Abstract
Adeno-associated virus type 2 (AAV2) is a human parvovirus that relies on a helper virus for efficient replication. Herpes simplex virus 1 (HSV-1) supplies helper functions and changes the environment of the cell to promote AAV2 replication. In this study, we examined the accumulation of cellular replication and repair proteins at viral replication compartments (RCs) and the influence of replicating AAV2 on HSV-1-induced DNA damage responses (DDR). We observed that the ATM kinase was activated in cells coinfected with AAV2 and HSV-1. We also found that phosphorylated ATR kinase and its cofactor ATR-interacting protein were recruited into AAV2 RCs, but ATR signaling was not activated. DNA-PKcs, another main kinase in the DDR, was degraded during HSV-1 infection in an ICP0-dependent manner, and this degradation was markedly delayed during AAV2 coinfection. Furthermore, we detected phosphorylation of DNA-PKcs during AAV2 but not HSV-1 replication. The AAV2-mediated delay in DNA-PKcs degradation affected signaling through downstream substrates. Overall, our results demonstrate that coinfection with HSV-1 and AAV2 provokes a cellular DDR which is distinct from that induced by HSV-1 alone.
INTRODUCTION
Adeno-associated virus type 2 (AAV2) is a small, nonenveloped parvovirus with a single-stranded DNA genome of 4.7 kb (52). In the absence of a helper virus, AAV2 establishes a latent infection characterized by site-specific integration of the viral genome into the AAVS1 site on human chromosome 19 (72). In the presence of a helper virus, AAV2 can replicate productively in the host cell nucleus. AAV2 DNA replication occurs at discrete sites in the nucleus, termed replication compartments (RCs). During the course of infection, several small RCs rapidly expand and fuse to large structures, which displace the cellular chromatin and fill the entire cell nucleus (28, 35, 37, 79, 91). AAV2 RCs contain AAV2 proteins, as well as defined helper virus proteins and cellular proteins (3, 35, 63, 65, 75, 79, 90, 91). Replicating AAV2 has inhibitory effects on both the host cell (9, 41, 68, 71, 73, 74, 100, 101) and the helper virus (5, 30, 31, 34, 40, 44, 61, 84, 100).
One of the helper viruses for AAV2 replication is herpes simplex virus 1 (HSV-1) (14). The minimal HSV-1 helper factors for AAV2 replication from plasmid substrates include the helicase-primase complex encoded by UL5, UL8, and UL52 and the major DNA binding protein ICP8 (3) (90). Besides viral helper factors, the fate of AAV2 replication also depends on cellular proteins. Recently, cellular proteins have been identified that interact with AAV2 Rep78/68 in adenovirus (Ad)- or HSV-1-supported AAV2 replication (63, 65). Of these, the largest functional categories correspond to cellular proteins which are involved in DNA metabolism, including DNA replication, repair, and chromatin modification.
There is accumulating evidence that the DNA damage response (DDR) pathways play central roles in viral replication (92). Control of DDR signaling may be a mechanism to prevent apoptosis and/or stop cell cycle progression (92). For example, DNA damage signaling has been shown to enhance the replication of the autonomous parvovirus minute virus of mice, perhaps in part by promoting cell cycle arrest (1). In response to DNA damage, a complex signaling network is activated that includes kinase regulation, transcriptional induction, and redistribution of a multitude of factors (33, 38). Depending on the extent of DNA damage, cell cycle progression is stopped to repair DNA breaks or apoptosis is induced. Two main pathways are classified for the repair of DNA double-strand breaks, homologous recombination and nonhomologous end joining (16, 36, 99). Proteins which are important for sensing of DNA double-strand breaks include H2AX and the Mre11/Rad50/Nbs1 (MRN) complex (for a review, see reference 47). The phosphatidylinositol-3-kinase-like kinases (PIKKs) ataxia telangiectasia mutated (ATM) and ATM and Rad3 related (ATR) are proximal signaling kinases that have key functions in signaling transduction in homologous recombination (24, 33, 60, 66, 69). ATM is recruited by the MRN complex (for a review, see reference 29) and catalytically activated through dimer dissociation and autophosphorylation at serine 1981 (S1981) (6, 103). Examination of ATR recruitment to sites of DNA damage revealed that binding of ATR to ATR-interacting protein (ATRIP) leads to colocalization of the ATR-ATRIP complex with replication protein A (RPA)-coated single-stranded DNA (7). It has been suggested that interaction of topoisomerase II-binding protein 1 with the ATR-ATRIP complex induces kinase activity of ATR (59). A third PIKK, DNA-dependent protein kinase (DNA-PK), belongs to the nonhomologous end-joining machinery and is composed of the Ku70/Ku80 heterodimer and the catalytic subunit of DNA-PK (DNA-PKcs). Ku70/80 directly recognizes DNA double-strand breaks and activates DNA-PKcs (for a review, see reference 15). Activity of DNA-PKcs is proposed to be regulated by autophosphorylation at several sites, including S2056 (19, 21). Investigation of downstream signaling via PIKKs suggests that checkpoint kinase 1 (Chk1) is mainly a substrate of ATR after the recognition of single-strand breaks and stalled-replication forks (22, 32, 53, 80, 83, 105), while Chk2 activation by ATM is more restricted to double-strand breaks, including those induced by ionizing radiation (2, 20, 42, 43, 56, 57). However, there is evidence that ATR (85, 87) and DNA-PK (50, 85) can also induce Chk2 phosphorylation. DNA-PK (49, 86, 88), ATR (46), and ATM (8) have all been reported to induce phosphorylation of p53.
HSV-1 induces the activation of a cellular DNA double-strand break response pathway involving the MRN complex, ATM, p53, RPA (nonphosphorylated), and Rad51 (13, 51, 76, 96), while the ATR response has been reported to be inhibited (58, 94). Similarly, signaling via DNA-PK is also inhibited by HSV-1 through ICP0-dependent proteasomal degradation of DNA-PKcs (48, 67). It has been shown that in the absence of the Ku70 subunit of the DNA-PK complex, HSV-1 replication is enhanced (81).
AAV2, although not replicating in the absence of a helper virus, induces a strong DDR mediated by ATR (41). A different DDR is induced upon coinfection with a helper virus that supports AAV2 replication; for example, during Ad-supported AAV2 replication, the DDR signaling is mediated primarily by DNA-PK and is independent of the MRN complex (25, 75). Furthermore, activation of ATM, Chk1, Chk2, RPA, and H2AX was also observed (25, 75). Given that DNA-PK is a key kinase in nonhomologous end joining, it seems that these events may play important roles not only in the (site-specific) integration of the AAV2 genome (18, 27, 77) but also in AAV2 genome replication (23). As opposed to Ad and AAV2 coinfection, the DDR induced by coinfection with HSV-1 and AAV2 has not previously been investigated. Thus, the goals of the present study were to identify cellular replication and repair proteins that accumulate at AAV2 RCs when HSV-1 is the helper virus and to determine the effect of AAV2 on the cellular DDR signaling pathways induced by HSV-1.
MATERIALS AND METHODS
Cells.
DNA-PKcs-positive (expressing one copy of DNA-PKcs) and DNA-PKcs-negative HCT116 cells were kindly provided by E. Hendrickson (University of Minnesota Medical School, Minneapolis) and maintained in growth medium containing Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin G, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (1% AB). DNA-PKcs-positive (Fus1) and DNA-PKcs-negative (Fus9) MO59J cells were kindly provided by T. Melendy (Department of Cellular and Molecular Biology, Roswell Park Cancer Institute, Buffalo, NY) and cultured in 50% F10 medium–50% DMEM supplemented with 10% FBS, 1% AB, and 250 μg/ml G418. AT22 IJE-T yZ5 (expressing ATM) and AT22 IJE-T pEBS7 (lacking ATM) fibroblast cells were kind gifts from Y. Shiloh (Department of Molecular Genetics and Biochemistry, Sackler School of Medicine, Tel Aviv University, Tel Aviv, Israel). These cells were maintained in DMEM supplemented with 10% FBS, 1% AB, and 100 μg/ml hygromycin B. U2OS GW33 cells were a kind gift from P. Nghiem (Department of Medicine/Dermatology, University of Washington, Seattle) and were cultured in DMEM containing 10% FBS, 1% AB, 200 μg/ml G418, and 50 μg/ml hygromycin B. Vero cells were maintained in DMEM supplemented with 10% FBS and 1% AB. All cells were maintained at 37°C in a 95% air–5% CO2 atmosphere.
Viruses.
HSV-1 strain F was kindly provided by B. Roizman (Marjorie B. Kovler Viral Oncology Laboratories, University of Chicago, Chicago, IL). rHSV-1 dl1403 (rHSV-1ΔICP0) was kindly provided by N. D. Stow (MRC Virology Unit, University of Glasgow, Glasgow, United Kingdom), and rHSV-1vEYFP-ICP4 and rHSV-1 vECFP-ICP4 were a kind gift R. D. Everett (MRC Virology Unit, University of Glasgow, Glasgow, United Kingdom). Viruses were grown on Vero cells. HSV-1 strain F titers were determined on Vero cells, and rHSV-1 dl1403 titers were determined on U2OS cells. AAV2 and Ad2 were kindly provided by H. Buening (University of Cologne, Cologne, Germany) and U. Greber (University of Zurich, Zurich, Switzerland), respectively. The recombinant AAV2CherryRep (rAAVCR) genome, containing the AAV2 inverted terminal repeats flanking the rep open reading frames fused at its 5′ terminus with the mCherry coding sequence, has been described previously (3). rAAVCR particles of AAV serotype 2 were produced by transient transfection of 2935Z with pDG and pAAVCR and purified on two successive CsCl gradients, and titers of genome-containing particles were determined by dot blot assay. rAAV2GFP was kindly provided by M. Linden (King's College London School of Medicine, London, United Kingdom).
Antibodies.
The following primary antibodies were used: anti-actin (Santa Cruz Biotechnology SC-10731; dilution for Western blotting [WB], 1:10,000), anti-ATM (Genetex 70107; dilution for WB, 1:1,000), anti-ATM-P-S1981 (Rockland Immunochemicals 200-301-400; dilution for WB, 1:500; dilution for immunofluorescence [IF] assay, 1:50), anti-ATR (SC-28901; dilution for WB, 1:1,000), anti-ATR-P-S428 (Cell Signaling Technology CST-2853; dilution for WB, 1:500; dilution for IF assay, 1:200), anti-ATRIP (Abcam Ab-19531; dilution for WB, 1:2,000; dilution for IF assay, 1:500), anti-Chk1 (SC-8408; dilution for WB, 1:1,000), anti-Chk1-P-S345 (CST-133D3; dilution for WB, 1:1,000), anti-Chk2 (SC-5278; dilution for WB, 1:1,000), anti-Chk2-P-T68 (SC-16297; dilution for WB and immunoprecipitation, 1:250; dilution for IF assay, 1:100), anti-DNA-PKcs (NeoMarkers M5370; dilution for WB, 1:500; dilution for IF assay, 1:50; dilution for flow cytometry, 1:250), anti-DNA-PKcs-P-S2056 (Ab-18192; dilution for WB, 1:2,000; dilution for IF assay, 1:500), anti-HSV-1 ICP8 (Ab-20193; dilution for WB, 1:1,000; dilution for IF assay, 1:200), anti-H2AX-P-S139 (Millipore 05-636; dilution for WB, 1:500; dilution for IF assay, 1:50), anti-Nbs1 (Novus Biologicals 100-143; dilution for WB, 1:1,000), anti-Nbs1-P-S343 (Ab-47272; dilution for WB, 1:500; dilution for IF assay, 1:100), anti-p53 (Ab-1101; dilution for WB, 1:1,000), anti-p53-P-S15 (Ab-38497 [purchased in June 2009 and no longer available]; dilution for WB, 1:500; dilution for IF assay, 1:500), anti-AAV2 Rep (Fitzgerald Industries 10R-A111A; dilution for WB, 1:200), anti-RPA32 (Bethyl Laboratories BL-A300-244A; dilution for WB, 1:2,000; dilution for IF assay, 1:500), anti-RPA32-P-S4/8 (BL-A300-245A; dilution for WB, 1:200; dilution for IF assay, 1:200), and anti-USP7 (CST-3277; dilution for WB, 1:750). The following secondary antibodies were used: rabbit anti-mouse IgG-horseradish peroxidase (HRP; Sigma A9044; dilution, 1:10,000), goat anti-rabbit IgG-HRP (Sigma A6154; dilution, 1:10,000), rabbit TrueBlot, goat anti-rabbit IgG (H+L)-Alexa Fluor 405 (AF405; Molecular Probes A31556; dilution, 1:500), goat anti-rabbit IgG (H+L)-AF594 (Molecular Probes A11012; dilution, 1:1,000), goat anti-mouse IgG (H+L)-AF594 (Molecular Probes A11005; dilution, 1:1,000), goat anti-mouse IgG (H+L)-fluorescein isothiocyanate (FITC; Southern Biotechnology 1031-02; dilution, 1:200), goat anti-rabbit IgG (H+L)-FITC (Southern Biotechnology 4050-02; dilution, 1:200), and goat anti-mouse IgG-Cy5 (Millipore AP181S; dilution, 1:500).
WB analysis.
A total of 106 HCT116 cells, 5 × 106 AT22 IJE-T, or 5 × 106 MO59J fusion cells were seeded into 6-cm plates. The following day, the cells were mock infected, infected with either AAV2 (multiplicity of infection [MOI], 2,000) or HSV-1 (MOI, 1.5), or coinfected with AAV2 (MOI, 2,000) and HSV-1 (MOI, 1.5) in DMEM supplemented with 2% FBS and 1% AB. Cells treated for 18 h with hydroxyurea (HU; 3 mM) served as a positive control for activation of DDR proteins. After 3 h, 6 h, 9 h, 12 h, 24 h, or 48 h, cells were trypsinized, washed once with phosphate-buffered saline (PBS), and resuspended in 200 μl EBC-170 lysis buffer (50 mM Tris-HCl, 170 mM NaCl, 0.5% Nonidet P-40, Complete Mini-EDTA-free protease inhibitor [Roche Diagnostics, Rotkreuz, Switzerland]). After 30 min of incubation at 4°C under constant agitation, the suspension was centrifuged (20 min at 13,200 × g) and the supernatant was collected, mixed with 2× loading buffer (4% SDS, 10% β-mercaptoethanol, 20% glycerol, 0.005% bromphenol blue, 0.125 M Tris-HCl, pH 6.8), and boiled for 10 min. Cell lysates were separated, depending on the molecular weight of the protein of interest, on 8%, 10%, or 12% SDS-polyacrylamide gels and transferred to Protran nitrocellulose membranes (Whatman, Bottmingen, Switzerland). For detection of γH2AX, proteins were separated on a 12% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane with a pore size of 0.45 μm (Amersham Hybond-P; GE Healthcare Bio-Sciences AB, Uppsala, Sweden). The membranes were blocked with PBS-T (PBS containing 0.3% Tween 20) supplemented with 5% nonfat dry milk for 1 h at room temperature (RT). Incubation with antibodies was carried out with PBS-T supplemented with 2.5% nonfat dry milk. Primary antibodies were incubated overnight at 4°C, while secondary antibodies were incubated for 1 h at RT. Membranes were washed three times with PBS-T for 10 min after each antibody incubation step. HRP-conjugated secondary antibodies were detected with ECL detection reagent (ECL WB blotting systems; GE Healthcare, Zurich, Switzerland). The membranes were exposed to chemiluminescence detection films (Roche Diagnostics, Rotkreuz, Switzerland). Detection of anti-actin served as a loading control for the lysate.
Fluorescence-activated cell sorting (FACS) and WB analysis.
A total of 6.6 × 106 AT22 IJE-T cells were seeded into 10-cm cell culture dishes. Cells were mock infected, infected with rHSV-1vECFP-ICP4 (MOI, 2 or 4), or coinfected with rHSV-1vECFP-ICP4 (MOI, 2 or 4) and rAAV2CR (MOI, 4,000). Cells positive for mCherry (rAAV2) or ICP4-ECFP (HSV-1) were sorted and prepared for WB analysis as described above. The same number of mock-infected and HU-treated cells (3 mM, 18 h postinfection [hpi]) was used as a control.
Immunoprecipitation.
AT22 IJE-T cells (90,000) were seeded into 6-well plates. The next day, cells were mock infected, infected with either AAV2 (MOI, 2,000) or HSV-1 (MOI, 1.5), or coinfected with AAV2 (MOI, 2,000) and HSV-1 (MOI, 1.5) in DMEM supplemented with 2% FBS and 1% AB. After 24 h, cells were trypsinized, washed once with PBS, and resuspended in 200 μl EBC-170 lysis buffer containing the primary antibody. After incubation for 2 h at 4°C under constant agitation, 30 μl protein A Sepharose CL-4B (GE Healthcare, Zurich, Switzerland) was added and the mixture was incubated for 2 h at 4°C under constant agitation. Then, Sepharose beads were pelleted and the supernatant was collected as loading and infection controls. The pellet was washed twice with PBS for 15 min at 4°C under constant agitation, and protein was eluted using 20 to 40 μl of a 4 M urea solution (pH 7.5). After 10 min of incubation at 4°C under constant agitation, beads were pelleted and the supernatant was collected. Samples were analyzed by WB assay as described above; however, for detection of immunoprecipitated proteins, the secondary antibody rabbit IgG TrueBlot (anti-rabbit IgG-HRP; eBioscience, San Diego, CA) was used, which preferentially detects the nonreduced form of rabbit IgG over the reduced, SDS-denatured form of IgG.
IF analysis.
HCT116 cells (5 × 104), AT22 IJE-T cells (7,5 × 104), or MO59J fusion cells (7,5 × 104) were seeded onto coverslips (12-mm diameter; Glaswarenfabrik Karl Hecht GmbH & Co. KG, Sondheim, Germany) in 24-well plates. The next day, cells were mock infected, infected with HSV-1 (MOI, 1.5), or coinfected with rAAV2CR (MOI, 250) and HSV-1 (MOI, 1.5), rHSV-1dl1403 (MOI, 0.9), or Ad2 (MOI, 12). UV-exposed cells (10 J/m2) or HU (3 mM)-treated cells served as a positive control for activation of DDR proteins. After 24 h, cells were washed once with cold PBS and fixed with 3.7% formaldehyde in PBS for 15 min at RT. The fixation process was stopped by incubation with 0.1 M glycine for 10 min at RT. Cells were washed twice with cold PBS. For permeabilization, cells were treated for 2 min with precooled (−20°C) acetone and washed three times with PBS. Cells were blocked for 30 min with 3% bovine serum albumin (BSA) in PBS. For staining, cells were incubated with antibodies diluted in PBS-BSA (3%) in a humidified chamber at RT in the dark. Coverslips were placed on droplets (40 μl) of primary antibody solution. After incubation for 1 h, cells were washed three times with PBS. DAPI (4′,6′-diamidino-2-phenylindole) staining was performed together with the secondary antibody staining. For this, DAPI (0.5 μg/ml) and the secondary antibody were diluted in PBS-BSA (3%). After incubation for 1 h at RT in the dark, cells were washed three times with PBS and once with H2O. Coverslips were embedded in Glycergel (DakoCytomation, Carpinteria, CA) containing DABCO (26 mg/ml; Fluka, Sigma-Aldrich Chemie GmbH, Munich, Germany), and cells were observed using a confocal laser scanning microscope (CLSM; Leica TCS SP2 AOBS; Leica Microsystems, Wetzlar, Germany). To prevent cross talk between the channels for the different fluorochromes, all channels were recorded separately and fluorochromes with longer wavelengths were recorded first. Images from the CLSM were deconvolved with Huygens Essential 2.6.0p1 software (Scientific Volume Imaging, Hilversum, Netherlands) and processed using Imaris 5.0.1 (Bitplane AG, Zurich, Switzerland).
Flow cytometry.
The day before infection, 106 HCT116 cells were seeded into 6-cm tissue culture plates. The cells were mock infected; infected with rHSV-1vEYFP-ICP4 (MOI, 1.5); or coinfected with AAV2 (MOI, 250), rAAV2GFP (MOI, 250), and HSV-1 (MOI, 1.5) in DMEM supplemented with 2% FCS and 1% AB; and incubated for 20 h at 37°C in a 5% CO2 atmosphere. Cells were trypsinized and washed once with PBS. For fixation and staining, the BD Cytofix/Cytoperm Plus kit (BD Biosciences) was used according to the manufacturer's instructions. Flow cytometry was performed on a FACScalibur (BD). DNA-PKcs contents were analyzed in HSV-1-infected cells positive for EYFP-ICP4 (rHSV-1 encoded) and in coinfected cells positive for enhanced green fluorescent protein (EGFP; rAAV encoded). A minimum of 80,000 events were scored for each sample. The mean fluorescence intensity of cell populations was determined using FlowJo software (FlowJo version 8.6.3; Stanford University, Stanford, CA).
RESULTS
Activation of primary DNA damage markers Nbs1 and H2AX upon virus infection.
We compared the DDRs of cells infected with HSV-1 alone or coinfected with AAV2. As we have previously observed that at low MOIs, HSV-1-supported AAV2 RCs are not detectable before approximately 18 hpi, we analyzed the cells for up to 48 hpi.
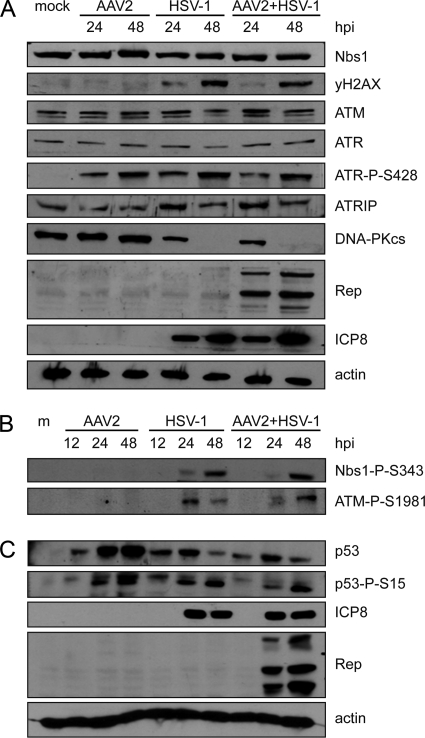
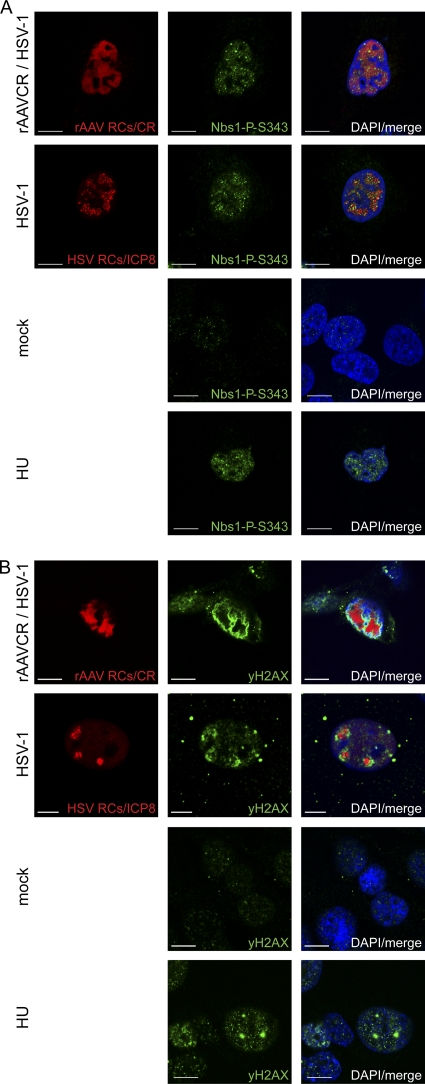
The overall levels of Nbs1 were not altered by virus infection (Fig. 1A). However, Nbs1 was phosphorylated upon infection with HSV-1 alone or coinfection with AAV2 (Fig. 1B); moreover, phosphorylated Nbs1-P-S343 colocalized with AAV2 RCs and with HSV-1 RCs (Fig. 2A). Nbs1 expression was also examined in AT22 IJE-T and HCT116 cells with similar results (data not shown). In HCT116 cells, HSV-1 infection- and coinfection-induced phosphorylation was also indicated by shifted Nbs1 bands (data not shown). In addition, H2AX was found to be phosphorylated (Fig. 1A) and to surround HSV-1-supported AAV2 RCs (Fig. 2B). As previously observed, H2AX also surrounded HSV-1 RCs (Fig. 2B) (94).
Fig 1.
DNA damage signaling induced by viral infection. (A to C) WB analysis of infected cell lysates. MO59J Fus1 cells were mock infected (m), infected with AAV2 alone (MOI, 2,000) or HSV-1 alone (MOI, 1.5), or coinfected with AAV2 (MOI, 2,000) and HSV-1 (MOI, 1.5) and harvested at the indicated times postinfection. Total proteins were extracted, separated by SDS-PAGE, blotted onto nitrocellulose membrane, and analyzed with the antibodies indicated. Actin served as a loading control; detection of HSV-1 ICP8 and AAV2 Rep was used as an infection control.
Fig 2.
Activation of primary DDR proteins. MO59J Fus1 cells were mock infected, infected with HSV-1 (MOI, 1.5), or coinfected with rAAVCR (MOI, 250) and HSV-1 (MOI, 1.5). After 24 h, cells were fixed and processed for IF analysis. rAAVCR RCs (AAV RCs) were visualized by binding of the rAAVCR-encoded mCherry-Rep68/78 fusion protein (CR) to AAV DNA (red). HSV-1 RCs were visualized with a primary antibody specific for the HSV-1 major DNA binding protein ICP8 and an AF594-labeled secondary antibody (red). Cells treated with HU (3 mM) served as a DDR control. To identify phosphorylated Nbs1 (A) and H2AX (B), cells were stained with antibodies specific for Nbs1-P-S343 or H2AX-P-S139 (γH2AX) and an FITC-labeled secondary antibody (green). DAPI was used to stain cellular DNA. Images were taken using a CLSM and represent a single optical z slice of the nuclei. Scale bars, 10 μm.
Coinfection with AAV2 and HSV-1 induces the phosphorylation of ATM and ATR.
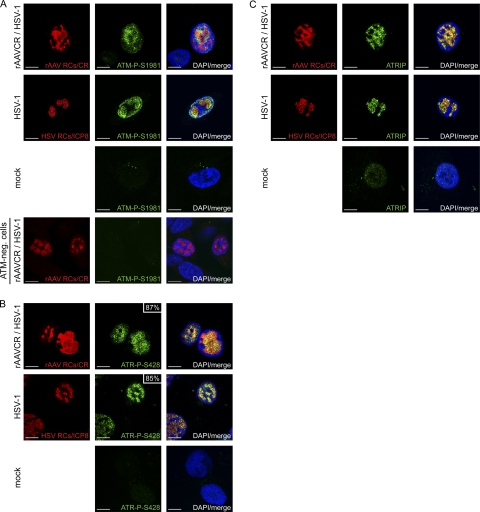
Previous studies revealed that ATM-mediated signaling is induced (51, 76, 96) and that ATR-mediated signaling is blocked in HSV-1-infected cells (58, 94). To examine the activity of these two PIKKs in cells coinfected with HSV-1 and AAV2, we first determined their phosphorylation status by WB and IF analyses. While total ATM levels were similar in virus-infected cells and mock-infected cells (Fig. 1A), ATM-dependent staining with an antibody generated to autophosphorylated ATM was detected only in cells infected with HSV-1 alone or coinfected with AAV2 (Fig. 1B) and in these cells it colocalized with HSV-1 and AAV2 RCs (Fig. 3A). As this antibody detects other phosphorylated targets of ATM such as 53BP1, this finding only demonstrates that the staining requires ATM activity but does not necessarily represent ATM itself.
Fig 3.
Recruitment of ATM-P-S1981, ATR-P-S428, and ATRIP into HSV-1 and AAV2 RCs. MO59J Fus1 cells were infected and processed for IF, and viral RCs (red) were visualized as described in the legend to Fig. 2. Cells were stained with an antibody specific for ATM-P-S1981 (A), ATR-P-S428 (B), or ATRIP (C) and an FITC-labeled secondary antibody (green). DAPI was used to stain cell nuclei. In panel A, cells deficient for ATM (AT-22 IJE T) served as a control for the phosphospecific ATM antibody. In panel B, the percentage of ATR-P-S428 in viral RCs is indicated. Scale bars, 10 μm.
Total ATR levels were also comparable in virus-infected cells and mock-infected cells (Fig. 1A), but phosphorylated ATR-P-S428 was present only in cells infected with either HSV-1 or AAV2 alone or coinfected with both viruses (Fig. 1A), and it colocalized with approximately 85% of the HSV-1 RCs and approximately 87% of the AAV2 RCs (Fig. 3B). The observed phosphorylation of ATR in cells infected with HSV-1 alone was surprising, as it appeared to be in contrast to previous studies which did not find HSV-1-induced activation of ATR (58, 94). We therefore further investigated the phosphorylation/activation of ATR by (i) testing the specificity of the ATR-P-S428 antibody (70), (ii) monitoring ATRIP, a protein that is important for the recruitment of ATR to sites of DNA damage (89, 106), and (iii) analyzing the phosphorylation status of the ATR target Chk1. The results of these experiments can be summarized as follows. (i) Immunoprecipitation with an ATR-specific antibody resulted in ATR-P-S428-specific bands in cells infected with HSV-1 and/or AAV2 but not in mock-infected cells (see Fig. S1A in the supplemental material). (ii) ATRIP was detected in both mock-infected cells and virus-infected cells (Fig. 1A), where it colocalized with AAV2 and HSV-1 RCs (Fig. 3C). (iii) WB analysis revealed no phosphorylation of the ATR target Chk1 at S345 (see Fig. S1B in the supplemental material) and S317 (data not shown) in infected cells. From the results described above, it seems that although ATR is phosphorylated in infected cells, the kinase is not activated and therefore not able to transmit DDR signaling via its downstream target, Chk1. Supporting this, Nam and Cortez (62) recently showed that phosphorylation of ATR on S428 is not indicative of ATR activity and cannot be inhibited by the commonly used ATR inhibitor caffeine. Moreover, Liu et al. reported that the mutation of S428 to A428 did not prevent the phosphorylation of Chk1 in response to replicative stress (54).
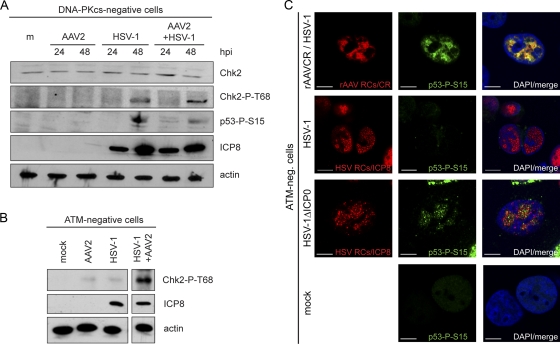
HSV-1 ICP0-dependent degradation of DNA-PKcs is delayed during coinfection with AAV2.
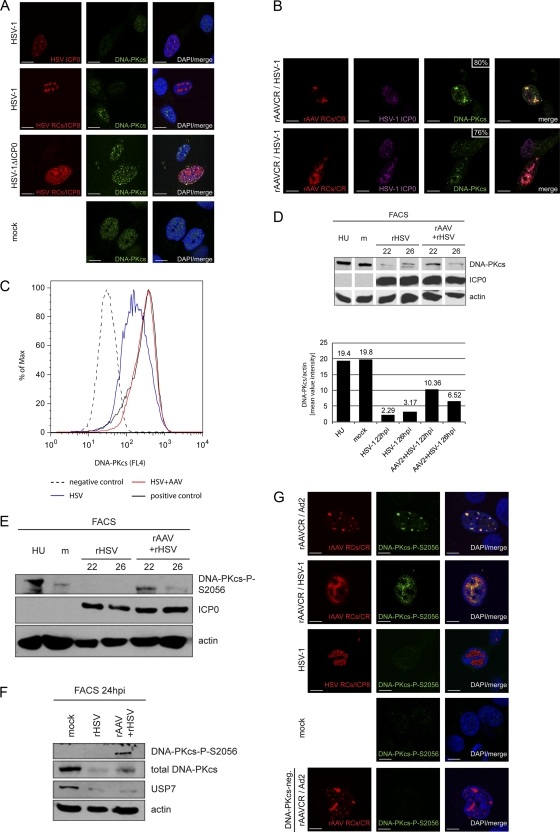
HSV-1 ICP0 is known to induce proteasomal degradation of DNA-PKcs (48, 67), which, together with Ku70 and Ku80, forms DNA-PK, the third main PIKK besides ATM and ATR (reviewed in reference 55). Consistent with these reports, our WB results displayed loss of DNA-PKcs by 48 h after infection with HSV-1 or coinfection with HSV-1 and AAV2 (Fig. 1A). Accordingly, at the single-cell level, DNA-PKcs was not detected simultaneously with either the HSV-1 immediate-early (IE) protein ICP0 or HSV-1 early (E) protein ICP8 in cells infected with HSV-1 alone. However, DNA-PKcs was readily detected in cells infected with rHSV-1ΔICP0 (Fig. 4A). Interestingly, IF analysis revealed that DNA-PKcs colocalized with HSV-1 proteins ICP0 and ICP8 in 80% and 76% of the AAV RCs, respectively (Fig. 4B). This indicates that HSV-1 ICP0-mediated DNA-PKcs degradation in coinfected cells is not as efficient as in cells that are infected with HSV-1 alone, even if the intensity of DNA-PKcs staining in large AAV RCs is lower than that in small AAV RCs (Fig. 4B). This possibility was further investigated by flow cytometry data. Cells infected with HSV-1 or coinfected with HSV-1 and AAV2 were identified using rHSV-1 and rAAV expressing ICP4-EYFP or EGFP, respectively. Similar to the results obtained with wild-type HSV-1 (strain F), DNA-PKcs was also degraded in cells infected with autofluorescent recombinant HSV-1 (strain17; Fig. 4C). In contrast, the fluorescence intensity of DNA-PKcs staining in cells coinfected with wild-type HSV-1 and AAV2 was the same as that in mock-infected cells (Fig. 4C). At the chosen MOIs, the ratio of cells that show AAV RCs at 24 hpi is only approximately 10%, while 24 h after infection with HSV-1 alone, more than 40% of the cells contain HSV-1 RCs. This may explain why in the WB analysis of total cell lysates shown in Fig. 1, the degradation of DNA-PKcs appeared equally efficient in HSV-1-infected cells and in coinfected cells while, on the single-cell level (Fig. 4B and C), DNA-PKcs degradation appeared to be less efficient in coinfected cells.
Fig 4.
Activation and delayed degradation of DNA-PKcs in cells coinfected with HSV-1 and AAV2. (A) MO59J Fus1 cells were infected with HSV-1 (MOI, 1.5) or HSV-1ΔICP0 (MOI, 0.9) or mock infected. After 24 h, cells were fixed and processed for IF analysis. HSV-1 infection was detected using an antibody specific for ICP0 or ICP8 and an AF594-labeled secondary antibody (red). Additionally, cells were stained with an antibody specific for DNA-PKcs and an FITC-labeled secondary antibody (green). DAPI was used to stain cellular DNA. Scale bars, 10 μm. (B) IF analysis of MO59J Fus1 cells at 24 h after coinfection with rAAVCR (MOI, 250) and HSV-1 (MOI, 1.5). rAAVCR RCs (red) were visualized as described in the legend to Fig. 2. Additionally, cells were stained with antibodies specific for ICP0 or ICP8 and an AF405-labeled secondary antibody (purple) and with an antibody specific for DNA-PKcs and an FITC-labeled secondary antibody (green). The percentage of DNA-PKcs in rAAVCR RCs is indicated. Scale bars, 10 μm. (C) Flow cytometric analysis of infected cells. DNA-PKcs-positive HCT116 cells were mock infected (positive control), infected with rHSV-1ICP4EYFP (MOI, 1.5), or coinfected with HSV-1 (MOI, 1.5), AAV2 (MOI, 250), and rAAVGFP (MOI, 250). HCT116 cells negative for DNA-PKcs served as a negative control. Cells were fixed at 20 h postinfection and stained with a DNA-PKcs-specific monoclonal antibody and a Cy5-labeled secondary antibody. DNA-PKcs was analyzed in HSV-1-infected cells positive for EYFP-ICP4 (HSV) and in coinfected cells positive for EGFP (HSV+AAV). A minimum of 80,000 events were scored for each sample. Graphs were overlaid to show the fluorescence shift of HSV-1-infected populations. (D and E) WB analysis of AT22 IJE-T cells sorted for productive HSV-1 and AAV2 infection at 22 and 26 hpi. Cells were mock infected, infected with rHSV-1vECFP-ICP4 (rHSV; MOI, 2), or coinfected (rHSV + rAAV) with rHSV-1vECFP-ICP4 (MOI, 2) and rAAV2CR (MOI, 4,000). Lysates of sorted cells were processed for WB analysis and stained with the antibodies indicated. Quantification of WB band intensities was done with a Gel Doc system using Quantity One software (version 4.6.1; Bio-Rad, Hercules, CA). (F) WB analysis of AT22 IJE-T cells sorted for productive HSV-1 and AAV2 infection at 24 hpi. Cells were mock infected, infected with rHSV-1vECFP-ICP4 (rHSV; MOI, 4), or coinfected (rHSV + rAAV) with rHSV-1vECFP-ICP4 (MOI, 4) and rAAV2CR (MOI, 4,000). Lysates of sorted cells were processed for WB analysis and stained with the antibodies indicated. (G) MO59J Fus1 cells were mock infected, infected with HSV-1 (MOI, 1.5), or coinfected with rAAVCR (MOI, 250) and either HSV-1 (MOI, 1.5) or Ad2 (MOI, 12.5). After 24 h, cells were fixed and processed for IF analysis. rAAVCR and HSV-1 RCs (red) were visualized as described in the legend to Fig. 2. DNA-PKcs phosphorylation was detected with an antibody specific for DNA-PKcs-P-S2056 and an FITC-labeled secondary antibody (green). As a negative control, MO59J Fus9 DNA-PKcs-negative cells were coinfected with rAAVCR (MOI, 250) and Ad2 (MOI, 12.5) and stained for pDNA-PKcs. DAPI was used to stain cellular DNA. Scale bars, 10 μm.
To further examine this observation, we performed WB analysis of sorted cells at 22 and 26 hpi (Fig. 4D and E). Cells were infected with rHSV-1 ICP4-ECFP or coinfected with rAAVCR and rHSV-1 ICP4-ECFP. HSV-1-infected cells were sorted based on the expression of ICP4-ECFP, and cells that contained rAAV RCs were sorted based on the expression of mCherry-Rep. Sorted mock-infected cells served as a control. Mock-infected and HU-treated cells served as an activation control for DNA-PKcs. Quantification of the WB assay shown in Fig. 4D revealed 3-times-higher levels of DNA-PKcs at 22 hpi during AAV replication than in cells infected with HSV-1 alone (Fig. 4D). At 26 hpi, the intensity of the DNA-PKcs staining was also decreased in coinfected cells compared to that seen at 22 hpi but still approximately 2 times as high as that in cells infected with HSV-1 alone at 26 hpi. Moreover, phosphorylation of DNA-PKcs at S2056 was detected only in cells coinfected with both viruses (Fig. 4E and F); however, pDNA-PKcs levels also decreased in coinfected cells over time (Fig. 4E). Staining for the cellular deubiquitinase ubiquitin-specific peptidase 7 (USP7) at 24 hpi, which is another known target of HSV-1 ICP0-induced proteasomal degradation (12), revealed that the AAV-mediated delay of the ICP0 function was specific for DNA-PKcs, as the degradation of USP7 was equally efficient in cells infected with HSV-1 alone and in coinfected cells (Fig. 4F).
Next, we analyzed the localization of DNA-PKcs-P-S2056 within infected cells. As positive and negative controls, we coinfected DNA-PKcs-positive cells and DNA-PKcs-deficient cells with Ad and AAV2, which are known to activate DNA-PKcs and recruit it into RCs (75). While DNA-PKcs-P-S2056 staining was readily detected in cells coinfected with AAV2 and either Ad2 or HSV-1, where it also colocalized with AAV2 RCs, we did not observe staining in cells that contained HSV-1 RCs (Fig. 4G).
Phosphorylation of Chk2 and p53.
To explore a potential DNA-PK-mediated signaling to downstream targets in coinfected cells, we assessed virus-induced phosphorylation of Chk2 and p53 in normal cells and in cells defective for either DNA-PKcs (MO59J Fus9) or ATM (AT22 IJE-T pEBS7). HSV-1 has previously been shown to induce the phosphorylation of Chk2 in an ATM-dependent manner (51, 76). Our results are in line with these reports, as in normal cells (data not shown) and in DNA-PKcs-deficient cells (Fig. 5A), Chk2 was phosphorylated at 24 h after infection with HSV-1, and this also occurred in cells coinfected with HSV-1 and AAV2. However, in ATM-deficient cells, Chk2 was phosphorylated only in cells coinfected with both viruses but not in cells infected with HSV-1 alone (Fig. 5B). This suggests that in these cells, Chk2 phosphorylation was possibly facilitated by the AAV2-mediated delayed degradation of DNA-PKcs. Also consistent with previous observations, we detected that HSV-1 (13) and AAV2 (68) induced the stabilization of p53 (Fig. 1C). Additionally, we observed that p53 was phosphorylated at S15 in cells infected with either HSV-1 or AAV2 or coinfected with both viruses (Fig. 1C) and that p53-P-S15 was recruited into both HSV-1 RCs and AAV2 RCs (data not shown). p53-P-S15 was also detected in DNA-PKcs-deficient cells infected with HSV-1 alone or coinfected with HSV-1 and AAV2 (Fig. 5A); in these cells, phosphorylation is likely ATM mediated. However, as opposed to the situation in normal cells infected with AAV2 alone, p53-P-S15 was not detected in DNA-PKcs-deficient cells infected with AAV2 alone (Fig. 5A), indicating that in the absence of the helper virus, AAV2-induced phosphorylation of p53 is DNA-PK dependent. In ATM-deficient cells, p53 was phosphorylated only when cells were infected with rHSV-1ΔICP0 or coinfected with AAV2 and HSV-1 (Fig. 5C) and not in cells infected with HSV-1 alone (Fig. 5C), indicating again that in coinfected cells, p53 phosphorylation was supported by the delayed degradation of DNA-PKcs.
Fig 5.
DNA-PKcs- and ATM-dependent activation of Chk2 and p53 upon viral infection. (A) WB analysis of DNA-PKcs-deficient MO59J Fus9 cells. Cells were mock infected (m), infected with AAV2 (MOI, 2,000) or HSV-1 (MOI, 1.5), or coinfected with AAV2 (MOI, 2,000) and HSV-1 (MOI, 1.5). Total proteins were extracted at the indicated times postinfection and subjected to WB analysis using antibodies specific for actin (loading control), ICP8 (HSV-1 infection control), Chk2, Chk2-P-T68, and p53-P-S15. (B) Immunoprecipitation and WB analysis of ATM-negative AT22 IJE-T cells at 24 h after mock infection or infection with AAV2 (MOI, 2,000), HSV-1 (MOI, 1.5), AAV2 (MOI, 2,000), and HSV-1 (MOI, 1.5). Lysates were analyzed using the antibodies indicated. (C) IF analysis of ATM-negative AT22 IJE-T cells at 24 h after infection with HSV-1 (MOI, 1.5) and rAAVCR (MOI, 250), HSV-1 (MOI, 1.5), or HSV-1ΔICP0 (MOI, 0.9) or mock infection. rAAVCR and HSV-1 RCs (red) were visualized as described in the legend to Fig. 2. p53 activation was detected with an antibody specific for p53-P-S15 and an FITC-labeled secondary antibody (green). DAPI was used to stain cellular nuclei. Scale bars, 10 μm.
Phosphorylation of RPA.
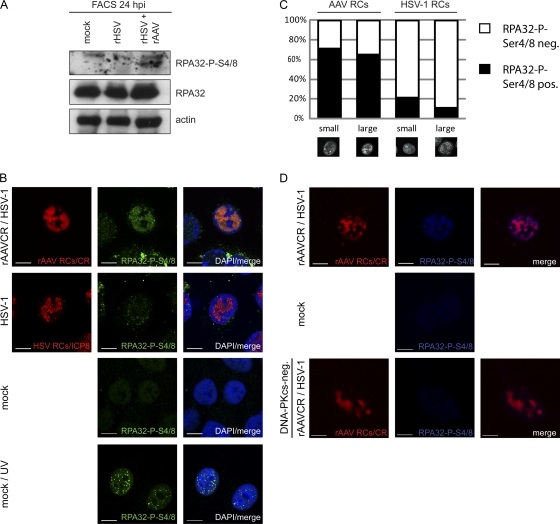
The trimeric RPA complex composed of RPA70, RPA32, and RPA14 is involved in DNA repair, as well as replication and transcriptional regulation of both cellular and viral DNAs (10, 97). RPA has a strong affinity for single-stranded DNA (98) and was shown to be a component of Ad-supported AAV2 RCs (79). Upon DNA damage, RPA32 becomes phosphorylated at several residues (reviewed in reference 107). It was suggested that phosphorylation at S33 is mediated by ATR, while phosphorylation at S4/8 is mediated in a DNA-PK-dependent manner (4, 104). Here we examined the spatial localization of the RPA subunit RPA32 and its phosphorylation at S4/8 in infected cells. Our results show that total RPA32 is recruited into HSV-1 and AAV2 RCs (data not shown) and that its expression levels were not altered by virus infection (Fig. 6A). WB analysis of cells productively infected with either HSV-1 or AAV2 revealed phosphorylation of RPA32 at Ser4/8 only upon AAV2 replication (Fig. 6A). More precisely, RPA32-P-S4/8 clearly colocalized with approximately 70% of the small and large AAV2 RCs but with only 21% of the small and 11% of the large HSV-1 RCs (Fig. 6B and C). UV-treated cells served as a positive control (Fig. 6B). Moreover, accumulation of RPA32-P-S4/8 was observed only in AAV2 RCs of DNA-PKcs-positive cells (Fig. 6D). In summary, these data show that coinfection with AAV2 and HSV-1 induces DNA-PKcs-dependent phosphorylation of RPA32 at S4/8.
Fig 6.
Phosphorylation of RPA32 upon AAV2 and HSV-1 replication. (A) WB analysis of sorted AT22 IJE-T cells at 24 hpi. Cells were mock infected, infected with rHSV-1vECFP-ICP4 (rHSV; MOI, 4), or coinfected (rHSV + rAAV) with rHSV-1vECFP-ICP4 (MOI, 4) and rAAV2CR (MOI, 4,000). Sorted cells were subjected to WB analysis and analyzed with the antibodies indicated. (B) IF analysis of U2OS cells after infection with HSV-1 (MOI, 1.5) or rAAVCR (MOI, 250) and HSV-1 (MOI, 1.5) or mock infection at 24 h. UV-treated cells (10 J/m2) served as a positive control. rAAVCR and HSV-1 RCs (red) were visualized as described in the legend to Fig. 2. To detect phosphorylated RPA32, cells were stained with an antibody specific for RPA32-P-S4/8 and an FITC-labeled secondary antibody (green). Cellular DNA was stained with DAPI. Scale bars, 10 μm. (C) Quantification of RPA32-P-S4/8 colocalization with small and large AAV2 or HSV-1 RCs in U2OS cells. Fifty cells per sample were counted. Black columns, RPA32-P-S4/8-positive viral RCs; open columns, RPA32-P-S4/8 negative viral RCs. (D) IF analysis of MO59J Fus1 or Fus9 (DNA-PKcs-negative) cells at 24 h after mock infection or coinfection with HSV-1 (MOI, 1.5) and rAAVCR (MOI, 250). rAAVCR RCs (red) were visualized as described in the legend to Fig. 2. Cells were stained with an antibody specific for RPA32-P-S4/8 and an AF405-labeled secondary antibody (blue). Scale bars, 10 μm.
DISCUSSION
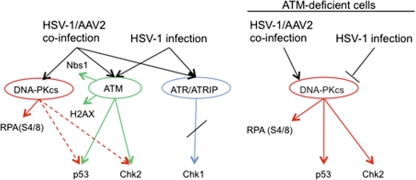
In this study, we compared the phosphorylation status and spatial organization of DDR proteins between cells infected with HSV-1 alone and cells coinfected with HSV-1 and AAV2. WB and IF analyses demonstrated that HSV-1 and AAV2 coinfection induced a strong DDR (Table 1 and Fig. 7). Specifically, HSV-1-supported AAV2 replication induced the phosphorylation of Nbs1 and H2AX, as well as ATM and its substrates Chk2 and p53. These responses are similar to those induced by HSV-1 infection alone, as observed here and in previous studies (13, 49, 67, 76, 81, 95). We also detected phosphorylation of ATR and recruitment of pATR along with its binding partner ATRIP into HSV-1-supported AAV2 RCs, as well as into HSV-1 compartments, although kinase activity of ATR was blocked upon HSV-1 and AAV2 replication, as we did not detect phosphorylation of Chk1 at ATR target sites S317 and S345. Our results are similar to those of a previous study showing that ATR, although recruited together with ATRIP into HSV-1 RCs, is unable to activate Chk1. In addition, Mohni et al. showed that both ATR and ATRIP, even if not activated, contribute to efficient HSV-1 infection (58).
Table 1.
DNA damage signaling in cells infected with HSV-1 or coinfected with HSV-1 and rAAV2a
| Protein | HSV-1–rAAV2 coinfection |
HSV-1 infection |
||||||
|---|---|---|---|---|---|---|---|---|
| Inhibition | Induction | Phosphorylation | Localization | Inhibition | Induction | Phosphorylation | Localization | |
| ATM | + | NUCc + RCs | + | RCs | ||||
| ATR | − | − | + | RCs | − | − | + | RCs |
| ATRIP | − | + | NDd | RCs | − | + | ND | RCs |
| DNA-PKcs | +b | − | + | RCs | + | − | − | − |
| USP7 | + | − | ND | RCs | + | − | ND | ND |
| H2AX | ND | ND | + | NUC | ND | ND | + | NUC |
| NBS1 | − | − | + | RCs | − | − | + | RCs |
| RPA32 (P-Ser4/8) | − | − | + | RCs | − | − | − | RCs |
| p53 | − | + | + | RCs | − | + | + | RCs |
| Chk1 | − | − | − | RCs | − | − | − | RCs |
| Chk2 | − | − | + | RCs | − | − | + | RCs |
Shown is a summary of data from WB analysis, IF analysis, and flow cytometry. Bold, AAV-induced modulation of DDR.
Inhibition delayed.
NUC, nucleus.
ND, not done.
Fig 7.
Models of the DDR signaling induced by HSV-1 replication and HSV-1-supported AAV replication. The analysis of the DDRs in ATM-deficient, DNA-PKcs-deficient, and normal cells supports the following models. Infection of cells with HSV-1 induces phosphorylation of ATM and ATR and signaling to their targets Nbs1, H2AX, Chk2, and p53 but not to the ATR target Chk1. In cells infected with HSV-1 alone, DNA-PKcs is rapidly degraded in an HSV-1 ICP0-dependent manner and no DNA-PKcs-mediated signaling occurs. In contrast, coinfection with HSV-1 and AAV2 induces the activation of DNA-PKcs, which enables the phosphorylation of RPA32 at S4/8. Further support for the activation of DNA-PKcs in coinfected cells comes from experiments performed with ATM-deficient cells, as in the absence of ATM, HSV-1 and AAV2 coinfection still induced the phosphorylation of p53, Chk2, and RPA32 at S4/8. In ATM-deficient cells infected with HSV-1 alone, the DDR signaling is broadly reduced, with undetectable levels of Chk2-P-T86 and p53-P-S15 (51, 76).
Due to the strong affinity of RPA32 for single-stranded DNA (102, 107) and its function in recruiting the ATRIP-ATR complex to sites of DNA damage (7, 26, 106), it is not surprising that we and others (65, 79) detected RPA32 in HSV-1-supported AAV2 RCs. In HSV-1-infected cells, RPA32 colocalizes with the HSV-1 major single-stranded DNA binding protein ICP8 (81) in HSV-1 RCs (82, 93). Replicating HSV-1 DNA contains stretches of single-stranded DNA (39) which may recruit RPA32 into HSV-1 RCs also independently of ICP8. Single-stranded DNA binding proteins such as RPA32 are proposed to stimulate AAV2 DNA replication (64, 79), e.g., by protecting the single-stranded replication products from nucleases (63) and by enhancing the binding and nicking of Rep proteins at the replication origins (79).
HSV-1 has been demonstrated to induce the degradation of DNA-PKcs in an ICP0-dependent manner (48, 67). Although WB analysis of total infected cells displayed a similar degradation of DNA-PKcs, in cells infected with HSV-1 or coinfected with HSV-1 and AAV2, a striking difference was observed at the single-cell level, as well as in cells productively infected with HSV-1 and AAV2 captured by FACS. Flow cytometry, IF analysis, and WB analysis of sorted cells revealed an AAV2-mediated delay in the HSV-1-dependent degradation of DNA-PKcs, as well as AAV2-induced phosphorylation of DNA-PKcs. The observed discrepancy in the results of WB assays of total cell lysates and analyses on the single-cell level is likely due to the fact that, under the chosen experimental conditions, only approximately 10% of the cells supported AAV2 replication. In order to estimate the extent of the delay in DNA-PKcs degradation, we covisualized DNA-PKcs and the HSV-1 IE and E proteins ICP0 and ICP8, respectively, which allowed monitoring of the progression of HSV-1 infection. While DNA-PKcs and ICP0 or ICP8 were not codetected in cells infected with HSV-1 alone, DNA-PKcs and ICP0 or ICP8 were consistently codetected in cells coinfected with HSV-1 and AAV2, and they colocalized with AAV2 RCs. As we did not observe an AAV2-mediated inhibition of ICP0 in this study (see Fig. S1B in the supplemental material) or in a previous study (31), we speculate that AAV2 replication can prevent the ICP0-dependent degradation of DNA-PKcs (48, 67), e.g., by shielding DNA-PKcs in viral RCs. We can exclude the possibility that AAV2 replication directly inhibits the E3 ubiquitin ligase activity of ICP0 because USP7, another target of ICP0-mediated proteasomal degradation (12), was rapidly degraded in coinfected cells. In addition, we can exclude a possible role for USP7 in preventing the degradation of DNA-PKcs by deubiquitinating (45) DNA-PKcs. Experiments performed with ATM-deficient cells revealed that the AAV2-mediated delay of DNA-PKcs degradation affected downstream signaling via Chk2 and p53. In these cells, coinfection with both viruses resulted in a DDR comparable to that induced by infection with HSV-1ΔICP0, including phosphorylation of p53. Consistent with previous data (51, 76), infection of ATM-deficient cells with HSV-1 alone resulted in a broad reduction of DDR signaling with undetectable levels of Chk2-P-T86 and p53-P-S15. We therefore hypothesize that in coinfected ATM-deficient cells, DNA-PK is responsible for the phosphorylation of Chk2 and p53 (Fig. 7). A role for DNA-PK in the phosphorylation of Chk2 and p53 has previously been reported (49, 50, 85, 86, 88). In fact, here we demonstrate that DNA-PK is essential for the activation of p53 in cells infected with AAV2 alone (Fig. 1C and 5A).
Recently, we reported hyperphosphorylation of RPA32 at the DNA-PK target site S4/8 in cells transfected with a plasmid encoding the AAV2 Rep68 or Rep78 protein (30). In the present study, we detected phosphorylation of RPA32 at S4/8 and recruitment of RPA32-P-S4/8 into HSV-1-supported AAV2 RCs. In contrast, HSV-1 infection alone did not induce phosphorylation of RPA32 at S4/8. Similar to the phosphorylation of Chk2 and p53 in coinfected cells, phosphorylation of RPA32 at S4/8 may also be a consequence of the delayed degradation of DNA-PKcs. Further support for this theory comes from the fact that DNA-PK has previously been demonstrated to phosphorylate RPA32 at S4/8 (4, 11, 104).
In a comparison of our results obtained with coinfected cells with those of previous studies examining the DDR signaling in cells infected with HSV-1 alone (13, 51, 76, 96) or Ad2 alone (17, 78) or coinfected with AAV2 and Ad2 (25, 75), it is remarkable that although Ad and HSV-1 by themselves induce very different DDRs, in the presence of AAV2, the induction and activation patterns of DDR proteins are more similar. It seems that during AAV2 replication, the cellular DDR is modulated toward DNA-PK-dependent signaling, which might have positive and/or negative impacts on AAV replication and transduction, depending on the cellular environment and the viral genome structure (23, 25, 75). Further experiments will address the question of whether this modulation can enhance HSV-1-supported AAV2 replication and/or inhibit helper virus replication.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Beard (ETH, Lausanne, Switzerland) for helpful discussions and A. Nicolas (INSERM, Lyon, France) for critically reading the manuscript. We are also grateful to E. Hendrickson, T. Melendy, Y. Shiloh, and P. Nghiem for providing cell lines and B. Roizman, N. Stow, R. D. Everett, H. Buening, U. Greber, and M. Linden for providing viruses.
This work was supported by Swiss National Science Foundation grant 31003A_124938 to C.F. D.L.G. is supported by fellowships from the Swiss National Science Foundation and the Swiss Foundation for Grants in Biology and Medicine (PBZHP3-122925 and PASMP3-132554). A.S. and N.J. are supported by INSERM and the Association Française contre les Myopathies (AFM). Work in the Weitzman laboratory was partially supported by a Pioneer Developmental Chair from the Salk Institute and by NIH grants CA97093 and AI43341 (M.D.W.).
Footnotes
Published ahead of print 19 October 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Adeyemi RO, Landry S, Davis ME, Weitzman MD, Pintel DJ. 2010. Parvovirus minute virus of mice induces a DNA damage response that facilitates viral replication. PLoS Pathog. 6:e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. 2000. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 60:5934–5936 [PubMed] [Google Scholar]
- 3. Alazard-Dany N, et al. 2009. Definition of herpes simplex virus type 1 helper activities for adeno-associated virus early replication events. PLoS Pathog. 5:e1000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anantha RW, Vassin VM, Borowiec JA. 2007. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 282:35910–35923 [DOI] [PubMed] [Google Scholar]
- 5. Antoni BA, et al. 1991. Adeno-associated virus Rep protein inhibits human immunodeficiency virus type 1 production in human cells. J. Virol. 65:396–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bakkenist CJ, Kastan MB. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499–506 [DOI] [PubMed] [Google Scholar]
- 7. Ball HL, Myers JS, Cortez D. 2005. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell 16:2372–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Banin S, et al. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674–1677 [DOI] [PubMed] [Google Scholar]
- 9. Berthet C, Raj K, Saudan P, Beard P. 2005. How adeno-associated virus Rep78 protein arrests cells completely in S phase. Proc. Natl. Acad. Sci. U. S. A. 102:13634–13639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Binz SK, Sheehan AM, Wold MS. 2004. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair 3:1015–1024 [DOI] [PubMed] [Google Scholar]
- 11. Block WD, Yu Y, Lees-Miller SP. 2004. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 32:997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boutell C, Canning M, Orr A, Everett RD. 2005. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J. Virol. 79:12342–12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boutell C, Everett RD. 2004. Herpes simplex virus type 1 infection induces the stabilization of p53 in a USP7- and ATM-independent manner. J. Virol. 78:8068–8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buller RM, Janik JE, Sebring ED, Rose JA. 1981. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 40:241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burma S, Chen BP, Chen DJ. 2006. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair 5:1042–1048 [DOI] [PubMed] [Google Scholar]
- 16. Cahill D, Connor B, Carney JP. 2006. Mechanisms of eukaryotic DNA double strand break repair. Front. Biosci. 11:1958–1976 [DOI] [PubMed] [Google Scholar]
- 17. Carson CT, et al. 2003. The Mre11 complex is required for ATM activation and the G[inf]2/M checkpoint. EMBO J. 22:6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cataldi MP, McCarty DM. 2010. Differential effects of DNA double-strand break repair pathways on single-strand and self-complementary adeno-associated virus vector genomes. J. Virol. 84:8673–8682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan DW, et al. 2002. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 16:2333–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chaturvedi P, et al. 1999. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene 18:4047–4054 [DOI] [PubMed] [Google Scholar]
- 21. Chen BP, et al. 2005. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 280:14709–14715 [DOI] [PubMed] [Google Scholar]
- 22. Chen Y, Sanchez Y. 2004. Chk1 in the DNA damage response: conserved roles from yeasts to mammals. DNA Repair 3:1025–1032 [DOI] [PubMed] [Google Scholar]
- 23. Choi YK, Nash K, Byrne BJ, Muzyczka N, Song S. 2010. The effect of DNA-dependent protein kinase on adeno-associated virus replication. PLoS One 5:e15073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clarke PR, Allan LA. 2009. Cell-cycle control in the face of damage—a matter of life or death. Trends Cell Biol. 19:89–98 [DOI] [PubMed] [Google Scholar]
- 25. Collaco RF, Bevington JM, Bhrigu V, Kalman-Maltese V, Trempe JP. 2009. Adeno-associated virus and adenovirus coinfection induces a cellular DNA damage and repair response via redundant phosphatidylinositol 3-like kinase pathways. Virology 392:24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dart DA, Adams KE, Akerman I, Lakin ND. 2004. Recruitment of the cell cycle checkpoint kinase ATR to chromatin during S-phase. J. Biol. Chem. 279:16433–16440 [DOI] [PubMed] [Google Scholar]
- 27. Daya S, Cortez N, Berns KI. 2009. Adeno-associated virus site-specific integration is mediated by proteins of the nonhomologous end-joining pathway. J. Virol. 83:11655–11664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fraefel C, et al. 2004. Spatial and temporal organization of adeno-associated virus DNA replication in live cells. J. Virol. 78:389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Giglia-Mari G, Zotter A, Vermeulen W. 2011. DNA damage response. Cold Spring Harb. Perspect. Biol. 3:a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Glauser DL, et al. 2010. Inhibition of herpes simplex virus type 1 replication by adeno-associated virus Rep proteins depends on their combined DNA-binding and ATPase/helicase activities. J. Virol. 84:3808–3824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Glauser DL, et al. 2007. Live covisualization of competing adeno-associated virus and herpes simplex virus type 1 DNA replication: molecular mechanisms of interaction. J. Virol. 81:4732–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo Z, Kumagai A, Wang SX, Dunphy WG. 2000. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14:2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harper JW, Elledge SJ. 2007. The DNA damage response: ten years after. Mol. Cell 28:739–745 [DOI] [PubMed] [Google Scholar]
- 34. Heilbronn R, Burkle A, Stephan S, Hzur Hausen. 1990. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol. 64:3012–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heilbronn R, et al. 2003. ssDNA-dependent colocalization of adeno-associated virus Rep and herpes simplex virus ICP8 in nuclear replication domains. Nucleic Acids Res. 31:6206–6213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Helleday T, Lo J, van Gent DC, Engelward BP. 2007. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair 6:923–935 [DOI] [PubMed] [Google Scholar]
- 37. Hunter LA, Samulski RJ. 1992. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J. Virol. 66:317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jackson SP, Bartek J. 2009. The DNA-damage response in human biology and disease. Nature 461:1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jacob RJ, Roizman B. 1977. Anatomy of herpes simplex virus DNA VIII. Properties of the replicating DNA. J. Virol. 23:394–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jing XJ, Kalman-Maltese V, Cao X, Yang Q, Trempe JP. 2001. Inhibition of adenovirus cytotoxicity, replication, and E2a gene expression by adeno-associated virus. Virology 291:140–151 [DOI] [PubMed] [Google Scholar]
- 41. Jurvansuu J, Raj K, Stasiak A, Beard P. 2005. Viral transport of DNA damage that mimics a stalled replication fork. J. Virol. 79:569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kastan MB, Lim DS. 2000. The many substrates and functions of ATM. Nat. Rev. Mol. Cell Biol. 1:179–186 [DOI] [PubMed] [Google Scholar]
- 43. Kastan MB, Lim DS, Kim ST, Xu B, Canman C. 2000. Multiple signaling pathways involving ATM. Cold Spring Harb. Symp. Quant. Biol. 65:521–526 [DOI] [PubMed] [Google Scholar]
- 44. Kleinschmidt JA, Mohler M, Weindler FW, Heilbronn R. 1995. Sequence elements of the adeno-associated virus rep gene required for suppression of herpes-simplex-virus-induced DNA amplification. Virology 206:254–262 [DOI] [PubMed] [Google Scholar]
- 45. Komander D, Clague MJ, Urbe S. 2009. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10:550–563 [DOI] [PubMed] [Google Scholar]
- 46. Lakin ND, Hann BC, Jackson SP. 1999. The ataxia-telangiectasia related protein ATR mediates DNA-dependent phosphorylation of p53. Oncogene 18:3989–3995 [DOI] [PubMed] [Google Scholar]
- 47. Lamarche BJ, Orazio NI, Weitzman MD. 2010. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 584:3682–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lees-Miller SP, et al. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70:7471–7477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW. 1992. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol. Cell. Biol. 12:5041–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li J, Stern DF. 2005. Regulation of CHK2 by DNA-dependent protein kinase. J. Biol. Chem. 280:12041–12050 [DOI] [PubMed] [Google Scholar]
- 51. Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 102:5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Linden RM, Berns KI. 2000. Molecular biology of adeno-associated viruses. Contrib. Microbiol. 4:68–84 [DOI] [PubMed] [Google Scholar]
- 53. Liu Q, et al. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 14:1448–1459 [PMC free article] [PubMed] [Google Scholar]
- 54. Liu S, et al. 2011. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol. Cell 43:192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lovejoy CA, Cortez D. 2009. Common mechanisms of PIKK regulation. DNA Repair 8:1004–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Matsuoka S, Huang M, Elledge SJ. 1998. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282:1893–1897 [DOI] [PubMed] [Google Scholar]
- 57. Matsuoka S, et al. 2000. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 97:10389–10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mohni KN, Livingston CM, Cortez D, Weller SK. 2010. ATR and ATRIP are recruited to herpes simplex virus type 1 replication compartments even though ATR signaling is disabled. J. Virol. 84:12152–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mordes DA, Cortez D. 2008. Activation of ATR and related PIKKs. Cell Cycle 7:2809–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morio T, Kim H. 2008. Ku, Artemis, and ataxia-telangiectasia-mutated: signalling networks in DNA damage. Int. J. Biochem. Cell Biol. 40:598–603 [DOI] [PubMed] [Google Scholar]
- 61. Nada S, Trempe JP. 2002. Characterization of adeno-associated virus rep protein inhibition of adenovirus E2a gene expression. Virology 293:345–355 [DOI] [PubMed] [Google Scholar]
- 62. Nam EA, Cortez D. 2011. ATR signalling: more than meeting at the fork. Biochem. J. 436:527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nash K, Chen W, Salganik M, Muzyczka N. 2009. Identification of cellular proteins that interact with the adeno-associated virus Rep protein. J. Virol. 83:454–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ni TH, et al. 1998. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J. Virol. 72:2777–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nicolas A, et al. 2010. Identification of Rep-associated factors in herpes simplex virus type 1-induced adeno-associated virus type 2 replication compartments. J. Virol. 84:8871–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ohnishi T, Mori E, Takahashi A. 2009. DNA double-strand breaks: their production, recognition, and repair in eukaryotes. Mutat. Res. 669:8–12 [DOI] [PubMed] [Google Scholar]
- 67. Parkinson J, Lees-Miller SP, Everett RD. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Raj K, Ogston P, Beard P. 2001. Virus-mediated killing of cells that lack p53 activity. Nature 412:914–917 [DOI] [PubMed] [Google Scholar]
- 69. Reinhardt HC, Yaffe MB. 2009. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr. Opin. Cell Biol. 21:245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Robinson K, Asawachaicharn N, Galloway DA, Grandori C. 2009. c-Myc accelerates S-phase and requires WRN to avoid replication stress. PLoS One 4:e5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rommelaere J, Cornelis JJ. 1991. Antineoplastic activity of parvoviruses. J. Virol. Methods 33:233–251 [DOI] [PubMed] [Google Scholar]
- 72. Samulski RJ, et al. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Saudan P, Vlach J, Beard P. 2000. Inhibition of S-phase progression by adeno-associated virus Rep78 protein is mediated by hypophosphorylated pRb. EMBO J. 19:4351–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schlehofer JR. 1994. The tumor suppressive properties of adeno-associated viruses. Mutat. Res. 305:303–313 [DOI] [PubMed] [Google Scholar]
- 75. Schwartz RA, Carson CT, Schuberth C, Weitzman MD. 2009. Adeno-associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase. J. Virol. 83:6269–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shirata N, et al. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 280:30336–30341 [DOI] [PubMed] [Google Scholar]
- 77. Song S, et al. 2004. DNA-dependent PK inhibits adeno-associated virus DNA integration. Proc. Natl. Acad. Sci. U. S. A. 101:2112–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stracker TH, Carson CT, Weitzman MD. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352 [DOI] [PubMed] [Google Scholar]
- 79. Stracker TH, et al. 2004. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 78:441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tapia-Alveal C, Calonge TM, O'Connell MJ. 2009. Regulation of chk1. Cell Div. 4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Taylor TJ, Knipe DM. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Taylor TJ, McNamee EE, Day C, Knipe DM. 2003. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology 309:232–247 [DOI] [PubMed] [Google Scholar]
- 83. Tibbetts RS, et al. 2000. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 14:2989–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Timpe JM, Verrill KC, Trempe JP. 2006. Effects of adeno-associated virus on adenovirus replication and gene expression during coinfection. J. Virol. 80:7807–7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tomimatsu N, Mukherjee B, Burma S. 2009. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 10:629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang S, et al. 2000. The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc. Natl. Acad. Sci. U. S. A. 97:1584–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang XQ, Redpath JL, Fan ST, Stanbridge EJ. 2006. ATR dependent activation of Chk2. J. Cell. Physiol. 208:613–619 [DOI] [PubMed] [Google Scholar]
- 88. Wang Y, Eckhart W. 1992. Phosphorylation sites in the amino-terminal region of mouse p53. Proc. Natl. Acad. Sci. U. S. A. 89:4231–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Warmerdam DO, Kanaar R. 2010. Dealing with DNA damage: relationships between checkpoint and repair pathways. Mutat. Res. 704:2–11 [DOI] [PubMed] [Google Scholar]
- 90. Weindler FW, Heilbronn R. 1991. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J. Virol. 65:2476–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Weitzman MD, Fisher KJ, Wilson JM. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70:1845–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Weitzman MD, Lilley CE, Chaurushiya MS. 2010. Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 64:61–81 [DOI] [PubMed] [Google Scholar]
- 93. Wilcock D, Lane DP. 1991. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349:429–431 [DOI] [PubMed] [Google Scholar]
- 94. Wilkinson DE, Weller SK. 2006. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J. Cell Sci. 119:2695–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wilkinson DE, Weller SK. 2005. Inhibition of the herpes simplex virus type 1 DNA polymerase induces hyperphosphorylation of replication protein A and its accumulation at S-phase-specific sites of DNA damage during infection. J. Virol. 79:7162–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wilkinson DE, Weller SK. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wold MS. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61–92 [DOI] [PubMed] [Google Scholar]
- 98. Wold MS, Kelly T. 1988. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc. Natl. Acad. Sci. U. S. A. 85:2523–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wyman C, Kanaar R. 2006. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 40:363–383 [DOI] [PubMed] [Google Scholar]
- 100. Yang Q, Chen F, Ross J, Trempe JP. 1995. Inhibition of cellular and SV40 DNA replication by the adeno-associated virus Rep proteins. Virology 207:246–250 [DOI] [PubMed] [Google Scholar]
- 101. Yang Q, Chen F, Trempe JP. 1994. Characterization of cell lines that inducibly express the adeno-associated virus Rep proteins. J. Virol. 68:4847–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. You JS, Wang M, Lee SH. 2000. Functional characterization of zinc-finger motif in redox regulation of RPA-ssDNA interaction. Biochemistry 39:12953–12958 [DOI] [PubMed] [Google Scholar]
- 103. You Z, Chahwan C, Bailis J, Hunter T, Russell P. 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 25:5363–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zernik-Kobak M, Vasunia K, Connelly M, Anderson CW, Dixon K. 1997. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem. 272:23896–23904 [DOI] [PubMed] [Google Scholar]
- 105. Zhao H, Piwnica-Worms H. 2001. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21:4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548 [DOI] [PubMed] [Google Scholar]
- 107. Zou Y, Liu Y, Wu X, Shell SM. 2006. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 208:267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.