Abstract
Cellular entry of Ebola virus (EBOV), a deadly hemorrhagic fever virus, is mediated by the viral glycoprotein (GP). The receptor-binding subunit of GP must be cleaved (by endosomal cathepsins) in order for entry and infection to proceed. Cleavage appears to proceed through 50-kDa and 20-kDa intermediates, ultimately generating a key 19-kDa core. How 19-kDa GP is subsequently triggered to bind membranes and induce fusion remains a mystery. Here we show that 50-kDa GP cannot be triggered to bind to liposomes in response to elevated temperature but that 20-kDa and 19-kDa GP can. Importantly, 19-kDa GP can be triggered at temperatures ∼10°C lower than 20-kDa GP, suggesting that it is the most fusion ready form. Triggering by heat (or urea) occurs only at pH 5, not pH 7.5, and involves the fusion loop, as a fusion loop mutant is defective in liposome binding. We further show that mild reduction (preferentially at low pH) triggers 19-kDa GP to bind to liposomes, with the wild-type protein being triggered to a greater extent than the fusion loop mutant. Moreover, mild reduction inactivates pseudovirion infection, suggesting that reduction can also trigger 19-kDa GP on virus particles. Our results support the hypothesis that priming of EBOV GP, specifically to the 19-kDa core, potentiates GP to undergo subsequent fusion-relevant conformational changes. Our findings also indicate that low pH and an additional endosomal factor (possibly reduction or possibly a process mimicked by reduction) act as fusion triggers.
INTRODUCTION
The filovirus Ebola virus (EBOV) is an extremely lethal pathogen for which there are currently no approved vaccines or antiviral agents (18). Entry of the virus into cells is mediated by GP, the sole glycoprotein species present on the virion surface (32). GP is a class I fusion protein (22, 53) that mediates binding to the cell surface and fusion with an intracellular membrane. Similar to many other class I fusion proteins (e.g., influenza virus hemagglutinin), native GP is a trimer of heterodimers composed of a receptor binding subunit (GP1) and a fusion subunit (GP2) (31). The two subunits arise by cleavage of a precursor polypeptide as GP transits the Golgi compartment and remain associated through a disulfide bond and noncovalent interactions. GP1 is a large subunit (apparent molecular mass of 130 kDa) that includes a glycan cap containing 4 N-linked carbohydrate addition sites and a heavily O-glycosylated mucin-like domain. The mucin-like domain is dispensable for infection of cells in culture; pseudovirions and virus-like particles bearing genetically engineered GP with a deletion of the mucin-like domain (GPΔ) are fully competent for entry and infection (17, 29, 46). The two carbohydrate-rich domains of GP1 sit atop a head domain, which contains a receptor-binding region (RBR) (5, 17, 29, 35, 39). In turn, the head domain of GP1 sits atop a base that interacts extensively with the fusion subunit (GP2), which houses the fusion loop (26).
The endosomal proteases cathepsin B (Cat B) and cathepsin L (Cat L) are required for EBOV to enter the cytoplasm and therefore to infect host cells (10, 29, 45). Cat B and Cat L cleave GP within the unstructured β13-β14 loop of GP1, generating a 20-kDa intermediate and then the key 19-kDa core (17, 25, 45). Processing appears to go through an ∼50-kDa intermediate that resembles GPΔ. During these massive cleavage events, GP2 is fully protected and remains disulfide bonded to GP1. Processing to 19-kDa GP removes the mucin-like domain, glycan cap, and outer β strand (β14) of the head domain (of GP1). The receptor-binding region (RBR) and base of GP1 as well as all of GP2 remain intact. We have designated cathepsin cleavage of GP1 a priming step, since we have proposed that GP2 remains clamped in the 19-kDa core (45). Evidence in support of this proposal includes the observation that entry by pseudovirions bearing 19-kDa GP is blocked by both bafilomycin and E64(d) (45, 54), suggesting that low pH and an E64-sensitive process are required to induce 19-kDa GP to mediate fusion and entry. A first non-mutually exclusive hypothesis (45) to explain these findings is that priming to the 19-kDa core potentiates GP to respond to a fusion trigger(s). A second is that cleavage to 19 kDa enhances the ability of GP to bind to an endosomal receptor (29).
Most viral fusion proteins reside on their respective virion surfaces in a metastable state. Exposure to a fusion trigger initiates conformational changes that drive the fusion protein to a lower energy state and promote fusion. For all viral fusion proteins, an important step of triggering is exposure and repositioning of the fusion peptide (or fusion loop) so that it can bind to the target membrane. Only when the fusion peptide (loop) has engaged the target membrane can the subsequent fold-back of the glycoprotein drive a merger of the viral and target bilayers (22, 53). In the present study, we first established an assay with which to measure a major early step of fusion triggering for EBOV GP: fusion loop-dependent binding to target membranes. We next used this assay to explore the role of cathepsin cleavage and low pH in triggering EBOV GP. Lastly, we obtained evidence that mild reduction can trigger 19-kDa GP. Our findings provide the first evidence for a triggered conformational change in EBOV GP as well as strong support for the hypothesis that a major role of cathepsin cleavage, specifically to 19-kDa GP, is to potentiate EBOV GP for fusion triggering. In addition, our results provide new insights into potential mechanisms of fusion triggering.
MATERIALS AND METHODS
EBOV GPΔ ectodomain production and purification.
The ectodomain of Zaire EBOV with the mucin-like domain deleted (GPΔ Ecto) was produced and purified essentially as described in reference 31. Briefly, a pDisplay vector encoding the GPΔ Ecto tagged with an HA epitope was transfected using CaPO4 into 293T cells, which were treated 24 h posttransfection with 10 mM butyric acid to enhance protein expression. Cell medium was harvested 48 h posttransfection, cleared of debris by two centrifugations at 2,500 rpm for 7 min at 4°C in an IEC Centra MP4R centrifuge, and concentrated by tangential flow over a PelliconXL Biomax 100-kDa membrane using a Labscale TFF System (Millipore). The retentate was then affinity purified over resin conjugated with antibody 3F10 (Roche), and fractions containing GPΔ Ecto were pooled and concentrated with Vivaspin concentrators (100-kDa molecular mass cutoff; Sartorius Stedim VS0241). Protein concentration was determined with a Nanodrop 1000 (Thermo Scientific).
Thermolysin cleavage of EBOV GPΔ Ecto to 19 kDa.
GPΔ Ecto (0.25 mg/ml) was incubated with thermolysin (Sigma P1512; 0.1 to 0.25 mg/ml; optimal concentration predetermined for different lots) in HM buffer (20 mM HEPES, 20 mM MES [morpholinepropanesulfonic acid], 130 mM NaCl [pH 7.5]) containing 2 mM CaCl2 at 37°C for 1 h. The cleavage was stopped by the addition of EDTA to 10 mM. The sample was concentrated, and the thermolysin and EDTA were removed by washing the sample 6 times with 500 μl of HM buffer following concentration through an Amicon Ultra spin concentrator (100-kDa cutoff; Millipore UFC510024). The final concentration of 19-kDa Ecto (in HM buffer) was adjusted to 0.2 mg/ml, determined as described above. All preparations were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Cathepsin L cleavage of EBOV GPΔ Ecto to 20 kDa.
GPΔ Ecto (0.25 mg/ml) was incubated with Cat L (5 μg/ml) in sodium acetate (NaOAc) buffer (100 mM NaOAc, 1 mM EDTA [pH 4.5]) containing 5 mM dithiothreitol (DTT) for 1 h at 37°C. The reaction mixture was then concentrated and washed, and the Cat L was removed as described above (for removing thermolysin and EDTA from the 19-kDa GP Ecto).
Preparation of biotinylated liposomes.
Mixtures of phosphatidylcholine (PC; Avanti 830051P), phosphatidylethanolamine (PE; Avanti 841118P), biotinyl-phosphatidylethanolamine (BtPE; Avanti 860562C), sphingomyelin (Sph: Avanti 860061P), and cholesterol (Chol; Sigma C-3137) in a ratio of 1.0:0.9:0.1:1.0:1.5 were dried in a glass tube with N2, lyophilized overnight, and resuspended in HM buffer such that the concentration of lipid was 4 mM. The liposome suspension was subjected to 5 cycles of freeze-thawing, extruded 30 times through a 100-nm Nuclepore filter (Fisher 800309), and stored at 4°C. Liposomes were used within 1 week.
Liposome binding assay.
19-kDa GP Ecto(0.4 μg) and 8 μl of biotinylated liposomes (4 mM) were incubated in a final volume of 40 μl of HM buffer at 4°C for 20 min. Samples were acidified to pH 5.5 or 5.0 with 3.5 to 5 μl of 100 mM citric acid and brought to a total volume of 50 μl with distilled water. For the pH 7.5 samples, the volume was brought to 50 μl with HM buffer. Samples were then incubated for various times at various temperatures, placed on ice, and the acidified samples were neutralized (to pH 7.5) by the addition of 4 to 5 μl of 0.5 M NaOH. An equivalent volume of HM buffer was added to pH 7.5 samples. Neutralized samples were added to 40 μl BcMag streptavidin-conjugated magnetic beads (Bioclone MMI-106; used within 2 months of purchase) for 1 h at room temperature. The solution above the beads was removed (and saved as the unbound fraction), and the beads were washed as follows: for Fig. 1, three times with HM buffer; for Fig. 3, twice with HM buffer and once with the indicated concentration of NaCl or urea (in HM buffer); for all other figures, twice with HM buffer and once with 1 M NaCl (in HM buffer). The washed beads were then resuspended in 50 μl HM buffer (bound fraction). The bound and unbound fractions were then separated by SDS-PAGE. Gels were transferred to nitrocellulose and probed with an anti-GP1 polyclonal antibody followed by an Alexa Fluor 680 anti-rabbit IgG secondary antibody (Invitrogen A-21109). Blots were imaged and quantified using a Li-Cor Odyssey gel imager and associated software. Except where indicated, results are displayed as the percent bound GP, calculated as [(bound GP)/(unbound GP + bound GP)] × 100. For Fig. 6, samples were set up with equivalent molar amounts (to 19-kDa Ecto) of 20-kDa GP Ecto (0.4 μg) and GPΔ Ecto (0.65 μg). For Fig. 3 and 5A, samples were set up with 0.8 μg of 19-kDa GP Ecto proteins and 16 μl of biotinylated liposomes and were incubated with 60 μl BcMag streptavidin-conjugated magnetic beads.
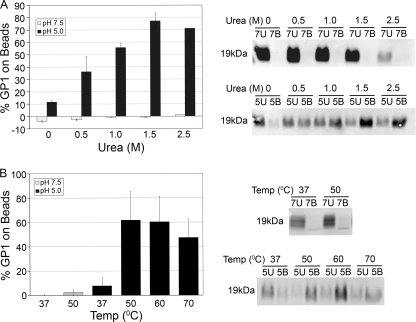
Fig 1.
Urea and elevated temperature each induce the 19-kDa GP Ecto to bind to liposomes at low pH. (A) GPΔ Ecto was cleaved with thermolysin to 19-kDa GP Ecto and incubated with liposomes in the presence of urea at pH 7.5 or pH 5.0 at 37°C for 10 min. The amount of 19-kDa GP Ecto bound to liposomes was then determined as described in Materials and Methods. The average percentage of total 19-kDa GP Ecto bound (% GP1 on beads) from duplicate samples is shown. Error bars indicate standard deviations (SD). The gels on the right are from one set of samples. (B) 19-kDa GP Ecto was incubated with liposomes at the indicated temperature and either pH 7.5 or pH 5.0, and the percentage of total 19-kDa GP Ecto bound to liposomes was measured as for panel A. Data are the averages from six to eight determinations. Error bars indicate SD. Representative gel samples are shown on the right.
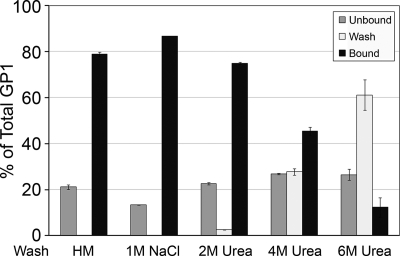
Fig 3.
Association of 19-kDa GP Ecto with liposomes is stable to treatment with 1 M NaCl or 2 M urea. The 19-kDa GP Ecto was incubated with liposomes at 50°C and pH 5.0 for 10 min and then processed as described in the legend to Fig. 1, except that after two washes with HM buffer, the liposome-bound beads were washed once with HM buffer containing 1 M NaCl or the indicated concentration of urea. The unbound, third-wash, and bound fractions were then analyzed for 19-kDa GP Ecto as described in the legend to Fig. 1. Data are the percentages of total GP1 detected in each fraction and are the averages from duplicate samples. Error bars indicate SD.
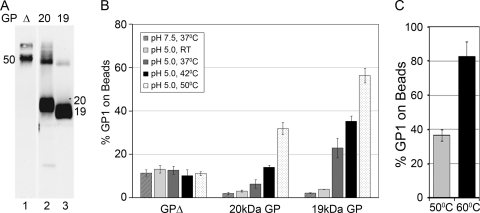
Fig 6.
Cathepsin cleavage of GPΔ Ecto to 19 kDa is necessary for optimal liposome binding. (A) GPΔEcto (Δ, lane 1) was digested with Cat L (20 kDa, lane 2) or thermolysin (19 kDa, lane 3), separated on an SDS–15% polyacrylamide gel, and analyzed by Western blotting for GP1. (B) Uncleaved GPΔ Ecto, 20-kDa GP Ecto, and 19-kDa GP Ecto were incubated as indicated. Liposome binding was then assessed as described in the legend to Fig. 1. (C) The 20-kDa GP Ecto was incubated with liposomes at pH 5.0 for 5 min at either 50°C or 60°C, and binding was measured as for panel B. Data are the averages from duplicate samples. Error bars indicate SD.
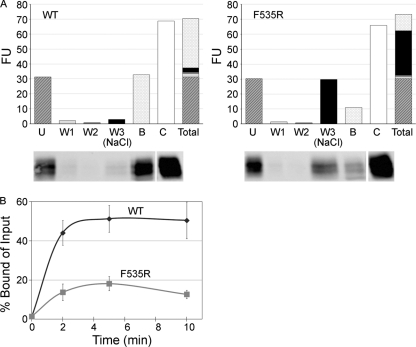
Fig 5.
The 19-kDa GP Ecto of a fusion loop mutant (F535R) is impaired in liposome binding. (A) Equivalent amounts (0.8 μg) of WT and F535R 19-kDa GP Ecto proteins were incubated with liposomes at pH 5.0 and 50°C for 10 min. Mixtures were then incubated with streptavidin magnetic beads as described in Materials and Methods and the following fractions analyzed by SDS-PAGE and Western blotting for GP1: unbound (U), 1st wash with HM buffer (W1), 2nd wash with HM buffer (W2), 3rd wash with HM buffer containing 1 M NaCl [W3 (NaCl)], and remaining bound protein (B). The fluorescence units (FU) of corresponding gel samples (below the graphs) are shown. The control (C) was the supernatant from a sample maintained for 10 min at 4°C and pH 7.5. “Total” shows a stacked column of the fluorescent units found in the U, W1, W2, W3, and B fractions (i.e., the sum equals all of the GP1 recovered). The control gel sample was repositioned from the far left on the gel to the far right of the image (indicated with a space) to align with the corresponding bars in the overlying graph. (B) Equivalent amounts of WT and mutant F535R 19-kDa GPΔ Ecto proteins were incubated with liposomes for the indicated times at 50°C and pH 5.0. The protein-liposome complexes were then collected on magnetic beads, which were washed twice with HM buffer and once with HM buffer containing 1 M NaCl. Samples were then separated on SDS gels and blotted for GP1 as described above. The results were calculated as [(bound GP)/(total input GP)] × 100, where total input GP is the unbound signal from parallel samples maintained at pH 7.5 and 4°C and processed in the identical manner. Data are the averages from three experiments, each performed in duplicate. Error bars indicate SD.
Infections with VSV GP pseudovirions.
Vesicular stomatitis virus (VSV) particles bearing GPΔ or GPΔ F535R and encoding green fluorescent protein (GFP) were produced as described previously (17, 45, 50). To generate VSV pseudovirions (for Fig. 8) bearing 19-kDa GP, pseudovirions with GPΔ (0.25 mg/ml) were incubated with thermolysin (0.25 mg/ml) at 37°C for 45 min. The reaction was quenched by the addition of EDTA (10 mM), and the thermolysin and EDTA were removed by centrifugation through a 1-ml cushion containing 20% sucrose (in HM buffer) at 32,000 rpm for 2 h in an SW55 rotor. The pseudovirion-containing pellet was resuspended in 100 μl of 10% sucrose (in HM buffer).
Fig 8.
Mild reduction inactivates the ability of 19-kDa GP pseudovirions to mediate infection. VSV GPΔ pseudovirions and VSV GPΔ pseudovirions that had been cleaved with thermolysin to generate 19-kDa GP (VSV-19kDa GP) were pretreated at pH 7.0 or at pH 5.5 with or without 5 mM TCEP for 10 min at 37°C. Samples were neutralized by adding 16 volumes of Optimem medium and added to Vero cells. After 24 h, the cells were detached and fixed, and the percentage of GFP-expressing infected cells was measured by flow cytometry. Data are from one of two experiments (performed in duplicate) with similar results. Error bars indicate SD.
RESULTS
Elevated temperature or urea induces 19-kDa GP Ecto to bind to liposomes.
We previously showed (45) that infection by pseudovirions bearing 19-kDa GP, produced by digesting GP pseudovirions either with Cat B plus Cat L or with thermolysin, is potently inhibited by bafilomycin, an agent that raises the pH of endosomes. This finding suggested that the fusion subunit (GP2) in 19-kDa GP is still clamped, i.e., that its fusion loop has not been exposed, and that 19-kDa GP is, therefore, in a metastable state. As a first test of this hypothesis, we expressed and purified the ectodomain of EBOV GP lacking its mucin-like domain (GPΔ Ecto) (31), cleaved it with thermolysin to produce 19-kDa GP Ecto, and asked if the addition of heat or urea could induce target membrane binding. This type of experiment has confirmed the metastability of many class I viral fusion proteins, including those of influenza virus (9), PIV5 (11), avian sarcoma and leukosis virus (ASLV) (37, 49), and Moloney murine leukemia virus (MoMLV) (51). The aforementioned studies monitored association of the fusion protein ectodomain with target liposomes using sucrose flotation gradients (7, 16, 20, 48). To increase the throughput of the analysis, we modified a method employing liposomes containing a biotinylated lipid (34). If a fusion protein ectodomain binds to the biotinylated liposomes, it will remain associated with the liposomes following incubation with magnetic beads coupled with streptavidin. We first tested the assay with the well-characterized low-pH-activated bromelain-released fragment of the influenza hemagglutinin (BHA). When BHA was exposed to pH 5 for 5 min at 37°C, the majority was pulled over by magnetic streptavidin beads (by virtue of binding to target biotinylated liposomes), whereas only trace amounts were bound to the beads in samples treated at pH 7 for 5 min at 37°C (data not shown). These behaviors are consistent with results obtained using liposome flotation gradients (16, 48).
We next used the magnetic bead liposome binding assay to ask if elevated temperature or urea could trigger the19-kDa GP Ecto to bind to liposomes. We performed the assays at both neutral and low pH, since GP-mediated fusion is thought to occur in late endosomes (10, 43, 45), which have a pH of ∼5, and since low pH is required for entry of pseudovirions bearing 19-kDa GP (45). As seen in Fig. 1A, treatment of the 19-kDa GP Ecto with urea at pH 5.0, but not at neutral pH, induced 19-kDa GP Ecto to bind to liposomes. Peak binding occurred upon exposure of the19-kDa GP Ecto to 1.5 M urea. Elevated temperature also induced the 19-kDa GP Ecto to bind to liposomes (Fig. 1B). At pH 5.0, a low level of binding was seen at 37°C and maximum binding was observed at 50°C to 60°C. No binding was observed at pH 7.5 at 37°C, with only a trace amount detected at 50°C. Our standard liposomes (see Materials and Methods) are composed of PC-PE-BtPE-Sph-Chol (1.0:0.9:0.1:1.0:1.5). In samples heated to 50°C at pH 5, the 19-kDa GP Ecto bound equally well to liposomes in which half of the (nonbiotinylated) PE was replaced with either POPG [1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)], LBPA (lysobisphosphatidic acid), or POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine) (data not shown).
Figure 2 shows a more detailed analysis of the temperature dependence of liposome binding. At pH 5, virtually no association was observed at 22°C, a sharp increase was seen between 37°C and 45°C, and maximal association was observed at ∼50°C (Fig. 2A). Temperature affected both the rate and extent of 19-kDa GP Ecto binding to liposomes (Fig. 2B). At 50°C, binding increased rapidly, reaching an apparent maximum at 2 min; at 42°C and 37°C, liposome binding occurred progressively more slowly and less extensively.
Fig 2.
Time and temperature dependence of 19-kDa GP Ecto binding to liposomes. (A) 19-kDa GP Ecto was incubated with liposomes at pH 5.0 (black diamonds) or pH 7.5 (gray circle) at the indicated temperatures for 10 min, and the percentage of total 19-kDa GP Ecto bound to liposomes was measured as described in the legend to Fig. 1. Results of one of two experiments (triplicate samples) with similar results are shown. (B) Samples of 19-kDa GP Ecto and liposomes were incubated at pH 5.0 for the indicated times at 50°C, 42°C, or 37°C and processed as for panel A. Data are the averages from duplicate samples. Error bars indicate SD.
19-kDa GP Ecto binds stably to liposomes through its fusion loop.
We next asked if the 19-kDa GP Ecto associates stably with liposomes. To do this, we incubated complexes of 19-kDa GP Ecto and liposomes, formed by incubation at 50°C and pH 5, with 1 M NaCl or various concentrations of urea; if the 19-kDa GP Ecto were only weakly (peripherally) bound, it should be removed by 1 M NaCl or low concentrations of urea. As seen in Fig. 3, the heat- and low-pH-induced interaction between the 19-kDa GP Ecto and liposomes was stable to treatments with 1 M NaCl or 2 M urea, suggesting a stable association in which a hydrophobic segment (e.g., the fusion loop in GP2) is embedded in the liposome membrane. Higher concentrations of urea (4 M and 6 M) caused some 19-kDa GP Ecto to elute from the liposomes. A wash with 1 M NaCl was included in all subsequent experiments to assess conditions that trigger the 19-kDa GP Ecto to bind stably to liposomes.
Based on the preceding result, we hypothesized that stable liposome association of 19-kDa GP is mediated by the fusion loop. To test this hypothesis, we compared the ability of the wild-type (WT) 19-kDa GP Ecto and a 19-kDa GP Ecto engineered to contain a mutation in its fusion loop (F535R) (26) to bind to liposomes. We first confirmed (Fig. 4A) that VSV pseudovirions bearing F535R are significantly less infectious than particles bearing WT GP (26), even though the mutant GP is well incorporated into pseudovirions (26) (data not shown). We also showed that the F535R mutant Ecto is efficiently cleaved by thermolysin (Fig. 4B). Purified WT and mutant 19-kDa GP Ecto proteins were then assessed for heat-induced liposome binding at pH 5. The mutant Ecto (Fig. 5A, right) is significantly impaired in its ability to bind stably to the liposomes (Fig. 5B), i.e., it binds ∼30% as well as the WT 19-kDa Ecto (Fig. 5A, left). The mutant Ecto binds initially but is eluted during the high-salt wash (Fig. 5A). For the fusion loop mutant, both the rate and extent of liposome association were significantly impaired compared to those for the WT Ecto (Fig. 5B). The behavior of the F535R EBOV fusion loop mutant is reminiscent of those of a classic influenza HA fusion peptide mutant (G1E) and a mutant (V30E) in the fusion loop of the ASLV Env glycoprotein, a fusion loop that shares many similarities with the fusion loop of EBOV (13, 21). Although severely debilitated with regard to fusion and infection, the HA (G1E) and ASLV Env (V30E) fusion peptide and loop mutants are only partially impaired in liposome binding (∼50% binding compared to WT) (19, 23, 24).
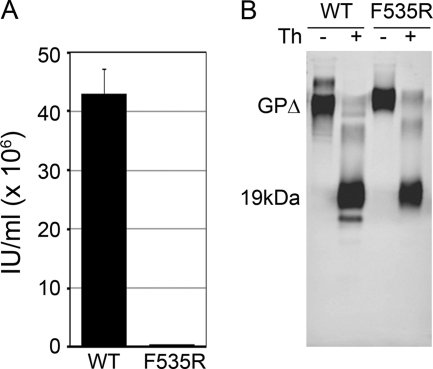
Fig 4.
Infectivity and thermolysin cleavage of the F535R fusion loop mutant. (A) Infectivity. Equivalent volumes of culture supernatants containing VSV pseudovirions bearing wild-type or F535R EBOV GPΔ were used to determine titers on Vero cells by serial dilution (triplicate samples). Infected cells were scored by visual inspection for GFP expression. Titers of wild-type and F535R virus were 4.30 × 107 IU/ml and 3.75 × 105 IU/ml, respectively. The data are from one experiment performed in triplicate. Bars indicate SD. (B) Thermolysin cleavage. WT or F535R GPΔ Ecto proteins (0.1 mg/ml) were treated with thermolysin (Th; 0.1 mg/ml) (+) or buffer (−) as described in Materials and Methods and analyzed by SDS-PAGE and Western blotting for GP1.
Sequential cleavage of GPΔEcto to the 19-kDa form progressively potentiates elevated temperature-induced liposome binding.
Cleavage of EBOV GP by cathepsins appears to occur in steps that trim the apparent mass of GP1 from 130 kDa to ∼50 kDa (with properties similar to those of the engineered GPΔ), then to 20 kDa, and finally to 19 kDa (17, 25, 29, 45, 54). Infection by pseudovirions bearing 19-kDa GP is less sensitive to inhibition by cathepsin antagonists than are infections by pseudovirions bearing either full-length GP or GPΔ (10, 45). Furthermore, there is a strong correlation between the relative amount of 20-kDa versus 19-kDa GP and the sensitivity of pseudovirion infection to antagonists of Cat B (45, 54). The 19-kDa GP is also more sensitive to subsequent cathepsin L digestion than 20-kDa GP (54) (data not shown). Given these differences between GPΔ, 20-kDa GP, and 19-kDa GP, we predicted that there would be a relationship between the degree of processing and the ease of triggering. To test this prediction, we prepared Ecto proteins corresponding to GPΔ, 20-kDa GP, and 19-kDa GP (Fig. 6A). We then compared the ability of the GPΔ, 20-kDa GP, and 19-kDa GP Ecto proteins to bind to liposomes in response to incubation at elevated temperatures at pH 5. As seen in Fig. 6B, only a small amount (∼10%) of the GPΔEcto bound to the liposomes, and this association occurred independently of temperature. This was in sharp contrast to the 19-kDa GP Ecto (Fig. 6B) which, as also shown in Fig. 1B and 2, associated strongly with liposomes in a highly temperature-dependent manner. Interestingly, the 20-kDa GP Ecto (Fig. 6B) bound to liposomes but required higher temperatures to achieve equivalent levels of binding compared to the 19-kDa GP Ecto. For example, at 50°C we observed ∼35% binding of the 20-kDa GP Ecto compared with ∼60% binding of the 19-kDa GP Ecto. Raising the temperature to 60°C increased the amount of the 20-kDa GP Ecto that bound (Fig. 6C). These findings support our hypotheses that GPΔ is refractory to fusion triggering and that 19-kDa GP is more readily triggered to bind to target membranes than 20-kDa GP.
Mild reduction triggers 19-kDa GP Ecto, but not GPΔ Ecto, to bind to liposomes.
We previously speculated that reduction of a disulfide bond(s) might trigger 19-kDa GP to initiate conformational changes leading to fusion (45). We therefore asked whether mild reduction triggers the 19-kDa GP Ecto to bind to liposomes (at 37°C). For this test we used Tris(2-carboxyethyl)phosphine hydrochloride (TCEP; Hampton Research), a reducing agent that has reducing activity comparable to that of DTT (at equivalent concentrations) but, unlike DTT, functions equally well over a wide pH range (information from manufacturer). The reducing potential of 5 mM DTT has been reported to be −215 mV (40). As seen in Fig. 7A, exposure to 1 or 5 mM TCEP at 37°C and pH 5.5 induced the 19-kDa GP Ecto to bind to liposomes. In contrast, no (1 mM TCEP) or only a low level (5 mM TCEP) of binding above background was observed with the unprimed GPΔ Ecto. As seen in Fig. 7B, TCEP induced greater binding of 19-kDa GP Ecto to liposomes at pH 5.5 than at pH 7.5, with the difference being most apparent in samples treated with 1 and 2.5 mM TCEP. Binding of the F535R fusion loop mutant (19-kDa Ecto) in response to TCEP (2.5 mM at 37°C) was greatly diminished compared to that of the corresponding WT protein (Fig. 7C). No other conditions that we tested for potential triggering at 37°C, including addition of 20 mM Ca2+, Mg2+, or Cs+ and chelation of cations with 20 mM EDTA or 20 mM EGTA, caused the 19-kDa GP Ecto to bind to liposomes at 37°C (data not shown). Collectively, the results in Fig. 7 indicate that mild reduction at physiological temperature (37°C) can trigger the primed 19-kDa GP Ecto, but not the unprimed GPΔ Ecto, to bind robustly to target liposomes, that TCEP-triggered liposome binding is dependent on the WT fusion loop sequence, and that reductive triggering occurs preferentially at low pH, i.e., at a pH found in endosomes.
Fig 7.
Mild reduction (preferentially at low pH) induces 19-kDa GP Ecto, but not GPΔ Ecto, to bind to liposomes; the F535R mutant Ecto is impaired in liposome binding in response to mild reduction. (A) GPΔ Ecto and 19-kDa GP Ecto were incubated with liposomes and the indicated concentration of TCEP for 10 min at pH 5.5 and 37°C, and liposome binding was assessed as described in the legend to Fig. 1. Data are the averages from duplicate samples. (B) The 19-kDa GP Ecto was incubated with liposomes and the indicated concentration of TCEP for 10 min at 37°C at pH 7.5 or pH 5.5, and liposome binding was assessed as for panel A. Data are the averages of duplicate samples. Similar results were observed in a second experiment. (C) Equivalent amounts of WT and F535R mutant 19-kDa Ecto proteins were incubated with liposomes, treated with 2.5 mM TCEP at 37°C at pH 7.5 or 5.5 as described above, and then analyzed for 19-kDa GP binding as described in the legend to Fig. 5B. Data are from one experiment performed in triplicate. Error bars indicate SD.
Pretreatment with TCEP strongly inactivates the ability of 19-kDa GP, but not GPΔ, to mediate infection.
Our results thus far suggest that the 19-kDa GP Ecto can be triggered by heat, urea, or mild reduction to insert its fusion loop into a target membrane. Conversely, none of the conditions tested robustly triggered the upstream, unprimed GPΔ Ecto to do so. For several enveloped viruses (e.g., influenza virus and SFV) pretreatment under conditions that expose the fusion peptide or fusion loop (in the absence of a target membrane) inactivates virions for subsequent fusion and infection (6, 52). To test if this is also the case for EBOV GP, we pretreated VSV pseudovirions bearing transmembrane domain-anchored forms of GPΔ or 19-kDa GP with 5 mM TCEP at pH 5.5. As seen in Fig. 8, this pretreatment significantly inactivated the ability of pseudovirions with 19-kDa GP to infect Vero cells while having only a modest effect on infections by pseudovirions bearing GPΔ. These results indicate that priming to 19-kDa GP also potentiates the ability of GP embedded in a pseudovirion membrane to change conformation in response to mild reduction at low pH.
DISCUSSION
Fusion of EBOV with host cells is mediated by GP, the sole glycoprotein on the surface of this filamentous virus. GP is exceptional among class I fusion proteins in that it undergoes a massive proteolytic priming event and is somewhat unusual in that priming is carried out by endosomal cathepsins (3, 4, 10, 38, 45). Priming of EBOV GP appears to occur in steps shifting the molecular mass of GP1 from 130 kDa to ∼50 kDa, then to 20 kDa, and finally to 19 kDa (10, 25, 45). The cleavage sites for the 20-kDa and 19-kDa species map within the unstructured β13-β14 loop in GP1 (17, 25). Entry of pseudovirions bearing 19-kDa GP, like that of pseudovirions bearing full-length GP or GPΔ (∼50 kDa), is dependent on low pH (45). These observations suggested that 19-kDa GP is in a clamped metastable conformation in which its fusion loop has not yet been exposed for interaction with the target membrane and that low pH plays a role in fusion triggering. In the present study, we identified conditions that enable the EBOV GP Ecto to undergo a major early step of fusion triggering: binding to target membranes. We report several new findings. (i) The 19-kDa GP is in a metastable state that can be triggered to bind to membranes by elevated temperature, urea, or mild reduction. (ii) Triggering occurs preferentially at low pH. (iii) The 19-kDa GP requires less energy to be triggered than 20-kDa GP. (iv) GPΔ cannot be triggered by any of the conditions tested. (v) A mutation (F535R) in the fusion loop impairs the ability of 19-kDa GP to bind to liposomes in response to elevated temperature or mild reduction. (vi) Mild reduction inactivates the ability of 19-kDa GP, but not GPΔ, embedded in a pseudovirus membrane to mediate infection.
Sequential cathepsin cleavage progressively lowers the energy barrier for a conformational change in GP.
We documented that 19-kDa GP is in a metastable state by showing that it can be triggered to bind to target membranes in a fusion loop-dependent manner by elevated temperature, urea, or mild reduction. In stark contrast, the ∼50-kDa GPΔ ectodomain (31) was not triggered to bind to liposomes in response to any of the treatments tested. This indicates that GPΔ and, by inference, full-length GP are “locked down” and, specifically, that the presence of the glycan cap and/or β-strand 14 (above the receptor binding region), which are removed by cathepsin priming, impede (most likely indirectly) conformational changes needed to expose the fusion loop for target membrane binding. Provocatively, we found that the 20-kDa GP Ecto requires more energy than the 19-kDa Ecto to achieve equivalent levels of liposome association. This difference suggests that removal of ∼10 residues (from the β13-β14 loop), which converts 20-kDa to 19-kDa GP, affects the conformation of GP such that it can respond more readily to a fusion trigger, perhaps by releasing a physical constraint on the fusion loop itself (15). This striking difference in ectodomain triggering correlates well with previously observed differences between 20-kDa and 19-kDa GP on pseudovirus particles (45, 54) (data not shown).
Role of low pH in fusion triggering.
Previous work suggested that low pH is necessary, but not sufficient, for EBOV fusion. While EBOV GP-mediated entry is inhibited by bafilomycin, exposure to low pH does not induce significant levels of fusion or entry (45). Moreover, because cell entry of pseudovirions bearing 19-kDa GP is blocked by bafilomycin (45), low pH likely provides a function for GP-mediated entry in addition to its requirement for optimal cathepsin activity. Low pH could alter the conformation of 19-kDa GP, and/or it could activate other endosomal factors required for entry. In this study, we provide the first demonstration that low pH affects the conformation of the trimeric 19-kDa GP ectodomain. Although we detected only minimal effects of low pH alone on 19-kDa GP (Fig. 1 and 6 to 8) (1), triggering of 19-kDa GP by elevated temperature or urea occurred only at low pH, and triggering by the reducing agent TCEP occurred preferentially at low pH. These observations, combined with our recent demonstrations that low pH induces a conformational change in the isolated EBOV fusion loop and is required for the fusion loop to mediate liposome-liposome fusion (21), support our contention (45) that low pH influences EBOV fusion per se, i.e., that low pH acts directly on 19-kDa GP and is needed beyond its requirement to maintain optimal cathepsin activity. It is also possible that low pH positively affects another endosomal factor needed to trigger GP.
Other factors required for fusion triggering.
A surprising finding of our study is that mild reduction is sufficient to trigger the 19-kDa GP Ecto to bind to liposomes (Fig. 7) and to inactivate the ability of 19-kDa GP (in a pseudovirus particle) to subsequently mediate infection (Fig. 8). In contrast, reduction does not trigger GPΔ and does not strongly inactivate pseudovirions bearing GPΔ for subsequent infection. A major question is how these findings relate to fusion in vivo. One possibility is that 19-kDa GP is triggered by a thiol isomerase, as is the case for several nonenveloped viruses (34, 44) and as has been proposed for several enveloped viruses (2, 27, 36). A second possibility is that cathepsin-primed GP is triggered by endosomal reductases or reducing potential. Endosomes have been reported to have a mild reducing potential (−220 mV) (28, 40), which is thought to be maintained by cysteine and cystine transporters (41, 42). Reducing conditions are necessary for optimal activity of cathepsins B, L, and K (28), and several cell types have been shown to possess endosomal reductases (47). Reduction of 19-kDa GP by endosomal reductases or reducing potential might serve a purpose similar to that of the action of the thiol isomerase motifs present in the MoMLV and HTLV Env glycoproteins (33, 51) but not present in the ASLV Env and EBOV GP (14). Another possibility is that the reductive triggering observed in vitro mimics the effects of another endosomal factor that triggers 19-kDa GP (at low pH). Possibilities include a tighter interaction between 19-kDa GP and a primary (cell surface) receptor (if still bound in endosomes) (17), a de novo interaction with an endosomal receptor (29), or effects of other endosomal proteins (8, 12) or environmental conditions. Interestingly, entry by pseudovirions bearing 19-kDa GP is completely blocked by E64d (45, 54), a sulfhydryl-modifying agent (30) known as a cysteine protease inhibitor. Perhaps E64(d) quenches a free sulfhydryl in a (nonproteolytic) factor involved in triggering 19-kDa GP.
Summary.
In conclusion, we provide the first evidence that the cathepsin-primed trimeric EBOV GP ectodomain can be triggered to bind to target membranes (in a process dependent on its WT fusion loop sequence). Most importantly, we found that the cleavages of GP to its 20-kDa form and then to its 19-kDa core progressively lower the energy barrier for triggering, supporting our previous model (45) that cleavage (specifically to the 19-kDa core) renders GP responsive for fusion triggering. A second major finding, coupled with our recent report (21), is that low pH plays an active role in fusion triggering. Based on these observations, we propose a model in which, following binding to the cell surface, EBOV is internalized and delivered to endosomes. Once in the proper endosomal compartment and primed by cathepsins, low pH and an E64 sensitive process, possibly reduction or possibly a process mimicked by reduction (e.g., interaction with other endosomal factors), act together to trigger conformational changes in 19-kDa GP that lead to fusion.
ACKNOWLEDGMENTS
We thank Edward Park and Elizabeth Nelson for technical assistance, Paul Bates for the anti-GP1 antibody, and Dusan Turk for purified cathepsin L.
This work was supported by RO1 AI22470 and U54 AI57168 (to J.M.W.) and R01 AI67927 and R01 AI081982, The Skaggs Institute for Chemical Biology, and an Investigator in the Pathogenesis of Infectious Disease award from The Burroughs Wellcome Fund (to E.O.S.). M.B. and K.S. were supported in part by, respectively, an Infectious Disease Training Grant (5T32 AI07046-27; NIAID) and a Biodefense Training Grant (5T32 AI055432-01; NIAID).
Footnotes
Published ahead of print 26 October 2011
REFERENCES
- 1. Bale S, et al. Ebolavirus glycoprotein needs additional trigger, beyond proteolytic priming for membrane fusion. PLoS Negl. Trop. Dis., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barbouche R, Miquelis R, Jones IM, Fenouillet E. 2003. Protein-disulfide isomerase-mediated reduction of two disulfide bonds of HIV envelope glycoprotein 120 occurs post-CXCR4 binding and is required for fusion. J. Biol. Chem. 278:3131–3136 [DOI] [PubMed] [Google Scholar]
- 3. Belouzard S, Chu VC, Whittaker GR. 2009. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 106:5871–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bosch BJ, Bartelink W, Rottier PJ. 2008. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 82:8887–8890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brindley MA, et al. 2007. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J. Virol. 81:7702–7709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bron R, Wahlberg JM, Garoff H, Wilschut J. 1993. Membrane fusion of Semliki Forest virus in a model system: correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 12:693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cairns TM, et al. 2011. Capturing the herpes simplex virus core fusion complex (gB-gH/gL) in an acidic environment. J. Virol. 85:6175–6184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carette JE, et al. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carr CM, Chaudhry C, Kim PS. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. U. S. A. 94:14306–14313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. 2006. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 103:17903–17908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cote M, et al. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delos SE, et al. 2008. Cysteines flanking the internal fusion peptide are required for the avian sarcoma/leukosis virus glycoprotein to mediate the lipid mixing stage of fusion with high efficiency. J. Virol. 82:3131–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delos SE, La B, Gilmartin A, White JM. 2010. Studies of the “chain reversal regions” of the avian sarcoma/leukosis virus (ASLV) and Ebola virus fusion proteins: analogous residues are important, and a His residue unique to EnvA affects the pH dependence of ASLV entry. J. Virol. 84:5687–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dias JM, et al. A shared structural solution for neutralizing ebolaviruses. Nat. Struct. Mol. Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doms RW, Helenius A, White J. 1985. Membrane fusion activity of the influenza virus hemagglutinin. The low pH-induced conformational change. J. Biol. Chem. 260:2973–2981 [PubMed] [Google Scholar]
- 17. Dube D, et al. 2009. The primed Ebola virus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J. Virol. 83:2883–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feldmann H, Geisbert TW. 2011. Ebola haemorrhagic fever. Lancet 377:849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gething MJ, Doms RW, York D, White J. 1986. Studies on the mechanism of membrane fusion: site-specific mutagenesis of the hemagglutinin of influenza virus. J. Cell Biol. 102:11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gibbons DL, et al. 2004. Multistep regulation of membrane insertion of the fusion peptide of Semliki Forest virus. J. Virol. 78:3312–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gregory SM, et al. 2011. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proc. Natl. Acad. Sci. U. S. A. 108:11211–11216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harrison SC. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hernandez LD, et al. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 139:1455–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hernandez LD, White JM. 1998. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J. Virol. 72:3259–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hood CL, et al. 2010. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J. Virol. 84:2972–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito H, Watanabe S, Sanchez A, Whitt MA, Kawaoka Y. 1999. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J. Virol. 73:8907–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jain S, McGinnes LW, Morrison TG. 2009. Role of thiol/disulfide exchange in Newcastle disease virus entry. J. Virol. 83:241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jordans S, et al. 2009. Monitoring compartment-specific substrate cleavage by cathepsins B, K, L, and S at physiological pH and redox conditions. BMC Biochem. 10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaletsky RL, Simmons G, Bates P. 2007. Proteolysis of the Ebola glycoproteins enhances virus binding and infectivity. J. Virol. 81:13378–13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim MJ, et al. 1992. Crystal structure of papain-E64-c complex. Binding diversity of E64-c to papain S2 and S3 subsites. Biochem. J. 287(Pt 3):797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee JE, et al. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee JE, Saphire EO. 2009. Neutralizing ebolavirus: structural insights into the envelope glycoprotein and antibodies targeted against it. Curr. Opin. Struct. Biol. 19:408–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li K, et al. 2008. Intersubunit disulfide isomerization controls membrane fusion of human T-cell leukemia virus Env. J. Virol. 82:7135–7143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Magnuson B, et al. 2005. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol. Cell 20:289–300 [DOI] [PubMed] [Google Scholar]
- 35. Manicassamy B, Wang J, Jiang H, Rong L. 2005. Comprehensive analysis of Ebola virus GP1 in viral entry. J. Virol. 79:4793–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Markovic I, et al. 2004. Thiol/disulfide exchange is a prerequisite for CXCR4-tropic HIV-1 envelope-mediated T-cell fusion during viral entry. Blood 103:1586–1594 [DOI] [PubMed] [Google Scholar]
- 37. Matsuyama S, Delos SE, White JM. 2004. Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. J. Virol. 78:8201–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matsuyama S, Taguchi F. 2009. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J. Virol. 83:11133–11141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mpanju OM, Towner JS, Dover JE, Nichol ST, Wilson CA. 2006. Identification of two amino acid residues on Ebola virus glycoprotein 1 critical for cell entry. Virus Res. 121:205–214 [DOI] [PubMed] [Google Scholar]
- 40. Okun I, et al. 2006. Screening for caspase-3 inhibitors: effect of a reducing agent on identified hit chemotypes. J. Biomol. Screen. 11:694–703 [DOI] [PubMed] [Google Scholar]
- 41. Pillay CS, Elliott E, Dennison C. 2002. Endolysosomal proteolysis and its regulation. Biochem. J. 363:417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pisoni RL, Acker TL, Lisowski KM, Lemons RM, Thoene JG. 1990. A cysteine-specific lysosomal transport system provides a major route for the delivery of thiol to human fibroblast lysosomes: possible role in supporting lysosomal proteolysis. J. Cell Biol. 110:327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. 2010. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 6:e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schelhaas M, et al. 2007. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131:516–529 [DOI] [PubMed] [Google Scholar]
- 45. Schornberg K, et al. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80:4174–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schornberg KL, et al. 2009. α5β1-integrin controls ebolavirus entry by regulating endosomal cathepsins. Proc. Natl. Acad. Sci. U. S. A. 106:8003–8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sinnathamby G, Maric M, Cresswell P, Eisenlohr LC. 2004. Differential requirements for endosomal reduction in the presentation of two H2-E(d)-restricted epitopes from influenza hemagglutinin. J. Immunol. 172:6607–6614 [DOI] [PubMed] [Google Scholar]
- 48. Skehel JJ, et al. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 79:968–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith JG, Mothes W, Blacklow SC, Cunningham JM. 2004. The mature avian leukosis virus subgroup A envelope glycoprotein is metastable, and refolding induced by the synergistic effects of receptor binding and low pH is coupled to infection. J. Virol. 78:1403–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takada A, et al. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 94:14764–14769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wallin M, Ekstrom M, Garoff H. 2005. The fusion-controlling disulfide bond isomerase in retrovirus Env is triggered by protein destabilization. J. Virol. 79:1678–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. White J, Kartenbeck J, Helenius A. 1982. Membrane fusion activity of influenza virus. EMBO J. 1:217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K. 2010. A forward genetic strategy reveals destabilizing mutations in the Ebola virus glycoprotein that alter its protease dependence during cell entry. J. Virol. 84:163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]