Abstract
It is well established that the Nef proteins of human and simian immunodeficiency viruses (HIV and SIV) modulate major histocompatibility complex class I (MHC-I) cell surface expression to protect infected cells against lysis by cytotoxic T lymphocytes (CTLs). Recent data supported the observation that Nef also manipulates CTLs directly by down-modulating CD8αβ (J. A. Leonard, T. Filzen, C. C. Carter, M. Schaefer, and K. L. Collins, J. Virol. 85:6867–6881, 2011), but it remained unknown whether this Nef activity is conserved between different lineages of HIV and SIV. In this study, we examined a total of 42 nef alleles from 16 different primate lentiviruses representing most major lineages of primate lentiviruses, as well as nonpandemic HIV-1 strains and the direct precursors of HIV-1 (SIVcpz and SIVgor). We found that the vast majority of these nef alleles strongly down-modulate CD8β in human T cells. Primate lentiviral Nefs generally interacted specifically with the cytoplasmic tail of CD8β, and down-modulation of this receptor was dependent on the conserved dileucine-based motif and two adjacent acidic residues (DD/E) in the C-terminal flexible loop of SIV Nef proteins. Both of these motifs are known to be important for the interaction of HIV-1 Nef with AP-2, and they were also shown to be critical for down-modulation of CD4 and CD28, but not MHC-I, by SIV Nefs. Our results show that down-modulation of CD4, CD8β, and CD28 involves largely overlapping (but not identical) domains and is most likely dependent on conserved interactions of primate lentiviral Nefs with cellular adaptor proteins. Furthermore, our data demonstrate that Nef-mediated down-modulation of CD8αβ is a fundamental property of primate lentiviruses and suggest that direct manipulation of CD8+ T cells plays a relevant role in viral immune evasion.
INTRODUCTION
Cellular immune responses, mediated by CD8+ cytotoxic T lymphocytes (CTLs), play a significant role in the control of viral replication (9, 39). However, human and simian immunodeficiency viruses (HIV and SIV) can evade CTL control by the acquisition of escape mutations in major histocompatibility complex class I (MHC-I)-restricted epitopes and have evolved elaborate mechanisms to counteract the host immune response (8, 29). As a consequence, CTLs are usually unable to suppress viral replication to harmless levels and, ultimately, almost invariantly fail to prevent progression to AIDS. In particular, the multifunctional accessory Nef protein of HIV and SIV is well known for its capability to manipulate multiple cellular functions to support viral immune evasion, replication, and pathogenesis (4, 30). Nef undermines the efficiency of the CTL response by several mechanisms. First, it down-modulates MHC-I molecules from the cell surface (2, 32, 33, 59). This Nef function reduces CTL lysis of HIV-1-infected cells (13) and is associated with a strong selective advantage in SIV-infected rhesus macaques (42, 66). Notably, Nef affects MHC-I cell surface expression in a selective manner, i.e., it down-modulates HLA-A and -B but not HLA-C or -E alleles, most likely to balance escape from CTL lysis with protection from attack by natural killer (NK) cells (11, 15, 62). Second, Nef impairs the function of CD4+ T cells and antigen-presenting cells (APCs) that are required to maintain proper CD8+ T cell responses. In virally infected CD4+ T cells, Nef reduces the levels of CD4, MHC-I, and (less efficiently) CD28 and CXCR4 (CXCL12) cell surface expression and alters signal transduction and trafficking pathways (4, 30). In APCs, Nef perturbs MHC-II-restricted antigen presentation by up-modulation of Ii cell surface expression (56, 64). Third, it has been proposed that Nef-mediated up-modulation of FAS-L induces apoptosis of “attacking” CTLs (47, 71).
Accumulating evidence suggests that HIV-1 may also infect CD8+ T cells and manipulate them directly. During development in the thymus, a significant proportion of T cells express both CD4 and CD8 (17, 21), and low levels of CD4 expression are induced on activated memory CD8+ T cells, thereby rendering them susceptible to HIV and SIV infection (10, 18, 31). Finally, it has been reported that some rare HIV-1 strains can use CD8 as a receptor for viral entry (49, 50, 72). In support of a relevant role in vivo, HIV-1-infected CD8+ T cells have been detected in infected individuals, particularly in late-stage AIDS patients (50). Thus, although CD4+ T cells are the main targets of HIV-1 infection, it may also be advantageous for the virus to manipulate infected CD8+ T cells. In fact, it has been reported that some Nef proteins interact specifically with the cytoplasmic tail (CT) of the β-chain of CD8αβ to down-modulate it from the cell surface (35, 63) and that a CD8+ T cell subpopulation with reduced CD8β chain expression emerges in HIV-1-infected individuals (57). CD8β cell surface expression is dependent on coexpression of the CD8α chain, and both form covalently linked CD8αβ heterodimers. Furthermore, CD8 also exists as an αα homodimer that is expressed mainly by intestinal γδ T cells and thymic T cell precursors, whereas the CD8αβ receptor is found mostly on thymocytes and mature cytotoxic/suppressor T cells (19). Since the latter T cell population is the one that attacks and eliminates HIV-infected cells, it is conceivable that the virus specifically targets the β-chain of the CD8 receptor.
The elegant study of Stove and coworkers established that some HIV-1 group M (major) Nef proteins, as well as the HIV-2 Rod and SIVmac239 Nef proteins, down-modulate CD8αβ heterodimers (63). More recent work by the Collins laboratory suggested that AP-1 is relevant for down-modulation of CD8β by the HIV-1 Nef protein (35). It remained largely elusive, however, whether Nef-mediated down-modulation of CD8β is functionally and mechanistically conserved between different lineages of primate lentiviruses and whether this Nef function changed after zoonotic transmission of SIVs from apes and monkeys to humans, during adaptation of HIV-1 and HIV-2 to the new host (60). To address this, we functionally analyzed a large panel of highly divergent Nef proteins. Our data demonstrate that the vast majority of primate lentiviral Nef proteins efficiently down-modulate human CD8αβ from the cell surface by specific interaction with the cytoplasmic tail of CD8β. We also show that CD8β down-modulation is generally dependent on the dileucine (E/DXXXLL/M) motif and on two adjacent acidic residues (D174D175 in HIV-1 NL4-3 Nef and D/ED/E in other Nef proteins), referred to herein as the “DD” motif, in the C-terminal flexible loop of primate lentiviral Nef proteins and generally involves domains highly similar to those employed by Nef to downregulate CD4 and CD28.
MATERIALS AND METHODS
Nef expression vectors.
The generation of pCGCG vectors (22) coexpressing enhanced green fluorescent protein (EGFP) and AU-1-tagged nef genes of HIV-1 NL4-3, NA7, JRCSF, YBF30, CK1.62, 8161K9, 13127K2, HJ162, and HJ736, SIVcpz MT145, MB897, GAB1, EK505, TAN1, TAN2, and TAN3, HIV-2 BEN, and SIVgor, -gsn, -mus, -mon, and -mac239 from a bicistronic RNA via an internal ribosome entry site (IRES) has been described previously (52, 56). Additional nef alleles from HIV-1 YBF116 and DJ131, SIVcpz US, GAB2, CAM5.1, Nok5, and Nik4, HIV-2 Cbl23, 60415K, and 310319, SIVsmm Fmm1, Fyr1, and Fwr1, and SIVrcm, -deb40, -syk51, -blue31, -sun, -tan1, and -sab1 were PCR amplified using primers introducing XbaI and MluI sites flanking the nef reading frame and were cloned into the pCGCG vector (56). An overview of these nef alleles is provided in Table S1 in the supplemental material. Site-directed mutagenesis was performed by splice overlap extension PCR. The pCGCG (nef−) control vector expressing only EGFP contains a nef gene with a mutation in the ATG initiation codon and two premature stop codons, at positions 3 and 40 of the reading frame (56). All constructs were verified by sequence analysis.
Proviral constructs.
Generation of HIV-1 (NL4-3-based) proviral constructs carrying functional nef genes followed by an IRES element and the eGFP gene has been described previously (1, 55, 56). Splice overlap extension PCR was used to replace the HIV-1 NL4-3 nef allele with the nef genes listed in Table S1 in the supplemental material, via the single restriction sites for HpaI and MluI. Further splice overlap extension PCR was used to introduce alanine substitutions in SIVagmSab Nef and to mutate the LL/M residues in the C-terminal flexible loop of various primate lentiviral Nef proteins as described elsewhere (55, 56). All constructs were verified by sequence analysis.
Cell culture.
293T and SupT1 cells were cultured as described elsewhere (35, 56). CD4+ and CD8+ T cells from healthy human donors were isolated using human CD4+ or CD8+ T cell RosetteSep kits (Stemcell Technologies) or CD4 or CD8 MicroBeads (MACS; Miltenyi Biotec) following the protocols provided by the manufacturers. The cells were stimulated for 3 days with human CD3/CD28 T-Activator Dynabeads (Invitrogen) at a cell-to-bead ratio of 1:1 and with 10 ng/ml interleukin-2 (IL-2) prior to transduction.
Preparation of CEM A2-CD8β fusion cell line.
A CEM T cell line stably expressing the HLA-A2-CD8β fusion was generated as previously described (35). Briefly, the construct expressing the HLA-A2-CD8β fusion molecule was introduced into CEM-SS cells by using a murine stem cell virus (MSCV) retroviral vector pseudotyped with vesicular stomatitis virus G protein (VSV-G). A uniform population was selected by culturing the cells in neomycin.
Virus stocks and transduction.
To generate viral stocks, 293T cells were cotransfected with the proviral HIV-1 constructs and a plasmid (pHIT-G) expressing VSV-G (1, 55). The latter was used to achieve comparably high initial infection levels for flow cytometric analysis. The medium was changed after overnight incubation, and virus was harvested 24 h later. All cell types were transduced with NL4-3-based proviral constructs coexpressing the various Nef proteins and EGFP as described previously (55). These constructs have the advantage that nef alleles are expressed from the wild-type HIV-1 long terminal repeat (LTR) promoter and via the regular splicing sites. For the detection of CD4 down-modulation, proviral HIV-1 constructs defective in vpu and env were used, since both gene products also reduce CD4 cell surface expression and thus mask the effect of Nef. Flow cytometric analysis was performed at 3 days posttransduction.
Electroporation of CEM and SupT1 cells.
Electroporation of CEM and SupT1 cells with pCGCG vectors expressing different Nefs was performed with a Gene Pulser Xcell apparatus (Bio-Rad) as recommended by the manufacturer. Briefly, 10 μg of vector DNA coexpressing EGFP and Nef was electroporated using 0.4-cm cuvettes (Bio-Rad) at 200 V and 950 μF. At 2 days posttransfection, receptor expression was determined by fluorescence-activated cell sorter (FACS) analysis.
Flow cytometric analysis.
CD4 (Invitrogen), CD28 (BD Biosciences), and MHC-I (Dako) expression in SupT1 or purified human CD4+ T cells transduced with HIV-1 (NL4-3) constructs coexpressing Nef and EGFP was measured as described previously (55). Flow cytometric analyses of A2-CD8β fusions in CEM cells or CD8β in SupT1 and primary CD8+ T cells were performed using the primary antibodies BB7.2 (Abcam) and CD8β (clone 2ST8.5H7; Acris), which recognize CD8αβ but not CD8αα, and a secondary allophycocyanin-conjugated goat anti-mouse antibody (Invitrogen). For quantification of Nef-mediated modulation of the specific surface molecules, the levels of receptor expression were determined for cells expressing EGFP. The extent of down-modulation (n-fold) was calculated by dividing the mean fluorescence intensity (MFI) obtained for cells transduced with the nef-negative NL4-3 control viruses by the corresponding values obtained for cells transduced with viruses coexpressing Nef and EGFP.
Western blot analysis.
To examine the expression of most Nef alleles, 293T cells were transfected as described above with 5 μg DNA of expression vectors coexpressing EGFP and AU-1-tagged Nefs. At 2 days posttransfection, cells were lysed with RIPA buffer (1% NP-40, 0.5% sodium desoxycholate, 3% SDS, 150 mM NaCl, 50 mM Tris-HCl, 5 mM EDTA, protease inhibitor cocktail). Lysates were boiled at 95°C for 10 min, separated in 4 to 12% Bis-Tris gradient acrylamide gels (Invitrogen), blotted onto nitrocellulose filters, and probed with anti-AU-1 (MMS-130P; Covance), anti-GFP (ab290; Abcam), and anti-β-actin (ab8227; Abcam). Subsequently, blots were probed with anti-mouse or anti-rabbit IRDye Odyssey antibodies, and proteins were revealed using a Li-Cor Odyssey scanner.
Statistical analysis.
All statistical calculations were performed with two-tailed Student's t test, using Graph Pad Prism, version 5.0. P values of <0.05 were considered significant. Correlations were calculated with the linear regression module of the software.
RESULTS
To determine whether down-modulation of CD8β is a conserved property of primate lentiviruses, we examined a total of 42 HIV and SIV nef alleles representing the majority of primate lentiviral lineages known to date (see Table S1 in the supplemental material for details). Western blot analysis of 293T cells with pCGCG vectors expressing AU-1-tagged versions of these Nef proteins showed that all of them were expressed at detectable, albeit variable, levels (see Fig. S1). To determine the reproducibility, specificity, and possible significance of the effect of Nef on CD8β, we used three different target cell lines. (i) The first cell line was a CEM T cell line stably expressing the ectodomain of HLA-A2 fused to the cytoplasmic tail (CT) of the CD8β chain (A2-CD8β fusion) (35). The A2-CD8β fusion helped to address whether the CT, which consists of only 17 amino acids (CCRRRRARLRFMKQMYK), is sufficient for Nef-mediated modulation. (ii) The human T cell lymphoblastic lymphoma cell line Sup T1, which expresses full-length CD8αβ, CD4, CD28, and MHC-I, allowed measurement of Nef-mediated modulation of unmodified Nef target proteins from the same HIV-1-infected cell culture. (iii) To exclude the possibility of in vitro artifacts, we verified all effects of Nef in primary CD8+ or CD4+ T cells.
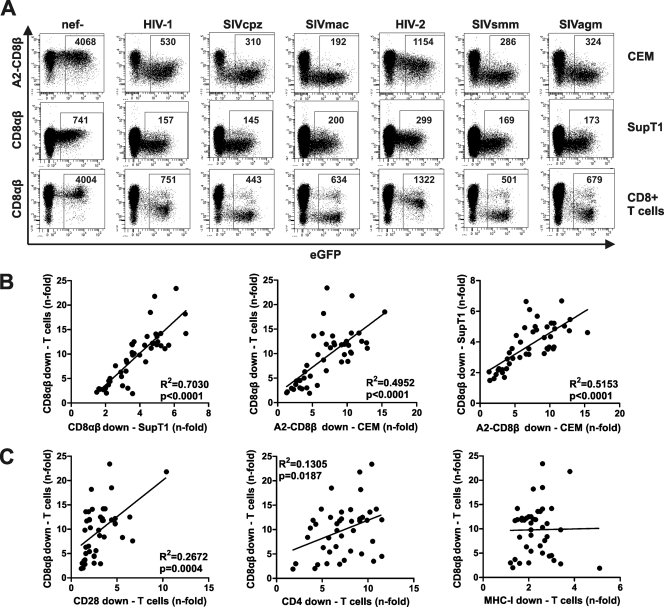
All cell types were transduced with HIV-1 NL4-3-based proviral constructs coexpressing the various Nef proteins and EGFP as described previously (55, 56). We found that HIV-1-infected (EGFP+) cells showed substantially lower levels of A2-CD8β fusion or CD8αβ surface expression than uninfected cells (examples are shown in Fig. 1A). This effect was not observed after infection with an otherwise isogenic HIV-1 reporter construct containing a disrupted nef gene and was thus Nef specific (Fig. 1A). Notably, some Nef proteins, e.g., those of SIVcpz, SIVmac, and SIVsmm, were highly effective at preventing CD8β surface expression in HIV-infected primary T cells at very low levels of EGFP (and thus Nef) expression (Fig. 1A). This is noteworthy because only CD4 and T cell receptor (TCR)-CD3 (in the case of HIV-2 and most SIVs) are down-modulated by Nef with similar efficacies, whereas significant modulation of MHC-I, CD28, and CXCR4 requires substantially higher levels of Nef expression (55). To quantify the efficiency of Nef-mediated receptor modulation, we divided the MFI for cells infected with the nef-defective control virus by the MFI for cells infected with viral constructs coexpressing Nef and EGFP. Statistical analyses showed that the effects of the various nef alleles on the surface expression of the A2-CD8β fusion on CEM cells correlated well with down-modulation of CD8αβ in SupT1 cells (R2 = 0.5153; P < 0.0001) and primary CD8+ T cells (R2 = 0.4952; P < 0.0001) (Fig. 1B). Thus, the CT of the human CD8β chain is sufficient for down-modulation by the vast majority of primate lentiviral Nefs, and down-modulation of the A2-CD8β fusion recapitulates the effect of Nef on unaltered CD8αβ molecules.
Fig 1.
Nef-mediated down-modulation of human CD8αβ is conserved between primate lentiviruses. (A) Flow cytometric analysis of CEM cells expressing A2-CD8β fusions, SupT1 cells, and primary CD8+ T cells transduced with HIV-1 recombinants expressing EGFP alone or together with the indicated nef alleles. The ranges of EGFP expression used to calculate the MFI of A2-CD8β fusion or CD8αβ expression are indicated. (B) Correlations between the efficiencies of Nef-mediated down-modulation of CD8αβ in primary CD8+ T cells and of A2-CD8β fusions in CEM cells or CD8αβ in SupT1 cells. (C) Correlations between the efficiencies of Nef-mediated down-modulation of CD8αβ in primary CD8+ T cells and of CD28, CD4, and MHC-I in primary CD4+ T cells. Each symbol represents average n-fold down-modulation (n = 3) of the indicated receptor molecule by one individual nef allele analyzed. An overview of the HIV and SIV nef alleles used is provided in Table S1 in the supplemental material.
It has been suggested previously that down-modulation of the human CD8β chain and CD4 involves common molecular interactions, i.e., Nef-mediated recruitment of AP-2 to the cytoplasmic domain of CD8β at the plasma membrane to promote clathrin-dependent endocytosis (63). In agreement with this assumption, we found that the potency of the 42 primate lentiviral Nef proteins in down-modulation of CD8αβ correlated with the effects on CD4 (R2 = 0.1305; P = 0.0187) and CD28 (R2 = 0.2672; P = 0.0004) in primary T cells but not with the modulation of MHC-I (Fig. 1C). However, the correlation between Nef-mediated modulation of CD4 and CD8αβ was imperfect, suggesting that both activities are mediated by overlapping domains in Nef but are functionally separable.
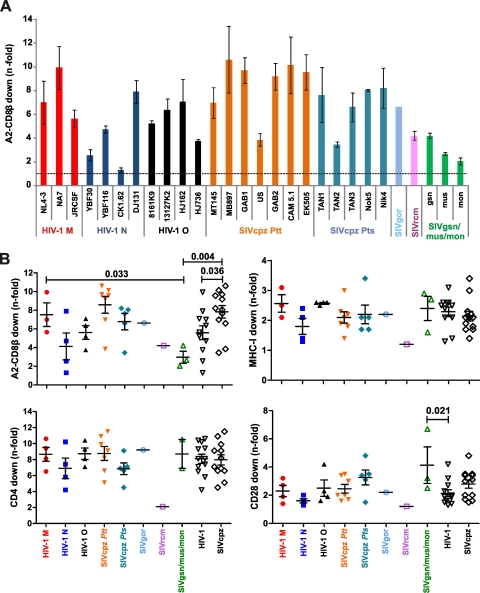
Next, we examined the effects of nef alleles on CD8αβ based on their species of origin and phylogenetic relationships. First, we examined 28 nef alleles from HIV-1 and its SIV precursors from apes and monkeys. This set encompassed HIV-1 M (major), the main causative agent of AIDS, and the nonpandemic or rare HIV-1 O (outlier) and N (non-M, non-O) groups, which arose from independent zoonotic transmissions of SIVcpz from the Pan troglodytes troglodytes subspecies of chimpanzees; SIVcpz from both P. t. troglodytes and Pan troglodytes schweinfurthii, which has not been found in humans; SIVgor from gorillas, which may be the original host of HIV-1 O; and SIVrcm, SIVgsn, SIVmus, and SIVmon, from red-capped mangabeys and greater spot-nosed, mustached, and mona monkeys, respectively (Cercocebus torquatus, Cercopithecus nictitans, Cercopithecus cephus, and Cercopithecus mona, respectively) (60). The latter are relevant for the emergence of HIV-1 because current data indicate that the precursor of SIVrcm recombined with the common ancestor of SIVgsn/mus/mon in chimpanzees to give rise to SIVcpz (6). Notably, most SIV nef alleles were obtained directly from uncultured ape or monkey material and thus did not contain adaptive changes in response to exposure to human cells (52, 55). For detailed quantitative analyses, we used the results obtained with CEM cells expressing A2-CD8β fusions because the effects of Nef showed the greatest dynamic range (Fig. 1A). Although all nef alleles that modulated A2-CD8β fusions also down-modulated unmodified CD8αβ molecules (Fig. 1B), the effects were often saturated in primary CD8+ T cells (Fig. 1A) and were (to some extent) dependent on the peripheral blood mononuclear cell (PBMC) donor (data not shown). We found that HIV-1, SIVcpz, and SIVgor nef alleles down-modulated A2-CD8β fusions with efficiencies ranging from 1.3- to 10.6-fold (Fig. 2A). On average, nef alleles from SIVcpz (7.8 ± 2.3; n = 12 [numbers give mean n-fold down-modulation ± standard deviation]) were significantly more active (P = 0.036) in modulating A2-CD8β fusions than HIV-1 nef alleles (5.6 ± 2.5; n = 11) (Fig. 2B). Our finding that SIVcpz Nefs are capable of targeting the CT of human CD8β is not surprising, because the amino acid sequence of this domain is identical in humans and chimpanzees (16). On average, nef alleles from the P. t. troglodytes subspecies of chimpanzees (which have transmitted the virus to humans) were more active in down-modulating A2-CD8β fusions than nef genes derived from the P. t. schweinfurthii subspecies (Fig. 2B). Furthermore, as a group, nef alleles from pandemic HIV-1 M strains reduced cell surface expression of A2-CD8β fusions more effectively than those from the nonpandemic N and O groups of HIV-1 (Fig. 2B). However, due to the limited sample size and relatively high variation of Nef function within each group, these differences failed to reach significance. In comparison to HIV-1 and SIVcpz Nefs, those of SIVgsn, SIVmus, and SIVmon showed only modest activity in down-modulating A2-CD8β fusions, although they potently kept MHC-I molecules away from the cell surface (Fig. 2B). The sequences of the CT domains of the CD8β molecules of greater spot-nosed, mustached, and mona monkeys are not available. Thus, it remains to be elucidated whether the SIVgsn, SIVmus, and SIVmon Nefs are more effective in down-modulating the CD8β variants found in their cognate host species. Most likely, SIVcpz obtained its nef gene from the precursor of SIVrcm, not from a possible common ancestor of SIVgsn/mus/mon (6, 55). Thus, it is noteworthy that the single SIVrcm Nef available for analysis was more active in modulating A2-CD8β fusions and CD8αβ than the SIVgsn, SIVmus, and SIVmon Nefs but was less effective than the majority of HIV-1 and SIVcpz Nefs (Fig. 2). Altogether, our results demonstrate that down-modulation of CD8αβ is a conserved property of the M, N, and O groups of HIV-1 and their simian counterparts, SIVcpz and SIVgor. Since the CT domains of CD8β are identical between chimpanzees and humans, SIVcpz Nefs were already capable of modulating human CD8αβ after zoonotic transmission, and this function was largely maintained in the new human host. Further studies are necessary to clarify whether more common infection of CD8+ T cells in SIVcpz-infected chimpanzees than in HIV-1-infected humans may explain why SIVcpz nef alleles are, on average, more active in modulating A2-CD8β fusions than those of HIV-1.
Fig 2.
Modulation of A2-CD8β fusions and other receptors by nef alleles from HIV-1 and its closest SIV counterparts. (A) Efficiencies of A2-CD8β fusion down-modulation by nef alleles from the indicated HIV-1 and SIV strains. Data shown are average values ± standard deviations (SD) derived from triplicate experiments. (B) Comparison of A2-CD8β fusion, MHC-I, CD4, and CD28 down-modulation by nef alleles from the indicated groups of primate lentiviruses. A2-CD8β fusion modulation was measured in CEM cells, and down-modulation of the remaining receptors was measured in primary CD4+ T cells. Each symbol represents the average for three independent measurements, and similar results were obtained in SupT1 cells.
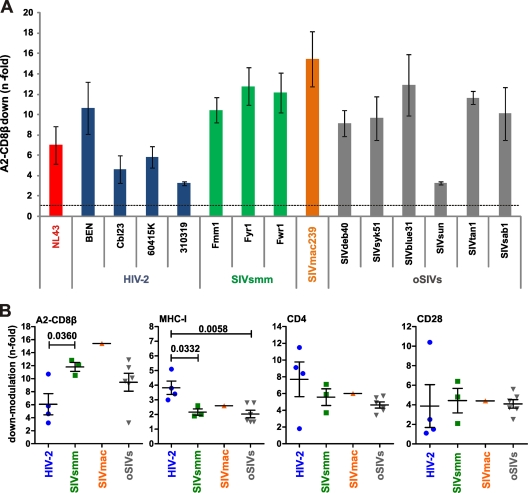
Next, we examined the capability of nef alleles from the SIVsmm/SIVmac/HIV-2 lineage to modulate A2-CD8β fusions in more detail. HIV-2 arose from multiple independent transmissions of SIVsmm from sooty mangabeys (Cercocebus atys) to humans (23, 60). The SIVsmm/SIVmac/HIV-2 lineage is of particular interest for studies on determinants of the pathogenesis of AIDS because SIVsmm is usually nonpathogenic in its natural monkey host but is highly pathogenic in experimentally infected rhesus macaques (SIVmac). The phenotype of HIV-2 in humans is intermediate: it can cause AIDS but is substantially less virulent than HIV-1, and many HIV-2-infected individuals show low viral loads and become long-term nonprogressors (24, 38, 43). All seven HIV-2 and SIVsmm nef alleles tested significantly reduced the levels of A2-CD8β fusions and CD8αβ cell surface expression, albeit with differential efficiencies, ranging from 3.2- to 12.8-fold in CEM cells and from 1.9- to 21.8-fold in primary T cells (Fig. 3A and data not shown). Unexpectedly, the SIVsmm Nefs were significantly more effective in down-modulating A2-CD8β fusions than those of HIV-2 (11.8- ± 1.2-fold versus 6.1- ± 3.2-fold; P = 0.0360), although HIV-2 Nefs were more effective at removing MHC-I molecules from the cell surface (3.8- ± 0.9-fold versus 2.2- ± 0.4-fold; P = 0.0332) (Fig. 3B). In agreement with published data (63), the nef allele of the molecular SIVmac239 clone, which is commonly used for studies on viral pathogenesis in the SIV-macaque model, was highly effective at down-modulating A2-CD8β fusions and CD8αβ from the cell surface (Fig. 3 and data not shown). Notably, it has been reported that activated intestinal double-positive CD4+ CD8+ memory T cells are a primary target of SIVmac infection in neonatal rhesus macaques (69) and are rapidly depleted in adult macaques with acute SIV infection (68). Our observation that nef alleles from SIVsmm and the macaque-adapted SIVmac derivative down-modulate CD8αβ with a high efficiency suggests a relevant role of this Nef function in viral immune evasion in both nonpathogenic natural and pathogenic experimental SIV infections.
Fig 3.
Modulation of A2-CD8β fusions and other receptors by nef alleles from HIV-2, their SIVsmm and SIVmac counterparts, and other highly divergent primate lentiviruses. (A) Efficiencies of A2-CD8β fusion down-modulation by nef alleles from the indicated HIV-2 and SIV strains. (B) Comparison of A2-CD8β fusion, MHC-I, CD4, and CD28 down-modulation by nef alleles from the indicated groups of primate lentiviruses. oSIVs, SIVs that were not involved in the evolution of HIV-1 and HIV-2. A2-CD8β fusion modulation was measured in CEM cells, and down-modulation of the remaining receptors was measured in primary CD4+ T cells. Refer to the legend to Fig. 2 for further details.
To further determine whether down-modulation of CD8β is a general property of primate lentiviral Nef proteins, we analyzed nef alleles from several additional highly divergent primate lentiviruses: SIVdeb from De Brazza monkeys (Cercopithecus neglectus), SIVsyk from Sykes monkeys (Cercopithecus albogularis), SIVblu from blue monkeys (Cercopithecus mitis), SIVsun from sun-tailed monkeys (Cercopithecus solatus), SIVtan from tantalus monkeys (Chlorocebus tantalus), and SIVsab from Sabaeus monkeys (Chlorocebus sabaeus). Together with the HIV and SIV nef alleles described above, these nef genes represented 5 of the 6 major primate lentiviral lineages known to date (23, 60). We found that all but the SIVsun sol-36 nef allele efficiently down-modulated A2-CD8β fusions and CD8αβ (Fig. 3 and data not shown). On average, these SIV nef alleles were more active in modulating A2-CD8β fusions than nef alleles derived from HIV-2 (9.5- ± 3.3-fold versus 6.1- ± 3.2-fold; P = 0.15), although the latter were more effective at modulating MHC-I (3.8- ± 0.9-fold versus 2.0- ± 0.6-fold; P = 0.0058) and CD4 (7.7- ± 4.1-fold versus 4.6- ± 0.9-fold; P = 0.11) cell surface expression (Fig. 3B). The effects of Nef on MHC-I, CD4, and CD28 correlated well in SupT1 and primary T cells, and these functional differences were observed in both cell types (data not shown).
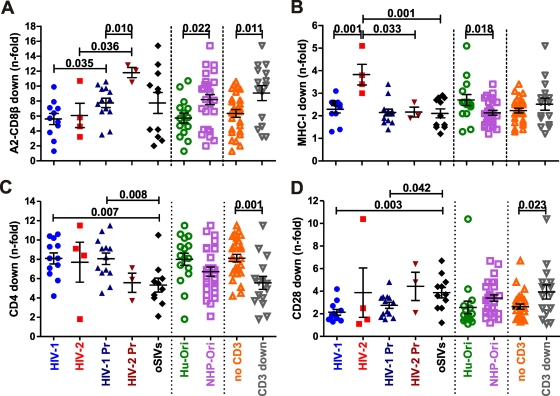
To further assess possible differences in Nef-mediated receptor modulation dependent on phylogenetic relationships, species of origin, and biological properties, we next compared the efficiencies of all nef alleles derived from HIV-1 and HIV-2, their direct SIV precursors, SIVs that (to our current knowledge) have not been transmitted to humans, viruses of human and nonhuman primate (NHP) origin, and viruses that do or do not down-modulate TCR-CD3 (55) in down-modulating A2-CD8β fusions, MHC-I, CD4, and CD28. HIV-1 Nefs were significantly less active than those of HIV-2 in down-modulating MHC-I (2.3- ± 0.6-fold versus 3.8- ± 0.9-fold; P = 0.001) but not in modulating A2-CD8β fusions or CD4 (Fig. 4). As a group, nef alleles derived from HIVs (Hu-Ori) were significantly (P = 0.022) less potent in modulating A2-CD8β fusions but more active in modulating MHC-I and CD4 than those derived from SIVs (NHP-Ori) (Fig. 4). Finally, we found that nef alleles from viruses that are unable to down-modulate TCR-CD3 (i.e., HIV-1 and its vpu-containing SIV precursors) were, on average, significantly less effective in down-modulating A2-CD8β fusions (6.3- ± 2.7-fold versus 9.1- ± 3.9-fold; P = 0.011) and CD28 (2.6- ± 1.2-fold versus 3.9- ± 2.4-fold; P = 0.023), but more active in down-modulating CD4 (8.1- ± 2.0-fold versus 5.6- ± 2.6-fold; P = 0.001), than nef genes from primate lentiviruses that down-modulate TCR-CD3 (Fig. 4).
Fig 4.
Comparison of Nef-mediated receptor modulation by human and nonhuman primate lentiviruses. Quantitative assessment was performed to measure down-modulation of the A2-CD8β fusion (A), MHC-I (B), CD4 (C), and CD28 (D) by nef alleles, based on the species of origin (humans versus nonhuman primates) and biological properties (unable or able to down-modulate TCR-CD3) of the nef alleles examined. A2-CD8β fusion modulation was measured in CEM cells, and down-modulation of the remaining receptors was measured in primary CD4+ T cells. Abbreviations: Pr, precursor; Hu, human; NHP, nonhuman primate.
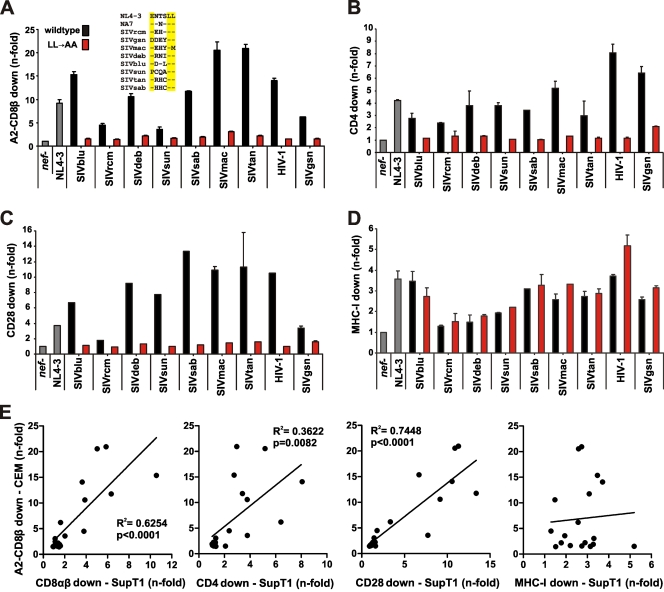
The data described above show that down-modulation of CD8β is a conserved property of the vast majority of primate lentiviral Nef proteins. Previous studies suggested that some conserved Nef functions, such as down-modulation of MHC-I, may involve different domains in different primate lentiviruses and thus may have evolved independently during primate lentiviral evolution (22). Thus, we next examined whether or not down-modulation of CD8β involves similar or distinct domains in highly divergent primate lentiviral Nef proteins. First, we focused on the E/DXXXLL motif in the C-terminal flexible loop of Nef because previous studies have shown that this motif is important for the ability of HIV-1 Nef to interact with different adaptor proteins (i.e., AP-1, AP-2, and AP-3) and to down-modulate CD4 and CD8β (12, 35, 63). The acidic residue and the first leucine are preserved in the great majority of HIV and SIV strains (12). In contrast, the second leucine is conserved in HIV-1 Nefs but is frequently replaced by a valine, threonine, or methionine residue in SIV and HIV-2 Nef proteins (Fig. 5A and data not shown). To further determine the functional relevance of the EXXXLL motif, we introduced alanine substitutions into the LL/M motif in eight highly divergent SIV Nefs and the control HIV-1 NA7 Nef. Notably, two SIV strains examined contained changes, to EXXXLM (SIVmac) and PXXXLL (SIVsun), in the otherwise conserved EXXXLL motif (Fig. 5A, inset). Mutations in the LL/M residues generally disrupted the capability of primate lentiviral Nefs to down-modulate CD4, CD28, and CD8β but had no significant effect on protein expression levels and MHC-I modulation (Fig. 5A to D and data not shown). Correlation analyses showed that the effects of the various wild-type and mutant nef alleles on the surface expression of A2-CD8β fusions on CEM cells correlated well with down-modulation of CD8αβ in CD8+ T cells (R2 = 0.6254; P < 0.0001) and with down-modulation of CD4 (R2 = 0.3622; P = 0.0082) and CD28 (R2 = 0.7448; P < 0.0001) but not MHC-I (Fig. 5E). The wild-type SIVsun Nef protein, which contains PXXXLL instead of EXXXLL, had little effect on CD8β but effectively down-modulated CD4 and CD28 from the cell surface (Fig. 5A to C). The latter is in agreement with the previous finding that the acidic residue plays a differential role in the modulation of dileucine-dependent Nef targets and is not generally required for Nef-mediated receptor modulation (12). Altogether, our results support the hypothesis that the E/DXXXLL motif is of general importance for the capability of primate lentiviral Nef proteins to down-modulate CD4, CD28, and CD8 from the cell surface. Our data also imply that some Nef proteins that contain changes in the acidic residue of the EXXXLL motif are still able to connect CD4 and CD28 to adaptor proteins.
Fig 5.
Importance of the EXXXLL motif in the C-terminal flexible loop for the modulation of cellular receptors by primate lentiviral Nef proteins. (A to D) CEM cells expressing the A2-CD8β fusion (A) or SupT1 cells (B to D) were transduced with VSV-G-pseudotyped vpu- and env-defective NL4-3-based constructs expressing the indicated nef alleles and assayed for surface expression of the A2-CD8β fusion (A), CD4 (B), CD28 (C), and MHC-I (D). Quantification was performed as described in Materials and Methods. Data shown are average values ± SD derived from two independent experiments. The inset in panel A shows an amino acid sequence comparison of the various EXXXLL-corresponding regions analyzed. The NL4-3 motif is shown as a reference at the top. Dots indicate amino acid identity. (E) Correlations between efficiencies of down-modulation of the A2-CD8β fusion on CEM cells and modulation of CD8αβ, CD4, CD28, and MHC-I on SupT1 cells.
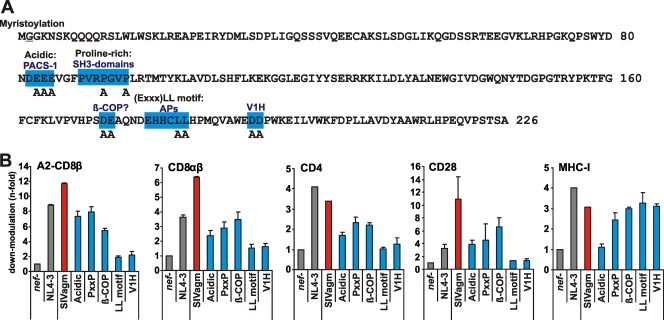
To further define the functional domains involved in the interaction of primate lentiviral Nefs with CD8β and other cellular receptors, we generated five mutant SIVagmSab nef constructs containing changes in the following sequence motifs known to be important for HIV-1 Nef function: (i) the acidic domain that interacts with the sorting protein PACS-2 to target Nef to the perinuclear region (5, 45) and with the CT of MHC-I molecules to support their interaction with AP-1 (70); (ii) the PXXP motif, involved in interactions with signaling molecules, such as the SH3 domains of various tyrosine kinases (51); (iii) two negatively charged residues (E154E155 in NL4-3 and D172E173 in SIVagmSab) that may interact with β-COP (27, 44, 53); (iv) the E/DXXXLL AP interaction site described above; and (v) the DD motif implicated in Nef binding to the V1H subunit of the vacuolar ATPase and AP-2 (36, 37) (Fig. 6A). SIVagmSab Nef was selected for detailed analysis because infection of African green monkeys with this virus represents one of the best-studied models of nonpathogenic natural SIV infection (61). We found that mutations in the dileucine-based and DD motifs in the C-terminal flexible loop disrupted the effects of SIVagm Nef on CD8β, CD4, and CD28 but had no effect on MHC-I down-modulation (Fig. 6B). In contrast, changes in the acidic domain disrupted MHC-I modulation but only partially impaired the other Nef functions (Fig. 6B). Changes in the PXXP motif and the DE residues preceding the EXXXLL motif resulted in intermediate phenotypes and reduced the modulation of CD8β, CD4, and CD28 (Fig. 6B). Our results show that both the dileucine-based motif and the DD residues in the C-terminal flexible loop of SIVagm Nef are required for down-modulation of CD8β, CD4, and CD28. Previous data have shown that analogous changes in the HIV-1 Nef protein also disrupt the modulation of these receptors (63), supporting the observation that the EXXXLL and DD motifs play similar roles in SIVagm and HIV-1 Nef function.
Fig 6.
Functional analysis of SIVagmSab Nef mutants. (A) The SIVagmSab Nef amino acid sequence is given in single-letter code. Mutated sequence motifs are highlighted by blue boxes, and positions of alanines are indicated. (B) CEM cells expressing A2-CD8β fusions or SupT1 cells were transduced with HIV-1 recombinants expressing EGFP alone or together with the indicated nef alleles and were assayed for surface expression of the A2-CD8β fusion, CD8αβ, CD4, CD28, and MHC-I. Quantification was performed as described in Materials and Methods. Data shown are average values ± SD derived from two independent experiments.
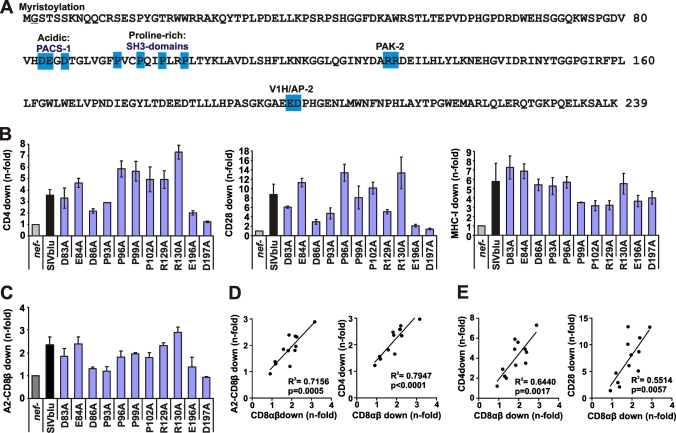
The results described above and previously published data (35, 63) suggest that down-modulation of CD8β, CD4, and CD28 may involve similar domains in Nef. To further examine whether this may generally be the case, we generated a set of 11 SIVblu mutant Nefs containing alanine substitutions at single amino acid residues. As shown in Fig. 7A, these point mutations affected the acidic domain, the proline-rich region, a dibasic motif reported to be critical for PAK-2 binding (46, 48), and the diacidic putative V1H–AP-2 interaction site in the C-terminal flexible loop of Nef. The Nef protein from SIVblu, which infects blue monkeys (Cercopithecus mitis), was selected for analysis because it is highly divergent from other Nef proteins that have been examined functionally in some detail (i.e., HIV-1, SIVmac, and SIVagm Nef proteins) and is active in most aspects of Nef function (Fig. 7B and data not shown). The mutations had differential effects on SIVblu Nef function. As expected from previous results on HIV-1 (56, 63) and SIVagm (Fig. 6) Nef function, the ED residues (V1H–AP-2 interaction site) were critical for down-modulation of CD4, CD8β, and CD28 (Fig. 7B and C). The D86A and P93A changes also markedly impaired these three Nef functions. This was unexpected, since the acidic domain and the PXXP motif are not critical for down-modulation of CD4 and CD28 by HIV-1 Nef (20). Notably, these changes did not reduce the efficiency of MHC-I down-modulation (Fig. 7B), and all SIVblu mutant Nefs were expressed at similar levels (see Fig. S2 in the supplemental material). The remaining mutations had little, if any, effect on modulation of CD4 and CD28, and in some cases (i.e., E84A, P96A, and R130A mutations) they even increased these Nef activities. The efficiencies of CD4 and CD28 down-modulation by this set of SIVblu Nefs correlated well with one another (R2 = 0.7718; P = 0.0002) (data not shown) but not with down-modulation of MHC-I (data not shown). The effects of wild-type and mutant SIVblu Nefs on CD8β were highly reproducible in CEM cells expressing A2-CD8β fusions and in SupT1 cells (R2 = 0.7156; P = 0.0005) (Fig. 7D). The efficiencies of the 12 SIVblu Nefs in down-modulating CD8αβ correlated very well with their potency in CD4 and CD28 modulation (Fig. 7D and E). These results thus further support the hypothesis that primate lentiviral Nefs use similar surfaces to modulate CD8β, CD4, and CD28 and further emphasize the important role of the DD motif in the C-terminal flexible loop of Nef in these activities. Our data also show, however, that residues in the acidic and proline-rich regions are more important for efficient modulation of CD8β, CD4, and CD28 by SIVblu Nef than by HIV-1 Nef proteins.
Fig 7.
Functional analysis of SIVblu Nef mutants. (A) The SIVblu Nef amino acid sequence is given in single-letter code. The positions of single alanine substitutions are highlighted by blue boxes, and the domains affected by these mutations are indicated. (B and C) Jurkat T cells (B) or CEM cells (C) were transfected with pCGCG vectors expressing EGFP alone or together with the indicated nef alleles and were assayed for surface expression of CD4, CD28, MHC-I, and the A2-CD8β fusion. Quantification was performed as described in Materials and Methods. Data shown are average values ± SD derived from two independent experiments. (D) Correlations between efficiencies of down-modulation of the A2-CD8β fusion on CEM cells and of CD8αβ (left) or CD4 (right) on SupT1 cells. (E) Correlations between Nef-mediated down-modulation of CD8αβ on SupT1 cells and of CD4 (left) or CD28 (right) on Jurkat T cells.
DISCUSSION
In the present study, we show that the ability of Nef to down-modulate CD8αβ from the surfaces of virally infected primary T cells is conserved in the vast majority of primate lentiviruses and that the 17-amino-acid CT sequence of CD8β is generally sufficient to confer sensitivity to Nef. Nef proteins of the SIV precursors of both human immunodeficiency viruses (i.e., SIVcpz, SIVgor, and SIVsmm) are highly effective in modulating human CD8αβ. Thus, like other Nef functions, such as modulation of CD4, CD3, CD28, and MHC-I and enhancement of viral infectivity and replication (3, 40, 41, 42, 55, 58, 62), and unlike Nef-mediated tetherin antagonism (28, 52, 74), the effect of Nef on CD8αβ is species independent, and SIVs can manipulate human CD8+ T cells without adaptive changes. This is not surprising, since the amino acid sequences of the cytoplasmic domains of human and macaque CD8β, encoded by the CY1 exon, which is present in all splice variants and targeted by Nef (63, 67), differ by only a single amino acid (CCRRRRARLRFMKQFYK to CCRRRRARLRFIKQFYK). Our finding that the effect of Nef on CD8αβ is conserved and usually highly effective supports a relevant role in direct HIV and SIV infections and manipulation of CD8+ CTLs in vivo.
Previous studies have shown that some Nef activities, i.e., the ability to down-modulate CD4, CD28, and MHC-I and also to enhance viral infectivity and replication, are conserved across most or all primate lentiviral lineages (4, 55). Thus, although primate lentiviral Nef proteins are highly variable and often share only about 30% amino acid identity (34), some functional interactions are obviously preserved. In some cases, however, the same activities may be mediated by different domains in Nef and thus evolved independently during primate lentiviral evolution (7, 25, 65). Our results support the hypothesis that the EXXXLL and (most likely) DD motifs in the C-terminal flexible loop of Nef are generally critical for Nef-mediated down-modulation of CD4, CD8β, and CD28. The finding that Nef proteins from several highly divergent primate lentiviruses use similar conserved motifs to perform these functions supports the idea that these motifs evolved early during primate lentiviral evolution.
Previous studies have demonstrated that the EXXXLL motif in the C-terminal flexible loop of Nef is critical for interaction with several clathrin adaptor proteins, i.e., AP-1, AP-2, and AP-3 (35, 36). In contrast, the adjacent DD motif is critical for AP-2 binding of HIV-1 Nef but dispensable for the interaction with AP-1 and AP-3 (36). Thus, our finding that changes in the DD motif disrupt the effects of HIV-1, SIVagm, and SIVblu Nefs on CD4, CD8β, and CD28 cell surface expression further supports an important role of AP-2 in the down-modulation of these receptors. Notably, recent data suggest that AP-1 is also important for down-modulation of CD8β and CD28 and (unlike the case with MHC-I) that this involves the EXXXLL motif in the HIV-1 Nef proteins (35). These results are not mutually exclusive, because accumulating evidence suggests that Nef may interact with various adaptor proteins in order to remove them from the cell surface and to induce their redistribution to the trans-Golgi network and endosomes and/or lysosomal degradation. The relative contributions of the different adaptors and pathways (e.g., endocytosis versus intracellular retention) need further study and seem to vary not only for different Nef functions but also between different primate lentiviruses. In either case, it is becoming evident that the EXXXLL and DD motifs in the C-terminal flexible loop of Nef both seem to be generally critical for down-modulation of CD4, CD8β, and CD28 as well as for up-modulation of the invariant chain (Ii) by Nef (56). A recent study showed that the EXXXLL and DD motifs are also critical for tetherin antagonism (73), further supporting their important role in primate lentiviral Nef function.
We observed striking correlations between the disruptive effects of mutations in the SIVagm and SIVblu Nefs on down-modulation of CD4, CD8β, and CD28 (Fig. 6 and 7). In agreement with published data on HIV-1 Nef function (35, 63), these results suggest that primate lentiviral Nef proteins use similar domains and largely overlapping surfaces to reduce the cell surface expression levels of these receptors. Nonetheless, the potencies with which primary HIV and SIV nef alleles modulate these receptors vary considerably and do not often correlate with one another. Furthermore, it is known that most of these Nef activities are genetically separable by specific point mutations (2, 14, 22, 26, 54, 65). One straightforward explanation for these results is that down-modulation of CD4, CD8β, and CD28 by Nef involves conserved interactions with cellular adaptor proteins and more variable interactions with the cytoplasmic tails of the various cellular receptors. However, alternative possibilities cannot be dismissed entirely. For example, recent data suggest that substitutions in and near the E/DXXXLL and DD motifs can have differential effects on the interaction of Nef with various adaptor proteins (12, 36). Thus, it is possible that some mutations may disrupt the interactions with specific adaptor proteins that are critical for Nef-mediated modulation of particular cellular receptors.
While mutations in the EXXXLL motif in Nef generally disrupted modulation of CD4, CD8β, and CD28, the effects of alanine substitutions in other conserved domains of Nef were context dependent. For example, the D86A and P93A changes, which affect the acidic domain and the proline-rich region of SIVblu Nef, severely impaired down-modulation of these receptors but not modulation of MHC-I (Fig. 7). In contrast, we and others have found that alanine substitutions in the acidic domain of HIV-1 Nef impair down-modulation of MHC-I much more severely than that of CD4 or CD8β (45, 56; data not shown). Furthermore, mutations in the PXXP motif of HIV-1 Nef disrupt the interaction with the SH3 domains of cellular kinases but not with CD4 (51). These findings further illustrate the enormous versatility and plasticity of Nef activities and suggest that some conserved domains may be involved in distinct functions in Nef proteins from different primate lentiviruses. We found that even analogous mutations in various HIV-1 Nefs may have entirely different effects on their functional activity (data not shown). Thus, generalizations based on functional analyses of single nef alleles should be avoided, which is noteworthy because the vast majority of studies have used the nef allele from the T cell line-adapted HIV-1 NL4-3 strain.
Our observation that Nef-mediated down-modulation of CD8β is a well-conserved and effective Nef function, together with the recent finding that the CT of CD8β represents a major target of HIV-1 Nef (35), supports a relevant role in viral immune evasion in vivo. This may seem surprising, since CD8+ T cells are not known to be major target cells of HIV or SIV infection. In fact, when we infected CD8+ T cells purified from PBMCs with HIV-1, we observed infection rates of only about 0.4% (data not shown), similar to published results (35). However, this may be different in vivo, because T cells frequently express both CD4 and CD8 during thymic development (17), and activated CD8+ T cells may express low levels of CD4 (10, 18, 31). In fact, virally infected CD8+ T cells have been detected in AIDS patients (50). Thus, further studies, particularly in appropriate animal models, are warranted and will help to elucidate the possible relevance of Nef-mediated down-modulation of CD8β in vivo. Infections of macaques with SIVmac and of African green monkeys with SIVagm currently represent the most common nonhuman primate models for studying the pathogenesis of AIDS and the mechanisms of viral immune evasion. Since the nef alleles from both SIVmac and SIVagm effectively down-modulate CD8β, in vivo studies on the importance of this function for primate lentiviral infection and manipulation of CD8+ T cells in vivo seem feasible, and our ongoing studies aim to define mutations in Nef that selectively impair down-modulation of CD8β.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jan Münch for support, Birgit Ott and Kerstin Regensburger for excellent technical assistance, and Daniel Sauter and Christine Goffinet for critical readings of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print 19 October 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Aiken C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871–5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akari H, et al. 2000. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J. Virol. 74:2907–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arhel N, et al. 2009. The inability to disrupt the immunological synapse between infected human T cells and APCs distinguishes HIV-1 from most other primate lentiviruses. J. Clin. Invest. 119:2965–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ariën KK, Verhasselt B. 2008. HIV Nef: role in pathogenesis and viral fitness. Curr. HIV Res. 6:200–208 [DOI] [PubMed] [Google Scholar]
- 5. Atkins KM, et al. 2008. HIV-1 Nef binds PACS-2 to assemble a multikinase cascade that triggers major histocompatibility complex class I (MHC-I) down-regulation: analysis using short interfering RNA and knock-out mice. J. Biol. Chem. 283:11772–11784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bailes E, et al. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 7. Bresnahan PA, Yonemoto W, Greene WC. 1999. Cutting edge: SIV Nef protein utilizes both leucine- and tyrosine-based protein sorting pathways for down-regulation of CD4. J. Immunol. 163:2977–2981 [PubMed] [Google Scholar]
- 8. Carlson JM, Brumme ZL. 2008. HIV evolution in response to HLA-restricted CTL selection pressures: a population-based perspective. Microbes Infect. 10:455–461 [DOI] [PubMed] [Google Scholar]
- 9. Chopera DR, et al. 2008. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 4:e1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cochrane A, et al. 2004. High levels of human immunodeficiency virus infection of CD8 lymphocytes expressing CD4 in vivo. J. Virol. 78:9862–9871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cohen GB, et al. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661–671 [DOI] [PubMed] [Google Scholar]
- 12. Coleman SH, et al. 2006. Modulation of cellular protein trafficking by human immunodeficiency virus type 1 Nef: role of the acidic residue in the ExxxLL motif. J. Virol. 80:1837–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. 1998. HIV-1 nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397–400 [DOI] [PubMed] [Google Scholar]
- 14. Craig HM, Pandori MW, Guatelli JC. 1998. Interaction of HIV 1 Nef with the cellular dileucine based sorting pathway is required for CD4 down regulation and optimal viral infectivity. Proc. Natl. Acad. Sci. U. S. A. 95:11229–11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeGottardi MQ, et al. 2008. Selective downregulation of rhesus macaque and sooty mangabey major histocompatibility complex class I molecules by Nef alleles of simian immunodeficiency virus and human immunodeficiency virus type 2. J. Virol. 82:3139–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delarbre C, Nakauchi H, Bontrop R, Kourilsky P, Gachelin G. 1993. Duplication of the CD8 beta-chain gene as a marker of the man-gorilla-chimpanzee clade. Proc. Natl. Acad. Sci. U. S. A. 90:7049–7053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Rossi A, et al. 1990. In vitro studies of HIV-1 infection in thymic lymphocytes: a putative role of the thymus in AIDS pathogenesis. AIDS Res. Hum. Retroviruses 6:287–298 [DOI] [PubMed] [Google Scholar]
- 18. Flamand L, et al. 1998. Activation of CD8+ T lymphocytes through the T cell receptor turns on CD4 gene expression: implications for HIV pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 95:3111–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gangadharan D, Cheroutre H. 2004. The CD8 isoform CD8alphaalpha is not a functional homologue of the TCR coreceptor CD8alphabeta. Curr. Opin. Immunol. 16:264–270 [DOI] [PubMed] [Google Scholar]
- 20. Geyer M, Fackler OT, Peterlin BM. 2001. Structure-function relationships in HIV-1 Nef. EMBO Rep. 2:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldrath AW, Hogquist KA, Bevan MJ. 1997. CD8 lineage commitment in the absence of CD8. Immunity 6:633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Greenberg ME, Iafrate AJ, Skowronski J. 1998. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 17:2777–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hahn BH, Shaw GM, De Cock KM, Sharp PM. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607–614 [DOI] [PubMed] [Google Scholar]
- 24. Hanson A, et al. 2005. Distinct profile of T cell activation in HIV type 2 compared to HIV type 1 infection: differential mechanism for immunoprotection. AIDS Res. Hum. Retroviruses 21:791–798 [DOI] [PubMed] [Google Scholar]
- 25. Hua J, Cullen BR. 1997. Human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus Nef use distinct but overlapping target sites for downregulation of cell surface CD4. J. Virol. 71:6742–6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iafrate AJ, Bronson S, Skowronski J. 1997. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 16:673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janvier K, et al. 2001. Nef-induced CD4 downregulation: a diacidic sequence in human immunodeficiency virus type 1 Nef does not function as a protein sorting motif through direct binding to beta-COP. J. Virol. 75:3971–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jia B, et al. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5:e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirchhoff F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8:55–67 [DOI] [PubMed] [Google Scholar]
- 30. Kirchhoff F, Schindler M, Specht A, Arhel N, Münch J. 2008. Role of Nef in primate lentiviral immunopathogenesis. Cell. Mol. Life Sci. 65:2621–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kitchen SG, Korin YD, Roth MD, Landay A, Zack JA. 1998. Costimulation of naive CD8+ lymphocytes induces CD4 expression and allows human immunodeficiency virus type 1 infection. J. Virol. 72:9054–9060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Le Gall S, et al. 2000. Distinct trafficking pathways mediate Nef-induced and clathrin-dependent major histocompatibility complex class I down-regulation. J. Virol. 74:9256–9266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Le Gall S, et al. 1998. Nef interacts with the μ subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC 1 molecules. Immunity 8:483–495 [DOI] [PubMed] [Google Scholar]
- 34. Leitner T, et al. 2003. HIV sequence compendium. LA-UR 04-7420. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 35. Leonard JA, Filzen T, Carter CC, Schaefer M, Collins KL. 2011. HIV-1 Nef disrupts intracellular trafficking of MHC-I, CD4, CD8, and CD28 by distinct pathways that share common elements. J. Virol. 85:6867–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lindwasser OW, et al. 2008. A diacidic motif in human immunodeficiency virus type 1 Nef is a novel determinant of binding to AP-2. J. Virol. 82:1166–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu X, Yu H, Liu SH, Brodsky FM, Peterlin BM. 1998. Interactions between HIV-1 Nef and vacuolar ATPase facilitate the internalization of CD4. Immunity 8:647–656 [DOI] [PubMed] [Google Scholar]
- 38. Marlink RG, et al. 1988. Clinical, hematological, and immunologic cross-sectional evaluation of individuals exposed to human immunodeficiency virus type-2 (HIV-2). AIDS Res. Hum. Retroviruses 4:137–148 [DOI] [PubMed] [Google Scholar]
- 39. Miura T, et al. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J. Virol. 83:2743–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Münch J, et al. 2007. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J. Virol. 81:13852–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Münch J, et al. 2005. Primary sooty mangabey simian immunodeficiency virus and human immunodeficiency virus type 2 nef alleles modulate cell surface expression of various human receptors and enhance viral infectivity and replication. J. Virol. 79:10547–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Münch J, Stolte N, Fuchs D, Stahl-Hennig C, Kirchhoff F. 2001. Efficient class I major histocompatibility complex down-regulation by simian immunodeficiency virus Nef is associated with a strong selective advantage in infected rhesus macaques. J. Virol. 75:10532–10536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pepin J, et al. 1991. HIV-2-induced immunosuppression among asymptomatic West African prostitutes: evidence that HIV-2 is pathogenic, but less so than HIV-1. AIDS 5:1165–1172 [PubMed] [Google Scholar]
- 44. Piguet V, et al. 1999. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of beta-COP in endosomes. Cell 97:63–73 [DOI] [PubMed] [Google Scholar]
- 45. Piguet V, et al. 2000. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat. Cell Biol. 2:163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pulkkinen K, Renkema GH, Kirchhoff F, Saksela K. 2004. Nef associates with p21-activated kinase 2 in a p21-GTPase-dependent dynamic activation complex within lipid rafts. J. Virol. 78:12773–12780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Quaranta MG, Mattioli B, Giordani L, Viora M. 2004. HIV-1 Nef equips dendritic cells to reduce survival and function of CD8+ T cells: a mechanism of immune evasion. FASEB J. 18:1459–1461 [DOI] [PubMed] [Google Scholar]
- 48. Renkema GH, Manninen A, Mann DA, Harris M, Saksela K. 1999. Identification of the Nef-associated kinase as p21-activated kinase 2. Curr. Biol. 9:1407–1410 [DOI] [PubMed] [Google Scholar]
- 49. Saha K, et al. 2001. Isolation of primary HIV-1 that targets CD8+ T lymphocytes using CD8 as a receptor. Nat. Med. 7:65–72 [DOI] [PubMed] [Google Scholar]
- 50. Saha K, Zhang J, Zerhouni B. 2001. Evidence of productively infected CD8+ T cells in patients with AIDS: implications for HIV-1 pathogenesis. J. Acquir. Immune Defic. Syndr. 26:199–207 [DOI] [PubMed] [Google Scholar]
- 51. Saksela K, Cheng G, Baltimore D. 1995. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 14:484–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sauter D, et al. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schaefer MR, Wonderlich ER, Roeth JF, Leonard JA, Collins KL. 2008. HIV-1 Nef targets MHC-I and CD4 for degradation via a final common beta-COP-dependent pathway in T cells. PLoS Pathog. 4:e1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schindler M, et al. 2004. Comprehensive analysis of nef functions selected in simian immunodeficiency virus-infected macaques. J. Virol. 78:10588–10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schindler M, et al. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055–1067 [DOI] [PubMed] [Google Scholar]
- 56. Schindler M, et al. 2003. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 77:10548–10556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schmitz JE, et al. 1998. Expression of the CD8alpha beta-heterodimer on CD8(+) T lymphocytes in peripheral blood lymphocytes of human immunodeficiency virus− and human immunodeficiency virus+ individuals. Blood 92:198–206 [PubMed] [Google Scholar]
- 58. Schmökel J, et al. 2011. The presence of a vpu gene and the lack of Nef-mediated downmodulation of T cell receptor-CD3 are not always linked in primate lentiviruses. J. Virol. 85:742–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338–342 [DOI] [PubMed] [Google Scholar]
- 60. Sharp PM, Hahn BH. 2010. The evolution of HIV-1 and the origin of AIDS. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:2487–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sodora DL, et al. 2009. Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat. Med. 15:861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Specht A, et al. 2008. Selective down-modulation of HLA-A and -B by Nef alleles from different groups of primate lentiviruses. Virology 373:229–237 [DOI] [PubMed] [Google Scholar]
- 63. Stove V, et al. 2005. Human immunodeficiency virus Nef induces rapid internalization of the T-cell coreceptor CD8αβ. J. Virol. 79:11422–11433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stumptner-Cuvelette P, et al. 2001. HIV-1 Nef impairs MHC class II antigen presentation and surface expression. Proc. Natl. Acad. Sci. U. S. A. 98:12144–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Swigut T, Iafrate AJ, Münch J, Kirchhoff F, Skowronski J. 2000. SIV and HIV Nef proteins use different surfaces to downregulate class I major histocompatibility antigen expression. J. Virol. 74:5691–5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Swigut T, et al. 2004. Impact of Nef-mediated downregulation of major histocompatibility complex class I on immune response to simian immunodeficiency virus. J. Virol. 78:13335–13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thakral D, Dobbins J, Devine L, Kavathas PB. 2008. Differential expression of the human CD8beta splice variants and regulation of the M-2 isoform by ubiquitination. J. Immunol. 180:7431–7442 [DOI] [PubMed] [Google Scholar]
- 68. Veazey RS, et al. 2000. Dynamics of CCR5 expression by CD4(+) T cells in lymphoid tissues during simian immunodeficiency virus infection. J. Virol. 74:11001–11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang X, Das A, Lackner AA, Veazey RS, Pahar B. 2008. Intestinal double-positive CD4+CD8+ T cells of neonatal rhesus macaques are proliferating, activated memory cells and primary targets for SIVMAC251 infection. Blood 112:4981–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wonderlich ER, Leonard JA, Collins KL. 2011. HIV immune evasion disruption of antigen presentation by the HIV Nef protein. Adv. Virus Res. 80:103–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xu XN, et al. 1997. Evasion of cytotoxic T lymphocyte (CTL) responses by Nef-dependent induction of Fas ligand (CD95L) expression on simian immunodeficiency virus-infected cells. J. Exp. Med. 186:7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zerhouni B, Nelson JA, Saha K. 2004. Isolation of CD4-independent primary human immunodeficiency virus type 1 isolates that are syncytium inducing and acutely cytopathic for CD8+ lymphocytes. J. Virol. 78:1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang F, et al. 2011. SIV Nef proteins recruit the AP-2 complex to antagonize tetherin and facilitate virion release. PLoS Pathog. 7:e1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang F, et al. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.