Abstract
Epstein-Barr virus (EBV) is a human herpesvirus which has been studied intensively for its role in certain human tumors. It also serves as a model of herpesviral latency because it establishes an immediate, latent infection in human B cells. When EBV infects quiescent, primary B cells it induces their continuous proliferation to yield growth-transformed B-cell lines in vitro. The lytic or productive phase of EBV's life cycle is induced by the expression of the viral BZLF1 gene in latently infected cells. The BZLF1 protein is a transactivator, which selectively binds to two classes of distinct DNA sequence motifs. One class is similar to the motifs that are bound by members of the AP-1 transcription factor family to which BZLF1 belongs. The second class, which contains CpG motifs, is predominant in viral promoters of early lytic genes and is BZLF1's preferred or exclusive target sequence when methylated. The BZLF1 gene is transiently expressed in newly infected B cells but fails to induce EBV's lytic cycle, potentially because the virion DNA is unmethylated. Here we report that the lack of 5-methylcytosine residues in CpG sites of virion DNA prevents the expression of essential lytic genes indispensable for viral DNA amplification during productive infection. This finding indicates that BZLF1 transactivates these promoters in a methylation-dependent fashion and explains how progeny virus synthesis is abrogated in newly infected B cells. Our data also reveal that viral lytic DNA synthesis precludes CpG methylation of virion DNA during EBV's lytic, productive cycle, which can be overcome by the ectopic expression of a prokaryotic cytosine methyltransferase to yield CpG-methylated virion DNA. Upon infection of B cells, randomly CpG-methylated virion DNA induces high expression of essential lytic genes in contrast to virion DNA free of 5-methylcytosine residues. Our data suggest that unmethylated virion DNA is part of EBV's strategy to prevent the viral lytic phase in newly infected B cells, allowing it to establish its characteristic latent infection in them.
INTRODUCTION
Upon infection, Epstein-Barr virus (EBV) delivers its linear genomic DNA of about 160 kps to human B cells. The viral genome is epigenetically naïve, i.e., free of histones and devoid of methylated CpG dinucleotides (14, 26, 29). When the linear viral DNA genome reaches the nucleus of these cells and forms a circular plasmid, it initiates a phase in the viral life cycle termed “prelatent.” This phase is characterized by the coexpression of two distinct sets of viral genes, which consist of the classical set of latent genes and a restricted number of the set of EBV's lytic genes. The expression of latent genes (Epstein-Barr nuclear antigens [EBNAs]), latent membrane proteins (LMPs), and viral noncoding RNAs and micro-RNAs activates the quiescent B lymphocytes, which become lymphoblasts and begin to proliferate. At this early stage, the concomitant expression of certain lytic genes, which encompass transcription factors and cytokines, protects the activated B lymphocytes from endogenous stress, immediate activation-induced apoptosis (1), and, presumably, DNA damage response signals (40). The prelatent phase is transient and ceases within 1 to 2 weeks postinfection (p.i.). No virus progeny is synthesized at this initial early stage, but nucleosomes become positioned onto the viral genome, histones acquire substantial epigenetic modifications over time, and the viral DNA becomes extensively methylated at CpGs (26).
The prelatent phase is replaced by a strictly latent phase, in which the virus establishes a stringent and stable virus-host relationship (25). Viral gene expression is entirely restricted to EBNAs, LMPs, and noncoding RNAs, the prevailing set of viral latent genes which support cellular proliferation and sustain lymphoblastoid cells lines (LCLs) in vitro. The EBNAs are now expressed from a different promoter than the prelatent phase, an important switch of viral gene regulation (1, 53). This switch reflects the gradual epigenetic modifications of viral DNA, which results in transcriptional repression and a high degree of CpG methylation of the viral genome (26). As a result, all lytic genes are efficiently repressed but latent viral genes are spared from epigenetic silencing. Much is known about their encoded gene products, which mimic central cellular functions and contribute to B-cell activation and proliferation (28).
Upon exogenous signals, such as antigen encounter or artificial cross-linking of the B-cell receptor (49, 51), cellular signaling pathways can activate a single viral gene, BZLF1. It is the molecular switch that can induce the lytic phase of viral gene expression in latently infected cells, leading to de novo virus synthesis (8, 50). BZLF1, a member of the family of cellular AP-1 transcription factors, binds viral and cellular promoters sequence specifically and induces their gene expression. Upon BZLF1's activation, a cascade of three classes of viral lytic genes begins, consisting of immediate-early (IE), early (E), and late (L) viral genes. BZLF1 and BRLF1 encode two viral factors of the immediate-early class; genes that directly or indirectly mediate lytic viral DNA amplification constitute the class of early genes; and genes encoding viral structural components form the late class of viral lytic genes. The sequential activation of the three classes of lytic genes is a hallmark of all herpesviruses. The lytically induced cells release viral progeny which contain viral DNA in its naïve state free of histones and with unmethylated CpGs (26).
Most herpesviruses initiate lytic infections on infecting cells. These productive infections are consistent with their genomes likely being free of methylated CpG dinucleotides and histones. This “naked” state of genomic DNA would be advantageous, since most herpesviruses exploit the transcription machinery of the cell to support their productive infections. CpG methylation and nucleosomal occupancy of virion DNA would interfere with the binding of cellular and viral transcription factors, a potential obstacle to immediate viral gene expression (35, 45). The peculiar epigenetically naïve state of herpesviral DNA is well-suited to promote de novo virus synthesis in most infected cells. It is therefore counterintuitive that EBV establishes only latent infections in infecting cells. EBV's genome is epigenetically naïve, but how then does EBV abrogate the onset of an initial lytic phase in newly infected cells?
The virion DNA of EBV is unmethylated but becomes heavily CpG methylated in latently infected cells over time (26). How EBV DNA acquires methylated cytosines is unclear, but once established, they are maintained in proliferating and latently infected B cells. In mammalian cells, semiconservative cellular DNA replication and DNA methylation are strictly coupled, and newly replicated daughter strands inherit the pattern of CpG methylation of parental DNA. The cellular DNA methyltransferase 1, DNMT1, which performs this task, and the critically involved factors, proliferating cell nuclear antigen (PCNA) and UHRF1 (also called Np95 or ICBP90), are known (6, 47). DNMT1, which is expressed during the early S phase of the cell cycle (30), and PCNA interact (7, 33, 52) and together with UHFR1 form complexes at sites where hemimethylated DNA is generated during DNA replication and converted into fully CpG-methylated DNA on both strands (2, 3, 21). Disrupting the interactions of DNMT1, PCNA, and UHRF1 promotes global DNA hypomethylation (23). Besides maintaining the pattern of DNA methylation, PCNA serves an additional critical function in the process of semiconservative DNA replication in eukaryotic cells. PCNA is a processivity factor of replicative DNA polymerases and is the sliding clamp on DNA, which ensures that the scanning polymerases are tethered to the DNA template during DNA synthesis.
Viral but not cellular factors mediate lytic herpesviral DNA replication (15, 20, 37). The function of PCNA in viral DNA replication is replaced by a structural homologue encoded by all herpesviruses, termed UL42 and BMRF1, in herpes simplex 1 and Epstein-Barr virus, respectively. PCNA seems to be excluded from lytic DNA replication of herpes simplex virus (see reference 36 and references therein), which is somewhat controversial in the case of EBV (9). We hypothesized that if herpesviral DNA replication is fully functional in the absence of PCNA, DNMT1 could not be recruited to the replication forks of the herpesviral DNA. As herpesviruses overreplicate, i.e., amplify their DNA several hundredfold during the lytic phase (37, 43), methylation marks at cytosine residues would be lost on both newly synthesized DNA strands, removing this epigenetic modification from EBV's genomic DNA, which had been acquired during latency.
This interesting hypothesis needs to be experimentally addressed. We have therefore asked “What happens when EBV virions contain genomic DNA with methylated CpGs? Would the infected B cell immediately support the lytic phase of EBV's life cycle, corroborating our hypothesis that EBV's latent program depends on its DNA first being unmethylated?”
MATERIALS AND METHODS
Cells.
HEK293, B95.8, and Raji cells have been described previously (18, 34, 44). The EBV-negative Daudi and Akata cells were a friendly gift from Kenzo Takada (38). The HEK293-based cell line for the production of recombinant wild-type 2089 EBV stocks has been described previously (11). Standard cell culture medium was high-glutamine RPMI 1640 (Gibco) supplemented with 10% fetal calf serum (PAA), 1% penicillin-streptomycin (Gibco), 1% sodium-pyruvate (Invitrogen), and 100 nM sodium selenite (ICN) at 37°C and 5% CO2. For continuous culture of EBV producer cell lines, 100 μg/ml hygromycin (PAA) was added to the culture medium.
Plasmids.
The wild-type maxi-EBV plasmid p2089 encompassing the complete EBV genome, the BZLF1 expression plasmid p509, and the BALF4 expression plasmid p2670 are described elsewhere (11, 19, 39). The reconstituted wild-type EBV 2089 encompasses the complete genome of the EBV prototype B95.8 strain (11). The coding region of the M.SssI de novo methyltransferase was cloned into the plasmid pRK5 and is constitutively expressed from the cytomegalovirus (CMV) promoter (p3663) as described previously (26). The tetracycline-inducible BZLF1 expression plasmid iBZLF1 (p3862) is based on the pRTS-2 plasmid (5), which contains tetO binding sites with two minimal CMV promoters oriented in opposite directions to support the simultaneous expression of the EBV gene BZLF1 and the reporter gene egfp upon addition of doxycycline (DOX). For selection in eukaryotic cells, hygromycin was added to the medium.
Stable transfection and establishment of cell lines.
Daudi and Akata cells (5 × 106) were electroporated with a capacity of 975 μF and 5 μg p3862 (iBZLF1) DNA in 250 μl RPMI1640 and 4-mm cuvettes at 190 V to 200 V and 185 V, respectively, using a Bio-Rad electroporation apparatus. Immediately after electroporation, cells were resuspended in 500 μl FCS and 5 ml cell culture medium was added. Cells were incubated in 6-well cluster plates at 37°C and 5% CO2 overnight. Afterwards, cells were plated into 96-well cluster plates in 200 μl cell culture medium supplemented with hygromycin (250 μg/ml and 200 μg/ml for Daudi and Akata cells, respectively). Within the next 4 to 5 weeks, resistant cells grew out, which were successively expanded into 48- and 24-well cluster plates, following cultivation in T-flasks under hygromycin selection.
Conditional expression of BZLF1 by fluorescence-activated cell sorter (FACS) and immunofluorescence analysis.
The conditional BZLF1 expression in iBZLF1-transfected Daudi and Akata cell lines was monitored by flow cytometry after addition of 1 μg/ml DOX for 24 h. The mean green fluorescence intensity of green fluorescent protein (GFP) in the viable cell population (defined by forward and side scatter) served as an indicator of the concomitant induction of BZLF1 expression. It was verified by staining the cells with a mouse monoclonal antibody specific for BZLF1 (24). For details, see results from Kalla et al. (26). B95.8 and EBV-negative Daudi and Akata cell lines served as positive and negative controls, respectively.
DNA transfection and generation of infectious viral particles.
For the production of recombinant EBV virus stocks, the lytic cycle was induced in HEK293 EBV producer cells carrying the maxi-EBV plasmid p2089 by transient transfection of an expression plasmid carrying BZLF1. DNA transfection was performed using polyethylenimine (PEI; Sigma-Aldrich). The day before transfection, 1 × 106 cells per well were seeded into 6-well cluster plates. During the preparation of the transfection mixture, cells were switched to Opti-MEM minimal medium (Invitrogen). For generation of wild-type 2089 EBV, 0.5 μg p509 carrying BZLF1 and 0.5 μg p2670 DNA carrying BALF4 were used per well of a 6-well cluster plate. For in vivo methylation of wild-type 2089 EBV (meEBV), 4 μg p3663 carrying the M.SssI gene was transiently cotransfected. The DNAs were mixed with 1 ml Opti-MEM and combined with 1 ml Opti-MEM containing 12 μl PEI dissolved in water (50%/50%, vol/vol). The mixture was incubated for 15 min at room temperature and added to the cells for 4 to 5 h. Virus-containing supernatants were harvested 3 days later and purified by pelleting debris of cells (300 × g, 5 min), followed by an additional centrifugation step (2,500 × g, 15 min). EBV virus stocks were stored at 4°C prior to use.
Quantification of EBV.
To adjust the virus stocks, quantitative PCR assays measured virion DNA after infection of EBV-negative B cells. The two EBV stocks were wild-type 2089 EBV (wtEBV) and 2089 EBV (meEBV). GFP expression in infected Raji cells was used as a surrogate marker to evaluate the concentration of infectious virus as described previously (12). In brief, 5 × 104 Raji cells per well of a 24-well cluster plate were infected with different amounts of virus stocks. After 3 days, the mean green fluorescence intensity of the viable cell population (defined by forward and side scatter) was quantified by flow cytometry to determine the concentration of infectious virus particles. GFP-positive Raji cells allow direct assessment of the concentration of infectious EBVs as “green Raji units” (GRUs), which were in the range of 0.5 to 10 × 104 GRU/ml for wtEBV stocks. meEBV stocks were produced in HEK293 cells expressing the de novo DNA methyltransferase M.SssI gene. Consistently, meEBV virus stocks had an apparently lower GRU concentration due to extensive CpG methylation of the CMV promoter driving expression of the egfp gene in 2089 wild-type EBV. To correct for this phenomenon, EBV-negative Daudi cells were infected with different amounts of titrated wtEBV and meEBV virus stocks for 3 days. Cells were washed extensively and genomic DNA was extracted and analyzed by quantitative PCR to assess the absolute molecular number of EBV DNA molecules. PCR primer pairs for the amplification of EBV DNA and housekeeping genes, which served as internal controls, are listed in Table 2. PCRs were performed as described in “Quantitative reverse transcription PCR” below.
Table 1.
PCR primers used for gene expression profiling
| Gene class | Gene | Function | Primer pair | Product size (nt) | Forward primer sequence (5′–3′) | Backward primer sequence (5′–3′) |
|---|---|---|---|---|---|---|
| IE | BZLF1 | Transcription factor | BZLF1/intr.spec./F1R1 | 104 | GCACATCTGCTTCAACAGGA | CCAAACATAAATGCCCCATC |
| IE | BRLF1 | Transcription factor | BRLF1/F2R2 | 100 | CCTGTCTTGGACGAGACCAT | AAGGCCTCCTAAGCTCCAAG |
| IE | BMLF1 | Mta | BMLF1/F1B1 | 212 | GGAACGGGTCTTGGATGGG | AAGCAACAGAGCAACCAGAGGTC |
| E | BBLF2 | Primase associated factor | BBLF2/F8B9 | 202 | GTCGGGAGTCTCGGTGGAATAG | AGCACAGGTGGTCTGCCAAAG |
| E | BBLF4 | DNA helicase | BBLF4/F6B3 | 170 | AAGCCTGCCTCATCCTTGACC | GACGAGCCTCTCCTTCACGG |
| E | BGLF5 | Alkaline exonuclease | BGLF5/nospan/F1R1 | 75 | TTCGGCCGCTATTAGCTTAG | GACGGGGGAATAATCAACCT |
| E | BMRF1 | DNA binding protein | BMRF1/nospan/F1R1 | 116 | CGTGCCAATCTTGAGGTTTT | CGGAGGCGTGGTTAAATAAA |
| E | BNFL2a | Immune evasion | BNLF2a/F1R1 | 98 | GGGAAAATTGTCACATTGGT | TGCTTTGCTAGAGCAGCAGT |
| E | BSLF1 | Primase | BSLF1/F3B4 | 199 | GCGGGTCCTCTGGATTAGATAGTC | CAGGGGCGGTGGTCTTAGC |
| L | BcLF1 | Major capsid protein | BcLF1/F10B10 | 202 | CCTCCCTGACCGTTCCCAG | GCAGTTTGAGACCGCCACATC |
| L | BDLF1 | Minor Capsid protein | BDLF1/F4B1 | 201 | GCACCTCCTCTGCTATGGGC | TGATACTCACCAAGATTGTTCCAGG |
| L | BLLF1 | Glycoprotein gp350/220 | Gp220/F23B25 | 164 | GTCACCTGTGTTATATTTTCACCACTTTC | CCTACCAACCTCACCGCACC |
| L | BLLF1 | Glycoprotein gp350/220 | Gp350/F69B81 | 228 | TGGCGAGTTTGCGTCCTCAG | CGTCCAGTGTCACGATTTCTTGG |
| L | BVRF2 | Capsid protein, protease | BVRF2/F2B4 | 170 | TTAGTGACCTGGAACCGTCTATCG | CCTGTCCTGCCTCAGAGTCTCC |
| HKG | CYC | Cytochrome c | ICYC/F1B1 | 264/178 | CCTGGTGGGCGTGTGCTAC | CAATGCTCCGTTGTTGGCAG |
| HKG | GUSB | β-glucuronidase | IGUSB/F29B35 | 196 | GTGCTGGGGAATAAAAAGGGG | ACTCGGGGAGGAAGGGACAC |
Quantification of CpG methylation of virion EBV DNA.
Southern blot hybridizations were performed to analyze the degree of CpG methylation of virion DNA in meEBV virus stocks, which were obtained by lytic induction of the HEK293 2089 producer cells and transient expression of the M.SssI gene. Viral particles were pelleted by centrifugation for 3 h at 19,000 rpm and 4°C in a TFA 20.250 rotor (Kontron) and resuspended in Tris-EDTA (TE) buffer. Sodium deoxycholate was added to a final concentration of 0.5%, and the suspension was incubated at 37°C with constant agitation for 1 h to dissolve viral envelopes, other microvesicles, or exomomes. DNase (100 μg/ml) and RNase (120 μg/ml) were added to the preparation in the presence of 1 mM MgCl2 and incubated at 37°C for 1 h to eliminate free RNA and DNA molecules. The free viral capsids were collected through a 36% (wt/vol) sucrose cushion in a Beckman SW32 rotor at 4°C at 28,000 rpm for 90 min. The capsids were resuspended in 600 μl TEN buffer, SDS was added to a final concentration of 1% together with 100 μg/ml RNase A, and the suspension was incubated at 37°C for 45 min. Proteinase K was added (500 μg/ml) and incubated at 50°C for 5 h. An additional RNase A digestion step followed at 37°C for 30 min. Nucleic acids were extracted with phenol-chloroform and precipitated with ethanol to recover DNA, which was dissolved in 25 μl TE buffer and cleaved with KpnI and the methylation-sensitive restriction enzyme AgeI as described previously (26). As controls, appropriate amounts of E. coli-derived, unmethylated, or in vitro-methylated p2089 EBV DNA were equally digested. The samples were separated by electrophoresis, transferred to charged nylon membranes by Southern blotting, and hybridized to a radioactive probe covering the BamW repeats as described in detail previously (26). After Southern blot hybridizations, the membranes were exposed to X-ray film.
A PCR approach was used to quantify the extent of CpG methylation in virion EBV DNA. Virions were generated in HEK293 2089 producer cells as described previously (39). Briefly, the cells were transfected with different amounts of the M.SssI gene expression plasmid p3663, together with the expression plasmids p509 and p2670, in order to induce the lytic phase in the cells and increase virus yield, respectively. EBV-negative Daudi cells were infected with defined amounts of wtEBV and meEBV virus stocks. Three days postinfection (p.i.), total cellular DNA was isolated and digested with the methylation-sensitive restriction enzyme HpaII. DNA is protected from HpaII cleavage if its recognition motif (5′-CCGG-3′) is CpG methylated. Subsequently, qPCR amplifications were performed with a primer pair specific for the BKRF4 gene locus. The DNA region of this PCR mixture contains a single HpaII cleavage site. As internal controls, additional qPCR primer pairs amplified parts of the viral BNLF1 locus and the cellular GUSB gene, which are both devoid of HpaII recognition sites. The qPCRs of the different samples were normalized with the GUSB qPCR product. For the list of primers, see Table 2.
Table 2.
PCR primers used for detection of DNA
| Gene class | Gene | Function | Name of primer pair | Product size (nt) | Forward primer sequence (5′-3′) | Backward primer sequence (5′-3′) |
|---|---|---|---|---|---|---|
| E | BMRF1 | DNA binding protein | EA-D/F2B2 | 233 | GCCGCCGTGTCATTTAGAAAC | TCGTTCCTCAGCCTGTCGC |
| E | BKRF4 | Virion protein | BKRF4/F3B9 | 220 | ACGAGGAGATTGATTTGGAGGAAG | CGGGAGAACGAAGGCGTG |
| L | BNLF1 | Tumor necrosis factor receptor | LMP1/F4B4 | 220 | GTGTCTGCCCTCGTTGGAGTTAG | CATCCTGCTCATTATTGCTCTCTATCTAC |
| HKG | GUSB | β-glucuronidase | GUSB/F1B1 | 105 | CACGACTACGGGCACCTGG | TGCTCCATACTCGCTCTGAATAATG |
Quantitative reverse transcription PCR.
Total RNA was extracted (RNeasy mini kit; Qiagen) according to the manufacturer's protocol. RNA was treated with DNase I (Invitrogen) at 37°C for 90 min to remove DNA, followed by inactivation of DNase for 10 min at 65°C. In order to monitor contaminating DNA in the RNA preparations, control PCRs were performed at this step. Reverse transcription of mRNA was performed at 55°C with the SuperScript III first-strand synthesis kit (Invitrogen) according to the manufacturer's protocol. One microliter of the cDNA product (final volume, 20 μl) was used as template for quantitative PCR amplification of selected EBV genes. Real-time PCR analysis was performed with a LightCycler instrument (Roche) according to the manufacturer's instructions. Quantifications of reverse-transcribed transcripts were monitored with the LightCycler FastStart reaction mix (SYBR green I; Roche). The amplification of PCR products was terminated after 40 cycles. The settings used were initial template denaturation at 95°C for 10 min. The cycles were 1 s at 95°C, 10 s at 62°C, and 10 s at 72°C. PCR primer pairs are listed in Table 1.
In vitro methylation of genomic EBV DNA.
p2089 EBV DNA was purified from Escherichia coli and methylated in vitro with the de novo methyltransferase M.SssI (New England BioLabs) as described previously (26). All CpG dinucleotides in p2089 DNA could be fully methylated with the recommended ratio of S-adenosylmethionine (SAM) and DNA according to the manufacturer's protocol. The extent of CpG methylation was controlled by limiting the methyl group donor SAM, which was monitored with the methyl-sensitive restriction enzymes AgeI or HpaII (NEB), followed by conventional agarose gel electrophoresis and ethidium bromide staining.
RESULTS
Generation and quantification of virions with unmethylated and CpG-methylated viral DNA.
We developed an inducible system to produce recombinant EBV in HEK293 cells, which are stably transfected with recombinant maxi-EBV DNA (11). To reconstitute infectious virus, clonal producer cell lines, which carry maxi-EBV DNAs, are transiently transfected with two expression plasmids carrying the lytic switch gene BZLF1 to induce EBV's lytic cycle and the viral glycoprotein BALF4 to boost virus synthesis (19, 39). The p2089 maxi-EBV DNA encompasses the complete wild-type genome of the EBV prototype strain B95.8 (11). Upon lytic induction of its producer cell line, 2089 EBV virus stocks, termed wtEBV in the manuscript, can be obtained with concentrations of up to 105 infectious units per ml. The recombinant EBVs are engineered to carry the egfp gene, and infectious units are defined with the aid of Raji cells (human Burkitt's lymphoma B-cell line), which turn GFP positive upon infection and allow the direct assessment of the concentration of infectious EBV virions as green Raji units (GRUs) per milliliter by FACS analysis (12). Like virions spontaneously released from the marmoset cell line B95.8, EBV virions generated in HEK293 producer cell lines contain viral genomic DNA free of methylated CpGs (14, 26, 29).
We wished to generate EBV virions that contain CpG-methylated genomic DNA to analyze and compare their phenotypes with “normal” wtEBV virions upon infection of human B cells. One of our sources of EBV DNA is recombinant maxi-EBV DNA propagated as plasmids in E. coli and therefore free of methylated CpGs. Previously, we made use of a prokaryotic de novo methyltransferase, M.SssI (41), to generate CpG-methylated genomic EBV DNA in vitro (26). Supernatants from HEK293 cells transiently cotransfected with CpG-methylated p2089 EBV DNA and BZLF1 contained infectious EBV, but supernatants from cells transfected with unmethylated p2089 EBV DNA did not (26). This finding indicated that methylation of viral DNA is a prerequisite for virus synthesis, which, however, yields unmethylated virion DNA exclusively (26).
It appears counterintuitive that methylated EBV DNA is required to support the lytic phase of EBV's life cycle because extensive DNA methylation of viral DNA is thought to contribute to gene silencing (14). However, BZLF1 transactivates certain early genes of EBV only when their promoters contain CpG-methylated BZLF1 binding sites (4). A number of early viral genes regulated in this manner code for viral factors and enzymes indispensable for lytic DNA replication of EBV (16). Therefore, methylation of viral DNA is a prerequisite for virus de novo synthesis.
We built on our previous findings and used the 2089 EBV producer cell line HEK293 (11), which was transiently cotransfected with expression plasmids carrying BZLF1 (p509), BALF4 (p2670), and the gene for M.SssI (p3663). We anticipated that the prokaryotic methyltransferase M.SssI would add methyl groups to cytosines in CpG dinucleotides of newly synthesized viral DNA during EBV's lytic phase. CpG-methylated EBV DNA might then become packaged and released as infectious virions.
Cell-free supernatants were harvested 3 days posttransfection, and the virus stocks were termed meEBV (Fig. 1). To our surprise, meEBV supernatants generated with this set of plasmids, including M.SssI, did not yield GFP-positive Raji cells in contrast to wtEBV virus stocks (Fig. 2). We hypothesized that coexpression of the M.SssI gene might interfere with virus production or yield virion DNA with a transcriptionally repressed egfp gene, which is controlled by the immediate-early promoter and enhancer of human cytomegalovirus. This cis-acting element encompasses multiple CpG dinucleotides and is prone to epigenetic silencing. We wished to address this problem and infected EBV-negative Daudi cells, a human Burkitt's lymphoma cell line (38), with discrete volumes of wtEBV or meEBV stocks. Prior to infection, the wtEBV stocks were quantified and their concentrations (GRUs/ml) were known (12). Daudi cells, which had been infected with titrated wtEBV or untitrated meEBV stocks, were washed intensively, and total genomic DNA was prepared 3 days p.i. We designed suitable primer pairs (Table 2) to detect EBV DNA and the housekeeping gene for β-glucuronidase (GUSB) as a reference for cellular DNA (10). Quantitative PCR analysis determined the absolute number of EBV DNA molecules per cellular genome equivalent in infected, EBV-negative Daudi B cells. Technical details of this quantification can be found in “Quantification of EBV” in Materials and Methods.
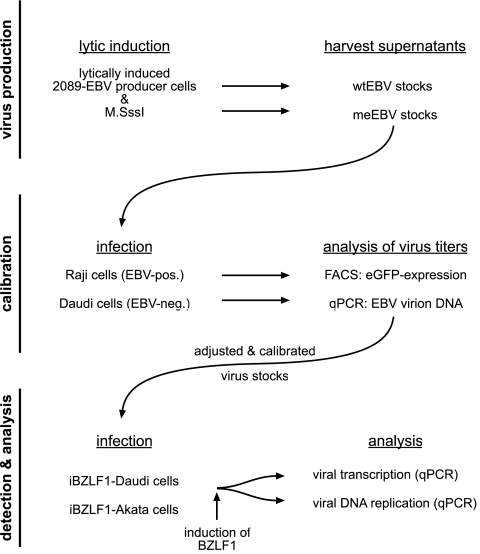
Fig 1.
Experimental flow chart. (Top) Production of wtEBV and meEBV virus stocks. HEK293 2089 producer cells (11) were transiently transfected with expression plasmids p509 and p2670 to induce EBV's lytic phase and increase virus yield, respectively (19, 39). Supernatants of meEBV stocks were generated by cotransfection of an additional expression plasmid carrying the methyltransferase M.SssI gene (26). After 3 days, the supernatants were harvested as virus stocks. (Middle) Titers of the virus stocks were determined by infecting Raji and Daudi cells. Three days p.i., the virus concentrations of wtEBV stocks were determined by assessing the number of GFP-positive Raji cells (GRU) per milliliters of virus stock as described elsewhere (Fig. 2) (1, 12). To calculate the molecular concentration of genomic EBV DNA in wtEBV and meEBV virus stocks, EBV-negative Daudi cells were infected with wtEBV at an MOI of 0.05 and equal volumes of meEBV stocks for 3 days. Total genomic DNA was prepared, and quantitative PCR analysis was used to assess the absolute number of virion DNA molecules. The analysis provided the coefficient to calculate and equilibrate meEBV virus stocks to obtain an equivalent concentration of infectious virus as wtEBV stocks. (Bottom) Daudi and Akata cells stably transfected with the conditional BZLF1 expression plasmid iBZLF1 were infected with the adjusted and calibrated virus stocks. Two days p.i., the cells were washed twice and DOX was added to the medium to induce expression of BZLF1 for two additional days. Genomic DNA and total RNA were extracted and analyzed by quantitative PCRs.
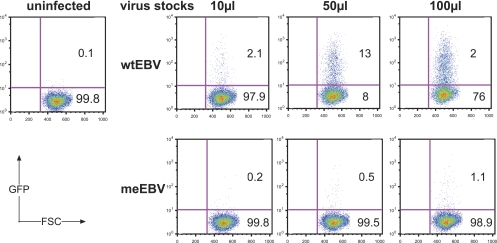
Fig 2.
Calibration of virus supernatants. Raji cells were infected with different volumes of wtEBV and meEBV stocks as indicated at the top of the panels. Recombinant EBV stocks carry the egfp gene (11). After 3 days, the percentage of GFP-positive Raji cells was determined by FACS analysis, followed by calculation of the virus titer as described previously (12). wtEBV stocks contained substantial concentrations of infectious units, which conferred GFP expression in wtEBV-infected Raji cells, but only a small fraction of cells infected with meEBV stocks became GFP positive. Quantification of meEBV virion DNA by PCR analysis indicated that meEBV stocks contained infectious virions at concentrations similar to those of wtEBV, suggesting that CpG methylation of the cytomegalovirus immediate-early promoter and enhancer driving the egfp gene interfered with its expression.
The analysis indicated that meEBV virus stocks contained considerable concentrations of virions, which were about a factor of two to four lower than wtEBV stocks (data not shown). This approach also revealed the number of infectious GRUs versus virion DNA molecules in wtEBV preparations, which was about 0.0025 because, on average, e.g., 104 GRUs contained about a total of 4 × 106 molecules of genomic EBV DNA. With this information at hand we could deduce the concentration of infectious virus also in meEBV stocks. Assuming that this correlation is valid, we adjusted the concentrations of our different virus stocks to use equal amounts of wtEBV and meEBV in the following experiments.
Quantification of methylated CpGs in virion DNAs of meEBV stocks.
We prepared large virus stocks of both wtEBV and meEBV as described in Materials and Methods in detail. Virions were purified by sequential rounds of sedimentation. Deoxycholate, an ionic detergent, was used to remove the virion envelopes and dissolve membranous vesicles, such as exosomes, that might contain EBV DNA, leaving capsids with virion DNA intact. Free nucleic acids were digested with DNase and RNase, and the virion capsids were again pelleted through a sucrose cushion by ultracentrifugation. After resuspension, addition of SDS, and digestion with proteinase K, virion DNA was extracted with phenol and recovered by ethanol precipitation.
Virion DNAs were digested with the restriction enzymes KpnI and AgeI. The latter does not cleave DNA if the CpG dinucleotide, which is in the center of its palindromic recognition sequence (Fig. 3A), carries a methyl group in the 5′ position of the pyrimidine ring of cytosine. The result of this analysis is shown in Fig. 3A and indicated that a substantial fraction of virion DNA molecules in meEBV stocks were CpG-methylated and, therefore, not cleaved by AgeI.
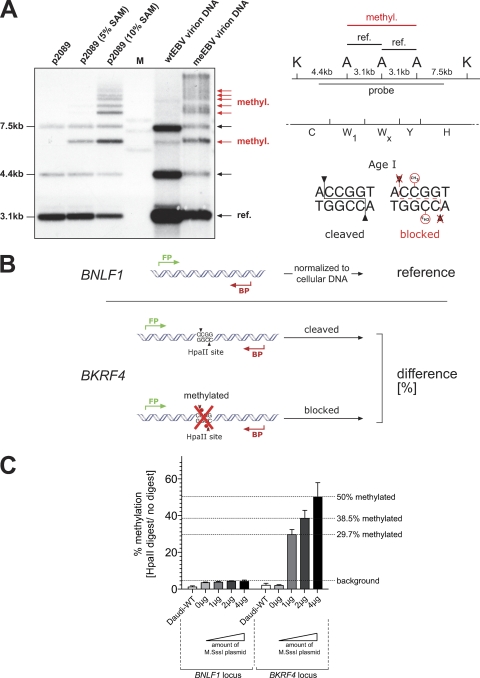
Fig 3.
CpG methylation of wtEBV and meEBV virion DNA. (A) In the left panel, an autoradiogram shows p2089 EBV DNA isolated from E. coli and EBV virion DNA after Southern blot hybridization with a radioactive probe. The DNAs were digested with two restriction enzymes, KpnI and AgeI. AgeI is blocked by CpG methylation as indicated in the right panel. E. coli-isolated p2089 DNA is free of methylated cytosine residues and cleaved by both enzymes to completion. After partial in vitro methylation of this DNA with limiting calculated amounts (5% and 10%) of the methyl group donor S-adenosyl methionine (SAM), the radioactive probe detected novel and slowly migrating DNA fragments (red arrows), which originated from partially cleavage-protected AgeI sites schematically depicted in the right panel. Virion DNA prepared from wtEBV stocks was cleaved by both restriction enzymes to near completion, revealing a lack of methylated CpG dinucleotides. In contrast, virion DNA prepared from meEBV stocks was mostly protected from AgeI cleavage, indicating the prevalence of methylated CpGs. (B) Overview of the experimental setup to determine the degree of CpG methylation in wtEBV and meEBV virion DNA. Daudi cells were infected, and 2 days p.i. genomic DNA was isolated and digested with the methylation-sensitive restriction enzyme HpaII, followed by quantitative PCR analysis with two different primer pairs specific for the viral BNLF1 and BKRF4 loci. The BKRF4 locus-specific PCR product spans a single HpaII site, which is protected from cleavage if the central CpG dinucleotide of the palindromic HpaII recognition sequence is methylated. Quantitative PCR analysis assessed the percentage of cleavage-protected virion DNA as a measure of CpG methylation. For normalization, quantitative PCRs of unrestricted genomic DNA of the housekeeping gene gusB served as an internal quantitative measure of cellular genome equivalents. The viral BNLF1 locus free of HpaII sites served as an internal reference of viral DNA molecules. (C) Transient transfections of different amounts of the M.SssI gene encoding expression plasmid into HEK293 2089 producer cells correlated with virion DNAs that were partially but variously CpG methylated.
We wanted to quantify the extent of CpG methylation of virion DNA and designed a suitable PCR-based approach, shown in Fig. 3B. PCR primer pairs were selected which amplify two different unique regions of the EBV genome: a part of the BKRF4 gene and a part of the BNLF1 gene containing a single or no HpaII recognition site, respectively. EBV-negative Daudi cells were infected with equal doses of wtEBV and meEBV stocks corresponding to a multiplicity of infection (MOI) of 0.05 GRUs, and total cellular DNAs were prepared 3 days p.i. The DNAs were left untreated or cleaved with the methylation-sensitive restriction enzyme HpaII to differentiate between unmethylated DNA, which HpaII cleaves, and CpG-methylated DNA, which HpaII does not cleave (Fig. 3B). Cellular DNA was quantified by qPCR with a primer pair that amplifies the housekeeping gene GUSB lacking HpaII sites for normalization of the DNA samples after HpaII restriction. The fraction of HpaII-sensitive and -resistant DNAs in cells infected with wtEBV or meEBV allowed calculation of the percentage of methylated viral DNA in Daudi cells infected with wtEBV or meEBV (Fig. 3B). The results are shown in Fig. 3C.
Virion DNA of wtEBV stocks contained very few CpG-methylated dinucleotides because the level set by the PCR probe of the BNLF1 locus compared to the BKRF4 locus indicated a degree of less than 4% methylated CpGs. Virion DNAs of meEBV generated with different amounts of transfected M.SssI gene-carrying p3663 plasmid DNA were protected from HpaII cleavage and contained up to 50% methylated CpG dinucleotides at this single HpaII site. Together the results clearly demonstrated that coexpression of a prokaryotic methyltransferase did yield EBV virion DNA, which can contain substantial proportions of methylated CpGs. The results also indicated that the extent of CpG methylation could be manipulated by altering the gene dosage of M.SssI (Fig. 3C). Next, we analyzed the phenotype of the two EBV stocks that contain unmethylated virion DNA (wtEBV) or virion DNA with up to 50% randomly methylated CpG residues (meEBV).
Generation of B-cell lines with a conditional BZFL1 gene.
We knew from previous experiments that methylated EBV DNA can yield virions, which transform B blasts to lymphoblastoid B-cell lines (26). We did not assess whether these virions contained methylated viral DNA that might spontaneously trigger viral lytic gene expression in infected cells to a degree that would support lytic amplification of viral DNA or even give rise to progeny virus. To our knowledge, in vitro no cell supports de novo synthesis of EBV upon infection, which is a major obstacle to generate high titer virus stocks to study the biology of EBV infection, or, for example, to produce attenuated vaccine strains in practically sufficient quantities. Rare reports suggest that certain primary epithelial cells might support EBV's lytic phase upon in vitro infection, but such experiments are difficult to generalize and viral gene expression in them has not been analyzed thoroughly (13, 46).
Latently infected, EBV-positive Akata cells support the lytic phase of EBV's life cycle upon B-cell receptor cross-linking (49). EBV-negative variants of this cell line as well as those of Daudi cells exist (38), and both EBV-negative cell lines can be infected with high efficiency to study viral gene expression.
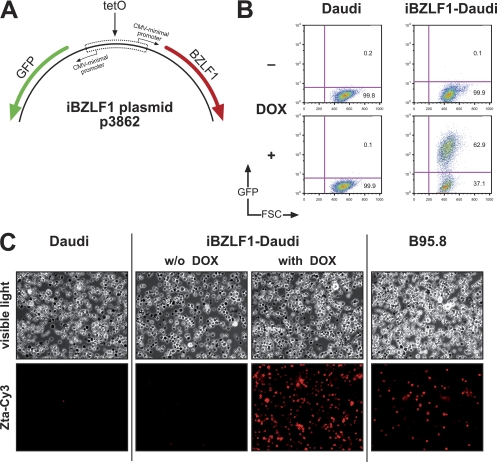
We constructed a plasmid that allows the conditional expression of BZLF1 upon addition of doxycycline (DOX). The plasmid is maintained extrachromosomally under selection in human cells and replicates via the EBV origin of plasmid DNA replication, oriP (54). Addition of DOX leads to the induced coexpression of BZLF1 and egfp, which serves a phenotypic marker to visualize gene expression at the level of individual cells by FACS analysis (5). We introduced the p3862 plasmid, iBZLF1 (Fig. 4A), into Daudi and Akata cells and selected them for stable plasmid maintenance. Addition of DOX readily induced GFP (Fig. 4B) and BZLF1 (Fig. 4C) in the majority of Daudi cells. Similar results were seen with Akata cells stably transfected with iBZLF1 (data not shown).
Fig 4.
Daudi cells with a doxycycline (DOX)-regulated conditional BZLF1 gene. (A) Construction of the iBZLF1 plasmid p3862 that carries a DOX-regulated bidirectional promoter. Schematically shown are the cluster of seven tetO binding sites (tetO) and the approximate positions of the two genes gfp and BZLF1. For selection in a eukaryotic cell, the plasmid confers resistance against hygromycin (not shown). (B) GFP expression by FACS analysis served as a phenotypic marker for DOX induction after 24 h. (C) Immunostaining of iBZLF1-Daudi cells with a BZLF1-specific monoclonal antibody. As negative and positive controls, the EBV-negative Daudi cell line and the spontaneously lytic B95.8 cell line were used, respectively.
Viral gene expression in cells infected with meEBV or wtEBV.
Next, we infected the iBZLF1-carrying Akata and Daudi cells with equal doses of wtEBV and meEBV stocks corresponding to an MOI of 0.05 GRUs per cell, divided the infected populations, and induced the BZLF1 gene in one fraction with DOX. The cell lines carrying the conditional iBZLF1 expression plasmid were readily infected with wtEBV stocks similar to the parental cells as indicated by GFP fluorescence and FACS analysis, and the meEBV stocks were confirmed to contain about 50% methylated CpGs at BKRF4 (Fig. 3C and data not shown). We divided the infected populations and induced the BZLF1 gene in one fraction with DOX. After 48 h, total RNAs were prepared and transcribed into cDNA libraries, which were analyzed by quantitative PCRs with primer pairs specific for immediate-early, early, and late viral transcripts. A possible contamination with viral DNA was carefully monitored to exclude background PCR signals and, where possible, the primer pairs spanned splice junctions to minimize this risk (Table 1). The stable transcript levels of the cellular housekeeping gene GUSB and cytochrome c (10) served as an internal reference for normalization of the data sets covering four different experimental conditions: cells infected with wtEBV or meEBV in the absence or presence of DOX for 48 h. The results are shown in Fig. 5.
Fig 5.
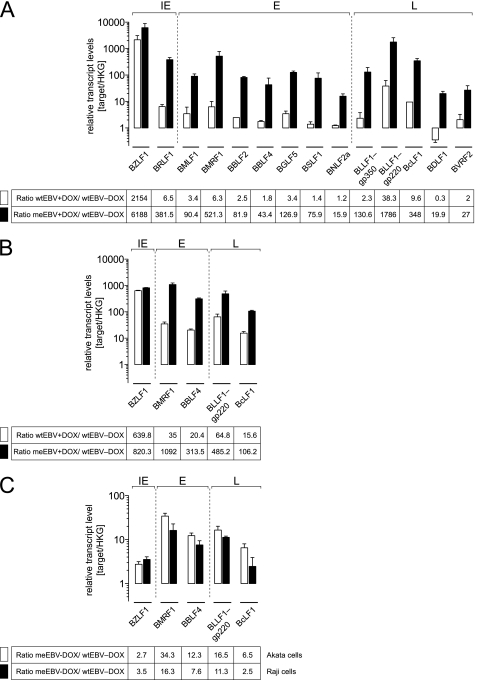
BZLF1 induces genes essential for viral replication in Daudi and Akata cells infected with meEBV stocks. EBV-negative Daudi and Akata cells carrying the conditional expression plasmid iBZLF1 were infected with wtEBV or meEBV for 2 days, when the cells were treated with DOX (+DOX) or left untreated (−DOX) and incubated for two additional days. After reverse transcription of cellular RNAs, relative levels of selected viral transcripts of the three classes of lytic viral genes, immediate early (IE), early (E), and late (L), were assessed by quantitative real-time PCRs. The results were normalized to the transcript levels of the constitutive housekeeping gene (HKG) GUSB and CYC. Results are shown for Daudi cells (A) and Akata cells (B). Ratios were calculated to analyze relative EBV transcript levels in uninduced or DOX-induced cell lines infected with either wtEBV or meEBV. White bars indicate wtEBV and DOX induction versus uninduced wtEBV; black bars indicate meEBV and DOX induction versus wtEBV without DOX. In panel C, ratios of viral transcripts levels are shown, which were calculated from Akata and Daudi cells infected with meEBV versus wtEBV in the absence of DOX 4 days p.i. The tables underneath the corresponding graphs show the mean values of three independent experiments.
Gene expression in infected Daudi cells was analyzed in detail. In DOX-treated wtEBV-infected cells, most viral genes were minimally induced in the order of 2- to 3-fold compared to cells without DOX. Exceptions were the transcripts of BRLF1, BMRF1, BLLF1-gp220, and BcLF1, which were induced in the range of 6-fold (BRLF1, BMRF1) to 38-fold (BLLF1-gp220) (Fig. 5A). The induction of the BZLF1 gene was dramatic but expected and served as a positive control since the BZLF1-specific primer pair amplified the viral transcript and the DOX-induced BZLF1 transcript of the conditional iBZLF1 expression plasmid.
With DOX induction, meEBV-infected Daudi cells showed substantial activation levels of all classes of viral transcripts compared to wtEBV-infected cells (Fig. 5A). The results indicated that viral transcription preferentially occurred from CpG-methylated virion template DNA as predicted (4). The ratio between DOX-treated meEBV-infected Daudi cells and wtEBV-infected cells without DOX corroborated this earlier finding at the level of individual genes, as can be seen with transcripts of four genes which were induced 400- to 500-fold (BRLF1, BMRF1), 350-fold (BcLF1), and more than 1,000-fold (BLLF1-gp220). Other lytic genes of all classes (BMLF1, BBLF2, BGLF5, BSLF1, and BLLF1-gp350) were induced by a factor of about 100, whereas BBLF4, BNLF2a, BDLF1, and BVRF2 were induced from 15-fold to more than 40-fold (Fig. 5A). Most likely, not all promoters of the induced lytic genes are direct targets of BZLF1 because late lytic genes were also upregulated by DOX, although they are not known to carry BZLF1-responsive elements (4).
DOX-treated wtEBV-infected Akata cells showed an induction of viral genes compared to cells without DOX treatment similar to Daudi cells but in a higher range, between 15-fold (BcLF1) and 65-fold (BLLF1-gp220) (Fig. 5B). The addition of DOX to meEBV-infected Akata cells led to the very impressive induction of all investigated transcripts from 100-fold up to 1,000-fold, exceeding levels seen with wtEBV-infected cells in the absence of DOX (Fig. 5B).
We also analyzed viral gene expression in the absence of DOX, i.e., without BZLF1 induction, and compared meEBV and wtEBV stocks in infected Daudi and Akata cells to comparatively assess spontaneous gene expression from unmethylated and partially CpG-methylated viral template DNA. Consistently, we found a clear upregulation of members of all classes of viral genes in Daudi and Akata cells infected with meEBV compared to those infected with wtEBV (Fig. 5C). Expression levels of two transcripts of early genes, BMRF1 and BBLF4, were induced about 10- to 30-fold, similar to the late gene transcript of BLLF1-gp220. In contrast, BZLF1 transcript levels were upregulated approximately 2- to 3-fold only. This finding is in line with the promoter of BZLF1, which is devoid of CpG dinucleotides, in contrast to the promoter of the BMRF1 or BBLF4 gene, for example. The paradoxical outcome of this experiment indicated that random CpG methylation of virion DNA led to spontaneously elevated levels of viral lytic transcripts in meEBV-infected Daudi and Akata cells (Fig. 5C). Additional experiments will be necessary to distinguish whether unknown cellular factors are responsible for methylation-dependent induction of viral genes or whether minimally elevated activation levels of the BZLF1 gene contribute to this effect in meEBV-infected B cells (Fig. 5C).
In summary, meEBV virion DNA led to the highest expression levels of viral transcripts in both B-cell lines when BZLF1 was inducibly expressed. The effect was more pronounced in Akata cells, but we also noted remarkable differences between this cell line and Daudi cells. Viral genes in Akata cells infected with wtEBV showed a considerably better inducibility upon BZLF1 expression compared to Daudi cells. The BZLF1-dependent transcription of almost all viral genes after infection with meEBV was expected and in line with our predictions (4). In addition, meEBV DNA but not wtEBV DNA readily acted as a template for viral transcription in Daudi and Akata cells when BZLF1 was not inducibly expressed. The results corroborated our working hypothesis and suggested that the BZLF1-induced transcript levels in meEBV-infected cells might suffice to amplify viral DNA or even yield progeny virus.
meEBV-infected B cells support limited amplification of viral DNA in Daudi cells.
A hallmark of the productive phase of herpesvirus infection is the asynchronous lytic DNA amplification of viral DNA, which can lead to a several hundredfold accumulation of newly synthesized EBV DNA (19, 20, 43). We asked whether Daudi or Akata cells infected with wtEBV or meEBV might support amplification of EBV DNA. Four days p.i. with wtEBV or meEBV stocks, we isolated whole genomic DNA of Daudi or Akata cells that had been incubated with DOX to induce the expression of BZLF1 for the last 2 days or left untreated. The copy number of viral DNA was analyzed with a primer pair that amplifies a unique viral locus (BMRF1) (Table 2). The cellular genome equivalents were also determined by quantitative PCR with a GUSB gene-specific primer pair. The results are shown in Fig. 6.
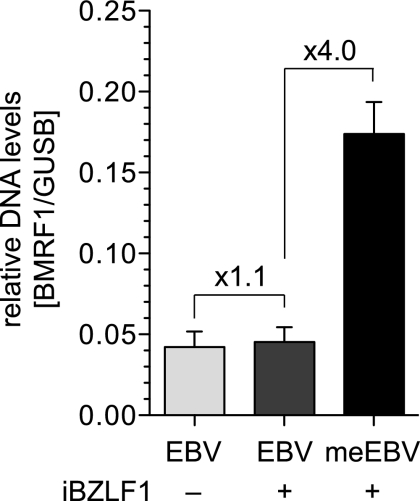
Fig 6.
Limited amplification of meEBV DNA in infected B cells. EBV-negative Daudi cells stably transfected with the DOX-regulated conditional expression plasmid iBZLF1 were infected with wtEBV and meEBV virus stocks for 2 days and incubated for two additional days with or without DOX. Genomic DNA was isolated and assessed by qPCR analysis with primers specific for the viral and cellular genes, BMRF1 and GUSB, respectively. BZLF1 expression did not lead to lytic DNA replication in DOX-induced Daudi cells infected with wtEBV, but infection with meEBV led to a detectable 4-fold increase in viral DNA on average in three independent experiments. Shown are relative levels of EBV DNA (BMRF1 locus) normalized to cellular DNA levels (GUSB locus).
Uninduced or DOX-induced Daudi cells infected with wtEBV did not differ in the copy number of viral DNA, but meEBV infections led to a clear DOX-dependent but small increase in viral DNA by only a factor of four. Similar results were obtained with Akata cells (data not shown). We tested the supernatants of the infected cells, similar to our previous experiments (26), for transforming EBV virus that the cells might release. No progeny virus could be detected.
DISCUSSION
As with other members of the herpesvirus family, EBV virion DNA is unmethylated upon infection, but in contrast to its many relatives, EBV must first establish a latent infection before its lytic, productive phase can ensue. In the latent phase, the viral DNA becomes methylated over time by the infected host B cell. Paradoxically, the viral transactivator BZLF1 prefers binding sites that encompass methylated CpG dinucleotides in the class of early lytic genes, which are essential for the synthesis of viral DNA (4), in order to induce the viral lytic cycle (26). This finding suggests that CpG methylation of viral DNA is a prerequisite for EBV in order to yield progeny virus. EBV has evolved its own time-dependent, epigenetic switch to control its biphasic life cycle and harnesses the host B cell's DNA methylation to modulate its own infection cycle (26). The dichotomous mode of rounds of infections, i.e., latent infection followed by the virus' lytic phase, is astounding but a major obstacle to propagating EBV in order to obtain large quantities of virus stocks.
Here, we attempted to generate virus stocks with 5-methylcytosine-containing virion DNA by inducing EBV's lytic phase in newly infected cells and bypassing the latent phase of EBV's life cycle. Our previous findings indicated that the bacterial de novo methyltransferase M.SssI is functionally expressed in HEK293 cells (26). The experiments in this report extend our initial observation and document 5-methylcytosine residues in virion DNAs of meEBV stocks generated in the HEK293 producer cell line 2089 (26).
In general, herpesviral virion DNA is free of 5-methylcytosines, which is surprising in the case of EBV because its lytic DNA replication relies on highly CpG-methylated viral DNA in latently infected cells (4, 26). In proliferating somatic cells, methylated cytosine residues are faithfully preserved during DNA replication and copied onto newly synthesized DNA strands, but our findings indicate that this rule does not apply to lytic replication of EBV DNA. EBV's lytic DNA synthesis eliminates the pattern of CpG methylation prevailing in the DNA templates of latent viral genomes. Several lines of evidence support our view. First is PCNA, which is critical for tethering the cellular DNA methyltransferase DNMT1 to newly replicating, hemi-methylated DNA but does not participate in herpesviral DNA amplification (36). Second, DNMT1 levels are regulated with the cell cycle and induced upon S-phase entry (48). Induction of EBV's lytic phase blocks cells at the G1/S boundary, presumably separating the temporal expression of DNMT1 from viral DNA amplification (31). Cellular DNA replication in lytically induced cells is blocked, suggesting that DNMT1 is not expressed. Conversely, EBV's lytic phase actively promotes an S-phase-like environment advantageous for viral lytic replication (32) that might include upregulation of DNMT1, a possibility that needs to be investigated. Third, lytic herpesviral DNA replication occurs in discrete nuclear compartments, which might exclude interfering factors or enzymes. It is therefore surprising to learn that the prokaryotic de novo methyltransferase M.SssI can gain access to herpesviral compartments of lytic DNA replication.
Upon infection of B cells, randomly CpG-methylated virion DNA readily induced expression of essential lytic genes in contrast to virion DNA free of 5-methylcytosine residues (Fig. 5A and B). Akata cells seemed to support this expression better than Daudi cells (Fig. 5B). In this series of experiments, we confirmed that the lack of 5-methylcytosine residues in CpG sites of virion DNA prevents the expression of lytic genes indispensable for viral DNA amplification during productive infection. However, even in Akata cells infected with meEBV stocks lytic viral DNA amplification was limited after the induced expression of BZLF1, suggesting that levels of expression of lytic genes were still inadequate. Lytic viral DNA amplification could also fail because randomly introduced 5-methylcytosines might prevent binding of cellular or viral factors to critical cis-acting elements in viral DNA, such as the lytic origin of DNA replication, oriLyt, or certain promoters of lytic genes. Random methylation of viral DNA does not reflect the profile of CpG methylation seen in latently infected cells because certain regions of genomic EBV DNA are spared from methylated CpGs during latency (26), an aspect which might also be important for EBV's lytic phase. Even Akata cells, which are known to support virus de novo synthesis if latently infected, do not synthesize new virus with randomly CpG-methylated EBV DNA (Fig. 6).
This report, together with our earlier findings (4, 26), suggests that EBV has developed a strategy that supports initial latent infections exclusively. In order to overcome this viral strategy, we have to consider alternative ideas. The key to success seems to lie in BZLF1 itself, because it has evolved to bind to and transactivate a certain class of viral DNA motifs in a methylation-dependent fashion. This strategy fosters latent infections, but an engineered version of BZLF1 that binds to the same but unmethylated DNA motifs of this class might overturn this strategy. The molecular structure of BZLF1 bound to a consensus AP-1 sequence motif is known (42), but how BZLF1 exactly binds sequence specifically to methylated DNA has not been resolved yet (27).
Assuming EBV could be propagated and amplified at will in vitro in consecutive rounds of infections like most other, conventional viruses, attractive goals spring to mind. For example, large scale, high-titer virus stocks would become feasible. They might serve as a prospective EBV vaccine based on genetically engineered, attenuated, and safe EBV mutants that lack viral oncogenes. Another example is safe viral vectors for human B cells, which are particularly recalcitrant to the transduction of therapeutic genes (17). Based on the highly specific B-cell tropism of EBV, such gene vectors could be engineered in sufficient quantities for the immune therapy of B-cell lymphomas or leukemias (22).
ACKNOWLEDGMENTS
We thank Bill Sugden for suggestions and critical reading of the manuscript and Dagmar Pich for her important experimental contributions. We acknowledge New England BioLabs for providing the M.SssI gene and Georg W. Bornkamm for the pRTS-2 plasmid.
Our work was supported by the Deutsche Forschungsgemeinschaft (SPP1230, SFB455, SFBTR5, SFBTR36) and the National Institutes of Health (grant no. CA70723).
Footnotes
Published ahead of print 26 October 2011
REFERENCES
- 1. Altmann M, Hammerschmidt W. 2005. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 3:e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. 2008. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455:818–821 [DOI] [PubMed] [Google Scholar]
- 3. Avvakumov GV, et al. 2008. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455:822–825 [DOI] [PubMed] [Google Scholar]
- 4. Bergbauer M, et al. 2010. CpG-methylation regulates a class of Epstein-Barr virus promoters. PLoS Pathog. 6:e1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bornkamm GW, et al. 2005. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 33:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bostick M, et al. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317:1760–1764 [DOI] [PubMed] [Google Scholar]
- 7. Chuang LS, et al. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277:1996–2000 [DOI] [PubMed] [Google Scholar]
- 8. Countryman J, Miller G. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. U. S. A. 82:4085–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daikoku T, et al. 2006. Postreplicative mismatch repair factors are recruited to Epstein-Barr virus replication compartments. J. Biol. Chem. 281:11422–11430 [DOI] [PubMed] [Google Scholar]
- 10. de Brouwer AP, van Bokhoven H, Kremer H. 2006. Comparison of 12 reference genes for normalization of gene expression levels in Epstein-Barr virus-transformed lymphoblastoid cell lines and fibroblasts. Mol. Diagn. Ther. 10:197–204 [DOI] [PubMed] [Google Scholar]
- 11. Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dirmeier U, et al. 2003. Latent membrane protein 1 is critical for efficient growth transformation of human B cells by Epstein-Barr virus. Cancer Res. 63:2982–2989 [PubMed] [Google Scholar]
- 13. Feederle R, et al. 2007. Epstein-Barr virus B95. 8 produced in 293 cells shows marked tropism for differentiated primary epithelial cells and reveals interindividual variation in susceptibility to viral infection. Int. J. Cancer 121:588–594 [DOI] [PubMed] [Google Scholar]
- 14. Fernandez AF, et al. 2009. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 19:438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fixman ED, Hayward GS, Hayward SD. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fixman ED, Hayward GS, Hayward SD. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Funke S, et al. 2009. Pseudotyping lentiviral vectors with the wild-type measles virus glycoproteins improves titer and selectivity. Gene Ther. 16:700–705 [DOI] [PubMed] [Google Scholar]
- 18. Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74 [DOI] [PubMed] [Google Scholar]
- 19. Hammerschmidt W, Sugden B. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427–433 [DOI] [PubMed] [Google Scholar]
- 20. Hammerschmidt W, Sugden B. 2006. Epstein-Barr Virus, p 687–705 In DePamphilis ML. (ed), DNA replication and human disease. Cold Spring Harbor Laboratory Press, New York, NY [Google Scholar]
- 21. Hashimoto H, et al. 2008. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455:826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hellebrand E, et al. 2006. Epstein-Barr virus vector-mediated gene transfer into human B cells: potential for antitumor vaccination. Gene Ther. 13:150–162 [DOI] [PubMed] [Google Scholar]
- 23. Hervouet E, et al. 2010. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLoS One 5:e11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Imbert-Marcille BM, et al. 2000. Sequential use of paraformaldehyde and methanol as optimal conditions for the direct quantification of ZEBRA and rta antigens by flow cytometry. Clin. Diagn. Lab. Immunol. 7:206–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalla M, Hammerschmidt W. 2011. Human B cells on their route to latent infection—early but transient expression of lytic genes of Epstein-Barr virus. Eur. J. Cell Biol. [Epub ahead of print.] doi:10.1016/j.ejcb.2011.01.014 [DOI] [PubMed] [Google Scholar]
- 26. Kalla M, Schmeinck A, Bergbauer M, Pich D, Hammerschmidt W. 2010. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc. Natl. Acad. Sci. U. S. A. 107:850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karlsson QH, Schelcher C, Verrall E, Petosa C, Sinclair AJ. 2008. Methylated DNA recognition during the reversal of epigenetic silencing is regulated by cysteine and serine residues in the Epstein-Barr virus lytic switch protein. PLoS Pathog. 4:e1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kieff E, Rickinson AB. 2007. Epstein-Barr virus and its replication, p 2603–2654 InKnipe DM, et al. (ed), Fields virology, 5th ed Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 29. Kintner C, Sugden B. 1981. Conservation and progressive methylation of Epstein-Barr viral DNA sequences in transformed cells. J. Virol. 38:305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kishikawa S, Murata T, Ugai H, Yamazaki T, Yokoyama KK. 2003. Control elements of Dnmt1 gene are regulated in cell-cycle dependent manner. Nucleic Acids Res. 3(Suppl):307–308 [DOI] [PubMed] [Google Scholar]
- 31. Kudoh A, et al. 2003. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high S-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. J. Virol. 77:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kudoh A, et al. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156–8163 [DOI] [PubMed] [Google Scholar]
- 33. Leonhardt H, Page AW, Weier HU, Bestor TH. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865–873 [DOI] [PubMed] [Google Scholar]
- 34. Miller G, Shope T, Lisco H, Stitt D, Lipman M. 1972. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc. Natl. Acad. Sci. U. S. A. 69:383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Minarovits J. 2006. Epigenotypes of latent herpesvirus genomes. Curr. Top. Microbiol. Immunol. 310:61–80 [DOI] [PubMed] [Google Scholar]
- 36. Muylaert I, Elias P. 2010. Contributions of nucleotide excision repair, DNA polymerase eta, and homologous recombination to replication of UV-irradiated herpes simplex virus type 1. J. Biol. Chem. 285:13761–13768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muylaert I, Tang KW, Elias P. 2011. Replication and recombination of herpes simplex virus DNA. J. Biol. Chem. 286:15619–15624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nanbo A, Inoue K, Adachi-Takasawa K, Takada K. 2002. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt's lymphoma. EMBO J. 21:954–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neuhierl B, Feederle R, Hammerschmidt W, Delecluse HJ. 2002. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc. Natl. Acad. Sci. U. S. A. 99:15036–15041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nikitin PA, et al. 2010. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8:510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nur I, et al. 1985. Procaryotic and eucaryotic traits of DNA methylation in spiroplasmas (mycoplasmas). J. Bacteriol. 164:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petosa C, et al. 2006. Structural basis of lytic cycle activation by the Epstein-Barr virus ZEBRA protein. Mol. Cell 21:565–572 [DOI] [PubMed] [Google Scholar]
- 43. Pfüller R, Hammerschmidt W. 1996. Plasmid-like replicative intermediates of the Epstein-Barr virus lytic origin of DNA replication. J. Virol. 70:3423–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pulvertaft JV. 1964. Cytology of Burkitt's tumour (African lymphoma). Lancet 39:238–240 [DOI] [PubMed] [Google Scholar]
- 45. Schaefer BC, Strominger JL, Speck SH. 1997. Host-cell-determined methylation of specific Epstein-Barr virus promoters regulates the choice between distinct viral latency programs. Mol. Cell. Biol. 17:364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shannon-Lowe C, et al. 2009. Features distinguishing Epstein-Barr virus infections of epithelial cells and B cells: viral genome expression, genome maintenance, and genome amplification. J. Virol. 83:7749–7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sharif J, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450:908–912 [DOI] [PubMed] [Google Scholar]
- 48. Szyf M, Detich N. 2001. Regulation of the DNA methylation machinery and its role in cellular transformation. Prog. Nucleic Acid Res. Mol. Biol. 69:47–79 [DOI] [PubMed] [Google Scholar]
- 49. Takada K. 1984. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt lymphoma lines. Int. J. Cancer 33:27–32 [DOI] [PubMed] [Google Scholar]
- 50. Takada K, Shimizu N, Sakuma S, Ono Y. 1986. Transactivation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J. Virol. 57:1016–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tovey MG, Lenoir G, Begon-Lours J. 1978. Activation of latent Epstein-Barr virus by antibody to human IgM. Nature 276:270–272 [DOI] [PubMed] [Google Scholar]
- 52. Vertino PM, et al. 2002. DNMT1 is a component of a multiprotein DNA replication complex. Cell Cycle 1:416–423 [DOI] [PubMed] [Google Scholar]
- 53. Woisetschlaeger M, Yandava CN, Furmanski LA, Strominger JL, Speck SH. 1990. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 87:1725–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yates JL, Warren N, Sugden B. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812–815 [DOI] [PubMed] [Google Scholar]