Abstract
The Epstein-Barr virus (EBV)-encoded immune evasion protein BNLF2a inhibits the transporter associated with antigen processing (TAP), thereby downregulating HLA class I expression at the cell surface. As a consequence, recognition of EBV-infected cells by cytotoxic T cells is impaired. Here, we show that sequence polymorphism of the BNLF2a protein is observed with natural EBV isolates, with evidence for positive selection. Despite these mutations, the BNLF2a variants efficiently reduce cell surface HLA class I levels. This conservation of BNLF2a function during evolution of EBV implies an important role for the viral TAP inhibitor in preventing T cell recognition during viral infection.
TEXT
Epstein-Barr virus (EBV) is a gamma-1 herpesvirus carried by over 90% of the adult human population worldwide. Whereas primary EBV infection of young children usually occurs unnoticed, primary infection of adolescents results in the development of infectious mononucleosis in at least 25% of the cases (25). In either scenario, primary infection is controlled by EBV-specific immunity. Despite this immunity, the virus persists for life in the infected host. Specific viral immune evasion strategies, eluding both innate and adaptive immunity, are thought to contribute to this persistence (24, 26).
Cytotoxic T lymphocytes (CTLs) play an important role in controlling EBV infection. Virus-infected cells are recognized by CTLs through the detection of virus-derived peptides presented at the cell surface in the context of HLA class I (HLA I) molecules. These peptides, generated by proteasomal degradation of viral proteins in the cytosol, are transported into the endoplasmic reticulum (ER) via the transporter associated with antigen processing (TAP) and subsequently are loaded onto newly synthesized HLA I molecules. To evade CTL recognition, the EBV lytic-phase protein BNLF2a inhibits TAP by interfering with the binding of both ATP and peptide to the TAP transporter (14, 16). As a result of diminished TAP-mediated peptide transport into the ER, peptide loading onto HLA I molecules is impaired and surface display of HLA I/peptide complexes is reduced (14, 16).
In humans, two EBV types (1 and 2) are distinguished based on sequence variation in the EBV latent proteins EBNA2, -3A, -3B, and -3C (10, 27). Additionally, considerable polymorphism is found among different strains belonging to the same type (7). Such mutations have been found to profoundly affect the T cell immunogenicity of EBV-encoded proteins. For instance, mutations in the HLA-A11-restricted immunodominant EBNA3B epitope allow escape from T cell recognition (11, 12). These mutant epitopes are more abundant in regions where HLA-A11 is highly prevalent than in regions where HLA-A11 is found at low frequencies, suggesting that these variants are selected by immunological pressure (11, 12). Similarly, polymorphisms in CTL epitopes of EBNA1 (3) and EBNA3A (1) affect T cell recognition. Sequence polymorphism has not been detected within the BNLF2a gene of the three EBV strains that have been fully sequenced, which are the type 1 strains B95.8 (infectious mononucleosis) and GD1 (Chinese nasopharyngeal carcinoma), and the type 2 strain AG876 (African Burkitt's lymphoma) (2, 13, 34).
In this study, we investigated the extent to which variation occurs for the immune evasion protein BNLF2a. The functional consequences of this variation were evaluated by measuring HLA I downregulation resulting from BNLF2a-mediated TAP inhibition.
Sequence variation among BNLF2a proteins of EBV isolates.
To investigate the frequency of genotypic variation within the EBV-encoded TAP inhibitor, the BNLF2a gene from EBV isolates from Australian Caucasians (3), Africans (4), and inhabitants of the Papua New Guinea highlands and lowlands (5) was sequenced. These viruses were isolated from lymphoblastoid cell lines raised “spontaneously” (without exogenous EBV addition). Furthermore, nasopharyngeal carcinoma (NPC) biopsy specimens from Hong Kong (4) were sequenced for the BNLF2a gene. These studies have been reviewed and approved by an appropriate institutional review committee, and the research has complied with all relevant federal guidelines and institutional policies.
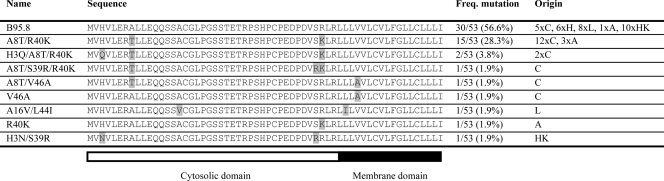
At the DNA level, multiple mutations were found; of these, two nucleotide substitutions found in the EBV isolates were silent, whereas all other mutations resulted in amino acid changes (see Fig. S1 in the supplemental material). The nucleotide polymorphisms observed for these 53 isolates resulted in nine different BNLF2a protein sequences (Table 1). In more than half of the EBV isolates, the BNLF2a amino acid sequence corresponded to that of the B95.8 reference strain (wild type [wt]). More than a quarter of the isolates encoded the same two amino acid substitutions, namely, an alanine-to-threonine replacement at position 8 combined with an arginine-to-lysine mutation at position 40. Other mutations were found only once or twice (Table 1).
Table 1.
BNLF2a protein sequences of EBV type 1 isolatesa
Freq. mutation, frequency of mutation; C, Caucasian (Australia) (spontaneous LCLs); H, Papua New Guinea highlands (spontaneous LCLs); L, Papua New Guinea lowlands (spontaneous LCLs); A, Africa (spontaneous LCLs); HK, Hong Kong (NPC biopsies).
Positive selection of the EBV BNLF2a protein during viral evolution.
The BNLF2a alignment was analyzed to identify individual residue positions likely to be under positive selection and also overall ratios of the rates of nonsynonymous to synonymous substitution (omega) using the codeml module of PAML 4.2b software (33). Of the 10 mutated positions in the DNA alignment, only two are synonymous. Positions 3, 8, and 39 in the alignment are identified as being under positive selection at a significance level (P value) of <0.05 (Table 2). At the other variable sites, statistical significance is not achieved.
Table 2.
Positive selection analysisa
| Residue | Pr(omega > 1) | Mean ± SE for omega |
|---|---|---|
| H3 | 0.980* | 7.314 ± 2.423 |
| A8 | 0.983* | 7.337 ± 2.399 |
| A16 | 0.804 | 6.003 ± 3.329 |
| S39 | 0.979* | 7.302 ± 2.436 |
| R40 | 0.869 | 6.514 ± 3.096 |
| L44 | 0.832 | 6.228 ± 3.239 |
| V46 | 0.839 | 6.282 ± 3.214 |
Pr(omega > 1), the probability that the ratio of nonsynonymous to synonymous substitutions (omega) is greater than 1 at that site, indicative of positive selection. The mean and standard error for omega are given for each site. Amino acids refer to the B95.8 EBV sequence. Positively selected sites (*, P > 95%); SE, standard error of the mean.
Conservation of BNLF2a function in different EBV isolates.
To examine whether the immune-evasive properties of BNLF2a are affected by the mutations as found in the EBV isolates, we transiently transfected Mel JuSo (MJS) cells (human melanoma cell line) (31) to express (mutant) BNLF2a using an Amaxa Nucleofector II. Two days after transfection, cells were analyzed by flow cytometry. The different BNLF2a variants of the EBV isolates from Australian Caucasians were cloned into the pLV-CMV-IRES-eGFP vector (32). This vector contains an internal ribosomal entry site (IRES) immediately downstream of BNLF2a, resulting in coexpression of enhanced green fluorescent protein (eGFP) together with the viral protein. Indeed, intracellular staining using a BNLF2a-specific antibody confirmed that expression levels of the BNLF2a protein correlated with GFP intensities (Fig. 1A). Therefore, in the experiments that follow, GFP levels are used as an indicator of the amounts of BNLF2a expressed.
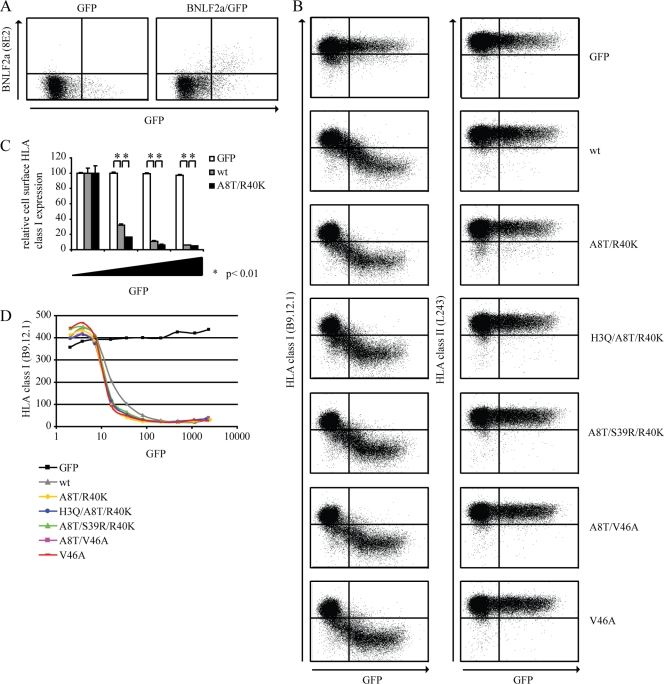
Fig 1.
EBV isolates retain BNLF2a-mediated HLA I downregulation. (A) MJS cells were transiently transfected to express the control protein GFP or to coexpress wild-type BNLF2a and GFP. After 48 h, cells were stained for intracellular expression of BNLF2a (monoclonal antibody [MAb] 8E2). Subsequently, the cells were analyzed by flow cytometry using CellQuest Pro software (BD Biosciences). (B) MJS cells were transiently transfected to express the control protein GFP, wild-type BNLF2a (wt), or one of the following BNLF2a mutants: A8T/R40K, H3Q/A8T/R40K, A8T/S39R/R40K, A8T/V46A, or V46A. After 48 h, cells were stained for cell surface expression of HLA I (MAb B9.12.1) and HLA II (MAb L243) and analyzed by flow cytometry using CellQuest Pro software (BD Biosciences). (C) Quantification of flow cytometry data. Cell surface expression levels of HLA I were correlated with GFP expression for cells transfected to express the control protein GFP, wild-type (wt) BNLF2a, or the A8T/R40K BNLF2a mutant. To this end, values were corrected for cell surface expression of HLA I in GFP-negative cells. The standard deviations are represented by the error bars. *, P < 0.01 as determined by a t test. (D) Graphical display of the results shown in panel B. The mean fluorescence index of HLA I expression is plotted against the mean fluorescence index of GFP expression. The results of one representative experiment out of at least three independent experiments are shown. For panel A, the experiment was performed in duplicate.
Surface expression of HLA I molecules was downregulated upon transfection of wild-type BNLF2a in a dose-dependent fashion, i.e., correlating with GFP expression (Fig. 1B, second panel on the left). This effect was specific, since surface HLA I display was not affected upon transfection of the control protein GFP (empty vector) (Fig. 1B, top left panel) and cellular HLA II levels remained unaltered in the presence of BNLF2a (Fig. 1B, second panel on the right). Expression of the BNLF2a mutants did not cause major alterations in HLA I surface expression compared to results for wild-type BNLF2a, indicating that they retained their ability to reduce the cell surface display of HLA I molecules (Fig. 1B, compare left panels).
To allow statistical analysis, the dot plot data were divided into small regions with comparable GFP expression levels. For each of these regions, the mean fluorescence intensity of HLA class I staining is depicted in Fig. 1C and D. This analysis revealed subtle differences between wild-type BNLF2a and the mutants. Cell surface HLA I expression is lower in BNLF2a-A8T/R40K-expressing cells than in wild-type BNLF2a-expressing cells; this difference is small but statistically significant (Fig. 1C). Also, the other mutant proteins were more efficient in downregulating HLA I expression than wild-type BNLF2a (Fig. 1D). No differences in the efficiency of reducing HLA I cell surface expression were observed among the various mutants (Fig. 1D).
N-terminal amino acid residues influence immune-evasive properties of BNLF2a.
Since the N-terminal domain of BNLF2a is required for its immune evasion properties (15), we focused on the N-terminal mutations that were found in the EBV isolates, especially the A8T mutation which is present at a high frequency in the EBV isolates (Table 1). However, the A8T mutation occurs in conjugation with other substitutions only, so its contribution to BNLF2a function cannot be inferred from these naturally occurring variants. Therefore, an A8T-only recombinant has been constructed and tested for TAP inhibition. Furthermore, we determined the effect of a double H3Q/A8T mutation in BNLF2a.
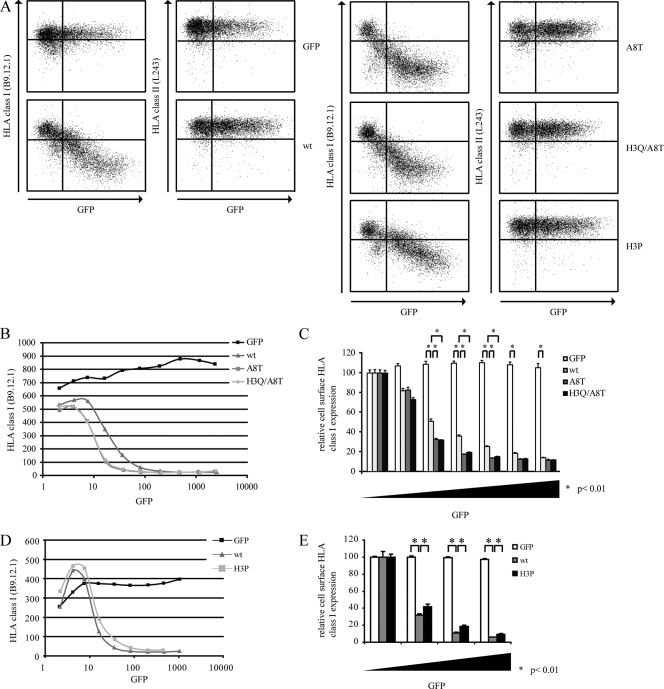
The BNLF2a-A8T and BNLF2a-H3Q/A8T mutants were slightly more efficient than wild-type BNLF2a in downregulating cell surface HLA I expression (Fig. 2A to C). No apparent differences were found between the two mutants (Fig. 2B and C), indicating that the alanine-to-threonine substitution at position 8 alone is sufficient to improve the HLA I-downregulating properties of BNLF2a, whereas the histidine-to-glutamine substitution at position 3 has no additional effect. Furthermore, our data confirm the important role of the N-terminal domain of BNLF2a in inhibiting TAP-mediated peptide transport.
Fig 2.
N-terminal amino acids of BNLF2a affect its immune evasion function. (A) MJS cells were transiently transfected to express the control protein GFP, wild-type BNLF2a (wt), BNLF2a-A8T (A8T), BNLF2a-H3Q/A8T (H3Q/A8T), or BNLF2a-H3P (H3P). After 48 h, cells were stained for cell surface expression of HLA I (MAb B9.12.1) and HLA II (MAb L243) and analyzed by flow cytometry using CellQuest Pro software (BD Biosciences). (B and D) Graphical display of flow cytometry data. The mean fluorescence index of HLA I expression is plotted against the mean fluorescence index of GFP expression. (C and E) Quantification of flow cytometry data. Cell surface expression levels of HLA I were correlated with GFP expression. To this end, values were corrected for cell surface expression of HLA I in GFP-negative cells. The standard deviations are represented by the error bars. *, P < 0.01 as determined by a t test. The results of one representative experiment out of three independent experiments are shown.
H3P mutation negatively affects BNLF2a-mediated HLA I downregulation.
Previously, we have demonstrated that BNLF2a prevents peptide binding to TAP (14). This might be achieved by binding of the cytosolic N terminus of BNLF2a, which is required for TAP inhibition (15), to the peptide-binding site of TAP, thereby directly competing with binding of peptides. For 7- to 11-mer peptide epitopes, a proline at position 1, 2, or 3 hampers interaction with TAP (20, 21, 28, 30). We hypothesized that the N terminus of BNLF2a associates with the peptide-binding domain of TAP; therefore, a histidine-to-proline substitution at position 3 of the BNLF2a protein might interfere with its ability to inhibit TAP and, consequently, to downregulate cell surface HLA I display.
Compared to wild-type BNLF2a, BNLF2a-H3P is impaired in its ability to reduce HLA I cell surface levels (Fig. 2A, D, and E). This observation indicates that the amino acid residue at position 3 contributes to BNLF2a function, possibly through perturbing the interaction between the N terminus of BNLF2a and the peptide-binding domain of TAP. However, other mechanisms by which the H3 residue influences BNLF2a function cannot be excluded.
In conclusion, the data presented in this study indicate that, whereas a single mutation within BNLF2a can impair its ability to reduce cell surface HLA I expression, the BNLF2a variants as identified in the natural isolates from Australian Caucasians retained their HLA I-downregulating properties despite the presence of multiple mutations in most cases. Furthermore, there is evidence for positive selection of the H3, A8, and S39 residues. One might expect the less-efficient BNLF2a variants to succumb during viral evolution. However, considering the polymorphism that is observed for TAP (6, 8, 22, 23, 29), it is not surprising to find variants of the TAP inhibitor BNLF2a as well. Whether these BNLF2a mutants have different efficiencies for inhibiting the various TAP alleles remains to be determined. Overall, these results suggest that BNLF2a has an important role in preventing antigen presentation to CTLs. In accordance, B cell lines transformed with a BNLF2a-deletion mutant of EBV display increased susceptibility toward CTLs targeting (immediate) early EBV antigens, compared to results for wild-type EBV-transformed cells (9).
Genetic variation, as is seen for BNLF2a, is also observed for other EBV lytic proteins that influence host-pathogen interactions. Sequencing of EBV isolates revealed mutations within the viral IL-10 protein (18), a cytokine that has immunomodulating properties (17). Similarly, amino acid sequence variation was observed for the antiapoptotic BHRF1 protein. These BHRF1 variants still confer protection against apoptosis, indicating conservation of the function of this viral protein (19).
Taken together, our present results demonstrate that the function of the TAP inhibitor BNLF2a is conserved during evolution of EBV, pointing toward an important contribution of immune evasion during the viral life cycle. By interfering with T cell recognition during productive EBV infection, BNLF2a is anticipated to create a window for the generation of viral progeny in the face of memory T cell immunity.
Nucleotide sequence accession numbers.
GenBank accession numbers for the BNLF2a sequences are JN703669 to JN703677, JN711406 to JN711425, and JN803886 to JN803909.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. J. Davison (University of Glasgow Centre for Virus Research, Glasgow, United Kingdom), J. M. Nicholls (University of Hong Kong, Hong Kong SAR, China), and R. C. Hoeben and A. Mulder (Leiden University Medical Center, Leiden, The Netherlands) for helpful advice and generously sharing reagents and constructs.
This work was supported by the Dutch Cancer Foundation (grant RUL 2005-3259 to D.H., M.E.R., and E.J.H.J.W.), The Netherlands Scientific Organization (NWO Vidi 917.76.330 to M.E.R.), the National Health and Medical Research Council of Australia (to S.R.B.) and the UK Medical Research Council (to D.G.).
The authors declare that they have no conflicts of interest.
Footnotes
Published ahead of print 19 October 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Apolloni A, et al. 1992. Sequence variation of cytotoxic T cell epitopes in different isolates of Epstein-Barr virus. Eur. J. Immunol. 22:183–189 [DOI] [PubMed] [Google Scholar]
- 2. Baer R, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211 [DOI] [PubMed] [Google Scholar]
- 3. Bell MJ, et al. 2008. Widespread sequence variation in Epstein-Barr virus nuclear antigen 1 influences the antiviral T cell response. J. Infect. Dis. 197:1594–1597 [DOI] [PubMed] [Google Scholar]
- 4. Burrows JM, et al. 2004. Selection pressure-driven evolution of the Epstein-Barr virus-encoded oncogene LMP1 in virus isolates from Southeast Asia. J. Virol. 78:7131–7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burrows JM, et al. 1996. Unusually high frequency of Epstein-Barr virus genetic variants in Papua New Guinea that can escape cytotoxic T-cell recognition: implications for virus evolution. J. Virol. 70:2490–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carrington M, Colonna M, Spies T, Stephens JC, Mann DL. 1993. Haplotypic variation of the transporter associated with antigen processing (TAP) genes and their extension of HLA class II region haplotypes. Immunogenetics 37:266–273 [DOI] [PubMed] [Google Scholar]
- 7. Chang CM, Yu KJ, Mbulaiteye SM, Hildesheim A, Bhatia K. 2009. The extent of genetic diversity of Epstein-Barr virus and its geographic and disease patterns: a need for reappraisal. Virus Res. 143:209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Colonna M, Bresnahan M, Bahram S, Strominger JL, Spies T. 1992. Allelic variants of the human putative peptide transporter involved in antigen processing. Proc. Natl. Acad. Sci. U. S. A. 89:3932–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Croft NP, et al. 2009. Stage-specific inhibition of MHC class I presentation by the Epstein-Barr virus BNLF2a protein during virus lytic cycle. PLoS Pathog. 5:e1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dambaugh T, Hennessy K, Chamnankit L, Kieff E. 1984. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc. Natl. Acad. Sci. U. S. A. 81:7632–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Campos-Lima PO, et al. 1993. HLA-A11 epitope loss isolates of Epstein-Barr virus from a highly A11+ population. Science 260:98–100 [DOI] [PubMed] [Google Scholar]
- 12. de Campos-Lima PO, et al. 1994. T cell responses and virus evolution: loss of HLA A11-restricted CTL epitopes in Epstein-Barr virus isolates from highly A11-positive populations by selective mutation of anchor residues. J. Exp. Med. 179:1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dolan A, Addison C, Gatherer D, Davison AJ, McGeoch DJ. 2006. The genome of Epstein-Barr virus type 2 strain AG876. Virology 350:164–170 [DOI] [PubMed] [Google Scholar]
- 14. Hislop AD, et al. 2007. A CD8+ T cell immune evasion protein specific to Epstein-Barr virus and its close relatives in Old World primates. J. Exp. Med. 204:1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horst D, et al. 2011. EBV protein BNLF2a exploits host tail-anchored protein integration machinery to inhibit TAP. J. Immunol. 186:3594–3605 [DOI] [PubMed] [Google Scholar]
- 16. Horst D, et al. 2009. Specific targeting of the EBV lytic phase protein BNLF2a to the transporter associated with antigen processing results in impairment of HLA class I-restricted antigen presentation. J. Immunol. 182:2313–2324 [DOI] [PubMed] [Google Scholar]
- 17. Hsu DH, et al. 1990. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science 250:830–832 [DOI] [PubMed] [Google Scholar]
- 18. Kanai K, et al. 2007. The vIL-10 gene of the Epstein-Barr virus (EBV) is conserved in a stable manner except for a few point mutations in various EBV isolates. Virus Genes. 35:563–569 [DOI] [PubMed] [Google Scholar]
- 19. Khanim F, et al. 1997. BHRF1, a viral homologue of the Bcl-2 oncogene, is conserved at both the sequence and functional level in different Epstein-Barr virus isolates. J. Gen. Virol. 78:2987–2999 [DOI] [PubMed] [Google Scholar]
- 20. Momburg F, et al. 1994. Selectivity of MHC-encoded peptide transporters from human, mouse and rat. Nature 367:648–651 [DOI] [PubMed] [Google Scholar]
- 21. Neefjes J, et al. 1995. Analysis of the fine specificity of rat, mouse and human TAP peptide transporters. Eur. J. Immunol. 25:1133–1136 [DOI] [PubMed] [Google Scholar]
- 22. Powis SH, et al. 1992. Polymorphism in a second ABC transporter gene located within the class II region of the human major histocompatibility complex. Proc. Natl. Acad. Sci. U. S. A. 89:1463–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Powis SH, et al. 1993. Alleles and haplotypes of the MHC-encoded ABC transporters TAP1 and TAP2. Immunogenetics 37:373–380 [DOI] [PubMed] [Google Scholar]
- 24. Ressing ME, et al. 2008. Epstein-Barr virus evasion of CD8+ and CD4+ T cell immunity via concerted actions of multiple gene products. Semin. Cancer Biol. 18:397–408 [DOI] [PubMed] [Google Scholar]
- 25. Rickinson AB, Kieff E. 2007. Epstein-Barr Virus, p 2655–2700 In Knipe D. M., et al. (ed), Fields virology, vol. 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 26. Rowe M, Zuo J. 2010. Immune responses to Epstein-Barr virus: molecular interactions in the virus evasion of CD8+ T cell immunity. Microbes Infect. 12:173–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sample J, et al. 1990. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 64:4084–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uebel S, et al. 1997. Recognition principle of the TAP transporter disclosed by combinatorial peptide libraries. Proc. Natl. Acad. Sci. U. S. A. 94:8976–8981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Endert PM, Lopez MT, Patel SD, Monaco JJ, McDevitt HO. 1992. Genomic polymorphism, recombination, and linkage disequilibrium in human major histocompatibility complex-encoded antigen-processing genes. Proc. Natl. Acad. Sci. U. S. A. 89:11594–11597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Endert PM, et al. 1995. The peptide-binding motif for the human transporter associated with antigen processing. J. Exp. Med. 182:1883–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Ham SM, et al. 1997. HLA-DO is a negative modulator of HLA-DM-mediated MHC class II peptide loading. Curr. Biol. 7:950–957 [DOI] [PubMed] [Google Scholar]
- 32. Vellinga J, Uil TG, de Vrij J, Rabelink MJ, Lindholm L, Hoeben RC. 2006. A system for efficient generation of adenovirus protein IX-producing helper cell lines. J. Gene Med. 8:147–154 [DOI] [PubMed] [Google Scholar]
- 33. Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591 [DOI] [PubMed] [Google Scholar]
- 34. Zeng MS, et al. 2005. Genomic sequence analysis of Epstein-Barr virus strain GD1 from a nasopharyngeal carcinoma patient. J. Virol. 79:15323–15330 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.