Abstract
Dengue is a mosquito-borne viral hemorrhagic disease that is a major threat to human health in tropical and subtropical regions. Here we report crystal structures of a peptide covalently bound to dengue virus serotype 3 (DENV-3) protease as well as the serine-protease inhibitor aprotinin bound to the same enzyme. These structures reveal, for the first time, a catalytically active, closed conformation of the DENV protease. In the presence of the peptide, the DENV-3 protease forms the closed conformation in which the hydrophilic β-hairpin region of NS2B wraps around the NS3 protease core, in a manner analogous to the structure of West Nile virus (WNV) protease. Our results confirm that flavivirus proteases form the closed conformation during proteolysis, as previously proposed for WNV. The current DENV-3 protease structures reveal the detailed interactions at the P4′ to P3 sites of the substrate. The new structural information explains the sequence preference, particularly for long basic residues in the nonprime side, as well as the difference in substrate specificity between the WNV and DENV proteases at the prime side. Structural analysis of the DENV-3 protease-peptide complex revealed a pocket that is formed by residues from NS2B and NS3; this pocket also exists in the WNV NS2B/NS3 protease structure and could be targeted for potential antivirus development. The structural information presented in the current study is invaluable for the design of specific inhibitors of DENV protease.
INTRODUCTION
Dengue virus (DENV) is a member of the flavivirus genus, which includes several viruses that are important human pathogens, including yellow fever virus (YFV), Japanese encephalitis virus (JEV), West Nile virus (WNV), and tick-borne encephalitis virus (TBEV). The four serotypes of DENV are estimated to cause 50 to 100 million human infections worldwide every year in tropical and subtropical regions (16). There are currently no clinically approved vaccines or therapeutics for DENV. Understanding the molecular details of DENV infection is essential for vaccine and antiviral development.
Flaviviruses contain a single strand of positive-sense RNA that encodes three structural and seven nonstructural (NS) proteins that are translated as a single polypeptide chain. NS3 contains two functions, an N-terminal serine protease and a C-terminal RNA helicase. The catalytic triad (His51, Asp75, and Ser135) is within the NS3 protease domain, but a region of NS2B is also required for catalytic activity (15). NS2B contains three predicted transmembrane helices, ensuring that the NS2B-NS3 protease is bound to the endoplasmic reticulum. The viral polyprotein is cleaved into the individual proteins by a combination of host proteases and the viral NS2B-NS3 protease (6, 30). The proteolytic activity of the viral protease is essential for viral replication, making it an excellent antiviral target (18).
Ligand-free structures of flavivirus proteases have been solved for DENV-1 and -2 and a WNV active-site mutant (1, 7, 13). In addition, the structure of Murray Valley encephalitis virus (MVEV) protease has been solved in the context of the full-length NS3 protein (2). The proteins used for all these structural studies are catalytically competent and include the approximately 40-amino-acid region of NS2B that is required for the protease catalytic activity. However, the NS2B in these structures adopts very different conformations, with the exception of a single β strand that extends the β-sheet core of NS3 (1, 2, 7, 13, 20, 25). In the MVEV NS3 structure, the rest of NS2B is disordered (2), whereas in the DENV-1 and -2 proteases, it adopts very different conformations that are likely to be dependent on crystal-packing contacts (7, 13). Several structures of WNV protease have been solved with either peptide-based inhibitors (13, 25) or protein inhibitors (1). In these WNV structures, the NS2B region forms a β hairpin that wraps around the NS3 core in the closed conformation that is not observed in the structures of the free enzyme. The β hairpin is part of the active site and directly interacts with the substrate, explaining why this region of NS2B is required for catalytic activity (1, 13, 25). However, it is not known whether the equivalent NS2B region in DENV and other flaviviruses also adopts a similar closed conformation during protease catalysis. Until now, in silico and rational approaches to design dengue virus-specific inhibitors have relied on homology models (17, 32), based on the assumption that dengue virus also forms a “closed” conformation that is similar to that of West Nile virus protease. It is therefore critical to solve the crystal structure of DENV protease to guide functional studies and drug discovery.
In this study, we report the crystal structure of DENV-3 NS2B-NS3 protease bound to a peptide-based inhibitor. The structure reveals, for the first time, the detailed interactions between the peptide substrate and the DENV protease. NS2B forms an ordered β hairpin that wraps around the NS3 protease to form an active, closed conformation. In addition, we have solved the crystal structure of the serine-protease inhibitor, aprotinin (also known as bovine pancreatic trypsin inhibitor), bound to DENV-3 protease, showing details of the interactions at the prime side of the active site. These are the first ligand-bound structures of the dengue virus protease and will be extremely important for a rational structure-based approach to design dengue virus-specific inhibitors.
MATERIALS AND METHODS
Cloning and protein production.
The cDNA of DENV-3 (strain S221/03), encoding NS2B residues 50 to 95 and NS3 residues 1 to 182 connected by a G4SG4 linker, was subcloned from a previously described construct (19) into the BamHI and XhoI sites of pGEX6P1 (GE Healthcare). The NS2B-G4SG4-NS3 gene was expressed in Escherichia coli BL21(DE3) using a standard protocol (19). The recombinant protein contained an N-terminal glutathione S-transferase (GST) and was purified from the clarified cell lysate by GST affinity chromatography. The GST fragment was removed using Prescission protease (GE Healthcare), which leaves the amino acid sequence GPLGS at the N terminus of the NS2B-NS3 fusion. The protease was further purified by size exclusion chromatography.
Protease inhibitors.
The peptide benzoyl-norleucine-Lys-Arg-Arg-aldehyde was synthesized as described previously (35), and aprotinin was purchased from Sigma.
Crystallization and structure solution.
DENV-3 protease in 20 mM Tris-HCl, pH 8.5, 150 mM NaCl was concentrated to 10 to 15 mg/ml and mixed with 2 to 3 mM benzoyl-norleucine-Lys-Arg-Arg-H from a 50 mM stock dissolved in water. The protein-peptide solution was crystallized by hanging-drop vapor diffusion by mixing it at a 1:1 ratio with a reservoir solution containing 0.1 M Tris-HCl, pH 6.8, and 1.8 M (NH4)2SO4. Crystals of approximately 30 by 50 by 100 μm grew within a week at 18°C. Crystals were transferred to the reservoir solution, supplemented with 20% glycerol, and flash frozen in liquid nitrogen. For cocrystallizing with aprotinin, DENV-3 protease at 13 mg/ml was mixed with 500 μM aprotinin (from a 50 mM stock dissolved in water). Hexagonal crystals of approximately 20 by 20 by 20 μm appeared after 4 weeks at 18°C from sitting-drop vapor-diffusion experiments in which 0.2 μl protein was mixed with 0.2 μl of the precipitant, 0.1 M morpholineethanesulfonic acid (MES), pH 6.5, plus 30% polyethylene glycol (PEG) 4000. Crystals were transferred to the reservoir solution, supplemented with 20% glycerol, and flash frozen in liquid nitrogen. Data were collected at beamline PXII of the Swiss Light Source. The data were integrated using MOSFLM and scaled using SCALA, part of the CCP4 suite of programs (4, 14, 33). The structures were solved by molecular replacement using 2FP7 and 2IJO as initial search models using MOLREP for the peptide- and aprotinin-bound structures, respectively (1, 13). The structures were rebuilt using COOT (12) and refined using REFMAC5 (22).
RESULTS
To understand whether NS2B adopts the same conformation in DENV as that observed in ligand-bound WNV protease structures (1, 13, 25), we solved two crystal structures of two distinct inhibitors bound to DENV-3 protease. The first structure is in complex with a peptide inhibitor, benzoyl-norleucine-Lys-Arg-Arg-aldehyde (Bz-nKRR-H), which was previously shown to inhibit DENV protease with a Ki (equilibrium inhibition constant) of 5.6 μM (35, 36). The second structure is in complex with aprotinin, a 58-amino-acid polypeptide that inhibits DENV protease with a Ki of 0.026 μM (21).
DENV-3 protease structure bound to Bz-nKRR.
Crystals of DENV-3 protease in the space group C2221 grew in the presence of a peptide-based aldehyde inhibitor, Bz-nKRR-H (13, 19, 35). The structure was solved by molecular replacement, which identified two molecules in the asymmetric unit. The model was refined to 2.3-Å resolution with an Rfactor of 21.8%, Rfree of 24.3%, and good stereochemistry (Tables 1 and 2). An essentially complete model for both monomers was built. Both molecules contain residues 50 to 88 of NS2B, while NS3 chains B and D contain residues 1 to 10 and 16 to 171 and residues 2 to 9 and 15 to 171, respectively. Both molecules in the asymmetric unit adopt a very similar conformation with a Cα root mean square difference (RMSD) of 0.47 Å for 190 of 203 residues. It should be noted that the peptide n-KRR, cocrystallized in the current structure, was the optimal tetrapeptide P1-P4 substrate identified for the DENV protease (19). Therefore, the cocrystal structure of DENV-3 NS2B-NS3 in complex with an aldehyde version of n-KRR should represent a trapped catalytically competent conformation of the protease complex, with NS2B wrapping around NS3 to complete the active site (Fig. 1A).
Table 1.
Data collection and phasing statistics
| Parameter | Value for |
|
|---|---|---|
| Bz-nKRR-H | Aprotinin | |
| Space group | C2221 | P321 |
| Cell parameters (a, b, c; Å) | 104.22, 133.12, 76.03 | 84.79, 84.79, 66.03 |
| Wavelength (Å) | 1.0 | 1.0 |
| Resolution range (Å) | 55.8–2.3 | 49.1–1.8 |
| No. of observed reflections | 133,915 | 194,151 |
| No. of unique reflectionsa | 23,827 (3,459) | 25,786 (3,703) |
| Completeness (%) | 99.7 (99.9) | 100 (100) |
| Multiplicity | 5.6 (5.5) | 7.5 (7.5) |
| Rmergeb | 0.077 (0.620) | 0.084 (0.585) |
| I/σ[I] | 12.4 (2.6) | 13.6 (3.1) |
| Solvent content (%) | 56.2 | 47.3 |
The numbers in parentheses refer to the last (highest-resolution) shell.
Rmerge = ∑h∑i|Ihi − <Ih>|/∑h,i Ihi, where Ihi is the ith observation of the reflection h, while <Ih> is its mean intensity.
Table 2.
Refinement statistics
| Parameter | Value for |
|
|---|---|---|
| Bz-nKRR-H | Aprotinin | |
| Resolution range (Å) | 30–2.3 | 30–1.8 |
| Completeness (%) | 99.6 | 99.9 |
| No. of reflections | ||
| Used for refinement | 22,563 | 24,442 |
| Used for Rfree calculation | 1,215 | 1,312 |
| No. of nonhydrogen atoms | 3,327 | 2,080 |
| No water molecules | 112 | 193 |
| Average B factors, Å2 | 50.6 | 29.2 |
| Rfactora (%) | 21.8 | 18.6 |
| Rfreeb (%) | 24.3 | 20.4 |
| Rms deviations from ideality | ||
| Bond lengths (Å) | 0.010 | 0.010 |
| Bond angles (°) | 1.33 | 1.26 |
| Ramachandran plot | ||
| Residues in most favored regions (%) | 97.2 | 98.3 |
| Residues in additional allowed regions (%) | 2.5 | 1.7 |
| Residues in disallowed regions (%) | 0.3 | 0.0 |
| Pdb code | 3U1I | 3U1J |
Rfactor = ∑ ||Fobs| − |Fcalc||/∑ |Fobs|.
Rfree was calculated with 5% of reflections excluded from the refinement.
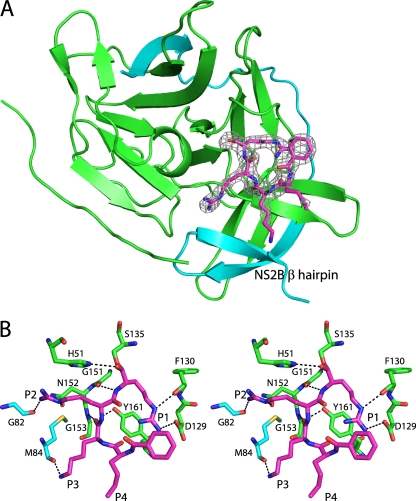
Fig 1.
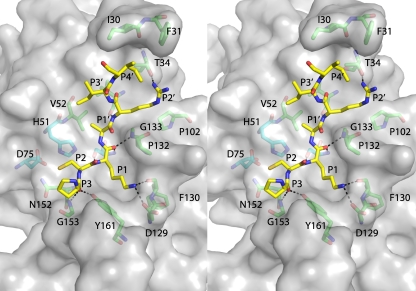
Structure of DENV-3 protease bound to Bz-nKRR-H. (A) The structure is shown in cartoon representation, with NS3 in green and NS2B in blue. The wrapped β hairpin is indicated. Bz-nKRR-H is shown in stick representation with the OMIT Fo-Fc density where the ligand is removed from the final round of refinement, contoured at 3σ. (B) Stereo image of the detailed interactions of Bz-nKRR-H. Potential hydrogen bonds are indicated by the dashed lines. The residues are colored as for the protein shown in panel A.
One NS2B-NS3 protease per asymmetric unit is bound to the inhibitor Bz-nKRR-H. As shown in Fig. 1B, Ser135 forms a covalent bond with the aldehyde carbon and the resulting hydroxyl of Bz-nKRR-H interacts with H51 of NS3. The product of this reaction is a tetrahedral hemiacetal that can result in either S or R stereochemistry, which cannot be distinguished at this resolution. The P1 side chain forms a charge-charge interaction with D129 of NS3 and an H bond with the backbone carbonyl of F130 of NS3. The P2 and P3 side chains interact with backbone carbonyls of G82 and M84 of NS2B, respectively; these interactions stabilize the β hairpin of NS2B in a closed conformation. In addition, P2 and P3 each make a single backbone interaction with G151 and N152 of NS3, respectively. It should be noted that the P2 and P3 Arg and Lys do not form any direct charge-charge interactions with acidic side chains of NS2B. This indicates that the basic residues at P2 and P3 positions are not required to stabilize the “closed” conformation. The preference for Arg at P2 likely also comes from interacting with Asp75, part of the catalytic triad, whereas Lys at P3 does not make any charge-charge interactions. This observation suggests that it may be possible to design peptidomimetics that contain synthetic polar side chains at P2 and/or P3 to mimic these interactions and reduce the requirement for charged groups in novel antivirals against dengue virus protease.
For the second monomer in the asymmetric unit, there is clear density for another smaller ligand (data not shown). A di-Arg peptide was modeled into the density because (i) the ligand likely originated from the Bz-nKRR-H sample and (ii) no other additives were present in the crystallization conditions that could account for this density. Crystal packing does not allow the second active site to contain a covalently bound Bz-nKRR-H peptide, and Ser135 does not have additional density indicating a covalent interaction. The remainder of Bz-nKRR-H is likely to be disordered. Alternatively, the di-Arg could be a breakdown product from prolonged storage of the peptide at −80°C or an impurity from the original synthesis. The Arg side chains form charge-charge interactions with D81 of NS2B and D129 of NS3 (data not shown). These two interactions are sufficient to transform the NS2B-NS3 into a closed conformation with NS2B in the active site.
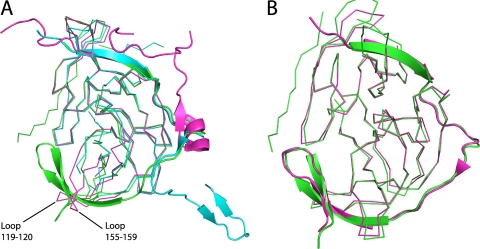
The overall structure of the DENV-3 protease is similar to that of the DENV-1 and -2 structures (7, 13) (RMSD of 0.80 and 1.11 to DENV-1 and DENV-2 for 158 of 186 and 147 of 174 residues, respectively). However, there is substantial movement in two loop regions of NS3 (residues 119 to 120 and 155 to 159), which are disordered in DENV-1 (7) and closer together in DENV-2 (13), so there is not enough space to accommodate the NS2B β hairpin (Fig. 2A). This agrees with results of previous nuclear magnetic resonance (NMR) spectroscopy experiments, which showed that the β hairpin of NS2B is largely disordered in solution for DENV and that substrate binding or small molecules are capable of inducing the closed conformation of the enzyme (5, 34).
Fig 2.
Comparison of DENV-3 protease with DENV-1 and -2 or WNV proteases. (A) DENV-3 protease in complex with Bz-nKRR-H (green) aligned with the proteases from DENV-1 (3L6P; blue) or DENV-2 (2FOM; magenta). NS3 is shown in ribbon representation and NS2B in cartoon representation. The flexible loops including residues 119 to 120 and 155 to 159 are indicated. (B) DENV-3 protease in complex with Bz-nKRR-H (green) aligned with WNV protease bound to the same ligand (2FP7; magenta). NS3 is shown as ribbons and NS2B as a cartoon.
The NS2B region of the protease adopts a very similar conformation to that seen in the ligand-bound WNV structures (1, 13, 25). The RMSD is 0.62 Å for 175 of 181 Cα atoms compared to WNV protease bound to the same ligand (13) (Fig. 2B), and many of the protein-ligand interactions are conserved, demonstrating the similarity of the WNV and DENV protease active sites. There are only two significant changes. One is the conformation of the oxyanion hole (discussed below), which is closed in the WNV protease (13), although it is open in the WNV protease bound to another peptide (25). The other difference is at the P3 position in the WNV structure, where the Lys forms two hydrogen bonds with NS2B: the backbone carbonyl of F85 and the side chain of Q86 (13). In the DENV-3 protease, the equivalent of Q86 is an Arg residue, and since this is positively charged, it points away from the P3 lysine. The only interaction the P3 residue makes in the DENV-3 protease is with the backbone carbonyl of M84 (Fig. 1B). There are no acidic residues found in the NS2B part of the S2-S3 pockets for any of the four serotypes of DENV. DENV-2 and -4 have Ser at position 85, which may make a direct interaction with the P3 basic side chain, while DENV-1 has a basic residue, K85. This again suggests that the preference for basic residues at P3 is to provide a long polar chain rather than to provide a positive charge.
WNV protease shows a preference for Lys at P2 (28), while DENV protease shows a preference for Arg (19, 24). It was proposed that this specificity was due in part to Asn84 in WNV protease (8), which is Ser or Thr83 in DENV proteases. This residue fits between the S2 and S3 pockets of the protease but does not make any specific interactions with either residue. The larger residue in WNV protease likely explains the preference for Lys at P2 in WNV (8).
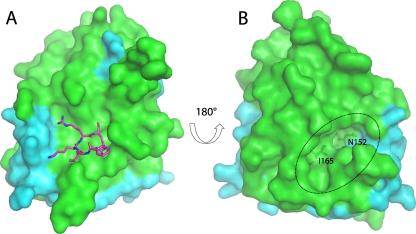
An analysis of the surface of the structure showed that the largest cavity is not the active site but a pocket on the opposite face of the molecule that is formed when NS2B is wrapped around the NS3 (Fig. 3) (10). This cavity is lined by residues from both NS2B (V78 and M84) and NS3 (73 to 74, 88, 89, 118, 120, 122 to 124, 147, 152, and 164 to 168). It is conserved in WNV protease structures where NS2B adopts the same closed conformation (1, 13, 25), suggesting that it is functionally important. Alanine mutagenesis of NS2B in WNV protease showed that this region within the β hairpin of NS2B was important for activity, and this cavity was proposed to be a potential binding site for antivirals (10). The cavity is not present in the structures of the free DENV protease structures because it requires NS2B to be in the closed conformation. It is likely that large conformational changes in the orientation of NS2B are required for catalytic activity, since removal of residues 11 to 20 within the flexible N terminus of DENV-4 protease reduced Vmax dramatically but had little effect on Km (7). Also, decreasing the length of the covalent linker between NS2B and NS3 dramatically reduces activity, indicating that flexibility is important (9). The identified cavity could potentially be targeted for antiviral drug development; compounds that bind to this pocket could lock the NS2B β hairpin in place, prohibiting the conformational change required for catalysis. Alternatively, they may prevent substrate binding (10). The functional importance of this pocket has been shown in dengue virus by mutagenesis of NS3 (26). Mutation of the conserved residue, N152 or I165 (Fig. 3), to Ala reduced activity dramatically, further indicating that targeting this cavity is a valid approach for antiviral development.
Fig 3.
Surface view of the DENV-3 protease. (A) The surface of the DENV protease shows the binding site of the Bz-nKRR-H peptide (as sticks; left). (B) The reverse face of the molecule shows the allosteric pocket (indicated by a dotted oval) that is lined by residues from both NS2B (cyan) and NS3 (green) and can potentially be targeted by low-molecular-weight inhibitors. N152 and I165 are shown as sticks.
DENV-3 protease structure bound to aprotinin.
In an attempt to gain more information about the prime side of the substrate-binding site, we solved the structure of DENV-3 protease bound to aprotinin (Fig. 4A), a serine-protease inhibitor. The structure was refined to 1.8 Å, with an Rfactor of 18.6% and Rfree of 20.4% (Tables 1 and 2). The structure was solved by molecular replacement using the structure of the WNV protease-aprotinin complex as a search model (2IJO) (1). The structure reveals, for the first time, the interactions between the DENV protease and residues at the prime side of the binding pocket.
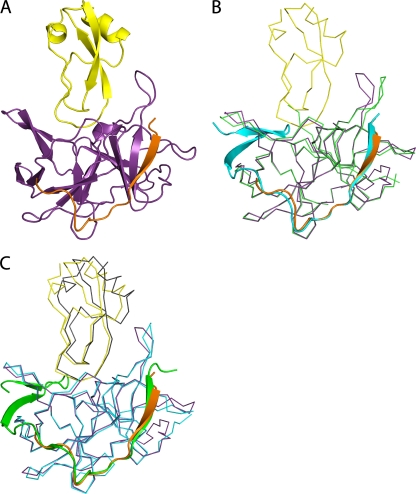
Fig 4.
DENV-3 protease-aprotinin structure and structural comparison with WNV protease-aprotinin and DENV-3 protease-Bz-nKRR-H structures. (A) Overview of the DENV-3 protease (NS3, purple; NS2B, orange) bound to aprotinin (yellow) shown in cartoon representation. (B) Alignment of the DENV-3 protease structures in complex with aprotinin (colored as in panel A) and in complex with Bz-nKRR-H (colored as in Fig. 1A). NS3 is shown in ribbon representation for clarity, with the rest in cartoon representation. (C) Comparison of the structures of DENV-3 protease-aprotinin (colored as in A) and WNV protease-aprotinin (NS3, blue; NS2B, green; aprotinin, gray), aligned on the Cα atoms of the protease only. NS2B is shown in cartoon representation, with the rest as ribbons.
The overall structure is similar to that of the WNV protease-aprotinin complex (1) with an RMSD of 0.62 Å for 157 of 177 Cα atoms when comparing the protease or 1.04 Å for 222 of 236 Cα atoms, including both the protease and aprotinin. In addition, the conformations of the NS3-NS2B protease in the Bz-nKRR-H- and aprotinin-bound structures are very similar, with an RMSD of 0.53 Å for 160 of 176 Cα atoms. The results further strengthen the conclusion that these structures represent an active conformation of the enzymes.
The major difference among the structures of DENV-3 protease-aprotinin, WNV protease-aprotinin (1), and DENV-3 protease-Bz-nKRR-H is that, in the DENV-3 protease-aprotinin structure, the β hairpin of NS2B is disordered (Fig. 4B and C). No direct contact between aprotinin and the NS2B β hairpin was observed in the DENV-3 protease-aprotinin structure (Fig. 4A), indicating that the binding of aprotinin to the protease active site does not require interactions with NS2B. This is in contrast to the protease-Bz-nKRR-H structure, in which the binding of Bz-nKRR-H to the protease active site requires interactions with NS2B. In the WNV protease-aprotinin structure, NS2B is in a closed conformation with one H-bond interaction with aprotinin (between Q86 of NS2B and R39 of aprotinin) (1). The conformational difference of NS2B observed in the DENV-3 and WNV protease-aprotinin structures is consistent with NMR data showing that the NS2B β hairpin is predominantly bound to NS3 in WNV protease (i.e., in the closed conformation), whereas it is largely disordered in DENV protease (31, 34). Since aprotinin is a large polypeptide, it is likely that its affinity is from multiple interactions and much of its inhibitory activity comes from steric hindrance of the active site.
DENV protease specificity.
WNV and DENV proteases have high sequence identity in the active site but still display significant differences in substrate specificity. Many of these differences can be rationalized by the current structures of DENV-3 protease. Figure 5 depicts the detailed interactions between aprotinin and DENV-3 protease. The P1 residue is Lys15, which makes a charge-charge interaction with D129, as well as an H bond with the backbone carbonyl of F130. Both DENV and WNV proteases have a strong preference for a basic residue at P1 (Arg is preferred) (19, 24, 28), which explains why aprotinin is a potent inhibitor of these proteases (21). Arg is preferred because it can interact with both D129 and F130 (as seen in the DENV-3-Bz-nKRR-H-structure) as well as make a π-cation interaction with Y161. The P2 residue is a Cys that forms a disulfide bond with another aprotinin residue, Cys38. The P2 Cys sits in a shallow pocket and forms Van der Waals interactions with the side chains of H51 and N152. In the WNV protease it also interacts with the side chain of N84 (1), but this interaction is not present in the DENV-aprotinin protease structure because the NS2B is in the open conformation. The P3 residue is Pro13 of aprotinin, and this sits deeper into the S3 pocket than in the WNV protease-aprotinin structure. This may be because WNV protease contains Ile155 in NS3, whereas in DENV protease Val155 is conserved and therefore slightly less bulky. As a result, the H bond between the backbone P3 carbonyl and the NH of Gly153 is lost in WNV protease. This will contribute to the lower potency of aprotinin for WNV protease than DENV protease (21).
Fig 5.
Detailed view of interactions between DENV-3 protease and aprotinin. Stereo diagram of the P3′-P4 residues of aprotinin (shown as yellow sticks) on the surface of the DENV-3 protease (gray). Potential hydrogen bonds are indicated by dashed lines. Other residues lining the pocket are shown as green sticks, with the catalytic triad shown as blue sticks.
The DENV-3 protease-aprotinin structure (Fig. 5) also provides information about the substrate specificity in the P1′ to P4′ positions. At P1′-P2′ positions, WNV shows a preference for Gly-Gly (28), whereas DENV prefers Ser over Gly at P1′ and shows a weak preference for acidic residues at P2′ (19). The preference for Ser at P1′ in DENV can be explained by the crystal structure where the Ala side chain of aprotinin points into a small pocket that would optimally fit a slightly larger Ser side chain. The P2′ Arg points into solvent. An acidic residue is preferred at this position, which may be explained by the presence of Thr34 in DENV-3 (instead of Tyr34 in WNV), with which an acidic residue could form an additional interaction. DENV also shows a stronger preference for Ser at P3′ (19). In the aprotinin structure, the P3′ Ile is pointing into solvent; a small polar residue, such as Ser, could potentially pick up additional H-bond interactions with the side chain of Gln27 (which is a Thr in WNV) or Van der Waals interactions with Val36. DENV also displays sequence preference for Gly at P4′ (19); the flexibility of P4′ Gly allows peptides to avoid steric clashes with I30 and F31 (Fig. 5).
The DENV-3 protease structures have an active conformation of the oxyanion hole.
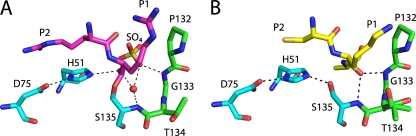
In both DENV-3 protease-Bz-nKRR-H (Fig. 6A) and protease-aprotinin structures (Fig. 6B), the oxyanion hole is formed by the NH groups of G133, T134, and S135. The oxyanion hole is in the productive conformation to accept the negatively charged tetrahedral intermediate. In the DENV-3 protease-Bz-nKRR-H structure, a water molecule and a sulfate occupy the oxyanion hole (Fig. 6A). In the WNV protease-Bz-nKRR-H structure (13), the G133 peptide bond is flipped, leading to the closure of the oxyanion hole (an inactive conformation); it was previously proposed that the closure of the oxyanion hole is due to the lack of a residue at the P1′ position (1). However, in the WNV protease structure with another peptide, naphthoyl-KKR-H (25), and in the current DENV-3 protease-Bz-nKRR-H structure, the substrates lack the P1′ residue, but the oxyanion hole is in the active conformation. These results indicate that a residue at P1′ is not required for this active conformation.
Fig 6.
Detailed view of the catalytically competent geometry in the DENV-3 protease structures. (A) Stick representation of the oxyanion hole and catalytic triad (H51, D75, and S135) from the DENV-3 protease-Bz-nKRR-H structure. The catalytic-triad residues are in blue, and other residues are in green. Only the P1 and P2 residues are shown for clarity. The water and sulfate by the oxyanion hole are shown as a sphere and sticks, respectively. Potential hydrogen bonds (up to 3.2 Å) are indicated by the dashed lines. (B) Sticks representation of the oxyanion hole of the DENV-3 protease-aprotinin structure. The protease residues are colored as in panel A. The P1′ to P2 residues of aprotinin are shown as yellow sticks, and the carbonyl of the P1 residue occupies the oxyanion hole.
In the DENV-3 protease-aprotinin structure, there are strong H-bond interactions among the catalytic triad residues (H51, D75, and S135); all hydrogen bonds between them are within 2.7 Å distance (Fig. 6). The geometry of the catalytic triad is in the productive conformation for catalysis. The carbonyl of the P1 Lys of aprotinin is inserted into the oxyanion hole (Fig. 6B), indicating that the oxyanion hole is in the active conformation. A similar conformation was observed in the WNV protease-aprotinin complex (1). These results strongly argue that the structures represent catalytically competent conformations.
DISCUSSION
We have solved ligand-bound crystal structures of the DENV-3 protease. The cocrystal structures in complex with an aldehyde peptide (Bz-nKRR-H) and aprotinin show catalytically competent conformations of the enzyme. In the Bz-nKRR-H structure, DENV-3 protease adopts the closed conformation in which the NS2B wraps around NS3, including the β hairpin that forms part of the active site. Our results demonstrate for the first time that the closed conformation previously observed in ligand-bound WNV structures is common to the DENV protease. It is likely that proteases of other flaviviruses catalyze reactions in a similar closed formation. The new structural information will be useful for rational structure-based design of DENV-specific inhibitors. Rational structure-guided programs looking for dengue antivirals have relied on homology modeling of the dengue virus protease structure (17, 32), based on the crystal structures of WNV protease (1, 13, 25). This study will enable those researchers to validate their models using experimental data and will enable new rational approaches.
It has previously been shown by NMR that WNV exists predominantly in the closed form in solution, whereas DENV protease is largely in the open form (31). This probably explains why similar peptides are typically >10-fold more potent inhibitors of WNV than DENV proteases (27). It also explains why previous attempts to crystallize DENV protease with bound inhibitors have been unsuccessful. It has been shown that increasing the number of clones from natural variants of an infectious agent greatly increases the probability of crystallization (3). Our success may be related to trying multiple DENV serotypes and isolates for crystallization trials. In addition, we designed new, shorter constructs for NS3 based on the optimal length for WNV protease activity (9).
Residues 50 to 61 of NS2B form a tight complex with NS3, and it has been proposed that W61 is the last residue that is tightly bound to NS3, after which the rest of the residues (residues 62 to 88) are flexible and able to adopt multiple conformations (1). However, the structures of both DENV-1 protease and DENV-3 protease-aprotinin show that more residues (beyond W61) from NS2B are interacting with NS3 (up to T68 and E66 of NS2B, respectively). These results suggest that the conformational change to the closed form is a dynamic and iterative process.
WNV protease shows a greater catalytic efficiency for the same small peptides than DENV protease (21), due to both a higher affinity for the peptides (lower Km) and a higher rate of catalysis (kcat). Since there are no obvious structural differences for this trend, it is likely that these differences are due to different propensities of the enzymes to form the closed conformation. It is reasonable to assume that formation of the closed form of the enzyme is entropically unfavorable such that this process is likely to reduce the affinity of substrates for the protease in vitro (i.e., increase the Km). Since WNV protease is more often in the closed form than DENV protease, the entropic penalty will be lower for WNV protease. Catalytic activity requires the closed form of the enzyme for turnover, as shown both by structural data and by the importance of amino acids in the β-hairpin region of NS2B for activity (10, 23).
In the WNV protease-aprotinin complex, the only direct interactions between NS2B and aprotinin are through Q86 of NS2B and R39 of aprotinin (1). Aligning the β-hairpin region of DENV-3 NS2B from the Bz-nKRR-H-bound structure onto the DENV-3 aprotinin-bound structure indicates that there would be similarly few direct interactions between aprotinin and the β hairpin of NS2B. It is unlikely that the NS2B β hairpin needs to bind to NS3 to form the catalytically competent active site before aprotinin can bind to the NS2B-NS3 complex. Aprotinin is a potent inhibitor of both WNV and DENV proteases, with a greater activity against DENV (21). In contrast, peptides typically show better potency against WNV protease than DENV protease (27). These results are in agreement with the observations that DENV protease with NS2B in the open conformation binds readily to aprotinin but needs to be induced to a closed conformation to bind peptide inhibitors. In contrast, the NS2B in WNV protease is more often in the closed conformation, favoring the binding of peptide inhibitors.
The crystal structures of DENV-3 and WNV proteases bound to aprotinin are remarkably similar, despite the difference in NS2B β-hairpin formation. Since most of the substrate-binding residues are conserved between the DENV-3 and WNV proteases, it is difficult to interpret subtle changes in sequence specificity. The specificity could derive from the flexibility and conformation of the peptide that is not immediately obvious from the structure. For an example, mutation of WNV P131 and T132 to the DENV sequence K and P, respectively, changes the P1′ and P2′ specificity to be more like those of DENV (29). T132 forms a hydrogen bond with the P1′ carbonyl in the WNV-aprotinin structure, and this interaction is lost in DENV, where the Thr is replaced by Pro. It is likely that this small change restricts the ability to accommodate slightly larger residues in the S1′ pocket and changes the preference from Ser to Gly.
One notable difference between the structures of WNV and DENV-3 proteases bound to aprotinin is the shape of the S4′ pocket. As shown in Fig. 5, the two hydrophobic residues (I30 and F31) of DENV-3 protease form a hydrophobic “lip” at the end of the binding pocket that could be used to design a peptide that terminates with a hydrophobic group at P4′ for increased affinity to the enzyme. This lip is flatter in the WNV protease-aprotinin structure (1), probably due to the presence of Y34 instead of T34. It is interesting to note that, in a study of synthetic peptides, the greatest catalytic efficiency was obtained with a P4-P3′ peptide that terminated with a 3-nitrotyrosine at P4′ (24). This peptide was cleaved more efficiently than longer or shorter peptides, raising the possibility that the nitrotyrosine itself was contributing to interactions in this pocket of the enzyme, as would be predicted from the DENV-3 protease structure. However, the hydrophobic residues at positions 30 and 31 have been proposed to interact with the membrane (2, 11), so it is not yet clear whether the P4′ pocket is available in vivo for interaction with inhibitors.
ACKNOWLEDGMENTS
We thank Siew Pheng Lim, Shahul Nilar, Qing Yin Wang, and other colleagues at Novartis Institute for Tropical Diseases for helpful discussions and support during the course of this study. We thank the beamline scientists at PXII of the Swiss Light Source for assistance with data collection.
Footnotes
Published ahead of print 26 October 2011
REFERENCES
- 1. Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. 2007. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 16:795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Assenberg R, et al. 2009. Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J. Virol. 83:12895–12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Au K, et al. 2006. Application of high-throughput technologies to a structural proteomics-type analysis of Bacillus anthracis. Acta Crystallogr. D Biol. Crystallogr. 62:1267–1275 [DOI] [PubMed] [Google Scholar]
- 4. Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67:271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bodenreider C, et al. 2009. A fluorescence quenching assay to discriminate between specific and nonspecific inhibitors of dengue virus protease. Anal. Biochem. 395:195–204 [DOI] [PubMed] [Google Scholar]
- 6. Cahour A, Falgout B, Lai CJ. 1992. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J. Virol. 66:1535–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandramouli S, et al. 2010. Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J. Virol. 84:3059–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. 2006. Insights to substrate binding and processing by West Nile Virus NS3 protease through combined modeling, protease mutagenesis, and kinetic studies. J. Biol. Chem. 281:38448–38458 [DOI] [PubMed] [Google Scholar]
- 9. Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. 2007. Generation and characterization of proteolytically active and highly stable truncated and full-length recombinant West Nile virus NS3. Protein Expr. Purif. 53:87–96 [DOI] [PubMed] [Google Scholar]
- 10. Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. 2008. Mutagenesis of the West Nile virus NS2B cofactor domain reveals two regions essential for protease activity. J. Gen. Virol. 89:1010–1014 [DOI] [PubMed] [Google Scholar]
- 11. Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. 2008. West Nile Virus NS2B/NS3 protease as an antiviral target. Curr. Med. Chem. 15:2771–2784 [DOI] [PubMed] [Google Scholar]
- 12. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 13. Erbel P, et al. 2006. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 13:372–373 [DOI] [PubMed] [Google Scholar]
- 14. Evans PR. 2011. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67:282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Falgout B, Pethel M, Zhang YM, Lai CJ. 1991. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 65:2467–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guzman MG, et al. 2010. Dengue: a continuing global threat. Nat. Rev. Microbiol. 8:S7–S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knehans T, et al. 2011. Structure-guided fragment-based in silico drug design of dengue protease inhibitors. J. Comput. Aided Mol. Des. 25:263–274 [DOI] [PubMed] [Google Scholar]
- 18. Lescar J, et al. 2008. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target. Antiviral Res. 80:94–101 [DOI] [PubMed] [Google Scholar]
- 19. Li J, et al. 2005. Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J. Biol. Chem. 280:28766–28774 [DOI] [PubMed] [Google Scholar]
- 20. Luo D, et al. 2008. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 82:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mueller NH, Yon C, Ganesh VK, Padmanabhan R. 2007. Characterization of the West Nile virus protease substrate specificity and inhibitors. Int. J. Biochem. Cell Biol. 39:606–614 [DOI] [PubMed] [Google Scholar]
- 22. Murshudov GN, et al. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67:355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niyomrattanakit P, et al. 2004. Identification of residues in the dengue virus type 2 NS2B cofactor that are critical for NS3 protease activation. J. Virol. 78:13708–13716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niyomrattanakit P, et al. 2006. Probing the substrate specificity of the dengue virus type 2 NS3 serine protease by using internally quenched fluorescent peptides. Biochem. J. 397:203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robin G, et al. 2009. Structure of West Nile virus NS3 protease: ligand stabilization of the catalytic conformation. J. Mol. Biol. 385:1568–1577 [DOI] [PubMed] [Google Scholar]
- 26. Salaemae W, Junaid M, Angsuthanasombat C, Katzenmeier G. 2010. Structure-guided mutagenesis of active site residues in the dengue virus two-component protease NS2B-NS3. J. Biomed. Sci. 17:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schuller A, et al. 2011. Tripeptide inhibitors of dengue and West Nile virus NS2B-NS3 protease. Antiviral Res. 92:96–101 [DOI] [PubMed] [Google Scholar]
- 28. Shiryaev SA, et al. 2007. Cleavage preference distinguishes the two-component NS2B-NS3 serine proteinases of Dengue and West Nile viruses. Biochem. J. 401:743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shiryaev SA, et al. 2007. Switching the substrate specificity of the two-component NS2B-NS3 flavivirus proteinase by structure-based mutagenesis. J. Virol. 81:4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stadler K, Allison SL, Schalich J, Heinz FX. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Su XC, et al. 2009. NMR analysis of the dynamic exchange of the NS2B cofactor between open and closed conformations of the West Nile virus NS2B-NS3 protease. PLoS Negl. Trop. Dis. 3:e561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wichapong K, Pianwanit S, Sippl W, Kokpol S. 2010. Homology modeling and molecular dynamics simulations of Dengue virus NS2B/NS3 protease: insight into molecular interaction. J. Mol. Recognit. 23:283–300 [DOI] [PubMed] [Google Scholar]
- 33. Winn MD, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu PSC, et al. 2007. Cell-free transcription/translation from PCR-amplified DNA for high-throughput NMR studies. Angew. Chem. Int. Ed. Engl. 46:3356–3358 [DOI] [PubMed] [Google Scholar]
- 35. Yin Z, et al. 2006. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg. Med. Chem. Lett. 16:40–43 [DOI] [PubMed] [Google Scholar]
- 36. Yin Z, et al. 2006. Peptide inhibitors of Dengue virus NS3 protease. Part 1: Warhead. Bioorg. Med. Chem. Lett. 16:36–39 [DOI] [PubMed] [Google Scholar]