Abstract
Stable HIV-1 replication requires the DNA repair of the integration locus catalyzed by cellular factors. The human RAD51 (hRAD51) protein plays a major role in homologous recombination (HR) DNA repair and was previously shown to interact with HIV-1 integrase (IN) and inhibit its activity. Here we determined the molecular mechanism of inhibition of IN. Our standard in vitro integration assays performed under various conditions promoting or inhibiting hRAD51 activity demonstrated that the formation of an active hRAD51 nucleofilament is required for optimal inhibition involving an IN-DNA complex dissociation mechanism. Furthermore we show that this inhibition mechanism can be promoted in HIV-1-infected cells by chemical stimulation of the endogenous hRAD51 protein. This hRAD51 stimulation induced both an enhancement of the endogenous DNA repair process and the inhibition of the integration step. Elucidation of this molecular mechanism leading to the restriction of viral proliferation paves the way to a new concept of antiretroviral therapy based on the enhancement of endogenous hRAD51 recombination activity and highlights the functional interaction between HIV-1 IN and hRAD51.
INTRODUCTION
DNA double-strand breaks (DSBs) arise during normal DNA metabolism [e.g., replication, meiosis, V(D)J recombination, etc.]. DNA-damaging agents, such as ionizing radiation and chemical compounds generating replication-blocking lesions like cis-platinum, also cause DSBs. Retroviral integration involving a cut of the genomic DNA can also be considered a potentially mutagenic event. The cellular DNA repair machinery could thus be required either for cell defense against viral infection or, inversely, for the stable integration of the viral genome. Since these machineries are not completely defined at present, they require better characterization. Retroviral integration is catalyzed by the virally encoded integrase (IN) in concert with other viral and cellular factors forming the preintegration complex (PIC). As a final step, stable integration of retroviral DNA requires the repair of the DNA gaps remaining after the reaction catalyzed by IN. Several cellular repair mechanisms, including base excision repair (3, 14, 43, 44), homologous recombination (HR) (19), and nonhomologous end-joining (NHEJ) (9–11, 26), are thought to be involved in the integration step. Direct interactions between HIV-1 IN and human RAD18 have been reported (31), and cell-based studies have indicated that RAD18 and RAD52 can inhibit HIV-1 infection (22, 27). These data suggest that cellular RAD DNA repair machineries can play a dual role by preventing or restricting retroviral viral replication/parasitism and/or participating directly in the stability of the integrated viral DNA, a crucial step of the infection process.
Human RAD51 (hRAD51), belonging to the HR DNA repair RAD52 epistasis group, has been previously shown to interact functionally with HIV-1 IN both in vitro and in a yeast integration model (12). A downregulation of HIV-1 IN activity by hRAD51 was also reported in in vitro integration assays, but the inhibition mechanism remained unsolved. Since the enhancement of this inhibition could constitute a new antiviral approach as well as signify an original restriction pathway, we focused our work here on the molecular mechanism underlying this process both in vitro and in vivo. We especially studied whether this hRAD51-mediated IN inhibition could be promoted in HIV-1-infected cells.
Using standard in vitro integration assays under conditions promoting or inhibiting hRAD51 activity (hRAD51 mutants, presence or absence of ATP, addition of a hRAD51 stimulatory compound), we show that the formation of an active hRAD51 presynaptic nucleofilament corresponding to the nucleocomplex formed on the two homologous DNA strands during HR is required for inhibition. An IN-DNA dissociation system was also set up in order to better determine the effect of the hRAD51 nucleofilament on the active integration complex. Using this system we show that this nucleofilament can dissociate IN from its substrate. Finally, the availability of RS-1, a chemical compound able to stimulate hRAD51 activity, allowed us to demonstrate that the stimulation of the hRAD51-mediated DNA repair process in HIV-1-infected cells can lead to the inhibition of retroviral replication by decreasing integration efficiency.
In addition to elucidating the molecular mechanism underlying IN inhibition by hRAD51, our data reveal an original intrinsic integration restriction property of hRAD51 that paves the way for a new concept of antiretroviral therapy based on the stimulation of the endogenous hRAD51 recombination activity in HIV-1-infected cells.
MATERIALS AND METHODS
In vitro procedures. (i) Proteins, antibodies, and chemical compounds.
HIV-1 IN was purified from a yeast expression system using the INHybrid methods described previously (24). Prototype foamy virus (PFV) integrase was purified according to the method reported before (41). Wild-type (wt), K133A, and K133R human hRAD51s were purified from an Escherichia coli expression system as described before (6). HIV-1 reverse transcriptase (RT) was purified according to the protocol previously described (1). Human HMG1A and bacterial RecA proteins were purchased from Sigma. RS-1 was previously selected from a 10,000-compound library (Chembridge DIVERSet); compounds were screened for their ability to affect the DNA binding property of hRAD51 (18). Cisplatin and monoclonal anti-FLAG and polyclonal anti-hRAD51 antibodies were purchased from Sigma. Anti-HIV-1 IN antibodies were purchased from Bio Products.
(ii) In vitro concerted integration assay.
Standard concerted integration reactions were performed as described previously (24) with some modifications. Briefly, purified HIV-1 IN (600 nM) was preincubated with both 5′-end-labeled donor DNA (15 ng) containing the unprocessed U3 and U5 long terminal repeat (LTR) sequences and the target DNA plasmid pBSK+ (150 ng) at 0°C for 20 min in a total volume of 5 μl. Then the reaction mixture (20 mM HEPES [pH 7.5], 10 mM dithiothreitol [DTT], 10 mM MgCl2, 15% dimethyl sulfoxide [DMSO], 8% polyethylene glycol [PEG], 30 mM NaCl) was added, and the reaction proceeded for 120 min at 37°C in a total volume of 10 μl. Purified wt or mutated hRAD51 was added at different concentrations and under different conditions, as reported in the text. Incubation was stopped by adding a phenol-isoamyl alcohol-chloroform mix (24/1/25 [vol/vol/vol]). The aqueous phase was loaded on a vertical 1% agarose gel in the presence of 1% bromophenol blue and 1 mM EDTA. After separation of the products, the gel was treated with 5% trichloroacetic acid (TCA) for 20 min, dried, and autoradiographed. All IN activities were quantified by scanning the bands (half-site plus full-site integration products) after gel electrophoresis and autoradiography using Image J software. Both target DNA and donor plasmids were kind gifts from K. Moreau (Université Claude Bernard-Lyon I, France). The target corresponds to the plasmid pBSK+ (Stratagene, La Jolla, CA) carrying the zeocin resistance-encoding gene. The 296-bp unprocessed donor was generated by cloning a donor containing ScaI ends into a pGEM-T vector (Promega). The resulting pGEM-T-SupFScaI vector was cleaved by ScaI, and the substrate fragment was purified and 5′ radiolabeled. For the PFV concerted integration assay, the 296-bp unprocessed donor DNA containing the PFV viral ends was generated by PCR using pGEM-T-SupFScaI as the template and primers PFV-SupF5′ (5′-ATTGTCATGGAATTTTGTATATTGATTATCCTTTAACGTTGCCCGGATCCGGTCGCGC) and PFV-SupF3′ (5′-ATTGTCATGGAATTTTGTATATTGATTATCCTGCGGCGCGTCATTTGATATGATGCG). The integration reaction was performed using purified PFV-IN (600 nM), 5′-end-labeled donor DNA (15 ng), and target DNA plasmid pBSK+ (150 ng) under the same conditions as those listed above.
(iii) hRAD51 in vitro activity.
hRAD51 activity was evaluated according to the previously reported strand exchange reaction (40) adapted to our concerted integration conditions. The 59-mer single-stranded DNA (ssDNA; 100 nM) was incubated with hRAD51 (0 to 0.5 μM) for 30 min at 37°C in the presence of 100 μM ATP and 20 mM HEPES (pH 7.5), 10 mM DTT, 10 mM MgCl2, 15% DMSO, 8% PEG, and 30 mM NaCl in a total volume of 10 μl. Then the homologous 32-mer double-stranded DNA (dsDNA; 10 nM) was added. The reaction products were separated by electrophoresis on nondenaturing 12% polyacrylamide gel. Strand exchange products were quantified after autoradiography of the gel using Image J software.
(iv) HIV-1 RT RNase H activity.
The RNA-DNA hybrid substrate was prepared as follows. Calf thymus DNA (10 μg) (Sigma) was incubated in 50 mM Tris-HCl (pH 7.8), 5 mM DTT, 5 mM Mg2+, 100 mM KCl, 500 μM (each) ATP, GTP, and CTP, 30 μCi [3H]UTP (31 Ci/mmol), and 1 unit of E. coli RNA polymerase for 1 h at 37°C (final volume, 50 μl). RT was preincubated with increasing amounts of RAD51 (or RS-1) for 15 min at 37°C in the presence of 100 μM ATP. Then the RNase H activity was measured by incubation in the presence of 50 mM Tris-HCl (pH 7.8), 5 mM DTT, 5 mM MgCl2, 100 mM KCl, and the RNA-DNA hybrid (20,000 cpm) for 20 min at 37°C in a 50-μl final volume. The reaction was stopped by addition of 1 ml cold 10% TCA. Samples were filtered on nitrocellulose filters and washed with 10% TCA, and the radioactivity was determined in a scintillation counter.
(v) HIV-1 RT DNA-dependent DNA polymerase activity.
RT was preincubated with increasing amounts of hRAD51 (or RS-1) for 10 min at 37°C in the presence of 100 μM ATP. Then the DNA-dependent DNA polymerase activity was measured for 15 min at 37°C (36). The reaction mixture contained, in a final volume of 50 μl, 50 mM Tris-HCl (pH 7.8), 10 mM DTT, 5 mM MgCl2, 50 μM (each) dATP, dCTP, and dGTP, 20 μM dTTP, 0.5 μCi [3H] TTP (31 Ci/mmol), and 20 μg/ml activated DNA. The reaction was stopped by addition of 1 ml cold 10% TCA. Samples were filtered on nitrocellulose filters and washed, and the radioactivity was determined in a scintillation counter.
(vi) In vitro IN-DNA dissociation assay.
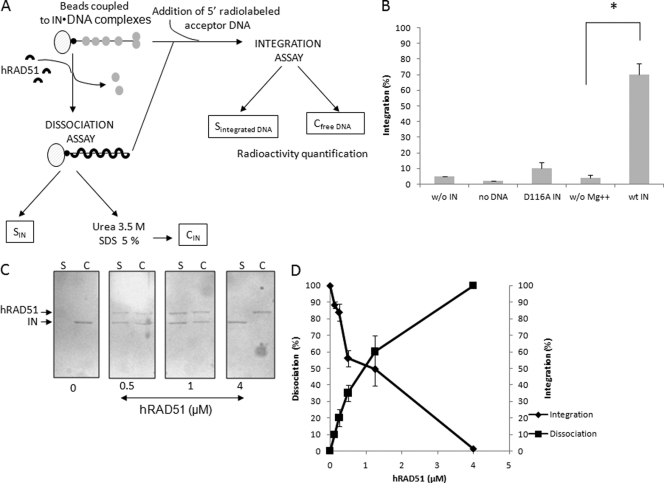
A schematic view of the method is provided in Fig. 4A. A 5′-biotinylated unprocessed donor DNA was generated by PCR using the PGEM-T-SupFScaI vector as the template and 5′-biotinylated U3 (5′-TATGGAAGGGCTAATTCACT-3′) and U5 (5′-TATGCTAGAGATTTTCCACA-3′) as the primers. The biotinylated fragments (100 ng) were coupled to streptavidin magnetic beads (10 μl; Ademtech) for 30 min at room temperature. After a washing with H2O, the amount of DNA bound to the beads was checked by SyberSafe-stained agarose gel analysis of the supernatant after magnetization of the beads. IN (40 pmol) was then added to the DNA-activated beads, and the coupling was performed for 20 min at 4°C in a total volume of 5 μl. After a washing, hRAD51 proteins (wt or mutated enzymes) were added for 60 min in a reaction buffer (20 mM HEPES [pH 7.5], 10 mM DTT, 7.5 mM MgCl2, 10% DMSO, 8% PEG, 30 mM NaCl) with or without ATP. Beads were then washed and subjected to magnetization. The bead pellet was treated with 3.5 M urea and 5% SDS and, in addition to the supernatant fraction, was subjected to SDS-PAGE analysis. After SDS-PAGE, HIV-1 IN and hRAD51 proteins were detected by Western blotting using a mixture of anti-IN and anti-hRAD51 polyclonal antibodies. For the integration assay on IN-DNA-coupled beads, a 32-bp double-stranded 5′ radioactively labeled acceptor oligodeoxynucleotide (ODN) (5′-CCATCCGCAAAAATGACCTCTTATCAAAAGGA-3′; hybridized to the homologous strand) was incubated with the activated beads for 1 h at 37°C under integration conditions (20 mM HEPES [pH 7.5], 10 mM DTT, 10 mM MgCl2, 15% DMSO, 8% PEG, 30 mM NaCl). The integration activity was measured by quantifying the radioactivity remaining covalently bound to the washed beads after their magnetization. The structure of the integration products was previously checked on polyacrylamide gels.
Fig 4.
Dissociation effect of hRAD51 on active IN-DNA integration complex. The 296-bp viral DNA used in the concerted integration assay and fused to biotin was coupled with magnetic beads. HIV-1 IN was then bound to the DNA (checked by SDS-PAGE). hRAD51 treatment was performed under different conditions promoting the formation of the active nucleofilament or not. IN-DNA dissociation and hRAD51 binding were checked by SDS-PAGE of the supernatant (C, lanes S). The proteins remaining bound to the DNA were analyzed after SDS and urea treatment of the bead pellet and SDS-PAGE followed by Western blotting (C, lanes C). The presence of IN and hRAD51 in the bead pellet and the supernatant was checked after SDS-PAGE of the corresponding fractions and Western blotting using a mixture of antibodies against each protein (strategy is shown in panel A). Coupling of active complexes was checked by integration assays using different conditions promoting (wt IN) or not (w/o Mg++; use of inactivated D116A enzyme) IN activity in the presence of a 5′-radiolabeled acceptor DNA. The activity of the coupled IN was measured by quantifying the radioactivity remaining covalently bound to the beads after repeated washings (B). A Student test was performed on serial values. *, P < 0.005. Treatment of the complexes with increasing concentrations of hRAD51 in the presence of 100 μM ATP led to the dissociation of IN from its substrate (C). (D) Quantification of the integration activity catalyzed by the IN coupled to the beads and IN dissociation after treatment with increasing concentrations of hRAD51.
Electron microscopy analysis.
Microscopy analysis was performed using HIV-1 IN (750 nM) (or the corresponding storage buffer) preincubated with the 296-bp donor DNA (1.5 μM) containing the 20 final bases of U3 and U5 LTR sequences at 0°C for 20 min in Tris-HCl (10 mM, pH 7.6), NaCl (30 mM), and EDTA (1 mM). Binding and control reaction mixtures were supplemented with the same volume of a solution containing hRAD51 (500 nM) (or the corresponding storage buffer) in HEPES-NaOH (40 mM, pH 7.6), NaCl (30 mM), magnesium acetate [Mg(OAc)2; 15 mM], DMSO (20%), DTT (1 mM), and ATP (100 μM). The reaction was carried out for 30 min at 37°C. Before the integration reaction buffer was added, 5 μl of binding reaction mixture was deposited on a 600-mesh copper grid (Delta Microscopies) covered with a thin carbon film activated by glow-discharge in the presence of pentylamine and positively stained with aqueous uranyl acetate (2% [wt/vol]), as previously described (2). Reaction mixtures with or without hRAD51 were diluted 10-fold in 20 mM HEPES (pH 7.5), NaCl (30 mM), and Mg(OAc)2 (7.5 mM) before positive staining carried out for binding or without further dilution on 200-mesh copper grids (Delta Microscopies) covered with a collodion-carbon film freshly glow-discharged and negatively stained with 2% uranyl acetate. Samples were observed in the annular dark-field mode for positive staining or in bright-field mode for negative staining using a Zeiss 902 transmission electron microscope. Images were captured at magnifications of ×85,000 and ×140,000 with a MegaviewIII charge-coupled device (CCD) camera and iTEM software for acquisition (Olympus Soft Imaging Solution).
Cellular procedures. (i) Viral infectivity.
Viral infectivity was assayed on peripheral blood mononuclear cells (PBMC), HeLa cells, or HeLa P4 cells expressing CD4 and CXCR4 receptors and carrying the stably integrated lacZ gene under the control of the HIV-1 LTR (8). Under this system the activity of β-galactosidase, whose expression is linked to the expression of the Tat protein, is proportional to HIV-1 integration. In the infection experiments, HeLa or HeLa P4 cells were plated in 48-well plates at 50,000 cells/well using 400 μl of Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) containing 10% (vol/vol) fetal calf serum (FCS; Invitrogen) and 50 μg/ml of gentamicin (Invitrogen). For RS-1 treatments, cells were incubated for 24 h prior to infection with increasing concentrations of RS-1. After overnight incubation at 37°C, medium was replaced with 400 μl of fresh DMEM containing either HIV-1 Lai (1 × 108 particles/ml; multiplicity of infection [MOI] = 0.4), produced as described in reference 29, or PFV at a MOI of 0.4 (pMD9 vector [16]) and the 4-plasmid system, as described in references 38 and 32, using codon-optimized expression constructs for PFV Gag, Pol, and Env. After 24 h at 37°C, cells were washed three times with 400 μl of 0.9% NaCl. For HeLa P4 cells each well was refilled with 400 μl of a reaction buffer containing 50 mM Tris-HCl (pH 8), 100 mM β-mercaptoethanol, 0.05% Triton X-100, and 5 mM 4-methylumbelliferyl-β-d-galactoside (4-MUG) (Sigma, St. Louis, MO). The level of the reaction was measured in a fluorescence microplate reader (Cytofluor II; Applied Biosystems, Foster City, CA) at 360/460 nm (excitation/emission wavelengths) after 24 h of incubation. PFV replication was measured by flux cytometry after infection of HeLa cells.
PBMC were isolated from blood samples using Ficoll-Hypaque gradient centrifugation. After separation, PBMC were pelleted by centrifugation. The cell culture medium consisted of RPMI 1640 supplemented with 20% heat-inactivated fetal bovine serum, 5% interleukin-2 (IL-2), and 50 μg gentamicin/ml. PBMC (2 × 106) isolated from whole blood were incubated with various concentrations of RS-1 (0, 15, 30, 75, and 100 μM) for 24 h at 37°C. Next, PBMC were harvested (7 min, 400 × g) and resuspended in 500 μl culture medium. Ten microliters of HIV-1 subtype B virus (MOI = 0.1) was added to PBMC, and cells were incubated at 37°C for 3 h. Then, the medium was removed, and 10 ml of RPMI 1640 was added to wash the cells. The cells were harvested at low speed (400 × g), and the washed-cell pellet was resuspended in 2 ml of supplemented RPMI 1640. The cell suspension was added to wells of a 24-well tissue culture plate and incubated at 37°C. HIV-1 RNA from plasma samples was determined 24, 48, and 72 h postinfection. Replication in PBMC was quantified by HIV-1 RNA determination in cellular supernatant using Amplicor HIV Cobas TaqMan, version 2 (Roche, Basel, Switzerland), with a lower limit of detection of 20 copies/ml of plasma.
(ii) Quantification of HIV-1 DNA population.
Cells were harvested 48 h postinfection by centrifugation of 2 × 106- to 10 × 106-cell aliquots, and cell pellets were kept frozen at −80°C until further analysis. Total DNA (including integrated HIV-1 DNA and episomal HIV-1 DNA) was extracted using the QIAmp blood DNA minikit (Qiagen, Courtaboeuf, France) according to the manufacturer's protocol. Elution was performed in 50 μl of elution buffer. The total HIV-1 DNA was amplified by quantitative real-time PCR using the Light Cycler instrument (Roche Diagnostics, Meylan, France). Amplification was performed in a 20-μl reaction mixture containing 1× Light Cycler Fast Start DNA master hybridization probes (Roche Diagnostics), 3 mM MgCl2, 500 nM forward primer LTR152 (5′-GCCTCAATAAAGCTTGCCTTGA-3′, and 500 nM reverse primer LTR131 (5′-GGCGCCACTGCTAGAGATTTT-3′), located in an LTR region with highly conserved fluorogenic hybridization probe LTR1 (50 nM; 5′-6-carboxyfluorescein [FAM]-AAGTAGTGTGTGCCCGTCTGTT[AG]T[GT]TGACT-3′-6-carboxytetramethylrhodamine [TAMRA]). After an initial denaturation step (95°C for 10 min), total HIV-1 DNA was amplified for 45 cycles (95°C for 10 s, 60°C for 30 s), followed by 1 cycle at 40°C for 60 s. The copy number of total HIV-1 DNA was determined using the 8E5 cell line. The 8E5/LAV cell line, used for a standard curve, was derived from a CEM cellular clone containing a single, integrated, defective (in the pol open reading frame), constitutively expressed viral copy. 8E5 DNA (5 to 5 × 104 copies) was amplified. Results were expressed as the copy number of total HIV-1 DNA per 106 cells.
The 2-LTR DNA circles were amplified with primers HIV-F and HIV-R1, spanning the LTR-LTR junction, as described elsewhere (4). Briefly, amplification was performed in a 20-μl reaction mixture containing 1× Light Cycler Fast Start DNA master hybridization probes (Roche Diagnostics), 4 mM MgCl2, 300 nM forward and reverse primers spanning the LTR-LTR junction, and 200 nM each fluorogenic hybridization probe. Copy number of 2-LTR circles was determined in reference to a standard curve prepared by amplification of quantities ranging from 10 to 1 × 106 copies of a plasmid comprising the HIVLAI 2-LTR junction (4) by using Light Cycler quantification software, version 4.1 (Roche Diagnostics). Results are expressed as copy number of 2-LTR circles per 1× 106 cells.
Integrated DNA in HeLa P4 cells was first amplified by Alu PCR performed in a 50-μl reaction mixture containing 200 ng total DNA, 1× HF Phusion mix, 200 nM deoxynucleoside triphosphates (dNTP), 500 nM primer PBS (5′-TTTCAAGTCCCTGTTCGGGCGCCA-3′), located in the PBS sequence of the viral genome, and 500 nM primer Alu-164 (5′-TCCCAGCTACTGGGGAGGCTGAGG-3′), located in the Alu sequence of the cellular genome. After an initial denaturation step (98°C for 30 s), the heterogeneously sized population of integrated DNA was amplified for 35 cycles (98°C for 10 s, 60°C for 20 s, 72°C for 2 min 30 s). A second nested PCR was then performed in a 50-μl reaction mixture containing 5 μl of Alu PCR product, 1× HF Phusion mix, 500 nM dNTP, 500 nM primer NI-1 (5′-CACACACAAGGCTACTTCCCT-3′), and 500 nM primer NI-2 (5′-GCCACTCCCCAGTCCCGCCC-3′); primer sequences match sequences localized in the viral genome. After an initial denaturation step (94°C for 12 min), the expected 351-bp fragment of integrated DNA was amplified for 42 cycles (94°C for 1 min, 60°C for 1 min, 72°C for 1 min). Results were analyzed on 1.2% agarose SYBR safe stain gel. A first-round PCR control was ran in the absence of polymerase in order to quantify any unspecific amplification during the second round. Quantification of the integrated DNA in PBMC was done by standard quantitative PCR as previously reported (5). Briefly, in a first round of PCR, integrated HIV-1 sequences were amplified with the two outward-facing Alu primers reported above and then the amplified products were subjected to quantitative PCR using the NI-1 and NI-2 primers mentioned above in the Light Cycler instrument (Roche Diagnostics, Meylan, France). The copy number of integrated HIV-1 DNA was determined in reference to a standard curve generated by concomitant two-stage PCR amplification of a serial dilution of the standard HeLa R7 Neo cell DNA mixed with uninfected-cell DNA to yield 50,000 cell equivalents.
(iii) Quantification of cisplatin resistance.
Determination of the effect of protein overexpression and RS-1 treatment on hRAD51-mediated homologous recombination in HeLa and HeLa P4 cells was performed by a standard cisplatin resistance assay described previously (18). Forty-eight hours after transfection with the expression vector or 24 h after RS-1 treatment, the cells were incubated for 24 h with increasing concentrations of cisplatin. After removal of the drug, the cells were allowed to grow in complete media for an additional 6 days. Cell survival experiments were then performed using a sulforhodamine B assay (34). Nine hundred cells were plated in a 96-well plate (one well per condition; each condition was tested in triplicate). Sulforhodamine B-stained cells were quantified with a Synergy (BioTek) plate reader. Survival was reported as optical density (OD) at 564 nM for the experimental wells divided by that for control well (cells without cisplatin treatment).
To quantify cisplatin resistance in the PBMC, an alternative procedure was chosen owing to poor viability of the cells. After 24 h of RS-1 treatment, the cells were incubated for 24 h with increasing concentrations of cisplatin. The viability was then directly evaluated by measuring the production of formazan catalyzed by the mitochondrial reductase in living cells from the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) substrate. Survival was reported as the OD at 492 nM (Synergy [BioTek] plate reader) for the cisplatin-treated cell fraction (preincubated of not with RS-1) divided by that for untreated cells.
Ethics statement.
Human PBMC were obtained from an HIV-1-seronegative donor from Etablissement Français du Sang (EFS) Aquitaine-Limousin. EFS has approved the study, and a written informed consent was provided by study participants.
RESULTS
hRAD51 specifically inhibits HIV-1 IN in vitro.
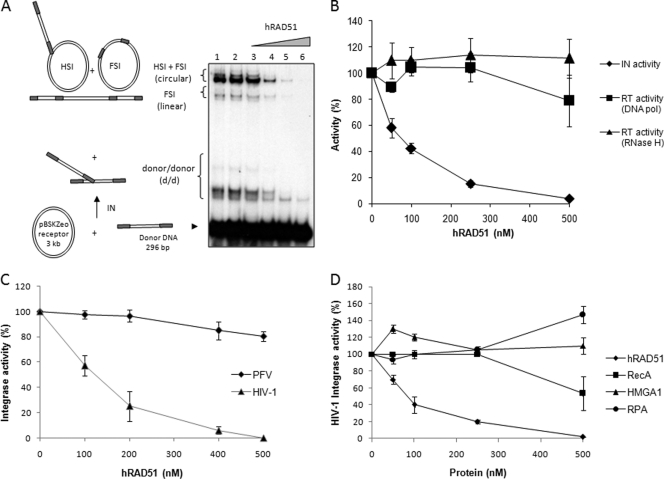
Increasing concentrations of purified human hRAD51 added to a concerted integration reaction mixture (24) under conditions allowing the formation of an active nucleofilament (presence of 100 μM ATP, as previously determined [6]) induce a strong inhibition of all the activities (autointegration and half-site and full-site concerted integration) catalyzed by the recombinant HIV-1 IN (50% inhibitory concentration [IC50] in the 75 to 125 nM range) (Fig. 1A; quantification in Fig. 1B). No direct inhibition of IN was observed with ATP (Fig. 1A, lane 2 compared to lane 1). The specificity of this in vitro inhibition was analyzed by testing the effect of hRAD51 on other HIV-1 enzymes acting on DNA. Since the core domain of IN and the RNase H domain of the HIV-1 reverse transcriptase (RT) have closely related structures (42), we assayed the effect of hRAD51 on both RNase H and DNA polymerase activities of recombinant purified RT. Reactions were performed under the standard conditions described in Materials and Methods, in the presence of increasing amounts of hRAD51. Results clearly showed that hRAD51 had no effect on in vitro RT activities within the IN inhibition concentration range (Fig. 1B). To better determine the specificity of this inhibition, we further tested the effect of hRAD51 on another retroviral integrase: the PFV integrase. A concerted integration reaction performed with the recombinant purified PFV integrase in the presence of hRAD51 showed that the DNA repair factor did not inhibit PFV in vitro integration compared to that of HIV-1 (Fig. 1C). Taken together these results suggested that inhibition by hRAD51 was due to the specific action of hRAD51 on the integrase protein in the HIV-1 active integration complex.
Fig 1.
HIV-1 integrase-specific inhibition by hRAD51. (A) Effect of hRAD51 on in vitro concerted integration catalyzed by HIV-1 integrase. The concerted integration assay was performed using 600 nM IN, 150 ng of acceptor plasmid, 15 ng of unprocessed donor DNA without ATP (lane 1), and 100 μM ATP in the absence (lane 2) or in the presence of 50, 100, 250, or 500 nM recombinant hRAD51 (lanes 3 to 6). The reaction products were loaded on 1% agarose gel. The positions and the structures of the different products obtained after half-site (HSI), full-site (FSI), and donor/donor integration are reported. (B) Comparison of hRAD51 effects on HIV-1 IN and reverse transcriptase in vitro activities. The concerted integration assay was performed as reported for panel A, and integration products (circular HSI and FSI plus linear FSI forms) were quantified. The hRAD51 in vitro effect on the HIV-1 reverse transcriptase activity was checked in standard DNA-dependent DNA polymerase (DNA pol) and RNase H reactions and compared to the IN inhibition. (C) Comparison of hRAD51 effect on HIV-1 and PFV retroviral integrase in vitro integration activity. The concerted integration assay and quantification of integration products were performed as reported for panel A. The PFV integration assay was performed under the same conditions as for HIV-1 but using specific 296-bp PFV donor substrate, as reported in Materials and Methods. (D) Comparison of effects of several DNA binding proteins on HIV-1 IN in vitro activity. The concerted integration assay was performed as for panel A but using increasing concentrations of either bacterial hRAD51 counterpart RecA, human HMGA1, human RPA, or hRAD51 proteins. Integration products were quantified on gel as reported before. All the quantifications shown in panels B, C, and D are reported as the means from at least three independent experiments ± standard deviations (error bars). In the quantification data reported in panels B, C, and D, 100% activity corresponds to the activity observed in the absence of hRAD51 or DNA binding proteins.
To determine whether IN inhibition by hRAD51 could be due to the nonspecific DNA binding property of the added cellular factor, we tested several eukaryotic and prokaryotic proteins also displaying unspecific interaction with DNA. We first assayed the bacterial RecA homolog of hRAD51, previously shown to interact in vitro with HIV-1 IN (33) and sharing close DNA binding properties with hRAD51 (39). As reported in Fig. 1D, RecA was able to inhibit IN but with a lower efficiency than its human counterpart. In contrast, when the human dsDNA binding protein HMG1A or the ssDNA binding protein RPA was tested, no inhibition was observed. This indicates that the IN inhibition observed with the RAD enzymes was specific to this type of DNA repair factor and not due to nonspecific DNA interactions.
The hRAD51 nucleofilament inhibits HIV-1 IN in vitro.
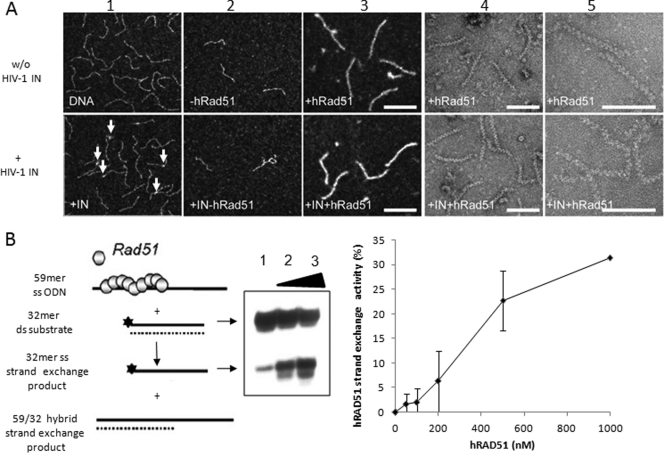
A prerequisite step in hRAD51-mediated recombination involves the ATP-dependent polymerization of hRAD51 on its single-stranded DNA substrate, following its initial binding to nucleotides (nucleation), leading to the formation of an active presynaptic nucleofilament (for a recent review see reference 37). Thus, we further studied whether this active nucleoprotein complex could also be formed on the double-stranded DNA integration substrate used under our IN inhibition conditions. For this purpose we first sought to detect this complex by electron microscopy in the presence or absence of IN. As reported in Fig. 2A, a typical hRAD51 nucleofilament was found on the viral DNA integration substrate both in the absence and presence of HIV-1 IN, strongly suggesting that hRAD51 could form this crucial recombination intermediate on the double-stranded integration substrate. Next, we investigated whether this nucleoprotein complex was active under our concerted integration reaction conditions. For this purpose we performed a standard hRAD51-mediated strand exchange reaction as previously described (40) but adapted to our conditions of concerted integration inhibition. As shown in Fig. 2B, hRAD51 was found to be active under these conditions, confirming that the protein could form a fully active nucleofilament in our concerted integration inhibition reactions.
Fig 2.
hRAD51 forms active nucleofilaments under in vitro integration conditions. (A) Transmission electron microscopy analysis of IN-hRAD51-DNA complexes. Electron microscopy analysis was performed as reported in Materials and Methods in the presence (+ HIV-1 IN) or absence (w/o HIV-1 IN) of HIV-1 IN (600 nM) (or the corresponding storage buffer). Preincubation was with the 296-bp donor DNA (1.5 μM) containing the 20 final nucleotides from U3 and U5 LTR sequences. (1) Control DNA before reaction in the presence (bottom) or absence (top) of IN after 20 min on ice. Samples were observed in the annular dark-field mode for positive staining. Magnification, ×85,000. Discrete nucleoprotein complexes could be observed (some examples are indicated by arrows). (2) DNA after reaction (top) and in the presence of IN (bottom) (a few figures corresponding to possible integration events could be observed, but binding could no longer be seen, as it seems quite dynamic and does not support dilution in our hands). Samples were observed in the annular dark-field mode for positive staining. Magnification, ×85,000. (3 to 5) DNA after reaction in the presence of hRAD51 (top) and in the presence of IN and RAD51 (bottom). (4 and 5) Samples were observed in the bright-field mode for negative staining. Magnifications, ×85,000 (3) and ×140,000 (4 and 5). Bars, 100 nm. (B) Strand exchange activity of hRAD51 nucleofilament. A standard strand transfer activity was performed in the presence of 100 μM ATP and in the absence (lane 1) or presence of 250 (lane 2) or 500 nM (lane 3) hRAD51 (a schema of the reaction is provided). Products were loaded on nondenaturing 12% polyacrylamide gel. The single-stranded (ss) strand-exchanged product is shown as well as the double-stranded (ds) 32-mer substrate. Quantifications from three representative strand exchange reactions performed with increasing concentrations of hRAD51 are shown at the right as means ± standard deviations (error bars) of percentages of 32-mer ss strand exchange product formed.
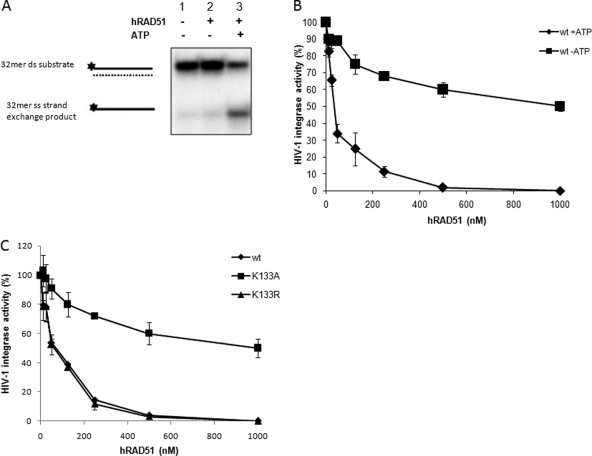
To determine the importance of this hRAD51 nucleofilament in the IN inhibition mechanism, we tested the hRAD51 efficiency for inhibiting integration under several well-defined hRAD51 activity-promoting or -restricting conditions. ATP was previously shown to be required for the formation of an active nucleofilament (6). hRAD51 activity was also found to require ATP under our conditions since strand exchange was observed only in the presence of ATP (Fig. 3A). Interestingly, the omission of ATP in the hRAD51-mediated IN inhibition reaction strongly decreased the negative effect of the cellular factor, confirming that the IN inhibition property of hRAD51 also required ATP (Fig. 3B). To confirm that the fully active protein was required for optimal IN inhibition and to define more precisely the role of hRAD51 polymerization on DNA in the IN inhibitory effect, we analyzed well-characterized ATP hydrolysis mutants of hRAD51 that were still able to bind IN (data not shown) but that presented different activities on DNA (see the analysis of the mutant activities on DNA in reference 6). As reported in Fig. 3C, the K133A mutant, which remains able to bind DNA but is unable to form an active nucleofilament due to the poor binding of ATP (6), was defective for IN inhibition. We also tested the K133R protein, which also carries a punctual mutation on the same amino acid, is unable to hydrolyze ATP but still able to form stable RAD51 filaments, and presents a level of recombination activity similar to that of the wild-type enzyme (6). In this case we observed a level of inhibition similar to that detected for the wild-type protein (Fig. 3C). The slight inhibition remaining due to the lack of ATP or the use of the K133A mutant indicates that a minor part of the inhibition process could involve the initial binding of hRAD51 on DNA and/or its interaction with IN. However, the observed difference in inhibition level between wt and K133A hRAD51 enzymes demonstrates that the active hRAD51-DNA complex is highly required for an optimal IN inhibition.
Fig 3.
Involvement of hRAD51 nucleofilament in inhibition of integration in vitro. (A) ATP-dependent strand exchange activity. The hRAD51 strand transfer reaction (left panel) was performed in the presence of 500 nM hRAD51 and in the absence or presence of 100 μM ATP. Products were loaded on nondenaturating 12% polyacrylamide gel. The single-stranded (ss) strand exchange product is shown, as well as the double-stranded (ds) 32-mer substrate. (B) ATP-dependent HIV-1 IN inhibition by hRAD51. Concerted integration reactions were performed under standard conditions with increasing concentrations of wild-type hRAD51 in the absence or presence of 100 μM ATP. Integration products were quantified on agarose gel. Values are percentages of donor DNA integrated as circular HSI and FSI plus linear FSI forms. (C) Inhibitory effect of hRAD51 mutants defective for DNA binding and/or nucleofilament formation on HIV-1 IN inhibition. Concerted integration reactions were performed under standard conditions in the presence of increasing concentrations of wild-type protein or K133R and K133A RAD51 mutants and 100 μM ATP. The reaction products were loaded on 1% agarose gel. Integration products were quantified on agarose gel. Values are percentages of donor DNA integrated as circular HSI and FSI plus linear FSI forms. The data reported are the means from at least three independent experiments ± standard deviations (error bars).
The mechanism of IN inhibition by hRAD51 involves dissociation of the retroviral enzyme from its complex with the viral DNA substrate by formation of an active nucleofilament.
A powerful in vitro property of protein-DNA displacement was reported for hRAD51, as demonstrated recently on nucleosomal templates (13). We thus wondered whether this mechanism could also occur on the IN-DNA complexes and be responsible for the IN inhibition. Electron microscopy analyses of the hRAD51 polymerization on DNA in the presence of IN (Fig. 2A) showed that preformation of IN-DNA complexes prior to hRAD51 incubation did not affect either the formation or the structure of the nucleofilament. This result supported the hypothesis of an IN displacement by hRAD51 polymerization on DNA. However, the method did not allow us to clearly show a direct dissociation of the IN-DNA complexes. Thus, in order to demonstrate this displacement, we set up the dissociation experiment described in Materials and Methods and Fig. 4A, derived from the hRAD51-DNA dissociation system reported by Chi et al. (6). Briefly, IN was bound to biotinylated DNA coupled to streptavidin magnetic beads prior to hRAD51 treatment. Coupling of functional IN-DNA complexes was checked by performing an integration reaction on the activated beads. This control showed that the 296-bp viral ends containing DNA bound to the beads and associated with wt IN could be integrated efficiently into a short acceptor DNA (Fig. 4B). The structure of the integrated product was further checked by polyacrylamide gel analysis (data not shown), confirming that integration was catalyzed by active complexes coupled to the beads.
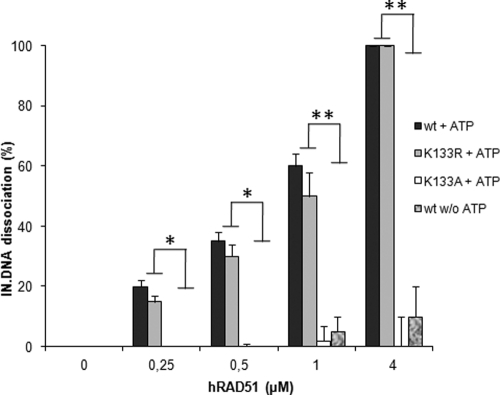
Experiments performed under conditions allowing the formation of the hRAD51 nucleoprotein filament (i.e., using wild-type hRAD51 and 100 μM ATP) showed that the addition of hRAD51 to the IN-DNA complex linked to the beads led to its dissociation and the release of the retroviral enzyme into supernatant fractions (Fig. 4C). An integration assay performed on the activated beads after hRAD51 dissociation showed that the reaction was also strongly inhibited, following a pattern inversely correlated with the IN-DNA dissociation efficiency (Fig. 4D). The involvement of the active hRAD51 nucleofilament in the dissociation process was further studied by using the two previously described K133R and K133A mutants and by omitting ATP in the reaction conditions. As reported in Fig. 5, a weak dissociation effect was observed in the absence of ATP or by using the hRAD51-defective K133A mutants, in contrast to the active wt and K133R proteins.
Fig 5.
The active hRAD51 nucleofilament dissociates IN-DNA complexes. Quantification of the released IN after treatment of the beads with wt hRAD51 and K133R and K133A mutants in the presence or absence of 100 μM ATP was performed by scanning the IN corresponding bands in the Western blot analyses reported in Fig. 4. The data shown are the means from three independent experiments ± standard deviations (error bars). A Student test was performed on serial values. *, P < 0.05; **, P < 0.005.
The observed close correlation between IN dissociation efficiency and the integration restriction property of hRAD51 strongly suggests that the cellular factor dissociates functionally relevant IN-DNA complexes and that this mechanism is probably responsible for the integration inhibition by the recombinase.
Stimulation of hRAD51 activity inhibits HIV-1 integration both in vitro and in infected cells.
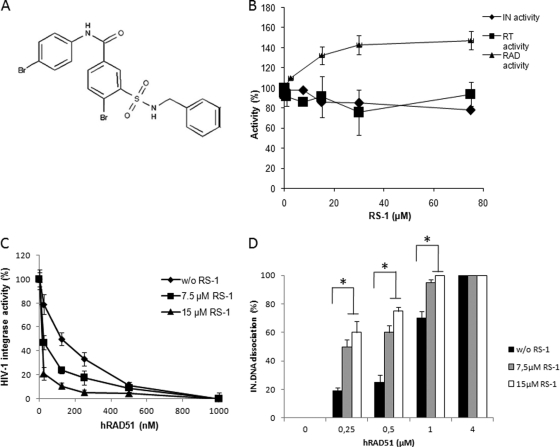
The above-mentioned results showing that hRAD51 was able to inhibit HIV-1 integration by dissociation of the IN-DNA complex may lead to a new therapeutic approach. Thus, it seemed interesting to investigate whether this observation was also true in infected cells and whether the stimulation of hRAD51 could intensify the restriction effect. Enhancement of hRAD51-DNA nucleofilament formation has been shown to be promoted by chemical compounds, such as the previously published hRAD51-stimulatory compound 1 RS-1 (18), whose structure is reported in Fig. 6A. This compound was shown to act presumably as an allosteric effector promoting the formation of the active nucleofilament and specifically stimulates hRAD51 activity both in vitro and in fibroblast cells (18). This hRAD51 stimulation was also observed under our in vitro conditions (Fig. 6B). In contrast, RS-1 did not affect either the HIV-1 IN or RT in vitro activity (Fig. 6B), allowing us to use this molecule as a molecular tool for dissecting hRAD51-mediated IN inhibition.
Fig 6.
In vitro effect of hRAD51 activity stimulation by RS-1 on its integration inhibition property. (A) Chemical structure of RS-1 {3-[(benzylamino)sulfonyl]-4-bromo-N-(4-bromophenyl)benzamide} molecule. (B) In vitro effect of RS-1 on HIV-1 IN, reverse transcriptase, and hRAD51 enzymes. The effect of RS-1 on in vitro integration was analyzed by using the concerted integration system with 600 nM IN, 150 ng of acceptor plasmid, and 15 ng of donor DNA. The effect of RS-1 on the RT enzyme was analyzed in a typical RNase H assay. The RS-1 effect on hRAD51 was measured in the strand exchange reaction shown in Fig. 2B in the presence of 500 nM hRAD51. The data reported are the means from at least two representative independent experiments ± standard deviations (error bars). One hundred percent corresponds to the activity observed in the absence of RS-1. (C) Effect of RS-1 on in vitro inhibition of HIV-1 IN activity by hRAD51. Increasing concentrations of wt hRAD51 were added in a standard concerted integration assay in the presence of 100 μM ATP and in the absence or presence of 7.5 or 15 μM RS-1. The data reported are the means from at least three independent experiments ± standard deviations (error bars). Integration is shown as the percentage of donor DNA integrated as circular HSI and FSI plus linear FSI forms. (D) Effect of RS-1 on hRAD51-mediated dissociation of the IN-DNA complex. The dissociation experiment was performed as shown in Fig. 4 with increasing concentrations of hRAD51 in the presence of 100 μM ATP and with or without 7.5 or 15 μM RS-1. The data reported are the means from at least three independent experiments ± standard deviations (error bars). A Student test was performed on serial values. *, P < 0.05.
Addition of RS-1 to hRAD51 in an in vitro integration assay increased its IN inhibition efficiency (Fig. 6C) (P < 0.05). This indicated that the inhibition property of hRAD51 can be promoted, at least in vitro, by RS-1. Dissociation experiments performed as reported in Fig. 5 but in the presence of RS-1 showed that the molecule also stimulated the hRAD51-mediated IN-DNA dissociation process (Fig. 6D), confirming that the molecule enhances IN inhibition by hRAD51 via the simulation of IN releasing from its substrate.
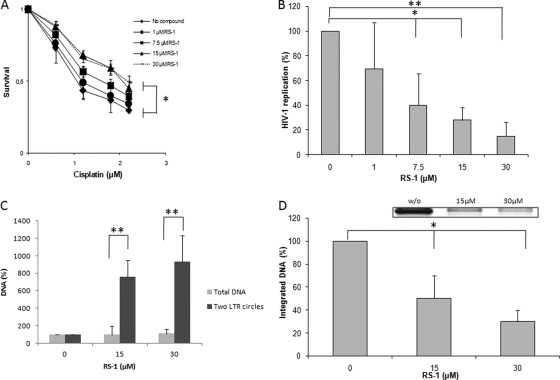
Since RS-1 was previously shown to be efficient for stimulating hRAD51 activity in human cells (18), we wondered whether it could also promote the hRAD51-mediated inhibition of IN in HIV-1-infected cells and hence retroviral replication. RS-1 treatment was first used to specifically stimulate the activity of the endogenous hRAD51 in the broadly used HeLa P4 cell model. To determine the effect of this RS-1 treatment on the cellular DNA repair processes mediated by HR catalyzed by hRAD51, we performed a standard cisplatin resistance assay as reported before using the sulforhodamine B method (18). Figure 7A indicates that a 24-h treatment with increasing concentrations of RS-1 ranging from 7.5 to 15 μM promoted an increased resistance to the cross-linking agent. No effect was observed at a concentration below 7.5 μM, and concentrations above 15 μM did not increase cisplatin resistance. These data indicate that RS-1 is capable of gaining intracellular access and increasing DNA repair efficiency in our model system, as previously reported in fibroblasts (18). Importantly, within the range of concentrations analyzed, RS-1 did not show any cytotoxicity, as measured by the standard MTT method (data not shown). We thus analyzed the effect of RS-1 treatment on HIV-1 replication. Increasing concentrations (0 to 30 μM) of RS-1 were added 24 h prior to infection under the same conditions as those inducing cisplatin resistance. As reported in Fig. 7B, RS-1 induced a dose-dependent inhibition of HIV-1 replication and 30 μM RS-1 treatments induced 80% inhibition of HIV-1 replication in this cell type.
Fig 7.
Effect of RS-1 on HIV-1 replication and integration in HeLa P4 cells. (A) Effect of RS-1 treatment on hRAD51 DNA repair endogenous activity. The ability of RS-1 to promote cisplatin resistance was checked in a standard survival analysis after 24 h of treatment with increasing concentrations of the compound. (B) Effect of RS-1 treatment on HIV-1 replication in HeLa P4 cells. The effect of RS-1 on early steps of HIV-1 replication was measured by 4-MUG quantification of the β-galactosidase activity 48 h postinfection (MOI = 0.4) after 24 h of pretreatment with increasing RS-1 concentrations. (C) Effect of RS-1 on amount of two-LTR circles. The total and two-LTR circle viral DNA populations were quantified by quantitative PCR as described in Materials and Methods for two effective concentrations of RS-1. Data are reported as the ratios between the percentages of two-LTR circles and total DNA. (D) Effect of RS-1 on integrated viral DNA amount. The integrated DNA was amplified by specific PCR as reported in Materials and Methods and quantified on agarose gel (an example of amplification results obtained in the absence [w/o] or presence of 15 or 30 μM RS-1 treatment is shown). One hundred percent corresponds to the integrated DNA quantification obtained without RS-1 treatment. All results are the means from at least three representative independent experiments ± standard deviations (error bars). A Student test was performed on serial values. *, P < 0.05; **, P < 0.005.
The retroviral replication step targeted by the treatment was next determined by quantification of the different viral DNA populations. As reported in Fig. 7C, the level of two-LTR DNA increased significantly at 15 μM or 30 μM RS-1 without affecting the amount of total viral DNA, strongly suggesting a specific inhibition of the HIV-1 integration step by RS-1 treatment. This is supported by the in vitro data reported in Fig. 6B showing that RS-1 did not induce any direct effect on reverse transcriptase. However, to demonstrate unambiguously that the integration step was specifically impaired by RS-1 treatment, the integrated DNA was amplified by nested PCR using primers specific to this form as described in Materials and Methods. As shown in Fig. 7D, RS-1 treatment induced a strong decrease in the amount of amplified integrated DNA.
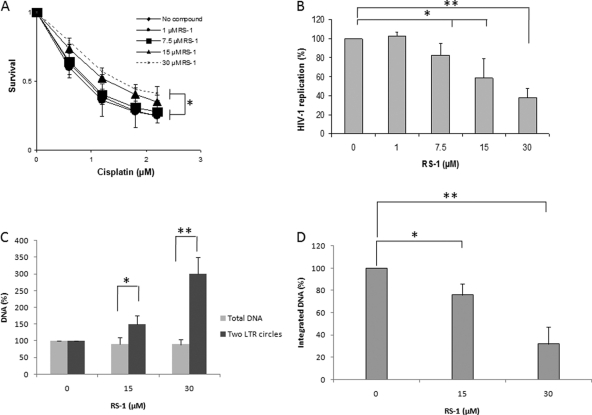
The cellular model used here is a transformed cell line where the endogenous hRAD51 activity could be different from that in naturally HIV-1-infected cells (17; reviewed in reference 20). In order to determine whether hRAD51-dependent restriction of viral replication could be promoted in natural HIV-1 infection conditions, we tested the RS-1 effect in primary peripheral blood mononuclear cells (PBMC) (Fig. 8). Analysis of the cisplatin resistance of cells treated with RS-1 showed that hRAD51 activity could also be stimulated in this cellular type but with a lower efficiency than in transformed HeLa P4 cells (compare Fig. 8A and 7A). This could be due to the difference in endogenous hRAD51 recombination levels existing in these cells (which appear more sensitive to cisplatin). As in HeLa P4 cells, no significant effect of RS-1 treatment on cell survival was detected (data not shown). The effect of RS-1 on HIV-1 replication in these cells was evaluated by addition of the molecule 24 h before infection and quantification of the release of genomic RNA using standard procedures described in Materials and Methods. As reported in Fig. 8B, inhibition of replication by RS-1 was also observed in these cells. The level of inhibition was lower than in HeLa P4 cells, since a 30 μM RS-1 treatment induced a 60% inhibition of HIV-1 replication. This is in agreement with the lower effect of the molecule on HR activity, as shown by the lower effect of RS-1 on cisplatin resistance in these cells (Fig. 8A). Accumulation of the two-LTR circles confirmed that the integration step was also impaired by the compound in these cells (Fig. 8C). To unambiguously determine the viral replication step impaired by the treatment, we quantified the integrated DNA by specific quantitative PCR. Results shown in Fig. 8D indicate that treatment with the RS-1 compound led to a decrease of the integrated DNA amount. These data, in addition to the 2-LTR accumulation, demonstrate that the integration step was also inhibited in PBMC by the molecule.
Fig 8.
Effect of RS-1 on HIV-1 replication and integration in PBMC. (A) Effect of RS-1 treatment on hRAD51 DNA repair endogenous activity. Cells were incubated for 24 h with increasing concentrations of RS-1. The effect of RS-1 on hRAD51 activity was then checked by quantifying its ability to promote cisplatin resistance in a standard survival analysis, as reported above, with some adjustment due to the property of the primary cells. (B) Effect of RS-1 treatment on HIV-1 replication in PBMC. PBMC were incubated for 24 h in media containing various concentrations of cisplatin in the presence of RS-1. Drugs were then removed, and cell survival was evaluated by an MTT assay as described in Materials and Methods. The effect on HIV-1 replication of a 24-hour treatment of the infected cells with RS-1 prior to infection (MOI = 0.1) was measured by quantifying the genomic RNA released in the medium 48 h postinfection by quantitative PCR. (C) Effect of RS-1 on the amount of two-LTR circles. Total DNA and two-LTR circles were quantified by quantitative PCR as described in Materials and Methods for two effective concentrations of RS-1. (D) Effect of RS-1 on integrated viral DNA amount. Integrated DNA was measured by quantitative PCR as reported in Materials and Methods. One hundred percent corresponds to the integrated DNA quantification obtained without RS-1 treatment. All results are the means from at least three representative independent experiments ± standard deviations (error bars). A Student test was performed on serial values. *, P < 0.05; **, P < 0.005.
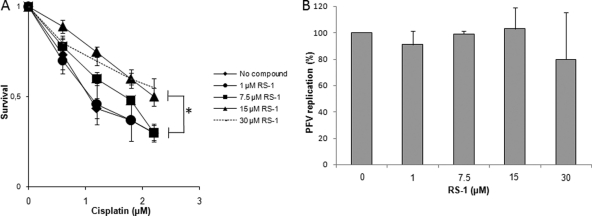
Finally, in order to determine the specificity of the effect of RS-1 treatment on HIV-1 replication, we tested the effect of the molecule on infection by PFV, whose IN was previously shown to be resistant in vitro to the restriction property of this cellular factor (cf. Fig. 1C). The infection assay was performed in HeLa cells; RS-1 treatment was also found to stimulate hRAD51-mediated DNA repair (Fig. 9A). As reported in Fig. 9B, no significant inhibition of PFV was observed with RS-1 compared to HIV-1 inhibition, confirming the specificity of integration inhibition by hRAD51 for HIV-1.
Fig 9.
Effect of RS-1 on PFV replication in HeLa cells. (A) Effect of RS-1 treatment on hRAD51 DNA repair endogenous activity. The ability of RS-1 to promote cisplatin resistance was checked in a standard survival analysis after 24 h of treatment with increasing concentrations of the compound. (B) Effect of RS-1 treatment on PFV replication in HeLa cells. HeLa cells pretreated or not with increasing concentrations of RS-1 were infected with PFV at a MOI of 0.4. Replication was quantified by fluorescence-activated cell sorting (FACS) 48 h postinfection. All results are the means from at least three representative independent experiments ± standard deviations (error bars). A Student test was performed on serial values. *, P < 0.05.
DISCUSSION
Our previous research using the yeast model system suggested a downregulation of HIV-1 IN by the DNA repair factor RAD51 (12). In the present work we used biochemical procedures to elucidate the molecular mechanism involved in this process. The effect of hRAD51 on IN was analyzed in vitro by using the concerted integration assay in the presence of purified recombinant hRAD51 protein. The in vitro IN inhibition assays performed under conditions promoting the formation of the hRAD51 nucleoprotein filament, required for efficient recombination activity, i.e., presence of ATP and hRAD51-stimulatory compound RS-1 addition, showed stimulated hRAD51 inhibition efficiency. In contrast, the reaction conditions that did not allow the formation of the active nucleoprotein filaments, i.e., lack of ATP or use of a K133A hRAD51 mutant defective for active polymerization on DNA, strongly impaired hRAD51 inhibition efficiency. Interestingly, the hRAD51 K133A mutant, which is still able to bind DNA and IN but is unable to form an active nucleoprotein filament, lost a significant fraction of inhibitory activity (Fig. 3C). These observations suggest that the IN binding property of hRAD51 is not sufficient for fully efficient inhibition and that, even if the initial nucleation of hRAD51 on DNA can induce a weak inhibition of IN activity, as observed with the K133A mutant, the formation of the active hRAD51 nucleoprotein filament is very necessary for fully efficient integration inhibition. This result was confirmed by the strong decrease of hRAD51 IN inhibition in the absence of ATP, a condition allowing the binding of the protein to DNA but impairing its polymerization (6). The remaining inhibition of IN by the K133A enzyme and in the absence of ATP suggests that this mechanism could also involve the initial nucleation of hRAD51 on DNA and/or its binding to IN. However, the poor inhibition efficiency found under these conditions indicates that the main component for optimal inhibition is the formation of the active nucleofilament. This requirement highlights the specificity of IN inhibition by hRAD51 and rules out a mechanism involving an unspecific coating of the dsDNA. This was confirmed by the fact that other DNA binding proteins tested in the integration assay did not show any inhibition of retroviral IN (Fig. 1D). Strikingly, hRAD51 was unable to inhibit the PFV retroviral integrase (Fig. 1C), again underlying the specificity of the action of this repair enzyme on the HIV-1 integration complex. Additionally, since no interaction between the PFV integrase and hRAD51 was detected (data not shown), and in view of the high structural and functional similarities between PFV and HIV-1 proteins (15, 21, 28), our data suggest that the physical interaction between HIV-1 IN and hRAD51 is important for inhibition. However, determination of the interaction domains between both proteins by direct dissociation approaches using mutants or peptide derivates from the association domains (under way in our laboratory) would be helpful for a better understanding of the role of this interaction.
Electron microscopy showed that the formation of the structured hRAD51 nucleoprotein filament was not affected by the presence of IN (Fig. 2A), suggesting that hRAD51 could polymerize on an IN-DNA complex. Treatment of magnetic beads coupled to the IN-DNA complex with hRAD51 under conditions promoting the formation of the active presynaptic filament led to a dose-dependent dissociation of IN from its complex with DNA (Fig. 4C). An integration reaction performed on the activated beads after hRAD51 treatment or without hRAD51 treatment indicated that active IN-DNA complexes were dissociated by the cellular enzyme (Fig. 4D). Owing to the constant correlation between the ability of hRAD51 to form an active nucleoprotein filament and its IN inhibition property, in addition to its capacity to dissociate the active IN-DNA complex, we conclude that hRAD51 can inhibit integration by remodeling the functional IN-DNA complex and releasing the retroviral enzyme from its substrate.
Since the HIV-1 integration inhibition property of hRAD51 could constitute a new antiviral approach, we then studied the possibility of promoting this inhibition in infected cells. For this purpose we used the RS-1 compound specifically stimulating the formation and stability of the hRAD51 nucleofilament (18). Under our conditions, RS-1 was also found to induce a strong increase in the in vitro hRAD51-mediated inhibition of IN activity, without having any direct effect on IN alone (Fig. 6B). Treatment of HeLa P4 HIV-1-infected cells induced both stimulation of hRAD51-mediated HR and inhibition of viral replication (Fig. 7A and B). PCR quantification of the different DNA species after hRAD51 overexpression or RS-1 treatment of HIV-1-infected cells showed an accumulation of two-LTR circles without changes in the total viral DNA amount. These data indicated that the enhancement of homologous recombination mediated by hRAD51 induced the inhibition of integration efficiency without affecting the entry of virions, RNA reverse transcription, or DNA nuclear import. This hypothesis was supported by in vitro RT assays indicating that neither the RNase H activity nor the DNA polymerase activity catalyzed by the recombinant RT was affected by hRAD51 (Fig. 1B). The detection and quantification of the integrated viral DNA unambiguously showed a strong decrease in this population after RS-1 treatment (Fig. 7D). Therefore, we conclude that stimulation of the recombination activity catalyzed by hRAD51 in HIV-1-infected cells specifically inhibited the integration steps via the formation of an active nucleofilament. Furthermore, this HIV-1 replication inhibition was also observed after RS-1 treatment in other HIV-1-infected cells such as 293T and MT4 systems (data not shown). More importantly the similar results obtained in primary PBMC confirm that the inhibition effect can be reproduced in more-physiological models independently from their transformation state (Fig. 8). Quantification of the integrated DNA clearly demonstrated that the integration step was also inhibited in these primary cells and, thus, prior to Tat-dependent LTR transcription that was previously reported to be activated by hRAD51 (7, 35). These data indicate that the hRAD51-mediated inhibition of integration is not cell dependent. However, the difference in inhibition efficiency between all those cell models suggests that inhibition efficiency could depend on the cell line.
Previous experiments performed in other mammalian cell lines showed that the overexpression of hRAD51 increased resistance to ionizing radiation via an enhancement of the homologous recombination mediated by this factor (see above). Our data strongly suggest that the same mechanism could occur in HIV-1-infected cells if hRAD51 is stimulated, leading to resistance to retroviral integration. This was supported by hRAD51 overexpression experiments performed in HeLa P4 and 293T HIV-1-infected cells showing both stimulation of the endogenous DNA repair mechanism and HIV-1 replication inhibition, as observed with RS-1 treatment (data not shown). Whether the IN inhibition property of hRAD51 under stimulation conditions reported here signifies a possible regulatory function of this factor under normal infection conditions remains unclear. Even though this point is under study in our laboratory, several hypotheses can be proposed based on the molecular mechanism elucidated here and the cellular function known for hRAD51. Early interaction of hRAD51 with the free incoming intasome and with the host DNA-bound intasome before integration could lead to the displacement of IN from its substrate by hRAD51 polymerization, thereby limiting integration and replication. This could be promoted by simulating the activity of the factor. In contrast, a later interaction of hRAD51 with the integration complex, i.e., after the catalysis of the strand transfer by HIV-1 IN, could lead to the release of IN from the integration loci via the formation of the nucleofilament and the dissociation mechanism reported in Fig. 4. It has been shown that the postintegration repair processes (action of FEN1 and polymerases) cannot occur in vitro when IN is bound to its integration product and may require the active disassembly of the IN complex (3, 43), as has been previously reported for other transposases (25). Additionally, the postintegration mechanism also requires the disassembly of the IN complex and its address to the proteasome in infected cells (30). Finally, recent data from our laboratory highly suggest that integration into chromatin could require dynamic nucleosomes at the integration locus (23). The chromatin remodeling property of hRAD51 previously reported (13) could, thus, also play a role during the interaction of the incoming integration complex with suitable chromosomal loci.
Such late action of hRAD51 at the postintegration step could thus improve the establishment of a stable insertion of the viral genome into the host DNA. Under physiological conditions, i.e., without hRAD51 stimulation, the function of hRAD51 could result from this equilibrium between its pro- and anti-integration properties. All these hypotheses remain to be studied under physiological in vivo conditions or with in vitro models reproducing more accurately the integration process. More especially, nucleosomal templates and LEDGF/p75 that could participate in or modulate hRAD51 activity on retroviral integration should be used. Indeed LEDGF/p75 may stabilize the intasome, modulating its dissociation by hRAD51 during native replication conditions.
Independently of the possible function of hRAD51 during HIV-1 replication, the promotion of its integration-inhibitory property reported and elucidated here could serve as a basis for a new antiviral therapy. The feasibility of this concept is strongly supported by our results obtained with the hRAD51-stimulatory agent RS-1 especially in PBMC including the physiological target of the HIV-1 infection. The mechanism of inhibition of HIV-1 replication illustrated in this work and the assay of new hRAD51 stimulation agents are under active study, which should lead to the improvement and optimization of such a therapeutic strategy in the future.
ACKNOWLEDGMENTS
We are deeply grateful to R. Cooke (University Bordeaux 2) for proofreading the manuscript.
No conflict of interest is declared.
This work was supported by the French Agence Nationale de Recherche contre le SIDA, the Centre National de la Recherche Scientifique, SIDACTION, and the University Bordeaux Segalen (to V.P. and M.A). A. De Cian is a recipient of a grant from the Association pour la Recherche contre le Cancer. P. Sung's grant number is RO1ES015252. P. P. Connell's NIH grant number is CA142642-02.
V. Parissi designed and performed the experiments, analyzed the results, and wrote the manuscript. O. Cosnefroy, A. Tocco, P. Lesbats, S. Thierry, O. Delelis, C. Calmels, S. Reigadas, A. De Cian, S. Desfarges, T. Wiktorowicz, and P. Bonot performed the experiments. Y. Kwon, J. San Filippo, A. Rethwilm, P. P. Connell, and P. Sung provided the reagents and analyzed the results. M. L. Andréola, E. Le Cam, H. Fleury, and S. Litvak analyzed the results.
Footnotes
Published ahead of print 19 October 2011
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Andreola ML, et al. 2001. DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry 40:10087–10094 [DOI] [PubMed] [Google Scholar]
- 2. Beloin C, et al. 2003. Contribution of DNA conformation and topology in right-handed DNA wrapping by the Bacillus subtilis LrpC protein. J. Biol. Chem. 278:5333–5342 [DOI] [PubMed] [Google Scholar]
- 3. Brin E, Yi J, Skalka AM, Leis J. 2000. Modeling the late steps in HIV-1 retroviral integrase-catalyzed DNA integration. J. Biol. Chem. 275:39287–39295 [DOI] [PubMed] [Google Scholar]
- 4. Brussel A, et al. 2003. Longitudinal monitoring of 2-long terminal repeat circles in peripheral blood mononuclear cells from patients with chronic HIV-1 infection. AIDS 17:645–652 [DOI] [PubMed] [Google Scholar]
- 5. Brussel A, Sonigo P. 2003. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J. Virol. 77:10119–10124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. 2006. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 5:381–391 [DOI] [PubMed] [Google Scholar]
- 7. Chipitsyna G, Sawaya BE, Khalili K, Amini S. 2006. Cooperativity between Rad51 and C/EBP family transcription factors modulates basal and Tat-induced activation of the HIV-1 LTR in astrocytes. J. Cell. Physiol. 207:605–613 [DOI] [PubMed] [Google Scholar]
- 8. Clavel F, Charneau P. 1994. Fusion from without directed by human immunodeficiency virus particles. J. Virol. 68:1179–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daniel R, et al. 2001. Wortmannin potentiates integrase-mediated killing of lymphocytes and reduces the efficiency of stable transduction by retroviruses. Mol. Cell. Biol. 21:1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daniel R, Katz RA, Skalka AM. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644–647 [DOI] [PubMed] [Google Scholar]
- 11. Daniel R, et al. 2004. Histone H2AX is phosphorylated at sites of retroviral DNA integration but is dispensable for postintegration repair. J. Biol. Chem. 279:45810–45814 [DOI] [PubMed] [Google Scholar]
- 12. Desfarges S, et al. 2006. Chromosomal integration of LTR-flanked DNA in yeast expressing HIV-1 integrase: down regulation by RAD51. Nucleic Acids Res. 34:6215–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dupaigne P, et al. 2008. Rad51 polymerization reveals a new chromatin remodeling mechanism. PLoS One 3:e3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Espeseth AS, et al. 2011. siRNA screening of a targeted library of DNA repair factors in HIV infection reveals a role for base excision repair in HIV integration. PLoS One 6:e17612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heinkelein M, et al. 2002. Improved primate foamy virus vectors and packaging constructs. J. Virol. 76:3774–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hine CM, Seluanov A, Gorbunova V. 2008. Use of the Rad51 promoter for targeted anti-cancer therapy. Proc. Natl. Acad. Sci. U. S. A. 105:20810–20815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jayathilaka K, et al. 2008. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc. Natl. Acad. Sci. U. S. A. 105:15848–15853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kilzer JM, et al. 2003. Roles of host cell factors in circularization of retroviral dna. Virology 314:460–467 [DOI] [PubMed] [Google Scholar]
- 20. Klein HL. 2008. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 7:686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krishnan L, et al. 2010. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. U. S. A. 107:15910–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lau A, Kanaar R, Jackson SP, O'Connor MJ. 2004. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 23:3421–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lesbats P, et al. 2011. Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLoS Pathog. 7:e1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lesbats P, et al. 2008. In vitro initial attachment of HIV-1 integrase to viral ends: control of the DNA specific interaction by the oligomerization state. Nucleic Acids Res. 36:7043–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levchenko I, Luo L, Baker TA. 1995. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 9:2399–2408 [DOI] [PubMed] [Google Scholar]
- 26. Li L, et al. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lloyd AG, et al. 2006. Effect of DNA repair protein Rad18 on viral infection. PLoS Pathog. 2:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maertens GN, Hare S, Cherepanov P. 2010. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468:326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Metifiot M, et al. 2007. Cellular uptake of ODNs in HIV-1 human-infected cells: a role for viral particles in DNA delivery? Oligonucleotides 17:151–165 [DOI] [PubMed] [Google Scholar]
- 30. Mousnier A, et al. 2007. von Hippel Lindau binding protein 1-mediated degradation of integrase affects HIV-1 gene expression at a postintegration step. Proc. Natl. Acad. Sci. U. S. A. 104:13615–13620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mulder LC, Chakrabarti LA, Muesing MA. 2002. Interaction of HIV-1 integrase with DNA repair protein hRad18. J. Biol. Chem. 277:27489–27493 [DOI] [PubMed] [Google Scholar]
- 32. Mullers E, Stirnnagel K, Kaulfuss S, Lindemann D. 2011. Prototype foamy virus Gag nuclear localization: a novel pathway among retroviruses. J. Virol. 85:9276–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parissi V, et al. 2003. The lethal phenotype observed after HIV-1 integrase expression in yeast cells is related to DNA repair and recombination events. Gene 322:157–168 [DOI] [PubMed] [Google Scholar]
- 34. Pauwels B, et al. 2003. Comparison of the sulforhodamine B assay and the clonogenic assay for in vitro chemoradiation studies. Cancer Chemother. Pharmacol. 51:221–226 [DOI] [PubMed] [Google Scholar]
- 35. Rom I, et al. 2010. Activation of HIV-1 LTR by Rad51 in microglial cells. Cell Cycle 9:3715–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sallafranque-Andreola ML, et al. 1989. Human immunodeficiency virus reverse transcriptase expressed in transformed yeast cells. Biochemical properties and interactions with bovine tRNALys. Eur. J. Biochem. 184:367–374 [DOI] [PubMed] [Google Scholar]
- 37. San Filippo J, Sung P, Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77:229–257 [DOI] [PubMed] [Google Scholar]
- 38. Stirnnagel K, et al. 2010. Analysis of prototype foamy virus particle-host cell interaction with autofluorescent retroviral particles. Retrovirology 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sung P, Klein H. 2006. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7:739–750 [DOI] [PubMed] [Google Scholar]
- 40. Takizawa Y, et al. 2004. Mutational analyses of the human Rad51-Tyr315 residue, a site for phosphorylation in leukaemia cells. Genes Cells 9:781–790 [DOI] [PubMed] [Google Scholar]
- 41. Valkov E, et al. 2009. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 37:243–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang W, Steitz TA. 1995. Recombining the structures of HIV integrase, RuvC and RNase H. Structure 3:131–134 [DOI] [PubMed] [Google Scholar]
- 43. Yoder KE, Bushman FD. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191–11200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yoder KE, et al. 2011. The base excision repair pathway is required for efficient lentivirus integration. PLoS One 6:e17862. [DOI] [PMC free article] [PubMed] [Google Scholar]