Abstract
Replication of all positive-strand RNA viruses is intimately associated with membranes. Here we utilize electron tomography and other methods to investigate the remodeling of membranes in poliovirus-infected cells. We found that the viral replication structures previously described as “vesicles” are in fact convoluted, branching chambers with complex and dynamic morphology. They are likely to originate from cis-Golgi membranes and are represented during the early stages of infection by single-walled connecting and branching tubular compartments. These early viral organelles gradually transform into double-membrane structures by extension of membranous walls and/or collapsing of the luminal cavity of the single-membrane structures. As the double-membrane regions develop, they enclose cytoplasmic material. At this stage, a continuous membranous structure may have double- and single-walled membrane morphology at adjacent cross-sections. In the late stages of the replication cycle, the structures are represented mostly by double-membrane vesicles. Viral replication proteins, double-stranded RNA species, and actively replicating RNA are associated with both double- and single-membrane structures. However, the exponential phase of viral RNA synthesis occurs when single-membrane formations are predominant in the cell. It has been shown previously that replication complexes of some other positive-strand RNA viruses form on membrane invaginations, which result from negative membrane curvature. Our data show that the remodeling of cellular membranes in poliovirus-infected cells produces structures with positive curvature of membranes. Thus, it is likely that there is a fundamental divergence in the requirements for the supporting cellular membrane-shaping machinery among different groups of positive-strand RNA viruses.
INTRODUCTION
Positive-strand RNA viruses infect eukaryotic hosts from phytoplankton algae to humans. A unique feature apparently contributing to the outstanding evolutionary success of this group of pathogens is their obligatory dependence on cellular membranes for viral genome replication. It has been postulated that this strategy may provide several advantages for the virus. First, association of viral replication proteins with membranes creates a high local concentration of viral products; second, assembly of the components of viral replication complexes on membranes may provide a structural scaffold facilitating their proper spatial organization; and third, the membrane enclosure of replication sites provides protection from cellular sensors of infection and defense mechanisms (47). Different positive-strand RNA viruses target different cellular membranes and induce various degrees of morphological alteration. Nevertheless, the relatively small genome size of these viruses and the considerable level of sequence conservation among functional domains of viral proteins (1, 32, 33) suggest that there may be common strategies for transformation of cellular membranes into viral replication sites shared by even evolutionarily distant groups of positive-strand RNA viruses.
Poliovirus is arguably one of the best-studied positive-strand RNA viruses that infect animal cells. It is the prototype member of the Picornaviridae family, which includes many important human and livestock pathogens, such as Coxsackie viruses, foot and mouth disease virus, rhinoviruses, and hepatitis A virus. The genome RNA of poliovirus is about 7,500 nucleotides (nt) in length and has the polarity of mRNA, thus defining it as positive stranded. Upon entrance into the cell, the viral RNA is directly translated into one long polyprotein that is cleaved in cis and in trans by three virus-specific proteases into structural and replication proteins. The proteolytic processing cascade generates about 10 mature proteins and a number of intermediate products, many of which perform their own independent functions in the viral life cycle. About half of the nonstructural proteins of poliovirus have been implicated in interference with cellular membrane metabolism, resulting in major rearrangements of pre-existing subcellular organelles. The precise mechanisms utilized by poliovirus to subvert normal cellular pathways remain unknown.
Massive virus-induced membrane remodeling is the most conspicuous morphological feature observed in images of poliovirus-infected cells. This phenomenon was described more than 50 years ago (29), and the morphology, nature, and mechanisms of formation of these replication structures have been a matter of investigation and debate ever since. It has been shown that development of poliovirus-induced membranous structures utilizes pre-existing cellular membranes and also requires de novo-synthesized lipids (25, 42). However, the origin of those membranes proved difficult to pinpoint. A significant body of evidence, including biochemical and structural data, suggests that the endoplasmic reticulum (ER) must play a major role in the formation of those structures (9, 17, 46). On the other hand, the earliest virus-induced replication complexes were observed to be associated with Golgi compartments (11, 25) whereas markers from the ER, Golgi compartments, and lysosomes were all found to be associated with poliovirus replication structures later in infection (48).
The very descriptions of the morphology of these virus-induced membrane rearrangements remain controversial. The first report identified them as U-bodies because of their horseshoe-shaped appearance in early transmission electron micrographs (TEM) of infected cells (29). Later, Bienz et al. described them as clusters of single-membrane vesicles (9, 12), while other reports (48, 51) brought to attention the double-membrane morphology of poliovirus-induced vesicles detected after the use of high-pressure freezing for sample processing. The single- versus double-membrane morphology of the vesicles was interpreted as indicating two different models of their formation. Rust et al. proposed that single-membrane vesicles in poliovirus replication structures were formed due to viral subversion of the cellular COPII-dependent process of vesicle budding from the ER membrane (46). COPII-coated vesicles normally transfer cargo from the ER to Golgi compartments in the cellular secretory pathway. The idea of the involvement of at least some components of the secretory pathway in poliovirus replication is supported by the sensitivity of poliovirus infection to brefeldin A (BFA) (26, 39), a specific inhibitor of cellular secretion, and the strong dependence of virus replication on the cellular guanine nucleotide exchange factor GBF1, a key component of the early steps of the secretory pathway (6, 8). On the other hand, an autophagy-like mechanism was postulated to explain the observation of the double-membrane vesicles clearly associated in infected cells with viral translation and replication products (48, 51). It was shown that the cellular autophagy pathway was indeed activated in infected cells and that stimulation or suppression of autophagy by pharmacological agents or small interfering RNAs (siRNAs) targeting its key components resulted in enhancement or inhibition of viral propagation, respectively (53, 54).
These conflicting observations prompted us to investigate the development of poliovirus membranous replication complexes in detail, using electron tomography to obtain better insight into the three-dimensional (3-D) architecture and mechanisms of formation of these structures. We found that membrane remodeling in poliovirus-infected cells starts by formation of a tight network of irregularly shaped single-membrane branching tubular structures. Often, the original sites of their formation are associated with a Golgi antigen. Later, they appear to be transformed by a membrane-wrapping process into assemblies of double-membrane structures. The highest rate of viral RNA synthesis is observed when the membranous replication structures are represented predominantly by single-membrane, convoluted tubules.
MATERIALS AND METHODS
Cells and viral infection.
All the experiments were performed on HeLa cells grown in Dulbecco modified Eagle medium (DMEM) supplemented with pyruvate and 10% heat-inactivated fetal bovine serum. Infections were performed with poliovirus type I (Mahoney strain) at a multiplicity of infection of 50 PFU/cell. Cells were grown overnight in 12-well plates on Thermanox coverslips (Nunc) at an initial density of 450,000 cells/well unless otherwise indicated. Before infection, the cells were washed once with serum-free DMEM and then incubated for 30 min in 500 μl of HEPES-buffered DMEM with the appropriate amount of virus at room temperature. The medium was then replaced with complete growth medium, and the cells were incubated at 37°C for the indicated times. Mock-infected cells were treated identically but without the addition of virus. Poliovirus mutant with a hemagglutinin (HA) tag in the 3A sequence have been previously described (55).
Antibodies.
Mouse monoclonal antibodies against poliovirus proteins 2C and 3A were a gift from K. Bienz, University of Basel, Switzerland. Monoclonal anti-double-stranded RNA (dsRNA) antibodies were from English & Scientific Consulting Bt (Szirák, Hungary). Monoclonal anti-BrdUTP and anticalnexin antibodies were from Sigma-Aldrich. Monoclonal anti-GM130 antibody was from Becton Dickinson. Monoclonal antiactin horseradish peroxidase (HRP)-conjugated antibodies were from Sigma-Aldrich. Sheep anti-mouse polyclonal HRP-conjugated Fab fragments were from Jackson ImmunoResearch Laboratory. Rabbit monoclonal anti-HA antibodies were from Cell Signaling.
Transmission electron microscopy (TEM).
Cell cultures were fixed overnight at 4°C in 4% paraformaldehyde–2.5% glutaraldehyde–0.1 M sodium cacodylate buffer. All subsequent processing was carried out using a Pelco Biowave laboratory microwave system (Ted Pella, Inc., Redding, CA) at 250 W and cycles of 2 min on, 2 min off, and 2 min on in a vacuum (Hg, 20 in) unless otherwise indicated. The specimens were postfixed with 0.5% osmium tetroxide–0.8% potassium ferricyanide in 0.1 M sodium cacodylate and then in 1% tannic acid–distilled water and stained en bloc with 1% aqueous uranyl acetate. Samples were rinsed with distilled water and dehydrated in a graded ethanol series for 45 s each time. Subsequently, they were infiltrated with ethanol and Spurr's mixture (1:1) and 100% resin for 5 min in a vacuum, embedded in resin, and cured overnight in a 68°C oven. Thin sections (90 nm) were cut using a UC6 ultramicrotome (Leica Microsystems, Vienna, Austria) and stained with 4% aqueous uranyl acetate and Reynold's lead citrate prior to viewing on a Hitachi H-7500 TEM (Hitachi, Tokyo, Japan) at 80 kV or a Tecnai BioTwin Spirit TEM (FEI, Hillsboro, OR) at 120 kV. Digital images were acquired with a Hammamatsu XR-100 digital camera system (AMT, Danvers, MA.)
Electron tomography.
Poliovirus-infected HeLa cells were processed as described above, 200-nm-thick sections were collected on glow-discharged carbon grids, and a mixture of 10- and 15-nm-thick colloidal gold fiducial markers was applied. Using a linear tilt scheme and a Tecnai BioTwin Spirit TEM (FEI) operated at 120 kV, a series of single-axis tilt images were collected. Images captured over a tilt range of ± 68° (1° increments) at a 1-μm defocus level were recorded using an UltraScan 1000 Gatan charge-coupled-device (CCD) camera (2,048 by 2,048 pixels) and automated tomography acquisition software (Xplore 3D; FEI). The resulting images had a binning factor of 1 and a pixel size of 0.46 nm or 0.57 nm. The images from the tilt series were aligned using either Inspect 3D (FEI) or an IMOD software package (version 4.2.5), and SIRT reconstructions of 35 iterations were performed. All 3-D surface models were created from unfiltered tomograms with inverted contrast by manually selecting areas of interest and smoothing the 3-D volumes by the use of the Amira visualization package (version 5.3.0; Visage Imaging, Carlsbad, CA).
Immunotransmission electron microscopy (IEM).
Cells were fixed in 4% paraformaldehyde–phosphate-buffered saline (PBS) for 20 min. After they were subjected to washing with PBS, they were labeled for an hour with primary and secondary antibodies in 0.05% saponin solution–PBS. All subsequent DAB labeling steps were carried out as mentioned elsewhere (41).Thin sections (90 nm) were cut using a UC6 ultramicrotome (Leica Microsystems) prior to viewing on a Hitachi H-7500 TEM (Hitachi) at 80 kV or a Tecnai BioTwin Spirit TEM (FEI) at 120 kV. Digital images were acquired with a Hammamatsu XR-100 digital camera system (AMT.)
Cryoscanning electron microscopy (cryo-SEM).
Infected HeLa cells fixed overnight at 4°C with 2% paraformaldehyde were washed with double-distilled water prior to suspension in Hanks-buffered saline solution–10% bovine serum albumin (BSA). For examination of alternative fracture planes across lipid bilayers, specimens were additionally postfixed for 1 h with 0.5% osmium tetroxide–0.8% potassium ferricyanide–0.1 M sodium cacodylate prior to washing. All specimens were divided into aliquots of “freeze fracture hats” (Leica Microsystems, Vienna, Austria) for cryoimmobilization in a Leica EMPact2 high-pressure freezer (Leica). The hats were transferred into a BAF 060 (Leica) freeze-etching device, using a vacuum at 1 × 10−6 mbar and a stage temperature of −145°C for fracturing and sputter coating. After the fracturing step, the specimens were sublimated at −95°C for 15 to 20 min and shadowed at −145°C by electron beam evaporation with 1.8 to 3.5 nm of platinum at a fixed angle of 45° followed by an additional 14 to 20 nm of carbon (rotary shadowed at a 90° angle). After a coating step, frozen samples were mounted in a Gatan CT-3500 cryo-holder (Gatan, Inc., Abingdon, UK) and observed on a Hitachi S-5200 in-lens microscope (Hitachi) at −150°C or colder after 15 min of further sublimation at −95°C within the microscope to remove ice contamination.
Confocal microscopy.
HeLa cells grown on glass coverslips were fixed with 4% paraformaldehyde–phosphate-buffered saline (PBS) for 20 min at the indicated time postinfection (p.i.), washed with PBS 3 times, and stored in PBS at 4°C. The cells were permeabilized with 0.2% Triton X-100–PBS for 5 min and washed 3 times with PBS. After that, the cells were incubated in 3% nonfat dry milk solution for 1 h to block nonspecific binding sites. The cells were incubated sequentially for 1 h with the primary and secondary antibodies prepared in the same blocking solution followed by three PBS washes after each incubation. Images were taken with a Zeiss LSM 510 confocal microscope. Digital images were processed with Adobe Photoshop software; equal adjustments were applied to all images.
BrUTP labeling.
Infected and mock-infected cells were transfected at 3.5 h postinfection (h.p.i.) with a BrUTP-lipofectin mix in the presence of actinomycin D (5 μg/ml) essentially as described in reference 14 for 1 h and then fixed and processed for immunoelectron microscopy with anti-BrdUTP antibodies as described above.
Viral RNA accumulation assay.
Total RNA from infected or mock-infected HeLa cells was isolated with QIAshredder and RNeasy columns (Qiagen) according to the manufacturer's recommendations. Material from 3 wells of a 12-well plate was combined for each time point. RNA was reverse transcribed using random primers and a high-capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR (qPCR) was performed using a TaqMan probe (Applied Biosystems) developed for poliovirus sequences and an 18S rRNA probe as a reference. Poliovirus RNA accumulation in infected cells was calculated relative to the signal from mock-infected cells.
RESULTS
Multistage remodeling of cellular membranes in poliovirus-infected cells.
The poliovirus replication cycle in HeLa cells occurs over a period of 6 to 7 h (Fig. 1A [inset]), at the end of which the internal morphology of the cell has become vastly reorganized. The nucleus is crenated and is usually pushed to the periphery of the cell, while clusters of viral replication structures occupy a large portion of the cytoplasm and usually coalesce in the perinuclear region (Fig. 1A). To investigate the progression of virus-induced membrane remodeling in poliovirus-infected cells, we followed the course of infection by sampling and observing cells at 1-h intervals during the course of a single-step growth cycle. In accordance with earlier reports (12, 17), the first clear virally induced membrane rearrangements were detectable between 2 and 3 h.p.i. At that time, virus-induced structures appeared as small clusters of associated round and elongated single-membrane compartments of about 100 to 200 nm in cross-section. The interior of the compartments usually appeared empty, although a light amorphous residue was sometimes visible (Fig. 1B). As infection progressed, those clusters grew in size and developed increasing complexity in shape, but they still maintained their mostly single-membrane morphology and lack of detectable inner content (Fig. 1C). At 4 h.p.i., however, intensive crowding and bending of those structures were observed, the round shapes almost disappeared, and occasional unmistakable double-membrane structures (arrow, Fig. 1C) were seen among the single-membrane ones. The apparent deflated, crumpled, “saggy” look of the single-membrane structures was not due to dehydration during the fixation procedure, since the samples processed for cryo-SEM imaging without the use of dehydrating agents also showed both the “round” and “saggy” shapes of virus-specific membranes (Fig. 1G). The double-membrane structures were about 100 to 300 nm in diameter and always contained some material that appeared indistinguishable from the surrounding cytoplasm, indicating that they may have developed by an autophagy-like mechanism, as was proposed previously (48). As the infection process continued, the number of such double-membrane compartments grew, while the number of single-membrane empty chambers continuously declined (Fig. 1D) until they had almost disappeared at ∼6 h.p.i. (Fig. 1E). While the progression of morphological changes in the samples observed at different times postinfection was clearly evident, the ratio of single- versus double-membrane compartments on the cellular scale could not be reliably quantified, given the limitations of the two-dimensional electron microscopy imaging. This temporal development of membrane remodeling can serve as a description of the majority of cells at a given time postinfection. However, even under these experimental conditions, when the cells were simultaneously infected with a multiplicity of infection of 50 viral PFU/cell, the cells did not display exactly the same degree of virus-induced membranous rearrangements. Rather, the cell population always contained a spectrum of cells with “early” and “late” features. Note that the regions of early empty-looking single-membrane clusters were present in cells even at 7 h.p.i. (Fig. 1F, arrowhead). Thus, membrane remodeling in poliovirus-infected cells showed sequential changes that proceeded from the appearance of “early” small clusters of single-membrane round compartments to “intermediate” bigger assemblies of irregular shaped single-membrane structures, which in turn later were replaced by either round or irregularly shaped “late” double-membrane formations.
Fig 1.
Virus-induced membrane remodeling in poliovirus-infected HeLa cells at various times postinfection. (A) Low-magnification transmission electron micrograph of a HeLa cell infected with poliovirus at 4 h.p.i. Inset: qPCR data showing kinetics of accumulation of viral RNA. (B, C, D, E, and F) Membranous structures in infected cells at 3, 4, 5, 6, and 7 h.p.i., respectively. Arrow, first detection of double-membrane structures in the middle of the infectious cycle (4 h.p.i.). Arrowhead, early single-membrane structures found at the late stage of infection (7 h.p.i.). (G) Cryo-SEM image showing clusters of early “saggy” membranous structures. Bars, 100 nm (unless otherwise indicated).
A Golgi marker but not an ER marker is associated with viral membrane structures.
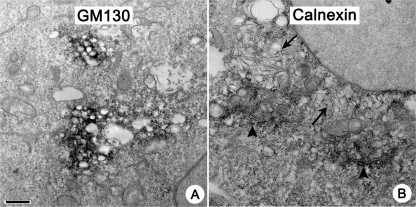
In order to investigate the origin of the membranes used to form the viral replication complexes, we performed IEM with antibodies against markers for cellular organelles. We concentrated on the early stage of infection because, first, this is a time of initial, active remodeling of cellular membranes into replication structures; however, such structures do not yet fill all the cytoplasmic space, and normal cellular organelles are still easily detectable. Second, the reliable staining of antigens hidden within big masses of virus-induced membranes found later in infection required stronger permeabilization conditions for IEM staining, resulting in severe destruction of the membrane morphology. Interestingly, permeabilization was much more detrimental for virus-induced structures than for cellular organelles, probably reflecting significant differences in membrane composition. We tested several antibodies to different cellular antigens, and obtained satisfactory staining results with antibodies for GM130, a cis-Golgi protein (43), and the ER-resident chaperone calnexin (45). We found that clusters of early single-membrane chambers are strongly associated with staining for GM130 (Fig. 2A). Staining for calnexin was never found on viral structures, even when calnexin-positive ER tubules (Fig. 2B, arrowhead) were located in close proximity to the developing clusters of virus-induced membranous chambers (Fig. 2B, arrows), arguing that the normal ER is not directly transformed into viral replication structures.
Fig 2.
Poliovirus replication structures and cellular organelles. Images were taken at 3 h.p.i. (A) Viral replication structures are strongly associated with staining for a Golgi antigen, GM130. (B) Staining for a resident ER protein calnexin (arrowheads) that is excluded from virus replication membranes (arrows). Bar, 500 nm.
The GM130 cis-Golgi marker is redistributed into multiple foci in poliovirus-infected cells.
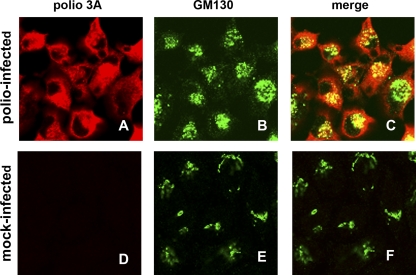
Association of clusters of poliovirus replication membranes with a GM130 cis-Golgi marker as identified by EM observations prompted us to look at the general distribution of this protein in infected cells. To simultaneously visualize GM130 and a poliovirus antigen, we took advantage of a poliovirus mutant that has an engineered HA tag in the 3A protein sequence. This mutant was previously reported to have the same growth characteristics in cell culture as the wild-type virus (55). 3A is a membrane-binding nonstructural protein, an important component of the poliovirus replication complex, and a modulator of cellular membrane trafficking pathways (5, 35, 51, 58). Whereas GM130 staining mostly showed elongated ribbons and big agglomerates consistent with normal Golgi localization in mock-infected cells (Fig. 3E), poliovirus infection resulted in clear redistribution of this antigen into multiple small foci concentrated in big assemblies in perinuclear areas (Fig. 3B). They often localized together with the signal for poliovirus protein 3A, but sometimes the signal for GM130 was separated from the areas positive for 3A in the confocal plane (Fig. 3C), suggesting that not all of the GM130 was associated with replication complexes. Thus, poliovirus infection induces complex reorganization of cellular membrane organelles that could be necessary for formation of viral replication membranes.
Fig 3.
Golgi antigen GM130 is redistributed into multiple dot-like structures in poliovirus-infected cells. HeLa cells were infected for 6 h with a poliovirus mutant with an HA tag within the 3A protein. Poliovirus-infected (A to C) and mock-infected (D to F) cells were stained simultaneously with anti-HA and anti-GM130 antibodies.
Electron tomography reveals a transition from single-membrane convoluted tubular structures to double-membrane vesicles during the course of poliovirus infection.
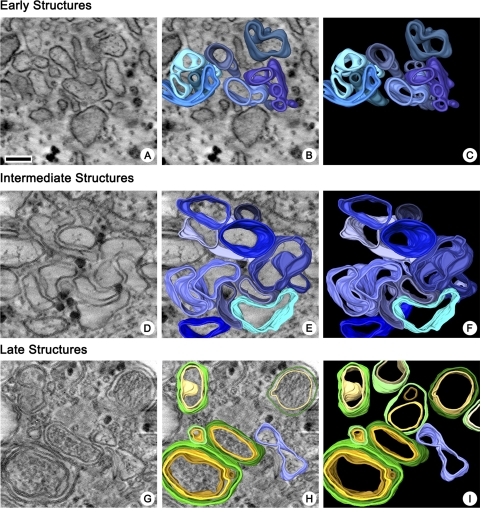
Whereas the conventional thin-section EM images show that a dramatic reorganization of cellular membranes occurred during the course of infection, they do not allow visualization of the 3-D structures of the viral replication complexes. To reveal their spatial organization, 200-nm-thick sections of cells fixed at different times after infection were examined by electron tomography (4, 37). At ∼2 to 3 h.p.i., early small clusters of single-membrane compartments were abundant in cells. 3-D reconstruction reveals that these structures represented irregularly shaped, branching tubular structures (Fig. 4A, B, and C; see also movie S1 in the supplemental material).
Fig 4.
Reconstructions showing the 3-D architecture of poliovirus membranous replication complexes at the early (3 h.p.i.), intermediate (4 h.p.i.), and late (7 h.p.i.) stages of development. (A, D, and G) Central slices in tomographic volumes. (B, E, and H) Central slices with segmented overlays. (C, F, and I) Segmented volumes, with blue indicating single-membrane structures and yellow and green indicating inner and outer membranes of double-membrane structures, respectively. Bar, 100 nm.
The intermediate single-membrane structures observed from 3 to 5 h.p.i. showed increased complexity of the membranous network; 3-D reconstructions of structures from areas with a large amount of tightly packed chambers were difficult, as the interpretation of the distinction between continuous versus merely adjacent membranes in the closely packed structures became highly subjective. Consequently, only peripherally located clusters with relatively well-separated individual membranous compartments were selected for reconstruction and 3-D analysis. These intermediate structures were more irregularly shaped, with frequent bending and wrapping around each other and more extensive branching of tubular protrusions, than the early structures (Fig. 4D to F; see also movie S2 in the supplemental material). ER and Golgi were readily distinguishable in infected cells at up to 4 h.p.i. However, we did not visualize transitions or connections between normal cellular organelles and clusters of viral replication structures, even when they were found in close proximity.
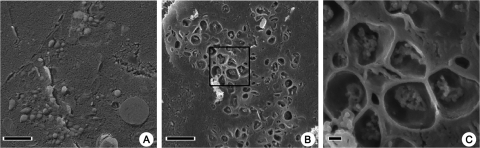
During the final stages of infection, from h 5 to 7, the cytoplasmic structures were almost exclusively represented by double-membrane and occasionally multilayered membrane compartments. Tomographic reconstructions revealed some long tubular structures and double-membrane vesicles that appeared to be spherical. Almost no branching of late double-membrane structures was observed (Fig. 4G to I; see also movie S3 in the supplemental material). The presence of sealed spherical structures was confirmed by freeze fracturing of poliovirus-infected HeLa cells and visualization by cryo-SEM (Fig. 5A); it is possible, though, that these structures contained small perforations in the membranes, especially at the early stages of their formation, that are not visible in cryo-EM images. Specimens osmicated prior to freezing to allow fractures across lipid bilayers (28, 44) revealed contents within the spherical structures that were consistent with the cytoplasmic material observed by TEM of the later double-membrane-bound vesicles (Fig. 5B and C).
Fig 5.
(A) A cryo-SEM image of freeze-fractured HeLa cells infected with poliovirus, showing late spherical structures at 6 h.p.i. (B) A cryo-SEM image of HeLa cells infected with poliovirus osmicated before freeze fracturing, revealing the inner contents of late spherical membrane structures. (C) Higher magnification of the region of interest demarcated in panel B. Bars, 1 μm (A and B) and 100 nm (C).
Double-membrane vesicles are formed by remodeling of single-membrane tubular structures.
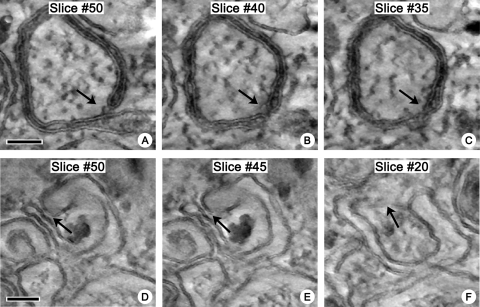
Our data show that poliovirus induces formation of single-membrane structures at the early stage of infection, whereas at the end of the replication cycle, the cytoplasm is packed with double-membrane tubules and vesicles filled with cytoplasmic material. This suggests either that there is a mechanism for conversion of the early single-membrane chambers into the late double-membrane ones or that the late structures may form de novo from the remaining cellular membranes that are not yet affected by virus-induced remodeling. The latter possibility seems unlikely, given the scarcity of available “normal” cellular membranes observed late in the virus growth cycle, when the double-membrane structures become prevalent. To determine the origin of the double-membrane vesicles and tubules, we performed electron tomography using the cells at the middle of the virus growth cycle, when the double-membrane structures start to appear. This analysis revealed multiple intermediate forms between the early single-membrane and the late double-membrane structures. The latter appeared to form by wrapping of the single-membrane sacks and tubules, which resulted in engulfment of cytoplasm during the formation of the double-membrane structure (Fig. 6 [arrows]; see also movies S4a and b in the supplemental material). It should be stressed that the figure does not show the time course of development of the double-membrane structures but presents different slices through a tomographic volume, demonstrating that open and sealed regions exist simultaneously in one continuous membranous structure. The darker intraluminal staining of the structure shown in Fig. 6A, B, and C probably reflects entrapment of the electron-dense material between closely opposed membranes during sample preparation, since the part of the same structure with more widely separated membranes (Fig. 6A [lower part of the image]) shows a level of luminal staining indistinguishable from that seen with other single-membrane compartments. Thus, the origin of the double-membrane structures in infected cells is most likely attributable to collapsing of the lumen and/or wrapping of the walls of early single-membrane chambers.
Fig 6.
Late double-membrane structures form by wrapping of early single-membrane membranous chambers. Slices (1.4 nm) through tomographic reconstructions are shown. Bead masking during tilt series alignment (IMOD) and 3-D noise-reduction median filtering (Amira) were applied for visualization. (A, B, and C) Slices from a single volume, showing coexistence of open single-membrane configurations and closed double-membrane regions in continuous membranous structures in infected cells. Cells were fixed at 4 h.p.i. (D, E, and F) Slices from a second volume, showing a potential region of closure that appears to have engulfed cytoplasmic granules. Cells were fixed at 6 h.p.i. Bar, 100 nm.
Poliovirus proteins and dsRNA are associated with both single- and double-membrane structures.
To investigate whether viral proteins and RNA are associated with single-membrane or double-membrane structures, we performed IEM imaging with antibodies against viral proteins and dsRNA, an indicator of viral RNA replication. The permeabilization procedure required for the penetration of antibodies into intracellular structures results in a marked deterioration of the fine structure of the membranous viral replication complexes; however, as shown in Fig. 7A, B, and C, viral proteins and dsRNA signals were found on both early single (arrows) and late double (arrowheads) membranes. We did not see differences between the viral replication proteins and dsRNA with respect to the patterns of their association with either single- or double-membrane structures. The labeling signal almost never encircled the entire membrane compartment outline but always appeared in the form of confined patches, indicating that viral proteins and RNAs are not evenly distributed on all the available membranous surfaces but are instead concentrated in distinct areas. These observations suggest that viral replication complexes form as distinct spatially separated loci.
Fig 7.
Poliovirus replication proteins and dsRNAs are associated with both single- and double-membrane structures. (A, B, and C) HeLa cells infected with poliovirus at 4 h.p.i. were stained with antibodies against poliovirus protein 3A (A) or 2C (B) or dsRNA (C). Arrows show single-membrane structures; arrowheads show double-membrane structures. (D) Background staining of infected cells processed without primary antibody. Bar, 100 nm.
The most active viral RNA replication occurs when the replication structures are in early single-membrane form.
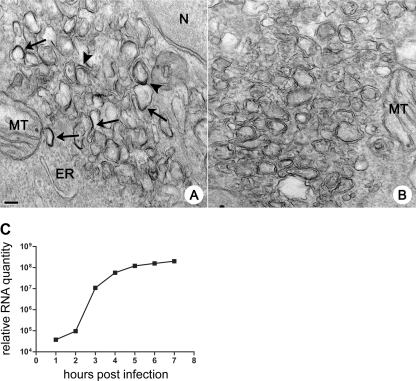
The observation of double- and single-membrane replication structures in poliovirus-infected cells has been a long-standing matter of controversy with respect to what structures are actually used for the replication of viral RNA. Although we detected dsRNA associated with both types of membranes, the presence of dsRNA does not necessarily indicate an active replication process. The RNAs may have formed earlier and then remained associated with membranes that are no longer involved in viral RNA replication. To detect active replication sites on membranes, we performed 1 h of pulse-labeling of cells in the middle of the infectious cycle with BrUTP that can be incorporated by the viral RNA-dependent RNA polymerase into nascent RNA chains. IEM with antibodies against BrUTP revealed that the label was associated with both single- and double-membrane structures, just as was observed for viral proteins and dsRNA, but not with the cellular organelle membranes of mitochondria and the ER (Fig. 8A). Positive labeling was visible only on a subset of membranous structures in the field, and the appearance of the BrU signal was patchy, as described earlier for labeling of viral proteins, supporting the idea that functional viral replication complexes occupy distinct localized sites within the membranous scaffold.
Fig 8.
Actively replicating poliovirus RNA is associated with single- and double-membrane structures. (A) HeLa cells infected with poliovirus were pulsed-labeled with BrUTP from 3.5 to 4.5 h.p.i. and stained with anti-BrdUTP antibodies. Arrows point to labeled single-membrane structures; arrowheads show labeled double-membranes structures. ER, endoplasmic reticulum; MT, mitochondrion; N, nucleus. (B) Background staining control. Infected cells processed without primary antibody are shown. (C) qPCR data reflecting kinetics of accumulation of viral RNA. Bar, 100 nm.
We assayed the kinetics of viral RNA accumulation in cells infected and incubated under the same conditions as used for preparation of samples for electron microscopy. qPCR analysis showed that the exponential phase of viral RNA accumulation occurred between 2 and 4 h.p.i. (Fig. 8C), when the replication-associated membranes are almost exclusively represented by single-membrane early and intermediate structures. Nevertheless, the absolute amount of viral RNA synthesized after 4 h.p.i. was greater than that synthesized during the exponential phase. At 4 h.p.i., the level of signal for relative RNA accumulation was 5.7 × 107, whereas the level of signal at 7 h.p.i. was 2 × 108 (Fig. 8C). Real-time PCR data reflect the total accumulated RNA, which showed that by 4 h.p.i., only ∼28% of total RNA was synthesized; therefore, the replication complexes existing after 4 h.p.i. must account for the majority of total viral RNA yield. Thus, our data demonstrate that the early and intermediate single-membrane structures were associated with the initial and most intensive rates of poliovirus RNA synthesis, while the late double-membrane structures appear in considerable quantities after the most active phase of viral RNA synthesis is over but may significantly contribute to overall viral RNA production.
DISCUSSION
Our research demonstrated that the transformation of cellular membranes into poliovirus replication complexes in infected cells is a well-coordinated multistage process. Previous reports of poliovirus replication structures described conflicting results and generated alternative models of their development. Nevertheless, these structures were generally visualized as “vesicles” whether surrounded by a single-membrane (11, 12) or a double-membrane (48) bilayer. It is indeed tempting to interpret the round membranous contours on 2-dimensional electron microscopy images as representing cross-sections of “vesicles,” but our data demonstrate that this perception is misleading. Electron tomography reconstruction shows that, until the final stages of infection, the membranous matrix associated with viral replication is represented by irregularly shaped branching convoluted single-membrane tubular structures that look more like a porous sponge rather than like clusters of spherical individual vesicles.
Other data supporting a vesicular organization of poliovirus replication complexes were obtained from transmission EM images of viral replication-associated membranes isolated from infected cells. They appeared as clusters of elongated vesicles with protruding interwoven narrow necks. It was suggested that the inflated and the narrow portions, called “compact membranes,” of the vesicles may be associated with different forms of viral replication complexes (10, 18). In our experiments, we were able to investigate samples that were 200 nm in thickness. We did not see the “rosette-like” arrangements of replication membranes previously described for isolated replication complexes (10); neither was a segregation of areas corresponding to “compact membranes” obvious. It is possible that the shapes of the structures isolated from infected cells disrupted by homogenization in buffered aqueous solution resulted from fragmentation of the continuous structures that we had directly observed in infected cells.
Many positive-strand RNA viruses form their replication complexes on specific cellular organelles. For example, the outer mitochondrial membrane (40), ER (38), chloroplasts (16), and plasma membrane (50) were previously shown to be sites of assembly of replication complexes for different viruses. The data available for poliovirus suggest that it utilizes membranes from multiple cellular sources (48). In agreement with some previous reports (10, 24), the first virus-induced membrane structures that we observed were associated with a cis-Golgi protein GM130, suggesting that Golgi membranes may be the initial site of poliovirus replication complex formation. The redistribution of this protein into multiple small foci in infected cells supports this idea. Other biochemical data have also pointed to the Golgi membrane as an important target of poliovirus replication. It has been shown that poliovirus proteins specifically engage components of the cellular secretory pathway, such as the small GTPase Arf and the Arf guanine nucleotide exchange factors GBF1, BIG1, and BIG2 (5–7), that are known to play a role in Golgi homeostasis. However, pre-existing Golgi membranes likely could not provide sufficient material for the masses of membranous replication complexes that can occupy most of the cytoplasmic space in infected cells later in infection. The ER is the most likely source of membranes for growth and expansion of viral replication complexes. Previous studies showed virus-induced structures and tubules of the ER in close proximity (9, 17). However, we believe it is unlikely that the ER is directly transformed into viral replication structures. The resident ER protein calnexin was excluded from the poliovirus replication structures, and the tomographic reconstruction did not reveal any apparent direct connections between the ER and the clusters of virus-induced membranous chambers. These data suggest that ER membranes must be modified somehow before they are used for virus replication.
The role of double-membrane structures in the infectious cycle of poliovirus remains uncertain. The exponential stage of viral RNA replication is essentially finished by 4 h.p.i., when these structures begin to form. It is possible that the wrapping of single-membrane early structures that generates the double-membrane tubules or vesicles is the result of the cell's response to virus infection and that they represent a dead-end development of the replication complexes. On the other hand, the amount of RNA synthesized after 4 h constitutes the major portion of the total viral RNA yield; thus, either double-membrane structures significantly contribute to the overall RNA production or single-membrane replication-active membranous complexes are being constantly generated throughout the course of infection. Our estimates of the intracellular constituents based on thin-section EM images are not quantitative, and there might be a sufficient source of membranes in infected cells that could contribute to the continuous formation of replication structures. Indeed, we saw regions of typical single-membrane early-looking membranous clusters in cells even at 7 h.p.i (Fig. 1F). Given the high level of RNA synthesis associated with the single-membrane chambers, their contribution to RNA production may be higher than that of more copious double-membrane structures. Unfortunately, analysis of thin-section EM images does not allow reliable statistical evaluation of the true distribution of different structures in cells. It is possible that the double-membrane structures are responsible not for viral RNA replication but for other stages in the infectious cycle such as RNA encapsidation or, as has been recently proposed, for nonlytic exit of virus from infected cells (52, 54). The latter phenomenon may be important for virus spread in an animal host, although the amount of virus released in the absence of cell lysis represents only a tiny fraction of the total amount of virus synthesized.
The process of forming double-membrane structures in poliovirus-infected cells has been suggested to involve at least the initial steps of the normal cellular autophagy pathway. This hypothesis was based on the morphological similarity of virus-induced double-membrane vesicles to early autophagosomes as well as on the stimulation of cleavage and modification of cellular LC3 protein in human 293T and MCF7 cells and its colocalization with the LAMP1 protein, an indicator of autophagosome development (27, 53). The data supporting the contribution of the autophagy pathway to enterovirus replication are mixed. It was demonstrated that inhibition or stimulation of autophagy results in modest inhibition or stimulation of poliovirus and coxsackie B3 virus yield, and data were also presented suggesting involvement of autophagy in replication of rhinoviruses 2 and 14 (27, 59). However, Brabec-Zaruba et al. reported that replication of rhinovirus 2 was insensitive to manipulation of autophagy with pharmacological agents and did not induce detectable modification of LC3 (13). This may indicate that different cells or cell types incubated under different conditions respond to poliovirus infection with different levels of activation of the autophagy program. It is also possible that the double-membrane structures in infected cells bear an only superficial morphological resemblance to those of autophagosomes. Our data indicate that the poliovirus double-membrane structures are formed by collapsing and/or wrapping of the early single-membrane structures, whereas recent advanced imaging studies of autophagosome formation demonstrate that they are formed by de novo synthesis of the isolation membrane and not by the simple wrapping of pre-existing organelle membranes (22, 23, 60). It is possible that changes in the concentration of intraluminal solutes result in luminal collapse due to change of osmotic pressure. Enterovirus 2B proteins are known viroporins (2, 56), and accumulation of this protein during the time course of infection could increase the permeability of these structures, contributing to their collapse at later times. Recently, the 3-D architecture of membranous replication complexes of several positive-strand RNA viruses was investigated by electron tomographic methods similar to those used in the studies reported here. Together with the previously accumulated data on membrane modifications induced by positive-strand RNA viruses, the images reveal that viruses from different families infecting different hosts demonstrate significant similarities in the overall organization of their replication complexes. Flock House virus of the Nodaviridae family (34), dengue virus of the Flaviviridae family (57), and Semliki Forest virus of the Togaviridae family (50) remodel membranes of mitochondria, ER, and plasma, respectively, to form invaginations harboring the viral replication machinery. Similar invaginated spherules were previously described as replication sites for other plant and animal viruses (21, 24, 30, 36). In those studies, invaginations were formed by inducing negative curvature in the pre-existing membrane bilayer. On the other hand, our study and previous data on membrane remodeling by picornaviruses and coronaviruses showed that these viruses shape membranes into tubular and/or spherical vesicle-like structures with a positive curvature (3, 15, 19, 31, 49). The total spectrum of positive-strand RNA viruses can be classified into three superfamilies of alpha-like, flavi-like, and picorna-like viruses on the basis of their genome sequences and organization (20, 32). These higher-order taxonomic units encompass diverse viruses infecting different hosts from almost all kingdoms of life. It appears that negative membrane curvature structures initiated by membrane invaginations are a predominant characteristic of the replication complexes of alpha- and flavi-like viruses, while picorna-like viruses almost exclusively develop complexes with positive membrane curvature. This division implies that two different subsets of supporting cellular membrane remodeling pathways are recruited by those groups of viruses and may suggest that there are two basic strategies evolved by these pathogens to develop their replication complexes.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Institute of Allergy and Infectious Diseases, NIH.
We thank Jeffrey Lengyel for technical assistance and critical review of the manuscript.
Footnotes
Published ahead of print 9 November 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ahlquist P, et al. 1985. Sindbis virus proteins nsP1 and nsP2 contain homology to nonstructural proteins from several RNA plant viruses. J. Virol. 53:536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agirre A, Barco A, Carrasco L, Nieva JL. 2002. Viroporin-mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem. 277:40434–40441 [DOI] [PubMed] [Google Scholar]
- 3. Amako K, Dales S. 1967. Cytopathology of mengovirus infection. II. Proliferation of membranous cisternae. Virology 32:201–215 [DOI] [PubMed] [Google Scholar]
- 4. Baumeister W, Grimm R, Walz J. 1999. Electron tomography of molecules and cells. Trends Cell Biol. 9:81–85 [DOI] [PubMed] [Google Scholar]
- 5. Belov GA, et al. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 81:558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. 2008. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 4:e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belov GA, Habbersett C, Franco D, Ehrenfeld E. 2007. Activation of cellular Arf GTPases by poliovirus protein 3CD correlates with virus replication. J. Virol. 81:9259–9267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belov GA, Kovtunovych G, Jackson CL, Ehrenfeld E. 2010. Poliovirus replication requires the N-terminus but not the catalytic Sec7 domain of ArfGEF GBF1. Cell Microbiol. 12:63–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bienz K, Egger D, Pasamontes L. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220–226 [DOI] [PubMed] [Google Scholar]
- 10. Bienz K, Egger D, Pfister T, Troxler M. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bienz K, Egger D, Rasser Y, Bossart W. 1983. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology 131:39–48 [DOI] [PubMed] [Google Scholar]
- 12. Bienz K, Egger D, Rasser Y, Bossart W. 1980. Kinetics and location of poliovirus macromolecular synthesis in correlation to virus-induced cytopathology. Virology 100:390–399 [DOI] [PubMed] [Google Scholar]
- 13. Brabec-Zaruba M, Berka U, Blaas D, Fuchs R. 2007. Induction of autophagy does not affect human rhinovirus type 2 production. J. Virol. 81:10815–10817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brass V, Gosert R, Moradpour D. 2009. Investigation of the hepatitis C virus replication complex. Methods Mol. Biol. 510:195–209 [DOI] [PubMed] [Google Scholar]
- 15. Dales S, Franklin RM. 1962. A comparison of the changes in fine structure of L cells during single cycles of viral multiplication, following their infection with the viruses of Mengo and encephalomyocarditis. J. Cell Biol. 14:281–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Graaff M, Coscoy L, Jaspars EM. 1993. Localization and biochemical characterization of alfalfa mosaic virus replication complexes. Virology 194:878–881 [DOI] [PubMed] [Google Scholar]
- 17. Egger D, Bienz K. 2005. Intracellular location and translocation of silent and active poliovirus replication complexes. J. Gen. Virol. 86:707–718 [DOI] [PubMed] [Google Scholar]
- 18. Egger D, Pasamontes L, Bolten R, Boyko V, Bienz K. 1996. Reversible dissociation of the poliovirus replication complex: functions and interactions of its components in viral RNA synthesis. J. Virol. 70:8675–8683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Friedmann A, Lipton HL. 1980. Replication of Theiler's murine encephalomyelitis viruses in BHK21 cells: an electron microscopic study. Virology 101:389–398 [DOI] [PubMed] [Google Scholar]
- 20. Goldbach R. 1987. Genome similarities between plant and animal RNA viruses. Microbiol. Sci. 4:197–202 [PubMed] [Google Scholar]
- 21. Grimley PM, Levin JG, Berezesky IK, Friedman RM. 1972. Specific membranous structures associated with the replication of group A arboviruses. J. Virol. 10:492–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hailey DW, et al. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayashi-Nishino M, et al. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437 [DOI] [PubMed] [Google Scholar]
- 24. Hrsel I, Brcak J. 1964. Ultrastructural changes in chloroplasts and cytoplasm caused by local infection of tobacco with tobacco mosaic virus and cucumber virus 4. Virology 23:252–258 [DOI] [PubMed] [Google Scholar]
- 25. Hsu NY, et al. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Irurzun A, Perez L, Carrasco L. 1992. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology 191:166–175 [DOI] [PubMed] [Google Scholar]
- 27. Jackson WT, et al. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. James R, Branton D. 1971. The correlation between saturation of membrane fatty acids and the presence of membrane fracture faces after osmium fixation. Biochim. Biophys. Acta 233:504–512 [DOI] [PubMed] [Google Scholar]
- 29. Kallman F, Williams RC, Dulbecco R, Vogt M. 1958. Fine structure of changes produced in cultured cells sampled at specified intervals during a single growth cycle of polio virus. J. Biophys. Biochem. Cytol. 4:301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim UH, Mukhajonpan V, Nii S, Kato S. 1977. Ultrastructural study of cell cultures infected with swinepox and orf viruses. Biken J. 20:57–67 [PubMed] [Google Scholar]
- 31. Knoops K, et al. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koonin EV, Dolja VV. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375–430 [DOI] [PubMed] [Google Scholar]
- 33. Koonin EV, Wolf YI, Nagasaki K, Dolja VV. 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6:925–939 [DOI] [PubMed] [Google Scholar]
- 34. Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lama J, Paul AV, Harris KS, Wimmer E. 1994. Properties of purified recombinant poliovirus protein 3ab as substrate for viral proteinases and as co-factor for RNA polymerase 3dPol. J. Biol. Chem. 269:66–70 [PubMed] [Google Scholar]
- 36. Lee WM, Ahlquist P. 2003. Membrane synthesis, specific lipid requirements, and localized lipid composition changes associated with a positive-strand RNA virus RNA replication protein. J. Virol. 77:12819–12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lucić V, Forster F, Baumeister W. 2005. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 74:833–865 [DOI] [PubMed] [Google Scholar]
- 38. Más P, Beachy RN. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147:945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maynell LA, Kirkegaard K, Klymkowsky MW. 1992. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 66:1985–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller DJ, Ahlquist P. 2002. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856–9867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mital J, Miller NJ, Fischer ER, Hackstadt T. 2010. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell Microbiol. 12:1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mosser AG, Caliguiri LA, Tamm I. 1972. Incorporation of lipid precursors into cytoplasmic membranes of poliovirus-infected HeLa cells. Virology 47:39–47 [DOI] [PubMed] [Google Scholar]
- 43. Nakamura N, et al. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131:1715–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nermut MV, Ward BJ. 1974. Effect of fixatives on fracture plane in red blood cells. J. Microsc. (Oxford) 102:29–39 [DOI] [PubMed] [Google Scholar]
- 45. Ou WJ, Cameron PH, Thomas DY, Bergeron JJ. 1993. Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature 364:771–776 [DOI] [PubMed] [Google Scholar]
- 46. Rust RC, et al. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808–9818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salonen A, Ahola T, Kaariainen L. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schlegel A, Giddings TH, Jr., Ladinsky MS, Kirkegaard K. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Skinner MS, Halperen S, Harkin JC. 1968. Cytoplasmic membrane-bound vesicles in echovirus 12-infected cells. Virology 36:241–253 [DOI] [PubMed] [Google Scholar]
- 50. Spuul P, Balistreri G, Kääriäinen L, Ahola T. 2010. Phosphatidylinositol 3-kinase, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 84:7543–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Suhy DA, Giddings TH, Jr., Kirkegaard K. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953–8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taylor MP, Burgon TB, Kirkegaard K, Jackson WT. 2009. Role of microtubules in extracellular release of poliovirus. J. Virol. 83:6599–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taylor MP, Kirkegaard K. 2007. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 81:12543–12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Taylor MP, Kirkegaard K. 2008. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy 4:286–289 [DOI] [PubMed] [Google Scholar]
- 55. Teterina NL, Pinto Y, Weaver JD, Jensen KS, Ehrenfeld E. 2011. Analysis of poliovirus protein 3A interactions with viral and cellular proteins in infected cells. J. Virol. 85:4284–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van Kuppeveld FJM, et al. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16:3519–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Welsch S, et al. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wessels E, Duijsings D, Notebaart RA, Melchers WJG, van Kuppeveld FJM. 2005. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-Golgi transport. J. Virol. 79:5163–5173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wong J, et al. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 82:9143–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5:1180–1185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.