Abstract
The envelope glycoprotein (Env) of human immunodeficiency virus type 1 (HIV-1) is composed of two noncovalently associated subunits: an extracellular subunit (gp120) and a transmembrane subunit (gp41). The functional unit of Env on the surface of infectious virions is a trimer of gp120/gp41 heterodimers. Env is the target of anti-HIV neutralizing antibodies. A considerable effort has been invested in the engineering of recombinant soluble forms of the virion-associated Env trimer as vaccine candidates to elicit anti-HIV neutralizing antibody responses. These soluble constructs contain three gp120 subunits and the extracellular segments of the corresponding gp41 subunits. The individual gp120/gp41 protomers on these soluble trimers are identical in amino acid sequence (homotrimers). Here, we engineered novel soluble trimeric gp140 proteins that are formed by the association of gp140 protomers that differ in amino acid sequence and glycosylation patterns (heterotrimers). Specifically, we engineered soluble heterotrimeric proteins composed of clade A and clade B Env protomers. The clade A gp140 protomers were derived from viruses isolated during acute infection (Q168a2, Q259d2.17, and Q461e2), whereas the clade B gp140 protomers were derived from a virus isolated during chronic infection (SF162). The amino acid sequence divergence between the clade A and the clade B Envs is approximately 24%. Neutralization epitopes in the CD4 binding sites and coreceptor binding sites, as well as the membrane-proximal external region (MPER), were differentially expressed on the heterotrimeric and homotrimeric proteins. The heterotrimeric gp140s elicited broader anti-tier 1 isolate neutralizing antibody responses than did the homotrimeric gp140s.
INTRODUCTION
At the end of 2009, an estimated 33.3 million people were living with human immunodeficiency virus type 1 (HIV-1) and an estimated 2.6 million people became infected with that virus in the same year (http://data.unaids.org/pub/Report/2009/JC1700_Epi_Update_2009_en.pdf). These statistics illustrate the urgent need for the development of effective prevention approaches, including the development of an effective vaccine. It is widely accepted that an effective vaccine against HIV-1 should elicit diverse antiviral immune responses, including neutralizing antibodies (NAbs) capable of preventing infection from diverse isolates (broadly neutralizing antibodies [bNAbs]) (40, 56).The expectation that vaccine-elicited bNAbs will contribute to protection from HIV infection is based on results from passive antibody-infusion studies conducted in nonhuman primates that demonstrated the protective potential of known anti-HIV-1 neutralizing monoclonal antibodies (MAbs) (1, 9, 12, 37, 38, 55, 58, 60, 65, 74).
The target of anti-HIV-1 NAbs is the viral envelope surface glycoprotein (Env) which has a molecular weight of approximately 160 (gp160). gp160 is encoded as a single polypeptide which during intracellular processing is cleaved by furin-like cellular proteases into two noncovalently associated subunits: the transmembrane subunit (gp41) and the extracellular subunit (gp120) (25, 27, 85). The functional unit of Env is a trimer of gp120/gp41 heterodimers. Both subunits are targeted by NAbs elicited during HIV infection, and as a result, recombinant soluble versions of Env have been generated and evaluated preclinically and clinically for their ability to elicit bNAbs against HIV (for a review of this topic, see references 40, 41, and 56).
The earliest soluble forms of Env tested preclinically and clinically as immunogens were based on the monomeric gp120 subunit (6, 33, 36, 43, 59, 82). These constructs were shown to elicit neutralizing antibody responses of very narrow breadth; i.e., they elicited antibodies that primarily targeted the homologous virus and a few “easy-to-neutralize” viruses (tier 1 viruses) but not primary viruses (tier 2 and 3 viruses) (30, 57, 59). Subsequently, soluble derivatives of the trimeric Env gp160 were engineered by introducing stop codons immediately upstream from the transmembrane region of gp41. These soluble proteins, commonly referred to as gp140s, elicit broader cross-reactive neutralizing antibody responses than do the corresponding monomeric gp120s, but the responses are of much narrower breadth than those that need to be elicited by vaccination to offer protection (3–5, 8, 20, 21, 26, 28, 34, 61, 67, 78–81, 86, 88).
Simian immunodeficiency virus (SIV)/HIV heterodimeric forms of Env can be formed on the cell surface of cells cotransfected with two plasmids, one expressing the HIV Env and the other one expressing the SIV Env (22). Whether the SIV and HIV Envs can associate into heterotrimeric fusion-competent spikes is unknown. Heterotrimeric forms of clade B Envs were shown to form in the context of a cell membrane-anchored Env (68, 83). However, it is unknown whether stable soluble forms of heterotrimeric gp140 can be produced. Here we designed, expressed, purified, and characterized antigenically and immunogenically stable, soluble gp140 heterotrimeric Envs whose protomers differ in amino acid sequencing and glycosylation patterns. Specifically, we generated heterotrimeric gp140 proteins between one of three clade A Envs (Q168a2, Q259d2.17, and Q461e2) (7) and the clade B Env SF162 (13, 75). The Q168 Env shares 80% amino acid sequence identity with that of SF162, whereas the Q259 and Q461 Envs are 76% identical in sequence to the SF162 Env (47).
We report that such novel constructs can be produced and are stable enough to be purified and to be characterized antigenically. The exposure of certain epitopes that are targets of known broadly neutralizing MAbs is enhanced on such heterotrimeric constructs, compared to their exposure on the corresponding clade A and B homotrimeric gp140 proteins. Interestingly, the clade A/B heterotrimeric gp140s elicit more potent anti-tier 1 neutralizing antibody responses than do the corresponding homotrimeric gp140s. Despite the lack of elicitation of broadly neutralizing antibody responses (i.e., against tier 2 or 3 viruses), the fact that the heterotrimeric gp140 proteins elicited more potent anti-tier 1 neutralizing antibody responses than did the corresponding homotrimeric gp140s is a promising finding.
MATERIALS AND METHODS
Plasmid constructions.
Soluble trimeric gp140s (homotrimeric or heterotrimeric) were generated using envelope sequences from the clade A Envs Q168, Q259, and Q461 (7) and the clade B Env SF162 (14). Stop codons were introduced immediately upstream from the transmembrane regions of gp41, and the gp120/gp41 cleavage site was eliminated by mutagenesis, as previously described (28, 29, 76, 77, 79). The gp140 sequences were inserted in the mammalian expression vectors pEMC* and COpTT3 (73).
The SF162 gp140 was 6× His tagged (HHHHHH) at its carboxy terminus, while the clade A gp140s were 3× FLAG tagged (DYKDHDGDYKDHDIDYDDDDK) at their carboxy termini. Two versions of the SF162 gp140 were constructed. One version, SF162gp140L (L, linker), contained a 14-amino-acid (aa) linker peptide (SILEVLFQGPLGSP) between the C terminus of gp140 and the His tag. The other version, SF162gp140NL (NL, no linker), did not have a linker peptide, and the His tag was added directly to the C terminus of SF162 gp140. In the case of the clade A gp140s, the FLAG tags were added to the C terminus of gp140 without a linker.
Transfection of 293F cells.
gp140 proteins were produced by transient transfection of 293F suspension cells (Invitrogen, Carlsbad, CA) in serum-free medium, using a high-density transfection protocol (2). In total, five homotrimeric (SF162L, SF162NL, Q168, Q259, and Q461) and four heterotrimeric (Q168/SF162L, Q461/SF162L, Q259/SF162NL, and Q461/SF162NL) gp140s were produced.
The transfection conditions for the production of the homotrimeric gp140 proteins were previously described (69, 73). For the generation of heterotrimeric gp140s, the plasmids expressing the clade A gp140 and the clade B gp140 were first mixed at a predetermined ratio that resulted in the equal expression of the two gp140s and then the mixtures of plasmids were added to the cells. For the Q168/SF162L heterotrimer, a 10:1 clade A/clade B ratio was used; for Q259/SF162NL, a clade A/clade B ratio of 1:5 was used; and for the Q461/SF162L and Q416/SF162NL heterotrimers, a 1:10 clade A/clade B ratio was used. A 6.25-mg amount of total DNA was added to 5 × 109 293 F cells in a volume of 250 ml (2 × 107 cells/ml) and swirled gently. A 12.5-mg amount of polyethylenimine Max (PEI Max) transfection reagent (Polysciences, Warrington, PA) was added, and the transfection reagents and the cells were placed on an orbital shaker at 125 rpm in a 37°C incubator. Following an incubation period of 3 h, the cell culture volume was expanded to 5 liters (1 × 106 cells/ml) and protein expression was carried out for 6 days, at which point the cell supernatants were collected and protease inhibitors [phenylmethylsulfonyl fluoride (PMSF) (0.5 mM), 4-(2-aminoethyl)-benzenesulfonyl fluoride (AEBSF) (0.5 mM), aprotinin (0.3 μM), E-64 (1 μM), pepstatin (1 μM), leupeptin (10 μM), and bestatin (1 μM) (Sigma, St. Louis, MO)] were added.
Purification of soluble trimeric gp140s.
The transfection supernatants were first clarified by centrifugation at 1,000 × g for 10 min at room temperature (RT). The clarified supernatants of the homotrimeric gp140 transfections were then concentrated and purified by Galanthus nivalis lectin affinity chromatography and gel filtration, as described previously (69, 73). The only variation from this protocol was with the SF162NL gp140 homotrimeric protein, which was not purified by lectin affinity chromatography but instead using Ni2+ Sepharose 6 Fast Flow (GE Healthcare, Waukesha, WI) chromatography followed by gel filtration on a Superdex 200PG 26/60 column (GE Healthcare).
Two different protocols were developed for the purification of the heterotrimeric gp140s (protocols I and II). Protocol I was used to purify heterotrimers containing SF162L, while protocol II was used to purify heterotrimers containing SF162NL. In protocol I, the clarified transfection supernatants were concentrated ∼20× using a 30-kDa-molecular-mass cutoff (MMCO) on a tangential flow filtration device (Ultrasette 30K; Pall Life Sciences, Ann Arbor, MI) and buffer exchanged 5× into 20 mM Tris (pH 7.4), 100 mM NaCl (G. nivalis agglutinin [GNA] binding buffer). The concentrated and buffer-exchanged samples (∼250 ml) were loaded at 3 ml/min onto an XK-26 column packed with 20 ml of GNA resin equilibrated with GNA binding buffer (20 mM Tris, pH 7.4, 100 mM NaCl) and washed until all unbound proteins were eliminated. The GNA-bound Env glycoproteins were eluted with 4 column volumes of GNA elution buffer (20 mM Tris, pH 7.4, 100 mM NaCl, 1 M methyl-α-d-mannopyranoside). Env-containing fractions were pooled and concentrated to ∼10 ml using a Centricon Plus 70 centrifugal concentrator (30-kDa MMCO; Millipore, Billerica, MA). They were then loaded onto the Superdex 200PG size exclusion chromatography (SEC) column equilibrated in 20 mM monobasic sodium phosphate, pH 7.5, 500 mM NaCl, and 10 mM imidazole (His tag binding buffer). The Env oligomer-containing fractions were pooled and loaded directly onto a 5-ml HisTrap FF cartridge (GE Healthcare) column at 1 ml/min. The column was washed until the absorbance at 280 nm returned to baseline. At this stage, gp140 proteins that are exclusively FLAG tagged and which do not bind to the HisTrap column are removed in the wash. Only the His-tagged proteins (homotrimeric and heterotrimeric) remain on the column. The bound Env proteins were eluted at 5 ml/min with elution buffer (20 mM sodium phosphate, pH 7.5, 500 mM NaCl, and 250 mM imidazole). The eluted Env protein-containing fractions were then buffer exchanged into 1× phosphate-buffered saline (PBS) using 10-kDa-MMCO SnakeSkin dialysis tubing (Pierce, Rockford, IL). The dialyzed elution pool from the Ni2+ column was loaded at 1 ml/min onto a 2.5-ml anti-FLAG M2 resin (Sigma, St. Louis, MO) packed in a Tricorn 10/50 column (GE Healthcare). The resin was washed until the absorbance at 280 nm returned to baseline. During this step, gp140s that are exclusively His tagged do not bind to the anti-FLAG M2 resin column and thus are removed in the wash. At this step, only the hetero-oligomeric FLAG-tagged proteins remain on the column. The bound Env proteins were eluted with PBS plus 0.5 mg/ml FLAG peptide (3× DYKDDDDK) (Sigma). The eluate was concentrated as described above (30-kDa MMCO, 4,000 × g) and loaded onto an SEC column using the same column as described above but equilibrated with PBS. This “polishing” SEC step isolates the heterotrimers while simultaneously removing the FLAG peptide. Protocol II was similar to protocol I, with the following modifications: GNA, HisTrap FF, and anti-FLAG followed by SEC.
Blue native (BN) PAGE was used to determine the fractions containing trimers (which at this point were all in a heterotrimeric configuration). These fractions were then pooled. All purified samples were analyzed by SDS-PAGE, blue native PAGE, and standard and native Western blotting. The bicinchoninic acid (BCA) assay (Pierce) was used to determine the protein concentration using bovine serum albumin (BSA) as a standard.
SDS-PAGE.
Two micrograms of purified homotrimeric gp140 Env and 4 μg of heterotrimeric gp140 Env were loaded per lane of a 4 to 12% NuPAGE gel (Invitrogen). Electrophoresis took place at 200 V for 60 min. Proteins were visualized with Simply Blue stain (Invitrogen).
Blue native PAGE.
BN PAGE electrophoreses were performed as follows: 2.5 μg/well of purified homotrimeric gp140 proteins and 5 μg/well of purified heterotrimeric gp140 proteins in PBS were mixed 3:1 (vol/vol) with 4× native PAGE loading buffer and loaded on a 3 to 12% native gel (Invitrogen). Electrophoresis assays were run at 150 V for 2 h. The gels were fixed for 15 min (40% methanol [MeOH], 10% acetic acid) and then destained (8% acetic acid) for 2 to 3 h.
Native Env Western blotting.
Following native PAGE and destaining, the proteins were transferred to polyvinylidene difluoride (PVDF) membranes (VWR International) at 70 V for 50 min in 1× transfer buffer (39 mM glycine, 48 mM Tris base, 0.037% SDS, 20% methanol). After the molecular weight markers were marked with pencil, the PVDF membranes were blocked in 5% nonfat milk (NFM)-0.2% Tween 20 for 1 h at room temperature (RT). Different primary antibodies were used to assess the composition of the heterotrimeric Envs: anti-FLAG M2-horseradish peroxidase (HRP) MAb (1:1,000) (Sigma), tetra-His MAb (1:2,500) (Qiagen, Valencia, CA), or purified IgG from several HIV-1-infected individuals (1:2,000). The PVDF membranes were incubated with primary antibody overnight (O/N) at 4°C. The membranes were then washed 5× for 5 min each in wash buffer (1× PBS, 0.6% Tween 20). In the case of anti-FLAG M2-HRP, development took place at this stage using ECL Western blotting detection reagent (GE Healthcare) mixed 1:1. For the purified anti-HIV+ IgG and anti-His blot analyses, the secondary antibodies, rabbit anti-human-HRP (1:3,000) and goat anti-mouse-HRP (1:3,000), respectively, were added and incubated for 60 min at RT with shaking. After a final 5× wash, the blots were developed as described above.
Apparent molecular mass determination.
The apparent molecular masses of the homotrimeric and heterotrimeric Env proteins were determined by analytical SEC using a Superdex 200 10/300 GL column (GE Healthcare) with the following proteins as standards: thyroglobulin (669 kDa), apoferritin (440 kDa), catalase (232 kDa), lactate dehydrogenase (140 kDa), and BSA (67 kDa). All standard proteins were from Sigma except lactate dehydrogenase (Lee BioSolutions, Inc., St. Louis, MO). The column was run at 0.75 ml/min in 1× PBS, and 100 μg of Env protein in 100 μl (1× PBS) was injected. A standard curve was generated by measuring the elution volumes of the standard proteins and then plotting their Kav values. Kav values were plotted against the logarithm of the corresponding molecular mass. Next, the molecular masses of the unknown Env proteins were determined from the calibration curve after calculating their respective Kav value from the measured elution volume by using the following equation: Kav = Ve − V0/Vt − V0, where Ve is the elution volume, V0 is the void volume of the column, and Vt is the column bed volume.
Relative binding of known neutralizing antibodies to gp140s.
The following anti-Env reagents were used: 2F5 and 4E10 (anti-gp41 membrane-proximal external region [MPER]; Polymun Scientific, Vienna, Austria), 2G12 (anti-terminal mannose residues on gp120; Polymun Scientific), VRC01 (anti-CD4 binding site [BS]; kindly provided by J. Mascola), b12 (anti-CD4-BS; Polymun Scientific), IgG-CD4 (anti-CD4-BS; Progenics Pharmaceuticals Inc., Tarrytown, NY), and 17b (anti-coreceptor binding site; kindly provided by J. Robinson). For these experiments, the same molar concentrations of homotrimeric and heterotrimeric gp140 were adsorbed on enzyme-linked immunosorbent assay (ELISA) plates. gp140 proteins were added in duplicate wells of 96-well high-binding polystyrene plates (Immulon 2Hb; Thermo Scientific) (10 nM in the first well) and then serially diluted 3-fold in NaHCO3 (pH 9.5). Following an O/N incubation at RT, the plates were washed 3× with wash buffer (imidazole-buffered saline with Tween 20; KPL, Gaithersburg, MD) using a Skan Washer 300 (Molecular Devices, Sunnyvale, CA) and then blocked with ELISA blocking buffer (PBS, 10% nonfat milk, and 0.3% Tween 20) for 1 h at 37°C. The plates were again washed, and monoclonal antibody (MAb) was added in each well in ELISA dilution buffer (PBS, 10% nonfat milk, and 0.03% Tween 20) and incubated for 90 min at 37°C. The plates were then washed, and the secondary antibody goat-anti-human-HRP (1:3,000) was added for 90 min at 37°C. ELISA 1-Step Ultra-TMB reagent (Thermo Scientific, Waltham, MA) was added to each well for a 4-min development. The reaction was then quenched with 1 M H2SO4. The graphed data compare the absorbances at 450 nm at the highest Env concentration (10 nM).
Rabbit immunizations with gp140s.
Immunizations took place at the Pocono Rabbit Farm (Canadensis, PA). Animals were immunized with one of the following four heterotrimeric gp140s: Q168/SF162L, Q461/SF162L, Q259/SF162NL, and Q461/SF162NL. In each case, a second group of animals was immunized with the corresponding mixture of homotrimeric gp140s. Rabbits (n = 3) were immunized with 0.2 ml of a mixture of 100 μg of protein and 1 mg of 25-kDa branched polyethylenimine (Polysciences) in PBS. An 0.1-ml amount of this formulation was administered intramuscularly in each hind leg. Immunizations took place every 4 weeks (0, 4, 8, 12, and 16 weeks) with the exception of the Q461/SF162L and Q461+SF162L constructs, which were administered eight times (0, 6, 9, 21, 28, 32, 52, and 57 weeks). Blood was collected at −1, 0, 6, 10, 14, and 18 weeks for the three groups with 5 immunizations. Blood was collected at −1, 0, 8, 11, 23, 30, 34, 54, and 59 weeks for the study with 8 immunizations.
Relative endpoint antibody titers.
The relative endpoint antibody titers against the corresponding Env gp140 were determined by ELISA. Briefly, 50 ng/well of Env antigen was added O/N at RT in 100 mM NaHCO3, pH 9.5, in 96-well plates. Following washing, 5-fold-serially diluted rabbit sera were added for 90 min at 37°C. After washing, secondary antibody (1:3,000; goat anti-rabbit) was added for 60 min at RT. Endpoint titers were reported as the dilution at which the titrated curve intersected the 3× prebleed values.
Luminex multiplex assay.
Peptides derived from the V1, V2, and V3 regions of each Env gp140 tested as immunogens were individually amine coupled through a lysine at their carboxyl termini to Bioplex beads (Bio-Rad, Hercules, CA), according to the manufacturer's instructions (12 μg of peptide per 100 μl of beads). Peptide sequences are found in Table 1. Serially diluted sera (pre- and postimmunization) were incubated with the cocktail of beads for 1 h at RT. Following 4 washes, the beads were incubated with anti-rabbit-phycoerythrin (PE)-conjugated secondary MAb (1:500) for 1 h at room temperature. A final wash step was performed prior to analysis on a Luminex 200 system (Invitrogen).
Table 1.
V1, V2, and V3 loop peptides used for epitope mapping of immune sera
| Clade A peptide | Sequence |
|---|---|
| Q168a2 V1 N terminus | CTNVNNNTTNVNNNTGWDEE |
| Q168a2 V1V2 junction | GWDEERKNCSFNITTELRDK |
| Q168a2 V2 crown | ELRDKRQKVYSLFYKLDVVQ |
| Q168a2 V2 C terminus | LDVVQIDNSSYRLINCNTSA |
| Q168a2 V3 N terminus | CTRPDNNTRTSIRIGPGQAF |
| Q168a2 V3 C terminus | PGQAFYATGIIGDIRQAYCT |
| Q259d2.17 V1 N terminus | CYNVTKSDKITKDMQEEIKN |
| Q259d2.17 V1V2 junction | EEIKNCSFNITTELRDKKQK |
| Q259d2.17 V2 crown | DKKQKVHSLFYRLDVVPMGG |
| Q259d2.17 V2 C terminus | VPMGGKNDSQYRLINCNTSA |
| Q259d2.17 V3 N terminus | CTRPNNNTRKSVRIGPGQAF |
| Q259d2.17 V3 C terminus | PGQAFYATDDIIGNIRQAYC |
| Q461e2 V1 N terminus | CTDWTNNATSTNQTTPATSE |
| Q461e2 V1V2 junction | PATSEETGVKNCSFNITTEL |
| Q461e2 V2 N-crown | ITTELRDKKQKVYSLFYKLD |
| Q461e2 V2 C-crown | FYKLDVVQISENNSSNSSNF |
| Q461e2 V2 C terminus | NSSNFTQYRLINCNTSAITQ |
| Q461e2 V3 N terminus | CIRPGNNTRKSVRIGPGQAF |
| Q461e2 V3 C terminus | PGQAFYATGDITGDIRNAHC |
| Q769h5 V1 N terminus | CSNINNIPNVNASSIPKDVK |
| Q769h5 V1V2 junction | PKDVKEEIKNCSFNMTTELK |
| Q769h5 V2 N-crown | TTELKDKKQNVYSLFYRLDV |
| Q769h5 V2 C-crown | YRLDVVPLETNLKQNSSHSR |
| Q769h5 V2 C terminus | NLKQNSSHSRYRLINCNTSA |
| Q769h5 V3 N terminus | CIRPGNNTRKSIHLGPGKVF |
| Q769h5 V3 C terminus | PGKVFYATNIIGDIRKAHCN |
MPER-biotinylated ELISA.
Streptavidin (SA; 50 ng/well) was applied as a coating onto high-binding polystyrene ELISA plates O/N at RT in 100 mM NaHCO3, pH 9.5. Plates were then washed prior to blocking with 3% BSA in PBS for 1 h at 37°C. Biotinylated peptides derived from the MPER of gp41 (2F5, EQELLELDKWASLWN, and 4E10, NWFDITNWLWYIRKKK) were added at 0.2 μg/well (100 μl of 2 μg/ml) for 1 h at 37°C in PBS plus 0.2% BSA. After washing, 5-fold-serially diluted rabbit serum was added for 1 h at 37°C in PBS plus 0.2% BSA. Goat anti-rabbit secondary antibody was added at 1:5,000 in PBS plus 0.2% BSA and incubated for 1 h at 37°C before development as described above.
AviTag ELISA.
AviTag peptides were synthesized by Genscript (Piscataway, NJ) so that targeted, residue-specific biotinylation could be accomplished in addition to presenting the affinity tag optimally in solution using an ELISA format. The peptides were designed with the AviTag at the N terminus adjacent to a 6-aa soluble linker and an alpha-helix (EAAAK)4 to control tag orientation, followed by a 6-aa soluble linker with the affinity tag comprising the C terminus. The AviTag peptides were enzymatically biotinylated (Genecopia, Rockville, MD), and the biotinylated peptides were purified using the appropriate affinity resin (FLAG-M2 for the biotinylated FLAG AviTag peptide or Sepharose 6 Fast Flow for the biotinylated His AviTag peptide). Purified peptides were then concentrated and buffer exchanged into PBS using a 3,000-molecular-weight-cutoff (MWCO) centrifugal concentrator. Peptide concentrations were determined by BCA assay (Pierce, Rockford, IL). ELISA plates were coated with biotinylated peptide as described above. Threefold-serially diluted rabbit sera were added and incubated at 37°C in PBS plus 0.2% BSA for 1 h. Secondary antibody treatment and development of ELISA plates were done as described above for the biotinylated ELISA.
Neutralization.
Neutralization assays were performed at the Stamatatos, Montefiori, and Seaman laboratories using single-round entry-competent viruses and TZM-bl cells as targets. The protocol used at the Stamatatos laboratory was previously described (20, 47, 70). Briefly, a predetermined amount of virus (2 × 105 relative luminescence units [RLU]) was mixed (1:1) with a single serum dilution (1:20, final) at 37°C for 1.5 h; total volumes in duplicate wells were 60 μl. Fifty microliters of the virus/serum mixture was transferred to wells of flat-bottomed 96-well tissue culture plates containing 3 × 103 Polybrene-treated TZM-bl cells. Following a 3-day incubation at 37°C, the cell supernatants were removed and 100 μl of SteadyGlo luciferase (Promega) was added to each well for 15 min at room temperature. The number of relative luminescence units associated with 75 μl of cell lysate was determined on a Fluoroskan Ascent FL (Thermo Labsystems). Percent neutralization at 1:20 serum dilution was determined for each immune serum against the corresponding serum collected prior to immunization, based on the following equation: (RLU preimmunization − RLU postimmunization/RLU preimmunization) × 100.
The Montefiori and Seaman laboratory neutralization protocols were described previously (49, 71). Briefly, 200 tissue culture infective doses (TCID) of virus was incubated with serial 3-fold dilutions of test sample in duplicate in a total volume of 150 μl for 1 h at 37°C in 96-well flat-bottomed culture plates. Freshly trypsinized cells (10,000 cells in 100 μl of growth medium containing 75 μg/ml DEAE dextran) were added to each well. One set of control wells received cells plus virus (virus control), and another set received cells only (background control). After a 48-h incubation, 100 μl of cells was transferred to a 96-well black solid plate (Costar) for measurements of luminescence using the Britelite luminescence reporter gene assay system (PerkinElmer Life Sciences). Neutralization titers are the dilution at which relative luminescence units (RLU) were reduced by 50% compared to those of virus control wells after subtraction of background RLU. Assay stocks of molecularly cloned Env-pseudotyped viruses were prepared by transfection in 293T cells and were titrated in TZM-bl cells as described previously (49, 71).
The following viruses (clade) were tested: tier 1, MN.3 (B), SF162.LS (B), HxB2 (B), W61D (TCLA) 0.71 (B), Bal.26 (B), BZ167.12 (B), Bx08.16 (B), SS1196.1 (B), MW965.26 (A), Q461d1 (A), Q168b23 (A), DJ263.8 (A), Q259w6 (A), Q769.b9 (A), TV1.21 (C), and 92BR025.9 (C); tier 2, 3988.25 (B), REJO4541.67 (B), TRO.11 (B), ADA (B), JRFL (B), YU2 (B), 89.6 (B), 6535.3 (B), 7165 (B), THRO4156.18 (B), AC10.0.29 (B), QH0692.42 (B), AC10.0.29 (B), RHPA4259.7 (B), WITO4160.33 (B), Q23.17 (A), Q842.D12 (A), 3365.v2.c20 (A), 3415.v1.c1 (A), 0260.v5.c1 (A), 191955_A11 (A), 191084 B7-19 (A), 9004SS_A3A4 (A), Q168a2 (A), Q461e2 (A), Q769h5 (A), Du422.1 (C), ZM197M.PB7 (C), ZM214M.PL15 (C), ZM233M.PB6 (C), ZM249M.PL1 (C), Q259d2.17 (A), CAP210.2.00.E8 (C), Du156.12 (C), Du172.17 (C), CAP45.2.00.G3 (C), and ZM53M.PB12 (C); and tier 3, PVO.4 (B).
Statistical analyses to differentiate the neutralizing antibody responses elicited by the heterotrimers and the homotrimers were performed with Welch's t test.
RESULTS
Optimizing transfection conditions for the efficient coexpression of clade B and A gp140s.
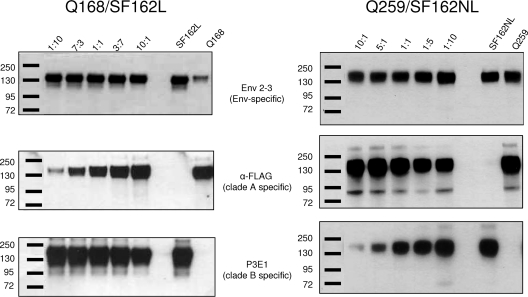
In preparation for large-scale expression of heterotrimeric gp140s, small-scale cotransfection experiments with different ratios of two plasmids, one expressing the clade A gp140 and the other expressing the clade B gp140, were performed. These pilot experiments aimed at defining the ratios of the two plasmids that resulted in similar levels of expression of the clade A and clade B gp140 proteins. During these experiments, the total amount of DNA added to the cells remained constant, while the ratio of clade A to clade B gp140-expressing plasmids varied from 1:10 to 10:1. The relative expression levels of the clade A and B gp140s were determined using MAbs recognizing the clade A or B gp140s. Two examples are shown in Fig. 1. For Q168/SF162L, a ratio of 10:1 was determined as optimal and used for all subsequent large-scale production experiments; for Q461/SF162L, a ratio of 10:1 was used; for Q259/SF162NL, a 1:5 ratio was used; and for Q461/SF162NL, a ratio of 1:10 was used.
Fig 1.
Optimizing the coexpression of clade A and B gp140s. 293 cells were cotransfected with the indicated ratios of two plasmids, one expressing the His-tagged clade B gp140 and the other expressing the FLAG-tagged gp140. The relative expression of the clade A and clade B gp140s was determined with the use of an anti-FLAG MAb, specific for the FLAG-tagged clade A gp140, or a clade B-specific anti-V3 MAb, P3E1, as discussed in detail in Materials and Methods. Anti-HIV polyclonal sera (Env 2-3) were used to monitor the overall Env expression. Molecular weight standards are shown at the left of each panel.
Purification of homotrimeric and clade B/A heterotrimeric gp140 proteins.
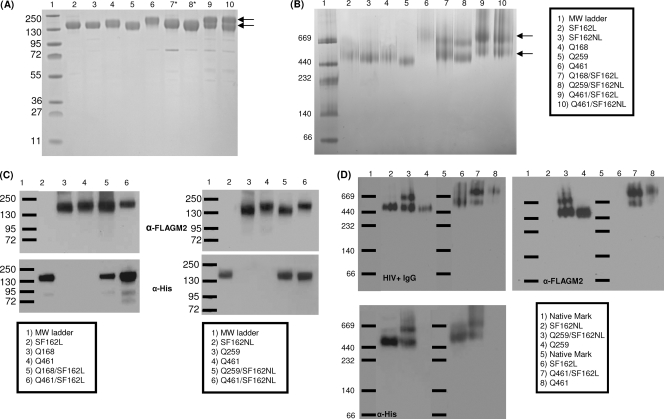
Our expression/purification protocols yielded between 0.8 and 4.5 mg/liter of purified homotrimeric gp140 and between 0.2 and 1.1 mg/liter of purified heterotrimeric gp140s. The purity of these preparations was estimated to be >95% for the homotrimers and >90% for the heterotrimers based on SDS-PAGE (Fig. 2A). Our initial designs included a 14-amino-acid linker sequence between the carboxy terminus of the SF162 gp140 and the 6His tags. We erroneously assumed that by placing the positively charged His tag away from gp140, a more efficient binding of the His tag to the nickel column would ensue and that this would facilitate the purification step of both the SF162 gp140 homotrimers and the clade A/B heterotrimers. Experimentally, however, we determined that this design had the opposite effect (data not shown), most likely because the presence of the linker sequence resulted in poor exposure of the 6His tag. Our subsequent designs did not include the 14-amino-acid linker, and instead the 6His tag was positioned directly at the carboxy terminus of the SF162 gp140 (SF162gp140NL). This resulted in improving by up to 5-fold the purification efficiency of both the homotrimeric SF162 gp140 and of the clade A/B heterotrimeric gp140s.
Fig 2.
Purification of heterotrimeric gp140s. (A) SDS-PAGE of the purified homotrimeric and heterotrimeric gp140 preparations. (B) Blue native PAGE of the same proteins as those in panel A. In both panels, the arrows indicate the presence of the 2 gp140s that migrate differently (heterotrimeric preparations) on the gel due to differences in molecular masses of the clade A and B gp140s (SDS-PAGE) or charge/mass ratios (native PAGE) (*, size differences between these clade A and clade B Envs determined by this method are negligible). Env visualization on both gels was done with Coomassie blue staining. (C) Detection of the His tag (clade B gp140) and/or FLAG tag (clade A gp140) by anti-His MAb or anti-FLAG MAbs of the indicated protein preparations. In the case of the heterotrimeric preparations, both the clade A and clade B gp140s are visualized by the use of both the anti-His MAb and anti-FLAG MAb. (D) Western blot assays of blue native gels of the indicated Env preparations. Envs were detected with either a purified pooled HIV+ IgG preparation (top left panel), an anti-FLAG MAb (top right panel), or an anti-His MAb (bottom left panel).
The migration patterns of the clade A and B gp140 protomers on SDS-PAGE gels differ because of differences in polypeptide length and extent of protein glycosylation (7, 47). As a result, two Env gp140s of different molecular masses are visualized in the purified preparation of heterotrimers, but only a single Env gp140 in the preparation of homotrimeric Envs is observed by Coomassie blue staining of SDS-PAGE gels (Fig. 2A and B). The presence of both clade A and B gp140s in the final purified preparation of heterotrimers was also shown by Western blotting and the use of specific MAbs (Fig. 2C). The anti-His specific MAb selectively recognizes the His-tagged clade B gp140s (both in the context of homotrimers and in the context of heterotrimers), and an anti-FLAG specific MAb selectively recognizes the FLAG-tagged clade A gp140 (both in the context of homotrimers and in the context of heterotrimers).
We note that two heterotrimeric gp140 species coexist in the final purified sample (arrows in Fig. 2A and 2B): one species consists of 2 clade A gp140 protomers and 1 clade B gp140 protomer and the other consists of 1 clade A gp140 protomer and 2 clade B gp140 protomers. The chromatographic properties of these two species are nearly identical, which precludes their separation with available methodologies, at least at this stage. The relative proportion of the two species was approximately 1:1 (based on results from several independent trimer preparations).
To confirm that both the clade A and B gp140 protomers exist on both the 2:1 and 1:2 heterotrimeric molecules, blue native gel electrophoresis and Western blotting methodologies were employed (Fig. 2D). Env gp140s were visualized by either a pooled HIV+ IgG preparation (top left panel), an anti-FLAG MAb (top right panel), or an anti-His MAb (bottom left panel). HIV+ IgG allows for the detection of both clade A and B gp140s simultaneously, while the anti-FLAG MAb is specific for the FLAG-tagged clade A gp140s and the anti-His MAb is specific for the His-tagged clade B gp140. A positive signal in both bands of the heterotrimeric preparations by both the anti-His and anti-FLAG MAbs indicates that both clade A and clade B protomers are present in each species. One species contains 2 clade A gp140 molecules and 1 clade B gp140 molecule, and the other species contains 2 clade B gp140 molecules and 1 clade A gp140 molecule.
The above results, combined with the fact that purified heterotrimeric preparations run as a single peak by analytical SEC (data not shown), provide evidence for the stable purification of soluble trimeric clade A/B gp140 proteins.
Analytical SEC was used to determine the apparent molecular mass of the HIV Env trimers (Table 2). Of note, the determined apparent molecular mass was greater than that predicted based on the primary amino acid sequence and the number of potential N-linked glycosylation sites (PNLGS) (assuming ∼2 kDa of sugars per NLGS) and using a globular molecular mass standard.
Table 2.
Molecular mass of homotrimeric and heterotrimeric gp140s
| Envelope trimer | Molecular mass (Da) |
|---|---|
| Q168FLAG | 534,880 |
| Q259FLAG | 509,806 |
| Q461FLAG | 661,791 |
| SF162NL | 488,006 |
| SF162L | 505,684 |
| Q259/SF162NL | 541,951 |
| Q461FLAG/SF162L | 642,970 |
| Q461FLAG/SF162NL | 625,989 |
Relative exposure of specific neutralization epitopes on heterotrimeric and corresponding homotrimeric gp140s.
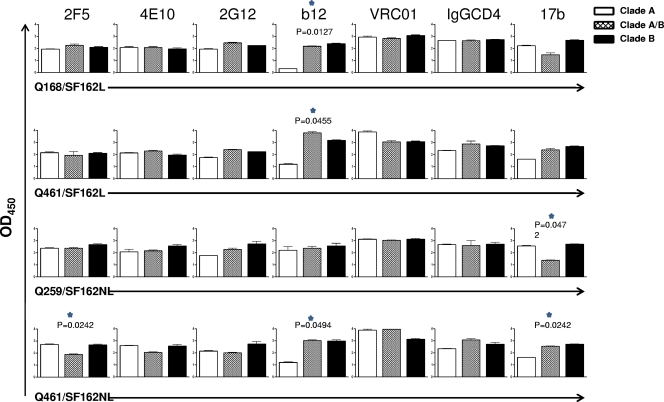
In order to determine whether the association of heterologous gp140 protomers into a heterotrimeric configuration results in changes in the exposure of neutralization epitopes, relative to the exposure of the same epitopes on the corresponding homotrimeric gp140 proteins, we compared the binding levels of several bNAbs to the heterotrimeric gp140s (clade A/B) and to the corresponding two homotrimeric gp140s (clade A and clade B) (Fig. 3). Since the molecular masses of the heterotrimeric and homotrimeric gp140s differ (Table 2), we performed ELISAs during which the wells were coated with identical molar concentrations of each gp140 to ensure that the same numbers of gp140 protomers were available for antibody recognition. Assuming a 1:1 ratio between the two heterotrimeric species (see above), the number of clade B gp140 protomer molecules in the wells coated with the clade A/B heterotrimers will be half the number of the clade B gp140 protomer molecules in the cells coated with the clade B homotrimers. The same will be the case for the clade A gp140 protomers. If a bNAb binds to the clade B gp140 but not the clade A gp140, one would expect that the maximum binding of this antibody to the clade A/B heterotrimer would be approximately half of that observed with the clade B homotrimer.
Fig 3.
Relative epitope exposure on homotrimeric and heterotrimeric gp140s. The binding of the indicated MAbs to the indicated heterotrimeric and homotrimeric gp140s was determined as described in Materials and Methods. The statistical analysis compares the measured value for MAb binding to the heterotrimers to the theoretical value expected if there were no change in relative epitope exposure. Unpaired t test with Welch's correction. OD450, optical density at 450 nm.
The anti-gp41 antibodies 2F5 and 4E10 bound similarly to the homotrimeric clade A and B gp140s tested here. As expected, therefore, 2F5 and 4E10 bound equally to the clade A and B Q168/SF162L, Q259/SF162NL, and Q461/SF162L heterotrimers and to the corresponding homotrimers. Overall, the epitopes of these two MAbs appear to be similarly exposed on these gp140s irrespective of whether they are in a homotrimeric or heterotrimeric form. In contrast, the binding of 2F5, but not of 4E10, to the Q461/SF162NL heterotrimer was significantly lower (P = 0.0242) than expected, based on its binding to the homotrimeric clade A and clade B homotrimeric gp140s. It appears, therefore, that the formation of the Q461/SF162NL heterotrimer decreases the exposure of the 2F5 epitope. Whether this decrease is occurring on the clade B gp140 component, on the clade A gp140 component, or on both components of the Q461/SF162NL heterotrimer is currently unknown.
No significant differences in the binding of MAb 2G12 to the homotrimeric clade A and B gp140s tested here were observed. As anticipated, no significant differences in the binding of 2G12 to the heterotrimeric gp140s and the corresponding homotrimeric gp140s were observed.
In contrast, MAb b12 recognized the clade A Q168 and Q461 gp140 homotrimers less efficiently than the clade B SF162 gp140 homotrimer. This is especially evident in the case of Q168 gp140. These results indicate that the b12 epitope is not as well exposed on these two clade A Envs (and in the case of the Q168 Env, potentially not present) as it is on the clade B SF162 gp140. The fact, however, that b12 binds to the Q168/SF162 and Q461/SF162 heterotrimers as efficiently as to the SF162 gp140 homotrimer suggests that either the b12 epitopes on clade SF162 gp140 protomers become more exposed in the context of the A/B heterotrimers or/and that, while the b12 epitope is occluded on the clade A homotrimer, it becomes exposed on the clade A protomers in the context of the heterotrimeric configurations.
To investigate whether the CD4-BS overall was more exposed on the Q168/SF162 and Q461/SF162 heterotrimers, we compared the binding of a second anti-CD4-BS MAb, VRC01, and that of IgG-CD4 to the homotrimers and heterotrimers. In contrast to what we observed for b12, we did not record any obvious changes in the relative epitope exposures of VRC01 and IgG-CD4 between the homotrimers and heterotrimers. Overall, the data indicate that certain elements of the CD4-BS become preferentially exposed in the context of some heterotrimeric gp140s but that the CD4-BS as a whole may not be more exposed on the heterotrimers than the homotrimers.
MAb 17b was utilized to investigate the relative exposure of the CCR5 coreceptor binding site on the homotrimers and heterotrimers. We note that these studies were performed in the absence of CD4 binding to gp140. Based on the relative binding of 17b to the homotrimeric clade B and A gp140s, our data indicate that the formation of heterotrimers affects the relative exposure of the 17b epitope (Q461/SF162L is the exception). In the cases of Q259/SF162NL and Q461/SF162NL, these changes in the relative exposure of the 17b epitope on the heterotrimer compared to the homotrimers were significant (P = 0.0472 for Q259/SF162 and P = 0.0242 for Q461/SF162). However, in the case of Q259/SF162 there was a decrease in the exposure of the 17b epitope (relative to what was expected based on the binding results with the corresponding homotrimers) while in the case of Q461/SF162 there was an increase in the 17b epitope (again, what was expected based on the binding results with the corresponding homotrimers). Therefore, the relative exposure of the CCR5 coreceptor binding site either cannot be affected by the formation of the heterotrimeric gp140 structure or it can increase or decrease, depending on the backbone of the clade A component of the heterotrimer. The relative binding of 17b to the Q461/SF162 heterotrimer was greater than expected (based on the binding results with the corresponding homotrimers) irrespective of whether the SF162 gp140 had a linker peptide at its carboxy terminus (SF162L) or not (SF162NL). The increase in 17b binding to the heterotrimer reached significance only when the nonlinker version was used. It is, therefore, possible that the presence of the linker peptide altered the position of the His tag in a way that may have changed the exposure of certain epitopes (Q461/SF162L versus Q461/SF162NL). One of these epitopes is the epitope recognized by MAb 17b, and another is the epitope recognized by MAb 2F5 (Fig. 3).
Immunogenicity of heterotrimeric and homotrimeric gp140s.
As discussed above, animals were immunized with a heterotrimer and control animals were immunized with the corresponding mixture of the homotrimers. Similar anti-clade A Env antibody titers were elicited by the heterotrimer and the mixture of the homotrimeric gp140s (Fig. 4). This indicates that the overall immunogenic properties of the three clade A Envs used here are not affected by the presence of the clade B SF162 gp140 in the context of either homotrimers or heterotrimers. In contrast, the immunogenicity of the clade B SF162 gp140 was greater in the case of the Q168/SF162L and Q461/SF162NL heterotrimers than in the case of the corresponding homotrimers (Fig. 5A and D). This was not the case with Q461/SF162L or Q259/SF162NL (Fig. 5B and C). In contrast, therefore, to what we observed for the clade A component of the heterotrimers, the association of clade B and A gp140s into stable heterotrimeric proteins can result in an enhancement of the clade B gp140 component.
Fig 4.
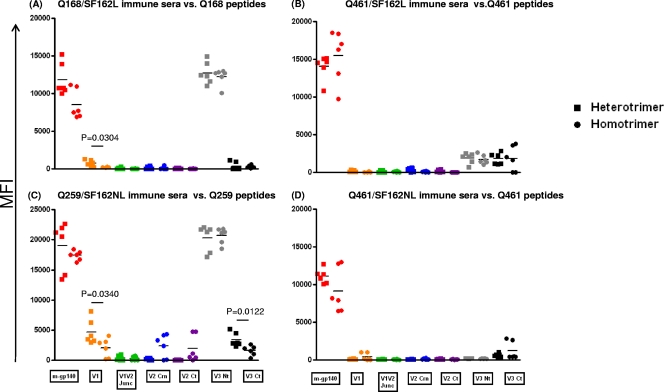
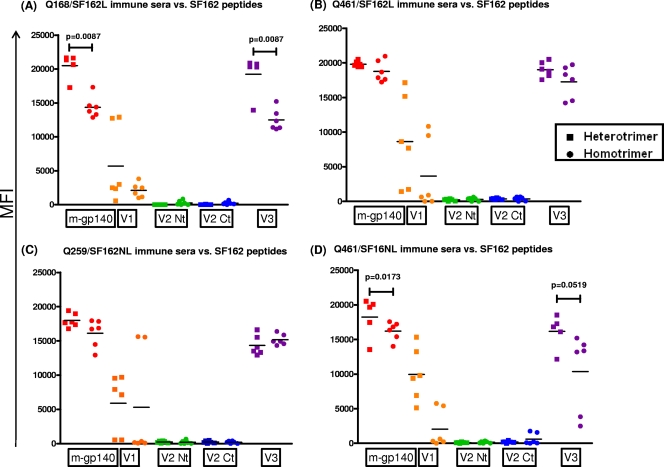
Immunogenicities of the V1, V2, and V3 regions from clade A Envs. Animals were immunized with Q168/SF162L (A), Q461/SF162L (B), Q259/SF162NL (C), or Q461/SF162NL (D). Antibody reactivities to peptides derived from the V1, V2, and V3 regions of each clade A Env were determined by multiplex Luminex assay. Antibody reactivity to monomeric gp140 (m-gp140) was also determined. ■, sera from animals immunized with heterotrimeric gp140; ●, sera from animals immunized with mixtures of homotrimers. MFI, mean fluorescent intensity. V1, peptide spanning the V1 loop; V1V2 Junc, peptide spanning the N-terminal side of the V2 loop; V2 Crn, peptide spanning the crown portion of the V2 loop; V2 Ct, peptide spanning the carboxy-terminal site of the V2 loop; V3 Nt, peptide spanning the amino-terminal region of the V3 loop; V3 Ct, peptide spanning the carboxy-terminal region of the V3 loop (Table 1 shows peptide sequences). Statistical analyses were performed with the unpaired t test with Welch's correction analysis.
Fig 5.
Immunogenicities of the V1, V2, and V3 regions from the clade B SF162 Env. The experiment was similar to that in Fig. 4, but the peptides were derived from the SF162 Env.
The fact that similar overall binding titers to the clade A gp140s were elicited by the homotrimeric and heterotrimeric proteins does not necessarily mean that the relative immunogenicity of specific Env regions was not different in the context of homotrimeric and heterotrimeric proteins. To address this point, we compared the antibody titers to specific clade A Env regions (Fig. 4). Similar analyses were performed for the SF162 gp140 (Fig. 5).
We focused on the variable regions 1, 2, and 3 and the MPER in gp41 because reagents derived from these Env regions are easily available and this allows for the determination of Ab binding to these regions. In contrast, appropriate reagents derived from structurally more complex Env regions, such as the CD4-BS or the coreceptor binding sites, are not available, and this does not allow for a direct determination of anti-CD4-BS or anti-receptor binding site Ab titers.
(i) Immunogenicity of clade A Env variable regions on homotrimeric and heterotrimeric gp140 proteins.
The relative antibody titers to the V1, V2, or V3 loop of gp120 on the heterotrimers and the corresponding mixtures of the homotrimers were determined using variable loop-derived peptides from the clade A and the clade B Env protomers (Fig. 4 and 5). Animals immunized with the Q168 and SF162L gp140s (Fig. 4A) or with Q259 and SF162L gp140s (Fig. 4C) (both as homotrimers and as heterotrimers) elicited high antibody titers to the corresponding clade A V3 N-terminal region. There was no significant difference in antibody binding titers between the heterotrimeric and homotrimeric sera. In contrast, very weak to undetectable antibody titers against the corresponding Q168 V3 carboxy-terminal region were observed (Fig. 4A). Higher antibody responses against the Q259 V3 carboxy-terminal region were observed in animals immunized with Q259 and SF162 (either as homotrimers or as heterotrimers) (Fig. 4C). The anti-Q259 V3 carboxy-terminal region antibodies were significantly higher in the case of the heterotrimeric sera than in the case of the homotrimeric sera (P = 0.0122). Similarly, weak or undetectable anti-V3 antibody titers were observed in the case of the Q461 or SF162 (either with the linker or without the linker) constructs (Fig. 4B and D).
The anti-V2 antibody titers were either undetectable or very low, although animals immunized with the Q259 and SF162 homotrimeric mixture developed very weak anti-V2 antibodies to the central and carboxy-terminal regions of the V2 loop (Fig. 4C).
Immunization with Q461 and SF162 did not result in the elicitation of detectable anti-V1 antibodies (Fig. 4B and D). In contrast, immunization with Q168 or Q259 (with or without SF162) resulted in the elicitation of measurable anti-V1 antibodies (Fig. 4A and C). In both cases, the anti-V1 antibodies were higher in the case of heterotrimeric sera than in the case of homotrimeric sera (P = 0.0304 in the case of Q168 and P = 0.0340 in the case of Q259).
(ii) Immunogenicity of clade B Env variable regions on homotrimeric and heterotrimeric gp140 proteins.
The V3 loop of SF612 was immunogenic in the context of homotrimeric and heterotrimeric proteins. However, higher levels of anti-V3 loop antibodies were present in the heterotrimeric Q168/SF162 (Fig. 5A) and Q461/SF162NL (Fig. 5D) sera than in the corresponding homotrimeric sera. The association, therefore, of the SF162 gp140 with certain clade A gp140s into stable trimers can lead to increases in the immunogenicity of the SF162 V3 loop. As discussed above, this was not the case for the clade A Envs. Similarly to what we observed for the clade A Envs, the immunogenicity of the V2 loop of the SF162 gp140 was poor. In accordance with our previous reports (15, 20), the V1 loop of SF162 was immunogenic in the context of SF162 gp140 homotrimers. The association of SF162 gp140 with the clade A gp140s into heterotrimeric conformations did not alter the immunogenicity of the SF162 V1 loop. This observation contrasts with that made with the immunogenicity of the clade A Q168 and Q259 V1 loops (Fig. 4A and C), which increased in the context of the heterotrimeric gp140s.
Overall, these results indicate that the immunogenic properties of the V1 and V3 loops from clade A and B Envs are differentially affected by the association of these Envs into heterotrimeric configurations.
(iii) Immunogenicity of the MPER on homotrimeric and heterotrimeric gp140 proteins.
Antibodies against the 2F5 and 4E10 peptides were detected in immune sera from several animals (Fig. 6). All three animals immunized with the Q461/SF162L heterotrimer gp140 elicited higher titers of both anti-4E10 (Fig. 6A; P = 0.0056) and anti-2F5 (Fig. 6B; P = 0.0063) antibodies than those in the matched groups immunized with the mixture of Q461 and SF162L homotrimers. Two of three animals immunized with either the Q461/SF162NL heterotrimers or the mixture of the corresponding homotrimers developed anti-4E10 antibodies (Fig. 6A). In contrast, the antibody responses to the 2F5 epitope were either undetectable or very weak (Fig. 6B), although it should be pointed out that two of three animals immunized with the mixture of the Q461and SF162NL homotrimers did develop weak titers of anti-2F5 antibodies (Fig. 6B). When an animal developed both anti-4E10 and anti-2F5 antibodies, the anti-4E10 antibody titers were higher than the anti-2F5 antibody titers.
Fig 6.
Immunogenicities of the 4E10 and 2F5 epitopes in the MPER of gp41. Sera were diluted at 1:100, and the antibody reactivities to the 4E10 (A) and 2F5 (B) peptides were determined (optical density at 450 nm [OD450]). Each point indicates an individual animal (3 animals per immunization group). The immunogens are indicated under the x axis. Heterotrimeric immunogens are indicated by a slash—for example, Q168/SF162L—while immunogens composed of the mixture of homotrimers are indicated by a plus sign—for example, Q168+SF162L. Statistical analyses were performed with the unpaired t test with Welch's correction analysis.
All three animals immunized with the Q259/SF162NL heterotrimer gp140 developed anti-4E10 antibodies while only one of the three animals immunized with the mixture of the corresponding homotrimers developed such antibodies (Fig. 6A). Two of three animals immunized with the Q259/SF162NL heterotrimer developed anti-2F5 antibodies while all three animals immunized with the mixture of the corresponding homotrimers developed weak anti-2F5 antibodies (see Fig. 6B).
At this stage, we do not know the reasons for the greater immunogenicity of the 2F5 and 4E10 epitopes in the context of the Q461/SF162L heterotrimer, but it does not appear to be linked to an increased exposure of these two epitopes on this protein (Fig. 3).
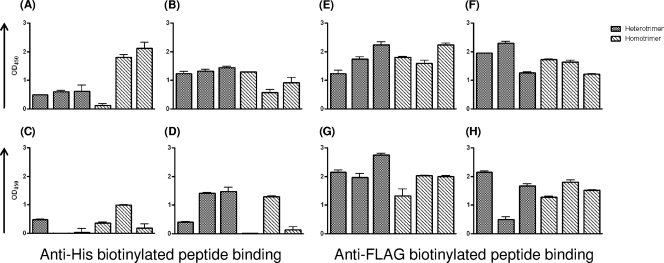
Immunogenicity of the His and FLAG tags.
As discussed in detail above, the His and FLAG peptide tags were added to the carboxy termini of the clade A and B gp140s, respectively, to facilitate purification of soluble clade A/B heterotrimeric gp140s. Anti-His antibodies were undetectable in immune sera from several animals (Fig. 7A to D) whereas all animals developed anti-FLAG antibodies (Fig. 7E to H). While very similar anti-FLAG antibody titers were recorded in all animals (with one exception, the Q461/SF162NL group [Fig. 7H]), the anti-His antibody titers varied not only among animals but also depending on the Env immunogen formulation.
Fig 7.
Immunogenicities of the His and FLAG tags. All clade A gp140s were tagged with FLAG, and the clade B gp140 was tagged with His. The anti-tag antibody responses in immune sera were determined at a 1:25 serum dilution, as described in Materials and Methods. Each bar indicates an individual animal. (A to D) Anti-His antibody responses. (E to H) Anti-FLAG antibody responses. (A and E) Sera from animals immunized with Q461 and SF162L; (B and F) sera from animals immunized with Q168 and SF162L; (C and G) sera from animals immunized with Q259/SF162NL; (D and H) sera from animals immunized with Q461/SF162NL.
Neutralizing activities in immune sera.
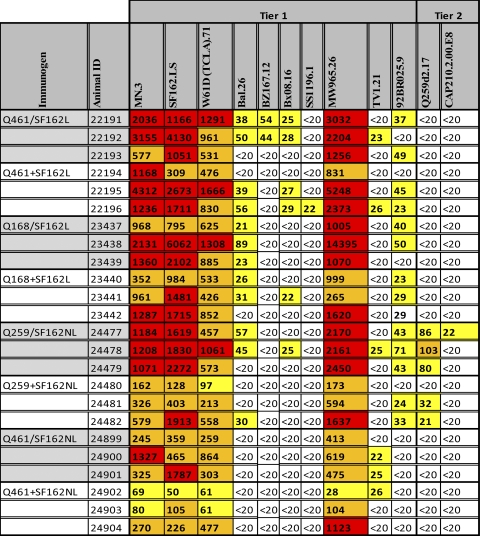
For the comparative analysis of the neutralizing activities of immune sera from animals immunized with heterotrimers or with the mixture of homotrimers, neutralization values of 50% or higher were considered (Table 3 and Fig. 8). Although several tier 1 viruses were susceptible to neutralization by both the heterotrimeric and homotrimeric sera, only 2/37 tier 2 viruses (the clade A Q259d2 and the clade C CAP210.2.00.E8) were susceptible to neutralization by these sera using TZM-bl cells as targets. We have not tested these sera using A3R5 cell lines or human peripheral blood mononuclear cells (PBMC) as targets. The Q259.d2 virus was neutralized only by sera from animals immunized with Env gp140 containing the autologous gp140 (3/3 heterotrimeric and 2/3 homotrimeric sera). The CAP210 virus was susceptible only to 1/3 heterotrimeric sera from animals immunized with the heterologous Q259/SF162 heterotrimers.
Table 3.
Neutralizing activities of immune seraa
Fifty percent inhibitory concentration neutralizing antibody serum titers are shown. Sera from rabbits immunized with the indicated immunogens were tested against the indicated viruses (tier 1 or tier 2). Heterotrimeric immunogens are indicated by a slash—for example, Q461/SF162L—while immunogens composed of the mixture of homotrimers are indicated by a plus sign—for example, Q461+SF162L. The results are presented as a heat map: white boxes, <20 (50% neutralization was not recorded at the lowest serum dilution tested [1:20]); yellow boxes, 50% inhibitory concentration titers below 1:100; orange boxes, 50% inhibitory concentration titers over 1:100 and below 1:1,000; red boxes, 50% inhibitory concentration titers above 1:1,000.
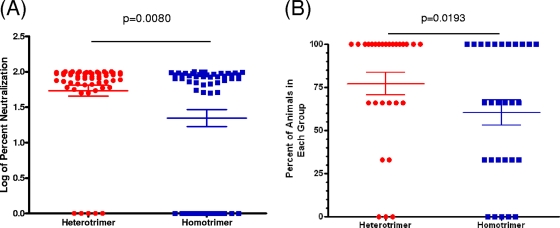
Fig 8.
Comparison of serum neutralizing activities in heterotrimeric and homotrimeric sera. (A) Each point indicates the log percent neutralization of a given serum against a given virus tested, irrespective of the immunogen. Only tier 1 viruses and one tier 2 virus (Q259) were included in this analysis. (B) The points represent the percentage of animals within each immunization group (n = 3) that neutralized a given virus. Red circles, sera from animals immunized with heterotrimeric gp140s; blue squares, sera from animals immunized with the mixture of homotrimeric gp140s. Statistical analyses were performed with Welch's t test.
Although no significant differences between the neutralizing potentials of the heterotrimeric and homotrimeric sera against tier 2 viruses were evident, the potency of neutralization of the heterotrimeric sera (at 1:20 dilution) against tier 1 isolates was greater than that of the homotrimeric sera (Fig. 8A; P = 0.008). Also, immunization with gp140 heterotrimers resulted in the more frequent elicitation of heterologous neutralizing antibody responses than did immunization with the corresponding mixture of gp140 homotrimers (Fig. 8B; P = 0.0193).
DISCUSSION
In an effort to enhance the immunogenicity of conserved neutralization epitopes on soluble Env-based immunogens, diverse approaches have been tested over the past 2 decades, such as prime-boost or cocktail immunizations with distinct envelope immunogens (16, 72, 84); deletion of variable but immunogenic Env regions (3, 20, 35, 42, 44, 53, 54, 87); addition or deletion of glycosylation sites either to occlude immunodominant but variable regions or to enhance the exposure of conserved neutralization epitopes, respectively (10, 17, 32, 50, 64, 66); repositioning of conserved epitopes within more variable, but also more immunogenic, regions of the HIV Env (48, 51); design of non-HIV Env scaffold proteins that express epitopes of broadly neutralizing antibodies (11, 18, 19, 39, 62, 63); molecular evolution-based approaches (24); generation of ancestral or consensus Env sequences (23, 31, 45, 46, 52); and others. Thus far, all of these approaches have had a very limited success in the elicitation of broad and potent neutralizing antibody responses against tier 2 HIV-1 viruses.
Here we tested a novel approach. We engineered soluble trimeric proteins composed of two distinct gp140 protomers that differed up to 24% from each other in amino acid sequence. It was previously reported that SIV and HIV gp160 can associate on the surface of cells to form heterodimers and that gp160s from different clade B Envs can form functional trimeric membrane-bound trimers (22, 68, 83). However, it was unknown whether soluble gp140 protomers belonging to two different HIV clades could associate in stable trimeric conformations. We hypothesized that the association of heterologous gp140s into a stable trimeric configuration would not be as “perfect” as the association of three identical gp140s and that, as a result, the relative exposure of epitopes on the heterotrimers would differ from that on the corresponding homotrimers. Potentially, differences in epitope exposure could be translated into differences in epitope immunogenicity.
By tagging the two heterologous gp140 proteins with two distinct tags (His and FLAG), we were able to devise a purification scheme that resulted in the isolation of three distinct clade A/B soluble gp140 heterotrimeric proteins. Using 5L transient-transfection protocols, we were able to purify several-milligram quantities of each heterotrimeric protein that were stable for several weeks (at 4°C at a concentration of 1 mg/ml in PBS).
The variable loops were not equally immunogenic on the three clade A Envs. For example, the amino-terminal side of the V3 loops of Q168 and Q259 Envs was immunogenic while that same region on the V3 loop of Q461 was weakly immunogenic. This was the case in the context of both the heterotrimeric and the homotrimeric forms. Therefore, the immunogenic properties of the amino-terminal side of the V3 loops of Q168 and Q259 were not altered by the presence of the neighboring SF162 gp140 molecules in the context of the homotrimers. The carboxy-terminal side of the V3 loop was weakly immunogenic in all cases. Interestingly, in the case of the Q259 Env, the antibody titers against the carboxy-terminal side of the V3 loop were higher in the homotrimeric mixture than in the heterotrimeric sera. This suggests that the association of the Q259 gp140 in heterotrimeric configurations with SF162 gp140 results in decreases in the immunogenic properties of the carboxy-terminal region of the V3 loop. Potentially, the formation of the Q259/SF162 heterotrimers results in the partial occlusion of the carboxy-terminal region of the clade A V3 loop.
The V2 loops of the three clade A Envs were not immunogenic (at least based on the assays used in our studies). We note, however, that weak antibody titers to the central and carboxy-terminal domains of the V2 loop were recorded in the case of the Q259/SF162 heterotrimers, while such responses were not evident in the homotrimeric mix (although the differences in titers were not significant). Therefore, the immunogenicity V2 loop of Q259 is increased in the context of the heterotrimer. Potentially, the central and carboxy-terminal regions of the clade A V2 loop are more exposed on the heterotrimers than on the homotrimers.
The V1 loop was not immunogenic in the Q461 Env, in the context of both the heterotrimeric and the homotrimeric configurations. In contrast, weak antibody responses to the V1 loop were observed in the cases of the Q168 and Q259 Envs. In these two cases, significantly higher antibody titers were present in the sera from animals immunized with the mixture of homotrimers than in sera from animals immunized with the heterotrimers. Thus, the immunogenicity of the V1 loops of Q168 and Q259 is decreased by the presence of neighboring heterologous SF162 gp140 molecules. Potentially, the V1 regions of the clade A could be more exposed on the homotrimers than on the heterotrimers.
In summary, the presence of heterologous clade A/B gp140s within single heterotrimeric molecules differentially alters the immunogenic properties of the V1, V2, and V3 loops. The effect is dependent on the backbone of the clade A Env. We note that the immunogenicities of the V1, V2, and V3 regions were determined by assessing the reactivities of serum antibodies to linear peptides but that antibodies to conformational V1, V2, or V3 epitopes may be present but are not detectable by the reagents used.
Our analysis of the antigenic structures of these heterotrimers and the corresponding homotrimers indicates that the association of gp140 protomers of distinct amino acid sequence and glycosylation patterns results in alterations in the exposure of epitopes located in the CD4-BS, the coreceptor binding site, and the gp41 subunit. Such global changes are of an extent that does not abrogate the association of the gp140 protomers into stable trimeric conformations.
Despite differences in the relative exposures of specific conserved neutralization epitopes on gp120, such as that recognized by MAb b12 (on the Q168/SF162 and Q461/SF162 heterotrimers), the sera from the animals that were immunized with the heterotrimers did not display greater breadth of neutralization than did the sera from animals immunized with the homotrimers. Although we did not specifically examine the presence of b12-like antibodies in those sera, we assume that such antibodies were not elicited or that they were of very low titers that did not affect the overall neutralizing activities of the sera. Most likely, the heterologous neutralizing activities in the homotrimeric and heterotrimeric sera were due to antibodies that recognize less conserved epitopes, such as those present in the V1, V2, and V3 regions. The greater potency of neutralization displayed by the heterotrimeric sera could be due to these types of antibodies.
The effect of heterotrimerization on the immunogenic properties of Env gp140 was also evident in the case of the MPER. Specifically, higher titers of anti-2F5 and anti-4E10 epitope antibodies were elicited by immunization with Q461/SF162 heterotrimers than by immunization with the corresponding mixture of homotrimers. Here, too, therefore, changes in the immunogenicity of MPER resulting from the formation of homotrimers were clade A Env dependent, since similar observations were not made with the Q168 or Q259 clade A Env.
In summary, our study reveals that clade A- and B-derived envelope gp140 protomers, which differ by up to 24% in amino acid sequence, can form stable heterotrimeric proteins that can be purified and characterized. The antigenic structures of these heterotrimeric proteins differ from those of the corresponding homotrimeric gp140 proteins, and when used as immunogens, the heterotrimeric gp140s elicit more potent anti-HIV neutralizing antibodies than do the homotrimeric gp140 proteins. Whether this finding is biologically relevant should be determined in virus-challenge studies in a relevant animal model.
ACKNOWLEDGMENTS
This study was supported by grant 38660 from the Bill and Melinda Gates Foundation, as part of the CAVD program.
We are indebted to all those who contributed reagents. We thank Noah Sather for many helpful suggestions and comments.
Footnotes
Published ahead of print 26 October 2011
REFERENCES
- 1. Baba TW, et al. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200–206 [DOI] [PubMed] [Google Scholar]
- 2. Backliwal G, Hildinger M, Hasija V, Wurm FM. 2008. High-density transfection with HEK-293 cells allows doubling of transient titers and removes need for a priori DNA complex formation with PEI. Biotechnol. Bioeng 99:721–727 [DOI] [PubMed] [Google Scholar]
- 3. Barnett SW, et al. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beddows S, et al. 2007. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology 360:329–340 [DOI] [PubMed] [Google Scholar]
- 5. Beddows S, et al. 2005. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 79:8812–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berman PW, et al. 1992. Neutralization of multiple laboratory and clinical isolates of human immunodeficiency virus type 1 (HIV-1) by antisera raised against gp120 from the MN isolate of HIV-1. J. Virol. 66:4464–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blish CA, Nedellec R, Mandaliya K, Mosier DE, Overbaugh J. 2007. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS 21:693–702 [DOI] [PubMed] [Google Scholar]
- 8. Broder CC, et al. 1994. Antigenic implications of human immunodeficiency virus type 1 envelope quaternary structure: oligomer-specific and -sensitive monoclonal antibodies. Proc. Natl. Acad. Sci. U. S. A. 91:11699–11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burton DR, et al. 2011. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc. Natl. Acad. Sci. U. S. A. 108:11181–11186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chakrabarti BK, et al. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 76:5357–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chakraborty K, et al. 2006. Design of immunogens that present the crown of the HIV-1 V3 loop in a conformation competent to generate 447-52D-like antibodies. Biochem. J. 399:483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen L, et al. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng-Mayer C, Liu R, Landau NR, Stamatatos L. 1997. Macrophage tropism of HIV-1 and utilization of the CC-CKR5 coreceptor. J. Virol. 71:1657–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng-Mayer C, Quiroga M, Tung JW, Dina D, Levy JA. 1990. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J. Virol. 64:4390–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ching L, Stamatatos L. 2010. Alterations in the immunogenic properties of soluble trimeric human immunodeficiency virus type 1 envelope proteins induced by deletion or heterologous substitutions of the V1 loop. J. Virol. 84:9932–9946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cho MW, et al. 2001. Polyvalent envelope glycoprotein vaccine elicits a broader neutralizing antibody response but is unable to provide sterilizing protection against heterologous simian/human immunodeficiency virus infection in pigtailed macaques. J. Virol. 75:2224–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cole KS, Steckbeck JD, Rowles JL, Desrosiers RC, Montelaro RC. 2004. Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J. Virol. 78:1525–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Correia BE, et al. 2011. Computational protein design using flexible backbone remodeling and resurfacing: case studies in structure-based antigen design. J. Mol. Biol. 405:284–297 [DOI] [PubMed] [Google Scholar]
- 19. Correia BE, et al. 2010. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure 18:1116–1126 [DOI] [PubMed] [Google Scholar]
- 20. Derby NR, et al. 2006. Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J. Virol. 80:8745–8762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dey B, et al. 2007. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J. Virol. 81:5579–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doms RW, Earl PL, Chakrabarti S, Moss B. 1990. Human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus env proteins possess a functionally conserved assembly domain. J. Virol. 64:3537–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doria-Rose NA, et al. 2005. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J. Virol. 79:11214–11224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Du SX, et al. 2011. A directed molecular evolution approach to improved immunogenicity of the HIV-1 envelope glycoprotein. PLoS One 6:e20927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Earl P, Moss LB, Doms RW. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 65:2047–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Earl PL, et al. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68:3015–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Earl PL, Doms RW, Moss B. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 87:648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Earl PL, Koenig S, Moss B. 1991. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J. Virol. 65:31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Earl PL, Moss B. 1993. Mutational analysis of the assembly domain of the HIV-1 envelope glycoprotein. AIDS Res. Hum. Retroviruses 9:589–594 [DOI] [PubMed] [Google Scholar]
- 30. Flynn NM, et al. 2005. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191:654–665 [DOI] [PubMed] [Google Scholar]
- 31. Gao F, et al. 2005. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J. Virol. 79:1154–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garrity RR, et al. 1997. Refocusing neutralizing antibody response by targeted dampening of an immunodominant epitope. J. Immunol. 159:279–289 [PubMed] [Google Scholar]
- 33. Gorse GJ, et al. 1999. HIV-1MN recombinant glycoprotein 160 vaccine-induced cellular and humoral immunity boosted by HIV-1MN recombinant glycoprotein 120 vaccine. National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. AIDS Res. Hum. Retroviruses 15:115–132 [DOI] [PubMed] [Google Scholar]
- 34. Grundner C, et al. 2005. Analysis of the neutralizing antibody response elicited in rabbits by repeated inoculation with trimeric HIV-1 envelope glycoproteins. Virology 331:33–46 [DOI] [PubMed] [Google Scholar]
- 35. Gzyl J, et al. 2004. Effect of partial and complete variable loop deletions of the human immunodeficiency virus type 1 envelope glycoprotein on the breadth of gp160-specific immune responses. Virology 318:493–506 [DOI] [PubMed] [Google Scholar]
- 36. Haigwood NL, et al. 1992. Native but not denatured recombinant human immunodeficiency virus type 1 gp120 generates broad-spectrum neutralizing antibodies in baboons. J. Virol. 66:172–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hessell AJ, et al. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 15:951–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hessell AJ, et al. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ho J, MacDonald KS, Barber BH. 2002. Construction of recombinant targeting immunogens incorporating an HIV-1 neutralizing epitope into sites of differing conformational constraint. Vaccine 20:1169–1180 [DOI] [PubMed] [Google Scholar]
- 40. Hoxie JA. 2010. Toward an antibody-based HIV-1 vaccine. Annu. Rev. Med. 61:135–152 [DOI] [PubMed] [Google Scholar]
- 41. Hu SL, Stamatatos L. 2007. Prospects of HIV Env modification as an approach to HIV vaccine design. Curr. HIV Res. 5:507–513 [DOI] [PubMed] [Google Scholar]
- 42. Jeffs SA, Shotton C, Balfe P, McKeating JA. 2002. Truncated gp120 envelope glycoprotein of human immunodeficiency virus 1 elicits a broadly reactive neutralizing immune response. J. Gen. Virol. 83:2723–2732 [DOI] [PubMed] [Google Scholar]
- 43. Keefer MC, et al. 1996. Safety and immunogenicity of Env 2-3, a human immunodeficiency virus type 1 candidate vaccine, in combination with a novel adjuvant, MTP-PE/MF59. NIAID AIDS Vaccine Evaluation Group. AIDS Res. Hum. Retroviruses 12:683–693 [DOI] [PubMed] [Google Scholar]
- 44. Kim YB, Han DP, Cao C, Cho MW. 2003. Immunogenicity and ability of variable loop-deleted human immunodeficiency virus type 1 envelope glycoproteins to elicit neutralizing antibodies. Virology 305:124–137 [DOI] [PubMed] [Google Scholar]
- 45. Kothe DL, et al. 2007. Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology 360:218–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kothe DL, et al. 2006. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology 352:438–449 [DOI] [PubMed] [Google Scholar]
- 47. Kraft Z, et al. 2008. Characterization of neutralizing antibody responses elicited by clade A envelope immunogens derived from early transmitted viruses. J. Virol. 82:5912–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Law M, Cardoso RM, Wilson IA, Burton DR. 2007. Antigenic and immunogenic study of membrane-proximal external region-grafted gp120 antigens by a DNA prime-protein boost immunization strategy. J. Virol. 81:4272–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li M, et al. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J. Virol. 80:11776–11790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Y, et al. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liang X, et al. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862–2872 [DOI] [PubMed] [Google Scholar]
- 52. Liao HX, et al. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353:268–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lorin C, et al. 2004. A single injection of recombinant measles virus vaccines expressing human immunodeficiency virus (HIV) type 1 clade B envelope glycoproteins induces neutralizing antibodies and cellular immune responses to HIV. J. Virol. 78:146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lu S, et al. 1998. Immunogenicity of DNA vaccines expressing human immunodeficiency virus type 1 envelope glycoprotein with and without deletions in the V1/V2 and V3 regions. AIDS Res. Hum. Retroviruses 14:151–155 [DOI] [PubMed] [Google Scholar]
- 55. Mascola JR, et al. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mascola JR, Montefiori DC. 2010. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 28:413–444 [DOI] [PubMed] [Google Scholar]
- 57. Mascola JR, et al. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340–348 [DOI] [PubMed] [Google Scholar]
- 58. Mascola JR, et al. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207–210 [DOI] [PubMed] [Google Scholar]
- 59. Nara PL, et al. 1988. Purified envelope glycoproteins from human immunodeficiency virus type I variants induce individual, type-specific neutralizing antibodies. J. Virol. 62:2622–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nishimura Y, et al. 2003. Transfer of neutralizing IgG to macaques 6 h but not 24 h after SHIV infection confers sterilizing protection: implications for HIV-1 vaccine development. Proc. Natl. Acad. Sci. U. S. A. 100:15131–15136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nkolola JP, et al. 2010. Breadth of neutralizing antibodies elicited by stable, homogeneous clade A and clade C HIV-1 gp140 envelope trimers in guinea pigs. J. Virol. 84:3270–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ofek G, et al. 2010. Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl. Acad. Sci. U. S. A. 107:17880–17887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ou W, Lu N, Yu SS, Silver J. 2006. Effect of epitope position on neutralization by anti-human immunodeficiency virus monoclonal antibody 2F5. J. Virol. 80:2539–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pantophlet R, Wilson IA, Burton DR. 2003. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J. Virol. 77:5889–5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Parren PW, et al. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Quinones-Kochs MI, Buonocore L, Rose JK. 2002. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J. Virol. 76:4199–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Richardson JTM, et al. 1996. Humoral response to oligomeric human immunodeficiency virus type 1 envelope protein. J. Virol. 70:753–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Salzwedel K, Berger EA. 2000. Cooperative subunit interactions within the oligomeric envelope glycoprotein of HIV-1: functional complementation of specific defects in gp120 and gp41. Proc. Natl. Acad. Sci. U. S. A. 97:12794–12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sather DN, et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Saunders CJ, et al. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 79:9069–9080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Seaman MS, et al. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Seaman MS, et al. 2007. Standardized assessment of NAb responses elicited in rhesus monkeys immunized with single- or multi-clade HIV-1 envelope immunogens. Virology 367:175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sellhorn G, Caldwell Z, Mineart C, Stamatatos L. 2009. Improving the expression of recombinant soluble HIV envelope glycoproteins using pseudo-stable transient transfection. Vaccine 28:430–436 [DOI] [PubMed] [Google Scholar]
- 74. Shibata R, et al. 1999. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat. Med. 5:204–210 [DOI] [PubMed] [Google Scholar]
- 75. Shioda T, Levy JA, Cheng-Mayer C. 1991. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature 349:167–169 [DOI] [PubMed] [Google Scholar]
- 76. Srivastava IK, et al. 2003. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J. Virol. 77:11244–11259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Srivastava IK, et al. 2002. Purification and characterization of oligomeric envelope glycoprotein from a primary r5 subtype B human immunodeficiency virus. J. Virol. 76:2835–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Srivastava IK, VanDorsten K, Vojtech L, Barnett SW, Stamatatos L. 2003. Changes in the immunogenic properties of soluble gp140 human immunodeficiency virus envelope constructs upon partial deletion of the second hypervariable region. J. Virol. 77:2310–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Stamatatos L, Lim M, Cheng-Mayer C. 2000. Generation and structural analysis of soluble oligomeric envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV-1 isolates. AIDS Res. Hum. Retroviruses 16:981–994 [DOI] [PubMed] [Google Scholar]
- 80. Stamatatos L, Wiskerchen M, Cheng-Mayer C. 1998. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV-1 isolate on viral envelope structure, cell-entry and replication. AIDS Res. Hum. Retroviruses 14:1129–1139 [DOI] [PubMed] [Google Scholar]
- 81. VanCott TC, et al. 1997. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J. Virol. 71:4319–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. VanCott TC, et al. 1999. Cross-subtype neutralizing antibodies induced in baboons by a subtype E gp120 immunogen based on an R5 primary human immunodeficiency virus type 1 envelope. J. Virol. 73:4640–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Walker LM, et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang S, et al. 2006. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology 350:34–47 [DOI] [PubMed] [Google Scholar]
- 85. Weiss CD, Levy JA, White JM. 1990. Oligomeric organization of gp120 on infectious human immunodeficiency virus type 1 particles. J. Virol. 64:5674–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yang X, Wyatt R, Sodroski J. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yang ZY, et al. 2004. Selective modification of variable loops alters tropism and enhances immunogenicity of human immunodeficiency virus type 1 envelope. J. Virol. 78:4029–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang PF, Jr, et al. 2007. Extensively cross-reactive anti-HIV-1 neutralizing antibodies induced by gp140 immunization. Proc. Natl. Acad. Sci. U. S. A. 104:10193–10198 [DOI] [PMC free article] [PubMed] [Google Scholar]