) is essential for the establishment of latent EBV infection since it replicates and segregates the viral genome in proliferating cells (reviewed in reference 17). Additionally, it is a transcriptional activator for other EBV latency genes. Aside from its roles in viral persistence, increasing numbers of observations suggest that EBNA1 alters cellular processes in ways that contribute to host cell survival and proliferation. First, EBNA1 is expressed in all latency forms in proliferating cells and is the only EBV protein expressed in all EBV-associated tumors (3, 38). In addition, downregulation of EBNA1 in Raji Burkitt's lymphoma (24) and NPC cells (55) using RNA interference has been found to decrease cell proliferation. Similarly, increased apoptosis was observed in Burkitt's lymphoma cells overexpressing a dominant-negative form of the EBNA1 mutant (28). Finally, EBNA1 expression in lymphoblastoid cell lines was shown to decrease apoptosis (34).
One mechanism by which EBNA1 contributes to cell survival is by lowering p53 levels. This occurs as a result of EBNA1 binding the cellular ubiquitin-specific protease USP7, which deubiquitinates and stabilizes p53 (31). EBNA1 was found to block the USP7-p53 interaction by competing for the same binding pocket on USP7 (22, 41, 44). In keeping with these findings, the expression of EBNA1 (but not a USP7-binding mutant of EBNA1) interfered with p53 stabilization after DNA damage, thereby decreasing apoptosis (41). In addition, EBNA1 limits p53 acetylation and apoptosis and interferes with DNA repair by inducing the loss of promyelocytic leukemia (PML) nuclear bodies, known to play important roles in these processes (46, 47). The loss of the PML bodies results from degradation of the PML tumor suppressor proteins. This process involves EBNA1-mediated recruitment of both USP7 and the cellular protein kinase CK2 (casein kinase 2) to the PML bodies.
Genome-wide analyses of the cellular effects of EBNA1 have been performed using microarray analyses, with various results (8, 48, 54). However, the ability of EBNA1 to affect the stability of p53 and PML proteins shows that EBNA1 can modulate the levels of some proteins without affecting RNA levels, due to the ability of EBNA1 to bind and reroute cellular-protein-modifying enzymes, including USP7, CK2, and protein methyl transferases PRMT1 and PRMT5 (23). Since CK2 and USP7 affect multiple cellular processes, it is likely that EBNA1 binding to these proteins will affect the posttranslational modifications and/or levels of cellular proteins in addition to p53 and PML. However, to date there are no comprehensive studies on the effects of EBNA1 on the human proteome. To gain greater insights into the host cell alterations in the presence of EBNA1, we used an unbiased proteomics-based approach, 2-dimensional difference gel electrophoresis (2-D DiGE), coupled with mass spectrometry to compare the proteomes of an EBV-negative NPC cell line in the presence and absence of stable EBNA1 expression. This approach identified several additional cellular proteins that are affected by EBNA1, and the results suggest roles for EBNA1 in modulating oxidative stress and metastases.
MATERIALS AND METHODS
Cell lines.
The EBV-negative nasopharyngeal carcinoma cell line CNE2 (also called CNE2Z) and the CNE2E cell line that stably expresses EBNA1 have been previously described (47, 49). Both cell lines were grown at 37°C in alpha minimal essential medium (αMEM; Gibco) supplemented with 10% fetal calf serum (Sigma), and 0.5 mg of hygromycin B/ml was added to CNE2E cells to stably maintain EBNA1 expression. The EBV-positive NPC cell line C666-1 has been previously described (12) and was grown in HEPES-modified RPMI 1640 (Sigma).
Sample preparation for 2-D DiGE.
Four 15-cm2 plates of CNE2 and CNE2E cells at ∼90% confluence were harvested by scraping and washed twice in phosphate-buffered saline (PBS). The cell pellet was resuspended in a 10:1 ratio (buffer/pellet [vol/vol]) of cold hypotonic buffer (20 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 1 mM dithiothreitol [DTT], and 10 μl/ml of protease inhibitor cocktail no. 8340 from Sigma-Aldrich) on ice for 10 min. The cells were then disrupted by hypotonic lysis using a Dounce homogenizer (10 strokes of pestle B), and nuclei were pelleted by centrifugation at 1,500 × g for 10 min at 4°C. The nuclear pellet was washed with hypotonic buffer to remove any cytosolic proteins and then resuspended in a 2:1 ratio (buffer/nuclear pellet) of buffer A (200 mM NaCl, 1% NP-40, 1 mM DTT, and protease inhibitors). After 15 min on ice, the extract was subjected to centrifugation at 15,000 × g for 30 min, and the soluble fraction (called the soluble nuclear fraction) was precipitated using a 2-D clean-up kit (GE Healthcare) to remove salts according to the manufacturer's recommendations and then resuspended in buffer B (8 M urea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 1 mM DTT). The pellet fraction of the nuclear extract was solubilized in buffer B to generate the chromatin-enriched fraction. Nucleic acids in this fraction were then removed by the addition of 10 mM spermine followed by ultracentrifugation at 200,000 × g for 1 h, and proteins were precipitated with the 2-D clean-up kit to remove spermine, as described for the soluble nuclear samples.
2-D DiGE.
Soluble nuclear and chromatin-enriched extracts were labeled with the fluorophores Cy3, Cy5, and Cy2 (GE Healthcare) according to the manufacturer's procedure. Two biological samples were used for each fraction to achieve statistical confidence. Amounts of 50 μg of lysates (∼7 mg/ml) were minimally labeled with 400 pmol of dye for 30 min in the dark at 4°C. Samples from each cell line (CNE2 and CNE2E) were labeled with either Cy3 or Cy5; dye swapping was also performed to minimize labeling-dependent bias. Equal amounts of protein from all four samples (for each fraction) were pooled to form an internal control, 50 μg of which was labeled with Cy2 to serve as a reference for spot normalization and comparison between gels. The reaction was stopped by the addition of 10 mM lysine and incubation for 10 min at 4°C. Labeled samples from each set were combined in 2-D DiGE rehydration buffer (8 M urea, 4% CHAPS, 0.002% bromophenol blue, 0.5% immobilized pH gradient [IPG] buffer [pH 3 to 11], and 40 mM DTT).
The 1st-dimension isoelectric focusing (IEF) of Cy3-, Cy5-, and Cy2-labeled protein samples was performed on an Ettan IPGPhor II electrophoresis system (GE Healthcare) using 13-cm Immobiline DryStrips, pH 3 to 11, nonlinear (GE Healthcare). IEF was performed at 20°C with the following parameters: 50 μA/strip for 14 h of rehydration, stepped to 500 V for 3.5 kV/h, gradient stepped to 1,000 V for 0.8 kV/h, gradient stepped to 8,000 V for 14 kV/h, and 8,000 V for 10 kV/h. Proteins in the IPG strips were reduced and alkylated with 1% DTT and 4% iodoacetamide, respectively, in equilibration buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, and 0.00125% bromophenol blue) for 15 min. The 2nd-dimension separation was done using 13% SDS-PAGE at 4°C with 15 min at 10 mA followed by 5 h at 30 mA. The 2-D gels were scanned on a Typhoon scanner (GE Healthcare) at a 100-μm resolution with λ excitation/λ emission values of 488/520, 532/580, and 633/670 nm for Cy2, Cy3, and Cy5, respectively, and were then analyzed using DeCyder software (version 6.5; GE Healthcare). Gels were matched, and protein spots were automatically and manually landmarked for determination of protein abundance. The average volume ratio of each spot was determined with normalization and statistical analysis (using analysis of variance [ANOVA] and Student's t test [P < 0.01]). Protein spots were considered to be of interest if there was a volume ratio change of ≤1.5 (decreased) or ≥1.5 (increased) between the CNE2 and CNE2E samples with Student's t test values at 99% statistical confidence (P < 0.01).
Protein identification by mass spectrometry.
Preparative 2-D gels for protein identification were used to analyze 300 μg of unlabeled protein samples as described above (150 μg each from CNE2 and CNE2E samples). The 2-D gels were then stained overnight using colloidal Coomassie dye (17% ammonium sulfate, 3% ortho-phosphoric acid, 34% methanol, 0.1% G-250). Spots of interest were manually excised by matching the preparative 2-D gels to analytical gels. These were subjected to overnight in-gel trypsin digestion using 13 ng of trypsin (Sigma) per μl of gel dissolved in 50% acetonitrile, 25 mM ammonium bicarbonate. Peptides were extracted in 16 μM ammonium bicarbonate, then 0.02% formic acid, then 40% acetonitrile for 10 min each (repeated twice). Mass spectrometry on extracted peptides was performed by the Advanced Protein Technology Centre (Hospital for Sick Children, Toronto, ON, Canada), using an on-line liquid chromatography (LC)-tandem mass spectrometry setup using an Agilent 1100 capillary LC system (Palo Alto, CA) fitted to a linear trap quadrupole (LTQ) ion trap mass spectrometer (Thermo Electron, San Jose, CA). A C18 precolumn (100-μm inner diameter by 5.0-cm length) and a micro-liquid chromatography analytical column (75 μm by 10 cm) that also served as a micro-electrospray ionization emitter was used to separate the digested proteins. The identity of peptides was determined using Mascot (version Mascot; Matrix Science, London, United Kingdom) and validated using Scaffold (version Scaffold_2_06_00; Proteome Software, Inc., Portland, OR). Some spots were also analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry after in-gel trypsin hydrolysis. The peptides were purified and analyzed by MALDI-TOF mass spectrometry using a cyano-4-hydroxycinnamic acid matrix (Sigma) on a Voyager DE-STR instrument (Applied Biosystems). Identification of the proteins using these mass fingerprinting data was carried out using ProFound software.
Western blotting.
Whole-cell lysates were generated by lysing cells in 8 M urea, 10 mM Tris, pH 8.0, followed by clarification by centrifugation. Amounts of 15 μg of soluble nuclear (obtained as described above) or 50 μg of whole-cell lysates were subjected to 15% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were blocked in 5% nonfat dry milk in PBS and then incubated with antibodies against stathmin 1/Op18 (sc-55531; Santa Cruz), maspin (sc-22762; Santa Cruz), superoxide dismutase 1 (SOD1) (sc-11407; Santa Cruz), Nm23-H1 (sc-343; Santa Cruz), Prx1 (ab15571; Abcam), NADPH oxidase 1 (NOX1) (NBP1-31546; Novus Biologicals), NOX2 (catalog no. ab80508; Abcam), EBNA1 (OT1X; kindly supplied by Jaap Middeldorp), or actin (Ab-1; Oncogene Research Products). After washing, blots were probed with goat anti-mouse peroxidase or goat anti-rabbit peroxidase (Santa Cruz) and developed using enhanced chemiluminescence/chemifluorescence reagents (ECL Plus; GE Healthcare). Bands were imaged on a Typhoon scanner and quantified using Image Quant software.
RNA interference.
Amounts of 1.5 × 105 CNE2E or C666-1 cells were transfected with 50 pmol small interfering RNA (siRNA) against green fluorescent protein (GFP) (GCAAGCUGACCCUGAAGUUCAU) or EBNA1 (GGAGGUUCCAACCCGAAAUTT) using 2 μl Lipofectamine 2000 (Invitrogen). Cells underwent three rounds of transfection separated by 24 h (CNE2E) or 48 h (C666-1).
Transient EBNA1 expression.
EBNA1 (with a C-terminal sequential peptide affinity [SPA] tag) was transiently expressed in CNE2 cells using an adenovirus delivery system. To generate the adenovirus, the coding sequence for the SPA tag (56) was first inserted between the EcoRI and NotI sites of the multicloning site of pENTR4 (Invitrogen), and then an EBNA1 or LacZ coding sequence was individually inserted between the BamHI and KpnI sites of the multicloning site to generate constructs expressing C-terminally tagged EBNA1 or LacZ. The Adenoviral Gateway Expression system (Invitrogen) was used to make the adenovirus carrying the EBNA1 or LacZ cassettes. CNE2 cells in 15-cm2 plates at 70% confluence were infected with the LacZ or EBNA1 adenovirus, and the expression of EBNA1 and LacZ in the majority of the cells was verified by immunofluorescence microscopy. Cells were harvested 48 h postinfection for Western blot analysis.
Intracellular ROS measurements.
Changes in intracellular reactive oxygen species (ROS) levels were determined by 2′,7′-dichlorodihydro-fluorescein diacetate (H2DCF-DA; Molecular probes) labeling. Cells at ∼70% confluence were loaded with 10 μM H2DCF-DA in PBS for 30 min at 37°C. Cells were then washed with PBS and scraped for analysis by flow cytometry using excitation and emission settings of 488 nm and 530 nm, respectively. The mean fluorescence peak was analyzed from the gated cell population of 10,000 cells. Experiments were performed with three biological replicates. Flow cytometry analysis was performed at the University of Toronto, Faculty of Medicine, Flow Cytometry Facility.
RNA extraction and real-time PCR analysis.
Total cellular RNA was isolated using TRIzol reagent (Invitrogen), following the manufacturer's instructions. Total cDNA synthesis was performed using a mixture of total cellular RNA (1 μg), random hexamers (200 ng), oligo(dT) (0.5 μg), and 1 mM deoxynucleoside triphosphates (dNTPs) in a final 12-μl volume. The mixture was incubated at 65°C for 5 min and quick chilled on ice, and then 8 μl of reaction mixture (4 μl 5× First Strand buffer, 2 μl 0.1 M DTT, 40 units RNaseOUT, and 100 units SuperScript II reverse transcriptase) was added to each sample. The samples were then incubated at room temperature for 10 min, followed by incubation at 42°C for 30 min and heat inactivation at 85°C for 5 min. The cDNA products were used for real-time quantitative PCR analysis in a final 10-μl volume consisting of 1 μl 10-fold-diluted cDNA products, 2 mM forward and reverse primer mix, and 5 μl SsoFast EvaGreen supermix (catalog no. 172-5201; Bio-Rad). A Rotor-Gene RG-3000A real-time PCR machine (Corbett Research) was used with the following cycling parameters: initial denaturation/enzyme activation at 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 55°C for 15 s, and 72°C for 25 s. The human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as the reference gene to normalize the expression level between samples, and expression variations were calculated using the cycle threshold (ΔΔCT) method. All primer sequences are shown in Table 1.
Table 1.
Primers used for mRNA quantification by real-time PCR
| Gene product | Forward primer | Reverse primer |
|---|---|---|
| GAPDH | 5′-GAGTCAACGGATTTGGTCGT-3′ | 5′-AATGAAGGGGTCATTGATGG-3′ |
| NOX1 | 5′-TAGGCGCCCTAAGTTTGAAG-3′ | 5′-AAACCGGAGGATCCTTTCAC-3′ |
| NOX2 | 5′-CCCTTTGGTACAGCCAGTGAAGAT-3′ | 5′-CAATCCCAGCTCCCACTAACATCA-3′ |
| PRX1 | 5′-AGCTAGCGAGATTTTCCAAAGCATATGGT-3′ | 5′-GAAGCTTCATGTTTCATCTACAGATCGTCC-3′ |
| SOD1 | 5′-CTGAAGGCCTGCATGGATTC-3′ | 5′-CCAAGTCTCCAACATGCCTCTC-3′ |
| STMN1 | 5′-GTTCCAGAATTCCCCCTTTC-3′ | 5′-TCTCGTGCTCTCGTTTCTCA-3′ |
| XDH | 5′-AGGTGGACCACTTCAGCAAT-3′ | 5′-GTTGGAGGGAAGGTTGGTTT-3′ |
RESULTS
2-D DiGE strategy for analysis of the effects of EBNA1 on the nuclear proteome.
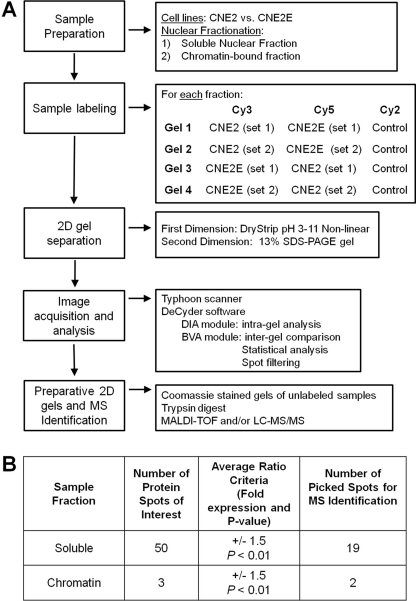
In order to more globally determine the effects of EBNA1 on the host cell proteome, we used 2-D DiGE to compare an EBV-negative NPC cell line that stably expresses EBNA1 (CNE2E) (47) to the parental CNE2 cell line lacking EBNA1 expression. CNE2E cells express EBNA1 from an integrated cassette at a low level similar to that of EBV-positive NPC cells. Since EBNA1 is a nuclear protein with several nuclear functions, we were particularly interested in surveying its effects on the nuclear proteome and, therefore, compared nuclear extracts of CNE2 and CNE2E cells. We chose the 2-D DiGE method, involving the labeling of protein samples with fluorescent Cy dyes prior to 2-D electrophoresis, because it allows for visualization and quantification of differences in protein abundance between samples and is conducive to statistical analysis. For better separation and visualization of abundant proteins, we further fractionated the nuclear extract into two parts, soluble and chromatin-enriched fractions, and subjected them individually to 2-D DiGE. In order to achieve high statistical confidence, two biological samples were used and each set of samples was analyzed twice, with dye swapping (Cy3 and Cy5). Additionally, internal controls in which an equal mixture of the samples was labeled with a third dye (Cy2) were used for accurate analysis between gels. In total, 4 gels were analyzed for each nuclear fraction. Figure 1A illustrates the strategy used in detail. For protein identification, preparative gels were run with unlabeled samples and stained with colloidal Coomassie dye, and protein spots of interest were excised and subjected to mass spectrometry.
Fig 1.
Experimental design and analysis. (A) Flow chart showing the details of the 2-D DiGE strategy used for identification of candidate proteins. LC-MS/MS, liquid chromatography-tandem mass spectrometry. (B) Summary of the number of candidate proteins identified using DeCyder analysis, selection criteria for protein spots of interest, and number of spots picked on preparative gels for identification via mass spectrometry.
Identification of differentially expressed proteins in the presence of EBNA1.
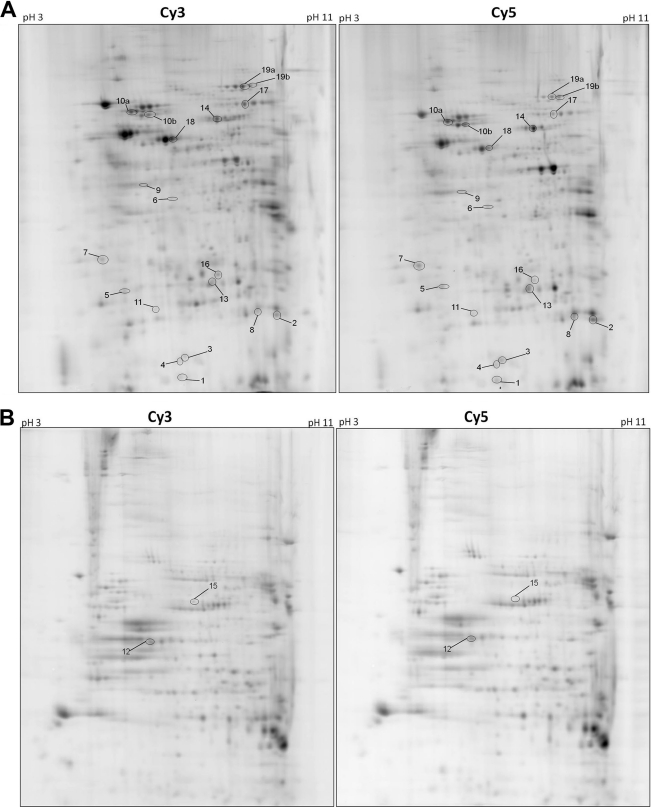
Comparison of normalized spot volumes of the two differentially labeled samples (from CNE2 and CN2E cells) identified spot volumes that were increased or decreased between the cell lines. Approximately 1,000 spots were detected in the gels for the soluble nuclear fraction, 50 of which showed an average volume ratio change of ±1.5-fold or greater with a P value of less than 0.01 (based on Student's t test). For the chromatin fraction, 3 out of 600 spots were differentially expressed with at least a 1.5-fold difference (Fig. 1B). Figure 2 shows representative 2-D gels generated from the soluble nuclear (Fig. 2A) and chromatin-enriched (Fig. 2B) fractions. Nineteen of the EBNA1-modulated protein spots were excised from the preparative gel of the soluble nuclear fraction for identification, while 2 EBNA1-modulated spots were excised from the chromatin-enriched gel (indicated by numbered spots in Fig. 2A and B). Table 2 summarizes the identity of the excised proteins as determined by mass spectrometry. Stathmin 1 (also called OP18), peroxiredoxin 1 (Prx1), and nonmetastatic protein 23 (Nm23-H1) were reliably identified and increased the most in abundance with EBNA1 expression. Of the proteins that decreased in abundance with EBNA1 expression, the most striking was the microRNA regulatory protein KSRP, followed by tapasin, which is involved in antigen presentation for the major histocompatibility complex (MHC) class 1 molecules. Interestingly, several proteins that were found to increase in the presence of EBNA1 are involved in oxidative stress, including the antioxidants superoxide dismutase 1 (SOD1), Prx1, Prx6, and glutathione S-transferase (GST). In addition, three proteins known to affect metastasis (Nm23-H1, maspin, and stathmin 1) were increased with EBNA1 expression, suggesting that EBNA1 may affect the ability of EBV-transformed tumor cells to metastasize. Interestingly, our finding of increased Nm23-H1 levels in the nuclear extracts with EBNA1 is consistent with a previous report that Nm23-H1 interacts with EBNA1 in B lymphocytes and is relocalized by EBNA1 to the nucleus (36).
Fig 2.
Analytical gels from 2-D DIGE. Representative 2-D DiGE Cy3 and Cy5 gel images from soluble nuclear (A) and chromatin-enriched (B) fractions are shown. Amounts of 50 μg of CNE2 (Cy3) and CNE2E (Cy5) protein samples, along with 50 μg of internal control samples, were run on 13-cm, pH 3 to 11, nonlinear Immobiline DryStrips gels (1st dimension) and 13% SDS-PAGE (2nd dimension). The positions of the differentially expressed protein spots picked for mass spectrometry are indicated by the numbers, which correspond to the protein spot numbers in Table 2.
Table 2.
Summary of proteins differentially expressed in CNE2 and CNE2E cells
| Spot no.a | Avg ratio (CNE2E/CNE2) | Accession no. | Protein name | Alternate name(s) | Molecular mass (kDa) | pI | No. of unique peptides | % Sequence coverage | Mascot score if identified by MALDI-TOF | Function |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4.1 | BAG70133 | Stathmin 1 | Op18, STMN1 | 17 | 5.75 | 12 | 59.10 | Regulator of microtubules | |
| 2 | 4.0 | NP_002565 | Peroxiredoxin 1 | Prdx1, Prx1 | 22 | 8.72 | 15 | 63.30 | Antioxidant | |
| 3 | 4.0 | CAA35621 | Nm23 protein | NM23 | 17 | 7.06 | 9 | 57.90 | Regulator of metastasis | |
| 4 | 3.8 | CAG46542 | Cu/Zn-superoxide dismutase 1 | SOD1 | 16 | 5.70 | 6 | 41.80 | Antioxidant | |
| 5 | 3.5 | NP_001036817 | Tropomyosin alpha-3 | 29 | 4.78 | 14 | 39.10 | Actin binding; muscle contraction | ||
| 6 | 3.3 | NP_002630 | Maspin | Serpin B5 | 43 | 5.72 | 16 | 52.40 | Regulator of metastasis | |
| 7 | 2.8 | BAG37888 | Unnamed protein product | 29 | 4.72 | 22 | 58.10 | |||
| 8 | 2.7 | NP_002558 | Phosphatidylethanolamine-binding protein 1 | PEBP1, RKIP | 21 | 7.01 | 12 | 66.30 | EGFR1 and NF-κB signaling | |
| 9 | 2.6 | 2JAI_A | Dimethylarginine dimethylaminohydrolase 1 | DDAH1 | 31 | 5.53 | 14 | 48.40 | Nitric oxide generation | |
| 10a, 10b | 2.3 (1.9) | 3FZF_A | Heat shock protein 70 | Hsp70 | 71 | 6.38 | 8 | 25 | 80 | Stress-induced heat shock protein |
| 11 | 2.3 | CAA30894 | Glutathione S-transferase | GST | 23 | 5.43 | 10 | 57.10 | Antioxidant | |
| 12 | 2.3 | Unnamed protein product | 48 | 7 | 32.00 | |||||
| 13 | 2.1 | NP_004896 | Peroxiredoxin 6 | Prxd6, Prx6 | 25 | 6.00 | 16 | 65.20 | Antioxidant | |
| 14 | 1.8 | XP_001163388 | Stress-induced phosphoprotein 1 | STIP1 | 63 | 7.80 | 45 | 58.00 | NAb | |
| 15 | −1.6 | EAW52998 | Lamin A/C | 87 | 8.91 | 38 | 46.00 | Nuclear structure | ||
| 16 | −1.7 | NP_004083 | Mitochondrial short-chain enoyl-coenzyme A hydratase 1 | 31 | 8.34 | 13 | 41.00 | Fatty acid beta oxidation | ||
| 17 | −1.9 | EAW64946 | IMP dehydrogenase 2 | IMPDH2 | 56 | 8.67 | 19 | 39.50 | Purine ribonucleoside monophosphate biogenesis | |
| 18 | −2.1 | 3F8U_A | Tapasin-ERP57 heterodimer | 54 | 5.60 | 29 | 57.60 | Antigen presentation | ||
| 19a, 19b | −3.1, −1.9 | AAH85004 | KH-type splicing regulatory protein | KHSRP, KSRP | 70 | 8.00 | 9 | 17 | 87 | microRNA biogenesis |
Positions of spots on 2-D DIGE analytical gels are shown in Fig. 2.
NA, not applicable.
Validation of EBNA1-mediated modulation of candidate proteins.
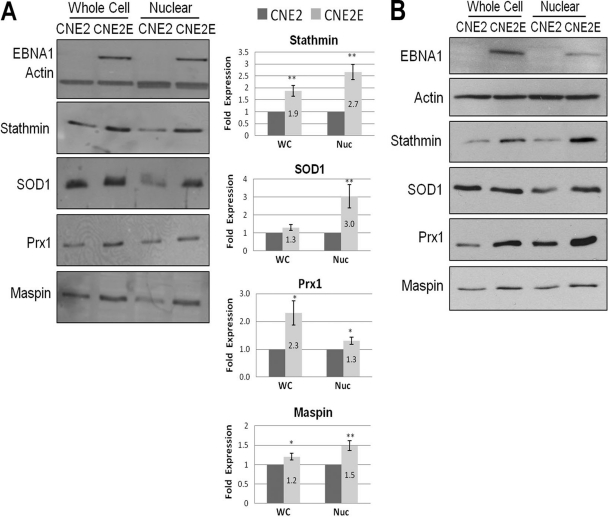
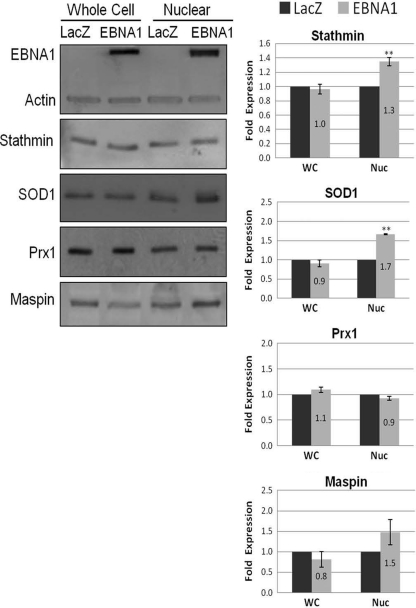
To validate the effects of EBNA1 expression on candidate proteins identified as described above, Western blots were performed on whole-cell and nuclear extracts from cells with and without EBNA1 expression. Since our DiGE analysis identified two major pathways (oxidative stress and metastasis) as being altered by EBNA1, validation was focused on proteins from these pathways. In particular, the antioxidants SOD1 and Prx1 were further investigated since they showed the greatest change by DiGE analysis. Validation of the metastasis proteins focused on stathmin 1 and maspin since studies validating the effect of EBNA1 on Nm23-H1 have been previously performed (36). Comparison of CNE2 and CNE2E extracts from 3 biological samples showed that stable expression of EBNA1 resulted in significant increases in stathmin 1 and Prx1 in both whole-cell and nuclear extracts (Fig. 3A). Smaller but still statistically significant increases were also seen for maspin in both extracts from EBNA1-expressing cells. For SOD1 levels, little difference was detected in whole-cell extracts from CNE2 and CNE2E cells; however, SOD1 levels were increased 3-fold in CNE2E nuclear extracts compared to the levels in CNE2 nuclear extracts. This suggests that EBNA1 increases the nuclear localization of SOD1. Overall, the results confirmed the findings of the 2-D DiGE that stable EBNA1 expression increased the nuclear levels of stathmin 1, maspin, SOD1, and Prx1. Since CNE2E cells are normally grown in hygromycin to maintain EBNA1 expression, we also repeated the experiments whose results are shown in Fig. 3A with CNE2E cells grown for a week without hygromycin to determine if hygromycin affected the results (Fig. 3B). However, the same protein changes in CNE2E cells relative to CNE2 cells were seen as in the experiments whose results are shown in Fig. 3A, indicating that hygromycin does not affect the levels of the proteins in question.
Fig 3.
Validation of candidate proteins by Western blotting. (A) A representative Western blot comparing CNE2 and CNE2E whole-cell (WC) and nuclear (Nuc) extracts probed with the antibodies indicated to the left of the gels and detected by enhanced chemifluorescence. Histograms show average values with standard errors of the bands from three independent experiments; values for CNE2 extracts (dark bars) are set to 1. P values are calculated using Student's t test and are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) CNE2E cells were grown for 1 week without hygromycin and compared to CNE2 cells by performing Western blots as described above.
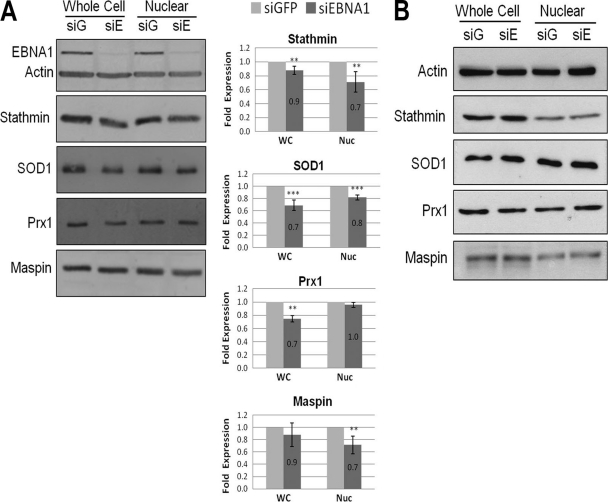
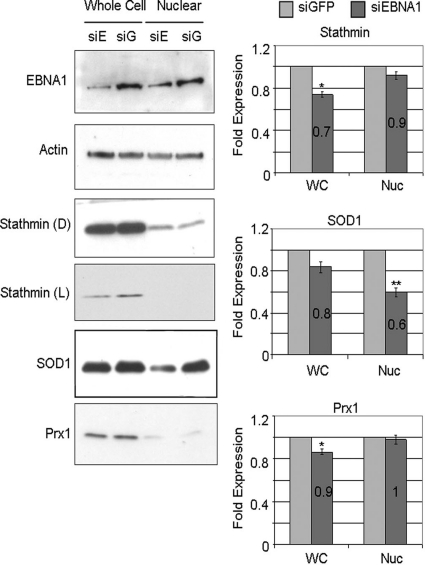
Next, we validated the effects of EBNA1 on these host proteins by comparing protein abundance in CNE2E cells after downregulation of EBNA1 with siRNA to protein abundance in CNE2E cells treated with nonspecific siRNA against GFP (Fig. 4A). Downregulation of EBNA1 resulted in small but statistically significant decreases in stathmin 1 and SOD1 in both the whole-cell and nuclear extracts. Consistent with the results shown in Fig. 3A, EBNA1 depletion decreased maspin levels in the nuclear extract more than in the whole-cell extract and decreased Prx1 more in the whole-cell extract than in the nuclear extract. Note that whole-cell extracts not only differ from nuclear extracts in containing cytoplasm but also contain additional nuclear proteins that leach out of the nucleus upon cell lysis. To ensure that the EBNA1 siRNA was not affecting proteins independent of EBNA1, we also compared the specific protein levels in CNE2 cells after GFP and EBNA1 siRNA treatments (Fig. 4B). However, in the absence of EBNA1, EBNA1 siRNA treatment had no effect on the whole-cell or nuclear levels of any of the proteins in question. Therefore, the EBNA1 depletion results further confirm the findings from the 2-D DiGE analysis.
Fig 4.
Effect of EBNA1 depletion on candidate protein expression. (A) CNE2E cells were transfected with siRNA against EBNA1 (siE/siEBNA1) or GFP (siG/siGFP), and cells were grown in the absence of hygromycin. A representative Western blot comparing whole-cell (WC) and nuclear (Nuc) extracts is shown. Western blots and quantification were performed as described for Fig. 3A except that values for extracts from GFP siRNA-treated samples (light bars) are set to 1. (B) CNE2 cells (lacking EBNA1 expression) were treated with siRNA against EBNA1 or GFP, and then whole-cell and nuclear lysates were analyzed by Western blotting.
Assessment of immediate effects of EBNA1.
We next asked whether the EBNA1-induced changes observed with stable EBNA1 expression occur gradually (as might be expected if EBNA1 affects the protein level indirectly) or whether these changes are due to more immediate/direct effects of EBNA1. To this end, we compared the whole-cell and nuclear extracts of CNE2 cells with and without transient expression of EBNA1. To ensure that EBNA1 expression occurred in the majority of the cells, an adenovirus-based system was used to deliver either EBNA1 or the LacZ negative control to the CNE2 cells, and cells were harvested 48 h postinfection for Western blot analysis (Fig. 5A). Consistent with the results from stable EBNA1 expression, transient EBNA1 expression resulted in increased nuclear levels of stathmin 1, SOD1, and maspin (although the increase in maspin from multiple experiments was not statistically significant due to higher variability). However, unlike the result for stable expression of EBNA1, we did not observe an increase in stathmin 1 expression in whole-cell lysates, nor did we detect any EBNA1-induced increase in Prx1 in either whole-cell or nuclear extracts. Therefore, the results suggest that the nuclear increases in stathmin 1, SOD1, and possibly, maspin are immediate effects of EBNA1 expression, while increases in Prx1 and in whole-cell levels of stathmin 1 only occur after more prolonged EBNA1 expression.
Fig 5.
Effect of transient EBNA1 expression on candidate proteins. A representative Western blot and quantification comparing CNE2 cells after delivery of EBNA1 or LacZ expression cassettes via adenovirus infection. Details are as described for Fig. 3A. Values for LacZ-transfected samples (dark bars) were set to 1 in the histograms.
Assessment of the effects of EBNA1 in EBV-positive NPC.
We also examined whether EBNA1-mediated changes in the levels of the candidate cellular proteins occurred in the context of EBV-infected NPC cells. For these experiments, we used the only naturally EBV-positive NPC cell line that stably maintains EBV genomes, C666-1 (12). EBNA1 expression was downregulated in C666-1 cells using siRNA against EBNA1, and cell extracts were compared to extracts of C666-1 cells treated with negative-control siRNA (against GFP). Western blots of whole-cell and nuclear extracts showed that EBNA1 downregulation modestly decreased the whole-cell levels of stathmin 1, SOD1, and Prx1 (Fig. 6) and had a greater effect on SOD1 nuclear levels (consistent with the results shown in Fig. 3A and 5). Note that EBNA1 was only partially depleted in these experiments and, therefore, large effects on protein levels were not expected. Interestingly, only a small fraction of the stathmin 1 in the C666-1 cells was found to be in the nucleus, in contrast to its largely nuclear localization in CNE2 and CNE2E cells. In addition, we found that C666-1 cells had very low levels of maspin, which precluded reliable detection and quantification of the effects of EBNA1 silencing (data not shown). The data suggest that, as in EBV-negative NPC cells, EBNA1 increases the levels of stathmin 1, Prx1, and SOD1 in EBV-positive NPC cells.
Fig 6.
Effect on candidate proteins of EBNA1 silencing in EBV-positive cells. A representative Western blot and quantification comparing EBV-positive C666-1 cells after transfection with siRNA against EBNA1 (siE/siEBNA1) and GFP (siG/siGFP) are shown. Details are as described for Fig. 3A. Values for GFP siRNA-treated samples (light bars) were set to 1. Both dark (D) and light (L) exposures of the same stathmin blot are shown.
Stable EBNA1 expression increases ROS levels and transcription of NOX1 and NOX2 oxidases.
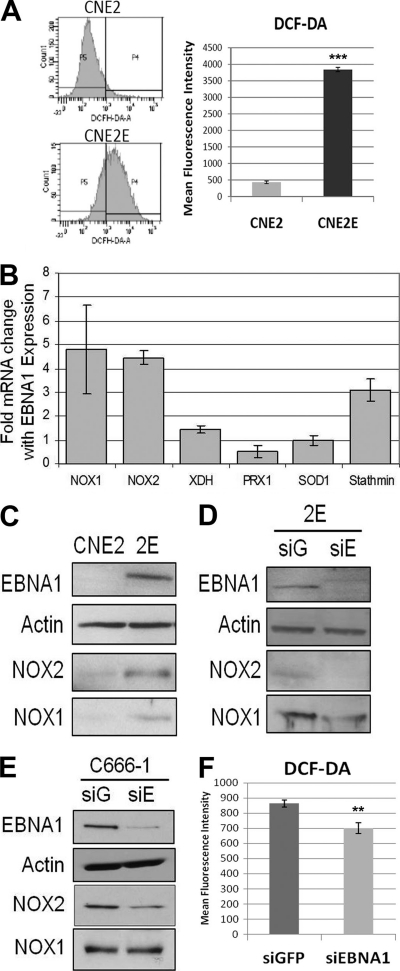
Since we observed an upregulation of several antioxidants (i.e., SOD1, Prx1, Prx6, and GST) in the presence of EBNA1 in our 2-D DiGE screen, we wondered if EBNA1 affected the balance of redox regulation in NPC cells. For this purpose, we compared the intracellular levels of reactive oxygen species (ROS) between CNE2 and CNE2E cells by labeling with DCF-DA, a membrane-permeant indicator that fluoresces upon oxidation (15). We observed that the EBNA1-expressing cells had about 7-fold-higher levels of ROS than the same cells lacking EBNA1 (Fig. 7A). Very similar results were obtained when CNE2E cells were grown for a week without hygromycin and compared to CNE2 cells (data not shown), showing that hygromycin is not responsible for the difference in ROS levels.
Fig 7.
Effect of EBNA1 on ROS, NOX1, and NOX2. (A) ROS levels were measured by monitoring changes in oxidized DCF fluorescence in CNE2 and CNE2E cells. Fluorescence-activated cell sorting data from one representative experiment are shown (left panels), along with quantification of results from multiple experiments shown in the histogram. Bars represent means ± standard errors of three independent experiments. ***, P < 0.0001. (B) Total RNA was extracted from CNE2 and CNE2E cells and subjected to quantitative reverse transcription-PCR for NOX1, NOX2, XDH, SOD1, PRX1, and stathmin transcripts. Data were normalized to values for GAPDH and expressed as fold change of results for CNE2E relative to results for CNE2 cells. Bars represent means ± standard errors of three independent experiments measured in triplicate. (C to E) Western blots were performed on whole-cell lysates with the indicated antibodies, comparing CNE2 and CNE2E (2E) cells (C), CNE2E cells with GFP (siG) and EBNA1 (siE) siRNA treatments (D), and C666-1 cells with GFP and EBNA1 siRNA treatments (E). Different exposure times were used for the different experiments to maximize protein detection over background. (F) ROS levels in C666-1 cells after GFP (siGFP) and EBNA1 (siEBNA1) siRNA treatments were measured as described for panel A.
The accumulation of ROS may be caused by enhanced oxidative metabolism or by downregulation of antioxidants (52). Since we observed increased ROS in CNE2E cells even with the increased abundance of several antioxidants, we investigated the cellular levels of other players in redox regulation, particularly oxidases involved in ROS generation. To this end, we extracted total mRNA from CNE2 and CNE2E cells and compared the transcript levels of NADPH oxidases (NOX1 and NOX2) and xanthine oxidase (XDH) (Fig. 7B). We consistently detected large increases in the levels of NOX1 and NOX2 transcripts (4.5-fold on average) and a slight increase in XDH transcripts (1.5-fold on average) in the presence of EBNA1. An increase in NOX1 and NOX2 proteins was also seen in CNE2E cells compared to the levels in CNE2 cells (Fig. 7C), and these levels were decreased by treating CNE2E cells with siRNA against EBNA1 (Fig. 7D). The data indicate that EBNA1 increases ROS production and NOX1 and NOX2 levels in CNE2 cells, suggesting that increased ROS may be due to increased levels of oxidases.
We also examined whether EBNA1 affected NOX1, NOX2, or ROS in C666-1 cells by comparing the levels of these proteins in cells treated with siRNA against GFP or EBNA1. EBNA1 downregulation in these cells was found to decrease NOX2 but not NOX1 levels (Fig. 7E) and to result in a small but statistically significant decrease in ROS levels (Fig. 7F). We conclude that EBNA1 affects ROS and NOX2 in NPC cell lines and that effects on NOX1 vary in different cell lines.
EBNA1 increases stathmin 1 transcripts.
As part of the transcript quantification shown in Fig. 7B, we also examined the effect of EBNA1 expression on Prx1, SOD1, and stathmin 1 transcripts in order to determine if the EBNA1-induced increased levels of these proteins in nuclei were due to effects on the RNA transcripts or occurred at the protein level. We observed 3-fold-increased levels of stathmin 1 transcripts in the presence of EBNA1, suggesting that the increase in protein abundance described above is due to effects on stathmin 1 transcripts. In contrast, we did not see a significant change in the level of SOD1 or Prx1 transcripts in the presence of EBNA1, suggesting that EBNA1 was not stimulating transcription of these mRNAs but, rather, might be affecting the stability or nuclear localization of these proteins.
DISCUSSION
Considerable evidence indicates that EBNA1 makes important contributions to EBV-induced cell immortalization and malignant transformation. Some of these EBNA1 effects stem from the ability of EBNA1 to bind several cellular protein-modifying enzymes, thereby affecting the stability of some host proteins. To gain a more comprehensive understanding of the cellular proteins affected by EBNA1, we used the 2-D DiGE proteomic approach coupled to mass spectrometry. 2-D DiGE technology is a powerful and quantitative tool to address proteomic changes in complex samples. This and related 2-D gel techniques have been used previously to provide insight into cellular changes induced by viral infection or by specific viral proteins (1, 2, 7, 9, 14, 42, 53). Using this approach, we have identified 19 cellular proteins whose abundance is altered in the presence of EBNA1 in NPC cells. These alterations include the upregulation of several proteins involved in metastasis and redox regulation. In addition, cells expressing EBNA1 had decreased levels of a few proteins, most notably KSRP, an interferon-responsive regulator of microRNA biogenesis (32, 51), and tapasin, a protein important for MHC class I antigen presentation (18, 21). Recent studies suggest that regulation of viral and host miRNA may be an important component of NPC (11, 13, 57), and hence, the effects of EBNA1 on KSRP could have biologically important outcomes for miRNA expression. The downregulation of tapasin suggests that EBNA1 may damp down MHC class I-mediated antigen presentation in NPC. In keeping with this possibility, NPC tumor biopsy specimens have been reported to have decreased tapasin and HLA class I antigens (37).
The DiGE results showed that EBNA1 upregulated the nuclear levels of three different proteins that contribute in various ways to metastases: stathmin 1, maspin, and Nm23-H1. These findings are in keeping with a previous report that EBNA1 expression in HONE-1 nasopharyngeal carcinoma cells increased primary tumor formation, as well as metastases, in nude mice (45). The ability of EBNA1 to increase nuclear levels of Nm23-H1, a negative regulator of metastases, has been previously reported in B lymphocytes and involves relocalization of Nm23-H1 to the nucleus (36). This interaction was found to counteract Nm23-H1-mediated suppression of cell migration and, thereby, increase the metastatic potential of EBNA1-expressing cells. In addition, a subsequent study showed that EBNA1 expression in a breast carcinoma cell line reversed the growth inhibitory effect of Nm23-H1 and increased lung metastases (27). The fact that our 2-D DiGE screen identified the nuclear enrichment of Nm23-H1 provides confidence in our method.
In addition to effects on Nm23-H1, we found that EBNA1 also increased the nuclear levels of maspin and stathmin 1, both of which have been found to contribute to metastases. Therefore, the published reports of increased metastases in the presence of EBNA1 may be due to multiple EBNA1 effects. Maspin is a serine protease inhibitor (also called serpin B5) involved in cell adhesion, motility, apoptosis, and angiogenesis (29). It is usually considered to be a tumor suppressor (4) but has been reported to both inhibit and increase metastatic potential in different systems (43, 50). By Western blotting, we confirmed the nuclear enrichment of maspin in NPC cells stably and transiently expressing EBNA1, as well as decreased nuclear levels upon EBNA1 silencing.
We also confirmed by Western blotting that EBNA1 upregulates stathmin 1 in NPC cell nuclei. This increase is detectable after transient EBNA1 expression but is more pronounced in cells stably expressing EBNA1. Stathmin 1 is a regulator of microtubule dynamics that stimulates cell proliferation, migration, and metastatic potential and is upregulated in many cancers (6, 25, 39). Interestingly, the transcription of stathmin 1 is negatively regulated by p53 (16) and we have previously shown that EBNA1 destabilizes p53 (41). This suggests that EBNA1 may increase stathmin 1 transcription due to its ability to decrease p53 levels. In keeping with this hypothesis, EBNA1 was indeed found to increase the level of stathmin 1 transcripts, suggesting that the increased levels of stathmin 1 in the nucleus result from a transcriptional effect. Stathmin 1 has also been reported to be aberrantly regulated by EBV latent-membrane protein 1 (LMP1) in NPC, thereby promoting cell proliferation (33). Our findings suggest that EBNA1 may also contribute to deregulation of stathmin 1 in NPC.
A second group of proteins that we found to be upregulated by EBNA1 are involved in the oxidative stress response, including the antioxidants SOD1 and Prx1. The EBNA1-induced increase in both of these proteins was validated by Western blotting, although only SOD1 was increased in cell nuclei after transient EBNA1 expression, suggesting that changes in Prx1 occur more gradually. The levels of transcripts encoding these proteins were unaffected by EBNA1, suggesting that EBNA1 affects these proteins at the protein level. In addition, our findings that SOD1 levels were preferentially increased in the nuclear fraction suggest that EBNA1 may increase the nuclear localization of SOD1, similar to its effects on Nm23-H1. SOD1 facilitates the conversion of free superoxide radicals into oxygen and hydrogen peroxide and is distributed in the cytoplasm, nucleus, and mitochondrial intermembrane space (35). SOD1 in the nucleus protects genomic integrity, as depletion of nuclear SOD1 increases DNA damage and reduces viability (26). Therefore, our finding that EBNA1 increases SOD1 levels in the nucleus may suggest a way of counteracting genome instability. Interestingly, high levels of autoantibodies against SOD1 have been identified in NPC patients, which is consistent with EBV-mediated SOD1 induction (19). SOD1 induction by viruses appears to be a conserved phenomenon, as various 2-D gel analyses comparing cells before and after infection with respiratory syncytial virus, African swine fever virus, rice yellow mottle virus, and tobacco mosaic virus all reported significant induction of SOD, although the other oxidative stress response proteins induced by EBNA1 were not commonly reported (1, 7, 9, 53).
The observation of EBNA1-induced antioxidants prompted us to ask if EBNA1 also induced ROS, as increased ROS could promote the expression of antioxidants to counteract the ROS. Indeed, we observed increased ROS levels in NPC cells stably expressing EBNA1. This effect may be due to EBNA1-mediated transcriptional activation of NADPH oxidases, as we detected increases in NOX1 and NOX2 transcripts and proteins in EBNA1-expressing cells. We also found that EBNA1 depletion in C666-1 cells decreased NOX2 and ROS, suggesting a link between NOX2 and EBNA1-induced ROS levels. These observations are consistent with those of Gruhne et al. (20), who found that EBNA1 increased ROS and NOX2 levels in B cells. In addition, primary EBV infection has been shown to increase oxidative stress in B cells, epithelial cells, and lymphoblastoid cell lines (30), as well as in EBV-positive tumor cells (5, 10). In contrast to our results, Gruhne et al. (20) reported that EBNA1 did not affect ROS or NOX2 transcripts in epithelial cells. However, this conclusion was based on EBNA1 expression in epithelial cells by transient transfection, which would require an immediate effect of EBNA1. We were also unable to detect EBNA1 induction of ROS after transient EBNA1 expression (data not shown), indicating that this EBNA1 effect occurs gradually and may be indirect. In contrast, increased levels of nuclear SOD1 were detected after transient EBNA1 expression, suggesting that ROS induction is not a prerequisite for the EBNA1-mediated SOD1 nuclear increase and that these two effects may occur independently. Nonetheless, the interplay between these counterbalancing effects may be important for cell survival.
In conclusion, our proteomics study showed that EBNA1 affects the levels of a small percentage of the host proteins and that these proteins are enriched for those associated with oxidative stress responses and those that affect cell motility and proliferation. The latter effects suggest that EBNA1 may increase the metastatic potential of EBV-positive NPC cells, while the gradual accumulation of ROS suggests a mechanism by which EBNA1 may contribute to the genomic instability observed in NPC. The results further strength the growing contention that EBNA1 directly contributes to the development of NPC.
ACKNOWLEDGMENTS
We thank Kathy Shire and Feroz Sarkari for cloning and generation of the EBNA1- and LacZ-expressing adenoviruses and Andy Wilde for the DC-FDA dye for ROS labeling. We also gratefully acknowledge Rick Collins and Andrew Keeping for helpful advice on DiGE methods and MALDI-TOF mass spectrometry and William Navarre for computer access for DiGE analysis.
This work was supported by Canadian Institutes of Health Research operating grant number 12477 awarded to L.F. L.F. is a tier 1 Canada Research Chair in Molecular Virology and therefore receives salary support from the Canada Research Chairs program.
Footnotes
Published ahead of print 19 October 2011
REFERENCES
- 1. Alfonso P, Rivera J, Hernaez B, Alonso C, Escribano JM. 2004. Identification of cellular proteins modified in response to African swine fever virus infection by proteomics. Proteomics 4:2037–2046 [DOI] [PubMed] [Google Scholar]
- 2. Antrobus R, et al. 2009. Proteomic analysis of cells in the early stages of herpes simplex virus type-1 infection reveals widespread changes in the host cell proteome. Proteomics 9:3913–3927 [DOI] [PubMed] [Google Scholar]
- 3. Babcock GJ, Hochberg D, Thorley-Lawson AD. 2000. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13:497–506 [DOI] [PubMed] [Google Scholar]
- 4. Bailey CM, Khalkhali-Ellis Z, Seftor EA, Hendrix MJ. 2006. Biological functions of maspin. J. Cell. Physiol. 209:617–624 [DOI] [PubMed] [Google Scholar]
- 5. Belfiore MC, et al. 2007. Involvement of 5-lipoxygenase in survival of Epstein-Barr virus (EBV)-converted B lymphoma cells. Cancer Lett. 254:236–243 [DOI] [PubMed] [Google Scholar]
- 6. Belletti B, et al. 2008. Stathmin activity influences sarcoma cell shape, motility, and metastatic potential. Mol. Biol. Cell 19:2003–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brasier AR, et al. 2004. Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected a549 cells identified by high-resolution two-dimensional gel electrophoresis. J. Virol. 78:11461–11476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Canaan A, et al. 2009. EBNA1 regulates cellular gene expression by binding cellular promoters. Proc. Natl. Acad. Sci. U. S. A. 106:22421–22426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Casado-Vela J, Selles S, Martinez RB. 2006. Proteomic analysis of tobacco mosaic virus-infected tomato (Lycopersicon esculentum M.) fruits and detection of viral coat protein. Proteomics 6(Suppl 1):S196–S206 [DOI] [PubMed] [Google Scholar]
- 10. Cerimele F, et al. 2005. Reactive oxygen signaling and MAPK activation distinguish Epstein-Barr virus (EBV)-positive versus EBV-negative Burkitt's lymphoma. Proc. Natl. Acad. Sci. U. S. A. 102:175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen SJ, et al. 2010. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One 5:e12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheung ST, et al. 1999. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer 83:121–126 [DOI] [PubMed] [Google Scholar]
- 13. Cosmopoulos K, et al. 2009. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J. Virol. 83:2357–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dallaire F, Blanchette P, Branton PE. 2009. A proteomic approach to identify candidate substrates of human adenovirus E4orf6-E1B55K and other viral cullin-based E3 ubiquitin ligases. J. Virol. 83:12172–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eruslanov E, Kusmartsev S. 2010. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol. Biol. 594:57–72 [DOI] [PubMed] [Google Scholar]
- 16. Fang L, et al. 2010. Downregulation of stathmin expression is mediated directly by Egr1 and associated with p53 activity in lung cancer cell line A549. Cell Signal. 22:166–173 [DOI] [PubMed] [Google Scholar]
- 17. Frappier L. 2010. EBNA1 in viral replication and persistence. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 18. Garbi N, Tan P, Momburg F, Hammerling GJ. 2001. Role of tapasin in MHC class I antigen presentation in vivo. Adv. Exp. Med. Biol. 495:71–78 [DOI] [PubMed] [Google Scholar]
- 19. Gargouri B, et al. 2009. High levels of autoantibodies against catalase and superoxide dismutase in nasopharyngeal carcinoma. South. Med. J. 102:1222–1226 [DOI] [PubMed] [Google Scholar]
- 20. Gruhne B, et al. 2009. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc. Natl. Acad. Sci. U. S. A. 106:2313–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansen TH, Bouvier M. 2009. MHC class I antigen presentation: learning from viral evasion strategies. Nat. Rev. Immunol. 9:503–513 [DOI] [PubMed] [Google Scholar]
- 22. Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. 2003. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J. Biol. Chem. 278:47753–47761 [DOI] [PubMed] [Google Scholar]
- 23. Holowaty MN, et al. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987–29994 [DOI] [PubMed] [Google Scholar]
- 24. Hong M, et al. 2006. Suppression of Epstein-Barr nuclear antigen 1 (EBNA1) by RNA interference inhibits proliferation of EBV-positive Burkitt's lymphoma cells. J. Cancer Res. Clin. Oncol. 132:1–8 [DOI] [PubMed] [Google Scholar]
- 25. Iancu-Rubin C, Atweh GF. 2005. p27(Kip1) and stathmin share the stage for the first time. Trends Cell Biol. 15:346–348 [DOI] [PubMed] [Google Scholar]
- 26. Inoue E, et al. 2010. SOD1 is essential for the viability of DT40 cells and nuclear SOD1 functions as a guardian of genomic DNA. J. Nucleic Acids 2010:795946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaul R, Murakami M, Choudhuri T, Robertson ES. 2007. Epstein-Barr virus latent nuclear antigens can induce metastasis in a nude mouse model. J. Virol. 81:10352–10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kennedy G, Komano J, Sugden B. 2003. Epstein-Barr virus provides a survival factor to Burkitt's lymphomas. Proc. Natl. Acad. Sci. U. S. A. 100:14269–14274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khalkhali-Ellis Z. 2006. Maspin: the new frontier. Clin. Cancer Res. 12:7279–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lassoued S, et al. 2008. Epstein-Barr virus induces an oxidative stress during the early stages of infection in B lymphocytes, epithelial, and lymphoblastoid cell lines. Mol. Cell. Biochem. 313:179–186 [DOI] [PubMed] [Google Scholar]
- 31. Li M, et al. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648–653 [DOI] [PubMed] [Google Scholar]
- 32. Lin W-J, et al. 2011. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol. Cell. Biol. 31:3196–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin X, et al. 2009. EBV-encoded LMP1 regulates Op18/stathmin signaling pathway by cdc2 mediation in nasopharyngeal carcinoma cells. Int. J. Cancer 124:1020–1027 [DOI] [PubMed] [Google Scholar]
- 34. Lu J, et al. 2011. Epstein-Barr virus nuclear antigen 1 (EBNA1) confers resistance to apoptosis in EBV-positive B-lymphoma cells through up-regulation of survivin. Virology 410:64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miao L, St Clair DK. 2009. Regulation of superoxide dismutase genes: implications in disease. Free Radic. Biol. Med. 47:344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murakami M, Lan K, Subramanian C, Robertson ES. 2005. Epstein-Barr virus nuclear antigen 1 interacts with Nm23-H1 in lymphoblastoid cell lines and inhibits its ability to suppress cell migration. J. Virol. 79:1559–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ogino T, et al. 2007. Association of immunoescape mechanisms with Epstein-Barr virus infection in nasopharyngeal carcinoma. Int. J. Cancer 120:2401–2410 [DOI] [PubMed] [Google Scholar]
- 38. Raab-Traub N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 12:431–441 [DOI] [PubMed] [Google Scholar]
- 39. Rana S, Maples PB, Senzer N, Nemunaitis J. 2008. Stathmin 1: a novel therapeutic target for anticancer activity. Expert Rev. Anticancer Ther. 8:1461–1470 [DOI] [PubMed] [Google Scholar]
- 40. Rickinson AB, Kieff KE. 2001. Epstein-Barr virus, p. 2575–2627 In Knipe DM, et al. (ed), Fields virology, 2nd ed,vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 41. Saridakis V, et al. 2005. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol. Cell 18:25–36 [DOI] [PubMed] [Google Scholar]
- 42. Schlee M, et al. 2004. Identification of Epstein-Barr virus (EBV) nuclear antigen 2 (EBNA2) target proteins by proteome analysis: activation of EBNA2 in conditionally immortalized B cells reflects early events after infection of primary B cells by EBV. J. Virol. 78:3941–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sheng S, et al. 1996. Maspin acts at the cell membrane to inhibit invasion and motility of mammary and prostatic cancer cells. Proc. Natl. Acad. Sci. U. S. A. 93:11669–11674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sheng Y, et al. 2006. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat. Struct. Mol. Biol. 13:285–291 [DOI] [PubMed] [Google Scholar]
- 45. Sheu LF, et al. 1996. Enhanced malignant progression of nasopharyngeal carcinoma cells mediated by the expression of Epstein-Barr nuclear antigen 1 in vivo. J. Pathol. 180:243–248 [DOI] [PubMed] [Google Scholar]
- 46. Sivachandran N, Cao JY, Frappier L. 2010. Epstein-Barr virus nuclear antigen 1 hijacks the host kinase CK2 to disrupt PML nuclear bodies. J. Virol. 84:11113–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sivachandran N, Sarkari F, Frappier L. 2008. Epstein-Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathog. 4:e1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sompallae R, Callegari S, Kamranvar SA, Masucci MG. 2010. Transcription profiling of Epstein-Barr virus nuclear antigen (EBNA)-1 expressing cells suggests targeting of chromatin remodeling complexes. PLoS One 5:e12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun Y, et al. 1992. An infrequent point mutation of the p53 gene in human nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. U. S. A. 89:6516–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Terashima M, et al. 2005. Gene expression profiles in human gastric cancer: expression of maspin correlates with lymph node metastasis. Br. J. Cancer 92:1130–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trabucchi M, et al. 2009. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459:1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Valko M, et al. 2007. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 39:44–84 [DOI] [PubMed] [Google Scholar]
- 53. Ventelon-Debout M, et al. 2004. Proteome analysis of cultivar-specific deregulations of Oryza sativa indica and O. sativa japonica cellular suspensions undergoing rice yellow mottle virus infection. Proteomics 4:216–225 [DOI] [PubMed] [Google Scholar]
- 54. Wood VH, et al. 2007. Epstein-Barr virus-encoded EBNA1 regulates cellular gene transcription and modulates the STAT1 and TGFbeta signaling pathways. Oncogene 26:4135–4147 [DOI] [PubMed] [Google Scholar]
- 55. Yin Q, Flemington EK. 2006. siRNAs against the Epstein Barr virus latency replication factor, EBNA1, inhibit its function and growth of EBV-dependent tumor cells. Virology 346:385–393 [DOI] [PubMed] [Google Scholar]
- 56. Zeghouf M, et al. 2004. Sequential peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3:463–468 [DOI] [PubMed] [Google Scholar]
- 57. Zhu JY, et al. 2009. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 83:3333–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]