Abstract
The incorporation of tegument proteins into the herpes simplex virus 1 (HSV-1) virion during virion assembly is thought to be a complex, multistage process occurring via numerous interactions between the tegument and the capsid, within the tegument, and between the tegument and the envelope. Here, we set out to examine if the direct interaction between two essential tegument proteins VP1/2 and VP16 is required for connecting the inner tegument with the outer tegument. By using glutathione S-transferase (GST) pulldowns, we identified an essential role of lysine 343 in VP16, mutation of which to a neutral amino acid abrogated the interaction between VP1/2 and VP16. When the K343A substitution was inserted into the gene encoding VP16 (UL48) of the viral genome, HSV-1 replicated successfully although its growth was delayed, and final titers were reduced compared to titers of wild-type virus. Surprisingly, the mutated VP16 was incorporated into virions at levels similar to those of wild-type VP16. However, the analysis of VP16 on cytoplasmic capsids by fluorescence microscopy showed that VP16 associated with cytoplasmic capsids less efficiently when the VP16-VP1/2 interaction was inhibited. This implies that the direct interaction between VP1/2 and VP16 is important for the efficiency/timing of viral assembly but is not essential for HSV-1 replication in cell culture. These data also support the notion that the incorporation of tegument proteins into the herpesviruses is a very complex process with significant redundancy.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is a large DNA virus in which the viral genome is enclosed in an icosahedral capsid, which is surrounded by a complex proteinaceous layer called the tegument. Lastly, the virion is enclosed in a lipid envelope bearing several viral envelope proteins.
HSV-1 assembly is an ordered process that begins with viral genome replication and packaging into preassembled capsids in the nucleus. These nucleocapsids then exit the nucleus and gain access to the cytoplasm through a process of budding at the inner nuclear membrane, followed by fusion with the outer nuclear membrane. Mature virus particles are then formed via budding of nucleocapsids with associated tegument into the lumen of membrane compartments thought to be derived from the trans-Golgi network (TGN) and/or endosomal compartments, a process called secondary envelopment (15 and 25).
It is thought that the incorporation of tegument proteins into the virion is an ordered, multistage process occurring via numerous tegument-capsid, tegument-tegument, and tegument-envelope interactions throughout the virus assembly process (15, 17, and 25). Although the study of tegument assembly is complicated by this large number of possible interactions and potential redundancy, many details concerning tegument structure and incorporation into the virions have been elucidated, primarily through studies on HSV-1 and the related porcine pathogen pseudorabies virus (PrV) (15, 17, and 25). It is generally considered that tegumentation can originate at two distinct sites: (i) at the nucleocapsid by interaction of tegument proteins with capsid (inner tegument) and (ii) at the future secondary envelopment site by binding of tegument proteins to cytoplasmic domains of viral envelope proteins (outer tegument).
Within the complex tegument network, two proteins that appear to be likely candidates to function in coordinating virus assembly are VP1/2 (pUL36) and VP16 (pUL48). VP1/2 is a large protein (>3,000 amino acids [aa]) encoded by the UL36 gene that is generally considered to be one of the first tegument proteins to associate with capsids and may act as an important scaffold during tegument assembly. This view is based on the observations that (i) VP1/2 is one of the few tegument proteins that stays associated with capsids migrating to the nuclear pores during entry (1, 11), (ii) VP1/2 remains associated with capsids after detergent solubilization/salt extraction of purified HSV-1 virions while most of the other tegument proteins are lost (33), (iii) VP1/2 interacts with other tegument proteins such as VP16 and pUL37 (18, 20, 26, 36) and has been shown to be essential for their recruitment to capsids (20), and (iv) VP1/2 directly interacts with the minor capsid protein pUL25 (4, 28). On the other hand, the tegument protein VP16, encoded by the UL48 gene, appears to be important for secondary envelopment in many alphaherpesviruses as (i) its deletion from the viral genome of HSV-1, PrV, and equine herpesvirus 1 (EHV-1) strongly impairs secondary envelopment (7, 27, 37) and as (ii) VP16 is able to interact with many viral proteins including tegument proteins VP1/2, Vhs (pUL41), VP11/12 (pUL46), VP13/14 (pUL47), and VP22 (pUL49) (6, 32, 36, 40) and the cytoplasmic domains of glycoproteins B, D, and H (12, 16, 41). However, it should be noted that VP16 may not be essential for all alphaherpesvirus assembly because the homologue in varicella-zoster virus ([VZV] open reading frame 10 [ORF10]) is dispensable for virus replication in cell culture (3). In addition to its structural role, VP16 forms a transcription complex with host cell factors HCF-1 and Oct-1 and drives the expression of immediate early (IE) genes (8, 14, 34, 38).
The aim of this study was to examine the role of the interaction between VP1/2 and VP16 in HSV-1 assembly. As both proteins are indispensable for HSV-1 replication (5, 17, 27) and as VP16 has been reported to bind directly to VP1/2 (36) and to require VP1/2 in order to be recruited to capsids, we hypothesized that their direct interaction could be an essential link connecting the inner tegument with the outer tegument. Using glutathione S-transferase (GST) pulldowns, we identified a single lysine in VP16, the mutation of which abrogated the interaction between VP1/2 and VP16. When this mutation was introduced into the viral genome, VP16 was less efficiently recruited to cytoplasmic nucleocapsids, and a minor inhibition of HSV-1 replication was observed. However, normal amounts of mutated VP16 were incorporated into mature extracellular virions, suggesting that the direct interaction between VP1/2 and VP16, although contributing to efficient virus assembly, is not essential for HSV-1 replication in cell culture.
MATERIALS AND METHODS
Cells and viruses.
Vero, HS30 (5), HaCaT, and HFF-hTERT (human telomerase reverse transcriptase immortalized human foreskin fibroblast) cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS), 100 U/ml penicillin G, 0.1 mg/ml streptomycin, and 2 mM glutamine.
HSV-1 containing a large deletion in the UL36 gene (KΔUL36; strain KOS) was grown, and titers in the UL36 complementing cell line HS30 were determined (5). All other viruses were propagated in Vero cells, and titers were determined.
Generation of VP1/2-specific monoclonal antibodies.
Female BALB/c mice were infected with HSV-1 (strain 17) by ear scarification, followed by an intraperitoneal boost 1 month later. Spleens were harvested 3 days later, and B-cell hybridomas were generated as previously described (10). Hybridoma supernatants were screened for reactivity in immunofluorescence assays using cells transfected with a green fluorescent protein (GFP)-VP1/2 expression plasmid. A cloned hybridoma line secreting an antibody with a strong reactivity to HSV-1 VP1/2 was selected and named CB4.
Plasmid construction.
Sequences of all primers used in this work are listed in Table 1. The 5′ end of the KOS UL36 ORF was amplified by PCR using primers COL162 and COL163 (Table 1) and cloned into the EcoRI and SalI sites of pEGFPC2 (Clontech). This plasmid expresses GFP fused to VP1/2 codons 1 to 581 (GFP-VP1/2NT; codon numbers correspond to the original strain 17 VP1/2 sequence; accession number NP_044638.1). Truncation mutants of VP16 tagged with GST were created by amplifying indicated regions of the UL48 gene from the HSV-1 KOS genome by PCR using specific sets of primers, followed by cloning into the EcoRI and XhoI sites of pCAG GST ENX (from J. Martin-Serrano, Kings College, London, United Kingdom) (Tables 1 and 2). Amino acid substitution mutants of VP16 in the context of GST-tagged VP16 consisting of residues 1 to 411 [GST-VP16(1–411)] were created by site-directed mutagenesis using specific primers and GST-VP16(1–411) as a template (Tables 1 and 3). All plasmids constructed using PCR products were verified by sequencing. Two universal transfer plasmids were used in the creation of recombinant viruses, pEP-mCherry-in and pEP-EYFP(A206K)-in. pEP-EYFP(A206K)-in contains the A206K mutation to reduce dimerization of enhanced yellow fluorescent protein (EYFP) (39) and was created by site-directed mutagenesis using primers SS15 and SS16 (Table 1) and pEP-EYFP-in as a template. pEP-mCherry-in and pEP-EYFP-in were gifts from N. Osterrieder, Freie Universität, Berlin, Germany.
Table 1.
Primers used in this work
| Primer name | Sequence |
|---|---|
| COL162 | GAAGCTTGAATTCATGGGTGGCGGAAACAAC |
| COL163 | GTCTAGAGTCGACTCAGGCGTAGTCGGGGACGTCATATGGGTAGAGCTCCAGCAGCCCC |
| COL281 | CTGAGAATTCGCCACCATGGACCTCTTGGTCGACG |
| COL282 | CTGACTCGAGTCACCCACCGTACTCGTCAATTC |
| COL307 | CTGACTCGAGTCACGACAGTCTGCGCGTGTG |
| SS07 | ACAGCCCTCCCGTCCGACACCCCCATATCGCTTCCCGACCTCCGGTCCCGATGGTGAGCAAGGGCGAGGAG |
| SS08 | CCAAGCGCCCGGACGCTATCGGTGGTAACGGTGCTGGGGCGGTGAAATTGCTTGTACAGCTCGTCCATGCC |
| SS09 | AGTTTGAGCAGATGTTTACCGATGCCCTTGGAATTGACGAGTACGGTGGGGTGAGCAAGGGCGAGGAG |
| SS10 | GGGACGGGAGGGGAAAACCCAGACGGGGGATGCGGGTCCGGTCGCGCCCCTTACTTGTACAGCTCGTCCATG |
| SS15 | CCTGAGCTACCAGTCCAAGCTGAGCAAAGACCCCA |
| SS16 | TGGGGTCTTTGCTCAGCTTGGACTGGTAGCTCAGG |
| SS19 | GATCGACTCGAGCTAAGATAGGAATTTACACTCCCGG |
| SS20 | GATCGACTCGAGCTACATTTCTCCCAGGTCGCGA |
| SS24 | GACTGAGAATTCGCCACCATGCGCGAGCACCTTAACCTCC |
| SS25 | GATCGACTCGAGCTACGACAGTCTGCGCGTGTG |
| SS26 | GATCGACTCGAGCTAGCCATGCAGGGTGGGAGG |
| SS27 | GATCGACTCGAGCTAGGAATACGAGTCCAACTTCG |
| SS28 | GATCGACTCGAGCTACGTACGCGCGCGGCTGTA |
| SS29 | GATCGACTCGAGCTAGTCGTCGTCCGGGAGATC |
| SS30 | GATCGACTCGAGCTACGGGAGGTTAAGGTGCTCG |
| SS33 | GACTGAGAATTCCCTCCCACCCTGCATGGC |
| SS34 | GACTGAGAATTCGCCTCTGGGTACTTTATGG |
| SS35 | GACTGAGAATTCGCGAAGTTGGACTCGTATTC |
| SS36 | GACTGAGAATTCCCCTCCGAGGCGGTCATG |
| SS37 | GACTGAGAATTCTACAGCCGCGCGCGTACG |
| SS44 | GATCGACTCGAGCTACATGACCGCCTCGGAGGG |
| SS45 | GATCGACTCGAGCTACATAAAGTACCCAGAGGCG |
| SS46 | GATCGACTCGAGCTACTCCTCCGTAGCCGCGCT |
| SS54 | CAACCAGGCCCGCGCCGCTGCGTACTTTATGGTGTTG |
| SS55 | CAACACCATAAAGTACGCAGCGGCGCGGGCCTGGTTG |
| SS56 | GCCCGCGCCTCTGGGGCCGCTGCGGTGTTGATTCGGGC |
| SS57 | GCCCGAATCAACACCGCAGCGGCCCCAGAGGCGCGGGC |
| SS58 | CTGGGTACTTTATGGCGGCGGCTCGGGCGAAGTTGGAC |
| SS59 | GTCCAACTTCGCCCGAGCCGCCGCCATAAAGTACCCAG |
| SS60 | CTTTATGGTGTTGATTGCGGCGGCGTTGGACTCGTATTC |
| SS61 | GAATACGAGTCCAACGCCGCCGCAATCAACACCATAAAG |
| SS62 | GTTGATTCGGGCGAAGGCGGCCGCGTATTCCAGCTTCAC |
| SS63 | GTGAAGCTGGAATACGCGGCCGCCTTCGCCCGAATCAAC |
| SS64 | GCGAAGTTGGACTCGGCTGCCGCCTTCACGACCTCGCC |
| SS65 | GGCGAGGTCGTGAAGGCGGCAGCCGAGTCCAACTTCGC |
| SS74 | TTTATGGTGTTGATTGCGGCGAAGTTGGACTC |
| SS75 | GAGTCCAACTTCGCCGCAATCAACACCATAAA |
| SS76 | GTGTTGATTCGGGCGGCGTTGGACTCGTATTC |
| SS77 | GAATACGAGTCCAACGCCGCCCGAATCAACAC |
| SS93 | GGCCCGCGCCTCTGGGTACTTTATGGTGTTGATTCGGGCGGCGTTGGACTCGTATTCCAGCTAGGATGACGACGATAAGTAGGG |
| SS94 | CTCGGAGGGCGAGGTCGTGAAGCTGGAATACGAGTCCAACGCCGCCCGAATCAACACCATAACAACCAATTAACCAATTCTGATTAG |
| SS102 | TGTTGATTCGGGCGAGGTTGGACTCGTATTC |
| SS103 | GAATACGAGTCCAACCTCGCCCGAATCAACA |
| SS104 | TGTTGATTCGGGCGATGTTGGACTCGTATTC |
| SS105 | GAATACGAGTCCAACATCGCCCGAATCAACA |
| SS106 | GTGTTGATTCGGGCGCAGTTGGACTCGTATT |
| SS107 | AATACGAGTCCAACTGCGCCCGAATCAACAC |
| SS108 | GTGTTGATTCGGGCGCATTTGGACTCGTATTCC |
| SS109 | GGAATACGAGTCCAAATGCGCCCGAATCAACAC |
Table 2.
GST-tagged VP16 truncation constructs
| Plasmida | Primers used |
|---|---|
| GST-VP16 (FL) | COL281 + COL282 |
| GST-VP16(1-411) | COL281 + COL307 |
| GST-VP16(1-387) | COL281 + SS29 |
| GST-VP16(1-369) | COL281 + SS28 |
| GST-VP16(1-359) | COL281 + SS44 |
| GST-VP16(1-348) | COL281 + SS27 |
| GST-VP16(1-337) | COL281 + SS45 |
| GST-VP16(1-327) | COL281 + SS26 |
| GST-VP16(1-314) | COL281 + SS46 |
| GST-VP16(1-305) | COL281 + SS30 |
| GST-VP16(1-206) | COL281 + SS20 |
| GST-VP16(1-106) | COL281 + SS19 |
| GST-VP16(299-411) | SS24 + SS25 |
| GST-VP16(322-411) | SS33 + SS25 |
| GST-VP16(332-411) | SS34 + SS25 |
| GST-VP16(342-411) | SS35 + SS25 |
| GST-VP16(354-411) | SS36 + SS25 |
| GST-VP16(364-411) | SS37 + SS25 |
| GST-VP16(332-348) | SS34 + SS27 |
Numbers in parentheses represent amino acids of VP16 encoded. FL, full-length.
Table 3.
VP16 mutants created by site-directed mutagenesis in the context of GST-VP16(1-411)
| Amino acid substitution(s) | Designated plasmid namea | Primers used |
|---|---|---|
| S333A G334A | M1 | SS54 + SS55 |
| Y335A F336A M337A | M2 | SS56 + SS57 |
| V338A L339A I340A | M3 | SS58 + SS59 |
| R341A K343A | M4 | SS60 + SS61 |
| L344A D345A S346A | M5 | SS62 + SS63 |
| Y347A S348A S349A | M6 | SS64 + SS65 |
| R341A | NA | SS74 + SS75 |
| K343A | NA | SS76 + SS77 |
| K343R | NA | SS102 + SS103 |
| K343M | NA | SS104 + SS105 |
| K343Q | NA | SS106 + SS107 |
| K343H | NA | SS108 + SS109 |
NA, not applicable
GST pulldown assays.
293T cells were seeded at 1.2 × 106 cells per well in six-well dishes and transfected several hours later using 1.5 μg of each plasmid and 12 μl of 1 mg/ml polyethylenimine (Polysciences). After 48 h, cells were harvested, pelleted, and lysed in 0.5 ml of lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 5% glycerol, 1% Triton X-100) supplemented with protease inhibitor cocktail (Roche) for 20 min at 4°C on a rotating wheel. Lysates were clarified at 17,000 × g for 10 min at 4°C and then incubated with glutathione-Sepharose 4B (Amersham Biosciences) at 4°C on a rotating wheel for 3 h. Sepharose beads were harvested at 2,400 × g for 3 min at 4°C and washed three times with lysis buffer. Bound protein complexes were eluted by heating in SDS-PAGE sample buffer at 98°C for 5 min and analyzed by Western blotting using antibodies specific for GST and GFP. All scanned blots were quantified using ImageJ, and signals in bead fractions of (GFP-VP1/2NT) were normalized to the amount of the GST-VP16 construct present in the same sample. These arbitrary values were used to classify GST-VP16 binding as follows: no binding, 0 to 0.05; weak, 0.05 to 0.5; moderate, 0.5 to 1; strong, 1 to 2; very strong, >2.
Coimmunoprecipitation.
HaCaT cells were grown to confluence in 100-mm dishes and infected with HSV-1 at a multiplicity of infection (MOI) of 5 PFU/cell. After 18 h, cell lysates were prepared as described above and precleared with protein A/G UltraLink resin (Thermo Scientific) for 1 h at 4°C on a rotating wheel. Samples were incubated with a VP16-specific monoclonal antibody (LP1; Abcam) (24) for a minimum of 1 h, followed by addition of protein A/G resin overnight on a rotating wheel at 4°C. Immunoprecipitated complexes were washed three times with lysis buffer, eluted, and analyzed by Western blotting as described below.
Purification of extracellular virions.
HaCaT cells grown in roller bottles were infected at 0.01 PFU/cell and incubated for 3 days. The culture medium was harvested and centrifuged at 2,000 rpm (Beckman GH3.8) for 20 min to remove cell debris. Supernatant virions were pelleted at 18,000 rpm (Beckman type 19 rotor) for 2 h, resuspended in 2 ml of 1% FCS–phosphate-buffered saline (PBS), and centrifuged through a 30-ml, 5 to 15% continuous Ficoll gradient at 12,000 rpm (Beckman SW 32Ti) for 1.5 h. The clear virus band in the middle of each gradient was harvested, and the virions were pelleted at 20,000 rpm (Beckman SW 32Ti) for 2 h. All centrifugations were performed at 4°C. The pellets were resuspended in PBS and stored at −70°C. Infectious virus titers were determined by plaque assay on Vero cells, and protein content was analyzed by Western blotting.
Western blotting.
Samples were boiled with SDS-PAGE sample buffer for 5 min, separated on polyacrylamide gels, and electrophoretically transferred to nitrocellulose membranes. Membranes were blocked and incubated with antibodies against GFP (JL-8; Clontech), GST (sc-459; Santa Cruz), VP1/2 (CB4), VP1/2 N terminus (anti-NT1 from P. O'Hare), VP16 (LP1; Abcam) (24), VP5 (ab6508; Abcam), pUL37 (from T. Mettenleiter), pUL41 (from B. Roizman), VP13/14 and VP22 (from G. Elliott), tubulin (MCA77G; AbD Serotec), gH (BBH1; Abcam), ICP0 (ab6513; Abcam), and ICP4 (58S) (31).
Construction of recombinant viruses.
All recombinant viruses were generated using a two-step red recombination technique (35) in Escherichia coli strain GS1783 (from G. Smith, Northwestern University) harboring bacterial artificial chromosome (BAC)-cloned HSV-1 genome (strain KOS; from D.A. Leib, Dartmouth Medical School) (9). First, singly modified viruses were constructed, as follows: HSV-VP16-Ch, where the coding sequence of mCherry was inserted in frame following codon 490 of VP16 (primers SS09 and SS10), and HSV-VP16(K343A) with lysine 343 in the UL48 gene mutated to alanine (primers SS93 and SS94). Subsequently, the BAC DNA of HSV-VP16-Ch was used as a template to insert the K343A substitution into the UL48 gene, creating HSV-VP16(K343A)-Ch, and to fuse EYFP(A206K) to codons 5 to 112 of the small capsid protein VP26 (primers SS07 and SS08), generating HSV-VP16-Ch/VP26-Y. Finally, the BAC DNA of HSV-VP16(K343A)-Ch was used as a template to fuse EYFP(A206K) to codons 5 to 112 of VP26, creating HSV-VP16(K343A)-Ch/VP26-Y. The sequences of primers used to create HSV-1 recombinants are listed in Table 1.
Virus reconstitution and BAC excision.
Vero cells were transfected with ∼100 ng of the BAC DNA together with 1 μg of pGS403 encoding Cre recombinase (from H. Coleman, University of Cambridge) using Fugene 6 (Roche) to reconstitute HSV-1 and excise the BAC backbone from the viral genome. After 3 to 4 days of incubation at 37°C, cell monolayers showing plaque formation were harvested, sonicated, and stored at −70°C. The efficiency of BAC backbone excision and the virus titers were determined by plaque assay on Vero cells followed by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining.
Growth analysis of recombinant viruses.
HaCaT cells were inoculated with recombinant viruses at 10 PFU/cell for 1 h at 37°C. The residual virus was inactivated by acid wash (40 mM citric acid, 135 mM NaCl, 10 mM KCl, pH 3.0) for 1 min at room temperature and three washes with PBS. At various times postinfection, cells were harvested and lysed by freezing/thawing and sonication. Virus yield was determined by plaque assay on Vero cells.
Immunofluorescence.
HFF-hTERT cells grown on 13-mm coverslips were infected at an MOI of 2 PFU/cell for 11 h. The cells were then fixed in 3% formaldehyde in PBS for 10 min and washed in PBS supplemented with 1% FCS. Samples were mounted in ProLong Gold antifade reagent with DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen). Images were acquired through the entire thickness of cells using the 60× (numerical aperture, 1.4) oil objective of an Olympus IX81 inverted wide-field microscope using Image-Pro Plus software (Media Cybernetics). The images were deconvolved with a blind deconvolution algorithm using AutoQuant software (Media Cybernetics). Using the “measure objects” function of the Image-Pro Plus software, mCherry emissions were determined for each EYFP punctum and plotted on histograms.
RESULTS
Identification of VP16 region required for binding VP1/2.
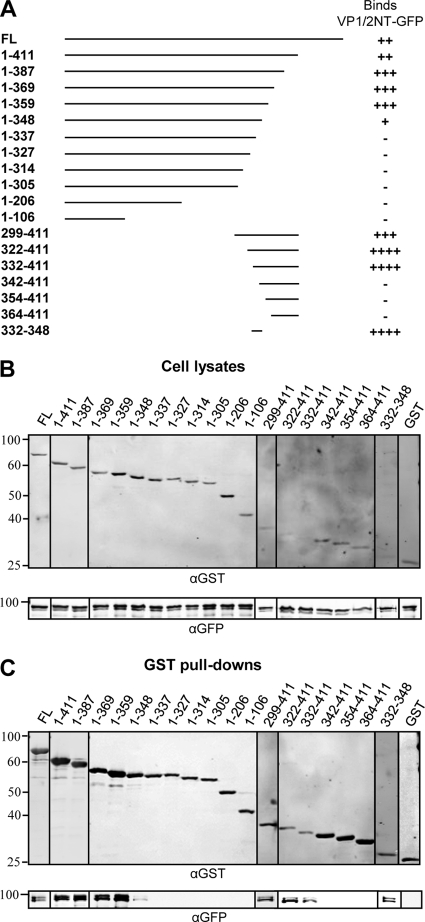
In order to determine the region of VP16 important for binding to VP1/2, a series of GST-tagged VP16 truncation mutants was generated and used in GST pulldown assays with the GFP-tagged N terminus of VP1/2 (GFP-VP1/2NT; residues 1 to 581) (Fig. 1A). The N terminus of VP1/2 was used for these assays as residues 124 to 511 of VP1/2 have been reported to be essential for binding VP16 (26). Western blotting confirmed that all GST-VP16 constructs were expressed and bound glutathione-Sepharose (Fig. 1B and C). Full-length VP16 efficiently bound GFP-VP1/2NT although removal of the activation domain of VP16 (residues 412 to 490) increased the association of GST-VP16 with GFP-VP1/2NT, suggesting a possible auto-inhibitory effect of this domain. Further truncation of the C terminus of VP16 to residue 359 had no effect on the interaction with VP1/2NT. However, removal of a further 11 amino acids reduced the efficiency of the interaction (residues 1 to 348), and any further truncation of the C terminus of VP16 abolished the interaction (Fig. 1C). Likewise, truncation of the N terminus of VP16 up to residue 331 did not prevent the interaction with VP1/2NT; however, any further N-terminal truncation abolished this interaction. A further construct (residues 332 to 348) containing the minimal overlapping region between the GST-VP16 constructs that demonstrated positive interaction with VP1/2NT was tested. This domain demonstrated a good interaction with GFP-VP1/2NT, suggesting that residues 332 to 348 of VP16 are both necessary and sufficient for the interaction with VP1/2.
Fig 1.
Mapping of the VP16 region involved in VP1/2 binding. (A) A schematic representation of GST-tagged VP16 truncation mutants and summary of results, indicating the GST-VP16 truncation mutants that bound very strongly (++++), strongly (+++), moderately (++), or weakly (+) or did not bind (−) to GFP-VP1/2NT. See Materials and Methods for details of quantification. FL, full-length. (B and C) 293T cells were cotransfected with GFP-VP1/2NT and the indicated GST-VP16 truncation mutants. Cell lysates were harvested at 48 h posttransfection and incubated with glutathione-Sepharose beads. Both cell lysates and bound protein complexes (GST pulldowns) were separated by SDS-PAGE and analyzed by Western blotting with anti-GST and anti-GFP antibodies. α, anti.
Substitution of lysine 343 in VP16 for a neutral amino acid inhibits binding to VP1/2.
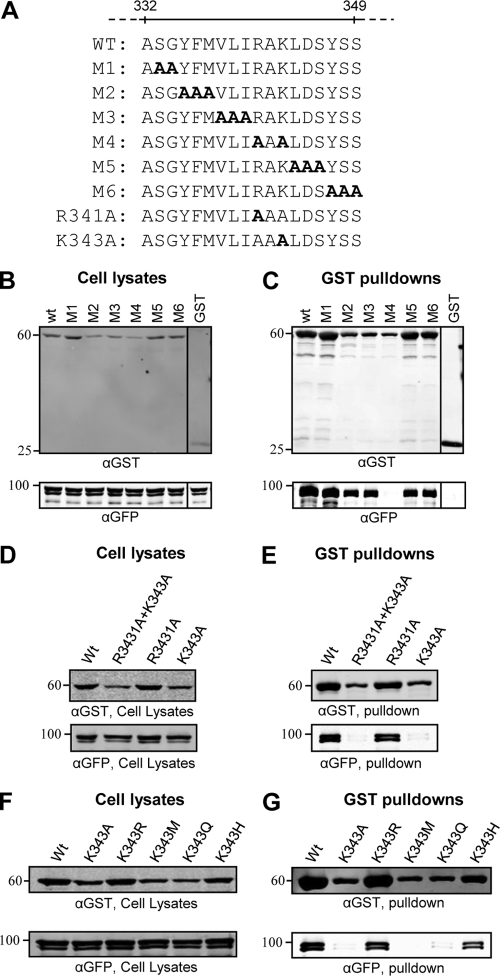
To further elucidate the residues of VP16 involved in binding to VP1/2, two or three amino acids spanning the region of residues 333 to 349 were mutated simultaneously to alanine [in the context of GST-VP16(1–411)] (Fig. 2A). GST pulldown assays demonstrated that VP1/2NT interacted with all mutants except M4, indicating that arginine 341 and/or lysine 343 is crucial for this interaction (Fig. 2C). Further mutation of the individual residues R341 or K343 to alanine demonstrated that K343 in VP16 is essential for the interaction with VP1/2 in GST pulldowns (Fig. 2E).
Fig 2.
Alanine scanning mutagenesis of VP16. (A) Amino acid sequence of the region spanning residues 332 to 349 of VP16 and mutants (M1 to M6) created in the context of GST-VP16(1–411). (B to G) 293T cells were cotransfected with GFP-VP1/2NT and indicated GST-VP16(1–411) mutants. Cell lysates were harvested at 48 h posttransfection and incubated with glutathione-Sepharose beads. Both cell lysates and bound protein complexes were separated by SDS-PAGE and analyzed by Western blotting with anti-GST and anti-GFP antibodies.
A specific role of K343 in VP1/2 interaction was surprising because the crystal structure of VP16 shows that this lysine residue has important intramolecular interactions within the VP16 core (23). Furthermore, a previous study demonstrated that replacement of K343 with alanine led to poor VP16 expression in E. coli, suggesting a major perturbation in the overall VP16 structure (21). In contrast, a more recent study found that VP16 containing a K343A substitution could still bind HCF-1 and activate transcription although its interaction with pUL41 was diminished (19). We also observed that VP16(K343A) reached lower expression levels than wild-type (wt) VP16 in 293T cells. While lower expression levels could be due to either reduced protein synthesis or increased turnover of protein, given that there is only a single amino acid change, it would seem most likely that the smaller amount of VP16(K343A) observed in cell lysates is due to a more rapid degradation of VP16(K343A) caused by greater instability of the protein structure rather than by any alteration to protein synthesis rates. We were therefore concerned that the inhibition of VP16-VP1/2 interaction was due to destabilizing the structure of VP16. However, other mutations (M2 and M3) demonstrated an equivalent level of expression/destabilization (Fig. 2B), and yet these constructs efficiently interacted with VP1/2 (Fig. 2C), suggesting a specific role of K343 in the VP16-VP1/2 interaction.
As lysine-to-alanine substitution is a relatively major side chain alteration, K343 was further mutated to arginine, methionine, glutamine, or histidine to investigate if it is possible to inhibit VP16-VP1/2 interaction while maintaining the stability of VP16. GST pulldown assays demonstrated that the presence of any positively charged side chain (K, R, or H) at residue 343 maintained both high levels of VP16 expression and VP1/2 interaction while mutation of K343 to uncharged residues (A, M, or Q) strongly inhibited the interaction with VP1/2 but also reduced VP16 expression (Fig. 2F and G). Overall, these data suggest that K343 has a direct role in the VP16-VP1/2 interaction, in addition to being important for the overall stability of VP16.
Generation of recombinant HSV-1 harboring VP16(K343A).
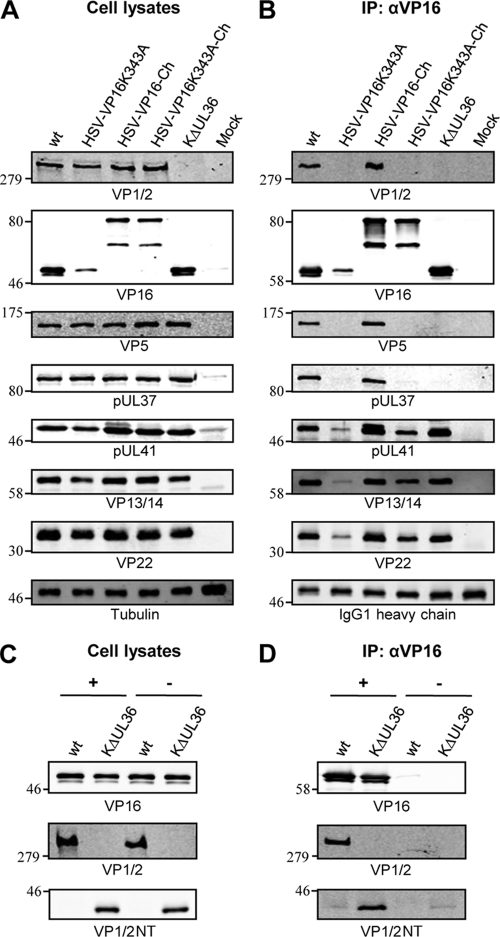
To examine if the VP16-VP1/2 interaction is essential for HSV-1 assembly, a recombinant HSV-1 expressing VP16(K343A) was created [HSV-VP16(K343A)]. Surprisingly, this virus was viable in cell culture and replicated to relatively high titers. Analysis of viral protein expression levels by Western blotting demonstrated that the K343A mutation had a more profound effect on the expression level/stability of virally expressed VP16 than that of the VP16 tagged with GST (compare Fig. 2D and 3A). These observations support a role for K343 in maintaining a stable core structure of VP16 and also suggest that the addition of a protein tag on VP16(K343A) increases its stability. Therefore, further recombinant viruses were constructed expressing VP16 tagged at the C terminus with mCherry (HSV-VP16-Ch) and additionally containing the K343A substitution [HSV-VP16(K343A)-Ch]. Analysis of infected cell lysates confirmed our notion that tagging of VP16(K343A) stabilizes the protein as mCherry-tagged VP16 and VP16(K343A) reached very similar levels (Fig. 3A). Two bands were consistently observed for VP16-mCherry in Western blots with an anti-VP16 antibody (LP1), suggesting a greater susceptibility to proteolytic cleavage than untagged VP16, although it is currently unclear whether this cleavage occurs normally within cells or during SDS-PAGE sample preparation. However, as the LP1 antibody binds an epitope near the N terminus of VP16, proteolytic cleavage is presumably occurring within the mCherry, and there was no apparent difference in proteolysis of wt or K343A VP16-mCherry.
Fig 3.
K343A substitution in VP16 inhibits interaction with VP1/2 during infection. (A and C) HaCaT cells were infected with the indicated viruses, and cell lysates were harvested at 18 hpi. (B and D) Cell lysates were subjected to immunoprecipitation (IP) using an anti-VP16 (αVP16) antibody. Cell lysates (A and C) and bound protein complexes (B and D) were separated by SDS-PAGE and analyzed by Western blotting using the indicated antibodies. Tubulin and the heavy chain of the anti-VP16 antibody were included as loading controls.
To investigate whether the K343A substitution in VP16 affects the interaction between VP16 and VP1/2 during infection, cells were infected with the HSV-VP16(K343A) mutants and their respective wt variants. After 18 h, cell lysates were used in immunoprecipitation assays with anti-VP16 (Fig. 3B). VP1/2 was efficiently coprecipitated with VP16 from cells infected with viruses expressing nonmutated VP16 (wt and HSV-VP16-Ch), as expected from published data (20), but not from cells infected with either VP16(K343A) virus or the KΔUL36 virus that contains a large deletion in the VP1/2 gene. Interestingly, both VP5 and pUL37 were also coprecipitated with VP16 from cells infected with wt and HSV-VP16-Ch but not from cells infected with HSV-VP16(K343A), HSV-VP16(K343A)-Ch, or KΔUL36, suggesting that these two proteins were coprecipitated via their interaction with VP1/2. VP1/2 is known to directly interact with pUL37 (26), and while no direct interaction has been shown with VP5, VP1/2 does interact with the minor capsid protein pUL25 (28), suggesting that the coprecipitation of VP5 could be indirect via this VP1/2-capsid association. We further tested the outer tegument proteins pUL41, VP13/14, and VP22, which have all been reported to directly interact with VP16 (6, 32, 36, 40). All three of these tegument proteins were coprecipitated with VP16 from all infected cells, although at much reduced levels from cells infected with HSV-VP16(K343A). This was expected as VP16(K343A) has significantly lower expression levels than wt VP16. However, smaller amounts of both pUL41 and VP22 were coimmunoprecipitated from cells infected with HSV-VP16(K343A)-Ch than with HSV-VP16-Ch, despite similar VP16 expression levels. This indicates that the K343A substitution in VP16 may also slightly inhibit the interactions of VP16 with VP22 and pUL41, which is in agreement with a previous report (19).
The KΔUL36 virus was used as a control for these assays as it lacks expression of full-length VP1/2. However, this virus does not harbor a full deletion of UL36, and an N-terminal fragment containing the first 361 amino acids of VP1/2, which overlaps substantially with the predicted VP16 binding site, is expressed by this virus (29). The monoclonal VP1/2 antibody that we have generated reacts with an epitope in the middle region of the protein and so does not detect this N-terminal fragment. To investigate whether the N-terminal fragment of VP1/2 expressed by KΔUL36 could still interact with VP16, cell lysates were used in immunoprecipitation assays in the presence or absence of anti-VP16, and samples were analyzed by Western blotting using an antibody specific for the N terminus of VP1/2 (2). Interestingly, the VP1/2 N-terminal fragment expressed by KΔUL36 was efficiently coprecipitated with VP16, suggesting that the interaction site lies within this domain of VP1/2 (Fig. 3C and D).
The K343A mutation in VP16 affects viral replication in cell culture and the expression of ICP4.
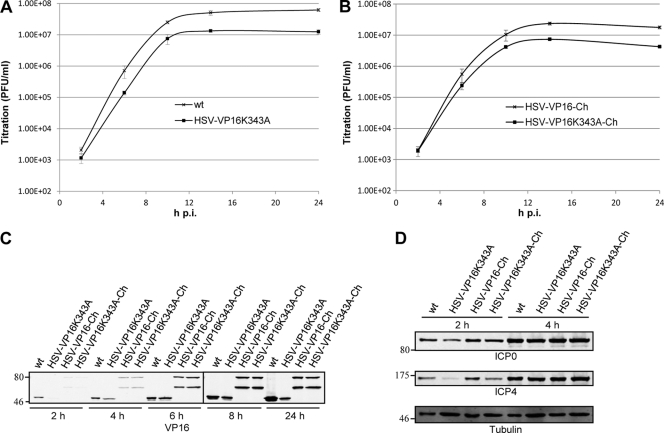
To assess if inhibiting the interaction between VP16 and VP1/2 has an effect on virus replication rates, single-cycle growth analyses were performed (Fig. 4A and B). Both mutant viruses replicated with slower kinetics than their parental strains, with final titers of HSV-VP16(K343A) and HSV-VP16(K343A)-Ch being 4- to 5-fold lower than those of wt and HSV-VP16-Ch, respectively. The observation that similar growth defects were detected with both K343A mutant viruses suggests that the slower kinetics of replication is due to the inhibition of VP16-VP1/2 interaction and not primarily caused by destabilization of VP16.
Fig 4.
K343A mutation in VP16 moderately inhibits virus replication. HaCaT cells were infected with the indicated viruses and harvested at various time points. (A and B) Total infectious virus yields were determined by plaque assay on Vero cells. The titers represent mean PFU/ml, and error bars represent standard errors of the means of triplicate samples. (C and D) Cell lysates were separated by SDS-PAGE and analyzed by Western blotting using the indicated antibodies. Tubulin was included as a loading control.
To examine the effect of the K343A mutation on VP16 expression levels during HSV-1 infection, cell lysates from cells infected with nonmutated and mutant viruses were analyzed at 2, 4, 6, 8, and 24 h postinfection (hpi). Interestingly, VP16(K343A) demonstrated similar levels of protein as wt VP16 at early time points compared to levels at 24 hpi, suggesting that the K343A mutation destabilizes VP16 only after it has passed a certain concentration in infected cells (Fig. 4C).
It has been reported that although the interaction of VP16(K343A) with HCF-1 is slightly diminished compared to wt VP16, VP16(K343A) is still able to activate transcription of a luciferase reporter gene linked to the promoter of the immediate-early (IE) gene ICP4 (19). To determine if the K343A substitution in VP16 affects expression of IE genes during infection, samples harvested at 2 and 4 hpi were also analyzed for the expression levels of two IE gene products, ICP0 and ICP4 (Fig. 4D). The expression levels of both ICP0 and ICP4 were diminished at 2 hpi in cells infected with HSV-VP16(K343A) compared to levels in cells infected with wt. In contrast, in cells infected with HSV-VP16(K343A)-Ch only, ICP4 levels were slightly reduced at 2 hpi compared to cells infected with HSV-VP16-Ch. No difference was seen in the expression level of ICP0 or ICP4 at 4 hpi. This suggests that the K343A mutation in VP16 slightly delays the transcription of IE genes, and this could be due to either the destabilization of VP16 or a reduced interaction with HCF-1. This may contribute to the delayed kinetics observed at the beginning of replication of both mutant viruses but seems unlikely to be the cause of reductions in final infectious titers.
VP16(K343A) is incorporated into virions at the same levels as wt VP16.
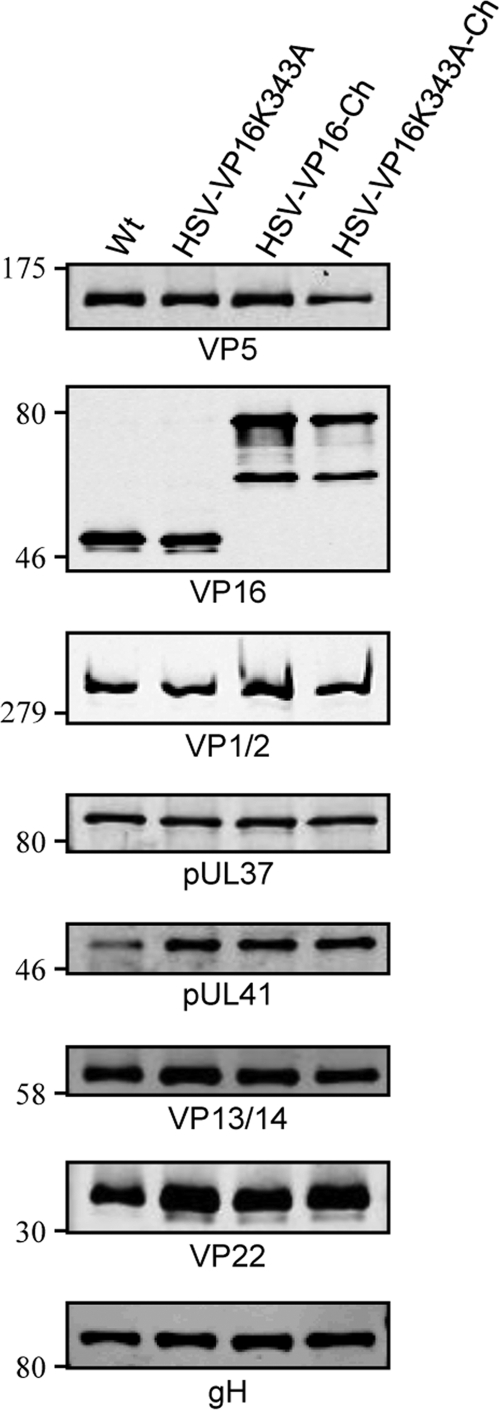
To examine if the inhibition of the VP16-VP1/2 interaction affects the incorporation of VP16 into virus particles, extracellular virions were purified from the medium of cells infected with both mutants and their wt variants. Western blotting of a range of PFU concentrations of the purified extracellular virion preparations and quantification of band intensities demonstrated 2- to 3-fold higher levels of VP5 in the K343A viruses than the equivalent PFU amounts of the wt variants, suggesting higher particle/PFU ratios for virions harboring the K343A mutation in VP16. Upon analysis of the purified viruses at similar levels of VP5 loading, it was apparent that similar levels of VP16 and VP16-Ch were incorporated into both wt and mutant viruses (Fig. 5). Similar levels of VP1/2, pUL37, VP13/14, and gH were also observed between all four purified viruses. However, a consistent increase of both pUL41 and VP22 in purified virions was observed in both K343A viruses. Quantification of several independent Western blots demonstrated that, when normalized to VP5 signals, VP16, VP1/2, pUL37, VP13/14, and gH levels all varied less than 2-fold between any sample. However, up to a 5-fold increase in pUL41 and up to a 3-fold increase in VP22 were observed in HSV-VP16(K343A) compared to wt levels, and 2- to 3-fold increases in both pUL41 and VP22 were observed in HSV-VP16(K343A)-Ch compared to HSV-VP16-Ch. Even though it is unclear why the K343A mutation would result in greater incorporation of pUL41 and VP22, these results indicate that the inhibition of VP16-VP1/2 interaction does not lead to any apparent reduction of VP16 in virions, suggesting alternative routes for recruiting VP16 into the tegument.
Fig 5.
VP16(K343A) is incorporated into virions in amounts similar to those of wt VP16. Extracellular virions were purified from the medium of infected HaCaT cells by Ficoll density gradient centrifugation, and VP5 levels in a range of PFU concentrations were analyzed by Western blotting. The appropriate PFU amounts to give equivalent levels of VP5 for each purified virion preparation were separated by SDS-PAGE and analyzed by Western blotting using the indicated antibodies. Numbers on the left represent molecular weight markers.
VP16(K343A) demonstrates a less efficient association with cytoplasmic capsids.
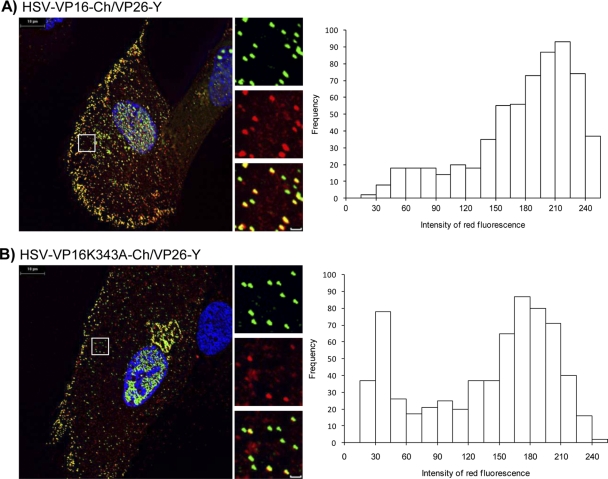
To test if the inhibition of the VP16-VP1/2 interaction may affect the efficiency of virus assembly, the capsid protein VP26 was labeled with EYFP in HSV-VP16-Ch and HSV-VP16(K343A)-Ch, resulting in HSV-VP16-Ch/VP26Y and HSV-VP16(K343A)-Ch/VP26Y, respectively. Cells were infected with these viruses, and the presence of VP16 on capsids was analyzed by measuring the red fluorescence intensity of all EYFP-labeled cytoplasmic particles. Histograms showing the red fluorescence profiles of VP16-Ch and VP16(K343A)-Ch formed two peaks, confirming that primarily two populations of cytoplasmic EYFP-VP26-labeled particles were observed in cells infected with the two viruses (Fig. 6), one emitting strong red fluorescence and one emitting very little red fluorescence. The right peak representing strong red fluorescence indicates EYFP-VP26-labeled capsids containing many VP16 molecules while the left peak with little or only background red fluorescence represents capsids with no or very few VP16 molecules. Interestingly, a far greater population of EYFP-VP26-labeled particles containing very little red fluorescence was observed in cells infected with HSV-VP16(K343A)-Ch/VP26-Y than for HSV-VP16-Ch/VP26-Y. This suggests that the inhibition of the VP16-VP1/2 interaction causes a less efficient and/or delayed association of VP16 with capsids in the cytoplasm of infected cells.
Fig 6.
Analysis of VP16-Ch and VP16(K343A)-Ch association with fluorescent capsids. Monolayers of HFF-hTERT cells grown on coverslips were infected with HSV-VP16-Ch/VP26-Y (A) or HSV-VP16(K343A)-Ch/VP26-Y (B). Cells were fixed and analyzed by fluorescence microscopy. Representative cells on right are shown with higher magnification (boxed area) slown along the right side of each large image. Scale bar of insets, 1 μm. Histograms at left show red fluorescence intensities of cytoplasmic EYFP-labeled particles.
DISCUSSION
Here, we have identified lysine 343 in VP16 as a crucial residue for the VP16-VP1/2 interaction. By mutating this residue in the HSV-1 genome and thus inhibiting the VP16-VP1/2 interaction during infection, we have further demonstrated that while the VP16-VP1/2 interaction does not appear to be essential for HSV-1 replication in tissue culture, it seems to be important for efficient HSV-1 assembly.
The finding that K343 is essential for VP16 to bind VP1/2 was surprising as, although this residue is highly conserved, the resolved crystal structure of the soluble VP16 core domain predicts that K343 is involved in a network of intramolecular polar interactions important for VP16 stability (23). This correlated with findings that the substitution of K343 for alanine resulted in complete inactivation of VP16 and poor VP16 synthesis in E. coli (21). In contrast, another study found that VP16 containing the substitution K343A was able to bind HCF-1 and activate transcription although its interaction with pUL41 was diminished (19). In our current study, VP16 carrying K343 substituted for a neutral amino acid demonstrated lower levels of protein expression than wt VP16, indicating that a positively charged residue at this position is indeed important for VP16 stability. We also found that any positively charged residue at position 343 maintains the interaction of VP16 with VP1/2. Therefore, it could be argued that an undetectable VP16-VP1/2 interaction caused by mutating K343 to a neutral amino acid is due to low expression levels/destabilization of VP16. However, this notion seems unlikely for several reasons. First, two VP16 mutants containing Y335A, F336A, and M337A or V338A, L339A, and I340A substitutions had protein expression levels similar to the level of VP16 carrying a K343A mutation but bound strongly to VP1/2. Also, VP16(K343A) tagged with mCherry appeared to have a level of stability similar to that of VP16-Ch and did not bind VP1/2 while VP16-Ch did. Furthermore, at around 6 to 8 hpi VP16(K343A) reaches protein levels similar to those of VP16 in cells and thus appears to be almost as stable as wt VP16. Therefore, a possible explanation could be that VP16(K343A) is destabilized only when free in solution while being prevented from degradation when it is bound to other proteins or fused to another protein domain. Early in infection the majority of VP16 molecules are likely to be in complexes with other proteins as they are needed for the assembly of new virions, while later in infection VP16 molecules may be overabundant and primarily free in the cytoplasm. Moreover, our observation that VP16(K343A) was incorporated into virions to the same extent as wt VP16 supports our theory that VP16(K343A) in protein complexes is as stable as wt VP16.
The fact that the crystal structure of VP16 shows the side chain of K343 projecting into the crystal core and forming a series of polar interactions would seem to preclude a direct interaction of this amino acid with VP1/2. However, the crystal structure of VP16 has been resolved only for free protein. It is conceivable that VP16 may adopt a different conformational state when bound to VP1/2, whereby K343 is exposed and available to mediate direct interactions with residues in VP1/2. If so, then it would appear that VP1/2 itself is sufficient to drive this conformational change as VP1/2 and VP16 interact in the absence of other viral proteins. This is currently speculation although it would be interesting in the future to investigate which VP16 and VP1/2 residues form direct contacts by determining the structure of a VP16-VP1/2 complex.
Based on the electrostatic surfaces of the VP16 core, it has been suggested that its concave surface, comprising residues around K343, forms a DNA-binding domain (23). After HSV-1 entry into the cell, VP16 is released from the virion, where it forms a VP16-induced transcription complex with two host cell factors, HCF-1 and Oct-1, on the viral IE promoters (8, 14, 34, 38). However, it is necessary for VP16 to perform this function only early in infection, and it is therefore tempting to speculate that both DNA and viral proteins compete for binding to this concave VP16 surface so that late in infection VP16 is biased toward assembling into virions rather than binding to IE promoters. In addition to VP1/2, pUL41 could be a potential candidate for binding to the concave VP16 surface as the residue L344 in VP16 has been shown to be crucial for the interaction of VP16 with pUL41 (19). In this way, binding of VP16 to at least two viral proteins could prevent its attachment to viral DNA and enhance its incorporation into virions. Given the proximity of K343 and L344, it is impossible to envisage how VP1/2 and pUL41 could bind to the same molecule of VP16, suggesting at least two pools of VP16 in the HSV-1 virion, one bound to VP1/2 and one bound to pUL41.
During our investigation into the VP16-VP1/2 interaction in infected cells, we utilized the KΔUL36 virus as a control that lacks expression of full-length VP1/2 (5). However, it is important to note that this virus contains only a partial deletion in UL36 and that an N-terminal fragment encoding the first 361 amino acids of VP1/2 is expressed in cells infected with KΔUL36 (29). Interestingly, this fragment of VP1/2 interacted efficiently with VP16 in our coimmunoprecipitation analysis. The interaction domain for VP16 within VP1/2 has previously been mapped to amino acids 124 to 511 (26), and so our data now add to this previous study and further narrow down the VP16 interaction site to residues 124 to 361 of VP1/2.
Unexpectedly, when the mutation K343A was inserted into the UL48 gene in the viral genome, despite potently inhibiting the VP16-VP1/2 interaction during infection, HSV-1 replication was largely unaffected, and the incorporation of VP16 into mature virus particles was not inhibited. This indicates that the VP16-VP1/2 interaction is dispensable for HSV-1 assembly and that VP16 does not require a direct interaction with VP1/2 in order to be incorporated into the virion. As there are thought to be only ∼100 copies of VP1/2 but >1,000 copies of VP16 in the virion (13), it would be unreasonable to expect that VP1/2 would be the only protein via which VP16 is normally incorporated into the virion. Furthermore, VP16 has been shown to interact with tegument proteins pUL41, VP11/12, VP13/14, and VP22 (6, 32, 36, 40) and the cytoplasmic domains of glycoproteins B, D, and H (12, 16, 41), offering VP16 a plethora of possible interactions for incorporation into virions. Interestingly, we did observe a consistent increase of both VP22 and pUL41 in purified VP16(K343A) virions compared to wt virions, and so it is possible that the increased levels of these two VP16-binding tegument proteins could compensate for the loss of VP16-VP1/2 interaction. We currently do not know the mechanism for this increased incorporation of VP22 and pUL41 in the absence of the VP16-VP1/2 interaction although it is conceivable that the delay in VP16 recruitment to cytoplasmic capsids we observe in VP16(K343A)-infected cells could lead to some deregulation of tegument assembly, allowing a greater association of VP22 and pUL41 with other tegument and/or envelope proteins and causing their enhanced incorporation. Such possibilities await further investigation.
Nevertheless, we had expected that the inhibition of the VP16-VP1/2 interaction would be severely attenuating for the virus as VP16 could act as a major link between the inner and the outer tegument/envelope via its interaction with VP1/2. Our observations that K343A-mutated viruses reached lower titers than wt and that VP16(K343A) associated with cytoplasmic capsids less efficiently suggest that the VP16-VP1/2 interaction is important for the efficiency of infectious HSV-1 assembly, although clearly not essential. It is possible that VP16 might interact with other inner tegument proteins in addition to VP1/2, such as pUL37 or pUS3, although such interactions have not been identified. Furthermore, several other interactions between outer tegument proteins and inner tegument or capsid proteins could serve as alternative routes for assembly, such as the recently reported pUL37-VP11/12 and pUL17-VP13/14 interactions (22, 30). In the future, it will be interesting to examine whether inhibiting the VP16-VP1/2 interaction has a greater effect on HSV-1 replication in the absence of VP11/12 and/or VP13/14 expression.
In conclusion, we have demonstrated that the interaction between two essential tegument proteins, VP1/2 and VP16, is important but not essential for HSV-1 replication in cell culture, further confirming the growing evidence that incorporation of tegument proteins into the virion is a complex process with significant redundancy. As all this work has been performed in tissue culture, it will be interesting in the future to determine whether VP16(K343A) viruses are viable in animal models as efficient assembly mediated through VP16-VP1/2 interaction may be important for virus replication in vivo.
ACKNOWLEDGMENTS
We thank N. Osterrieder (Freie Universität, Berlin, Germany), G. Smith (Northwestern University), D. A. Leib (Dartmouth Medical School), P. Desai (Johns Hopkins University), P. O'Hare (Imperial College London), T. Mettenleiter (Friedrich Loeffler Institutes), B. Roizman (University of Chicago), G. Elliott (Imperial College London), J. Martin-Serrano (King's College London), and H. Coleman (University of Cambridge) for reagents; S. Efstathiou, B. Bruun, and S. Colaco for help with monoclonal antibody generation; and other members of the Crump lab and D. Glauser for helpful discussions.
This work was funded by Wellcome Trust (Ph.D. studentship to S.S.), the Royal Society (UF090010), and the Medical Research Council UK (G0700129).
Footnotes
Published ahead of print 19 October 2011
REFERENCES
- 1. Antinone SE, Smith GA. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J. Virol. 84:1504–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bolstad M, Abaitua F, Crump CM, O'Hare P. 2011. Autocatalytic activity of the ubiquitin-specific protease domain of herpes simplex virus 1 VP1-2. J. Virol. 85:8738–8751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cohen JI, Seidel K. 1994. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J. Virol. 68:7850–7858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coller KE, Lee JI, Ueda A, Smith GA. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Desai PJ. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608–11618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elliott G, Mouzakitis G, O'Hare P. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fuchs W, Granzow H, Klupp BG, Kopp M, Mettenleiter TC. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerster T, Roeder RG. 1988. A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc. Natl. Acad. Sci. 85:6347–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gierasch WW, et al. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135:197–206 [DOI] [PubMed] [Google Scholar]
- 10. Gill MB, et al. 2006. Murine gammaherpesvirus-68 glycoprotein H-glycoprotein L complex is a major target for neutralizing monoclonal antibodies. J. Gen. Virol. 87:1465–1475 [DOI] [PubMed] [Google Scholar]
- 11. Granzow H, Klupp BG, Mettenleiter TC. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gross ST, Harley CA, Wilson DW. 2003. The cytoplasmic tail of Herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1–12 [DOI] [PubMed] [Google Scholar]
- 13. Heine JW, Honess RW, Cassai E, Roizman B. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14:640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herr W. 1998. The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harb. Symp. Quant. Biol. 63:599–607 [DOI] [PubMed] [Google Scholar]
- 15. Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9:382–394 [DOI] [PubMed] [Google Scholar]
- 16. Kamen DE, Gross ST, Girvin ME, Wilson DW. 2005. Structural basis for the physiological temperature dependence of the association of VP16 with the cytoplasmic tail of herpes simplex virus glycoprotein H. J. Virol. 79:6134–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. 2009. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 145:173–186 [DOI] [PubMed] [Google Scholar]
- 18. Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Knez J, Bilan PT, Capone JP. 2003. A single amino acid substitution in herpes simplex virus type 1 VP16 inhibits binding to the virion host shutoff protein and is incompatible with virus growth. J. Virol. 77:2892–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ko DH, Cunningham AL, Diefenbach RJ. 2010. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J. Virol. 84:1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lai JS, Herr W. 1997. Interdigitated residues within a small region of VP16 interact with Oct-1, HCF, and DNA. Mol. Cell. Biol. 17:3937–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee JH, Vittone V, Diefenbach E, Cunningham AL, Diefenbach RJ. 2008. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378:347–354 [DOI] [PubMed] [Google Scholar]
- 23. Liu Y, Gong W, Huang CC, Herr W, Cheng X. 1999. Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16. Genes Dev. 13:1692–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McLean C, et al. 1982. Monoclonal antibodies to three non-glycosylated antigens of herpes simplex virus type 2. J. Gen. Virol. 63:297–305 [DOI] [PubMed] [Google Scholar]
- 25. Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 26. Mijatov B, Cunningham AL, Diefenbach RJ. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26–31 [DOI] [PubMed] [Google Scholar]
- 27. Mossman KL, Sherburne R, Lavery C, Duncan J, Smiley JR. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roberts APE, et al. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scholtes LD, Yang K, Li LX, Baines JD. 2010. The capsid protein encoded by UL17 of herpes simplex virus 1 interacts with tegument protein VP13/14. J. Virol. 84:7642–7650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Showalter SD, Zweig M, Hampar B. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smibert CA, Popova B, Xiao P, Capone JP, Smiley JR. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spear PG, Roizman B. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9:143–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stern S, Tanaka M, Herr W. 1989. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature 341:624–630 [DOI] [PubMed] [Google Scholar]
- 35. Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634:421–430 [DOI] [PubMed] [Google Scholar]
- 36. Vittone V, et al. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. von Einem J, Schumacher D, O'Callaghan DJ, Osterrieder N. 2006. The α-TIF (VP16) homologue (ETIF) of equine herpesvirus 1 is essential for secondary envelopment and virus egress. J. Virol. 80:2609–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wysocka J, Herr W. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28:294–304 [DOI] [PubMed] [Google Scholar]
- 39. Zacharias DA, Violin JD, Newton AC, Tsien RY. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913–916 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y, Sirko DA, McKnight JL. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhu Q, Courtney RJ. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590–599 [DOI] [PubMed] [Google Scholar]