Abstract
We previously reported finding the RNA of a type K human endogenous retrovirus, HERV-K (HML-2), at high titers in the plasma of HIV-1-infected and cancer patients (R. Contreras-Galindo et al., J. Virol. 82:9329–9236, 2008.). The extent to which the HERV-K (HML-2) proviruses become activated and the nature of their activated viral RNAs remain important questions. Therefore, we amplified and sequenced the full-length RNA of the env gene of the type 1 and 2 HERV-K (HML-2) viruses collected from the plasma of seven HIV-1-infected patients over a period of 1 to 3 years and from five breast cancer patients in order to reconstruct the genetic evolution of these viruses. HERV-K (HML-2) RNA was found in plasma fractions of HIV-1 patients at a density of ∼1.16 g/ml that contained both immature and correctly processed HERV-K (HML-2) proteins and virus-like particles that were recognized by anti-HERV-K (HML-2) antibodies. RNA sequences from novel HERV-K (HML-2) proviruses were discovered, including K111, which is specifically active during HIV-1 infection. Viral RNA arose from complete proviruses and proviruses devoid of a 5′ long terminal repeat, suggesting that the expression of HERV-K (HML-2) RNA in these patients may involve sense and antisense transcription. In HIV-1-infected individuals, the HERV-K (HML-2) viral RNA showed evidence of frequent recombination, accumulation of synonymous rather than nonsynonymous mutations, and conserved N-glycosylation sites, suggesting that some of the HERV-K (HML-2) viral RNAs have undergone reverse transcription and are under purifying selection. In contrast, HERV-K (HML-2) RNA sequences found in the blood of breast cancer patients showed no evidence of recombination and exhibited only sporadic viral mutations. This study suggests that HERV-K (HML-2) is active in HIV-1-infected patients, and the resulting RNA message reveals previously undiscovered HERV-K (HML-2) genomic sequences.
INTRODUCTION
Human endogenous retroviruses (HERVs) are an integral part of the cellular DNA. These viruses entered hominid species over millions of years and have been transmitted in a Mendelian fashion (31). The HERV-K family constitutes the most recent entrant form of these viruses (40). HERV-K has replicated during the evolution of humans by infection rather than retrotransposition (4). During this process, these viruses have increased in copy number throughout the genome and today account for approximately 3,000 proviral fragments (35). A vast majority of these elements have accumulated lethal mutations in the viral genes through DNA replication. In addition, internal viral genes have been removed by recombination of the 5′ and 3′ long terminal repeats (LTRs), producing numerous solo HERV-K LTRs (20).
Phylogenetic analysis within the pol region indicates that the HERV-K family is subdivided into 10 subfamilies, termed HML-1 to HML-10 (27). HERV-K (HML-2) contains the most recent proviral forms and appears to be transcriptionally active, with proviruses with intact open reading frames (ORFs) for all viral genes. The HERV-K (HML-2) subfamily is subdivided into type 1 and type 2 elements on the basis of a 292-bp fragment that preserves the coding capacity of the env gene and is deleted in type 1 proviruses (31). Eight full-length HERV-K (HML-2) members have been shown to be polymorphically inserted among humans (3, 19, 43). HERV-K (HML-2) viruses also appear able to produce virus-like particles (VLPs) that are similar to exogenous retroviruses, but are widely held to be noninfectious (3, 7, 31, 40).
We previously detected HERV-K (HML-2) viral RNA in the plasma of human immunodeficiency virus type 1 (HIV-1)-infected patients at high titers (106 to 1010 RNA copies/ml in many cases), suggesting viral activation (5–7). Similar viral loads were detected in lymphoma and breast cancer patients. In the plasma of lymphoma patients, high HERV-K (HML-2) viral RNA titers, reverse transcriptase (RT) activity, and HERV-K (HML-2) viral proteins were found in blood fractions with HERV-K (HML-2) VLPs (7). However, which proviruses are expressed, the extent to which that expression varies, and the nature and characteristics of the viral RNA found in the plasma of patients with HIV-1 has remained unknown.
To address these issues, we amplified full-length HERV-K (HML-2) env RNA from plasma samples of HIV-1-infected patients collected over a period of 1 to 3 years, as well as from single plasma samples of breast cancer patients whose viral load was previously found to be elevated. Analysis of the HERV-K env gene sequences found in HIV-1 patients revealed evidence of active recombination, accumulation of synonymous mutations, and the preservation of encoded glycosylation sites. In contrast, in breast cancer patients who, like HIV-1-infected individuals, have high titers of HERV-K (HML-2) env RNA in their blood, the viral RNA sequences did not show evidence of recombination. Therefore, at least in some HIV-1-infected humans, HERV-K (HML-2) RNA found in the blood shows signs of having undergone reverse transcription and purifying selection. Further, we show that by studying these viral RNA messages in the blood of living patients, we can identify previously undiscovered HERV-K (HML-2) genomic sequences.
MATERIALS AND METHODS
Study subjects.
Plasma samples were obtained following protocols approved at Ponce School of Medicine, the University of Michigan, and the North Shore University Hospital. Plasma samples were collected from seven anonymous HIV-1-infected patients at different time points over 1 to 3 years. Six of these HIV-1 patients received highly active antiretroviral therapy (HAART) from 2001 to 2004, and the plasma samples were taken during two or three time points over the course of infection. One other HIV-1 patient had the CCR5-Δ-32 genomic mutation and did not receive HAART during the years 1994 to 2002. This patient developed aggressive and fatal Hodgkin's lymphoma in March 2003 and was started on HAART and chemotherapy just prior to his demise. Plasma samples were taken before and just at the onset of Hodgkin's lymphoma. The HIV-1-infected patients all had HIV-1 RNA viral loads of greater than 5,000 copies/ml. This study also included five HIV-1-negative breast cancer patients. These included one man and one woman with widely metastatic breast cancer who were on chemotherapy at the time of the study. In addition, plasma samples came from two women with invasive ductal carcinoma (IDC) and one woman with IDC plus intralobular carcinoma. The latter three patients had regional lymph node metastases and were on chemotherapy as well. Breast cancer samples were collected and analyzed at a single time point. Unlike healthy individuals, who typically have HERV-K (HML-2) viral loads of <102 copies/ml, these breast cancer and HIV-1 patients had HERV-K (HML-2) viral loads within a range of 108 to 1010 copies/ml (7).
DNA samples from people of diverse origins were obtained from the blood of Caucasian people (HRC2 Human Random Control DNA panel 2; Sigma-Aldrich) and from the peripheral blood mononuclear cells of diverse cancer patients recruited at the University of Michigan and the North Shore University Hospital. No additional patient information was available.
Iodixanol gradients and particle purification.
Five hundred microliters of plasma sample was diluted in 2 ml phosphate-buffered saline (PBS) and centrifuged at 3,000 rpm for 10 min. Supernatants were overlaid on 20% iodixanol cushions (Optiprep density gradient medium; Sigma, St. Louis, MO) and centrifuged at 45,000 × g for 2 h. Pellets were resuspended in 500 μl PBS and overlaid in discontinuous 10% to 50% iodixanol solutions in 0.85% NaCl and centrifuged in a swinging-bucket rotor at 350,000 × g for 6 h. Fractions of 400 μl were collected, and their density was calculated by measuring the absorbance at 340 nm.
Quantitation of HERV-K (HML-2) and HIV-1 RNAs by real-time RT-PCR.
Viral RNA was extracted from 100 μl of DNase-treated iodixanol fractions, and the HERV-K (HML-2) and HIV-1 RNAs were quantified by real-time RT-PCR as described previously (7, 30). Primers were designed to amplify and quantify the HERV-K (HML-2) gag (KgagRTF, 5′-AGC AGG TCA GGT GCC TGT AAC ATT-3′; KgagRTR, 5′-TGG TGC CGT AGG ATT AAG TCT CCT-3′), and the HERV-K (HML-2) env transmembrane (TM) (KenvTMF, 5′-GCT GTA GCA GGA GTT GCA TTG-3′; KenvTMR, 5′-TAA TCG ATG TAC TTC CAA TGG TC-3′) sequences. Quantitation of HIV-1 gag was performed as described previously (30), using primers SK462 (5′-ACA TCA AGC AGC CAT GCA AAT-3′) and SK431 (5′-CTA TGT CAC TTC CCC TTG GTT CTC T-3′) and the probe 5′-6-FAM-ACC ATC AAT GAG GAA GCT GCA GAA TGG G-TAMRA-3′.
Western blotting.
Western blotting was performed as described previously (7). Briefly, proteins were precipitated from the fractions with chloroform-methanol, separated on 15% sodium dodecyl sulfate-polyacrylamide gels, and blotted onto polyvinylidene difluoride membranes. The membranes were blocked in 10% dry milk for 1 h and incubated with primary mouse anti-HERV-K Env (HERM-1811-5) or mouse anti-HERV-K Gag (HERM-1841-5) monoclonal antibody (Austral Biologicals) in blocking solution. For immunodetection of HIV-1 proteins, the membranes were incubated with primary anti-HIV-1 immunoglobulin (HIV-Ig; catalog no. 3957; NIH AIDS Research and Reference Reagent Program, Bethesda, MD). The membranes were washed five times in PBS containing 1% Tween. The bound primary antibody was then detected with a horseradish peroxidase-conjugated goat anti-mouse or goat anti-human secondary antibody using the Super Signal West Pico system (Pierce Chemical Co., Rockford, IL). In addition, the membranes were incubated with secondary antibodies alone for 1 h to control for cross-reactivity with plasma proteins.
Electron microscopy (EM).
Viral particles purified from plasma using an iodixanol gradient, which sedimented in a fraction found at a density of 1.16 g/ml, were visualized using conventional transmission EM (TEM) and immuno-EM as described previously (7). Briefly, a 5-μl aliquot of a purified particle preparation with a final concentration of 0.052 μg/μl was applied to 200-square mesh copper-nickel grids coated with a thin carbon film. The grids were washed several times and subsequently negatively stained with 1% buffered uranyl acetate, pH 4.5. The grids were washed several times in double-distilled water and observed using a TEM FEI Tecnai G2 spirit microscope operating at 80kV and equipped with an Olympus SIS Morada charge-coupled device camera. For analysis by immunogold staining, particles were adsorbed to 300-mesh nickel grids, blocked in Tris-buffered saline–1% bovine serum albumin, and incubated with primary mouse anti-HERV-K Env or anti-HERV-K Gag monoclonal antibody (HERM-1811-5 or HERM-1841-5, diluted 1:10 in blocking solution; Austral Biologicals, San Ramon, CA) for 1 h. In addition, another set of grids was incubated with purified mouse IgG for 1 h to control for cross-reactivity of mouse serum with plasma proteins. Grids were washed 5 times and incubated with 5- or 10-nm gold-labeled anti-mouse secondary antibody for 1 h. In the experiments done with 5-nm gold particles, the secondary antibodies came from BB International (Madison, WI), and in the experiments done with 10-nm gold particles, the secondary antibodies came from Sigma (St. Louis, MO). Immunostained particles were fixed in 2% glutaraldehyde, negatively stained in 2% uranyl acetate, and visualized with a Philips CM-100 transmission electron microscope (Department of Internal Medicine, and Programs in Cancer Biology, Immunology, and Cellular and Molecular Biology, University of Michigan, Ann Arbor, MI) or a Philips CM-10 transmission electron microscope (Department of Cellular Microbiology and Informatics, Novartis Vaccines & Diagnostics, Siena, Italy) operating at 80 kV.
Viral RNA extraction, RT-PCR, cloning, and sequencing.
Plasma was treated with 200 U RNase-free DNase for 1 h at 37°C. Viral RNA was extracted using the QIAamp viral RNA minikit following the manufacturer's instructions (Qiagen, Valencia, CA). PCR was performed to verify the absence of genomic HERV-K (HML-2) DNA. The env gene was amplified using the One-Step RT-PCR kit (Qiagen, Valencia, CA) with primers that amplify the env SU (surface) and TM regions. Primers ES1 (5′-AGA AAA GGG CCT CCA CGG AGA TG-3′) and ES2 (5′-ACT GCA ATT AAA GTA AAA ATG AA-3′) expand the full-length env SU region and generate PCR products of 1,159 bp for type 1 HERV-K (HML-2) elements and 1,451 bp for type 2 HERV-K (HML-2) elements. The KenvTMF and KEnvTMR primers described above amplify a nearly full-length env TM region and generate an ∼464-bp product. The amplification products were cloned and sequenced.
Single-genome PCR sequencing (SGS).
The HERV-K (HML-2) viral load was determined in one HIV-1 patient as described previously (7). The SGS reaction was adapted from a protocol provided by John Coffin (36). Briefly, a sample containing 10,000 copies of HERV-K (HML-2) viral RNA was denatured in the presence of 5 μl of 10 mM deoxynucleoside triphosphates (dNTPs) and 5 μl of 50 ng/μl ES3 primer (5′-CAA TGC AAC TCC TGC TAC AGC-3′) in a 50-μl reaction volume at 65°C for 10 min, followed by chilling on ice. The RNA was reverse transcribed by adding 50 μl of reaction mixture to the reagents (final concentrations in PCR grade water) MgCl2 (5 mM), RNase OUT (20 U), dithiothreitol (1 mM), 1× RT buffer (Invitrogen, Carlsbad, CA), and RT (100 U; Superscript II; Invitrogen, Carlsbad, CA). The reaction was carried out for 50 min at 45°C, followed by heating for 10 min at 85°C and cooling to 4°C.
The cDNA was serially diluted 1:3 in 2 mM Tris-HCl (ph 8.0) to a final dilution of 1:6,561, in which there should be ∼1 single cDNA molecule, in theory. cDNA molecules from the two final dilutions were distributed into 96-well plates and amplified by PCR using primers ES1 and ES3 at 200 nM, 6 mM MgCL2, 200 mM dNTPs, 1× PCR buffer (Invitrogen, Carlsbad, CA), and Platinum Taq High Fidelity DNA polymerase (Invitrogen, Carlsbad, CA) in a final 10-μl reaction volume. According to the Poisson distribution, the cDNA dilution yielding less than 30% positive amplifications contains one copy of cDNA about 80% of the time (36). PCR products were directly sequenced using overlapping internal primers. No sequencing reactions identifying more than one amplified genome were obtained.
In silico sequence analysis.
The env sequences obtained in plasma from patients were BLASTed to the HERVd and NCBI databases (35). Most of the viral RNA sequences matched >99% to 34 provirus sequences, which were used for phylogenetic reconstruction. The sequences were aligned in BioEdit and exported to the MEGA4 matrix. Phylogenetic trees were constructed and corroborated by different methods (neighbor joining, maximum parsimony, and maximum likelihood), using the statistical bootstrap test (1,000 replicates) of inferred phylogeny and the Kimura two-parameter model (14, 18). ORFs were calculated using translated BLAST in the NCBI database. Highlighter plots were generated using the highlighter tool of the Los Alamos HIV sequence database (www.hiv.lanl.gov).
Test for recombination.
We evaluated sequences for potential recombination events using several methods. First, we inspected the neighbor-joining tree for each data set. Recombination of large portions of different elements may lead to branches with unresolved topology, resulting in taxonomic units (TUs) that either protrude far beyond the other taxa or fall far short in comparison. The potential recombinant sequences were verified and the parent sequences identified using RIP 3.0. This program used a sliding window (200 bp in this study) that moves over an alignment containing the query sequence and all of the possible parental proviruses. Best matches are marked if they are significant by using an internal statistical test. Recombination analyses were corroborated using the recombination analysis tool (RAT) program (13). We verified the sequence similarity between the putative parent and query sequences on each side of the recombination spot. On several occasions, recombinant sequences were more than 99% similar to each parental sequence. Recombination plots are available upon request.
Calculation of dN/dS ratios.
The ratios of nonsynonymous substitutions per site (dN) to synonymous substitutions per site (dS) were calculated using SNAP (synonymous/nonsynonymous analysis program) from the Los Alamos HIV sequence database (www.hiv.lanl.gov) and corroborated with PAL2NAL, a program that uses the PAML interface for phylogenetic analyses by maximum likelihood (22, 41, 44). dN/dS ratios were calculated using alignments for all of the viral sequences, for sequences that cluster in each major branch of the trees, and finally for each HERV-K (HML-2) viral element obtained in plasma of each patient during a period of time (1 to 3 years). Statistical significance was determined using the log likelihood Z test, assuming a null hypothesis of dN/dS = 1, which corresponds to neutral evolution.
N-glycosylation analyses.
Potential N-linked glycosylation sites were determined using the N-glycosite software from the Los Alamos HIV sequence database (www.hiv.lanl.gov), which searches for the N-glycosylation motif or sequon N(X)[ST], where X can be any amino acid.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database and assigned accession numbers DQ360503 to DQ360809, EU308642 to EU308718, GU476554, and JN656288 to JN656292.
RESULTS
Detection of HERV-K (HML-2) RNA titers, proteins, and VLPs in plasma from patients.
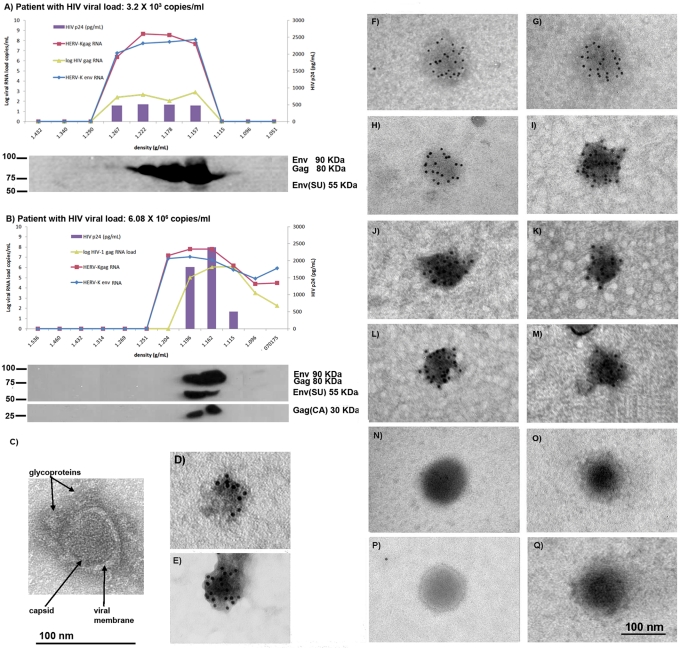
Plasma specimens from HIV-1-infected patients with high and low HIV-1 loads were subjected to iodixanol gradient ultracentrifugation in order to fractionate the plasma by density. HERV-K (HML-2) gag, env, and the HIV-1 gag RNA genes were quantified in different-density fractions by real-time RT-PCR, and the HIV-1 p24 antigen was quantified using a standard enzymatic assay. We also looked for the HERV-K (HML-2) Gag and Env proteins and HIV-1 proteins present in the different-density fractions by Western blotting (Fig. 1; see Fig. S1 in the supplemental material). We previously reported finding no HERV-K (HML-2) viral RNA or viral protein, and very little RT activity, in plasma fractions from control individuals (7). In contrast, as shown in Fig. 1A and B, in the plasma fractions of HIV-1-infected patients not only were HIV-1 RNA and HIV-1 p24 protein detected, but HERV-K (HML-2) RNA and immature and correctly processed proteins were all seen in each case in the fractions that were at the appropriate density (1.15 to 1.17 g/ml) for putative HERV-K (HML-2) viruses (11–12, 17, 23). In the patient with a low HIV-1 load (3.2 × 103 copies/ml), HIV-1 p24, unprocessed HERV-K (HML-2) Gag (80 kDa), unprocessed Env (90 kDa), and correctly processed Env (55 kDa) were seen by Western blotting (Fig. 1A; see Fig. S1A in the supplemental material). In the patient with a high HIV-1 load (6.08 × 106 copies/ml), unprocessed Gag (80 kDa) and Env (90 kDa) and correctly processed HERV-K (HML-2) Gag (30 kDa) and Env (55 kDa) were seen at the correct density for a HERV-K (HML-2) or an HIV-1 particle (∼1.16 g/ml) (Fig. 1B; see Fig. S1B in the supplemental material). HIV-1 p24 (capsid), p41 (matrix plus capsid), and p51 (RT) were additionally detected in Western blot assays incubated with the HIV-1 immunoglobulin in the same fractions where HERV-K antigens were detected (see Fig. S1B in the supplemental material). The maturation of the HERV-K (HML-2) proteins indicates that the HERV-K (HML-2) protease may be functional in this setting. These observations suggest that immature and mature HERV-K (HML-2) viruses may coexist with HIV-1 in the blood of patients.
Fig 1.
HERV-K (HML-2) RNA, proteins, and VLPs are found in the plasma of HIV-1 and breast cancer patients. Plasma samples from one HIV-1 patient with a low HIV-1 RNA load (A) and another patient with a high HIV-1 RNA load (B) were fractionated by density in iodixanol gradients. The density of each gradient is given on the x axis. The HIV-1 p24 antigen levels were measured by enzyme-linked immunosorbent assay using the HIV-1 p24 Antigen Capture kit (Science Applications International Corporation, NCI-Frederick, Frederick, MD) in each fraction, and the values are represented by purple bars given on the y axis on the right side in pg/ml. The HERV-K (HML-2) gag and env and the HIV-1 gag RNA titers (lines) were assessed by real-time RT-PCR (viral load is indicated on the y axis on the left side). HERV-K (HML-2) viral proteins were detected by Western blotting of protein extracts from each fraction. A protein size ladder is included at the left of each blot. (C) Transmission electron micrograph of a viral particle visualized in a plasma fraction at a density of 1.16 g/ml obtained from the patient in panel A. Arrows indicate viral structures, such as the viral membrane, the capsid, and glycoprotein spikes. (D to Q) Viral particles found in the plasma fraction at a density of 1.16 g/ml were visualized by EM. (D to M) Immuno-EM images of particles labeled with an antibody specific to HERV-K Env (D to I) or Gag (J to M) and a secondary antibody linked to 5-nm (D) or 10-nm (E to M) gold particles. All viral particles detected have a size of ∼100 nm, as expected for HERV-K. Some particles (E and I) show condensed cores and spikes symmetrically distributed around the viral membrane, suggesting mature viruses. Immunolabeled HERV-K VLPs were detected in preparations from both breast cancer (D and E) and HIV-1-infected (F to M) patients. Some condensed cores, presumably unenveloped viruses resulting from the preparation procedure, were immunolabeled with an antibody specific for Gag and had a size of ∼70 nm (J to M). (N to Q) No clustering of gold particles was observed in the negative controls in which the same preparations were labeled with purified mouse IgG antibody and detected with a 5-nm gold particle-labeled secondary antibody.
To ensure the specificity of our antibodies, the HERV-K monoclonal antibodies used in this study were tested for possible cross-reactivity with HIV-1 proteins. 293T cells were transfected with the pNL4.3 HIV-1 expression vector, and cellular extracts were incubated with the anti-HERV-K (HML-2) antibodies in Western blot assays (see Fig. S1C in the supplemental material). No apparent reactivity of HERV-K (HML-2) antibodies with HIV-1 proteins was observed in the Western blot assays, suggesting that the HERV-K (HML-2) antigens detected in this study are, in fact, HERV-K (HML-2) proteins. Furthermore, the Western blot assays from plasma fractions revealed that the size of HERV-K (HML-2) proteins is different from the size of HIV-1 proteins detected by the antibodies used in this study.
Detection of pathogenic viral particles in the blood of patients by EM is generally very difficult; in fact, such detection has never been done successfully for HIV-1 particles in plasma of infected patients. However, a few RNA viruses found at very high titers in the blood, such as hepatitis C virus, can be seen directly in the blood by EM (21). We detected HERV-K (HML-2) VLPs in the blood of patients with lymphoma for the first time (7). These viral particles were immunolabeled with HERV-K antibodies and were found in plasma fractions at a density of 1.16 g/ml, where HERV-K viral RNA, RT activity, and HERV-K proteins colocalized. As some of our HIV-1 patients had HERV-K (HML-2) RNA titers as high as 1010 copies/ml, and breast cancer patients had titers as high as 108 copies/ml, we searched for evidence of viral particles in the fractionated plasma of these patients. Indeed, we were able to detect VLPs in the blood of the patients by EM in the density fraction (1.16 g/ml) containing HERV-K (HML-2) viral RNA and proteins (Fig. 1C to Q). The ultrastructure of these VLPs was seen by TEM. The viral membrane, a symmetric capsid, and prominent glycoprotein spikes characteristic of a retroviral envelope were recognized (Fig. 1C). The morphology of these VLPs is different from that of HIV-1 particles cultured in vitro, which have been shown to have a cone-shaped capsid structure. In order to test whether antibodies against HERV-K structural proteins can recognize these VLPs, we used immuno-EM (Fig. 1D to Q). In spite of the difficulty of preserving virions from envelope shearing as a result of ultracentrifugation in gradients such as sucrose (21), we were able to detect clusters of gold-labeled Env proteins around VLPs with an anti-HERV-K Env mouse monoclonal antibody (Fig. 1D to I). Some particles showed condensed cores and spikes symmetrically distributed around the viral membrane, suggesting mature viruses (Fig. 1E and G). Some VLPs stained with anti-HERV-K Env antibodies were detected in preparations of breast cancer (Fig. 1D and E) and HIV-1 patients (Fig. 1F to I). We then performed immuno-EM using an antibody against the HERV-K Gag protein (Fig. 1J to M). We were able to detect clusters of gold-labeled Gag proteins around the viral cores of some virions, the envelopes having presumably been peeled off during the ultracentrifugation process. These viral cores are electron dense, suggesting that they are mature. No similar clustering of gold particles was seen in viral preparations probed with purified control mouse IgG antibody and detected with gold-labeled secondary antibody (Fig. 1N to Q). These EM findings, confirmed in two independent laboratories (Ann Arbor, MI, and Siena, Italy), further demonstrate that the viral particles, some mature and some immature, seen in the blood of the HIV-1 and breast cancer patients are indeed HERV-K.
Rationale for the study of the nature of HERV-K (HML-2) env RNA in the blood of patients.
Over the millions of years of endogenous retrovirus evolution, HERV-K has been under continuous purifying selection, suggesting that the expansion of HERV-K has been almost entirely due to germ-cell reinfection (4). However, no evidence of modern replication-competent HERV-K has yet been presented. Despite the presence of significantly elevated HERV-K (HML-2) RNA titers in the plasma of patients with HIV-1 infection, breast cancer, and lymphoma, whether HERV-K RNA sequences found in the plasma of modern humans retain signatures of purifying selection has been unknown. Lack of RNA mutations in the env sequences suggests that the presence of HERV-K (HML-2) sequences in the blood are simply the result of proviral induction. If RNA mutations are present, accumulation of synonymous mutations in the viral RNA sequences might be evidence of purifying selection, as these mutations retain the capacity to correctly code for the amino acids of the viral envelope necessary for cell attachment and infection (4). In contrast, RNA sequences would accumulate nonsynonymous mutations if the viruses evolve in such a way as to escape immune surveillance, as is the case of HIV-1 (22, 32). Viral RNA mutations showing a balanced number of synonymous and nonsynonymous mutations might indicate that the env gene has been influenced by a process other than purifying selection (i.e., segmental duplication, homologous recombination). As envelope glycosylation is an important feature of infectious capacity, a virus evolving in such a way as to retain infectious capability would be expected to conserve glycosylation sites. In addition, viral RNA genomes that have undergone reverse transcription, perhaps in a process related to reinfection, might be expected to show evidence of RNA recombination (26, 33). In view of these considerations, we examined the genetic evolution of the HERV-K (HML-2) env gene in HIV-1-infected individuals. For comparison, we examined plasma samples collected from five breast cancer patients in whose blood we have also detected high levels of HERV-K (HML-2) viral RNA (7).
Identification and coding capacity of HERV-K (HML-2) proviruses activated in HIV-1-infected and breast cancer patients.
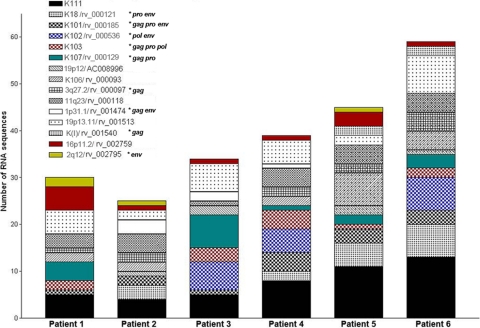
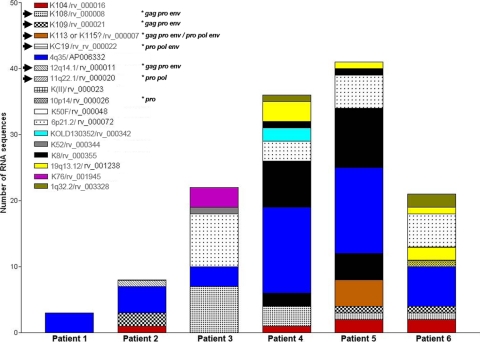
To characterize the HERV-K (HML-2) proviruses activated in HIV-1 and breast cancer patients, we BLASTed the sequences obtained by RT-PCR against all of the proviruses identified in the NCBI and HERVd databases. A sequence similarity of more than 99% was used as a cutoff for proviral identification, as has been described previously (10). The identification of the HERV-K (HML-2) elements and their chromosomal location were determined for each provirus by BLASTing the human genome databases, as well as by cross-referencing between the HERV-K (HML-2) proviruses identified in this study and the HERV-K (HML-2) proviruses reported in the literature (Table 1) (3, 8, 19, 25, 34, 38, 40, 42, 43). Of the 34 proviruses reported in this study, 6 have not been previously described and were named after their chromosomal location, except for K111 (see below). The relative abundance of the HERV-K (HML-2) viral RNA sequences derived from HERV-K (HML-2) proviruses was calculated. We found 15 different type 1 (Fig. 2) and 18 different type 2 (Fig. 3) HERV-K (HML-2) elements out of a total of 171 type 1 and 115 type 2 clone sequences in the plasma of HIV-1 patients. We detected 13 type 1 elements and 1 type 2 element in breast cancer patients, in whom it was rather difficult to amplify and clone type 2 transcripts. RNA transcripts from the proviruses K111 and 19p13.11 were found only in HIV-1 patients. RNA transcripts from the HERV-K type 2 provirus 4q35 predominated in all of the patients. The HERV-K (HML-2) proviruses activated in HIV-1 patients have ORFs and functional retroviral motifs in the translated sequences for 11 gag, 12 pro, 6 pol, and 7 type 2 env genes.
Table 1.
Identification of HERV-K RNA transcripts found in plasma of HIV-1 patients
| Name | HERVd fida | Accession no. | Location | Typeb | Reference(s) |
|---|---|---|---|---|---|
| K115 | rv_000007 | AY037929 | 8p23.1 | 2 | 43 |
| K108 | rv_000008 | Y17832 | 7p22.1 | 2 | 3, 42, 43 |
| 12q14.1 | rv_000011 | AC074261 | 12q14.1 | 2 | 8, 25 |
| K104 | rv_000016 | AF164612 | 5p14.3 | 2 | 3 |
| 11q22.1 | rv_000020 | AC007326 | 11q22.1 | 2 | 8 |
| K109 | rv_000021 | AL590785 | 6q14.1 | 2 | 3 |
| KC 19 | rv_000022 | Y17833 | 19p12-q12 | 2 | 25, 42 |
| K(2) | rv_000023 | AB047209 | 3q21.2 | 2 | 39 |
| 10p14 | rv_000026 | AL392086 | 10p14 | 2 | 8, 25 |
| K50F | rv_000048 | DQ112151 | 19q13.3 | 2 | 38 |
| 6p22.1 | rv_000072 | AL671879 | 6p22.1 | 2 | 25 |
| K106 | rv_000093 | AC024108 | 3q13.2 | 1 | 3, 25 |
| 3q27.2 | rv_000097 | AC069420 | 3q27.2 | 1 | 19 |
| 11q23.2 | rv_000118 | AP002954 | 11q23.2 | 1 | 8 |
| K18/K110 | rv_000121 | Y18890 | 1q23.3 | 1 | 34 |
| K107 | rv_000129 | AF164613 | 5q33.3 | 1 | 3, 25 |
| K101 | rv_000185 | AF164609 | 22q11.2 | 1 | 3 |
| KOLD 130352 | rv_000342 | DQ112134 | 8p23.1 | 2 | 38 |
| K52 | rv_000344 | DQ112101 | 19p12 | 2 | 38 |
| K8 | rv_000355 | DQ112094 | 5p12 | 2 | 38 |
| K102 | rv_000536 | AF164610 | 1q21 | 1 | 3, 25 |
| 19q13.13 | rv_001238 | AC012309 | 19q13.13 | 2 | 25 |
| 1p31.1 | rv_001474 | AC093156 | 1p31.1 | 1 | 19 |
| 19p13.11 | rv_001513 | AC011467 | 19p13.11 | 1 | 19 |
| K(I) | rv_001540 | AB047240 | 3q12.1 | 1 | 40 |
| K76 | rv_001945 | DQ112121 | 1p36.21 | 2 | 38 |
| 1q32.2 | rv_003328 | AL137789 | 1q32.2 | 2 | This study |
| 16p11.2 | rv_002759 | AC135776 | 16p11.2 | 1 | This study |
| 2q1 | rv_002795 | AC079776 | 2q1 | 1 | This study |
| K103 | AF164611 | 10p12.1 | 3, 25 | ||
| K113 | AY037928 | 19p13.11 | 2 | 43 | |
| 4q35 | AP006332 | 4q35 | 2 | This study | |
| 19p12 | AC008996 | 19p12 | 1 | This study | |
| K111 | GU476554 | 22cen | 1 | This study |
Identification number of the HERV-K sequence found in the HERVd database.
Type 2 viruses possess a 292-bp fragment, deleted in type 1 viruses, at the pol-env boundary.
Fig 2.
Cumulative frequency of type 1 HERV-K (HML-2) viral RNA sequences found in plasma of HIV-1 patients. The distribution and number of type 1 HERV-K (HML-2) viral RNA sequences identified for each patient are represented by cumulative bars. A total of 15 type 1 viral RNA sequences were found in the plasma of HIV-1 patients, of which K111 (black bars) is most highly represented in all HIV-1 patients but patient 3, in whom the frequency of K107 viral RNA predominates. Stars indicate an ORF for the designated gene. Accordingly, the majority of type 1 recombinant sequences contain a K111 viral sequence. Some viral RNA sequences, such as K102 (blue square bars) and 2q1/rv_002795 (yellow bars), are present in some patients but not in others. A sequence similarity of more than 99% was used as the cutoff for proviral identification as described previously (10). We detected 13 type 1 elements and 1 type 2 element (4q35/AP006332) in breast cancer patients.
Fig 3.
Cumulative frequency of type 2 HERV-K (HML-2) viral RNA sequences found in plasma of HIV-1 patients. The distribution and number of type 2 HERV-K (HML-2) viral RNA sequences identified for each patient are represented by cumulative bars. A total of 18 type 2 viral RNA sequences were found in the plasma of HIV-1 patients, of which 4q/35/AP006332 (blue bars) is most heavily represented in all HIV-1 patients, except patient 3. Arrows indicate polymorphic proviruses. Stars indicate an ORF for the designated gene. Some viral RNA sequences, such as K109 (black square bars) and K76/rv_001945 (purple bars), are present in some patients but not in others. An unidentified viral RNA sequence was found in patient 5 that shows 98.6% similarity to K113 and K115 (brown bars; accession no. DQ360585). Coding capacity for functional envelopes was found in seven type 2 env sequences (K104, K108 [two env sequences from the tandem repeat], K109, K115, 12q14.1, and KC19).
Importantly, intact type 2 env sequences were found in all HIV-1 patients, representing 22% of the type 2 env sequences. Similar sequences were not found in breast cancer patients. Nine type 2 env sequences detected in the plasma of HIV-1-infected individuals were less than 99% similar to reported HERV-K (HML-2) proviruses (clones 3e1, 3e2, 3e3, 3e5, 4d9, 5c7, 5b7, 5b12, and 5c5). Twenty-two percent of type 1 env sequences were less than 95% similar to reported HERV-K (HML-2) proviruses. These new viral RNA sequences might have originated from unfixed and/or insertionally polymorphic proviruses. These HERV-K (HML-2) viruses could be part of a pool of HERV-K (HML-2) viruses that can still be activated in modern humans (4). Full-length K113, often thought to be the HERV-K (HML-2) virus most likely to be capable of replication in modern humans, was not detected in this study (43).
Genomic organization of novel HERV-K proviruses.
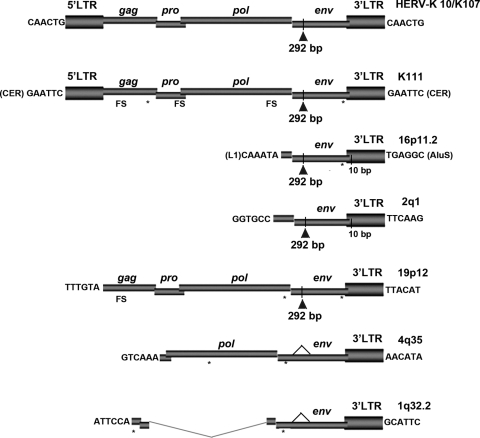
RNA transcripts derived from five previously undiscovered HERV-K (HML-2) proviruses were identified in this study by BLASTing human genome databases. Another novel HERV-K (HML-2) RNA transcript was discovered by phylogenetic reconstruction and termed K111 (see below). The five HERV-K (HML-2) proviruses were termed 16p11.2, 2q1, 19p12, 4q35, and 1q32.2 after their chromosomal location in humans and are not found in the chimpanzee genome sequence, suggesting that these viruses were inserted into the primate genome after the divergence of humans and chimpanzees. The genomic organization of the novel HERV-K (HML-2) proviruses identified in this study is described in Fig. 4. The viral genes from all of these viruses are characterized by stop codons and frameshift mutations, except for the env gene of provirus 2q1. Five of these proviruses are incomplete at the 5′ end and retain the 3′ LTR. In contrast to a HERV-K integration event that creates a 5- or 6-bp duplication at each end of the target site (19), the 6-bp “repeat” at the 3′ flanking sequence of these five proviruses is not repeated at the 5′ flanking sequence. The deletion of the 5′ genomic structure in these proviruses might have originated by genomic rearrangements or recombinational deletion as previously described (19–20). We did not recognize cellular promoters flanking the 5′ regions of these proviruses. When the integrity of the 5′ LTR is disrupted, the 3′ LTR could act as a promoter (9). The existence of RNA transcripts derived from proviruses devoid of a 5′ LTR may suggest that they were produced by 3′ LTR-mediated antisense transcription rather than 5′-linked cellular promoters. The mechanism of transcription of these proviruses requires further characterization.
Fig 4.
Genomic organization of novel HERV-K (HML-2) proviruses. The genomic organization of the six novel HERV-K (HML-2) proviruses reported in this study is illustrated relative to the provirus HERV-K10/K107. The proviruses were named after their chromosomal location (right), except for K111, which resides in centromeric areas unannotated in the human genome. The gag, pro, pol, and env genes are illustrated, together with the 5′ and 3′ LTRs. Stop codons (asterisks) and frameshift (FS; insertion or deletion) mutations, including the 292-bp deletion common to type 1 proviruses (vertical line with a triangle), are illustrated. An additional 10-bp deletion in the 3′ LTR of 16p11.2 and 2q1 is shown (vertical line). The 6-bp target site duplications flanking the 5′ and 3′ LTRs of K10 and K111 are depicted. The 6-bp sequences preceding and following the flanking sequences of the other proviruses are shown. Indicated in parentheses in the flanking sequence are repetitive elements that appear to be disrupted by the HERV-K (HML-2) provirus.
Phylogenetic reconstruction of HERV-K env sequences and discovery of a new HERV-K lineage.
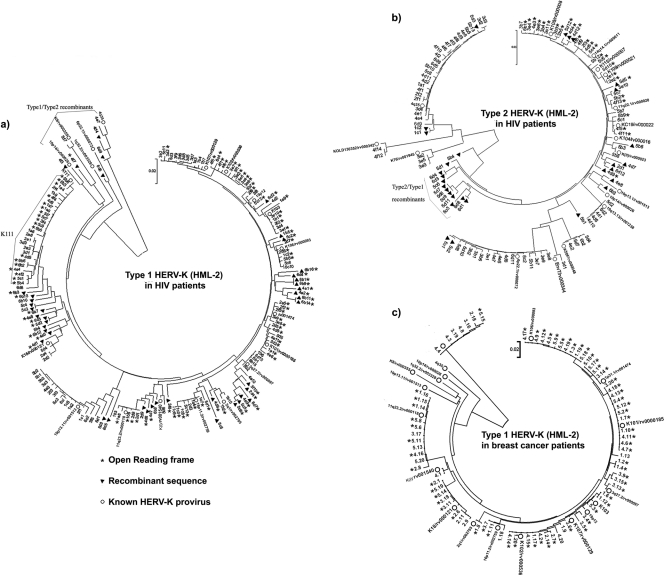
We constructed phylogenetic trees to show the evolutionary relationship of the HERV-K (HML-2) sequences found in the plasma of patients with HIV-1 infection and those with breast cancer. Figure 5 shows an overall picture of the viral sequences amplified from multiple patients, whereas Fig. 6 shows an analysis of env sequences from an individual HIV-1 patient. As expected, phylogenetic trees of the HERV-K (HML-2) env region showed several monophyletic lineages or major branches corresponding to previously identified HERV-K (HML-2) proviruses and their counterpart RNA sequences. However, we noted that in the type 1 and type 2 trees in HIV-1 patients (Fig. 5A and B), but not in the tree of breast cancer patients (Fig. 5C), a moderate number of TUs have unresolved topologies (branches that protruded far beyond or fell far short in comparison to the other taxa). These TUs represent recombinant HERV-K (HML-2) sequences present in HIV-1-infected patients but not in those with breast cancer.
Fig 5.
Phylogenetic neighbor-joining (NJ) tree of type 1 (a) and type 2 (b) HERV-K (HML-2) env SU sequences amplified from HIV-1 and breast cancer patients (c). The sequences were obtained from six HIV-1 patients (a and b; three samples each collected over a period of 1 to 3 years) and five breast cancer patients (c). The trees are unrooted, with taxa arranged for a balanced shape. The NJ trees were constructed using the Kimura 2-parameter model. The stability of branches was evaluated by bootstrap tests with 1,000 replications. White circles represent HERV-K (HML-2) proviruses reported in the HERVd and NCBI databases. Black triangles represent recombinant sequences. Clones with ORFs are depicted by stars. A cluster of sequences related to the novel HERV-K (HML-2) virus K111 (left) and putative type 1-type 2 recombinants and type 2-type 1 recombinants are indicated. The scale bars represent the nucleotide substitutions per sequence. Neither recombinant sequences nor the novel HERV-K111 sequence were identified in the breast cancer tree (c). The confidence values for the original trees we generated are available on request.
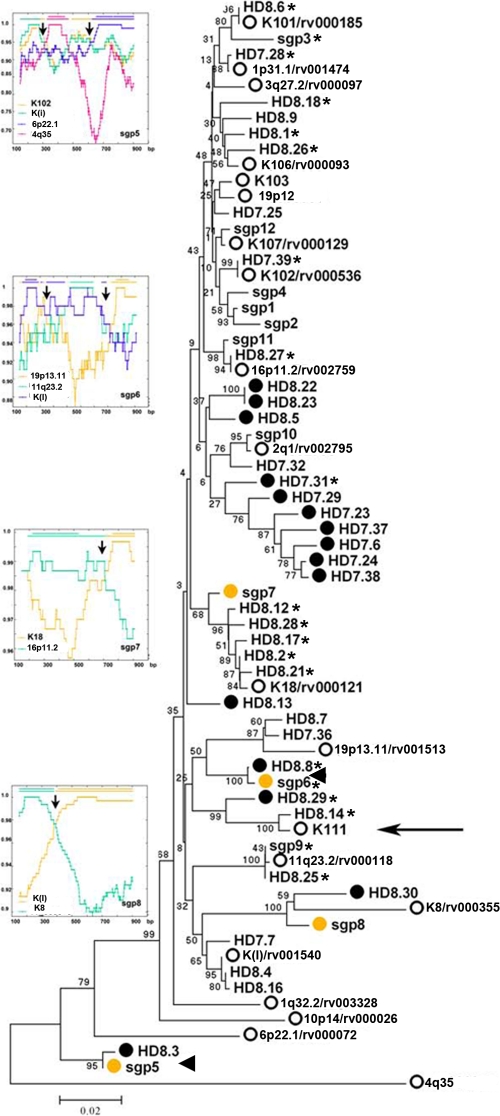
Fig 6.
Phylogenetic neighbor-joining (NJ) tree of type 1 HERV-K (HML-2) env SU sequences amplified from one HIV-1 patient by RT-PCR and SGS. The tree is unrooted, with taxa arranged for a balanced shape. The NJ tree was constructed using the Kimura 2-parameter model. The stability of branches was evaluated by bootstrap tests with 1,000 replications. Sequences labeled with the letters HD and the clone number represent clone sequences obtained by RT-PCR from one HIV-1-infected patient with Hodgkin's lymphoma. Sequences labeled with the letters sgp represent sequences obtained by SGS. White circles represent HERV-K (HML-2) proviruses reported in the HERVd and NCBI databases. Black circles represent recombinant sequences obtained by RT-PCR. Yellow circles represent recombinant sequences obtained by SGS. Similar recombinant sequences obtained by both amplification methods are indicated by arrowheads. ORFs are indicated by stars. A cluster of sequences related to the novel HERV-K (HML-2) virus K111 is indicated by an arrow. The scale bar represents 2% nucleotide substitutions per sequence. Recombination plots for individual env sequences obtained by SGS are shown at the left, and the arrows in the plots indicate the recombination spots.
In the HERV-K (HML-2) type 1 evolutionary tree derived from the plasma of HIV-1 patients (Fig. 5A), several sequences grouped in a major monophyletic branch that stands apart from other identified HERV-Ks. These elements cluster to a consensus sequence we termed K111. BLASTing of K111 to human genome databases retrieved only 95% sequence similarity to reported HERV-Ks, supporting the idea that these elements are sufficiently divergent to represent a novel HERV-K (HML-2) RNA sequence. In light of the fact that most HERV-K (HML-2) proviruses are found in our closest evolutionary relative, Pan troglodytes, we BLASTed the K111 sequence to the chimpanzee database. This allowed us to retrieve a chimpanzee counterpart, or CERV-K111, located in chromosome 7, which is ∼98% identical to the K111 query sequence. To corroborate the presence of K111 in the human genome, we designed primers strategically positioned near the integration site of CERV-K111 and internal HERV-K (HML-2) gag and env primers and detected full-length K111 proviruses in 129 DNA samples from people of diverse origin tested (96 DNA samples obtained from the blood of Caucasian people [HRC2 human random control DNA panel 2; Sigma-Aldrich] and 33 DNA samples obtained from the peripheral blood mononuclear cells of diverse cancer patients recruited at the University of Michigan and North Shore hospital).
The human full-length K111 proviral sequence was amplified, cloned, and sequenced in five human DNA samples, and the consensus sequence was deposited in the NCBI database (GenBank accession no. GU476554). Alignment of the LTRs of K111 revealed sequence similarity to the LTRs of provirus K105 (3). Both proviruses have the same 6-bp target site duplication flanking the LTRs and are inserted into centromeric repeats (CER). However, the 5′ LTR of K111 is 27 bp different from the corresponding LTR of the provirus K105, as is the 3′ LTR by 15 bp. Sequences flanking the 5′ and 3′ LTRs of K111 and K105 are only 90% similar, suggesting that these viruses are integrated at different locations and hence are separate viruses. K111 is integrated into the tandemly repetitive element D22Z3, which is found exclusively in the centromere of chromosome 22 in humans, and for this reason, the D22Z3 sequences are termed CER elements (29). The absence of published information about K111 in the human genome project may be due to the difficulty in sequencing and assembling centromeric regions. K111 and 19p13.11 RNA sequences were detected in the plasma of all of the HIV-1 patients and were absent in the plasma of all of the breast cancer patients (Fig. 5A and C, respectively), suggesting that these proviruses are activated upon HIV-1 infection.
Verification of recombinant sequences.
Unresolved topology in phylogenetic trees may indicate recombinant sequences (26). To assure ourselves initially that recombinant sequences are not artifacts of the RT-PCR method (15), we synthesized RNA transcripts in vitro from 12 different HERV-K (HML-2) proviruses activated in HIV-1 and breast cancer patients as previously described (5). The RNA transcripts were mixed and diluted serially down to 101 copies. A DNA-free RNA transcript mixture of different dilutions (102, 104, and 106) was amplified by RT-PCR using the env primers described. env sequences were cloned and sequenced. Of the 89 clones obtained by RT-PCR, only 4 showed one single base pair mutation, and no recombinant artifacts were generated (data not shown).
To confirm that the recombinant sequences obtained from HIV-1 patients are not the result of artifactual PCR recombination, we performed SGS (36) using plasma from one additional HIV-1 patient who had shown recombinant sequences as measured by RT-PCR. A phylogenetic reconstruction of the sequences generated by SGS and RT-PCR revealed that recombinant sequences are indeed detected using both methods (Fig. 6). The SGS method detected 4 (33%) of 12 recombinant sequences. Similarly, 15 (40%) of 37 sequences obtained by the RT-PCR method were found to be recombinant (Fig. 6). This information suggests that the percentage of recombinant sequences obtained by SGS and RT-PCR in plasma from this HIV-1 patient is very similar and supports the idea that most of recombinant sequences generated by the RT-PCR method are not likely induced by PCR-mediated recombination. In fact, two of the recombinant sequences generated by SGS (sgp 5 and sgp 6) were also amplified by the standard RT-PCR method, further supporting the idea that these recombinant HERV-K (HML-2) sequences are indeed present in this patient. These sequences are also likely to be the result of multiple recombinations, or several cycles of recombination, because their sequences match three distinct parental HERV-K (HML-2) proviruses. If these recombinant viruses were present due to artifacts of RT-PCR, then we would expect that they might be present in the plasma of patients with breast cancer who also had high levels of HERV-K (HML-2) viral RNA in their plasma. However, no recombinant (HML-2) sequences were detected in 77 sequences analyzed from the plasma of patients with breast cancer (Fig. 5C). This suggests that the process of activation of HERV K (HML-2) is different in HIV-1 infection and breast cancer.
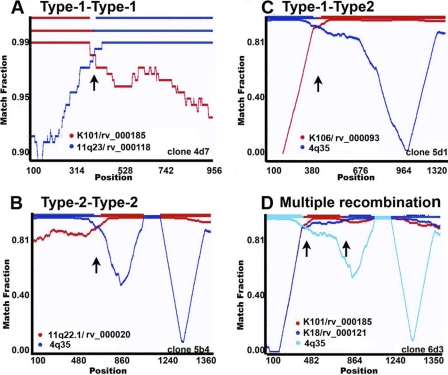
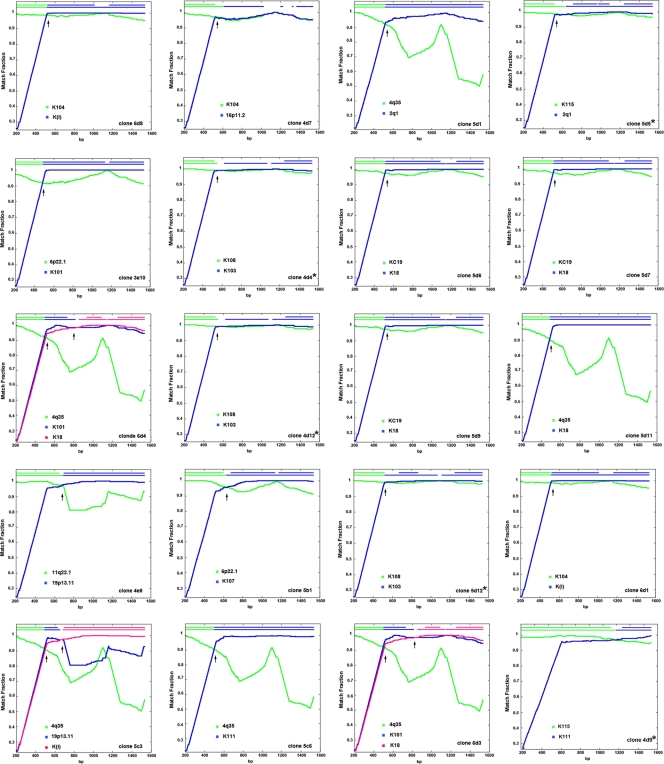
To verify the nature of putative recombinant HERV-K (HML-2) sequences that appear to have unresolved topology in the phylogenetic trees, we used the RIP 3.0, RAT, and nucleotide BLAST programs. Recombination analyses revealed that 19% and 25% of the HERV-K (HML-2) sequences detected in HIV-1 patients arose from intratype (type 2-type 2 or type 1-type 1) or intertype (type 1-type 2) recombination. Some examples are shown in Fig. 7. Approximately 93% of the putative recombinant sequences showed two distinct regions, each one highly (>99%) similar to a different parental HERV-K (HML-2) provirus. Six type 1 recombinant sequences displayed homologies to more than two proviruses (clones 4e1, 6b1, 6b9, 6b16, 6d2, and 6d7). Intertype recombinants (type 1-type 2) can be observed in the type 1 HERV-K (HML-2) phylogenetic tree (Fig. 5A and 6), matching with known type 2 viruses (10p14/rv_000026, K8/rv_000355, 1q32.2/rv_003328, 6p22.1/rv_000072, and 4q35/AP005332). The 20 type 1 sequences that acquired the 292-bp fragment from type 2 HERV-K (HML-2) genomes were considered type 2 sequences (clones 5b4, 5d1, and 6d3, among others) and, in the majority of the cases, clustered separately from other type 2 sequences in the type 2 phylogenetic tree (Fig. 5B and 8). Five (25%) of 20 recombinant type 1 env sequences that regain the 292-bp type 2 fragment reestablished the coding capacity for a functional envelope, i.e., clones 4d4, 4d9, 4d12, 5d5, and 5d12 (Fig. 8). While intratype recombination in the HERV-K (HML-2) family occurred randomly, in most of the type 1 recombinant sequences that regain the 292-bp fragment from type 2 viruses (16 [80%] of 20 recombinant sequences), recombination breakpoints were seen mostly in a region between 10 and 120 bp downstream of the type 1 292-bp deletion (Fig. 8). The prevalence of recombinant sequences containing the type 2 provirus 4q35 (6 of 20 sequences) might be explained by the predominant expression of this provirus in HIV-1 patients. Recombination plots for all of the recombinant clones are available upon request.
Fig 7.
Recombination plots of the different types of recombinant HERV-K (HML-2) env sequences found in HIV-1 patients. The similarity between the query recombinant sequence and each parental HERV-K (HML-2) virus is plotted for each position of an ∼1,000- or 1,350-bp sliding window. The y axis represents the match fraction of each query sequence to each parental sequence (red and blue lines, respectively). A match fraction of 1 means 100% identity. The representation of the recombinant query sequence is illustrated on the x axis (upper red/blue line at the top). Representative type 1-type 1 recombinant (A), type 2-type 2 recombinant (B), intersubtype recombinant (C), and multiply recombinant (D) sequences are illustrated. A third recombinant parental virus is indicated in clone 6d3 by light blue coloring. The identification number of the clone is shown in the lower right corner. Arrows indicate recombination spots.
Fig 8.
Recombination plots and breakpoints of type 1 envelope sequences that acquired the 292-bp type 2 fragment. The similarity between the query recombinant sequence and each parental HERV-K (HML-2) virus is plotted for each position of an ∼1,350-bp sliding window. The y axis represents the match fraction of each query sequence to each parental sequence (green and blue lines, respectively). A match fraction of 1 means 100% identity. The representation of the recombinant query sequence is illustrated in the top x axis (upper green/blue line at the top). A third recombinant parental virus can be seen in clones 5c3, 6d3, and 6d4 in fuchsia. The identification number of the clone is shown in the lower right corner. Arrows indicate recombination breakpoints. Stars indicate ORFs.
Evidence of HERV-K (HML-2) RNA sequences in HIV-1 patients that are under purifying selection.
To understand whether HERV-K RNA sequences found in the plasma of HIV-1 patients retain signatures of purifying selection we calculated the nonsynonymous (dN)-to-synonymous (dS) change (dN/dS) ratio in the HERV-K (HML-2) env sequences using SNAP and PAL2NAL (22, 41, 44). Frequent synonymous mutations occurred overall in the env gene, and the dN/dS ratio for the entire HERV-K (HML-2) family was low (0.53). In addition, the dN/dS ratios in the HERV-K (HML-2) env sequences found in plasma from each individual HIV-1 patient were also <1 (Table 2; P < 0.001), suggesting that these sequences remain under purifying selection in modern times as previously described (4).
Table 2.
Intrapatient dN/dS ratios of type 1 and type 2 HERV-K (HML-2) RNA sequences over three time points in plasma from HIV-1-positive patients
| Patient | Type 1 |
Type 2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of sequences | dN | dS | dN/dS ratio | P valuea | No. of sequences | dN | dS | dN/dS ratio | P valuea | |
| 1 | 27 | 0.0325 | 0.0730 | 0.44 | <0.001 | 3 | 0.0068 | 0.0424 | 0.16 | <0.001 |
| 2 | 30 | 0.0378 | 0.0606 | 0.62 | <0.001 | 6 | 0.0975 | 0.1494 | 0.65 | <0.001 |
| 3 | 40 | 0.0318 | 0.0618 | 0.51 | <0.001 | 22 | 0.0920 | 0.1977 | 0.46 | <0.001 |
| 4 | 27 | 0.0461 | 0.1180 | 0.39 | <0.001 | 34 | 0.1104 | 0.1684 | 0.65 | <0.001 |
| 5 | 28 | 0.0371 | 0.0651 | 0.56 | <0.001 | 33 | 0.0768 | 0.1511 | 0.50 | <0.001 |
| 6 | 31 | 0.0548 | 0.0964 | 0.56 | <0.001 | 17 | 0.0819 | 0.1960 | 0.41 | <0.001 |
Statistical significance taken from the log likelihood Z test. P values reflect comparisons of the dN/dS ratios obtained in this study to the null hypothesis (dN/dS = 1).
We observed accumulation of synonymous mutations (low dN/dS ratio) for most phylogenetically related HERV-K (HML-2) RNA sequences found in the blood of HIV-1 patients (Table 3). An exception is K111, which showed a dN/dS ratio of ∼1, indicating that K111 has been influenced by a process other than purifying selection. In contrast, in the vast majority of the sequences obtained from breast cancer patients, we did not observe substitutions at all. The very limited number of mutations found in phylogenetically related HERV-K (HML-2) RNA sequences found in breast cancer patients makes the calculation of dN/dS ratios unfeasible for four of the proviruses (K101, K106, 1p31.1, and 16p11.2; see Table S1 in the supplemental material). When mutations were present, the dN/dS ratios for many proviruses were not significantly different from 1, suggesting proviral induction. The dN/dS ratios of 5 of 11 (11q23.2, K18, K103, K107, and 3q27.2) were below 1 in breast cancer patients, suggesting some limited degree of purifying selection. However, we observed that the same proviruses have much lower dN/dS ratios in HIV-1 patients. The very sporadic mutations seen in the HERV-K (HML-2) sequences found in the blood of patients with breast cancer are likely to be the result of transcriptional errors. Therefore, the RNA sequences in breast cancer show only limited and infrequent evidence of purifying selection.
Table 3.
Interpatient dN/dS ratios of env (SU) genes of specific type 1 and type 2 HERV-K (HML-2) RNA sequences from the plasma of patients with HIV-1 infection
| Accession no. of HERV-K virus | No. of sequences | dN | dS | dN/dS ratio | P valuea |
|---|---|---|---|---|---|
| 11q23.2/rv_000118 | 6 | 0.0027 | 0.0077 | 0.35 | 0.008 |
| K106/rv_000093 | 8 | 0.0060 | 0.0143 | 0.41 | <0.001 |
| 19p13.11/rv_001513 | 16 | 0.0066 | 0.0106 | 0.62 | <0.001 |
| K18/rv_000121 | 4 | 0.0030 | 0.0131 | 0.22 | <0.001 |
| K101/rv_000185 | 8 | 0.0047 | 0.0117 | 0.40 | <0.001 |
| K103/AF164611 | 8 | 0.0072 | 0.0203 | 0.35 | <0.001 |
| K111/DQ360784 | 24 | 0.0122 | 0.0097 | 1.25 | 0.12b |
| 1p31.1/rv_001474 | 6 | 0.0020 | 0.0176 | 0.11 | <0.001 |
| 16p11.2/rv_002759 | 6 | 0.0065 | 0.0124 | 0.52 | <0.001 |
| K102/rv_000536 | 11 | 0.0040 | 0.0148 | 0.27 | <0.001 |
| K107/rv_000129 | 13 | 0.0037 | 0.0072 | 0.51 | <0.001 |
| 12q14.1/rv_000011 | 6 | 0.0029 | 0.0060 | 0.48 | 0.005 |
| 11q22.1rv_000020 | 4 | 0.0084 | 0.0208 | 0.40 | <0.001 |
| K50F/rv_000048 | 4 | 0.0049 | 0.0068 | 0.72 | 0.017 |
| 6p21.2/rv_000072 | 12 | 0.0063 | 0.0103 | 0.61 | 0.002 |
| 19q13.12rv_001238 | 6 | 0.0024 | 0.0038 | 0.63 | 0.011 |
| 4q35/AP006332 | 30 | 0.0109 | 0.0190 | 0.57 | <0.001 |
| K108/rv_000008 | 10 | 0.0048 | 0.0112 | 0.42 | <0.001 |
| K115/rv_000007 | 4 | 0.0211 | 0.0313 | 0.67 | 0.001 |
Statistical significance taken from the log likelihood Z test. P values reflect comparisons of the dN/dS ratios obtained in this study to the null hypothesis (dN/dS = 1). Note that, in contrast to the other viral elements, K111 shows similar numbers of nonsynonymous and synonymous mutations.
Not a significant value.
An idea of the nature and extent of the diversification observed by the dN/dS calculation can be seen in phylogenetic trees of all of the sequences obtained from all HIV-1 patients that are derived from the same type 1 or type 2 provirus, as well as their corresponding highlighter plots (see Fig. S2 in the supplemental material). To further analyze the above observations and confirm that these viruses keep accumulating synonymous mutations over the years in each single HIV-1 patient, the dN/dS ratio was calculated for HERV-K (HML-2) isolates from individual patients over the period of study. We found markedly low dN/dS ratios for the type 1 and 2 HERV-Ks when comparing the sequences at two or more time points over 1 to 3 years of viral evolution, further suggesting that HERV-K (HML-2) continues under purifying selection over time (Table 4, P < 0.05) in HIV-1 patients. As the Env TM protein is more functionally restricted than Env SU, the dN/dS ratio was calculated for 25 env TM sequences obtained from the plasma of six HIV-1 patients in this study and also showed accumulation of synonymous mutations (dN, 0.0227; dS, 0.0482; dN/dS, 0.47; P < 0.001), suggesting that Env SU, TM, or both are under purifying selection.
Table 4.
Intrapatient dN/dS ratios of the env (SU) gene of specific type 1 and type 2 HERV-K (HML-2) RNA sequences found in the plasma of patients with HIV-1 infection at different time points
| Accession no. of HERV-K provirus | Patient no. (time points) | No. of sequences | dN | dS | dN/dS ratio | P valuea |
|---|---|---|---|---|---|---|
| K111/DQ360784 | 5 (1–3) | 6 | 0.0083 | 0.0071 | 1.16 | 0.593b |
| K107/rv_000129 | 3 (1–3) | 7 | 0.0029 | 0.0096 | 0.30 | <0.001 |
| K102/rv_000536 | 3 (2–3) | 6 | 0.0037 | 0.0164 | 0.22 | <0.001 |
| K108/rv_000008 | 3 (1–3) | 7 | 0.0051 | 0.0138 | 0.36 | <0.001 |
Statistical significance taken from the log likelihood Z test. P values reflect comparisons of the dN/dS ratios obtained in this study to the null hypothesis (dN/dS = 1).
Not a significant value.
N-linked glycosylation studies of HERV-K (HML-2) Env proteins.
The presence of N-glycosylation sites (sequons) in the envelope protein of viruses is important for proper folding, functionality, and immunogenicity (28, 45). A sequon is defined by the amino acid sequence N-X-[S or T], where X can be any amino acid. A sequon is not glycosylated if it contains or is followed by a proline (28). Changes in the location of sequons may be advantageous to the virus to escape the immune system (1). Compared to exogenous lentiviruses such as HIV-1 and simian immunodeficiency virus (SIV) that generally have >30 sequons, few sequons are found in env sequences in the HERV-K (HML-2) viruses. There are seven conserved sequons in type 2 viruses (except K108, with eight sequons), and six in type 1 viruses, a finding that mirrors other nonlentiviral retroviruses. We searched for putative N-glycosylation sites in five coding-competent type 2 envelopes to determine whether immune pressure has influenced the sequon locations of HERV-K (HML-2) elements in our HIV-1 patients. N-linked glycosylation plots showed conserved locations of sequons in all type 2 elements, suggesting that HERV-K (HML-2) preserves glycosylation sites in its envelope, consistent with the low dN/dS ratios. Similarly, conserved sequon motifs are observed in type 1 viruses (data not shown).
DISCUSSION
The potential for replication of members of the HERV-K family in modern humans has been much debated over the past 2 decades. The existence of lethal mutations in most HERV-K genes and the apparent lack of infectivity of HERV-K particles produced by teratocarcinoma and mammary carcinoma cells are the major observations that support the idea that HERV-K particles lack replication capacity (2, 39). However, recent evidence exists to support the idea that HERV-K (HML-2) has expanded until recent times by reinfection, and therefore, a pool of replication competent viruses may still exist in humans (4, 43). Methods to assess the infectivity and mobilization of HERV-K particles remain a major problem in understanding the biology of these and other endogenous retroviruses. The presence of a large number of proviruses in the human genome makes it difficult to differentiate between recently inserted new virus and endogenous ancient virus. Two groups have recently shown that reanimated versions of HERV-K (HML-2), made from cloned constructs in which mutations have been corrected, are able to reinfect human and nonhuman cells (12, 23). These experiments demonstrate that ancestral HERV-K could have indeed replicated in the past, but they do not directly address whether activated HERV-K sequences in modern humans under certain circumstances can still be mobilized and perhaps replicate.
We have analyzed HERV-K (HML-2) RNA env sequences from the blood of patients to identify which proviruses are expressed, to study the extent to which the expression of these proviruses varies, and to track changes in individual viruses over time to see to what degree HERV-K (HML-2) might remain active in modern humans. This study provides information on the type 1 and 2 HERV-K (HML-2) proviruses expressed in HIV-1 and breast cancer patients. We observed that the nature of the expression of these proviruses varies in different diseases. Type 2 HERV-K (HML-2) RNA sequences were not easily detected in the plasma of breast cancer patients, suggesting some restriction of expression of functional viral envelopes. In stark contrast to HIV-1-infected patients, breast cancer patients show no evidence of recombination of the env gene. Expression of K111 and 19p13.11 was detected only in HIV-1-infected patients and not in breast cancer patients, again suggesting that specific activation of certain HERV-K (HML-2) proviruses varies in different diseases.
The methodology employed in these studies allowed the identification of six HERV-K (HML-2) proviruses that had not been discovered previously by genome sequencing and bioinformatic approaches (25, 38). Interestingly, the 5′ LTR of five of these viruses is deleted yet they retain the 3′ LTR. We did not observe cellular promoters close to sequences upstream of these proviruses. The transcriptional mechanism of these proviruses deserves further investigation. However, 3′ LTR-mediated antisense transcription may explain the finding of RNA transcripts having arisen from these proviruses, as previous investigators have also found expression of other retroviruses when the proviral sequences are devoid of the 5′ LTR (9). While well described in recent years for gammaretroviruses, HTLV-1, and HIV-1 (16, 24, 37), this transcriptional mechanism has not been described for the HERV-K (HML-2) family.
One other provirus, which we term HERV K111, is found in the plasma of all HIV-1-infected patients but in none of the plasma samples from the breast cancer patients tested. This endogenous virus has not been previously observed, and while we find it to be present in the DNA of all people tested, it was not identified in the human genome project. With the sequencing of the chimpanzee genome, we were able to localize a chimpanzee counterpart of K111. Using this information, we were able to locate a K111 provirus in the human genome. Through our studies examining the expression of HERV-K (HML-2) transcripts in the blood of living people, we have thus been able to uncover the existence of previously unreported human genomic elements.
We show that the expression of HERV-K (HML-2) in HIV-1 patients, but not in breast cancer patients, shows signatures of ongoing variability of env sequences, viral recombination, the accumulation of primarily synonymous substitutions over time, and preservation of glycosylation sites, strongly suggesting that HERV-K (HML-2) elements have been reverse transcribed and are under purifying selection. Not only type 2 but also type 1 env sequences retain N-glycosylation sites and accumulate predominantly synonymous substitutions, suggesting that they are also under purifying selection, perhaps to preserve the envelope structure.
Interestingly, we found that some type 1 env sequences can regain the deleted 292-bp fragment by recombination with type 2 sequences, reestablishing intact ORFs for the viral genome. Likewise, we found intact type 2 sequences that inherited stop codons from mutated sequences after recombination. While recombination breakpoints between HERV-K (HML-2) RNA sequences occurs randomly, a major recombination “hot spot” was detected in type 1-type 2 recombinants in sequences downstream of the 292-bp fragment from type 2 proviruses. This recombination hot spot increases the possibility that intact type 1 env genes restore coding capacity after regaining the deleted 292-bp fragment from type 2 viruses. During recombination, the inherited lethal mutations observed in certain HERV-K (HML-2) elements can be replaced with intact reading frames. Furthermore, in some instances, we saw recombinants originating from more than two proviruses, which may potentially be the result of viruses that have gone through several rounds of recombination, perhaps due to reinfection or retrotransposition.
Of course, it is reasonable to raise the question of whether these recombinants may have arisen from artifactual recombination during RT-PCR processing (15). However, we have shown that this is not likely the case. First, using a mixture of in vitro HERV-K (HML-2) RNA transcripts, we do not see such recombination by RT-PCR. In addition, using SGS from selected plasmas, we are able to demonstrate the existence of recombinant sequences. Remarkably, some of the recombinants seen by SGS are the same as the ones obtained by standard RT-PCR, strongly suggesting that these recombinant RNAs, found in the same HIV-1-infected patient, are not artifactual. The absence of recombinants in the plasma of breast cancer patients, in which the same methods of blood collection and RT-PCR were used, further argues that these recombinant HERV-K (HML-2) RNAs in the plasma of HIV-1 patients are real and arose in vivo. Although the above findings are consistent with reverse transcription and perhaps mobilization of activated HERV-K (HML-2) sequences in the somatic cells of people with HIV-1 infection, proof that these modern HERV-K (HML-2) viruses are competent to replicate awaits the demonstration of direct passaging of HERV-K (HML-2) viruses in the laboratory and discovery of reintegration sites of newly inserted HERV-K (HML-2) viruses into the genome, an inherent aspect of mobilization of genetic elements. As we did not observe RNA recombination and accumulation of synonymous mutations in the HERV-K (HML-2) viruses found in the plasma of breast cancer patients, perhaps replication-like mobilization of HERV-K (HML-2) may require coinfection with an exogenous retrovirus such us HIV-1.
Our results provide genetic evidence for activation of the HERV-K (HML-2) family in HIV-1-infected patients by a mechanism with similarities to reinfection, but further studies are needed to better understand these observations. Whether HERV-K (HML-2) plays a role in HIV-1 pathogenesis remains to be investigated. The evidence presented here suggests that HERV-K (HML-2) can be activated by diverse mechanisms in present-day humans, and the possibility that these viruses are not always fixed in the somatic cells of the modern human population requires further investigation.
Supplementary Material
ACKNOWLEDGMENTS
We thank John Coffin, Richard Gaynor, Michael Emerman, and Gil Omenn for their thoughtful comments; Michael Khodadoust for significant contributions to the manuscript; Antonello Covacci for comments on the methods; and Donna Gschwend and Jen Lewis for manuscript preparation.
This work was supported by the Research Center for Minority Institutions (RCMI) program (grant G12RR03050), by a generous grant (05-5089) to M.H.K. from the Concerned Parents for AIDS Research, and by grants RO1 AI062248 and RO1 CA144043 to D.M.M. from the National Institutes of Health. D.M.M. was the recipient of a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research.
Footnotes
Published ahead of print 26 October 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Bai XF., Liu J, Li O, Zheng P, Liu Y. 2003. Antigenic drift as a mechanism for tumor evasion of destruction by cytolytic T lymphocytes. J. Clin. Invest. 111:1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bannert N, Kurth R. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. U. S. A. 101:14572–14579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barbulescu M, et al. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861–868 [DOI] [PubMed] [Google Scholar]
- 4. Belshaw R, et al. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. U. S. A. 101:4894–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Contreras-Galindo R, et al. 2006. A new real-time-RT-PCR for quantitation of human endogenous retroviruses type K (HERV-K) RNA load in plasma samples: increased HERV-K RNA titers in HIV-1 patients with HAART non-suppressive regimens. J. Virol. Methods 136:51–57 [DOI] [PubMed] [Google Scholar]
- 6. Contreras-Galindo R, Kaplan M, Markovitz D, Lorenzo E, Yamamura Y. 2006. Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res. Hum. Retroviruses 22:979–984 [DOI] [PubMed] [Google Scholar]
- 7. Contreras-Galindo R, et al. 2008. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J. Virol. 82:9329–9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costas J. 2001. Evolutionary dynamics of the human endogenous retrovirus family HERV-K inferred from full-length proviral genomes. J. Mol. Evol. 53:237–243 [DOI] [PubMed] [Google Scholar]
- 9. Cullen BR, Lomedico PT, Ju G. 1984. Transcriptional interference in avian retroviruses—implications for the promoter insertion model of leukaemogenesis. Nature 307:241–245 [DOI] [PubMed] [Google Scholar]
- 10. de Parseval N, et al. 2005. Comprehensive search for intra- and inter-specific sequence polymorphisms among coding envelope genes of retroviral origin found in the human genome: genes and pseudogenes. BMC Genomics 6:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dewannieux M, Blaise S, Heidmann T. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 79:15573–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dewannieux M, et al. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 16:1548–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Etherington G, Dicks J, Roberts IN. 2005. Recombination analysis tool (RAT): a program for the high-throughput detection of recombination. Bioinformatics 21:278–281 [DOI] [PubMed] [Google Scholar]
- 14. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 15. Flockerzi A, et al. 2007. Expression pattern analysis of transcribed HERV sequences is complicated by ex vivo recombination. Retrovirology 4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gaudray G, et al. 2002. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 76:12813–12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanke K, et al. 2009. Reconstitution of the ancestral glycoprotein of human endogenous retrovirus K and modulation of its functional activity by truncation of the cytoplasmic domain. J. Virol. 83:12790–12800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hills D, Bull J. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182–192 [Google Scholar]
- 19. Hughes JF, Coffin JM. 2001. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat. Genet. 29:487–489 [DOI] [PubMed] [Google Scholar]
- 20. Hughes JF, Coffin JM. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. U. S. A. 101:1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaito M, et al. 1994. Hepatitis C virus particle detected by immunoelectron microscopic study. J. Gen. Virol. 75:1755–1760 [DOI] [PubMed] [Google Scholar]
- 22. Kils-Hütten L, Cheynier R, Wain-Hobson S, Meyerhans A. 2001. Phylogenetic reconstruction of intrapatient evolution of human immunodeficiency virus type 1: predominance of drift and purifying selection. J. Gen. Virol. 82:1621–1627 [DOI] [PubMed] [Google Scholar]
- 23. Lee Y, Bieniasz P. 2007. Reconstruction of an infectious human endogenous retrovirus. PLoS Pathog. 3(1):e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lefebvre G, et al. 2011. Analysis of HIV-1 expression level and sense of transcription by high-throughput sequencing of the infected cell. J. Virol. 85:6205–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Macfarlane C, Simmonds P. 2004. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J. Mol. Evol. 59:642–656 [DOI] [PubMed] [Google Scholar]
- 26. Martin DP, Williamson C, Posada D. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260–262 [DOI] [PubMed] [Google Scholar]
- 27. Medstrand P, Blomberg J. 1993. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J. Virol. 67:6778–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mellquist JL, Kasturi L, Spitalnik SL, Shakin-Eshleman SH. 1998. The amino acid following an Asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 37:6833–6837 [DOI] [PubMed] [Google Scholar]
- 29. Metzdorf R, Göttert E, Blin N. 1988. A novel centromeric repetitive DNA from human chromosome 22. Chromosoma 97:154–158 [DOI] [PubMed] [Google Scholar]
- 30. Moroney SM, Loree C, Widen R. 2006. Evaluation of two TaqMan PCR assays for the detection of HIV-1 proviral DNA in blood samples. J. Microbiol. Methods 65:350–353 [DOI] [PubMed] [Google Scholar]
- 31. Nelson PN, et al. 2003. Demystified. Human endogenous retroviruses. Mol. Pathol. 56:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nielsen R. 1999. Changes in ds/dn in the HIV-1 env gene. Mol. Biol. Evol. 16:711–714 [DOI] [PubMed] [Google Scholar]
- 33. Onafuwa-Nuga A, Telesnitsky A. 2009. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 73:451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ono M, Yasunaga T, Miyata T, Ushikubo H. 1986. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J. Virol. 60:589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paces J, et al. 2004. HERVd: the Human Endogenous RetroViruses Database: update. Nucleic Acids Res. 32(Database issue):D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palmer S, et al. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43:406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rasmussen MH, et al. 2010. Antisense transcription in gammaretroviruses as a mechanism of insertional activation of host genes. J. Virol. 84:3780–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Romano CM, Ramalho RF, Zanotto PM. 2006. Tempo and mode of ERV-K evolution in human and chimpanzee genomes. Arch. Virol. 151:2215–2228 [DOI] [PubMed] [Google Scholar]
- 39. Seifarth W, et al. 1998. Proviral structure, chromosomal location, and expression of HERV-K-T47D, a novel human endogenous retrovirus derived from T47D particles. J. Virol. 72:8384–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sugimoto J, et al. 2001. Transcriptionally active HERV-K genes: identification, isolation, and chromosomal mapping. Genomics 72:137–144 [DOI] [PubMed] [Google Scholar]
- 41. Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tönjes RR, Czauderna F, Kurth R. 1999. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J. Virol. 73:9187–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Turner G, et al. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531–1535 [DOI] [PubMed] [Google Scholar]
- 44. Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555–556 [DOI] [PubMed] [Google Scholar]
- 45. Zhang M, et al. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.