Abstract
Papillomavirus E2 protein is required for the replication and maintenance of viral genomes and transcriptional regulation of viral genes. E2 functions through sequence-specific binding to 12-bp DNA motifs—E2 binding sites (E2BS)—in the virus genome. Papillomaviruses are able to establish persistent infection in their host and have developed a long-term relationship with the host cell in order to guarantee the propagation of the virus. In this study, we have analyzed the occurrence and functionality of E2BSs in the human genome. Our computational analysis indicates that most E2BSs in the human genome are found in repetitive DNA regions and have G/C-rich spacer sequences. Using a chromatin immunoprecipitation approach, we show that human papillomavirus type 11 (HPV11) E2 interacts with a subset of cellular E2BSs located in active chromatin regions. Two E2 activities, sequence-specific DNA binding and interaction with cellular Brd4 protein, are important for E2 binding to consensus sites. E2 binding to cellular E2BSs has a moderate or no effect on cellular transcription. We suggest that the preference of HPV E2 proteins for E2BSs with A/T-rich spacers, which are present in the viral genomes and underrepresented in the human genome, ensures E2 binding to specific binding sites in the virus genome and may help to prevent extensive and possibly detrimental changes in cellular transcription in response to the viral protein.
INTRODUCTION
Human papillomaviruses (HPVs)are small DNA viruses that infect cutaneous or mucosal epithelium and are associated with cervical carcinoma and other anogenital cancers, as well as head, neck, and nonmelanoma skin cancers, in humans. The viral E2 protein is the main regulator of the papillomavirus life cycle. E2 is a modular sequence-specific DNA-binding protein with an N-terminal transactivation domain, a central hinge region, and a C-terminal DNA-binding and dimerization domain (DBD) (18). E2 recognizes the palindromic DNA motif ACCGN4CGGT, which is present in multiple copies within the upstream regulatory region (URR) of the viral genome (3, 21, 34). Interaction with these motifs enables the E2 protein to recruit viral helicase E1 to the origin during the initiation of viral DNA replication (10, 53) and tether viral episomes to mitotic chromosomes or other cellular structures in order to ensure nuclear retention during cell division (5, 23). In addition, E2 functions as a transcription factor and regulates papillomavirus early promoter activity in concert with cellular proteins (11, 43, 50).
E2 binds to DNA as a dimer with an antiparallel β-barrel structure; a surface-exposed α-helix from each of the monomers makes sequence-specific contacts with the E2 binding site (E2BS) half-site (ACCG) (19). The 4-nucleotide spacer—N4—separating the half-sites is conserved in length and influences E2-binding affinity, although the protein does not make direct contacts with these nucleotides. E2BSs in HPV genomes have A/T-rich spacers (45), and the corresponding E2 proteins generally bind to such sites with a higher affinity due to sequence-dependent conformational characteristics and flexibility of the DNA (20, 28). Altering the spacer sequence can decrease the affinity for E2BSs an order of magnitude in the case of HPV type 11 (HPV11) and HPV16 E2 (2, 12), 2 orders of magnitude in the case of HPV18 E2 (28), and even up to 3 orders of magnitude in the case of HPV6 E2 (12).
E2 proteins associate with the cellular chromatin throughout the cell cycle. Association with mitotic chromosomes ensures nuclear retention of the viral episomes and has been characterized extensively for different papillomaviruses; however, E2 interaction with chromatin in interphase cells has only been studied in the case of a few PV types. Previous studies in our lab have shown that a fraction of the bovine papillomavirus type 1 (BPV1) E2 protein in cells associates with active chromatin regions and that the N-terminal transactivation domain of E2 is responsible for this association (30). Similarly, Jang and others have demonstrated by chromatin immunoprecipitation (ChIP)-on-chip approach that BPV1 and HPV1a E2 proteins associate with active cellular promoters together with cellular Brd4 protein (24). This interaction enables the virus to avoid transcriptional silencing by targeting the viral genome to functionally active nuclear regions.
A few reports indicate that E2 proteins can also associate with cellular chromatin directly, in a sequence-specific manner, without the mediation of cellular proteins. Horner and DiMaio have demonstrated that an endonuclease with a BPV1 E2 DBD targets the integrated viral URR in HeLa cells, and an additional genomic locus on human chromosome 13 that contains an E2BS (22). In addition, HPV8 E2 has been shown to regulate two cellular genes that contain short E2BS consensus sites in their promoter region—ITGB4 and MMP9 (1, 41, 42). E2 binds to the corresponding DNA sequences in vitro and is able to repress ITGB4 and activate MMP9 in human keratinocytes. Intact transactivation and DNA-binding domains of HPV8 E2 are necessary for MMP9 activation, indicating that the E2BSs are involved in cellular promoter regulation by E2.
In this study, we have analyzed the occurrence and functionality of E2BSs in the human genome. Our computational analysis indicates that most E2BSs in the human genome are found in repetitive DNA regions and have G/C-rich spacer sequences. Experiments targeting a subset of the cellular E2BSs show that HPV11 E2 interacts with E2BSs within transcriptionally active chromatin regions and that E2 sequence-specific DNA-binding activity is necessary for this interaction. However, exogenous expression of the viral E2 protein does not change the cellular transcript levels of genes located in the proximity of E2BSs. E2BSs with A/T-rich spacers, which are present in the HPV genomes, occur rarely in the human genome compared to E2BSs with G/C-rich spacers. We suggest that the preference of HPV E2 proteins for E2BSs with A/T-rich spacers is useful for long-term maintenance of papillomavirus episomes, as it ensures preferential E2 binding to high-affinity viral E2BSs rather than abundant nonoptimal cellular E2BSs.
MATERIALS AND METHODS
Chromosome sequences and repeat masking.
For the main part of our analysis, we used the reference genome sequence obtained from the Ensembl database, version 60.37e (16). FASTA files with assembled chromosomes and chromosomes masked by RepeatMasker (http://www.repeatmasker.org) were obtained from the NCBI FTP server (46). Genome versions Human_37 and Btau_4.2 were used. Sequence regions around double binding site locations (1,000 nucleotides upstream and downstream) were analyzed with Tandem Repeats Finder (7).
Counting the observed and expected occurrences of E2BS.
We searched for the occurrence of E2BS in chromosome sequences with a Python script. Consensus sequence 5′-ACCGnnnnCGGT-3′ regular expression was used to find the observed number of sites in the genome. The distribution of distance between two successive sites was obtained by measuring nucleotide distance from the beginning of one site to the beginning of another. If a poly-N stretch longer than 10,000 nucleotides was found in the sequence between two sites, the sites were considered nonsuccessive.
The expected frequency μi of a given motif in sequence i was calculated using Markov model order 1 (44). Markov transition probabilities were obtained from dinucleotide frequencies in the genomic sequence. Genome length li was calculated from chromosome sequences discarding unknown N nucleotides. The expected motif count λi in sequence i is liμi. The statistical significance of the difference between observed and expected counts was evaluated using Poisson compound distribution with the mean value of λi.
Plasmids.
HPV11 E2 expression plasmid pQM11E2co encodes E2 open reading frame (ORF) with a codon-optimized N terminus allowing for a higher E2 expression level in human cell lines and has been described earlier (31). pQM11E2co_N294A encodes HPV11 E2 with a single amino acid substitution in the E2 DNA recognition helix. An asparagine-to-alanine substitution at position 294 was introduced into the plasmid by primer-directed mutagenesis using primer CAAGGTGATTCCGCTTGTTTAAAATGT. pQM11E2co_R37A/I73A encodes HPV11 E2 with an arginine-to-alanine substitution at position 37 (GAAGTGCATCGCGCTGGAAAGCGTGC) and an isoleucine-to-alanine substitution at position 73 (ACAACGCCGCCGAGATGCAGATGCAC), which were introduced into the plasmid using the primers listed above (the mutated nucleotides are in bold).
Cell lines and transfection.
SiHa and HaCaT cells were grown at 37°C in a 5% CO2 atmosphere in Iscove's modified Dulbecco's medium supplemented with 10% fetal calf serum and 100 U/ml penicillin and 50 μg/ml streptomycin. Cells were transfected by the electroporation method according to a previously described protocol (53). Electroporation was carried out with a Bio-Rad Gene Pulser at a capacitance setting of 975 μF and voltages of 220 and 210 V for SiHa and HaCaT cells, respectively.
ChIP.
SiHa cells were electroporated with 500 ng and HaCaT cells with 1,000 ng of empty vector, pQM11E2co, or pQM11E2co_N294A. Twice the amount of pQM11E2co_R37A/I73A plasmid was used in order to achieve similar expression levels of the wild-type (WT) and mutated E2 proteins. Twenty-four hours laters cells were fixed and ChIP was performed according to the Upstate ChIP assay protocol. Chromatin was immunoprecipitated from ∼2 × 106 to 3 × 106 cells with 4 μg of antibody. E2 was immunoprecipitated with 2 μg each of mouse monoclonal anti-HPV11 E2 antibody 10E12 (Institute of Technology, University of Tartu) and mouse monoclonal anti-E2Tag antibody 5E11 (Icosagen) (29). Histones were immunoprecipitated with antibodies for H3 (ab1791; Abcam), H3Ac (06-599; Millipore), and H3dimeK4 (07-030; Millipore). Brd4 was immunoprecipitated with 2 μg each of rabbit polyclonal antibodies Brd4B and Brd4C (47). DNA was extracted from immunoprecipitated and input material and analyzed in triplicate by quantitative PCR (qPCR) on the 7900HT real-time PCR system (Applied Biosystems). The PCRs were set up in 10 μl and contained 1/75 of the immunoprecipitated DNA or 1/300 of the input DNA and primers at a concentration 300 nM. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), HMGB2, PCDHA, and ARHGAP44 reaction mixtures were set up with 5 μl of Maxima SYBR green/ROX qPCR Master Mix (2×; Fermentas) and run with the following PCR program: 95°C for 10 min; 40 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s; and a dissociation stage consisting of 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s. The rest of the reaction mixtures contained 2 μl of 5× HOT FirePol EvaGreen qPCR Mix (Solis Biodyne) and were amplified with a PCR program of 95°C for 15 min; 40 cycles of 95°C for 15 s and 60°C for 60 s; and a dissociation stage consisting of 95°C for 15 s, 60°C for 15 s, and 95°C for 15 s. No-template controls were included in each assay, and amplification specificity was confirmed by dissociation curve analysis. The enrichment in immunoprecipitates was expressed as a percentage of the input material. Primer sequences are provided in Table S3 in the supplemental material.
Coimmunoprecipitation.
SiHa cells were electroporated with 500 ng of empty vector pQM11E2co or pQM11E2co N294A or 1,000 ng of pQM11E2co R37A/I73A. Twenty-four hours later, cells were lysed in lysis buffer (0.1 M KCl, 20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 10% glycerol, 0.1% Tween 20, 0.5 mM dithiothreitol, protease inhibitors) and sonicated briefly. Lysates were incubated on ice for 15 min and cleared by centrifugation. One-twentieth of the supernatant was saved for input control; the rest was incubated with anti-Brd4 antibodies Brd4B and Brd4C (47) (2 μg of each) for 2 h at 4°C with end-over-end rotation. Immunocomplexes were collected by the addition of protein G-Sepharose (Amersham Biosciences) beads. The samples were incubated for 2 h with end-over-end rotation at 4°C, washed four times with lysis buffer, resuspended in SDS loading buffer, and heated for 10 min at 95°C.
Western blotting.
Coimmunoprecipitation samples were separated electrophoretically in a 12% polyacrylamide–SDS gel; 1/40 of the input and half of the immunoprecipitated material were used. For Western blot analysis of the cells used in the ChIP assay, total protein from ∼105 cells was analyzed. Following electrophoresis, the samples were transferred to a polyvinylidene difluoride membrane (Millipore) by a semidry blotting method. Membranes were incubated with mouse monoclonal anti-HPV11 E2 antibody 10E12 (2,5 μg/ml), mouse monoclonal anti-β-actin (1/104; A2228; Sigma-Aldrich), or anti-α-tubulin antibody (1/104; B-512; Sigma-Aldrich); primary antibodies were visualized with secondary anti-mouse IgG antibodies conjugated with horseradish peroxidase (1/104; Icosagen). Signals were detected using an ECL detection kit following the recommendations of the provider (Amersham Biosciences).
mRNA analysis.
RNA was isolated from 106 cells using TRIzol reagent (Invitrogen) according to the protocol provided by the manufacturer. One microgram of total RNA was used in a reverse transcription reaction with oligo(dT)18 primers using a First Strand cDNA synthesis kit and following the manufacturers recommendations (Fermentas). cDNA was analyzed by qPCR on a 7900HT real-time PCR system (Applied Biosystems). Each reaction was set up in triplicate in a 10-μl reaction volume that contained 2 μl of 5× HOT FirePol EvaGreen qPCR Mix (Solis Biodyne), 1/100 of the synthesized cDNA, and primers at a concentration 300 nM. No-reverse-transcriptase and no-template control samples were included in each assay to check for genomic DNA and contamination in buffers. Amplification specificity was further confirmed by PCR product dissociation curve inspection. The threshold cycle values were recorded, and cDNA levels were determined by the relative quantitation method using the cellular HPRT1 as the reference and setting the sample transfected with an empty vector as the calibrator.
RESULTS
Papillomavirus E2BSs are found mainly in repetitive human genome regions and have G/C-rich spacer sequences.
We examined the occurrence of E2BS consensus sequence ACCGN4CGGT in the human reference genome and found that it occurs 3,388 times. Next, we compared the observed number of E2BSs with the number of binding sites that are expected to occur by chance. The expected number of E2BSs was calculated using the order 1 Markov model, which takes into account dinucleotide frequencies. This is particularly important for DNA motifs that contain CpG dinucleotides, such as E2BSs, since CpG dinucleotides are significantly underrepresented in the human genome. According to this model, the expected number of E2BSs in the human genome is only 1,094, while the observed number—3,388—is significantly higher.
To further elucidate the source of such abundance, the distribution of E2BSs in respect to repetitive DNA regions was studied. The human genome was masked with two repeat detection software programs—RepeatMasker and Tandem Repeat Finder. Fifty-two percent of the entire human genome sequence was marked as repetitive DNA by RepeatMasker. Analysis of E2BSs revealed that 79% of the E2BSs are located in the repeated regions of the human genome while only 21% are in nonrepeated regions (Table 1). Comparison of the observed and expected numbers of E2BSs in nonrepeated genome regions confirmed that E2BSs appear approximately at the expected frequency when repetitive DNA regions are excluded from the analysis (1.4-fold overrepresentation).
Table 1.
Observed and expected frequencies of E2BSs in the human reference genome
| Region | Binding site | Observed frequency | % of total | Expected frequencya | Overrepresentation (fold) | P valueb |
|---|---|---|---|---|---|---|
| All | HPV E2 | 3,388 | 100 | 1,094 | 3.1 | 0 |
| Nonrepeated | HPV E2 | 697 | 21 | 489 | 1.4 | 6E-19 |
| Repeated | HPV E2 | 2,691 | 79 | 632 | 4.3 | 0 |
| All | HPV E2 ×2 | 16 | 100 | 2.00 | 8.0 | 5E-10 |
| Nonrepeated | HPV E2 ×2 | 3 | 19 | 0.18 | 16.7 | 8E-04 |
| Repeated | HPV E2 ×2 | 13 | 81 | 1.82 | 7.5 | 4E-08 |
The expected count was calculated using dinucleotide frequencies.
The P value indicates whether the observed count of E2BSs is significantly different from the expected number of E2BSs. Significance was evaluated using Poisson compound distribution.
Multiple E2BSs allow cooperative E2 binding and can serve as E2-dependent enhancers (49, 51). Further analysis of E2BSs in the human genome revealed that there are 13 genomic locations where at least two E2BSs are found within 500 nucleotides. In human chromosome 17, there are 6 consecutive E2BSs separated by 26 nucleotides, making the number of double sites 16 (Table 1; see Table S1 in the supplemental material). Comparing this number with the expected frequencies of double sites indicates that the double E2BSs are present in the human genome more frequently than expected. It would be highly unlikely to find any double E2BSs in nonrepeated regions of the genome, yet we detected 3 such sites.
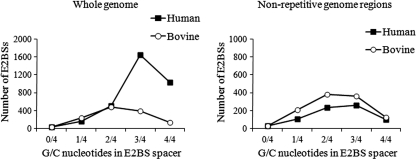
The affinity of E2 protein for E2BSs depends on the spacer sequence that separates the two half-sites of E2BS. HPV E2 proteins bind with a higher affinity to sites with A/T-rich central spacers, while BPV1 E2 has no such preference (12). Analysis of E2BSs present in the human genome showed that most of these sites have spacer sequences with high G/C content (Fig. 1). There are only 34 E2BSs in the human genome that contain only A and T nucleotides in the spacer region (see Table S2 in the supplemental material). For comparison, we analyzed the distribution of E2BSs in the bovine (Bos taurus) genome. The bovine genome contains 1,263 E2BSs, and only 13% of these sites are located in repeated regions while 40% of the bovine genome is masked by the RepeatMasker. The analysis of E2BS spacer sequences revealed that 41% of the E2BSs in the bovine genome have a high G/C content (3 or 4 G/C nucleotides), while the respective number in human genome is 80% (Fig. 1).
Fig 1.
E2BSs in the human genome have G/C-rich spacer sequences. Analysis of E2BS spacer sequences in the whole genomes and nonrepetitive regions of the human and bovine genomes.
In summary, there are over 3,000 E2BSs in the human genome; however, most of these are nonoptimal for HPV E2 protein binding because these E2BSs are either located in repetitive DNA regions or have G/C-rich spacer sequences.
HPV11 E2 protein associates with a subset of E2BSs present in the human genome in vivo.
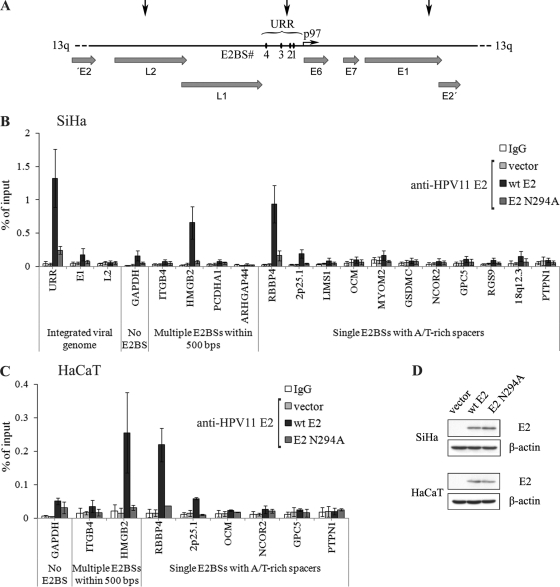
Our computational study detected 34 high-affinity E2BSs with A/T-only central spacers and 13 genomic sites where there are multiple E2BSs within 500 nucleotides. In order to study HPV E2 protein binding to cellular E2BSs determined by in silico analysis, ChIP analysis in transiently transfected human cell lines was performed. HPV11 E2 was used for this analysis due to the availability of ChIP-compatible antibodies against HPV11 E2 protein. First, the ChIP analysis was performed with the HPV-positive human cervical carcinoma-derived cell line SiHa, which contains 1 or 2 copies of HPV16 genome in the long arm of chromosome 13 (39) (Fig. 2A). In SiHa cells, the E2 ORF is disrupted; however, the viral URR and the ORFs encoding E6 and E7 are intact and the early promoter is active (4), as expression of viral oncogenes is essential for cell viability (25). The integrated viral sequences provide excellent controls for our ChIP assay, and E2 binding to three different HPV16 genome regions was analyzed, i.e., the viral URR, which contains E2BSs; and sequences located ∼1.5 kb downstream and upstream in the E1 and L2 coding regions, respectively. SiHa cells were transfected with either an HPV11 E2 expression plasmid or the same amount of an empty vector and fixed with formaldehyde 24 hours later. ChIP was performed with nonspecific IgG or anti-E2 antibodies, and immunoprecipitated DNA was analyzed by qPCR along with input DNA. As shown in Fig. 2B, the integrated HPV16 genome URR, but not E1 and L2 coding regions, was immunoprecipitated with anti-E2 antibodies, as expected. These data confirmed that E2 is able to bind specifically to E2BSs located in the viral early promoter in the context of chromatin and that our anti-E2 antibodies are suitable for use in a ChIP assay.
Fig 2.
HPV11 E2 associates with the integrated viral URR in SiHa cells and additional cellular chromatin regions that contain E2BSs in SiHa and HaCaT cells. (A) Schematic representation of the integrated HPV16 genome copy in chromosome 13. Three viral genome regions indicated with black arrows—URR and coding regions of E1 and L2—were used as controls in our ChIP assay. (B) SiHa and (C) HaCaT cells were electroporated with an empty vector or an HPV11 WT E2 or an HPV11 N294A E2 expression vector. Twenty-four hours later, cells were fixed and chromatin immunoprecipitated with nonspecific IgG or anti-E2 antibodies. DNA was extracted from immunoprecipitates and input fractions and analyzed by qPCR. Enrichment in immunoprecipitated fractions was calculated as a percentage of the input material. The data represent the average of three independent experiments and the standard deviation. The nonspecific IgG values of vector, WT, and N294A samples were similar, and the corresponding data reflect the average of all three samples. (D) Western blot analysis of E2 levels in the cells used in ChIP assays. Cells (105) were lysed, separated electrophoretically, and immunoblotted with a primary mouse monoclonal antibodies for HPV11 E2 and β-actin and an anti-mouse IgG secondary antibody conjugated with horseradish peroxidase.
Next, E2 binding to cellular DNA loci that contain one or two E2BSs was analyzed. We included 3 chromatin regions with multiple E2BSs within 500 nucleotides and 11 regions, out of a total of 34, that contain a single E2BS with an A/T-only spacer. These E2BSs, their locations, and their distances from the closest gene transcription start site (TSS) are shown in Table 2. Most of these sequences are located within coding or promoter regions of cellular genes and are referred to according to the closest gene symbol. Two sites—2p25.1 and 18p12.3—are present in intergenic regions and were named after the corresponding chromatin bands. In addition, the promoter regions of the genes for GAPDH and ITGB4 were included for comparison. The promoter of the gene for GAPDH is an active cellular promoter that does not contain any E2BSs, whereas the ITGB4 promoter region contains three short E2BS consensus sequences that, in some cell lines, are involved in ITGB4 transcriptional repression by the HPV8 E2 protein (41, 42). The ChIP assay revealed that HPV11 E2 protein binds to two genomic sites that contain E2BSs—HMGB2 and RBBP4—in SiHa cells (Fig. 2B). The HMGB2 locus contains two E2BSs separated by 126 nucleotides, and RBBP4 contains a single high-affinity E2BS. The rest of the sites displayed only a low level of enrichment in the HPV11 E2 sample compared to the control samples. Similar results were obtained in another human cell line—spontaneously immortalized human keratinocyte cell line HaCaT—which does not contain any HPV sequences (Fig. 2C).
Table 2.
Viral and genomic DNA regions analyzed by ChIP assay
| Chromosome band | Gene name (HGNC) | Protein | E2BS within locus | E2BS distance (bp) from TSS |
|---|---|---|---|---|
| 13q | HPV16 URR | aACCGatttTGGGt-82 bp-aACCGaaatCGGTt-15 bp-aACCGaaacCGGTt | −21 | |
| 13q | HPV16 E1 | HPV16 E1 | ||
| 13q | HPV16 L2 | HPV16 L2 | ||
| 12p13.31 | GAPDH promoter | Glyceraldehyde-3-phosphate dehydrogenase | ||
| 17q25.1 | ITGB4 | Integrin beta-4 | gACCtccctgGGTt-457 bp-gACCaggaaaGGTg-163 bp-cACCaggcatGGTg | −1,469 |
| 4q34.1 | HMGB2 | High-mobility group protein B2 | cACCGagtcCGGTg-126 bp-aACCGggccCGGTc | +275 |
| 5q31.3 | PCDHA12 | Protocadherin alpha 12 | tACCGcggtCGGTg-93 bp-aACCGgcggCGGTc | +1,869 |
| 17p12 | ARHGAP44 | Rho GTPase-activating protein | cACCGctgtCGGTg-355 bp-aACCGcagtCGGTc | +397 |
| 1p35.1 | RBBP4 | Retinoblastoma binding protein 4 | gACCGatttCGGTc | +273 |
| 1p35.1 | ZBTB8OS | Zinc finger and BTB domain containing 8 opposite strand | gACCGatttCGGTc | −980 |
| 2p25.1 | Intergenic | gACCGaattCGGTc | ||
| 2q12.3 | LIMS1 | LIM and senescent cell antigen-like-containing domain protein 1 | aACCGaaatCGGTg | +77,891 |
| 7p22.1 | OCM | Oncomodulin (parvalbumin beta) | aACCGaaaaCGGTg | −836 |
| 8p23.3 | MYOM2 | Myomesin 2 | aACCGatttCGGTa | +3,504 |
| 8q24.21 | GSDMC | Gasdermin C | tACCGttatCGGTa | +38,732 |
| 12q24.31 | NCOR2 | Nuclear receptor corepressor 2 isoform 1 | tACCGaaaaCGGTa | +184,407 |
| 13q31.3 | GPC5 | Glypican 5 | tACCGaaaaCGGTa | +798,339 |
| 17q24.1 | RGS9 | Regulator of G protein signaling 9 isoform 2 | tACCGattaCGGTa | +23,147 |
| 18q12.3 | Intergenic | aACCGaaaaCGGTa | ||
| 20q13.13 | PTPN1 | Protein tyrosine phosphatase, nonreceptor type | tACCGtaatCGGTt | +46,285 |
Sequence-specific DNA-binding activity of HPV11 E2 protein is necessary for binding to cellular E2BSs.
BPV1 and HPV1a E2 proteins have been shown to associate with active cellular promoters without the presence of E2-specific motifs in cellular DNA (24). In order to determine whether HPV11 E2 binding to cellular chromatin depends on E2 sequence-specific interaction with E2BSs within the cellular DNA loci, we generated an expression plasmid encoding N294A mutant E2 with a single amino acid substitution in the DNA recognition helix. Mutating asparagine at position 294 of HPV11 E2 to alanine has been shown to abolish HPV11 E2 DNA-binding activity (37), and the corresponding mutation in HPV16 E2 results in a 20-fold decrease in DNA-binding activity (15). Both the WT and N294A mutant E2 proteins localized in the nucleus of the cell, as determined by immunofluorescence analysis (data not shown). N294A mutant E2 was inefficient in supporting initiation of HPV11 origin-containing DNA replication in transient replication assay in U2OS cells (data not shown), indicating that N294A is defective in DNA binding.
In the ChIP assay, N294A mutant E2 bound to the E2BSs within the viral URR in SiHa cells less efficiently than WT E2 (Fig. 2B), although these E2 proteins were expressed at similar levels (Fig. 2D). Similarly, cellular E2BSs within HMGB2 and RBBP4 loci were immunoprecipitated less efficiently with N294A E2 than with WT E2 in both the SiHa and HaCaT cell lines (Fig. 2B and C), indicating that E2 binds directly to specific binding sites within cellular DNA.
HPV11 E2 protein binds E2BSs located in open chromatin regions.
The availability of regulatory sequences in the cellular genome is determined by the chromatin organization surrounding these elements. Access to specific DNA motifs depends on several chromatin structural features, such as nucleosome positioning and density, and posttranslational modifications of the constituent histones. In order to characterize the chromatin structure of the genomic DNA surrounding the E2BSs, we have analyzed the amount of H3, which indicates nucleosomal occupancy, and two different histone modifications associated with transcriptionally active chromatin—H3ac and H3dimeK4—in the corresponding chromatin regions in SiHa and HaCaT cells. ChIP analysis with anti-H3 antibodies showed that the H3 signal varied drastically between the chromatin regions investigated in this study (Fig. 3A). The promoter region of the actively transcribed cellular GAPDH-encoding gene exhibited a very low H3 signal level in both cell lines tested, in contrast to intergenic region 2p25.1, where the H3 signal was significantly higher. Low H3 occupancy was also observed in the URR, HMGB2, and RBBP4 loci (Fig. 3A), where E2BSs are located within regulatory regions—21 nucleotides upstream from the TSS in the case of the URR and 275 and 273 nucleotides downstream from the TSS in the cases of HMGB2 and RBBP4, respectively. The genomic DNA loci that exhibited low H3 occupancy had high levels of histone modifications indicative of transcriptionally active chromatin (Fig. 3B). Interestingly, the viral E1 region carries higher levels of open chromatin markers than the L2 region does, although these sites are located within the same distance from the TSS of the viral early promoter that controls the expression of oncogenes E6 and E7. The remaining genomic loci showed high H3 occupancy and very low levels of H3ac and H3dimeK4 modifications.
Fig 3.
HPV11 E2 protein interacts with cellular E2BSs that are present in transcriptionally active chromatin regions. (A) SiHa or HaCaT cells were fixed, and chromatin was immunoprecipitated with H3 antibodies. DNA was extracted from immunoprecipitates and input fractions and analyzed by qPCR. Enrichment in immunoprecipitated fractions was calculated as a percentage of the input material. The data represent the average of three independent experiments and the standard deviation. (B) SiHa or HaCaT cells were fixed, and chromatin was immunoprecipitated with H3Ac and H3diMeK4 antibodies. DNA was extracted from immunoprecipitates and input fractions and analyzed by qPCR. Enrichment is presented relative to the H3 signal for the corresponding chromatin regions. The data represent the average of three independent experiments and the standard deviation.
These data suggest that E2 interaction with cellular E2BSs correlates with transcriptionally active chromatin sites. E2 binds to cellular E2BSs in chromatin regions that exhibit low nucleosomal occupancy and have high levels of histone modifications indicative of transcriptionally active chromatin.
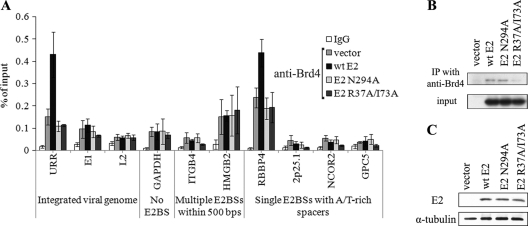
Chromatin adaptor proteins that associate with specific histone modifications can recruit sequence-specific DNA-binding factors to the corresponding chromatin regions. For papillomavirus E2 proteins, the bromodomain protein Brd4, which binds to acetylated lysines, has been identified as the major cellular interaction partner (54, 55). All papillomavirus E2 proteins interact with Brd4, and this interaction is important for E2 transcription regulatory function (38). Association with Brd4 helps to recruit HPV11 E2 to viral chromatin, enhances site-specific DNA recognition of E2, and stabilizes the viral protein (32, 55). Our ChIP assay with anti-Brd4 antibodies showed that Brd4 is bound to genomic loci positive for acetylated H3 in SiHa cells (Fig. 4A). Brd4 association with chromatin was further enhanced in the viral URR and RBBP4 region by the expression of WT E2 but not by that of N294A E2. Both the WT E2 and N294A E2 proteins were coimmunoprecipitated with endogenous Brd4 in SiHa cells (Fig. 4B), indicating that this amino acid substitution does not affect E2 interaction with Brd4, which is mediated by the N-terminal transactivation domain of E2.
Fig 4.
E2 binding enhances the chromatin association of Brd4. (A) SiHa cells were electroporated with an empty vector or an HPV11 WT E2, an HPV11 N294A E2, or an HPV11 R37A/I73A E2 expression vector. Twenty-four hours later, cells were fixed and chromatin immunoprecipitated with nonspecific IgG or anti-Brd4 antibodies. DNA was extracted from immunoprecipitates and input fractions and analyzed by qPCR. Enrichment in immunoprecipitated fractions was calculated as a percentage of the input material. The data represent the average of three independent experiments and the standard deviation. The nonspecific IgG values represent the average of vector, WT, N294A, and R37A/I73A samples. (B) SiHa cells were electroporated with an empty vector or an HPV11 WT E2, an HPV11 N294A E2, or an HPV11 R37A/I73A E2 expression vector. Twenty-four hours later, cells were lysed and E2 was coimmunoprecipitated (IP) with anti-Brd4 antibodies. Immunoprecipitates and input fractions were separated electrophoretically and immunoblotted with primary mouse monoclonal antibodies for HPV11 E2 and an anti-mouse IgG secondary antibody conjugated with horseradish peroxidase. (C) Western blot analysis of E2 levels in the cells used in ChIP assay. Cells (105) were lysed, separated electrophoretically, and immunoblotted with primary mouse monoclonal antibodies for HPV11 E2 and an anti-mouse IgG α-tubulin and secondary antibody conjugated with horseradish peroxidase.
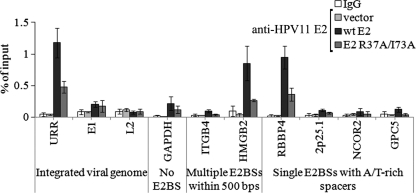
In order to further investigate Brd4 involvement in HPV11 E2 protein interaction with consensus E2BSs, we generated an E2 expression plasmid encoding HPV11 E2 mutant R37A/I73A. Residues R37 and I73 are important for E2 interaction with Brd4, and replacing these amino acids with alanines is known to reduce the Brd4-binding efficiency of E2 proteins from different papillomavirus types (38). R37A/I73A E2 was coimmunoprecipitated with endogenous Brd4 less efficiently than WT E2 (Fig. 4B) and was unable to enhance Brd4 binding to the URR and RBBP4 region in SiHa cells (Fig. 4A). ChIP analysis with R37A/I73A E2 and WT E2 in SiHa cells using anti-E2 antibodies revealed that disrupting the E2 interaction with Brd4 reduces E2 binding to both viral and cellular E2BSs (Fig. 5). The URR and the RBBP4 and HMGB4 regions were immunoprecipitated in R37A/I73A E2 samples less efficiently than in WT E2 samples, although the proteins were expressed at similar levels in these cells (Fig. 4C). However, this reduction was less severe than in the case of N294A E2, which is deficient in sequence-specific DNA-binding activity (compare Fig. 2B and 5). Our results indicate that both E2 activities—interaction with cellular Brd4 and sequence-specific DNA-binding activity—are important for binding to E2BSs within the chromatin context.
Fig 5.
Disruption of E2 interaction with Brd4 reduces E2 binding to E2BSs within the chromatin context. SiHa cells were electroporated with an empty vector or an HPV11 WT E2 or an HPV11 R37A/I73A E2 expression vector. Twenty-four hours later, cells were fixed and chromatin immunoprecipitated with nonspecific IgG or anti-E2 antibodies. DNA was extracted from immunoprecipitates and input fractions and analyzed by qPCR. Enrichment in immunoprecipitated fractions was calculated as a percentage of the input material. The data represent the average of three independent experiments and the standard deviation. The nonspecific IgG values represent the average of vector, WT, and R37A/I73A samples.
E2 binding to cellular E2BSs does not alter cellular transcription.
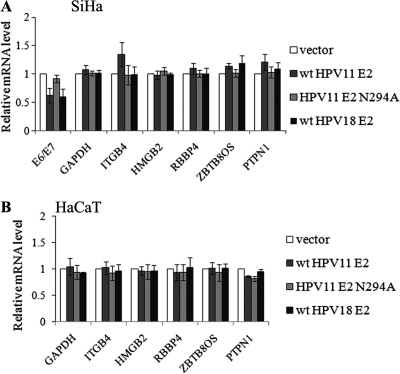
Our next goal was to determine whether direct binding of E2 to cellular E2BSs causes changes in cellular transcript levels. In SiHa cells, E2 expression leads to repression of early viral promoters (13, 14, 48), and transcripts for viral oncogenes E6 and E7 provided controls in our mRNA analysis. Cellular transcripts of the genes for GAPDH, ITGB4, HMGB2, RBBP4, ZBTB8OS, and PTPN1 were analyzed; the rest of the genes corresponding to the genomic loci analyzed in the ChIP assay are expressed at a very low level or not at all in these cell lines, since we could not detect their transcripts in either SiHa or HaCaT cells (data not shown). The GAPDH coding region does not contain any E2BSs, the gene for HMGB2 has two E2BSs separated by 126 nucleotides in the first intron, and the gene for RBBP4 contains a single E2BS in the first intron. The gene for ZBTB8OS is located just upstream from that for RBBP4 and is in the reverse strand. The E2BS located in the first intron of the gene for RBBP4 is situated 980 nucleotides upstream from the ZBTB8OS TSS. PTPN1 contains a single E2BS in the last exon. mRNA analysis was carried out with SiHa and HaCaT cells 24 h after transfection with expression vectors for the low-risk HPV11 WT and N294A E2 proteins and high-risk HPV18 E2 protein. Twenty-four hours later, RNA was extracted, reverse transcribed, and analyzed by qPCR. The cellular mRNA levels were normalized against HPRT1 and presented in relation to the sample transfected with empty vector. As shown in Fig. 6A, the viral E6/E7 transcript was suppressed by the WT HPV11 and HPV18 E2 proteins but not by HPV11 N294A E2, which is defective in sequence-specific DNA binding. The expression of E2 did not alter any cellular transcript levels in SiHa or HaCaT cells (Fig. 6). The cellular transcript levels were not affected over a wide range of E2 concentrations in SiHa or HaCaT cells, nor did we observe changes in cellular transcript levels in a U2OS cell line stably expressing E2 (data not shown). These data indicate that E2 sequence-specific binding to these cellular E2BSs has no effect on the expression of cellular genes.
Fig 6.
HPV11 E2 association with E2BSs does not result in changes in cellular transcript levels. SiHa and HaCaT cells were transfected with an empty vector or an HPV11 WT E2, an HPV11 N294A E2, or an HPV18 WT E2 expression vector. Twenty-four hours later, total RNA was isolated, reverse transcribed, and analyzed by qPCR. mRNA levels were calculated relative to the empty-vector sample, and HPRT1 was used as the reference. The data represent the average of three independent experiments and the standard deviation.
DISCUSSION
Papillomaviruses are able to establish persistent infection by replicating and maintaining their DNA as episomes in host cells. During this period, the viral E2 protein, which binds sequence specifically to DNA with a high affinity, is expressed at low levels. E2 protein is required for the viral genome replication and maintenance and has to function without damaging the host cell by causing global changes in cellular gene expression in the persistent phase of the viral life cycle. In order to better understand the molecular basis of such cooperation, we have analyzed the occurrence and functionality of specific E2BSs in the human genome.
Our computational analysis revealed that there are over 3,300 E2BSs—palindromic 12-bp DNA sequences ACCGN4CGGT—in the human reference genome. Only about 1,000 such motifs are expected to occur by chance, which indicates that E2BSs are three times overrepresented in the human genome. A similar analysis of the bovine genome, which is comparable in size to the human genome, revealed that the number of E2BSs is much lower and close to expected in this species. Furthermore, most of the E2BSs in the human genome have spacer sequences with high G/C content, while the spacer sequences of E2BSs in the bovine genome do not have such a distribution. We suggest that these two factors—the high number of cellular E2BSs and the high G/C content of these sites—may explain the HPV E2 protein preference for A/T-rich E2BSs. It is possible that E2BSs in HPV genomes have changed during viral evolution so that they would differ from those of the host cell. This difference may guarantee that E2 protein binds to the viral high-affinity E2BSs, rather than abundant cellular low-affinity E2BSs, and thereby ensures the replication and maintenance of the HPV genomes.
In the present study, we have investigated the binding of the HPV11 E2 protein to cellular E2BSs and its ability to discriminate between viral and cellular E2BSs in the context of chromatin. Using a ChIP approach, we demonstrated that the HPV11 E2 protein binds sequence specifically to at least 2 of the 15 cellular E2BSs tested. E2 binds to E2BSs within the integrated viral URR in SiHa cells and to cellular E2BSs located in transcriptionally active chromatin regions within the highly expressed cellular genes for HMGB2 and RBBP4. Jang et al. have previously shown that BPV1 E2 associates with transcriptionally active cellular chromatin, including the HMGB2 and RBBP4 promoter regions (24). However, Jang et al. focused on E2 association with cellular chromatin rather than direct binding to DNA. They showed that the BPV1 E2 protein binds to cellular chromatin independently of the presence of E2BSs (24). E2 binding to DNA and chromatin is mediated by different interactions and through different protein domains; E2 binds to DNA through its DNA-binding domain but associates with cellular chromatin through its activation domain. We show that HPV11 E2 protein binding to HMGB2 and RBBP4 is dependent on the sequence-specific DNA recognition activity of E2. Furthermore, HPV11 E2 binds to these sites with notably higher efficiency than to the cellular promoter region of GAPDH, which does not contain E2BSs. In the work of Jang et al., the level of BPV1 E2 binding to E2 consensus sites was very similar to that of the active promoters without consensus sites (24). We show that two E2 activities mediated by different E2 domains—sequence-specific binding to DNA and interaction with the cellular Brd4 protein—are both important for HPV11 E2 protein binding to E2BSs within the chromatin context and suggest that these interactions contribute additively to strong, sequence-specific interaction of HPV11 E2 with E2 consensus sites.
Bioinformatic approaches can be useful for finding all of the potential recognition motifs for a given factor; however, usually only a subset of these represent binding sites in vivo. In our study, E2 was able to bind to only one of the high-affinity E2BSs with A/T-rich spacers in vivo and to at least one of the two E2BSs within the HMGB2 intron that have AGTC and GGCC spacers, indicating that only a few of the cellular E2BSs are accessible to the viral factor. This is also true for cellular transcription factors. For example, in a systematic whole-genome study of p63-binding sites in the human genome, only 8% of the best and 1 to 3% of the typical motifs were bound by p63 in vivo and only 10 to 20% of the bound targets were involved in p63-dependent transcription regulation (56). Our data show that E2 binding to cellular E2BSs in vivo correlates with open chromatin regions. E2 binds to E2BSs in chromatin regions that exhibit low histone occupancy and have high levels of acetylated and K4-dimethylated histone H3. E2 binding to E2BSs within cellular chromatin depends on several factors. First, the accessibility of E2 consensus site within the cellular chromatin influences E2 binding. Second, DNA methylation, which was not investigated in the present study, could also explain poor E2 binding to at least some of the cellular high-affinity E2BSs. E2 binding motif contains CpG nucleotides, which are methylated in 70 to 80% of the cases in mammalian genomes (35), and methylation is known to inhibit E2 binding (52). Third, the availability and interaction with cellular adaptor proteins is important for E2 binding to consensus sites within cellular chromatin.
Brd4, the cellular bromodomain protein that binds to acetylated histones, has been demonstrated to recruit HPV11 E2 to HPV chromatin (32, 55). Our data show that Brd4 is prebound to all chromatin sites that are also bound by E2 and that E2 binding to specific binding sites in cellular chromatin recruits additional Brd4 molecules to these loci, demonstrating once again the tight interaction and cross talk between these two proteins. However, in spite of E2 binding and the recruitment of additional Brd4 molecules to these chromatin regions, the mRNA levels of genes expressed from cellular promoters containing E2BSs were not affected by the expression of HPV11 and HPV18 E2 proteins. This is consistent with previous studies (24, 26) and demonstrates that papillomaviruses have developed a long-term relationship with the host cell in order to guarantee the propagation of the virus. It is beneficial for the virus to utilize the host cell components without causing global changes in cellular gene expression, which could be detrimental to host cells.
The HPV11 E2 protein binds to cellular E2BSs with an efficiency similar to that of viral E2BSs within the integrated early promoter in HPV-positive SiHa cells. This region contains the origin of virus DNA replication. Kadaja and others have demonstrated that papillomavirus replication proteins E1 and E2 utilize the integrated viral replication origin when expressed from viral episomes at low levels (27). This indicates that in the presence of viral episomes, the E2BSs within the context of cellular chromatin compete for E2 binding, which provides further support for our hypothesis that the viral protein needs to distinguish between viral and abundant cellular E2BSs.
Nuclear viruses often encode their own transcription factors, and these can interfere with normal cellular transcription in virus-infected cells. This enables the virus to directly alter cellular physiology during the course of the viral life cycle and represents an intriguing aspect of virus-host interaction. For example, the EBNA1 protein, encoded by Epstein-Barr virus, binds sequence specifically to a large number of cellular promoters and regulates their transcriptional activity (9, 36). Some studies indicate that E2 is also involved in the modulation of the cellular environment during the course of the viral life cycle (8, 17); however, in most cases, these effects are caused by protein-protein interactions between E2 and host factors rather than direct interaction with cellular DNA (6, 33, 40), and our results are in agreement with this. We suggest that HPV E2 proteins and the E2BSs within viral genomes have adapted to the host cell in order to ensure the replication and maintenance of the viral DNA and at the same time prevent extensive and possibly detrimental changes in cellular transcription in response to the viral protein.
Supplementary Material
ACKNOWLEDGMENTS
We thank Aare Abroi, Ivar Ilves, and Piia Uusen for useful discussions, Ants Kurg for support, and Märt Möls for advice in statistical methods.
This study was supported by a grant from the Estonian Science Foundation (ETF7556) to R.K., by long-term research program SF0180175 of the Estonian Ministry of Education and Research to M.U., and by the European Regional Development Fund through the Center of Excellence in Chemical Biology. A.S. and M.R. were supported by grant SF0180026s09 from the Estonian Ministry of Education and Research and by the European Regional Development Fund through the Estonian Centre of Excellence in Genomics.
Footnotes
Published ahead of print 26 October 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Akgül B, et al. 2011. The E2 protein of human papillomavirus type 8 increases the expression of matrix metalloproteinase-9 in human keratinocytes and organotypic skin cultures. Med. Microbiol. Immunol. 200(2):127–135 [DOI] [PubMed] [Google Scholar]
- 2. Alexander KA, Phelps WC. 1996. A fluorescence anisotropy study of DNA binding by HPV-11 E2C protein: a hierarchy of E2-binding sites. Biochemistry 35:9864–9872 [DOI] [PubMed] [Google Scholar]
- 3. Androphy EJ, Lowy DR, Schiller JT. 1987. Bovine papillomavirus E2 trans-activating gene product binds to specific sites in papillomavirus DNA. Nature 325:70–73 [DOI] [PubMed] [Google Scholar]
- 4. Baker CC, et al. 1987. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J. Virol. 61:962–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bastien N, McBride AA. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124–134 [DOI] [PubMed] [Google Scholar]
- 6. Behren A, et al. 2005. Papillomavirus E2 protein induces expression of the matrix metalloproteinase-9 via the extracellular signal-regulated kinase/activator protein-1 signaling pathway. Cancer Res. 65:11613–11621 [DOI] [PubMed] [Google Scholar]
- 7. Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burns JE, Walker HF, Schmitz C, Maitland NJ. 2010. Phenotypic effects of HPV-16 E2 protein expression in human keratinocytes. Virology 401:314–321 [DOI] [PubMed] [Google Scholar]
- 9. Canaan A, et al. 2009. EBNA1 regulates cellular gene expression by binding cellular promoters. Proc. Natl. Acad. Sci. U. S. A. 106:22421–22426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiang CM, Dong G, Broker TR, Chow LT. 1992. Control of human papillomavirus type 11 origin of replication by the E2 family of transcription regulatory proteins. J. Virol. 66:5224–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chin MT, Hirochika R, Hirochika H, Broker TR, Chow LT. 1988. Regulation of human papillomavirus type 11 enhancer and E6 promoter by activating and repressing proteins from the E2 open reading frame: functional and biochemical studies. J. Virol. 62:2994–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dell G, et al. 2003. Comparison of the structure and DNA-binding properties of the E2 proteins from an oncogenic and a non-oncogenic human papillomavirus. J. Mol. Biol. 334:979–991 [DOI] [PubMed] [Google Scholar]
- 13. Demeret C, Desaintes C, Yaniv M, Thierry F. 1997. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J. Virol. 71:9343–9349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dong G, Broker TR, Chow LT. 1994. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J. Virol. 68:1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferreiro DU, Dellarole M, Nadra AD, de Prat-Gay G. 2005. Free energy contributions to direct readout of a DNA sequence. J. Biol. Chem. 280:32480–32484 [DOI] [PubMed] [Google Scholar]
- 16. Flicek P, et al. 2010. Ensembl's 10th year. Nucleic Acids Res. 38:D557–D562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frattini MG, Hurst SD, Lim HB, Swaminathan S, Laimins LA. 1997. Abrogation of a mitotic checkpoint by E2 proteins from oncogenic human papillomaviruses correlates with increased turnover of the p53 tumor suppressor protein. EMBO J. 16:318–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giri I, Yaniv M. 1988. Structural and mutational analysis of E2 trans-activating proteins of papillomaviruses reveals three distinct functional domains. EMBO J. 7:2823–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hegde RS, Grossman SR, Laimins LA, Sigler PB. 1992. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature 359:505–512 [DOI] [PubMed] [Google Scholar]
- 20. Hines CS, et al. 1998. DNA structure and flexibility in the sequence-specific binding of papillomavirus E2 proteins. J. Mol. Biol. 276:809–818 [DOI] [PubMed] [Google Scholar]
- 21. Hirochika H, Hirochika R, Broker TR, Chow LT. 1988. Functional mapping of the human papillomavirus type 11 transcriptional enhancer and its interaction with the trans-acting E2 proteins. Genes Dev. 2:54–67 [DOI] [PubMed] [Google Scholar]
- 22. Horner SM, DiMaio D. 2007. The DNA binding domain of a papillomavirus E2 protein programs a chimeric nuclease to cleave integrated human papillomavirus DNA in HeLa cervical carcinoma cells. J. Virol. 81:6254–6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ilves I, Kivi S, Ustav M. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jang MK, Kwon D, McBride AA. 2009. Papillomavirus E2 proteins and the host BRD4 protein associate with transcriptionally active cellular chromatin. J. Virol. 83:2592–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang M, Milner J. 2002. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene 21:6041–6048 [DOI] [PubMed] [Google Scholar]
- 26. Johung K, Goodwin EC, DiMaio D. 2007. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J. Virol. 81:2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kadaja M, et al. 2007. Genomic instability of the host cell induced by the human papillomavirus replication machinery. EMBO J. 26:2180–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim SS, Tam JK, Wang AF, Hegde RS. 2000. The structural basis of DNA target discrimination by papillomavirus E2 proteins. J. Biol. Chem. 275:31245–31254 [DOI] [PubMed] [Google Scholar]
- 29. Kurg R, et al. 1999. Effect of bovine papillomavirus E2 protein-specific monoclonal antibodies on papillomavirus DNA replication. J. Virol. 73:4670–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kurg R, Sild K, Ilves A, Sepp M, Ustav M. 2005. Association of bovine papillomavirus E2 protein with nuclear structures in vivo. J. Virol. 79:10528–10539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurg R, Uusen P, Vosa L, Ustav M. 2010. Human papillomavirus E2 protein with single activation domain initiates HPV18 genome replication, but is not sufficient for long-term maintenance of virus genome. Virology 408:159–166 [DOI] [PubMed] [Google Scholar]
- 32. Lee AY, Chiang CM. 2009. Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J. Biol. Chem. 284:2778–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee D, et al. 2002. Human papillomavirus E2 down-regulates the human telomerase reverse transcriptase promoter. J. Biol. Chem. 277:27748–27756 [DOI] [PubMed] [Google Scholar]
- 34. Li R, Knight J, Bream G, Stenlund A, Botchan M. 1989. Specific recognition nucleotides and their DNA context determine the affinity of E2 protein for 17 binding-sites in the Bpv-1 genome. Genes Dev. 3:510–526 [DOI] [PubMed] [Google Scholar]
- 35. Lister R, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu F, et al. 2010. Genome-wide analysis of host-chromosome binding sites for Epstein-Barr virus nuclear antigen 1 (EBNA1). Virol. J. 7:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matsumoto T, Nakashima N, Takase K, Hirochika H, Mizuno H. 1997. A mutation study of the DNA binding domain of human papillomavirus type11 E2 protein. J. Biochem. 121:138–144 [DOI] [PubMed] [Google Scholar]
- 38. McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. 2006. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 80:9530–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meissner JD. 1999. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J. Gen. Virol. 80(Pt. 7):1725–1733 [DOI] [PubMed] [Google Scholar]
- 40. Mole S, Milligan SG, Graham SV. 2009. Human papillomavirus type 16 E2 protein transcriptionally activates the promoter of a key cellular splicing factor, SF2/ASF. J. Virol. 83:357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oldak M, et al. 2010. Human papillomavirus type 8 E2 protein unravels JunB/Fra-1 as an activator of the beta4-integrin gene in human keratinocytes. J. Virol. 84:1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oldak M, et al. 2004. The human papillomavirus type 8 E2 protein suppresses beta4-integrin expression in primary human keratinocytes. J. Virol. 78:10738–10746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Phelps WC, Howley PM. 1987. Transcriptional trans-activation by the human papillomavirus type 16 E2 gene product. J. Virol. 61:1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Robin S, Schbath S, Vandewalle V. 2007. Statistical tests to compare motif count exceptionalities. BMC Bioinformatics 8:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sánchez IE, Dellarole M, Gaston K, de Prat Gay G. 2008. Comprehensive comparison of the interaction of the E2 master regulator with its cognate target DNA sites in 73 human papillomavirus types by sequence statistics. Nucleic Acids Res. 36:756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sayers EW, et al. 2011. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 39:D38–D51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Silla T, Mannik A, Ustav M. 2010. Effective formation of the segregation-competent complex determines successful partitioning of the bovine papillomavirus genome during cell division. J. Virol. 84:11175–11188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smith JA, et al. 2010. Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl. Acad. Sci. U. S. A. 107:3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Spalholz BA, Byrne JC, Howley PM. 1988. Evidence for cooperativity between E2 binding sites in E2 trans-regulation of bovine papillomavirus type 1. J. Virol. 62:3143–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spalholz BA, Yang YC, Howley PM. 1985. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell 42:183–191 [DOI] [PubMed] [Google Scholar]
- 51. Tan SH, Leong LE, Walker PA, Bernard HU. 1994. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J. Virol. 68:6411–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thain A, Jenkins O, Clarke AR, Gaston K. 1996. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J. Virol. 70:7233–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ustav M, Stenlund A. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu SY, Chiang CM. 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem. 282:13141–13145 [DOI] [PubMed] [Google Scholar]
- 55. Wu SY, et al. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 20:2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang A, et al. 2006. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol. Cell 24:593–602 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.