Abstract
Mouse mammary tumor virus (MMTV) encodes a Rev-like protein, Rem, which is involved in the nuclear export and expression of viral RNA. Previous data have shown that all Rev-like functions are localized to the 98-amino-acid signal peptide (SP) at the N terminus of MMTV Rem or envelope proteins. MMTV-SP uses endoplasmic reticulum-associated degradation (ERAD) for protein trafficking. Rem cleavage by signal peptidase in the ER is necessary for MMTV-SP function in a reporter assay, but many requirements for trafficking are not known. To allow detection and localization of both MMTV-SP and the C-terminal cleavage product, we prepared plasmids expressing green fluorescent protein (GFP) tags. N-terminal Rem tagging led to protein accumulation relative to untagged Rem and allowed signal peptidase cleavage but reduced its specific activity. C-terminal tagging also led to Rem accumulation yet dramatically reduced cleavage, GFP fluorescence, and activity relative to N-terminally tagged Rem (GFPRem). Substitutions of an invariant leucine at position 71 between the known RNA-binding and nuclear export sequences interfered with GFPRem accumulation and activity but not cleavage. Similarly, deletion of 100 or 150 C-terminal amino acids from GFPRem dramatically reduced both Rem and MMTV-SP levels and function. Removal of the entire C terminus (203 amino acids) restored both protein levels and activity of MMTV-SP. Only C-terminal GFP tagging, and not other modifications, appeared to trap Rem in the ER membrane. Thus, Rem conformation in both the ER lumen and cytoplasm determines cleavage, retrotranslocation, and MMTV-SP function. These mutants further characterize intermediates in Rem trafficking and have implications for all proteins affected by ERAD.
INTRODUCTION
Rem is a 301-amino-acid protein encoded by the betaretrovirus mouse mammary tumor virus (MMTV) from a doubly spliced mRNA (24, 31). Rem affects nuclear export of unspliced viral RNA as well as postexport functions (30, 31), as was previously seen with the Rev protein encoded by human immunodeficiency virus (HIV) (12, 17). These functions are localized to the 98-amino-acid signal peptide (SP) (see Fig. 1) (31). MMTV-SP contains the nuclear localization signal (NLS), an arginine-rich motif (ARM) RNA-binding domain, and a nuclear export sequence (NES) (31). Rem is an internally truncated version of the envelope (Env) protein, and Rem and Env share the same open reading frame, including the MMTV-SP (24, 31). The betaretroviruses Jaagsiekte sheep retrovirus (JSRV) and human endogenous retrovirus type K (HERV-K) both express regulatory proteins using sequences that include the signal peptides from their Env proteins (1, 8, 23).
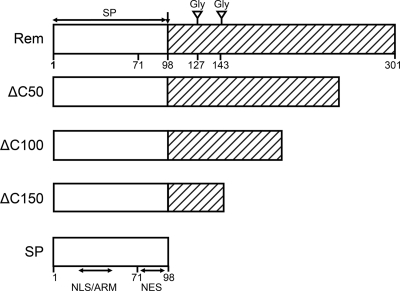
Fig 1.
Diagram of Rem and mutant constructs. The full-length Rem protein is depicted, with the white box representing the signal peptide and the hatched box representing the C-terminal domain. The inverted triangles indicate the positions of glycosylation sites (Gly) in the C terminus. The C-terminal truncation mutants are shown relative to their inclusion of Rem features. The domains indicated with horizontal double-headed arrows below MMTV-SP (SP) include the nuclear localization sequence (NLS)/arginine-rich motif (ARM) and the nuclear export sequence (NES). The relative position of mutations at amino acid 71 is also shown.
Our recent experiments revealed that MMTV-SP is generated from either the Rem or the Env proteins by signal peptidase cleavage in the endoplasmic reticulum (ER) (7). Mutation of the signal peptidase cleavage site in either rem or env cDNA was shown to prevent Rem cleavage and activity in an assay based on a reporter plasmid derived from the 3′ end of the MMTV genome (7, 31). In addition, green fluorescent protein (GFP)-tagged Rem with mutations in the signal peptidase cleavage site were unstable and exhibited very low level fluorescence compared to wild-type GFP-tagged Rem (7), which is consistent with misfolding and targeting to the proteasome. Removal of the NLS also prevents Rem activity (31), indicating that MMTV-SP must function in the nucleus. Thus, MMTV-SP has a unique trafficking pattern in virally infected cells involving retrotranslocation from the ER lumen prior to nuclear function (7, 13).
Retrotranslocation is part of the ER-associated degradation (ERAD) process used by eukaryotic cells for identification and disposal of misfolded or inappropriately assembled proteins (2, 6, 33). Defects in ERAD are known to result in a variety of diseased states, including cardiovascular disorders, neurodegenerative diseases, and cancer (19, 49). Furthermore, many viruses use ERAD to avoid the immune response, e.g., by downregulation of MHC class I (20, 47). Alternatively, hepatitis E virus uses retrotranslocation to produce cytosolic capsid protein (45), whereas simian virus 40 (SV40) and polyomaviruses use retrotranslocation to facilitate uncoating and virion entry (26, 39).
The extraction of incorrectly folded proteins from the ER requires energy, which is provided for some substrates by p97/VCP, the AAA ATPase (9, 10), as well as by the 19S subunit of the proteasome (46). Inhibition of proteasomal activity led to preferential accumulation of the Rem precursor relative to the cleaved MMTV-SP product, indicating that full-length Rem is an unstable intermediate (7). Rem activity was partially inhibited by expression of a dominant-negative p97 protein (48), which is consistent with a role for this ATPase in extraction of MMTV-SP from the ER membrane (7).
To further determine the requirements for correct Rem processing and function, we prepared tagged versions of Rem carrying mutations in either the SP or the C terminus. GFP tagging of Rem on the N terminus allowed protein accumulation, cleavage by signal peptidase, and activity in an MMTV-SP-dependent reporter assay, whereas GFP tagging on the C terminus had the opposite effect. Using N-terminally GFP-tagged Rem, mutations in either the MMTV-SP or the C terminus increased proteasomal degradation without affecting cleavage, but rescue from the proteasome did not restore activity. These results suggest a critical requirement for Rem folding and cleavage to allow retrotranslocation and escape from degradation prior to MMTV-SP function in the nucleus.
MATERIALS AND METHODS
Cell lines and transfections.
The conditions for growth of human embryonic kidney 293 and HC11 mouse mammary cells have been previously described (30). The 293 cells (5 × 105) were transfected using 6-well tissue culture plates and a modification of the calcium-phosphate precipitation method (30). HC11 cells were transfected by electroporation (30). All DNA samples for transfections were purified by cesium-chloride gradient centrifugation and quantitated by absorbance at 260 nm. Calcium phosphate transfections were performed in triplicate and used 250 ng of pGL3-Control firefly luciferase (Promega) and 250 ng of pHMRluc (Renilla luciferase) reporter plasmids in addition to the indicated amounts of rem or other expression constructs. The total amount of DNA was adjusted to 6 μg by the use of pcDNA3 control vector (Invitrogen). For electroporation of HC11 cells, 16 to 20 μg of total DNA was used for each electroporation of 1 × 107 cells in a volume of 200 μl of RPMI medium. All transfections in the same experiment were performed in triplicate, and all reaction mixtures contained the same amount of DNA. A BTX ECM600 electroporator was used with a 2-mm-gap cuvette at 140 V, 1,750 μF, and 72 Ω. Cells were harvested for luciferase assays and Western blot analysis after 48 h. In some instances, MG132 (Boston Biochem) or tunicamycin (Sigma) was added as indicated. Cells from each transfection were pelleted and washed once in phosphate-buffered saline. Half of each triplicate cell sample was resuspended in passive lysis buffer, subjected to three freeze-thaw cycles, and clarified by centrifugation at 5,000 × g. The supernatant was analyzed for protein levels prior to determination of luciferase values using a Dual-Luciferase assay kit (Promega). The other half of each of the triplicate samples was pooled for Western blotting. The protein amount for each pool was determined by a Bradford assay prior to lysis of the remaining cell pool in sodium dodecyl sulfate (SDS)-containing loading buffer for Western blotting. All transfections were repeated at least twice with similar results.
Plasmid constructs.
Expression vectors for RemGFP, GFPRem, untagged Rem, and their L71P mutants have been previously described (7, 31). The signal peptidase cleavage site (GFP-V96RG98R) and glycosylation site (GFP-N127QN143Q) mutants have also been previously reported (7). Constructs for the L71 or C-terminal truncations were prepared using rem cDNA in the enhanced GFP-C1 (EGFP-C1) vector (Promega) and site-directed mutagenesis with a Stratagene QuikChange site-directed mutagenesis kit. The EGFP sequences were removed for experiments involving untagged Rem. Details are available upon request. All plasmid constructs were confirmed by sequencing at the University of Texas Institute for Cellular and Molecular Biology (ICMB) sequencing facility.
Luciferase assays.
Luciferase assays were performed essentially as described previously (29). Cell lysates were used for the Dual-Luciferase reporter assays (Promega), and results for firefly and Renilla luciferase activity per 100 μg of protein were determined. Each of the Renilla assays that contained the pHMRluc vector with the Rem-responsive element (RmRE) was then normalized for nonspecific effects on transfection and expression by using the results of firefly luciferase assays as described by Mertz et al. (29). The averages and standard deviations of the results of triplicate experiments were calculated and expressed relative to the results seen with pHMRluc in the absence of Rem (assigned a relative value of 1).
Fluorescence microscopy.
Cells were transfected as described above for luciferase assays and Western blotting. In some cases, MG132 was added to achieve a concentration of 10 μM for 12 h to stabilize Rem expression. At approximately 48 h posttransfection, cells were fixed in 4% paraformaldehyde for 15 min, treated with 0.05% Triton X-100 for 30 min to maximize membrane permeability, and incubated with 150 nM DAPI (4′,6-diamidino-2-phenylindole) to stain nuclear DNA. Cells were examined using an Olympus IX70 fluorescence microscope as described previously (7).
Western blotting.
In most cases, cells were lysed in 0.125 M Tris-HCl (pH 6.8)–10% glycerol–1% SDS–2.5% β-mercaptoethanol–0.1% bromophenol blue. In a few cases, a similar buffer lacking β-mercaptoethanol was used. The same amounts of transfected cell extracts were analyzed on denaturing polyacrylamide gels. Separated proteins were transferred to nitrocellulose membranes. Blots were incubated with antibodies specific for MMTV-SP (7), GFP (mouse monoclonal; Clontech), or actin (Calbiochem) followed by horseradish peroxidase-conjugated donkey anti-rabbit or goat anti-mouse antibody (Jackson ImmunoResearch). Antibody binding was detected using a Western Lightning Plus ECL chemiluminescence kit (Perkin-Elmer).
Statistical analysis.
Student's two-tailed t test was used to determine whether luciferase values from triplicate transfections expressing pHMRluc and rem expression vectors were statistically different from the values from triplicate transfections performed with the reporter and pcDNA3 empty vector. Values of <0.05 were considered to be significant.
RESULTS
GFP tagging reduces Rem activity.
Previous experiments suggested that several factors influence the processing and activity of MMTV-SP generated from constructs expressing rem or env cDNA (7, 29). For example, C-terminal tagging of Rem significantly blocked cleavage by signal peptidase (29). To directly test the effect of GFP tagging, we compared rem constructs that expressed GFP at the N terminus (GFPRem) or the C terminus (RemGFP) relative to a construct expressing the untagged rem gene. Extracts from transfected cells were analyzed for Rem cleavage on Western blots using antibody specific for MMTV-SP (7). As in our published results (7), we could detect MMTV-SP after transfection of untagged rem constructs and GFP-SP from N-terminally tagged constructs (Fig. 2A). MMTV-SP from RemGFP-expressing plasmids was not observed (data not shown). Extracts from transfected cells were also used for Western blotting with GFP-specific antibody. Constructs carrying C-terminal tags showed little or no detectable GFP-tagged C-terminal product of the expected size (∼49 kDa) (Fig. 2B, lanes 7 to 9). Incubation of Western blots with actin-specific antibody confirmed similar protein loading from different extracts. These results indicated that the N-terminal GFP tag increased levels of both Rem and MMTV-SP without affecting Rem cleavage, whereas C-terminal GFP tags inhibited cleavage by signal peptidase.
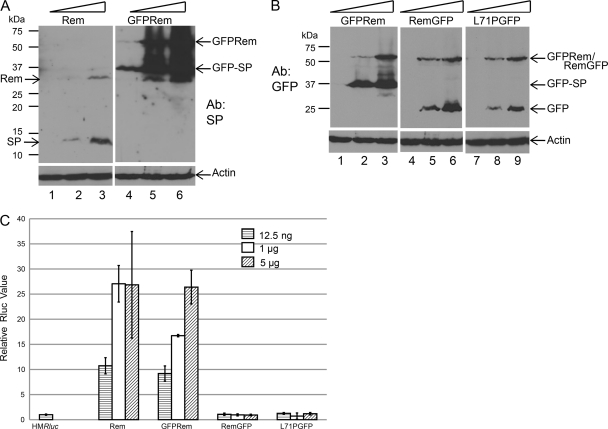
Fig 2.
GFP tagging affects Rem stability, cleavage, and function. (A) N-terminal GFP tagging stabilizes Rem and MMTV-SP expression. Expression constructs were transfected into 293 cells, and extracts from transfected cells (100 μg of protein) were subjected to Western blotting with SP-specific antibody (Ab) (upper panel). The total amount of DNA for each transfection was constant, but the amounts of rem expression construct differed (see triangles) as follows: 12.5 ng (lanes 1 and 4); 1 μg (lanes 2 and 5); and 5 μg (lanes 3 and 6). The lower panel shows Western blots of the same extracts after incubation with actin-specific antibody. (B) C-terminal tagging inhibits Rem cleavage. Western blots prepared with transfected cell extracts were incubated with GFP- or actin-specific antibodies (upper and lower panels, respectively). The total amount of DNA for each transfection was constant, but the amounts of rem expression construct differed as follows: 12.5 ng (lanes 1, 4, and 7), 1 μg (lanes 2, 5, and 8), and 5 μg (lanes 3, 6, and 9). (C) Both N-terminal and C-terminal GFP tagging inhibit Rem function. Extracts from the experiment described in the panel B legend were assayed for luciferase activity as described in Materials and Methods. Average levels of luciferase activity/100 μg of protein extract were determined. The activity of triplicate transfections performed using the pHMRluc reporter vector in the absence of Rem expression was assigned a relative value of 1 after normalization to the activity of a cotransfected firefly reporter plasmid that lacks the Rem-responsive element. Values determined in the presence of Rem are shown as the averages of the results of triplicate transfections with standard deviations. The amounts of rem expression construct differed as indicated. No significant differences between the results of experiments using pcDNA3 control vector and the C-terminally tagged rem expression plasmids were detected.
Interestingly, a band of ∼25 kDa was detectable on Western blots using GFP-specific antibody after expression of rem constructs with C-terminal, but not N-terminal, GFP tags (Fig. 2B; compare lanes 2 and 3 with lanes 5, 6, 8, and 9). The same band was also detected with GFP-specific rabbit antibody from another source (Santa Cruz) (data not shown). These data suggest that free GFP is readily generated from C-terminally but not N-terminally tagged Rem protein, presumably due to the presence of the C-terminal tag in the ER lumen (see Fig. 8B).
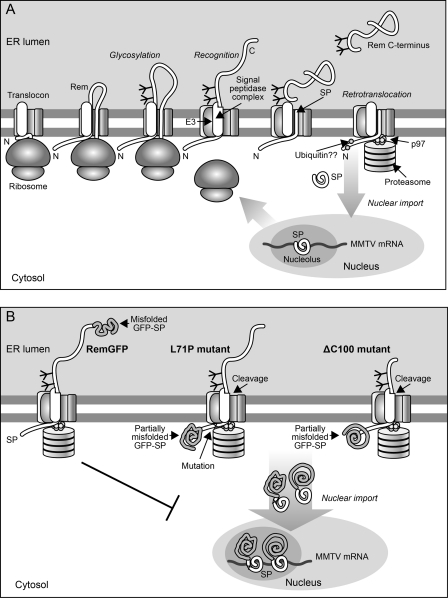
Fig 8.
Model for Rem processing. (A) Rem processing and trafficking pathway. The nascent Rem protein is recruited to the translocon by SRP to allow translation across the ER membrane. The hydrophobic domain predicted to bind SRP is adjacent to the signal peptidase cleavage site (13), suggesting that the Rem N terminus localizes to the cytoplasm. Since uncleaved Rem can be glycosylated, addition of sugars may occur prior to signal peptidase cleavage. Multiple cellular factors, including PDI proteins, likely bind to the Rem C terminus to assist glycosylation and folding. The misfolded state of Rem triggers association with the retrotranslocon, which may represent a translocon with additional components or a separate channel (25, 46). Components of the retrotranslocon include an E3 ligase, an enzyme required for the polyubiquitination of misfolded substrates (3, 34). Many substrates, including Rem, require the p97 ATPase to assist their extraction from the ER to the cytosol (7, 9, 10). Cleaved MMTV-SP then escapes proteasomal degradation to allow nuclear entry and nucleosomal targeting prior to MMTV RNA binding. The NES within MMTV-SP provides a bridge between the viral RNA and the Crm1 export protein for nuclear export of viral RNA (17, 36, 42). (B) Model for defects in Rem mutants. After Rem tagging at the C terminus, signal peptidase cannot cleave MMTV-SP and GFP does not fold properly. Failure to achieve cleavage prevents nuclear import and MMTV-SP function. In the L71 mutants with an N-terminal GFP tag, signal peptidase cleaves MMTV-SP, but a change in folding (as detected by lower fluorescence in the absence of MG132) prevents proper function after nucleolar localization. The C-terminal deletion mutants have a defect similar to the L71 mutants, except that GFP fluorescence (and folding) are further reduced, leading to defective nuclear function of MMTV-SP with a wild-type sequence.
GFP-tagged and untagged constructs were cotransfected into 293 cells with pHMRluc reporter vector. This vector contains the 3′ end of the MMTV genome, including the RmRE, yet lacks MMTV-SP sequences. Previous data showed that Renilla luciferase activity resulting from translation of unspliced RNA in the cytoplasm is responsive to MMTV-SP expression in trans (31). As expected, cotransfection of both N-terminally tagged and untagged versions of rem expression plasmids with pHMRluc increased activity relative to that observed with the reporter vector alone (Fig. 2C). Nevertheless, the specific activity of the untagged protein was higher than that of the GFP-tagged version, since the levels of the untagged protein were much lower (see Fig. 2A). Although the N-terminal GFP tag allows Rem cleavage and function in the reporter assay, the C-terminally tagged Rem protein had no detectable activity under these conditions with or without the originally described L71P mutation (31) (Fig. 2C), consistent with a defect in cleavage by signal peptidase. Although both N-terminal and C-terminal tagging with GFP negatively affected MMTV-SP activity, the effect of C-terminal tagging was much more dramatic.
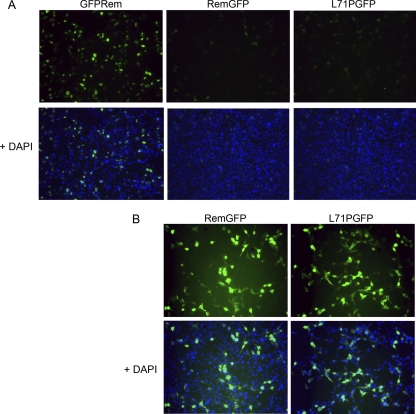
GFP fluorescence has been used as a measure of protein folding (11, 36, 38). Our previous experiments indicated that fluorescence associated with N-terminally GFP-tagged Rem carrying a proline variant at amino acid 71 was primarily localized to the nucleolus of transfected XC rat fibroblast cells (31). We then verified that the fluorescence of wild-type GFPRem in 293 cells was mostly nucleolar (Fig. 3A). In contrast, C-terminally GFP-tagged Rem (either wild-type L71 or L71P) had very low fluorescence, suggesting that the GFP was not properly folded. A 10-fold increase in the exposure time for transfected cells expressing C-terminally tagged wild-type or L71P proteins allowed better detection of the low-level fluorescent signal of these proteins, which was primarily localized outside the nucleus (Fig. 3B). Since the Western blotting results indicated that C-terminally tagged Rem is poorly cleaved to MMTV-SP and a GFP-tagged Rem C terminus, the uncleaved Rem may be retained within the ER, where the C-terminal GFP tag is partially misfolded.
Fig 3.
GFP tagging at the Rem C terminus reduces fluorescence and prevents MMTV-SP nuclear localization. (A) C-terminally GFP-tagged Rem proteins have reduced fluorescence. N-terminally or C-terminally tagged expression constructs (1 μg) were transiently transfected into 293 cells. Relatively larger amounts of plasmid DNA were transfected in this experiment to allow detection of fluorescent signals from proteins with C-terminal GFP tags. C-terminally tagged Rem proteins containing either the wild-type (L71) or mutant (L71P) sequences were analyzed. Fluorescence was recorded for GFP alone (top panels) or after combining the signals obtained from nuclear staining with DAPI (bottom panels). (B) C-terminally tagged RemGFP fusion proteins are localized primarily in the cytosol. The exposure time was increased 10-fold over that used in the experiments whose results are shown in panel A. The fluorescence of the N-terminally tagged GFPRem is greatly overemphasized under this condition (not shown).
The leucine at position 71 is required for optimal Rem stability and function.
Previous experiments indicated that GFP-tagged rem constructs encoding a proline at position 71 of MMTV-SP showed greatly decreased cleavage compared to constructs encoding a leucine at that position, resulting in a dramatic loss of activity (29). Examination of Rem sequences from different MMTV strains revealed that L71 is invariant (24) despite its location between the two functional domains, NLS/ARM and the NES (Fig. 1). To further examine the effect of L71, we prepared rem expression constructs with other nonpolar (proline, alanine, and glycine) amino acids or acidic (aspartic acid) or basic (histidine) amino acid substitutions. The mutant and wild-type constructs (250 ng) were cotransfected into 293 cells with pHMRluc. Western blots verified that each of the Rem mutants was cleaved to produce GFP-tagged SP, although the mutants exhibited lower expression than that obtained with the wild-type construct (data not shown; see below).
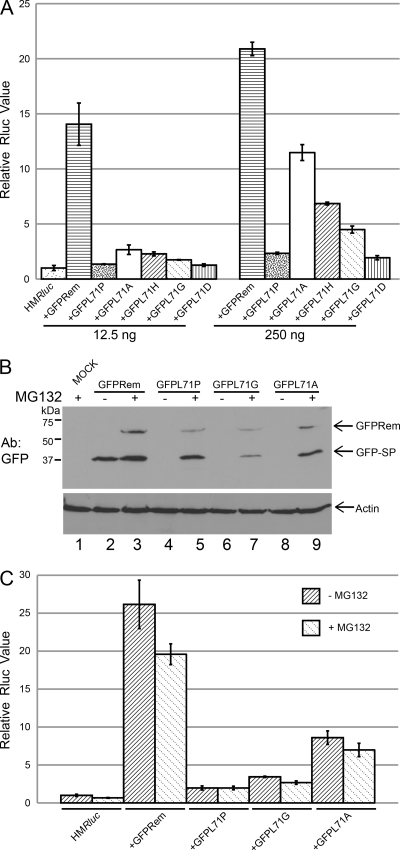
The activity of the L71 mutant constructs was assessed using reporter assays after transfection with two different amounts of DNA. In experiments using 12.5 ng of DNA, the rem construct expressing wild-type MMTV-SP (GFPRem) gave an approximately 14-fold increase in luciferase expression (Fig. 4A). Substitution with either nonpolar or polar residues at position 71 dramatically decreased MMTV-SP activity. An alanine substitution affected SP function less than the other mutations and resulted in 5- to 6-fold lower activity than was seen with GFPRem, whereas the proline and histidine substitution mutants had activities only slightly above the basal level of the activity of the luciferase reporter with the vector control. At higher DNA concentrations (250 ng), rem constructs showed a similar pattern, but the effect of the defect caused by the L71 substitution was diminished. This result appears to have been due to a plateau in the results of the assay performed using 250 ng of the wild-type rem expression construct in 293 cells (unpublished data). Nonetheless, each of the mutants retained some activity, albeit greatly reduced compared to that seen with wild-type Rem. Therefore, the leucine residue at position 71 in the signal peptide appears to be required for optimal Rem activity as well as stability.
Fig 4.
Amino acid substitutions at position 71 reduce Rem stability and activity. (A) Substitutions at amino acid 71 inhibit MMTV-SP activity. Two different amounts of N-terminally GFP-tagged rem expression constructs were transfected as indicated. The values determined for the vector control and 12.5 ng of the L71P mutant were not statistically different (P = 0.055). The other values were statistically different from the control vector values (P = <0.05). (B) Mutation of amino acid 71 in the MMTV-SP affects protein stability. Expression constructs (250 ng) and the reporter vector pHMRluc were transfected into 293 cells and incubated for 36 h before addition of 10 μM MG132 for 12 h. Transfected cell extracts (50 μg) were used for Western blotting with GFP-specific or actin-specific antibody (upper and lower panels, respectively). (C) Luciferase assays of MG132-treated cells transfected with L71 mutants. Cells transfected with 12.5 ng of rem expression constructs were incubated in the absence or presence of MG132, and then extracts were tested for luciferase activity. Values are reported as described for Fig. 2.
To further examine the significance of position L71 for Rem stability, we performed similar transfection experiments using wild-type N-terminally GFP-tagged Rem and representative mutant expression constructs. Half of the transfected 293 cells were treated with the MG132 proteasome inhibitor for 12 h prior to preparation of cell extracts for Western blotting using GFP-specific antibody. As observed previously (7), expression of wild-type Rem in the absence of MG132 primarily yielded the GFP-SP cleavage product, whereas the presence of MG132 preferentially stabilized the precursor (Fig. 4B; compare lanes 2 and 3). In contrast, each of the mutants analyzed (GFPL71P, GFPL71G, and GFPL71A) showed little or no detectable Rem or MMTV-SP expression in the absence of MG132. Both mutant Rem precursor and cleavage product levels increased in the presence of the proteasome inhibitor (lanes 5, 7, and 9). These data indicated that the MMTV-SP cleavage product carrying a mutation at L71 is more susceptible to proteasomal degradation than the wild-type protein.
The same transfections were also analyzed for Rem activity. Transfection of the wild-type rem construct gave approximately 26-fold-increased levels of luciferase activity; the levels were slightly decreased in the presence of MG132, as were baseline levels of the reporter vector alone (Fig. 4C). Although the levels of the mutant proteins were elevated in the presence of the proteasome inhibitor (Fig. 4B), no increases were observed in the activity of these proteins. The results suggest that function is compromised even after rescue from the proteasome.
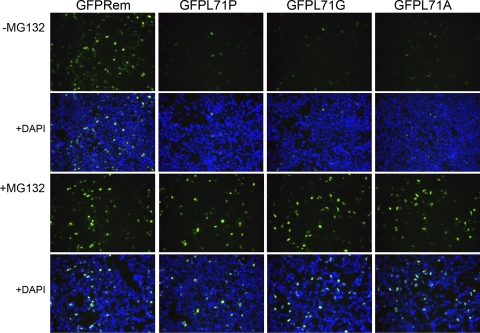
To determine whether the L71 mutants were mislocalized, wild-type rem and mutant constructs were transfected into 293 cells. After 36 h, one portion of the cells was treated with MG132, whereas the other portion was left untreated. As expected, each of the mutant Rem proteins showed lower fluorescence than the wild-type protein in the absence of the proteasome inhibitor (Fig. 5). This result could be explained by the lower level of expression of the L71 mutants (Fig. 4B). However, mutant expression and fluorescence recovered in the presence of MG132 (Fig. 4B and 5). In each case, mutants showed fluorescent localization in the nucleoli. These data suggested that, unlike the C-terminal GFP tagging results, the L71 mutants allowed both correct GFP folding at the N terminus and intracellular localization. Therefore, L71 mutations compromised both Rem and MMTV-SP stability as well as function in the reporter assay after nucleolar targeting.
Fig 5.
L71 substitutions reduce fluorescence of N-terminally tagged MMTV-SP. 293 cells were transfected with rem expression constructs (100 ng) encoding amino acid substitutions at L71 and an N-terminal GFP tag. Half of the cells were treated with the proteasome inhibitor MG132 for 12 h prior to fixation and fluorescence microscopy. Cells are shown with GFP fluorescence alone or combined with DAPI staining of the nuclei.
C-terminal truncations destabilize Rem.
Previous data indicate that both rem and env cDNAs produce MMTV-SP (7), which contains the Rev-like functional motifs (15). Furthermore, transfection of an MMTV-SP expression vector that does not express the Rem C terminus (see Fig. 1) increases luciferase expression from the pHMRluc reporter vector. Nevertheless, our previous results indicated that the C terminus influences MMTV-SP function (31). Therefore, we engineered constructs expressing Rem proteins that removed 50, 100, or 150 amino acids from the C terminus. Since our previous results indicated that the characteristics of Rem processing and trafficking are similar in different cell lines (7, 31), each mutant was transiently transfected into HC11 mouse mammary cells. After growth in the presence or absence of MG132, extracts were used for Western blotting performed with GFP-specific antibody (Fig. 6A).
Fig 6.
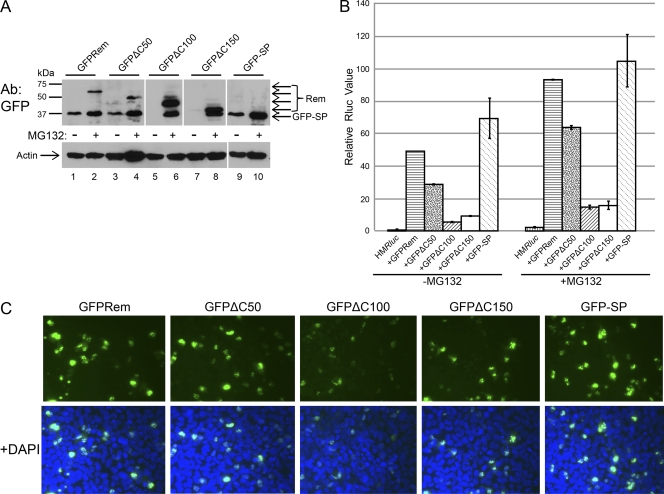
The Rem C terminus affects SP cleavage and activity. (A) Loss of 100 or 150 amino acids of the C terminus destabilizes Rem. Transfected HC11 cells expressing constructs for N-terminally GFP-tagged wild-type Rem or C-terminal mutants (25 ng) and the reporter vector pHMRluc were incubated in the absence or presence of MG132. Extracts (50 μg) were analyzed by Western blotting with GFP-specific (upper panel) or actin-specific (lower panel) antibodies. (B) Stabilization of C-terminal Rem mutants with proteasome inhibitors does not rescue wild-type SP activity. Extracts from cells transfected with GFPrem expression constructs (25 ng) were used for luciferase assays. Values are reported as described for Fig. 2. (C) Truncation of the Rem C terminus affects fluorescence of the N-terminal GFP tag. Expression constructs for wild-type Rem or C-terminal mutants (75 ng) were transfected into 293 cells. After 36 h, cells were treated with 10 μM MG132 for 12 h to stabilize mutant proteins. Cells were fixed, and nuclei were stained with DAPI prior to examination by fluorescence microscopy. Upper panels show GFP fluorescence alone, and lower panels show combined GFP and DAPI signals.
Removal of 50 amino acids from the C terminus showed little effect on Rem cleavage or the amount of total Rem produced (Fig. 6A; compare lanes 1 and 3). In contrast, removal of 100 or 150 C-terminal amino acids (GFPΔC100 or GFPΔC150) completely destabilized protein expression. Deletion of the entire C terminus (203 amino acids) yielded GFP-tagged SP and rescued wild-type protein levels (compare lane 1 with lanes 5, 7, and 9). MG132 treatment of cells transiently transfected with GFPΔC100 or GFPΔC150 rescued expression to levels comparable to those seen with wild-type Rem (Fig. 6A, lanes 2, 6, and 8). Each of the mutants was cleaved to yield GFP-tagged SP with the same mobility as the SP expression construct, although the GFPΔC100 and GFPΔC150 mutants exhibited a higher ratio of precursor to SP. These results indicated that the C-terminal amino acids influence both Rem stability and processing.
The C-terminal truncation mutants were also analyzed to determine functional activity in reporter assays in HC11 mouse mammary cells (Fig. 6B). With this cell type, vectors expressing wild-type Rem gave a 50-fold increase in luciferase activity. The activity obtained using a vector expressing only MMTV-SP (lacking all C-terminal amino acids) was slightly increased. The GFPΔC50 construct exhibited approximately 2-fold lower activity, whereas both GFPΔC100 and GFPΔC150 exhibited 5- to 10-fold lower activity than wild-type Rem. The function of both the GFPΔC100 and GFPΔC150 constructs was easily detectable in this assay, although Western blotting could not detect the proteins in the absence of MG132. These results confirm our previous observations indicating that Western blotting is significantly less sensitive than detection of MMTV-SP activity by the pHMRluc reporter assay (31).
Because levels of GFPΔC100 and GFPΔC150 increased relative to those observed with wild-type constructs in the presence of a proteasome inhibitor, we also tested extracts of transfected cells treated with MG132 for Rem activity (Fig. 6B). Similar to the results observed with untreated cells, wild-type Rem and MMTV-SP exhibited comparable levels of activity. In contrast, the mutants GFPΔC50, GFPΔC100, and GFPΔC150 all exhibited significantly reduced activity compared to wild-type Rem, despite rescue of wild-type levels of mutant proteins in the presence of MG132. As observed for reporter function in the absence of MG132, GFPΔC50 exhibited higher activity than either the GFPΔC100 or GFPΔC150 mutant. Together, these data suggest that the C-terminal mutants missing 100 or 150 amino acids are unstable yet can be cleaved by signal peptidase. This instability may be due to improper folding, signaling their rapid disposal by the proteasome.
Expression constructs of N-terminally GFP-tagged wild-type Rem and C-terminal mutants were transfected into 293 cells prior to examination by fluorescence microscopy. As expected, fluorescence of the GFPΔC100 or GFPΔC150 mutant in the absence of MG132 was not observed. Transfection of constructs expressing wild-type GFPRem, GFPΔC50, or GFP-SP revealed GFP fluorescence primarily localized to nucleoli as previously described (13, 24, 31) (data not shown). Transfected cells were also treated with MG132 before microscopy was performed (Fig. 6C). Under conditions in which similar amounts of mutant and wild-type protein were detectable by Western blotting, the GFPΔC100 and GFPΔC150 mutants showed less-intense fluorescence and fewer GFP-positive nucleoli than either wild-type Rem or SP. Combined with results from previous experiments, these data suggest that C-terminal truncations prevent proper folding of the GFP tag and GFP-SP localization to nucleoli. Instead, such mutants are degraded by the proteasome. Nevertheless, rescue from the proteasome is not sufficient to restore wild-type MMTV-SP function.
Formation of a disulfide-bonded protein complex correlates with a lack of Rem cleavage.
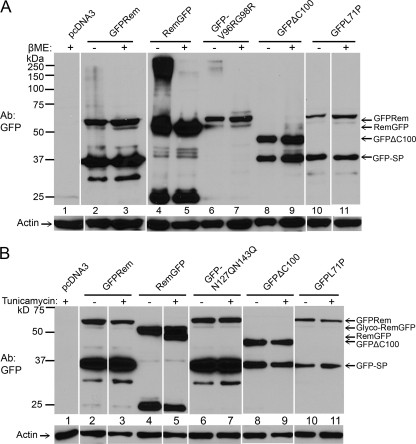
One known determinant of protein folding in the ER is interaction with members of the protein disulfide isomerase (PDI) family, which catalyzes formation and breakage of disulfide bonds, leading to protein isomerization (14, 18). PDI levels can increase after ER stress caused by the unfolded protein response (16, 18, 27). To determine whether disulfide bond formation is affected by Rem mutations, we transfected 293 cells with wild-type GFPrem expression plasmids as well as with constructs that are known to affect Rem cleavage and/or protein stability, including plasmids expressing RemGFP, GFP-V96RG98R, GFPΔC100, and GFPL71P. The GFP-V96RG98R mutant has a mutation in the consensus site for signal peptidase cleavage between Rem-SP and the C terminus. The cleavage site mutant is poorly expressed compared to wild-type Rem, has primarily cytosolic localization, and lacks activity in the pHMRluc reporter assay (7). Relatively large amounts of plasmid DNA were transfected to allow detection of Rem mutant proteins in the absence of proteasome inhibitors. After 48 h, cells were lysed in SDS-containing loading buffer in the presence or absence of β-mercaptoethanol to allow the detection of disulfide-bonded proteins. Equal amounts of protein extract were then used for Western blotting with GFP-specific antibody (Fig. 7A).
Fig 7.
GFP tagging at the Rem C terminus leads to the accumulation of glycosylated Rem precursor in a disulfide-bonded protein complex. (A) C-terminal GFP tagging allows accumulation of a disulfide-bonded protein complex with Rem. The 293 cells were transfected with 2.5 μg of GFP-tagged rem constructs as indicated. Cells were transfected with relatively large amounts of expression plasmids to allow detection of mutant proteins by Western blotting. After 48 h, cells were lysed in SDS-containing loading buffer in the presence or absence of β-mercaptoethanol (βME). Extracts (30 μg) were used for Western blotting with GFP-specific (upper panel) or actin-specific (lower panel) antibody. The positions of specific bands are indicated by arrows. (B) C-terminally GFP-tagged Rem is glycosylated. The 293 cells were transfected with 1 μg of the indicated rem expression constructs. After 24 h, half of the transfected cells were treated with tunicamycin (5 μg/ml). After an additional 24-h incubation, cells were lysed in SDS-containing loading buffer containing βME and used for Western blotting as described for panel A. The partial change in RemGFP mobility presumably resulted from protein synthesis prior to addition of tunicamycin or a partially effective drug concentration.
The results of this experiment revealed a high-molecular-mass complex with C-terminally tagged, but not N-terminally tagged, wild-type Rem protein in the absence of β-mercaptoethanol. Under the same conditions, lower levels of a similar complex were observed in extracts from cells expressing the GFP-V96RG98R mutant that lacks a consensus cleavage site for signal peptidase (7). Expression of mutants (e.g., GFPΔC100) that are cleaved efficiently by signal peptidase and expressed at similar levels had no detectable high-molecular-mass complex in the absence of a reducing agent. These results suggest that Rem carrying a C-terminal GFP tag or mutations to the signal peptidase cleavage site form a disulfide-bonded complex within the ER membrane prior to proteasomal degradation.
To further document whether the large protein complex formed by RemGFP involves trafficking through the ER membrane, constructs expressing N-terminally or C-terminally GFP-tagged Rem proteins were transfected into 293 cells. Half of each of the cell samples was treated with tunicamycin, an N-glycosylation inhibitor (44), prior to extract preparation and Western blot analysis (Fig. 7B). Although we and others have shown that a portion of wild-type GFPRem is glycosylated (7, 13), tunicamycin had no demonstrable effect on the mobility of the precursor under these conditions (Fig. 7B, lanes 2 and 3). The double-glycosylation mutant (GFP-N127QN143Q) was used as a control to reveal the position of the nonglycosylated Rem precursor (lanes 6 and 7). Previous results also indicated that the Rem precursor is unstable and not detectable in MMTV-infected cells (7), leading to the conclusion that Rem is rapidly glycosylated and cleaved, whereas uncleaved and nonglycosylated protein is retrotranslocated and degraded by the proteasome. Examination of the GFPL71P and GFPΔC100 mutants revealed the same results as those observed with wild-type GFPRem (compare lanes 8 to 11 with lanes 2 and 3). In contrast, the C-terminally tagged RemGFP protein showed a clear mobility shift in the presence of tunicamycin, consistent with the loss of glycosylation (lanes 4 and 5). Together with previous data, these results suggest that RemGFP is trapped in the ER membrane in a large protein complex that contains a glycosylated Rem C terminus.
DISCUSSION
Multiple viruses, including members of the Retroviridae, Hepeviridae, Polyomaviridae, and Herpesviridae families, use ERAD and retrotranslocation to facilitate their replication (7, 13, 20, 26, 28, 39, 41, 45, 47). MMTV-encoded Rem protein has a unique trafficking and processing pathway that involves ERAD and an unusually long signal peptide (7, 13). Two MMTV precursor proteins, Rem and Env, have an N-terminal SP that specifies the Rev-like domains, including the NLS/RNA-binding domain and NES (Fig. 1) (7, 31). The normal functions of cellular signal peptides are to bind the signal recognition particle (SRP) and direct mRNAs to the ER membrane for translation (22). Therefore, Rem must escape SRP binding for direct import into the nucleus or follow the typical routing of transmembrane proteins to the ER. Our recent published data indicate that Rem is a 33-kDa precursor protein that is processed by signal peptidase in the ER lumen to yield an 11-kDa protein (MMTV-SP), which must be retrotranslocated out of the ER membrane prior to nuclear import (7). Nevertheless, many details of this process are unknown. The results presented here indicate that changes to the N-terminal or C-terminal portions of the Rem precursor have dramatic effects on the folding (as judged by GFP fluorescence), cleavage, trafficking, and activity of the protein.
Our initial identification of the MMTV rem gene included development of a reporter assay that requires MMTV-SP function, including nuclear localization (31). We measured the ability of C-terminally tagged Rem protein with an L71P mutation (L71PGFP) to increase the luciferase activity of a reporter plasmid expressing an unspliced mRNA carrying the Rem-responsive element (29, 32). Despite transfections of much larger amounts of expression plasmids than were used in the current experiments, only 4- to 6-fold increases in reporter activity were detected (31). In HC11 mouse mammary cells, we now can achieve at least 50-fold increases with 1,000 times less plasmid DNA expressing N-terminally GFP-tagged Rem (GFPRem) rather than L71PGFP (31), i.e., at least a 10,000-fold increase in activity (not shown). Although the L71P mutation has a dramatic effect on Rem function (Fig. 4), the effect of C-terminal tagging is greater (Fig. 2C) due to inefficient Rem cleavage by signal peptidase (Fig. 2B).
Previous reports indicated that the differences seen in the activity of GFPRem and RemGFP are due to the efficient cleavage of the former but not the latter (29). Our current experiments confirm that either MMTV-SP-specific or GFP-specific antibody can detect cleavage of N-terminally GFP-tagged Rem (Fig. 2). The highly inefficient cleavage of C-terminally tagged Rem protein was also verified using the highly sensitive pHMRluc reporter assay (32) (Fig. 2C). Surprisingly, RemGFP showed cleavage of GFP (Fig. 2 and 7) despite the absence of a consensus cleavage site for signal peptidase between the Rem C terminus and GFP. Microscopy indicated that RemGFP had very low fluorescence compared to N-terminally tagged GFPRem (Fig. 3A). The low level of fluorescence of RemGFP was also absent from the nucleoli, which is typical of GFPRem and other Rev-like proteins (21, 37, 43) (see Fig. 3B) and reminiscent of the behavior of Rem cleavage site mutants (7). Misfolding of RemGFP is also consistent with the results seen after lysis of transfected cells in the absence of reducing agents, a condition allowing detection of a high-molecular-mass complex (Fig. 7A). Therefore, these data suggest that the C-terminal GFP tag leads to Rem misfolding that cannot be corrected by binding and refolding by members of the PDI family (27). Protein misfolding then leads to failure of signal peptidase cleavage and trapping of RemGFP in the ER membrane. Stable glycosylation of C-terminally tagged Rem precursor suggests ER membrane trapping (Fig. 7B). Cell fractionation experiments have also confirmed that RemGFP is detectable within the fraction containing ER proteins, whereas GFPRem is not (data not shown). Mislocalization of RemGFP may lead to blocks in normal cellular protein trafficking in the ER similar to those seen with the Rem proteins that carry mutations in the signal peptidase cleavage site (7).
Our previous experiments also identified a Rem variant containing a proline substitution for a highly conserved leucine at position 71 of the MMTV-SP (29). We further investigated the effects of amino acid substitutions of L71. The results indicated that all substitutions resulted in 5- to 10-fold decreases in reporter activity in the dose-dependent range of the assay, although proline and aspartic acid had the most dramatic effects (Fig. 4A). Western blotting indicated that the mutant proteins were unstable compared to wild-type GFPRem. Addition of MG132 to transfections could rescue the proteins from proteasomal degradation as well as allow correct localization of fluorescence to nucleoli (Fig. 5). Unlike RemGFP, L71 substitutions did not affect SP cleavage by signal peptidase. Examination of known motifs within Rem-SP indicated that position 71 is localized between the NLS/RNA-binding domain and the NES (see Fig. 1). Binding of cellular proteins to the L71 region may stabilize MMTV-SP after Rem cleavage. Since other Rev-like proteins contain multimerization domains (4, 5, 35), mutations at L71 could also affect assembly on the RmRE, leading to their defective activity in the reporter assay.
The highly active GFPRem construct was used to test several C-terminal truncation mutants. A mutant lacking 50 C-terminal amino acids showed slightly reduced MMTV-SP levels by Western blotting and about 40% lower activity than was obtained using wild-type constructs expressing GFPRem (Fig. 6). In contrast, deletion of 100 or 150 amino acids from the Rem C terminus greatly decreased the stability of the protein but could be rescued in the presence of MG132. Despite the rescue of Rem C-terminal mutants by proteasome inhibitors, the fluorescence and activity of these mutants, particularly GFPΔC100, were diminished even in the presence of MG132, likely due to defects in MMTV-SP folding. These mutations did not affect the known glycosylation sites, which are outside the region deleted from GFPΔC100 and GFPΔC150 (Fig. 1), and cleavage of the C-terminal mutants appeared normal. Strikingly, protein stability, fluorescence, and activity were completely restored by the deletion of the entire Rem C terminus, even in the absence of MG132. Although MMTV-SP is functional in the absence of the Rem C terminus, this protein is always expressed by the virus as part of the Rem or Env precursors (7). Mutations within the C terminus (particularly between amino acids 150 and 250) likely affect folding, protein stability, and interactions with ER-resident cellular factors, such as chaperones and PDI proteins. C-terminal mutations can alter MMTV-SP conformation in the cytosol with resulting decreases in viral replication (see Fig. 8B).
Experiments performed by Dultz et al. (13) indicated that Rem is cleaved into two gene products: an 11-kDa SP and a 22-kDa C terminus (Rem-CT). We have also detected Rem-CT by the use of an antibody specific for a C-terminal epitope (unpublished data), although our results were not consistent with localization of the uncleaved Rem product in the nucleus. Experiments performed using the signal peptidase cleavage site mutants (7, 13) and the results obtained with RemGFP presented here strongly argue that failure to get cleavage results in functional inactivation of MMTV-SP. Our previous data indicated that RemGFP and SP-GFP (both carrying the L71P mutation) had low specific activity but that SP-GFP was marginally more active (31), data similar to that obtained with N-terminally tagged Rem (Fig. 6B). Although the Rem C terminus can serve a regulatory role for MMTV-SP, the primary role during MMTV infection is likely to be formation of a separate gene product, Rem-CT.
Our results suggest the following model (Fig. 8A). On the basis of the appearance of a disulfide-linked complex of high molecular mass containing uncleaved Rem (either RemGFP or signal peptidase cleavage site mutants) in the absence of β-mercaptoethanol (Fig. 7A), we predict that PDI family members interact with the Rem precursor in the ER prior to cleavage. PDI proteins serve as chaperones and catalysts during protein isomerization (16, 18, 27, 28) and likely interact with the Rem C terminus, as judged by the absence of these complexes in cells expressing GFPΔC100 (Fig. 7A). Glycosylation of the Rem precursor prior to rapid signal peptidase cleavage in the ER is predicted (7, 13). Uncleaved mutants as well as wild-type Rem precursor are very unstable and are quickly delivered to the proteasome, as indicated by their rescue in the presence of MG132. Recognition of the Rem precursor and MMTV-SP retrotranslocation to the cytosol are likely to involve multiple proteins. Published results have shown that a dominant-negative p97/VCP, a cytoplasmic AAA ATPase associated with the retrotranslocon, blocks MMTV-SP activity but not cleavage. Although we have not observed Rem or MMTV-SP ubiquitination, E3 ligases associated with the retrotranslocon are widely believed to add polyubiquitin chains to substrates prior to their disposal by the proteasome (3, 34, 40, 42).
In summary, our results indicate that Rem precursor conformation is critical for cleavage to MMTV-SP. Both Rem C-terminal sequences and position 71 of MMTV-SP are important for protein stability and folding to allow efficient viral replication. Current knowledge of cellular protein quality control and disposal is limited. Viruses, which have evolved methods for using ERAD for viral protein trafficking or selective degradation of cellular proteins (7, 41, 45), are useful tools for understanding this protein degradation system. Results presented here predict cellular protein interactions of Rem and MMTV-SP during retrotranslocation. Our data clearly show that N- or C-terminal mutations or tagging outside the known Rev-like functional domains or both contribute to Rem folding, signal peptidase cleavage, retrotranslocation, and function. In addition, Rem mutants with various blocks impeding this process (Fig. 8B) can stabilize intermediates in protein processing and retrotranslocation. These mutants should be useful in screenings to identify cellular proteins involved in ERAD.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant R01 CA116813 from the National Cancer Institute.
We thank Jon Huibregtse for helpful suggestions.
Footnotes
Published ahead of print 9 November 2011
REFERENCES
- 1. Armezzani A, et al. 2011. The signal peptide of a recently integrated endogenous sheep betaretrovirus envelope plays a major role in eluding Gag-mediated late restriction. J. Virol. 85:7118–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bagola K, Mehnert M, Jarosch E, Sommer T. 2011. Protein dislocation from the ER. Biochim. Biophys. Acta 1808:925–936 [DOI] [PubMed] [Google Scholar]
- 3. Bernardi KM, et al. 2010. The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retro-translocation. Mol. Biol. Cell 21:140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boese A, Galli U, Geyer M, Sauter M, Mueller-Lantzsch N. 2001. The Rev/Rex homolog HERV-K cORF multimerizes via a C-terminal domain. FEBS Lett. 493:117–121 [DOI] [PubMed] [Google Scholar]
- 5. Bogerd H, Greene WC. 1993. Dominant negative mutants of human T-cell leukemia virus type I Rex and human immunodeficiency virus type 1 Rev fail to multimerize in vivo. J. Virol. 67:2496–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brodsky JL. 2007. The protective and destructive roles played by molecular chaperones during ERAD (endoplasmic-reticulum-associated degradation). Biochem. J. 404:353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Byun H, et al. 2010. Retroviral Rem protein requires processing by signal peptidase and retrotranslocation for nuclear function. Proc. Natl. Acad. Sci. U. S. A. 107:12287–12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caporale M, et al. 2009. The signal peptide of a simple retrovirus envelope functions as a posttranscriptional regulator of viral gene expression. J. Virol. 83:4591–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carlson EJ, Pitonzo D, Skach WR. 2006. p97 functions as an auxiliary factor to facilitate TM domain extraction during CFTR ER-associated degradation. EMBO J. 25:4557–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chapman E, Fry AN, Kang M. 2011. The complexities of p97 function in health and disease. Mol. Biosyst. 7:700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Craggs TD. 2009. Green fluorescent protein: structure, folding and chromophore maturation. Chem. Soc. Rev. 38:2865–2875 [DOI] [PubMed] [Google Scholar]
- 12. Cullen BR. 2003. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28:419–424 [DOI] [PubMed] [Google Scholar]
- 13. Dultz E, et al. 2008. The signal peptide of the mouse mammary tumor virus Rem protein is released from the endoplasmic reticulum membrane and accumulates in nucleoli. J. Biol. Chem. 283:9966–9976 [DOI] [PubMed] [Google Scholar]
- 14. Feige MJ, Hendershot LM. 2011. Disulfide bonds in ER protein folding and homeostasis. Curr. Opin. Cell Biol. 23:167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475–483 [DOI] [PubMed] [Google Scholar]
- 16. Gauss R, Kanehara K, Carvalho P, Ng DTW, Aebi M. 2011. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol. Cell 42:782–793 [DOI] [PubMed] [Google Scholar]
- 17. Groom HCT, Anderson EC, Lever AML. 2009. Rev: beyond nuclear export. J. Gen. Virol. 90:1303–1318 [DOI] [PubMed] [Google Scholar]
- 18. Haefliger S, et al. 2011. Protein disulfide isomerase blocks CEBPA translation and is up-regulated during the unfolded protein response in AML. Blood 117:5931–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hebert DN, Molinari M. 2007. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 87:1377–1408 [DOI] [PubMed] [Google Scholar]
- 20. Hegde NR, et al. 2006. The role of BiP in endoplasmic reticulum-associated degradation of major histocompatibility complex class I heavy chain induced by cytomegalovirus proteins. J. Biol. Chem. 281:20910–20919 [DOI] [PubMed] [Google Scholar]
- 21. Heger P, Rosorius O, Hauber J, Stauber RH. 1999. Titration of cellular export factors, but not heteromultimerization, is the molecular mechanism of trans-dominant HTLV-1 rex mutants. Oncogene 18:4080–4090 [DOI] [PubMed] [Google Scholar]
- 22. Hiss JA, Schneider G. 2009. Architecture, function and prediction of long signal peptides. Brief. Bioinformatics 10:569–578 [DOI] [PubMed] [Google Scholar]
- 23. Hofacre A, Nitta T, Fan H. 2009. Jaagsiekte sheep retrovirus encodes a regulatory factor, Rej, required for synthesis of Gag protein. J. Virol. 83:12483–12498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Indik S, Günzburg WH, Salmons B, Rouault F. 2005. A novel, mouse mammary tumor virus encoded protein with Rev-like properties. Virology 337:1–6 [DOI] [PubMed] [Google Scholar]
- 25. Johnson AE, Haigh NG. 2000. The ER translocon and retrotranslocation: is the shift into reverse manual or automatic? Cell 102:709–712 [DOI] [PubMed] [Google Scholar]
- 26. Lilley BN, Gilbert JM, Ploegh HL, Benjamin TL. 2006. Murine polyomavirus requires the endoplasmic reticulum protein Derlin-2 to initiate infection. J. Virol. 80:8739–8744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Määttänen P, Gehring K, Bergeron JJM, Thomas DY. 2010. Protein quality control in the ER: the recognition of misfolded proteins. Semin. Cell Dev. Biol. 21:500–511 [DOI] [PubMed] [Google Scholar]
- 28. Magnuson B, et al. 2005. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol. Cell 20:289–300 [DOI] [PubMed] [Google Scholar]
- 29. Mertz JA, Chadee AB, Byun H, Russell R, Dudley JP. 2009. Mapping of the functional boundaries and secondary structure of the mouse mammary tumor virus Rem-responsive element. J. Biol. Chem. 284:25642–25652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mertz JA, Lozano MM, Dudley JP. 2009. Rev and Rex proteins of human complex retroviruses function with the MMTV Rem-responsive element. Retrovirology 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mertz JA, Simper MS, Lozano MM, Payne SM, Dudley JP. 2005. Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J. Virol. 79:14737–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Müllner M, Salmons B, Günzburg WH, Indik S. 2008. Identification of the Rem-responsive element of mouse mammary tumor virus. Nucleic Acids Res. 36:6284–6294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakatsukasa K, Brodsky JL. 2008. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9:861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. 2008. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132:101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olsen HS, Cochrane AW, Dillon PJ, Nalin CM, Rosen CA. 1990. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 4:1357–1364 [DOI] [PubMed] [Google Scholar]
- 36. Reeder PJ, Huang Y-M, Dordick JS, Bystroff C. 2010. A rewired green fluorescent protein: folding and function in a nonsequential, noncircular GFP permutant. Biochemistry 49:10773–10779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruggieri A, et al. 2009. Human endogenous retrovirus HERV-K(HML-2) encodes a stable signal peptide with biological properties distinct from Rec. Retrovirology 6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanders JKM, Jackson SE. 2009. The discovery and development of the green fluorescent protein, GFP. Chem. Soc. Rev. 38:2821–2822 [DOI] [PubMed] [Google Scholar]
- 39. Schelhaas M, et al. 2007. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131:516–529 [DOI] [PubMed] [Google Scholar]
- 40. Shimizu Y, Okuda-Shimizu Y, Hendershot LM. 2010. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol. Cell 40:917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soetandyo N, Ye Y. 2010. The p97 ATPase dislocates MHC class I heavy chain in US2-expressing cells via a Ufd1-Npl4-independent mechanism. J. Biol. Chem. 285:32352–32359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stagg HR, et al. 2009. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J. Cell Biol. 186:685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stauber R, Gaitanaris GA, Pavlakis GN. 1995. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology 213:439–449 [DOI] [PubMed] [Google Scholar]
- 44. Strickler JE, Patton CL. 1980. Trypanosoma brucei brucei: inhibition of glycosylation of the major variable surface coat glycoprotein by tunicamycin. Proc. Natl. Acad. Sci. U. S. A. 77:1529–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Surjit M, Jameel S, Lal SK. 2007. Cytoplasmic localization of the ORF2 protein of hepatitis E virus is dependent on its ability to undergo retrotranslocation from the endoplasmic reticulum. J. Virol. 81:3339–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wahlman J, et al. 2007. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell 129:943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang X, Ye Y, Lencer W, Hansen TH. 2006. The viral E3 ubiquitin ligase mK3 uses the Derlin/p97 endoplasmic reticulum-associated degradation pathway to mediate down-regulation of major histocompatibility complex class I proteins. J. Biol. Chem. 281:8636–8644 [DOI] [PubMed] [Google Scholar]
- 48. Ye Y, Meyer HH, Rapoport TA. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414:652–656 [DOI] [PubMed] [Google Scholar]
- 49. Yoshida H. 2007. ER stress and diseases. FEBS J. 274:630–658 [DOI] [PubMed] [Google Scholar]