Abstract
Human noroviruses bind with their capsid-protruding domains to histo-blood-group antigens (HBGAs), an interaction thought to direct their entry into cells. Although human noroviruses are the major cause of gastroenteritis outbreaks, development of antivirals has been lacking, mainly because human noroviruses cannot be cultivated. Here we use X-ray crystallography and saturation transfer difference nuclear magnetic resonance (STD NMR) to analyze the interaction of citrate with genogroup II (GII) noroviruses. Crystals of citrate in complex with the protruding domain from norovirus GII.10 Vietnam026 diffracted to 1.4 Å and showed a single citrate bound at the site of HBGA interaction. The citrate interaction was coordinated with a set of capsid interactions almost identical to that involved in recognizing the terminal HBGA fucose, the saccharide which forms the primary conserved interaction between HBGAs and GII noroviruses. Citrate and a water molecule formed a ring-like structure that mimicked the pyranoside ring of fucose. STD NMR showed the protruding domain to have weak affinity for citrate (460 μM). This affinity, however, was similar to the affinities of the protruding domain for fucose (460 μM) and H type 2 trisaccharide (390 μM), an HBGA shown previously to be specifically recognized by human noroviruses. Importantly, competition STD NMR showed that citrate could compete with HBGA for norovirus binding. Together, the results suggest that citrate and other glycomimetics have the potential to block human noroviruses from binding to HBGAs.
INTRODUCTION
Human noroviruses, family Caliciviridae, are the dominant cause of outbreaks of gastroenteritis. Many aspects of human norovirus replication, however, remain unclear, mainly because these viruses cannot be grown in cell culture. Transmission predominately occurs through ingestion of contaminated foods, airborne transmission, and person-to-person contact. Medical treatment usually involves orally administered fluids and electrolyte replacement therapy. Currently, there is no effective vaccine.
Human noroviruses can be divided into 2 main genogroups (GI and GII), which can be further subdivided into at least 25 different genotypes (GI.1 to -8 and GII.1 to -17) (26, 57). The norovirus genome has three open reading frames (ORFs) that encode nonstructural, capsid, and small structural proteins, respectively. The capsid of human norovirus is composed of two domains, shell and protruding (P) domains. The shell forms a scaffold around the RNA, and the dimeric P domain contains determinants for both antigenicity and receptor binding (25, 43, 51). The P domain is further subdivided into P1 and P2 subdomains, where the P1 subdomain interacts with the shell domain and is buried under the outermost P2 subdomain.
Human noroviruses bind to histo-blood group antigens (HBGAs), with recognition occurring in the P domain. HBGAs are complex carbohydrates present on mucosal epithelial cells or free antigens in blood, saliva, and other fluids (32). X-ray crystal structures of norovirus P domains in complex with different HBGAs have defined distinct binding sites for GI and GII viruses (8, 11, 12, 21); in particular, the HBGA binding site of GII is located at the dimeric interface of two P domains, whereas the HBGA binding site in GI is located within a single P domain (8, 11, 12, 21).
A number of recent studies have shown that natural fruits or their constituents, including orange juice, pomegranate juice, cranberry juice, and grape seed extract, can inhibit and/or reduce feline calicivirus and murine norovirus infectivity (23, 48–50, 54). Although there have been no studies to support the idea that natural fruits or their constituents can prevent human norovirus infections, and data on the mode of inhibition of fruits have been lacking, the stability of human norovirus virus-like particles over a pH range of 3 to 7 (3) suggested that the effect might be related to a specific interaction with compounds in fruits rather than a pH effect. In this study, we used X-ray crystallography and saturation transfer difference nuclear magnetic resonance (STD NMR) to provide atomic-level details on the interaction of citrate and GII human noroviruses. We show that citrate specifically binds at the HBGA recognition site of GII noroviruses, and this inhibits P domain binding of both fucose and HBGA.
MATERIALS AND METHODS
Protein expression, purification, and crystallization of the norovirus P domain.
The norovirus Vietnam026 GII.10 P domain (GenBank accession no. AF504671) (22) was expressed in Escherichia coli as previously described (21). Briefly, a truncated form of the GII.10 P domain was optimized for E. coli expression, cloned in a modified pMal-c2x vector at the BamHI and NotI sites (New England BioLabs), and transformed into E. coli BL21 cells (Invitrogen), and expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 18 h at 22°C. After a series of purifications and cleavage steps, the P domain was concentrated to 2 mg/ml and stored in gel filtration buffer (0.35 M NaCl, 2.5 mM Tris [pH 7.0], 0.02% NaN3) before crystallization. Crystals of the P domain were obtained by the hanging-drop vapor diffusion method, with the mother solution containing citric acid triammonium (0.66 M [pH 6.5]) and isopropanol (1.65% [vol/vol]).
Data collection, structure solution, and refinement.
X-ray diffraction data at a 1.000-Å wavelength were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamline 22-BM at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL, and processed with HKL2000 (41). Prior to data collection, crystals were transferred to a cryoprotectant solution consisting of the mother liquor in 30% ethylene glycol, loop mounted, and flash-cooled in a nitrogen cryostat to 100°K. Structures were solved by molecular replacement with PHASER (35) by using Protein Data Bank (PDB) code 2OBR (11) as a search model. Structures were refined in multiple rounds of manual model building in COOT (16) and positional together with TLS refinement in REFMAC (13) and PHENIX (1).
Structure analysis and figures.
Citrate and H type 2 interactions were determined using Discovery Studio (Accelrys, version v2.5.5.9350). Figures were rendered using PyMOL (Schroedinger, LLC, version 1.2r3) and ChemDraw Ultra (Cambridgesoft, version 12.0.2.1076).
STD NMR.
All NMR data were recorded at 298°K on a Bruker Avance 600 NMR spectrometer equipped with a cryogenically cooled z-shielded gradient probe. One-dimensional (1D) STD NMR spectra were acquired with selective irradiation at −1 and +40 ppm (on and off resonance, respectively) using a train of 50-ms Gaussian-shaped radio frequency pulses separated by 1-ms delays and an optimized power level of 57 db. Water suppression was achieved with a binomial 3-9-19 pulse sequence. Samples were prepared in 20 mM sodium phosphate buffer containing 50 mM sodium chloride at pH 6.8. The NMR data were processed and analyzed with Topspin 2.1. STD enhancements were expressed as the STD amplification factor, ASTD, defined as ASTD = (I0 − ISAT) I0−1 ([Lt]/[P]), where Lt and P are the total ligand and protein concentrations, respectively (34). HBGAs, H type 2 disaccharide [α-l-fucose-(1-2)-β-d-galactose], and H type 2 trisaccharide [α-l-fucose-(1-2)-β-d-galactose-(1-4)-2-N-acetyl-β-d-glucosamine] were purchased from V-labs, and l-fucose was purchased from Sigma-Aldrich. For the citrate experiments, sodium citrate dihydrate (Sigma-Aldrich) was added to sodium phosphate buffer and then titrated at pH 6.85 ± 0.1.
Computational citrate docking studies of other saccharide-binding proteins.
Citrate docking analyses were performed against six different saccharide-binding proteins, including Anguilla anguilla agglutinin (PDB identification no. 1K12) (5), Aleuria aurantia lectin (PDB identification no. 1IUC) (19), Streptococcus pneumoniae virulence factor SpGH98 (PDB identification no. 2J1S) (7), Pseudomonas aeruginosa PA-IIL lectin (PDB identification no. 2JDH) (33), parainfluenza virus 5 hemagglutinin-neuraminidase (PDB identification no. 1Z4X) (56), and porcine adenovirus type 4 galectin domain (PDB identification no. 2WSV) (20). Water molecules and ligands were removed from the PDB files, with the exception of one water molecule (HOH 935) in 1Z4X, which is present in both ligand-free and sialyllactose-bound hemagglutinin-neuraminidase structures. For 2JDH, the two calcium ions in the fucose binding site were kept, and the partial charges for the calcium ions were assigned to 1.5 as suggested by previous studies (38). AutoDock4.2 (39) was used as the docking engine, with the grid files generated by Autogrid4.2 using default parameters and centered on the cocrystallized ligands. The citrate molecule was docked to the three structures using default parameters (ga_pop_size = 150, ga_num_evals = 2,500,000, and ga_run = 50). For each structure, the docking pose with the lowest estimated free energy of binding among the 50 docking runs was selected as the predicted binding pose. For comparison, for each complex, the cocrystallized ligand (or the terminal monosaccharide having the largest contact area with the binding site, if the cocrystallized ligand was not a monosaccharide) was docked in the saccharide binding site using the same procedure. Fucose and citrate molecules were also docked to the fucose-bound GII.10 P domain (PDB identification no. 3ONY) and the citrate-bound GII.10 P domain, respectively, for comparison. The water molecule (HOH 135) mediating the interaction between citrate and the protein was present during the citrate docking analysis.
Protein structure accession number.
Atomic coordinate and structure factors for the citrate-bound GII.10 P domain have been deposited in the Protein Data Bank under accession no. 3RY8.
RESULTS
X-ray crystal structure of citrate bound to the GII.10 P domain.
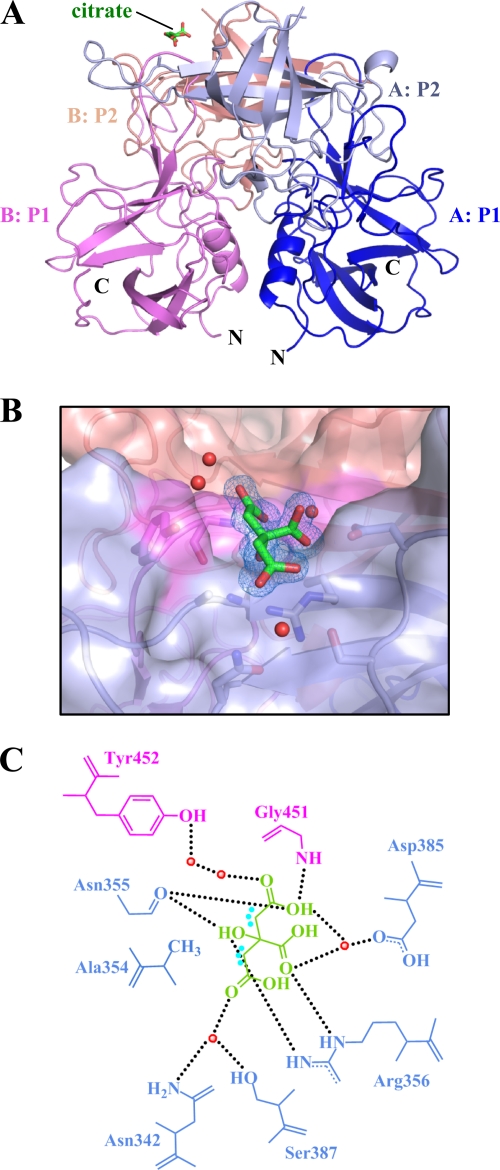
The GII.10 P domain protein could be expressed in E. coli to 2 mg/liter and was purified and prepared for crystallization as previously described (21). To obtain a GII.10 P domain-citrate complex, we chose a crystallization condition that was similar to our previously reported GII.10 P domain-HBGA complex conditions (21), though with the addition of citrate. The GII.10 P domain-citrate complex formed rectangular plate crystals, and X-ray diffraction data revealed a space group of P21, the same as the previous GII.10 P domain-HBGA complexes (21), and strong diffraction to 1.4 Å. Structure solution by molecular replacement revealed one dimer per asymmetrical unit (Fig. 1A), and refinement led to an Rvalue of 0.139 (Rfree = 0.151), with well-defined density for most of the P domain dimer (Table 1). Electron density for residues 296 to 299 (chain A) and 296 to 300 and 344 to 351 (chain B) was poor, and these residues were not modeled. Extra electron density was observed at the HBGA binding site, where a single citrate molecule was clearly distinguished and refined (Fig. 1B; see Fig. S1 in the supplemental material). The structure of the GII.10 P domain in complex with citrate was highly reminiscent of the other known structures (GI.1, GII.4, and GII.12), where the P1 subdomain contains a single α-helix and the P2 subdomain contains six anti-parallel β-strands that form two anti-parallel β-sheets (21).
Fig 1.
Citrate binding to the GII.10 P domain. (A) X-ray crystal structure of the GII.10 P domain dimer (ribbon structure) and the bound citrate (green sticks). Each P subdomain is colored differently: i.e., chain A, P1, blue; chain A, P2, light blue; chain B, P1, violet; and chain B, P2, salmon. (B) Surface representation of the GII.10 P domain (colored as in panel A) showing the residues (sticks) and water molecules (red spheres) interacting with the citrate molecule (green sticks). The 2Fo-Fc density was contoured at 1.0 σ. (C) Residues interacting with the citrate molecule were contributed by both monomers (colored as in panel A), where the black dotted lines represent the hydrogen bonds, the cyan dots near the citrate represent the hydrophobic interactions with Ala354, and the red spheres represent water molecules. For simplicity, only the backbone is shown for residues that were backbone mediated. Hydrogen bond distances were less than 3.1 Å, though the majority were ∼2.8 Å.
Table 1.
Data collection and refinement statistics for structures of the GII.10 Vietnam026 norovirus P domaina
| Parameter | Value(s) for citrate (026_citrate; PDB accession no. 3RY8)b |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 63.76, 79.81, 69.60 |
| α, β, γ (°) | 90, 96.84, 90 |
| Resolution (Å) | 50-1.40 (1.45-1.40) |
| Rsym | 7.3 (30.5) |
| I/σI | 18.7 (3.2) |
| Completeness (%) | 99.9 (99.6) |
| Redundancy | 3.7 (3.2) |
| Refinement | |
| Resolution range (Å) | 31.98-1.399 |
| No. of reflections | 131,576 |
| Rwork/Rfree | 0.1388/0.1506 |
| No. of atoms: | |
| Total | 5,587 |
| Protein | 4,722 |
| Ligand/ion | 57 |
| Water | 808 |
| B-factors | |
| Protein | 18.3 |
| Ligand/ion | 19.2 |
| Water | 30.2 |
| RMSD | |
| Bond length (Å) | 0.011 |
| Bond angle (°) | 1.393 |
Each data set was collected from a single crystal.
Values in parentheses are for the highest-resolution shell.
Citrate was highly coordinated by the GII.10 P domain.
At a 1.4-Å resolution, detailed interactions between citrate and the P domain could be defined. Seven residues of the P domain, many of which are conserved and located at the dimer interface, are involved in hydrogen bonding interactions with citrate (Fig. 1B and C). These include the side chain of Tyr452 and main chain of Gly451 from one P domain subunit as well as side chains Arg356 and Asp385 and the main chain of Asn355 of a second P domain subunit. Unique to citrate binding, side chains of Asn342 and Ser387 make a water-mediated hydrogen bond with the C-5 CO group of citrate. Superposition of citrate-bound and apo GII.10 P domain structures indicated that the citrate interaction did not cause any conformation changes in the GII.10 P domain.
Comparisons of citrate and HBGA interaction with the GII.10 P domain.
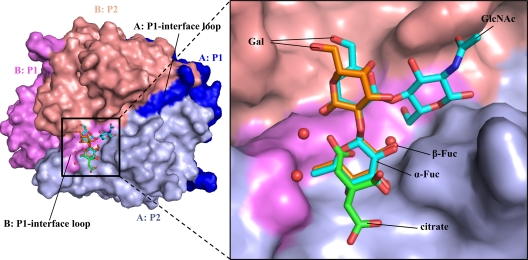
Compared with GII.10 P domains in complex with HBGAs, we found that citrate essentially mimics the fucose unit of HBGAs. By using H type 2 di- and trisaccharides as examples, superposition of the P domains revealed that three carbon atoms, including C-2, C-3, and the C-3 carboxy carbon, and three oxygen atoms, including the C-1 and C-3 carboxy oxygens and the C-3 hydroxyl group of citrate closely overlapped with C-5/C-4/C-3/O-5/O-4/O-3 of the terminal α-fucose ring (Fig. 2). In addition, a water molecule, present in the citrate-bound structure but absent from the HBGA-bound structures, occupied the site of the C-2 hydroxyl of fucose (Fig. 2). In this configuration, the citrate and associated water molecule formed a ring-like structure, mimicking the pyranoside ring of fucose. Finally, the comparisons showed that of the seven residues involved in hydrogen bonding interactions with citrate, five made almost identical interactions with their comparable atoms in fucose.
Fig 2.
The HBGA and citrate binding site overlapped on the top of the GII.10 P domain. (A) The citrate molecule (green sticks) bound at the HBGA binding site; shown here are the bound H type 2 di- and trisaccharides (orange and cyan sticks, respectively). (B) Close-up of the black square in panel A, showing the H type 2 disaccharide [α-l-fucose(1-2)-β-d-galactose] and H type 2 trisaccharide [β-l-fucose(1-2)-β-d-galactose(1-4)-2-N-acetyl-β-d-glucosamine].
Characterization of binding of citrate, H type 2 trisaccharide, and fucose to GII.10 P domain by STD NMR.
Given the remarkable similarities observed for citrate and fucose binding to the GII.10 P domain by crystallography, we sought to characterize in solution by NMR the binding of GII.10 P domain with citrate, HBGAs, and fucose and ultimately to determine their relative binding affinities and whether they bind competitively.
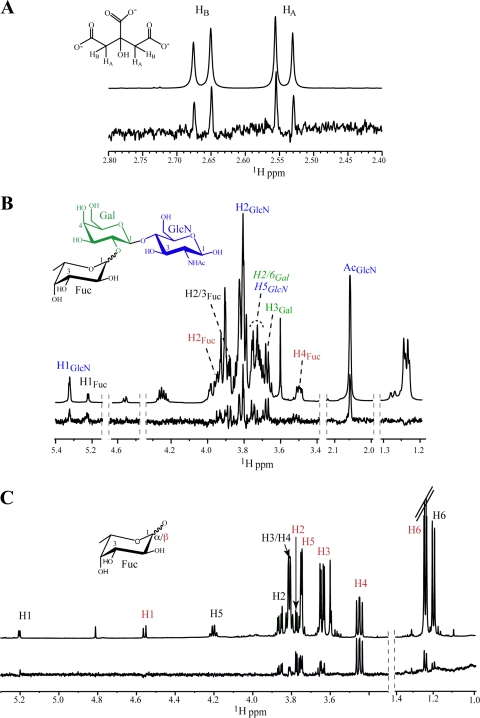
STD enhancements were observed for methylene protons H2A and H2B of citrate, consistent with their close proximity to the protein in the bound state (Fig. 3A). In the crystal structure, these hydrogens are within van der Waals contact of the methyl of Ala354 (Fig. 1C). With H type 2 trisaccharide, the most prominent STD signals that could be assigned corresponded to H-1, H-2, and H-4 of α-fucose; H-3 of galactose; and H-1, H-2, and N-acetyl of glucosamine (Fig. 3B). We also characterized binding of monosaccharide α/β-fucopyranose, as it also would be used in competition STD NMR experiments. As seen in Fig. 3C, binding of both anomers was observed, with H-1, H-2, and H-4 of α-fucose versus H-2, H-4, and H-5 of β-fucose showing the strongest enhancements. Although natural H type 2 HBGAs contain α-Fuc(1-2)Gal and not β-Fuc, it is interesting that the HBGA binding site of norovirus can bind both. By NMR, we observed binding of α and β forms of the monosaccharide (Fig. 3C) as well as synthetic H type 2 trisaccharide α/β-Fuc(1-2)β-Gal(1-4)β-GlcN (Fig. 3B) and H type 2 disaccharide α-Fuc(1-2)β-Gal(1-4) (data not shown), and by crystallography, binding of synthetic H type 2 trisaccharide β-Fuc(1-2)β-Gal(1-4)β-GlcN was observed, in addition to binding of other HBGAs containing the α-Fuc(1-2)β-Gal linkage (21). Finally, it is interesting to note that a similar mode of citrate binding was observed for the soluble GII.12 P domain (see Fig. S2 in the supplemental material).
Fig 3.
STD NMR spectra for citrate, H type 2 trisaccharide, and l-fucose bound to the GII.10 P domain. STD (lower) and reference (upper) spectra of (A) citrate (1.2 mM), (B) H type 2 trisaccharide (1.2 mM), and (C) l-fucose (mixture of α and β anomers) (1.2 mM) in the presence of the GII.10 P domain (15 μM). Nonoverlapping protons that exhibit STD enhancements are labeled and color coded by sugar residue, and signals for β-Fuc are red. One group of overlapping signals appears in italics.
Affinity of citrate, H type 2 tri- and disaccharides, and l-fucose to the GII.10 P domain.
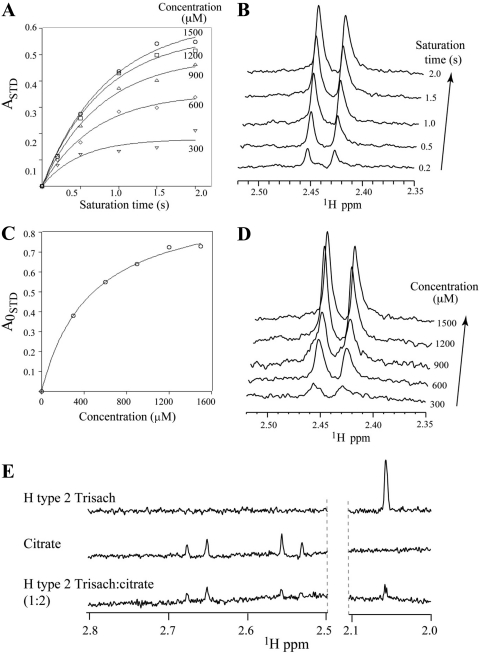
We used single-ligand titration STD NMR experiments to determine the KD (equilibrium dissociation constant) of citrate binding to the GII.10 P domain (Fig. 4A) (2). STD amplification factors (ASTD) (34) were calculated by integrating the signal at δH 2.54 ppm in difference and corresponding reference spectra. Initial growth rates (A0STD) were obtained by measuring the effect on ASTD as a function of various saturation time (tsat) and fitting the data to the equation ASTD = Amax STD[1 − exp(−ktsat)] for each concentration (300, 600, 900, 1,200, and 1,500 μM) of the ligand. The KD of citrate was in turn measured as 460 ± 80 μM by fitting A0STD values as a function of ligand concentration using the equation y = Bmax/(KD + x), where x is the ligand concentration and Bmax represents the plateau of the curve (Fig. 4) (2, 37). For H type 2 trisaccharide, the STD enhancements for the N-acetyl signal were sufficiently strong to allow for accurate integration, even at very short saturation times (0.1 s); thus, a KD value of 390 ± 70 μM could be determined directly by fitting ASTD values as a function of ligand concentration (40) (Fig. 5). KD values for fucose and H type 2 disaccharides were in turn obtained from single point competition STD experiments as described previously (36) to give values of 460 ± 10 and 420 ± 40 μM, respectively (see Fig. S3 in the supplemental material).
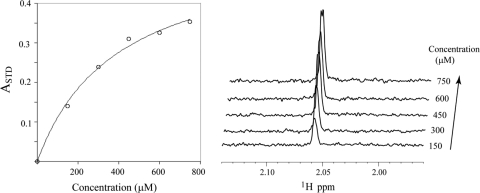
Fig 4.
Binding affinity of citrate and HBGAs to GII.10 P domain by STD NMR. Data were used to obtain the KD for citrate and H type 2 trisaccharide (Trisach) binding to GII.10 P by single-ligand titration STD NMR experiments (2). (A) Effect on STD enhancement (expressed as ASTD) (34, 37) as a function of saturation time (tsat) and ligand concentration; (B) stacked plots of spectra for 1.5 mM citrate as a function of tsat (y axis); (C) Langmuir binding curve used to obtain the KD from the initial slope of ASTD as a function of citrate concentration. (D) Stacked plot of various citrate concentrations (tsat, 2 s, 15 μM protein); (E) competition STD spectra of H type 2 trisaccharide (top), citrate (middle), and 1:2 H type 2 trisaccharide-citrate (0.75:1.5 mM; bottom) used to calculate the Ki of citrate (36).
Fig 5.
Binding affinity of H type 2 trisaccharide to the GII.10 P domain (left) effect on STD enhancement, expressed as ASTD, as a function of trisaccharide concentration in the presence of 15 μM GII.10 P domain. tsat = 0.1 s. Curve fitting (described in the text) provides the KD value. (Right) Stacked STD NMR spectra showing the change in enhancement of the nonoverlapped N-acetyl proton signals as a function of increasing concentration of H type 2 trisaccharide (40).
Competition of HBGAs and citrate with the GII.10 P domain.
To confirm the overlapping mode of binding observed in the crystal structure of citrate and fucose of H type 2 ligands, ASTD values of l-fucopyranose and H type 2 trisaccharide were monitored while titrating citrate to the samples. As seen in Fig. 4E, addition of citrate to a sample of P domain-H type 2 trisaccharide diminishes the trisaccharide signals in a concentration-dependent manner, indicating that citrate directly competes with the trisaccharide for P domain binding, giving a Ki of 600 ± 20 μM. Upon addition of citrate, the pH of the solutions was found to remain constant (pH 7.2 ± 0.1), indicating that the competition was a direct result of citrate binding rather than pH. The same effect was observed in STD competition experiments with l-fucose (data not shown). Importantly, the reverse set of experiments showed that HBGAs can compete with citrate for P domain binding (data not shown), indicating that the P domain is unaffected by the presence of citrate. Together, these results conclusively demonstrate molecular mimicry between citrate and fucose of HBGAs.
DISCUSSION
Despite the discovery of human norovirus nearly 40 years ago (27), little is known about the capsid interaction with ligands (18, 44) other than HBGAs (8, 11, 12, 15, 21, 45). Our finding that citrate binds at the terminal fucose binding site was somewhat unexpected, given that the structure of citrate is unlike the structure of fucose and considering that the GII.10 P domain could not bind HBGAs having an α-fucose1-3/4 saccharide (21). In an earlier enzyme immune assay study, Feng et al. screened ∼5,000 compounds (the Diversity screening set; Timtec, Inc.) for their ability to block GI and GII norovirus virus-like particles (VLPs) from binding to saliva samples of known HBGA type (18). They found 14 compounds that had strong inhibition; however, the mode of action was not determined. In a more recent NMR study, Rademacher et al. screened ∼500 compounds (the Maybridge Ro5 fragment library; Thermo Fisher Scientific, Inc.) for their ability to bind to a GII.4 VLP HBGA binding site (44). They showed that both univalent and multivalent compounds were capable of binding to the HBGA binding site. Interestingly, for both studies, the compounds that showed the highest affinities included compounds with at least one ring component. Taken together, these studies indicated that the HBGA binding site was capable of binding numerous compounds other than HBGAs, ranging from the small (smallest) citrate molecule to larger multivalent compounds.
For over a decade, the GII.4 noroviruses have remained as the dominant genotype of outbreaks of gastroenteritis around the world and as such the most well studied. Most studies agreed that a dominant GII.4 norovirus was replaced the following year or next by a new GII.4 “variant” norovirus that had ∼5% amino acid change in the capsid gene (6, 9, 10, 30, 31, 47). The reason that the GII.4 variants dominated and not some other genotype was unknown, but studies have shown specific mutations at or surrounding the HBGA binding site were capable of altering the HBGA binding patterns (15, 30, 31, 52). These small changes were thought to lead to new GII.4 variants capable of causing pandemics, analogous to influenza A virus evolution (14, 29). Despite these amino acid changes, few if any occurred at the fucose-binding site, thus highlighting the common site of vulnerability for GII noroviruses, especially for the pandemic GII.4 variant noroviruses. It is not known if the GI noroviruses will bind citrate given that the GI and GII P domain interactions with HBGAs were different, but since GI.1 P domain interacted with α-fucose1-2 and it was reported that the HBGA binding site was conserved among GI noroviruses (12), we suspect that GI noroviruses may also bind citrate, although further structural studies are needed.
Our unexpected finding that citrate and fucose have similar binding modes to the norovirus GII.10 P domain raises the question of whether such citrate mimicry of monosaccharide binding could be a general phenomenon or whether it is specific to norovirus and other caliciviruses. To investigate this, we performed in silico docking studies of citrate against four different fucose-binding proteins (Anguilla anguilla agglutinin, Aleuria aurantia lectin, Streptococcus pneumoniae virulence factor SpGH98, and Pseudomonas aeruginosa PA-IIL lectin) and two other saccharide-binding proteins (parainfluenza virus 5 hemagglutinin-neuraminidase and porcine adenovirus type 4 galectin domain), for which fucose or other saccharide-bound crystal structures were available (see Table S1 in the supplemental material). Computational docking analyses reveal different levels of citrate mimicry of monosaccharide binding for other saccharide-binding proteins. For Anguilla anguilla agglutinin, citrate, in its predicted binding pose, overlapped with the C-5, C-4, C-3, O-5, O-4, and O-3 atoms of fucose in a similar way to what was observed in the GII.10 P domain (Table S1), while forming hydrogen bonds with the same sets of protein residues as fucose (see Fig. S4 in the supplemental material). Citrate was thus predicted to show a high degree of mimicry to fucose, similarly to our experimental findings for the GII.10 P domain. For the other three fucose-binding proteins, citrate, in its predicted binding poses, did not overlap with the cocrystallized fucose, although it still formed the same sets of polar interactions as the cocrystallized fucose (see Fig. S5 to S7 in the supplemental material). Hence, our docking studies suggest that the mimicry between citrate and fucose binding observed for the GII.10 P domain could be a common, although not universal, phenomenon across other fucose-binding proteins. For all six fucose- and other saccharide-bound proteins for which docking was performed, the predicted citrate binding poses were able to form polar interactions with the same sets of protein residues as the cocrystallized ligand see (Fig. S4 to S9 in the supplemental material), indicating that citrate might be generally useful as a scaffold for designing glycomimetic inhibitors against these and other saccharide-interacting pathogens. Furthermore, a search of the ZINC database (4) revealed that there are more than three thousand compounds with at least 50% similarity to citrate. Thus, in silico screening of this database may present a promising approach for identifying small molecules that bind to saccharide-binding proteins. We note, however, that the predicted binding pose of citrate docked to fucose-bound GII.10 P domain had a root mean square deviation (RMSD) of 3.60 Å, while the predicted binding pose of citrate docked to citrate-bound GII.10 P domain with the cocrystallized water molecule had an RMSD of 1.87 Å. This indicates that the resulting docking modes could be error prone. Given that calculating small molecule-receptor binding energies is a difficult and error-prone task (24, 46), ultimately experimental validation would be necessary to confirm the generality of the citrate-saccharide mimicry predicted here.
The STD NMR data provided strong evidence that the integrity of the GII.10 P domain remained unchanged in the presence of different concentrations of citrate buffer and since the pH of the citrate buffer remained more or less the same during the titration, a specific effect of citrate was responsible for the reduction in HBGA attachment. Although the KD values of citrate and H type 2 trisaccharide for the GII.10 P domain are in the range of 360 to 490 μM, these relatively weak affinities are typical for univalent protein-carbohydrate interactions (17, 28). Given that 90 copies of dimeric P domains are present on norovirus capsid, it is plausible that a multivalent version of citrate- or fucose-like ligands would greatly enhance affinities and provide a starting point for norovirus inhibitors. Indeed, Rademacher et al. show that multivalent fucose-like compounds have increased avidity over their univalent counterparts (44).
In conclusion, we have described the structural basis by which citrate binds to the HBGA binding site of the norovirus GII.10 P domain and can in turn inhibit HBGA binding. Natural compounds, such as juice from lemons and limes, which contain ∼300 mM citric acid (42), may already reduce or inhibit norovirus infections, as suggested by a number of recent studies (23, 48–50, 54). In regard to this, it is tempting to speculate that a few drops of lemon juice with one's oysters might reduce norovirus infection. Epidemiological studies on the ingestion of foods high in citrate and norovirus infection may be illuminating, as may be correlations with related glycomimetics—e.g., with ascorbic acid (vitamin C). Controlled possibly volunteer studies should also provide an accurate assessment of norovirus inhibition. Additional compound screening will likely be required to identify a universal norovirus inhibitor with high potency and broad reactivity, and the structural basis for norovirus interaction with citrate as revealed here may be helpful in such efforts.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Structural Biology Section and Structural Bioinformatics Core at the NIH Vaccine Research Center for comments on the manuscript and J. Stuckey for assistance with figures.
G.S.H., S.H., K.K., C.A.B., and P.D.K. designed the research, G.S.H., S.H., J.S.M., and T.S. performed the research, G.S.H., S.H., J.S.M., G.-Y.C., I.G., C.A.B., and P.D.K. analyzed the data, and G.S.H., S.H., G.-Y.C., I.G., C.A.B., and P.D.K. wrote the paper, on which all authors commented.
Support for this work was provided by the Intramural Research Program of the National Institutes of Health (NIDDK, C.A.B; NIAID, P.D.K); the Intramural AIDS Targeted Antiviral Program, Office of the Director, NIH (C.A.B.); a grant from The Japan Health Science Foundation; and by grants from the Ministry of Health, Labor, and Welfare of Japan. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38.
Footnotes
Published ahead of print 26 October 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Adams PD, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angulo J, Enriquez-Navas PM, Nieto PM. 2010. Ligand-receptor binding affinities from saturation transfer difference (STD) NMR spectroscopy: the binding isotherm of STD initial growth rates. Chemistry 16:7803–7812 [DOI] [PubMed] [Google Scholar]
- 3. Ausar SF, Foubert TR, Hudson MH, Vedvick TS, Middaugh CR. 2006. Conformational stability and disassembly of Norwalk virus-like particles. Effect of pH and temperature. J. Biol. Chem. 281:19478–19488 [DOI] [PubMed] [Google Scholar]
- 4. Berman HM, et al. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bianchet MA, Odom EW, Vasta GR, Amzel LM. 2002. A novel fucose recognition fold involved in innate immunity. Nat. Struct. Biol. 9:628–634 [DOI] [PubMed] [Google Scholar]
- 6. Bok K, et al. 2009. Evolutionary dynamics of GII. 4 noroviruses over a 34-year period. J. Virol. 83:11890–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boraston AB, Wang D, Burke RD. 2006. Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J. Biol. Chem. 281:35263–35271 [DOI] [PubMed] [Google Scholar]
- 8. Bu W, et al. 2008. Structural basis for the receptor binding specificity of Norwalk virus. J. Virol. 82:5340–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bull RA, Tu ET, McIver CJ, Rawlinson WD, White PA. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44:327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bull RA, White PA. 2011. Mechanisms of GII.4 norovirus evolution. Trends Microbiol. 19:233–240 [DOI] [PubMed] [Google Scholar]
- 11. Cao S, et al. 2007. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81:5949–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choi JM, Hutson AM, Estes MK, Prasad BV. 2008. Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U. S. A. 105:9175–9180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collaborative Computational Project N 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 14. De Jong JC, Rimmelzwaan GF, Fouchier RA, Osterhaus AD. 2000. Influenza virus: a master of metamorphosis. J. Infect. 40:218–228 [DOI] [PubMed] [Google Scholar]
- 15. de Rougemont A, et al. 2011. Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens. J. Virol. 85:4057–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang M, Agha S, Lee R, Culpepper-Morgan J, D'Souza A. 2000. Perforation of jejunal diverticulum: case report and review of literature. Conn. Med. 64:7–10 [PubMed] [Google Scholar]
- 18. Feng X, Jiang X. 2006. Library screen for inhibitors targeting norovirus binding to their histo-blood group antigen receptors. Antimicrob. Agents Chemother. 51:324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fujihashi M, Peapus DH, Kamiya N, Nagata Y, Miki K. 2003. Crystal structure of fucose-specific lectin from Aleuria aurantia binding ligands at three of its five sugar recognition sites. Biochemistry 42:11093–11099 [DOI] [PubMed] [Google Scholar]
- 20. Guardado-Calvo P, et al. 2010. Crystallographic structure of porcine adenovirus type 4 fiber head and galectin domains. J. Virol. 84:10558–10568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansman GS, et al. 2011. Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability. J. Virol. 85:6687–6701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hansman GS, et al. 2004. Detection of norovirus and sapovirus infection among children with gastroenteritis in Ho Chi Minh City, Vietnam. Arch. Virol. 149:1673–1688 [DOI] [PubMed] [Google Scholar]
- 23. Horm KM, D'Souza DH. 2011. Survival of human norovirus surrogates in milk, orange, and pomegranate juice, and juice blends at refrigeration (4 degrees C). Food Microbiol. 28:1054–1061 [DOI] [PubMed] [Google Scholar]
- 24. Huey R, Morris GM, Olson AJ, Goodsell DS. 2007. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 28:1145–1152 [DOI] [PubMed] [Google Scholar]
- 25. Jiang X, Wang M, Graham DY, Estes MK. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kageyama T, et al. 2004. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to norovirus in Japan. J. Clin. Microbiol. 42:2988–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kapikian AZ, et al. 1972. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 10:1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kiessling LL, Young T, Gruber TD, Mortell KH. 2008. Glycoscience 12. 4:2483–2523 [Google Scholar]
- 29. Levin SA, Dushoff J, Plotkin JB. 2004. Evolution and persistence of influenza A and other diseases. Math. Biosci. 188:17–28 [DOI] [PubMed] [Google Scholar]
- 30. Lindesmith LC, Donaldson EF, Baric RS. 2011. Norovirus GII.4 strain antigenic variation. J. Virol. 85:231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lindesmith LC, et al. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marionneau S, et al. 2001. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 83:565–573 [DOI] [PubMed] [Google Scholar]
- 33. Marotte K, et al. 2007. X-ray structures and thermodynamics of the interaction of PA-IIL from Pseudomonas aeruginosa with disaccharide derivatives. Chem. Med. Chem. 2:1328–1338 [DOI] [PubMed] [Google Scholar]
- 34. Mayer M, Meyer B. 2001. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 123:6108–6117 [DOI] [PubMed] [Google Scholar]
- 35. McCoy AJ, et al. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meinecke R, Meyer B. 2001. Determination of the binding specificity of an integral membrane protein by saturation transfer difference NMR: RGD peptide ligands binding to integrin alphaIIbbeta3. J. Med. Chem. 44:3059–3065 [DOI] [PubMed] [Google Scholar]
- 37. Meyer B, Peters T. 2003. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem. Int. ed. Engl. 42:864–890 [DOI] [PubMed] [Google Scholar]
- 38. Mitchell EP, et al. 2005. High affinity fucose binding of Pseudomonas aeruginosa lectin PA-IIL: 1.0 A resolution crystal structure of the complex combined with thermodynamics and computational chemistry approaches. Proteins 58:735–746 [DOI] [PubMed] [Google Scholar]
- 39. Morris GM, et al. 1998. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 19:1639–1662 [Google Scholar]
- 40. Neffe AT, Bilang M, Meyer B. 2006. Synthesis and optimization of peptidomimetics as HIV entry inhibitors against the receptor protein CD4 using STD NMR and ligand docking. Org. Biomol. Chem. 4:3259–3267 [DOI] [PubMed] [Google Scholar]
- 41. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 42. Penniston KL, Nakada SY, Holmes RP, Assimos DG. 2008. Quantitative assessment of citric acid in lemon juice, lime juice, and commercially-available fruit juice products. J. Endourol. 22:567–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prasad BV, et al. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290 [DOI] [PubMed] [Google Scholar]
- 44. Rademacher C, et al. 2011. Targeting norovirus infection-multivalent entry inhibitor design based on NMR experiments. Chemistry 17:7442–7453 [DOI] [PubMed] [Google Scholar]
- 45. Rademacher C, Krishna NR, Palcic M, Parra F, Peters T. 2008. NMR experiments reveal the molecular basis of receptor recognition by a calicivirus. J. Am. Chem. Soc. 130:3669–3675 [DOI] [PubMed] [Google Scholar]
- 46. Shoichet BK. 2004. Virtual screening of chemical libraries. Nature 432:862–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Siebenga JJ, et al. 2010. Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants. PLoS Pathog. 6:e1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Su X, Howell AB, D'Souza DH. 2010. Antiviral effects of cranberry juice and cranberry proanthocyanidins on foodborne viral surrogates—a time dependence study in vitro. Food Microbiol. 27:985–991 [DOI] [PubMed] [Google Scholar]
- 49. Su X, Howell AB, D'Souza DH. 2010. The effect of cranberry juice and cranberry proanthocyanidins on the infectivity of human enteric viral surrogates. Food Microbiol. 27:535–540 [DOI] [PubMed] [Google Scholar]
- 50. Su X, Sangster MY, D'Souza DH. 2010. In vitro effects of pomegranate juice and pomegranate polyphenols on foodborne viral surrogates. Foodborne Pathog. Dis. 7:1473–1479 [DOI] [PubMed] [Google Scholar]
- 51. Tan M, Hegde RS, Jiang X. 2004. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J. Virol. 78:6233–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tan M, et al. 2009. Conservation of carbohydrate binding interfaces: evidence of human HBGA selection in norovirus evolution. PLoS One 4:e5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reference deleted.
- 54. Whitehead K, McCue KA. 2010. Virucidal efficacy of disinfectant actives against feline calicivirus, a surrogate for norovirus, in a short contact time. Am. J. Infect. Control. 38:26–30 [DOI] [PubMed] [Google Scholar]
- 55. Reference deleted. [Google Scholar]
- 56. Yuan P, et al. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13:803–815 [DOI] [PubMed] [Google Scholar]
- 57. Zheng DP, et al. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.