Abstract
Here we report a novel viral glycoprotein created by replacing a natural receptor-binding sequence of the ecotropic Moloney murine leukemia virus envelope glycoprotein with the peptide ligand somatostatin. This new chimeric glycoprotein, which has been named the Sst receptor binding site (Sst-RBS), gives targeted transduction based on three criteria: (i) a gain of the use of a new entry receptor not used by any known virus; (ii) targeted entry at levels comparable to gene delivery by wild-type ecotropic Moloney murine leukemia virus and vesicular stomatitis virus (VSV) G glycoproteins; and (iii) a loss of the use of the natural ecotropic virus receptor. Retroviral vectors coated with Sst-RBS gained the ability to bind and transduce human 293 cells expressing somatostatin receptors. Their infection was specific to target somatostatin receptors, since a synthetic somatostatin peptide inhibited infection in a dose-dependent manner and the ability to transduce mouse cells bearing the natural ecotropic receptor was effectively lost. Importantly, vectors coated with the Sst-RBS glycoprotein gave targeted entry of up to 1 × 106 transducing U/ml, a level comparable to that seen with infection of vectors coated with the parental wild-type ecotropic Moloney murine leukemia virus glycoprotein through the ecotropic receptor and approaching that of infection of VSV G-coated vectors through the VSV receptor. To our knowledge, this is the first example of a glycoprotein that gives targeted entry of retroviral vectors at levels comparable to the natural capacity of viral envelope glycoproteins.
INTRODUCTION
The lack of methods for targeting entry to achieve gene delivery in specific cell types and tissues in vivo remains a major technical barrier to in vivo human gene therapy (13, 53). Retroviral and lentiviral vectors for gene delivery have most commonly used vesicular stomatitis virus (VSV) G protein or envelope Env glycoprotein (Env) from gammaretroviruses, including amphotropic murine leukemia virus (MLV) and gibbon ape leukemia virus. Since these glycoproteins use widely expressed entry receptors, they mediate transduction of virtually all cells encountered after in vivo injection of vectors. The broad tropism seen when using these viral glycoproteins presents challenges to in vivo gene delivery, including the need to administer many times more vectors than would be needed if entry could be limited to the cells that would benefit from transduction of therapeutic genes. In addition, several important target cell types, including those of the lung epithelium, do not transduce well, because they display natural virus receptors on the poorly accessible basolateral side.
Strategies for entry targeting of retroviral and lentiviral vectors have primarily been based on creating chimeric envelope glycoproteins by inserting peptide ligand sequences within or at the beginning or end of viral envelope glycoproteins. Early efforts used insertion of ligands into the glycoprotein surface subunit (SU), the subunit of the retroviral envelope protein (Env) that binds the natural virus receptor. These chimeras reproducibly exhibited specific vector attachment but very poor or no transduction in the cells bearing the target receptor and lacking the natural virus receptor (2, 13). More recent efforts used nonretroviral glycoproteins. A targeting system for replicating measles virus was developed by inserting ligand sequences into the C terminus of measles virus hemagglutinin (HA) H, a type II membrane protein (8, 50). Once truncation of their cytoplasmic tails was shown to enable the use of measles virus H and fusion F proteins to pseudotype lentiviral vectors (19), the retargeting system was applied to lentiviral vectors, but the tendency of the chimeric scFv-H protein dimers to aggregate resulted in inconsistent expression and assembly into lentiviral particles (41). When titers were determined for cells that naturally express the target receptor and the CD46 measles virus receptor, seven different targeted measles virus H chimeras showed a capacity for entry that was 10- to 100-fold lower than that of their parent H and F proteins (41). The entry capacity of these chimeras versus that of VSV G has not yet been established using cells that express both the target and VSV G receptors (3, 19, 20, 22, 41). However, given that they have a 10- to 100-fold lower entry capacity than the parent measles virus glycoproteins, it is unlikely that their actual capacity for entry is comparable to that of VSV G. In addition, immunity due to vaccination or natural infection is likely to interfere with the in vivo use of modified measles virus glycoproteins for entry-targeted gene delivery in humans (32). An antibody conjugation system based on insertion of the Fc-binding ZZ domain from protein A into the E1 glycoprotein from Sindbis virus also targeted lentiviral vector entry (40), but the instability of the vector-antibody conjugates and their rapid inactivation by complement severely limited their usefulness in vivo (39), where entry targeting is most critical.
With respect to chimeric retroviral SU efforts, the most frequently used insertion sites include the amino terminus of the Moloney MLV (MoMLV) glycoprotein and the proline-rich region (PRR) (2). Vectors coated with ligand-inserted Env chimeras were internalized but failed to escape endosomal vesicles (61), evidently because they could not mediate membrane fusion (6, 47). Amino-terminal insertion failed to initiate fusion, whereas insertion into the PRR initiated but did not complete fusion (47). This evidence suggested that ligand insertion strategies fail largely because binding to the target receptors does not activate the correct conformational changes required to complete virus-cell membrane fusion (47).
Since these conformational changes are normally activated by interaction of a receptor binding site on Env with its natural receptor, we proposed that a reasonable new strategy would be to replace receptor binding residues with a peptide ligand sequence (47). In this type of Env chimera, binding of the ligand to its cognate, or target, receptor should mimic the interactions with the natural virus receptor and so induce the required set of conformational changes to complete membrane fusion and result in targeted transduction. Here, we report the construction and characterization of the first chimeric Env based on this novel design concept. The results establish a proof of principle that this type of chimeric glycoprotein can give specific, targeted infection of retroviral vectors comparable to the efficiency of infection using natural retroviral glycoproteins and approaching that of VSV G glycoprotein infection. We also report Sst receptor binding site (Sst-RBS)-pseudotyped self-inactivating (SIN) lentiviral vector transduction of cells.
MATERIALS AND METHODS
Construction of plasmids for chimeric envelope protein expression.
A cDNA encoding the chimeric Sst-RBS envelope glycoprotein and a codon-optimized version of the cDNA (designated Sst-RBSopti) were produced using oligonucleotide-directed mutagenesis and an ExSite mutagenesis kit (Stratagene). The nucleotide sequences of the original cDNA and the optimized cDNA encoding the receptor binding domains (RBD) were deposited in the GenBank database; sequences outside the RBD are identical to those of the previously published wild-type Moloney murine leukemia virus Env (J02255). Control chimeras Sst-NH2 and Sst-PRR were made by ligating an Sst-encoding fragment into the NotI restriction site in two previously described plasmids, pcDNA-MolMLV-Nflex and pcDNA-MoMLV-PRR, respectively (44). The Sst-230 chimeric sequence was constructed by ligating an Sst-encoding fragment into the EcoO109I site at codon 230 of the MoMLV Env cDNA. The sequence of each chimeric gene was verified by DNA sequence analysis.
Somatostatin receptor cDNAs and stable cell lines.
The mammalian expression vector pcDNA-hSSTR2a, encoding human Sst receptor (SSTR) subtype 2a, was constructed by subcloning the SSTR2a-encoding sequences from a pBluescript plasmid (gift of Graeme Bell) (55) into pcDNA3 (Invitrogen) under the control of the cytomegalovirus (CMV) promoter. pcDNA3 also contains a linked G418 resistance gene cassette. The expression plasmid 12CA5+/SSTR5, encoding amino-terminal HA epitope-tagged human SSTR subtype 5 (SSTR5-HA) linked to a G418 resistance gene cassette, was purchased from Affymax, Inc. (Palo Alto, CA). Cell lines stably expressing SSTR2a or SSTR5-HA were generated by selection of transfected 293 cells for growth in medium containing G418 (1 mg/ml) for 4 weeks posttransfection. After a 4-week selection in the medium containing G418, the G418-resistant SSTR5-HA cells were further selected for moderate surface expression of SSTR5-HA by three consecutive fluorescence-activated cell sorter (FACS) experiments using mouse anti-HA monoclonal antibody (MAb) HA.11 (Covance) and secondary goat anti-mouse antibody conjugated to fluorescein isothiocyanate (FITC) (Sigma).
Virus productions and titrations.
Pseudovirions were produced by transient transfection of H1-BAG cells (60), a clonal population of 293 cells stably expressing a lacZ-transducing retroviral genome in which the lacZ gene is under the control of a murine leukemia virus intact long-terminal-repeat promoter (42), with plasmids encoding the MoMLV gag and pol genes (32 μg) and either the parental wild-type MoMLV env gene or the chimeric env gene (20 μg) or VSV G protein cDNA (4 μg), by the use of a T75 flask at 80% to 85% confluence and standard calcium phosphate coprecipitation and harvested exactly as previously described (60). All transductions were performed by overnight exposure of quadruplicate cell samples to virus in the presence of Polybrene (4 μg/ml) in 24- or 48-well plates. 293 cells transiently transfected with SSTR2a cDNA were exposed overnight to undiluted virus stocks (see Fig. 2A). The transduction value determinations were performed by standard endpoint dilution using 10-fold serial dilutions of pseudovirus stocks containing comparable numbers of pseudovirions as judged by the relative amounts of capsid protein on Western blots (see Fig. 2C, 4A and C, 5A, and 7) (data not shown, except in Fig. 4C).
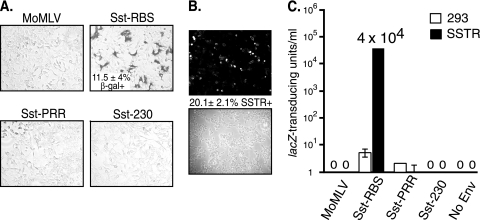
Fig 2.
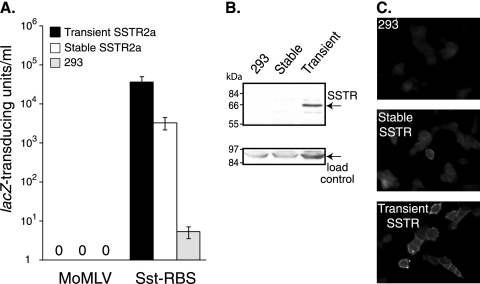
Sst-RBS gave transduction of human 293 cells transiently transfected with an SSTR2a cDNA. (A) Cells were exposed to unconcentrated MLV vectors coated with the indicated Env and then fixed and stained for transduced β-galactosidase (β-Gal+) activity 48 h later. The mean percentage of transduced cells ± standard deviation is shown for the Sst-RBS virus from an experiment that represents three that were performed; there were no lacZ-positive cells in the other cell samples in any of the experiments. (B) Quantification of SSTR-positive cells. The mean percentage of cells immunostained with anti-SSTR2a antiserum ± standard deviation is shown between representative fluorescence and phase micrographs from one of three experiments performed. (C) Quadruplicate wells of parent 293 cells or 293 cells transiently transfected with SSTR2a cDNA were exposed to serial 10-fold dilutions of vectors, and the endpoint dilution titration was calculated after staining for transduction of lacZ. Values shown represent the means ± standard deviations of the results of four independent titration experiments.
Fig 4.
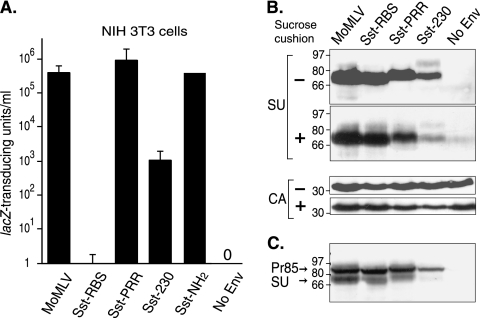
Sst-RBS loses the use of the ecotropic receptor and shows a stable virion association. (A) Murine NIH 3T3 cells expressing endogenous ecotropic receptors were exposed to serial 10-fold dilutions of the indicated virus. Values shown represent the means ± standard deviations of the results of four independent titrations. Sst-RBS gave 0 to 8 lacZ TU/ml in the four experiments. No infection was seen in cells exposed to vectors lacking Env (No Env). (B) Assembly and subunit association of chimeric Env into pseudovirions. Viral proteins in vectors pelleted from supernatants by ultracentrifugation in the absence or presence of a sucrose cushion were separated by SDS-PAGE, and the top portion of the membrane was immunoblotted to antiserum to the surface subunit (SU) of Env and the bottom portion to antiserum to the capsid (CA). (C) Equal amounts (50 μg) of whole-cell lysates from producer cells were separated by SDS-PAGE, transferred to nitrocellulose, and probed with anti-SU antiserum. Values shown to the left of the immunoblots represent molecular mass markers in kilodaltons. Pr85, 85-kDa Env precursor.
Fig 5.
(A) 293 cells transiently transfected with SSTR2a cDNA, 293 cells selected for stable expression of the G418-resistance gene linked to SSTR2a in pcDNA-hSSTR2a, and untreated 293 cells were exposed to serial dilutions of Sst-RBS or Moloney MLV pseudovirions and then fixed and stained for lacZ transduction. Values shown represent the means ± standard deviations of the results of four independent titrations. (B) Plasma membrane proteins were subjected to affinity purification using avidin-agarose after biotinylation of the proteins on the surface of live cells and then separated by SDS-PAGE. Replicate immunoblots of the samples were probed with anti-SSTR2a antiserum (top panel) or with anti-Na+K+ ATPase, a ubiquitous membrane protein, as an internal control for recovery and loading of total plasma membrane proteins (bottom panel). (C) Cells grown on glass coverslips were fixed, permeabilized, and then incubated with anti-SSTR2a antiserum. Micrographs representative of each cell type are shown.
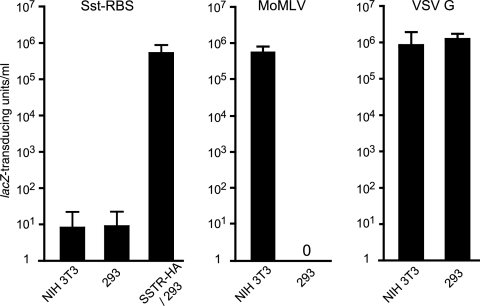
Fig 7.
Sst-RBS glycoprotein targets entry at levels comparable to those of entry mediated by wild-type Moloney murine leukemia virus glycoprotein and approaching those of VSV G pseudovirions. Quadruplicate wells of the host cells indicated on the x axes were exposed to 10-fold serial dilutions of independently produced vectors pseudotyped with the envelope glycoprotein indicated over each graph, and the titers were calculated from the endpoint dilutions. Values shown represent the numbers of mean lacZ-transducing units per milliliter ± standard deviations for four Sst-RBS (left panel), three wild-type MoMLV Env (middle panel), and four VSV G (right panel) pseudovirion stock titers.
Indirect immunofluorescence.
Untreated human 293 cells, stable SSTR2a/293 cells, and 293 cells transiently transfected with pcDNA-hSSTR2a were cultured on triplicate glass coverslips for 24 h and then fixed, permeabilized, incubated in blocking buffer (10% horse serum–phosphate-buffered saline [PBS]) for 1 h at room temperature, incubated with rabbit anti-SSTR2a antiserum (Novus Biologicals, Littleton, CO) in blocking buffer at 1:200 overnight at 4°C, and then incubated with donkey anti-rabbit FITC (Jackson Laboratory, 1:400 dilution) for 45 min at room temperature. Coverslips were examined under a Zeiss Axioplan2 epifluorescence microscope, and phase-contrast and fluorescent images were captured. The total numbers of immunostained cells (positive for SSTR) and negative cells were counted in micrographs of four contiguous fields per coverslip, and the percentages of positive cells were calculated by dividing the number of positive cells by the total number of cells for each sample.
Inhibition of infection by Sst-14 peptide.
293 cells transiently transfected with pcDNA-hSSTR2a were seeded into 24-well tissue culture plates and grown overnight. The next morning, quadruplicate wells were incubated with regular culture medium containing 0, 0.01, 0.1, 1, or 10 μM synthetic Sst-14 (Sigma) for 30 min at 22°C or 37°C. Sst-RBS vector stock was diluted 1:2 in HEPES-buffered DMEM–8% fetal bovine serum and brought to a final concentration of 0, 0.01, 0.1, 1, or 10 μM synthetic Sst-14 peptide. At the end of 30 min of preabsorption with Sst-14, the medium was replaced with the Sst-14-containing vector dilutions and cultures were incubated at 22°C or 37°C for 1 h, after which cells were washed once with regular growth medium and fed with fresh medium. At 48 h later, cells were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) for determinations of β-galactosidase activity. Transduction was quantified by counting the total number of blue (lacZ-positive) cells and the total number of cells. The average percentages of transduction were calculated as the number of blue cells divided by the total number of cells times 100 and then normalized to the average percentages of infected cells in the absence of Sst-14 (taken as representing 100% transduction).
Surface biotinylation.
The relative levels of surface expression of SSTR2a on transiently transfected and stable cell lines were determined as previously described (44) with the following modifications. SSTR2a protein was detected using rabbit anti-SSTR2a antiserum (1:500 dilution) incubated overnight at 4°C. As a control for total recovery of membrane proteins and loading of samples, duplicate blots were probed with mouse monoclonal anti-Na+K+-ATPase α clone M7-PB-E9 (Affinity Bioreagents) (1:250 dilution) overnight at 4°C. Subsequent incubation with goat anti-rabbit or goat anti-mouse antibody conjugated to horseradish peroxidase (HRP; Sigma) (1:10,000) was performed, and immunoblots were developed by detection of HRP with SuperSignal (Pierce).
Western blot analysis of pseudovirions.
Virus particles were purified from cell-free supernatant by centrifugation at 30,000 rpm for 90 min at 4°C (Beckman SW-40 rotor), and retroviral pellets were resuspended in PBS. In some cases, cell-free supernatant was layered over a 25% sucrose cushion prior to centrifugation. Producer cells were lysed using radioimmunoprecipitation (RIPA) buffer (20 mM Tris [pH 7.0], 0.1% Triton X-100, 0.05% sodium deoxycholate, 150 mM NaCl, 2% protease inhibitor). Nuclei were pelleted and discarded, and then the protein concentration in the lysates was determined using a Bio-Rad protein assay. Samples were separated using 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Protran; Schleicher & Schuell), and the envelope and capsid proteins were detected using goat anti-Rauscher gp70 (Quality Biotech Inc.) (1:100 dilution) and goat anti-Rauscher p30 (Quality Biotech Inc.) (1:10,000 dilution), respectively. Blots were incubated with HRP-conjugated mouse anti-goat antibody (Sigma) (1:10,000 dilution) and developed using SuperSignal (Pierce).
Equilibrium virus binding assay.
Assays were performed as previously described (58, 59). Briefly, stocks of Sst-RBS and control ecotropic Moloney murine leukemia virus Env-pseudotyped retrovirus were concentrated 20-fold by low-speed centrifugation using Centricon-100 devices (Millipore), incubated with NIH 3T3, SSTR-HA/293, or 293 cells and then with goat anti-Rauscher SU (a pan-ecotropic SU antiserum from Quality Biotech) (1:100 dilution), and cleared by preincubation with receptor-negative 293 cells and secondary mouse anti-goat FITC (Molecular Probes) (1:400 dilution). Binding assays were initially performed at 4°C; however, the signal for Sst-RBS pseudovirion binding was not above background levels and was barely detected for Moloney murine leukemia virus pseudotypes (data not shown). These results are consistent with those reported by two other groups (28, 57), who found that Moloney murine leukemia virus pseudovirion binding to NIH 3T3 cells was almost undetectable at 4°C but increased substantially at 37°C. Control experiments performed at 4°C and 37°C using synthetic somatostatin peptide instead of Sst-RBS pseudovirions showed that internalization of ligand-bound receptors was inhibited by 0.01% sodium azide at 37°C (data not shown). For this reason, 0.01% sodium azide was added to the assay buffers in the binding experiments reported here.
Nucleotide sequence accession numbers.
The nucleotide sequences of the original cDNA and the optimized cDNA encoding the receptor binding domains were deposited in the GenBank database under accession no. JN122323 and JN122324, respectively.
RESULTS
Receptor binding sequences.
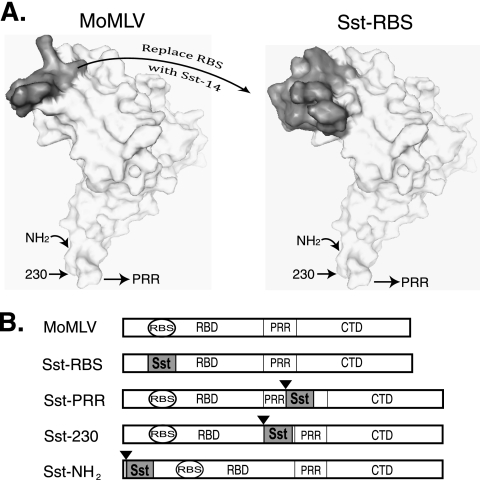
The crystal structure of residues 9 to 236, comprising the receptor binding domain from the highly homologous ecotropic Friend 57 MLV (17), has been solved, but no structure for an Env receptor complex is available due to the integral-membrane nature of the ecotropic receptor (1). However, receptor binding residues have been identified and oriented with respect to other subunits of the Env trimer by the use of genetic approaches (4, 5, 11, 15, 26, 35, 45, 59). Two key studies provided very strong genetic evidence that, of all the known binding site residues, aspartate 84 has the single greatest influence on receptor binding (35, 45). The corresponding residue in Friend 57 MLV, aspartate 86, showed similar importance (15). Since Friend 57 and Moloney MLV use the same cell surface receptor, their receptor binding sequences should have homologous structures. We submitted to SwissModel (24) the sequence of the receptor binding domain of Moloney MLV Env aligned to the Friend 57 1AOL template structure (17). The molecular surface of the predicted structure is shown in Fig. 1A (left panel). Aspartate 84 lies within a disulfide-bonded loop encompassing cysteine 72 to cysteine 85 (shown in dark gray) that is referred to here as the putative RBS.
Fig 1.
Design of the Sst-RBS and control glycoproteins. (A) Left panel, location of the putative ecotropic receptor binding sequences (shown in gray) on the predicted molecular surface of wild-type ecotropic Moloney MLV glycoprotein. Right panel, predicted location of the somatostatin Sst-14 ligand sequences (shown in gray) on the molecular surface of the Sst-RBS model structure. Arrows point to the positions of ligand insertion in traditional designs for chimeric Env. (B) Schematic diagrams of the SU proteins of Env chimeras. Black triangles indicate sites of insertion of the Sst ligand in control chimeras. RBS, putative ecotropic receptor binding sequences. RBD, receptor binding domain. PRR, proline-rich region. CTD, carboxy-terminal domain. Sst, somatostatin.
Choice of the ligand somatostatin.
Extensive searches of the structure databases were performed by visual comparisons to identify a ligand that was similar to the putative RBS in size and structure. The premise was that replacement of RBS with a similarly sized and structured ligand would increase the likelihood that the chimeric Env would fold correctly and be assembled into vector particles. One search identified a candidate molecule called octreotide, a peptide mimetic of mature somatostatin (Sst-14). The sequence of octreotide, FCFWKTCT, contains the core binding sequence FWKT (37), which is underlined here in the Sst-14 sequence AGCKNFFWKTFTSC. The nuclear magnetic resonance (NMR) structure of octreotide (1SOC) showed that the tryptophan and lysine residues form a turn and that the flanking cysteines (shown in bold) form a disulfide bridge (37, 38). Since the affinity of octreotide for Sst receptors (SSTR) is comparable to that of Sst-14 (9, 21, 27), we reasoned that the structure of the active FWKT sequence in Sst-14 is likely to be similar. In sum, we concluded that Sst-14 is similar to the putative RBS in size, contains a pair of disulfide bonded cysteines near each end, and is likely to have a similar structure.
Design of the Sst-RBS Env chimera and control chimeras.
A cDNA encoding Sst-RBS was constructed by swapping nucleotides encoding the Sst ligand for the putative RBS-encoding ones by the use of oligonucleotide-directed mutagenesis. The amino acid sequence of the first 240 residues of Sst-RBS was submitted to SwissModel (24) using an alignment to the Friend 57 1AOL template structure (17). The side chains of the critical FWKT binding sequence for the target SSTR are predicted to lie on the surface at a location similar to that of the natural RBS, and the cysteine residues are predicted to form a disulfide bond (Fig. 1A, right panel).
Despite strong evidence that, because binding does not properly activate conformational changes, ligand-inserted chimeras fail to give transduction, characteristics of the target receptor may also be important to membrane fusion. To control for this possibility, three additional chimeras were made. Two controls, Sst-NH2 and Sst-PRR, were traditional designs in which the Sst ligand was inserted between residues 6 and 7 and between residues 264 and 265, respectively. In the third control, Sst-230, the ligand was inserted between glycine 230 and proline 231, a design reported to give a low level of targeted infection (23). Diagrams of the chimeric Env are shown in Fig. 1B, and the positions of insertion sites are indicated in the molecular models in Fig. 1A.
Sst-RBS mediates infection of human cells expressing SSTR cDNA.
SSTR is expressed robustly in only a few cell types in vivo, and while neuroblastoma cell lines have been reported to express SSTR mRNA, they do not express detectable SSTR protein on their plasma membranes (data not shown). In vitro studies of SSTR function typically use cell lines transfected with SSTR cDNA. Consequently, a target cell line expressing SSTR was generated by transiently transfecting human 293 cells with a plasmid encoding human SSTR subtype 2a cDNA. Transfected cells were divided into two portions; the cells in the larger portion were seeded directly on 24-well plates for exposure to vectors, and some cells were seeded on glass coverslips for immunofluorescence staining of SSTR2a expression.
In the first experiment, transduction of the lacZ transgene was observed with the Sst-RBS pseudotype MLV, with a mean of 11.5% ± 4.0% of cells transduced (Fig. 2A). No lacZ transduction was observed in cells exposed to vectors coated with the control Sst-PRR and Sst-230 chimeras or the parent Moloney murine leukemia virus Env (Fig. 2A). At the time of virus exposure, the aliquots of transiently transfected cells that had been seeded onto glass coverslips were fixed, permeabilized, and stained with anti-SSTR2a antiserum against a peptide derived from its cytoplasmic tail. A mean of 20.1% ± 1.9% of transfected cells showed immunostaining for the target SSTR (Fig. 2B). Taken together, these results suggest that a single exposure of unconcentrated Sst-RBS-coated vector transduced approximately half of the cells transiently expressing the target SSTR.
Subsequent endpoint dilution titration of four independently produced Sst-RBS-coated virus stocks gave a range of 2 × 104 to 4 × 104 lacZ-transducing units (lacZ-TU)/ml on 293 cells transiently transfected with SSTR2a (Fig. 2C), whereas control Sst-PRR and Sst-230 showed almost no transduction of these cells. In each experiment, no more than two of the four replicate wells exposed to undiluted Sst-PRR or Sst-230 vectors contained a lacZ-positive focus, a level comparable to the infection seen in the control 293 cells.
Sst-RBS-mediated transduction was specific to the target SSTR.
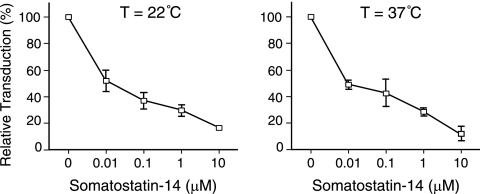
To determine whether transduction occurred via the target SSTR, the results of infection in the presence and absence of synthetic Sst-14 peptide were compared. Since the natural Sst-14 peptide hormone reportedly has a short half-life of just a few minutes in vivo (9) and higher binding affinity at 22°C than at 37°C (36), experiments were performed at both temperatures. Addition of Sst-14 decreased infection in a dose-dependent manner under both sets of conditions (Fig. 3), which is consistent with transduction specific to the target SSTR.
Fig 3.
Dose-dependent inhibition by somatostatin peptides is consistent with entry through the target SSTR. Human 293 cells transiently transfected with SSTR2a cDNA were exposed to Sst-RBS pseudovirions at 22°C (left panel) or 37°C (right panel) in the presence or absence of synthetic somatostatin-14 peptide at the indicated concentrations. Values shown represent the means ± standard deviations of the results of three independent experiments.
Sst-RBS lost use of the ecotropic receptor.
We asked whether use of the natural Moloney murine leukemia virus receptor was retained on rodent cells, since loss of this ability is one trait of targeted entry. A very low level of infection, ranging from 0 to 8 lacZ TU/ml in four independent titrations, was observed (Fig. 4A). Sst-PRR- and Sst-NH2-mediated transduction was comparable to that seen with Moloney murine leukemia virus MLV, while Sst-230 showed infection of the mouse fibroblasts that was more than 2 logs lower (Fig. 4A). Gollan and Green also observed lower infection of mouse cells by the use of chimeras containing a gastrin-releasing peptide inserted at glycine 230 (23). To determine whether chimeric Env assembled stably into virus particles, pseudovirions were purified from vector stocks by the use of ultracentrifugation to pellet virus directly or through a sucrose cushion for comparison. The latter method places greater mechanical stress on virions and thus can reveal differences in the strength of chimeric Env subunit associations (60). Comparable amounts of the SU from parent MoMLV, Sst-RBS, and Sst-PRR Envs were present on virions, while the Sst-230 SU appeared less abundant but was clearly detectable (Fig. 4B, upper panel). Sst-230 also appeared to have a weaker subunit association, since ultracentrifugation through a dense sucrose cushion decreased the level of virion-associated Sst-230 (Fig. 4B, lower panel). Expression of Sst-230 in producer cells was lower as well (Fig. 4C). The weaker subunit association, along with the lower levels of expression and incorporation, may explain the lower infection of Sst-230 seen in NIH 3T3 cells.
Establishing a stable SSTR-positive host cell line.
To assess the true capacity of Sst-RBS to mediate targeted entry, we needed a cell population with two important characteristics: (i) expression of the target receptor on the surface of every cell and (ii) display of a realistic level of receptors, specifically, at levels no greater than those seen with endogenous expression of natural virus receptors. 293 cells were transfected with a human SSTR2a expression plasmid, and stable cell lines were selected for growth by the use of G418 selection. Two independently generated stable cell lines showed 5- to 10-fold-lower transduction than was seen with transiently transfected 293 cells; the titration results for one of the lines are shown in Fig. 5A.
No binding of Sst-RBS-coated vector was detected using the stable cell lines in an equilibrium virus binding assay (data not shown), suggesting that either a small number of cells had substantial SSTR2a expression or, alternatively, the Sst-RBS Env does not bind the target receptor tightly enough to be detected in the virus binding assay. Cell surface levels of SSTR2a were then examined. First, membrane proteins on the surface of live cells were biotinylated using a membrane-impermeable reagent, and the biotin-labeled proteins were affinity purified using avidin-agarose beads, separated by SDS-PAGE, and immunoblotted using anti-SSTR2a antiserum and separately using anti-Na+K+ATPase antibody as a loading control. Samples from transiently transfected cells contained a prominent 65-kDa species as well as minor 72- and 55-kDa species that were absent from parent 293 cell membrane proteins (Fig. 5B). These sizes are consistent with the relative molecular masses previously reported for SSTR2a (16). The anti-SSTR2a reactive species were also present in the stable SSTR2a cell line, but their intensity was much lower and almost undetectable. Since the carboxy-terminal residues recognized by the antiserum lie on the cytoplasmic side of the plasma membrane, indirect immunofluorescence was used as the second method of assessing cell surface expression. Substantially fewer cells of the stable cell line showed anti-SSTR staining at their periphery compared to the transiently transfected cells (Fig. 5C). Taken together, these results suggest that human 293 cells may not tolerate prolonged expression of SSTR2a.
Next, we asked whether one of the other SSTR subtypes could be used to make a stable SSTR-positive host cell line. Since no antibody specific to the other subtypes was available at the time of experimentation, we obtained amino-terminal HA-tagged cDNAs of human subtype 5. As members of the seven-transmembrane G protein-coupled receptor (GPCR) family, SSTRs present their amino termini to the extracellular space such that the HA tag can be detected on live cells by the use of anti-HA MAb. Addition of an HA epitope tag to the amino terminus does not affect SSTR binding to Sst-14 or internalization (33). After 4 weeks of G418 selection, the majority of resistant cells exhibited surface expression SSTR5, as judged by flow cytometry of anti-HA MAb-stained live cells. This population was submitted to three sequential rounds of FACS analysis at 2-week intervals. The resulting cell population was named SSTR-HA/293.
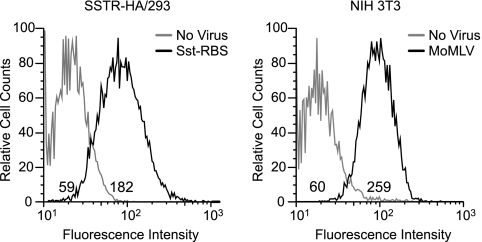
In generating the stable host line, each sorting procedure included gating at a medium level of fluorescence intensity with the goal of obtaining a population whose SSTR expression did not exceed that of the endogenous ecotropic receptor on murine NIH 3T3 cells. To compare the relative numbers of SSTR on the SSTR-HA/293 cells with the relative numbers of ecotropic receptors on NIH 3T3 cells, equilibrium binding assays were performed using Sst-RBS pseudovirions and wild-type Env-coated vectors. The mean fluorescence intensity (MFI) can be used as a measurement of the relative numbers of pseudovirion binding sites on cells of the two cell lines, assuming that the affinity of the antiserum is the same for both glycoproteins. Given that the anti-SU antiserum appeared to recognize the Sst-RBS and wild-type Moloney murine leukemia virus Env similarly on immunoblots (Fig. 4B), this assumption is reasonable. Representative histograms from one of three independent binding assays are shown in Fig. 6. The MFI of Sst-RBS Env bound to SSTR-HA/293 cells was 3-fold higher than the background MFI, whereas the MFI of MoMLV Env bound to NIH 3T3 cells was 4.3-fold higher than the background MFI. These results support the supposition that the density of Sst-RBS binding sites on the surface of SSTR-HA/293 cells is comparable to the density of endogenous ecotropic receptor on NIH 3T3 cells. This interpretation assumes that differences in affinity, particularly in comparisons of the off rates of the two pseudotypes, do not influence the quantification.
Fig 6.
Generation of a host cell line stably expressing levels of target receptor SSTR comparable to the levels of ecotropic receptor on NIH 3T3 cells. 293 cells were transfected with a cDNA-encoding human SSTR5 with an amino-terminal HA epitope tag (SSTR-HA), selected for G418 resistance, and then further selected for surface expression of the target receptor by three rounds of FACS experiments using anti-HA antibody, yielding a population of cells designated SSTR-HA/293. Left panel, aliquots of this cell line were incubated at 37°C with Sst-RBS pseudovirions or mock incubated (No virus), and then both samples were incubated with goat anti-SU antiserum followed by a mouse anti-goat FITC-conjugated secondary antibody plus propidium iodide and analyzed by flow cytometry. Values shown represent FITC intensities determined with live (propidium iodide-negative) cells. Right panel, aliquots of murine NIH 3T3 cells were incubated with MoMLV Env-pseudotyped virus or mock infected (No Virus), and virus binding was performed as described for the left panel. The histograms shown are representative of the results of three independent binding experiments.
Sst-RBS-mediated transduction at levels comparable to transduction using wild-type Moloney murine leukemia virus and VSV G glycoproteins.
To determine the relative capacities of the chimeric Sst-RBS Env to mediate targeted entry, quadruplicate wells of cells were exposed to serial dilutions of Sst-RBS-, Moloney murine leukemia virus Env-, or VSV G-pseudotyped vector stocks overnight and then fixed and stained for lacZ transduction 48 h later. Figure 7 shows the mean values of titers calculated from the endpoint dilutions. In four independent experiments, titers for unconcentrated Sst-RBS pseudovirion stocks were 2.5 × 105, 2.5 × 105, 7.5 × 105, and 1 × 106 TU/ml on SSTR-HA/293 cells, while titers for Moloney murine leukemia virus Env pseudotypes were 2.5 × 105, 5.0 × 105, and 1 × 106 TU/ml on NIH 3T3 cells and titers for VSV G-pseudotyped vectors were 1.0 × 105, 2.5 × 106, 1.0 × 106, and 7.5 × 105 TU/ml on 293 cells.
Sst-RBS-pseudotyped lentiviral vectors.
To further characterize the usefulness of Sst-RBS, human immunodeficiency virus (HIV)-based SIN lentiviral vectors transducing the mKate2 red fluorescent protein and firefly luciferase and pseudotyped with Sst-RBS (Sst-RBS-LV) or VSV G (VSV G-LV) were produced by the University of Tennessee Health Science Center (UTHSC) Viral Vector Core. The Viral Vector Core concentrated the vector stocks by the use of a standard ultracentrifugation method, certified them to be negative for replicating lentivirus, and quantified transducing units by endpoint dilution titration of SSTR-HA/293 and 293 cells by the use of a standard luciferase activity assay. The VSV G-LV stock contained 2.3 × 109 luciferase-transducing units (LTU)/ml, and the Sst-RBS-LV stock contained 9.2 × 106 LTU/ml. The 200-fold-lower titer seen with the Sst-RBS-LV stock compared to the VSV G-LV stock is similar to the titers of other retroviral glycoprotein pseudotypes of lentiviral vectors and is thought to result from a lower level of assembly of other glycoproteins than of VSV G due to the presence of cytoplasmic tail sequences (10, 49).
DISCUSSION
These results establish a proof of principle that replacement of the natural RBS with a heterologous ligand sequence represents a method to target retroviral vector entry to a clinically relevant receptor. The transduction was specific to the target SSTR, since synthetic Sst-14 peptide inhibited infection in a dose-dependent manner. In addition, the ability to infect murine cells was almost abolished, which is consistent with a loss of ecotropic receptor usage. A single exposure of unconcentrated virus stock coated with Sst-RBS glycoprotein gave targeted transduction of up to 106 lacZ TU/ml on human cells expressing moderate levels of SSTR, levels comparable to transduction of NIH 3T3 cells by ecotropic Moloney murine leukemia virus Env and approaching those seen with VSV G-pseudotyped vectors.
It has been proposed that target receptor characteristics represent the most important parameter for targeted entry (14, 29). Although SSTRs are not entry receptors for any known virus, they are seven-transmembrane, G protein-coupled receptors (GPCR) with membrane topology similar to that of the chemokine coreceptors used by human immunodeficiency viruses (HIV) (7) and the GPCR Xpr1 used by the xenotropic MLV (5). The natural Moloney murine leukemia virus receptor also traverses the membrane multiple times (1). Noting that the known gammaretrovirus receptors are multiple-membrane-spanning proteins, Cosset and colleagues tested the importance of this receptor characteristic by examining two amino-terminal insertion chimeras (14). Insertion of the receptor binding domain of amphotropic MLV gave titers of 103 TU/ml, whereas chimeras containing insertion of epidermal growth factor (EGF) bound but did not give targeted entry (14). In a later study, Katane and colleagues showed that insertion of stroma-derived factor 1α (SDF-1α) at proline 79 gave titers of 103 to 104 TU/ml on cells overexpressing the CXCR4 SDF-1α receptor, whereas insertion of EGF at the same position failed to result in infection (29). Since the natural amphotropic MLV receptor and CXCR4 are polytopic membrane proteins whereas the EGF receptor has a single transmembrane protein, Cosset and colleagues and Katane and colleagues concluded that receptor characteristics were most important for targeted entry. Katane and colleagues pointed out that polytopic proteins may be favored, because a virus bound to them would be close to the cell membrane (29). In addition, we note that polytopic membrane proteins are often located in detergent-resistant membrane domains rich in cholesterol and glycosphingolipids thought to be important for membrane fusion (43).
Alternatively, characteristics of the targeting glycoprotein may also represent an important parameter. Viral glycoproteins naturally assemble into virions in a metastable conformation that is not competent for membrane fusion. In the case of gammaretroviral Env, the transition from a metastable to a fusion-competent conformation is induced by receptor binding and begins with fusion peptide exposure (12) and subsequent isomerization of the disulfide bond between the SU and TM (48). It follows that the Env sequences that bind natural retrovirus receptors possess the ability to initiate and sustain these critical conformation changes. We propose that this capacity derives from the position of the RBS at the top of the receptor binding domain, and while this may not be the only location with the capacity to initiate and sustain membrane fusion, it is the most effective.
While characteristics of both the target receptor and chimeric Env likely influence targeted entry, replacement of the RBS represents the most important parameter contributing to the highly efficient targeted entry observed with Sst-RBS. Our reasoning is as follows: if properties of the SSTR represent the most important parameter, then any Sst chimera should give substantial infection. However, the Sst-NH2, Sst-PRR, and Sst-230 chimeras give almost no infection of SSTR-expressing human cells, and such infection as they exhibit occurs at levels no higher than those seen with 293 cells. Yet these control glycoproteins retain the ability to induce membrane fusion, since they mediate entry into NIH 3T3 cells expressing the ecotropic receptor. Thus, we favor the interpretation that Sst-RBS gives highly efficient targeted entry, primarily because the ligand Sst is placed in the unique position normally occupied by the natural RBS.
Early efforts to design ligand-inserted chimeras for targeted entry were made without knowledge of the secondary structure of the retroviral glycoproteins. Domain analysis of an avian retroviral Env serendipitously led to discovery of the amino-terminal insertion site (56), while linker insertion analysis was used to discover the insertion site in the PRR (54) and, later, in the site after residue 230 (23). Peptide ligand sequences were inserted most frequently in the amino terminus and the PRR, because these two sites reproducibly tolerated insertion of ligand sequences in the sense that chimeras were typically expressed on the cell surface and assembled into pseudovirions.
The crystal structure of ecotropic Friend 57 Env residues 1 to 236 (17) and the identification of the putative RBS within the crystallized fragment (15, 35, 45) provide several insights into the characteristics of the ligand-inserted chimeras. First, the amino-terminal and glycine 230 insertion sites are near each other in the secondary structure even though they are quite distant in the primary sequence (Fig. 1A). Second, both are located on the opposite end from the putative RBS. A ligand inserted in either location would be in a very different position within the Env spike than the sequences that bind the ecotropic receptor. This position is nonetheless accessible, since amino-terminal insertion chimeras typically bound their target receptors.
Most importantly, none of the insertion sites lie within the core structure of the glycoprotein's receptor binding domain, which consists of a slightly skewed β-barrel that begins at residue 9 and ends at residue 232 of Friend Env (residue 230 of Moloney murine leukemia virus Env) and is followed by a series of turns leading into the PRR (17). Ligands inserted just before (e.g., between residues 6 and 7) or immediately after (e.g., between residues 230 and 231) the core β-barrel likely fold separately and outside the receptor binding domain. In agreement with this possibility, addition of a flexible linker to amino-terminal insertion chimeras increased their expression, stable association with virions, and ability to bind target receptor (52). The commonly used PRR insertion site (between residues 264 and 265) lies downstream of the crystallized fragment, and so its location relative to the RBS is not known. It is apparent, however, that the PRR sequences comprise a separate folding domain (5, 26); therefore, insertion at this location would also place a ligand outside the core receptor binding domain. These insights suggest that ligand-inserted chimeras bind but do not induce membrane fusion, because the ligands were not an integral part of the natural receptor binding domain and consequently were not in a position allowing induction of the initial conformational changes or in a position that sterically interferes with the conformation changes needed for membrane fusion.
It was expected that Sst-RBS would completely lose the ability to interact with the ecotropic receptor, since the critical aspartate 84 was replaced, and would give no infection of the parental 293 cells, which lack an ecotropic receptor. However, NIH 3T3 and 293 cells consistently showed infection at levels ranging from 0 to 25 TU/ml. Since human and murine Sst-14 sequences are identical, it is possible that the background level of infection is attributable to the low levels of endogenous SSTR previously reported to be found on the surface of 293 cells (31). ras-transformed NIH 3T3 cells also display low levels of endogenous SSTR (46), and this may also be the case for NIH 3T3 cells not transformed by ras. Alternatively, NIH 3T3 infection may result from a residual capacity to bind and enter via the ecotropic receptor.
Do the size and structure of the ligand matter in considering the general applicability of this strategy? In designing Sst-RBS, we carefully considered the structures of the ligand and of the receptor binding sequences being replaced and, in particular, the overall similarities between them. This aspect of the design may have contributed to the success of the chimera, because similarities in the two structures increased the possibility that Sst-RBS would fold into the correct metastable conformation with the ligand Sst residues exposed at the surface. Given that the natural ecotropic RBS is relatively small (∼14 residues), the ecotropic Env may not accommodate the replacement of larger targeting ligands. If the size and structure of the ligand are limited to roughly those of the natural RBS, then it may be possible to extend the RBS replacement strategy to other ligands by using parent glycoproteins whose natural RBS are larger than and similar in structure to the ligand sequences of interest.
In vivo SSTR are abundant in a number of clinically relevant target tissues, specifically, the subthalamic nucleus, striatum, substantia nigra, amygdala, and hippocampus in the brain, the endocrine glands of the pancreas, an unknown cell type in the lungs, and the ileum, but not in other tissues (18, 25, 34, 51). However, the primary cells from these SSTR-positive tissues are extremely difficult to culture, and some, like cells from the subthalamic nucleus, have not been cultured. Therefore, in order to determine whether Sst-RBS transduces cells endogenously expressing SSTR and because the capacity for in vivo transduction is the next critical issue, we have begun in vivo studies. In a preliminary study, two immunocompetent mice (one mouse with VSV G-LV and the other with Sst-RBS-LV-transducing mKate2) were subjected to microinjection in the subthalamic nucleus of one hemisphere. The uninjected hemispheres showed no red fluorescence, and numerous red-fluorescing mKate2-transduced cells were observed along the lower length of the needle track in the VSV G-LV-injected mouse, whereas the needle track of the Sst-RBS-LV-injected brain showed very few red-fluorescing cells, but a burst of transduction at the end of the needle track in the subthalamic nucleus was apparent (preliminary data not shown). The regions directly above the subthalamic nucleus are reportedly negative for SSTR (18, 25, 34) but are positive for VSV G receptors (30). Interpretation of this preliminary result must be cautious, as further extensive in vivo studies are needed to determine which cell types were transduced and whether in vivo targeting to SSTR occurred.
This preliminary observation of in vivo Sst-RBS-mediated transduction suggests potential clinical relevance, as the subthalamic nucleus is involved in Parkinson's disease and has also been implicated in Alzheimer's disease. If the SSTR-positive substantia nigra and striatum are also transduced in vivo by Sst-RBS-pseudotyped vectors, then this new chimeric glycoprotein may be useful as a therapeutic and investigational tool for Alzheimer's and Huntington's disease as well as for other neurodegenerative diseases involving these regions of the brain. Additionally, the amino acid sequences of the targeting ligand Sst in humans, mice, dogs, and primates are identical; thus, Sst-RBS-pseudotyped vectors can be used in preclinical studies in most animal models of disease and would not have to be altered for use in clinical studies.
ACKNOWLEDGMENTS
These studies were supported by PHS grant CA81171 from the National Cancer Institute to L.M.A. and by PHS grant MH073389 from the National Institute of Mental Health and an NARSAD award from the Brain & Behavior Research Foundation to S.A.H.
We thank Graeme Bell (University of Chicago) for the gift of an SSTR2a-encoding plasmid, Krishnakumar Kizhatil for suggesting somatostatin as a ligand, Zhaohui Qian for helpful discussions and construction of pcDNA-SSTR2a, Xiaofei Wang for frozen section preparation, and Carolyn Fields, Marie Selders, Li Lu, and Sathyaramya Balasubramaniam for technical assistance. HIV-based replication-negative SIN lentiviral vectors were produced by the UTHSC Viral Vector Core.
Footnotes
Published ahead of print 19 October 2011
REFERENCES
- 1. Albritton LM, Tseng L, Scadden D, Cunningham JM. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659–666 [DOI] [PubMed] [Google Scholar]
- 2. Anderson WF. 1998. Human gene therapy. Nature 392:25–30 [DOI] [PubMed] [Google Scholar]
- 3. Anliker B, et al. 2010. Specific gene transfer to neurons, endothelial cells and hematopoietic progenitors with lentiviral vectors. Nat. Methods 7:929–935 [DOI] [PubMed] [Google Scholar]
- 4. Bae Y, Kingsman SM, Kingsman AJ. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Battini JL, Heard JM, Danos O. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 66:1468–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benedict CA, et al. 1999. Targeting retroviral vectors to CD34-expressing cells: binding to CD34 does not catalyze virus-cell fusion. Hum. Gene Ther. 10:545–557 [DOI] [PubMed] [Google Scholar]
- 7. Berson JF, Doms RW. 1998. Structure-function studies of the HIV-1 coreceptors. Semin. Immunol. 10:237–248 [DOI] [PubMed] [Google Scholar]
- 8. Bossow S, et al. 2011. Armed and targeted measles virus for chemovirotherapy of pancreatic cancer. Cancer Gene Ther. 18:598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bruns C, Lewis I, Briner U, Meno-Tetang G, Weckbecker G. 2002. SOM230: a novel somatostatin peptidomimetic with broad somatotropin release inhibiting factor (SRIF) receptor binding and a unique antisecretory profile. Eur. J. Endocrinol. 146:707–716 [DOI] [PubMed] [Google Scholar]
- 10. Christodoulopoulos I, Cannon PM. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J. Virol. 75:4129–4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chung M, Kizhatil K, Albritton LM, Gaulton GN. 1999. Induction of syncytia by neuropathogenic murine leukemia viruses depends on receptor density, host cell determinants, and the intrinsic fusion potential of envelope protein. J. Virol. 73:9377–9385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colman PM, Lawrence MC. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309–319 [DOI] [PubMed] [Google Scholar]
- 13. Cosset FL, Lavillette D. 2011. Cell entry of enveloped viruses. Adv. Genet. 73:121–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cosset FL, et al. 1995. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J. Virol. 69:6314–6322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davey RA, Zuo Y, Cunningham JM. 1999. Identification of a receptor-binding pocket on the envelope protein of friend murine leukemia virus. J. Virol. 73:3758–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dournaud P, et al. 1996. Localization of the somatostatin receptor SST2A in rat brain using a specific anti-peptide antibody. J. Neurosci. 16:4468–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fass D, et al. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 277:1662–1666 [DOI] [PubMed] [Google Scholar]
- 18. Fehlmann D, et al. 2000. Distribution and characterisation of somatostatin receptor mRNA and binding sites in the brain and periphery. J. Physiol. Paris 94:265–281 [DOI] [PubMed] [Google Scholar]
- 19. Frecha C, et al. 2008. Stable transduction of quiescent T cells without induction of cycle progression by a novel lentiviral vector pseudotyped with measles virus glycoproteins. Blood 112:4843–4852 [DOI] [PubMed] [Google Scholar]
- 20. Frecha C, Levy C, Cosset FL, Verhoeyen E. 2010. Advances in the field of lentivector-based transduction of T and B lymphocytes for gene therapy. Mol. Ther. 18:1748–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Froidevaux S, Eberle AN. 2002. Somatostatin analogs and radiopeptides in cancer therapy. Biopolymers 66:161–183 [DOI] [PubMed] [Google Scholar]
- 22. Funke S, et al. 2008. Targeted cell entry of lentiviral vectors. Mol. Ther. 16:1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gollan TJ, Green MR. 2002. Selective targeting and inducible destruction of human cancer cells by retroviruses with envelope proteins bearing short peptide ligands. J. Virol. 76:3564–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guex N, Diemand A, Peitsch MC. 1999. Protein modelling for all. Trends Biochem. Sci. 24:364–367 [DOI] [PubMed] [Google Scholar]
- 25. Hannon JP, et al. 2002. Somatostatin sst2 receptor knock-out mice: localisation of sst1-5 receptor mRNA and binding in mouse brain by semi-quantitative RT-PCR, in situ hybridisation histochemistry and receptor autoradiography. Neuropharmacology 42:396–413 [DOI] [PubMed] [Google Scholar]
- 26. Heard JM, Danos O. 1991. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J. Virol. 65:4026–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hofland LJ, Lamberts SW. 2003. The pathophysiological consequences of somatostatin receptor internalization and resistance. Endocr. Rev. 24:28–47 [DOI] [PubMed] [Google Scholar]
- 28. Kadan MJ, Sturm S, Anderson WF, Eglitis MA. 1992. Detection of receptor-specific murine leukemia virus binding to cells by immunofluorescence analysis. J. Virol. 66:2281–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katane M, Takao E, Kubo Y, Fujita R, Amanuma H. 2002. Factors affecting the direct targeting of murine leukemia virus vectors containing peptide ligands in the envelope protein. EMBO Rep. 3:899–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuroda H, Kutner RH, Bazan NG, Reiser J. 2008. A comparative analysis of constitutive and cell-specific promoters in the adult mouse hippocampus using lentivirus vector-mediated gene transfer. J. Gene Med. 10:1163–1175 [DOI] [PubMed] [Google Scholar]
- 31. Law SF, Yasuda K, Bell GI, Reisine T. 1993. Gi alpha 3 and G(o) alpha selectively associate with the cloned somatostatin receptor subtype SSTR2. J. Biol. Chem. 268:10721–10727 [PubMed] [Google Scholar]
- 32. Liu C, Russell SJ, Peng KW. 2010. Systemic therapy of disseminated myeloma in passively immunized mice using measles virus-infected cell carriers. Mol. Ther. 18:1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Q, et al. 2005. Receptor signaling and endocytosis are differentially regulated by somatostatin analogs. Mol. Pharmacol. 68:90–101 [DOI] [PubMed] [Google Scholar]
- 34. Ludvigsen E, Olsson R, Stridsberg M, Janson ET, Sandler S. 2004. Expression and distribution of somatostatin receptor subtypes in the pancreatic islets of mice and rats. J. Histochem. Cytochem. 52:391–400 [DOI] [PubMed] [Google Scholar]
- 35. MacKrell AJ, Soong NW, Curtis CM, Anderson WF. 1996. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J. Virol. 70:1768–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maggi M, et al. 1994. Identification, characterization, and biological activity of somatostatin receptors in human neuroblastoma cell lines. Cancer Res. 54:124–133 [PubMed] [Google Scholar]
- 37. Marbach P, et al. 1988. Structure-function relationships of somatostatin analogs. Horm. Res. 29:54–58 [DOI] [PubMed] [Google Scholar]
- 38. Melacini G, Zhu Q, Goodman M. 1997. Multiconformational NMR analysis of sandostatin (octreotide): equilibrium between beta-sheet and partially helical structures. Biochemistry 36:1233–1241 [DOI] [PubMed] [Google Scholar]
- 39. Morizono K, et al. 2010. Redirecting lentiviral vectors pseudotyped with Sindbis virus-derived envelope proteins to DC-SIGN by modification of N-linked glycans of envelope proteins. J. Virol. 84:6923–6934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morizono K, et al. 2009. A versatile targeting system with lentiviral vectors bearing the biotin-adaptor peptide. J. Gene Med. 11:655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Münch RC, et al. 2011. DARPins: an efficient targeting domain for lentiviral vectors. Mol. Ther. 19:686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Price J, Turner D, Cepko C. 1987. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc. Natl. Acad. Sci. U. S. A. 84:156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Puri A, et al. 1998. The neutral glycosphingolipid globotriaosylceramide promotes fusion mediated by a CD4-dependent CXCR4-utilizing HIV type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 95:14435–14440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qian Z, Donald R, Wang H, Chen Q, Albritton LM. 2003. Identification of a critical basic residue on the ecotropic murine leukemia virus receptor. J. Virol. 77:8596–8601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qian Z, Wang H, Empig C, Anderson WF, Albritton LM. 2004. Complementation of a binding-defective retrovirus by a host cell receptor mutant. J. Virol. 78:5766–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reardon DB, Dent P, Wood SL, Kong T, Sturgill TW. 1997. Activation in vitro of somatostatin receptor subtypes 2, 3, or 4 stimulates protein tyrosine phosphatase activity in membranes from transfected Ras-transformed NIH 3T3 cells: coexpression with catalytically inactive SHP-2 blocks responsiveness. Mol. Endocrinol. 11:1062–1069 [DOI] [PubMed] [Google Scholar]
- 47. Ryu BY, Zavorotinskaya T, Trentin B, Albritton LM. 2008. The block to membrane fusion differs with the site of ligand insertion in modified retroviral envelope proteins. J. Gen. Virol. 89:1049–1058 [DOI] [PubMed] [Google Scholar]
- 48. Sanders DA. 2000. Sulfhydryl involvement in fusion mechanisms. Subcell. Biochem. 34:483–514 [DOI] [PubMed] [Google Scholar]
- 49. Sandrin V, et al. 2002. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 100:823–832 [DOI] [PubMed] [Google Scholar]
- 50. Schneider U, Bullough F, Vongpunsawad S, Russell SJ, Cattaneo R. 2000. Recombinant measles viruses efficiently entering cells through targeted receptors. J. Virol. 74:9928–9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ten Bokum AM, et al. 2002. Tissue distribution of octreotide binding receptors in normal mice and strains prone to autoimmunity. Nucl. Med. Commun. 23:1009–1017 [DOI] [PubMed] [Google Scholar]
- 52. Valsesia-Wittmann S, et al. 1996. Improvement of retroviral retargeting by using amino acid spacers between an additional binding domain and the N terminus of Moloney murine leukemia virus SU. J. Virol. 70:2059–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Verhoeyen E, Cosset FL. 2009. Engineering the surface glycoproteins of lentiviral vectors for targeted gene transfer. Cold Spring Harb. Protoc. 2009:pdb.prot5276. doi:10.1101/pdb.top59 [DOI] [PubMed] [Google Scholar]
- 54. Wu BW, Lu J, Gallaher TK, Anderson WF, Cannon PM. 2000. Identification of regions in the Moloney murine leukemia virus SU protein that tolerate the insertion of an integrin-binding peptide. Virology 269:7–17 [DOI] [PubMed] [Google Scholar]
- 55. Yamada Y, et al. 1992. Cloning and functional characterization of a family of human and mouse somatostatin receptors expressed in brain, gastrointestinal tract, and kidney. Proc. Natl. Acad. Sci. U. S. A. 89:251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Young JA, Bates P, Willert K, Varmus HE. 1990. Efficient incorporation of human CD4 protein into avian leukosis virus particles. Science 250:1421–1423 [DOI] [PubMed] [Google Scholar]
- 57. Yu H, Soong N, Anderson WF. 1995. Binding kinetics of ecotropic (Moloney) murine leukemia retrovirus with NIH 3T3 cells. J. Virol. 69:6557–6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zavorotinskaya T, Albritton LM. 1999. Failure to cleave murine leukemia virus envelope protein does not preclude its incorporation in virions and productive virus-receptor interaction. J. Virol. 73:5621–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zavorotinskaya T, Albritton LM. 1999. A hydrophobic patch in ecotropic murine leukemia virus envelope protein is the putative binding site for a critical tyrosine residue on the cellular receptor. J. Virol. 73:10164–10172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zavorotinskaya T, Albritton LM. 2001. Two point mutations increase targeted transduction and stabilize vector association of a modified retroviral envelope protein. Mol. Ther. 3:323–328 [DOI] [PubMed] [Google Scholar]
- 61. Zhao Y, et al. 1999. Identification of the block in targeted retroviral-mediated gene transfer. Proc. Natl. Acad. Sci. U. S. A. 96:4005–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]