Abstract
CD8+ T cell responses rapidly select viral variants during acute human immunodeficiency virus (HIV)/simian immunodeficiency virus (SIV) infection. We used pyrosequencing to examine variation within three SIV-derived epitopes (Gag386-394GW9, Nef103-111RM9, and Rev59-68SP10) targeted by immunodominant CD8+ T cell responses in acutely infected Mauritian cynomolgus macaques. In animals recognizing all three epitopes, variation within Rev59-68SP10 was associated with delayed accumulation of variants in Gag386-394GW9 but had no effect on variation within Nef103-111RM9. This demonstrates that the entire T cell repertoire, rather than a single T cell population, influences the timing of immune escape, thereby providing the first example of conditional CD8+ T cell escape in HIV/SIV infection.
TEXT
Escape from CD8+ T cell responses is a hallmark of human immunodeficiency virus (HIV)/simian immunodeficiency virus (SIV) infection (6, 16, 24). Within days of the first detectable CD8+ T cell responses in blood, viral variants that abrogate binding to major histocompatibility complex class I (MHC class I) molecules, impair T cell receptor recognition, or diminish the efficiency of epitope processing begin to emerge (reviewed in references 15 and 29). In SIV-infected macaques, where the timing of infection is known, recognizable CD8+ T cell epitopes can be largely lost by 17 days postinfection in replicating viruses isolated from plasma (2, 11) and potentially even earlier in lymph nodes (26). Such responses are thought to be highly potent but of marginal protective value because they are short-lived.
In studies of CD8+ T cell escape during acute HIV/SIV infection, some individuals exhibit markedly less epitope variation than others, despite the presence of detectable CD8+ T cell responses (7, 20, 22). It has been proposed that delayed immune escape is a consequence of the CD8+ T cell immunodominance hierarchy, a characteristic of immunity that is influenced by an individual's set of MHC class I alleles (1, 13, 19, 21, 27). An alternate hypothesis for explaining the differential timing of immune escape is that the HIV/SIV genome can tolerate only a limited amount of simultaneous variation, such that variants accumulate in epitopes sequentially. Pyrosequencing has enabled investigators to quantify genome-wide variation within CD8+ T cell epitopes and explore the progression of immune escape with unprecedented resolution (2, 3, 11, 26). Using this approach, we tested the hypothesis that there is an upper limit to the number of SIV genomic sites in which simultaneous variation can be tolerated, such that escape within a given epitope may be delayed by the selection of variants in another epitope.
We studied a cohort of 18 MHC class I defined Mauritian cynomolgus macaques (MCM) infected intrarectally with 7,000 50% tissue culture infectious dose (TCID50) pathogenic SIVmac239 viruses. MCM have limited MHC genetic diversity, such that nearly all of their MHC alleles can be explained by seven common haplotypes, termed M1 to M7 (9, 23). Six MCM were homozygous for the M1 MHC haplotype (CY0320 to CY0325), six were M1/M3 heterozygotes (CY0326 to CY0331), and six were M3 homozygotes (CY0332 to CY0337). Both haplotypes carry Mafa-A1*063, an allele that restricts two epitopes (Gag386-394GW9 and Nef103-111RM9) targeted by acute-phase immunodominant CD8+ T cell responses (2, 8). The M3 haplotype also carries Mafa-B*075, an allele that restricts a strong acute-phase CD8+ T cell response directed against Rev59-68SP10 (8).
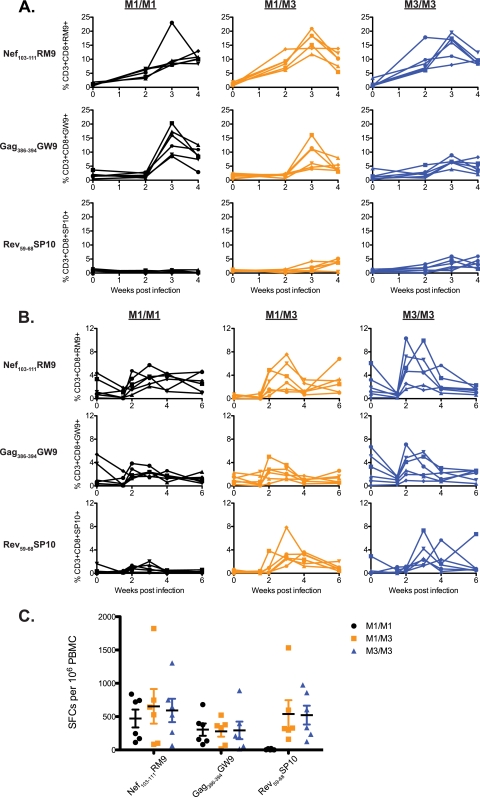
We measured CD8+ T cell responses against all three epitopes in cells from bronchoalveolar lavage (BAL) fluid and peripheral blood samples using MHC-peptide tetramers during acute infection. Strong responses against Nef103-111RM9 and Gag386-394GW9 were detected in BAL fluid samples of all 18 animals (Fig. 1A; see also supplemental Table 1 at go.wisc.edu/44305k/). Rev59-68SP10 responses were detected in M3+ animals, but their magnitude was more modest than that of the responses targeting the other two epitopes. Responses in BAL fluid samples were more robust than those in the blood samples (Fig. 1B and Table S1), consistent with earlier studies of SIV-specific CD8+ T cell responses at this mucosal site (17).
Fig 1.
Acute immunodominant CD8+ T cell responses in cells isolated from BAL fluid and peripheral blood samples. (A) MHC-peptide tetramers representing three SIVmac239-derived epitopes (Nef103-111RM9, Gag386-394GW9, and Rev59-68SP10) were used to quantify the percentages of antigen-specific CD3+ CD8+ T cells in BAL fluid samples at 0, 2, 3, and 4 weeks after SIVmac239 infection. (B) The same tetramers were used to quantify the percentages of antigen-specific CD3+ CD8+ T cells at 0, 1.5, 2, 3, 4, and 6 weeks after infection in peripheral blood samples. For panels A and B, six animals with each MHC genotype were followed: M1/M1 MCMs (black), M1/M3 MCMs (orange), and M3/M3 MCM (blue). Each shape represents a different animal. (C) T cell responses targeting the three SIV-derived peptide epitopes were measured by an IFN-γ ELISPOT assay 3 weeks after SIVmac239 infection. The average numbers of spot-forming colonies (SFCs) per 106 PBMC and the standard errors are shown. Six animals with each MHC genotype are shown as indicated in the legend.
At 3 weeks postinfection, we explored all three epitope-specific immune responses in the peripheral blood samples by a gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISPOT) assay. Although there are limitations to this assay, it can be used as a proxy for T cell functionality (25, 28). We found that the frequencies of IFN-γ-secreting T cells specific for Nef103-111RM9 and Gag386-394GW9 were similar among all three groups (Fig. 1C). Responses targeting Rev59-68SP10 were robust in M3+ MCM but absent in M1/M1 MCM. This assay suggests that the presence of peripheral IFN-γ-secreting T cells specific for Rev59-68SP10 did not curb the frequency of IFN-γ-secreting T cells specific for Gag386-394GW9 or Nef103-111RM9.
To examine the kinetics of CD8+ T cell selection, we isolated and pyrosequenced plasma virus from the SIVmac239 inoculum and all 18 animals at 4 weeks postinfection, using methods described previously (3). Briefly, four overlapping reverse transcription-PCR (RT-PCR) amplicons spanning all SIV genes were generated and purified. Amplicons were fragmented with a Nextera DNA sample prep kit (Epicentre, Madison, WI), and then multiplex identifier (MID) tags and adapter sequences for Roche/454 pyrosequencing were added. Purified fragments were pyrosequenced using a Roche/454 GS Junior. We obtained an average of 46,889 reads per genome, with an average coverage of 1,126 reads at each position (see supplemental Table 2 at go.wisc.edu/4430sk/). In viral populations from all animals except CY0330, the theoretical number of templates was greater than the average coverage, thus alleviating concerns about resampling.
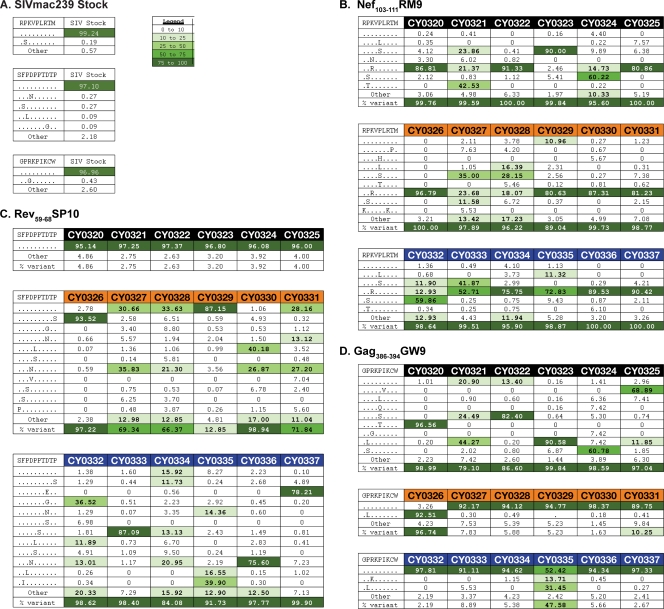
Amino acid variation within the three CD8+ T cell epitopes was examined using a custom data analysis pipeline (see supplemental Fig. 1 at go.wisc.edu/4430sk/) in Galaxy, an open-source system for processing next-generation sequence data (4, 5, 14). Sequence reads were translated in all six reading frames and aligned against SIVmac239 using BLAT (BLAST-Like Alignment Tool) (18). Reads containing low-quality bases within epitopes were discarded. The inoculum showed no substantial variation at any of the three epitopes (Fig. 2A). As expected, Nef103-111RM9 rapidly accumulated sequence variation in viruses from all 18 animals (Fig. 2B) (2). The wild-type sequence comprised a mean of 1.7% in all animals, with only one animal having more than 10% wild-type virus (CY0329). The majority of epitope sequences in Rev59-68SP10 were variantin 11 of 12 M3+ MCM (Fig. 2C), but negligible variation was observed in M1/M1 MCM.
Fig 2.
Amino acid variation within three epitope sequences at 4 weeks after SIVmac239 infection. (A) The SIVmac239 inoculum was subjected to full-genome Roche/454 pyrosequencing. No significant variation was detected in the three epitope sequences Nef103-111RM9, Rev59-68SP10, and Gag386-394GW9 in the SIVmac239 inoculum. Variants were included in the table if they were also present in panel B, C, or D. (B, C, and D) Viruses replicating in the plasma samples of all 18 animals 4 weeks after SIVmac239 infection were subjected to full-genome Roche/454 pyrosequencing. Amino acid variants in the three epitopes Nef103-111RM9, Rev59-68SP10, and Gag386-394GW9 in M1/M1 animals (black), M1/M3 animals (orange), and M3/M3 animals (blue) were interrogated. Sequence variation is shown for Nef103-111RM9 (B), Rev59-68SP10 (C), and Gag386-394GW9 (D). The wild-type sequence is shown at the top of each table. Masked identical bases are shown with a dot, and amino acid replacements are shown with a capital letter. The frequency of a given epitope variant sequence is shown. A variant sequence had to be present in at least one animal at a frequency of 5% or greater to be included in the table. The legend indicates the color associated with a given variant frequency. A “0” indicates that no reads were detected with that sequence.
In contrast to the widespread sequence variation detected in Nef103-111RM9 and Rev59-68SP10, sequence variation within Gag386-394GW9 was largely restricted to M1/M1 MCM (10) (Fig. 2D). All 18 animals possessed two copies of the restricting Mafa-A1*063 allele, which is carried on both the M1 and the M3 haplotypes. Extensive variation within Gag386-394GW9 was observed in all 6 M1/M1 MCM but only 2 of the 12 M3+ animals. On average, 83.34% of reads were wild type in M3+ MCM, while only 6.64% of reads were wild type in M1/M1 MCM. Virus isolated from an M1/M3 (CY0328) animal and an M3/M3 (CY0336) animal had high frequencies of nonsynonymous substitutions (23.1% and 82.5%, respectively) at the arginine codon immediately upstream of the epitope, but it is unclear whether this mutation affects epitope presentation. Different variants predominated in each M1/M1 animal, suggesting that multiple distinct variants can be tolerated within this epitope.
We sequenced Gag386-394GW9 from provirus in genomic DNA of lymph node cells isolated at 3 weeks postinfection, as a recent report described that the earliest immune escape variants are present at this site (26). Using amplicon-based pyrosequencing (2), we found that variants in Gag386-394GW9 were detectable in M1/M1 MCM but absent in M3+ MCM (see supplemental Fig. 2 at go.wisc.edu/4430sk/), a pattern consistent with our observations in the plasma virus at 4 weeks (Fig. 2D).
To determine whether variants eventually accumulate in the Gag386-394GW9 epitope in M3+ MCM, we sequenced the plasma virus in 15 of the animals at 12 weeks postinfection, including 11 M3+ MCM. Although viral loads were markedly lower, Gag386-394GW9 sequences were nearly completely variant (range, 96 to 100%) in the M3+ animals (see supplemental Fig. 3 at go.wisc.edu/4430sk/). These results demonstrate that Gag386-394GW9-specific CD8+ T cell responses do eventually select for sequence variants in M3+ animals.
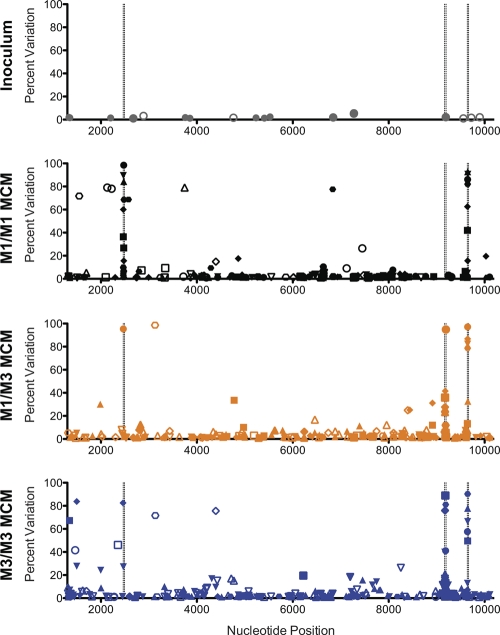
Given the lack of nonsynonymous substitutions within Gag386-394GW9 in the majority of M3+ animals at 4 weeks postinfection, we asked whether other sites in the viral genome were accumulating variants. We performed nucleotide alignments using the LASTZ short read mapper as implemented in Galaxy (see supplemental Fig. 4 at go.wisc.edu/4430sk/). Our qualifications for considering a variant authentic were quite conservative relative to those in other studies (11, 26); we required that the percentage of variants was ≥1% and greater than three times the percentage of N and indel sequences. We applied this strategy to the SIVmac239 inoculum and the viral populations replicating in all 18 animals at 4 weeks postinfection (Fig. 3). We detected low-frequency variants in the inoculum (see supplemental Table 3 at go.wisc.edu/4430sk/), but this was not entirely unexpected, as it was passaged for 5 days on rhesus macaque peripheral blood mononuclear cells (PBMC). Variants at the same positions in the sequences from the 18 animals were relatively rare (Table S4). Across the 18 viral genomes, we detected 659 variants, including 82 nonsynonymous mutations at a frequency of 10% or greater (Table S5). Of these high-frequency variants, 66% were located within the three immunodominant CD8+ T cell epitopes described above.
Fig 3.
Limited genome-wide nucleotide variation at 4 weeks after SIVmac239 infection. The percent variation at each nucleotide position (from site 1300 to site 10200) is shown for the inoculum, six M1/M1 MCM (CY0320 to CY0325; black), six M1/M3 MCM (CY0326 to CY0331; orange), and six M3/M3 MCM (CY0332 to CY0337; blue). Each symbol shape represents a different animal. Nonsynonymous mutations are shown as filled characters, and synonymous mutations are shown as open characters. The nucleotide positions corresponding to the N and C termini of Gag386-394GW9, Rev59-68SP10, and Nef103-111RM9 are shown as dashed vertical lines. Only variants present at a frequency of 1% or greater are shown. For an alternate visualization of these data with the Layercake visualization tool (12), please go to http://goo.gl/6uaDY.
In this study, we show that accumulation of sequence variants in CD8+ T cell epitopes during the acute phase is predictable and reproducible when both viral and host genetics are controlled, but this is not the case among animals sharing only a single MHC class I allele. We provide evidence that acquisition of sequence variants within one epitope targeted by a shared CD8+ T cell response is delayed when an additional discrete CD8+ T cell response is present. We found that the accumulation of variants in Rev59-68SP10 preceded the development of variation in Gag386-394GW9 when CD8+ T cell responses targeting both epitopes were present. One explanation for this “conditional” CD8+ T cell escape during acute SIV infection is that there is a limited amount of simultaneous variation tolerated by the HIV/SIV genome. Alternately, it is possible that variants in Rev59-68SP10 are simply tolerated better than those in Gag386-394GW9, and therefore variants in Rev59-68SP10 accumulate more rapidly when CD8+ T cell responses targeting both epitopes are present. Nonetheless, these data suggest that variants generally accumulate in a finite number of epitopes immediately after CD8+ T cell responses emerge, even when there are additional CD8+ T cell responses present that also have the potential to rapidly select escape variants.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01 AI084787, R01 AI077376-04A1, and R21 AI082880. S.L.O. is a CHAVI/HVTN Early Stage Investigator. D.A.P. is a Medical Research Council (United Kingdom) Senior Clinical Fellow. M.C. and M.G. were supported by NSF awards IIS-0946598 and CMMI-0941013. The research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grants RR15459-01 and RR020141-01.
We also thank members of the Wisconsin National Primate Research Center, a facility supported by grant P51 RR000167, and members of the O'Connor and Friedrich laboratories for their review of the manuscript.
Footnotes
Published ahead of print 19 October 2011
REFERENCES
- 1. Betts MR, et al. 2000. Putative immunodominant human immunodeficiency virus-specific CD8(+) T-cell responses cannot be predicted by major histocompatibility complex class I haplotype. J. Virol. 74:9144–9151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bimber BN, et al. 2009. Ultradeep pyrosequencing detects complex patterns of CD8+ T-lymphocyte escape in simian immunodeficiency virus-infected macaques. J. Virol. 83:8247–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bimber BN, et al. 2010. Whole-genome characterization of human and simian immunodeficiency virus intrahost diversity by ultradeep pyrosequencing. J. Virol. 84:12087–12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blankenberg D, et al. 2010. Manipulation of FASTQ data with Galaxy. Bioinformatics 26:1783–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blankenberg D, et al. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19:Unit 19.10.1-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boutwell CL, Rolland MM, Herbeck JT, Mullins JI, Allen TM. 2010. Viral evolution and escape during acute HIV-1 infection. J. Infect. Dis. 202(Suppl. 2):S309–S314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brumme ZL, et al. 2008. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J. Virol. 82:9216–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Budde ML, et al. 2011. Transcriptionally abundant major histocompatibility complex class I alleles are fundamental to nonhuman primate simian immunodeficiency virus-specific CD8+ T cell responses. J. Virol. 85:3250–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Budde ML, et al. 2010. Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics 62:773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burwitz BJ, et al. 2009. Mauritian cynomolgus macaques share two exceptionally common major histocompatibility complex class I alleles that restrict simian immunodeficiency virus-specific CD8+ T cells. J. Virol. 83:6011–6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cale EM, et al. 2011. Epitope-specific CD8+ T lymphocytes cross-recognize mutant simian immunodeficiency virus (SIV) sequences but fail to contain very early evolution and eventual fixation of epitope escape mutations during SIV infection. J. Virol. 85:3746–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Correll M, Ghosh S, O'Connor D, Gleicher M. 2011. Visualizing virus population variability from next generation sequencing data. In Proceedings of BioVis 2011 IEEE, New York, NY [Google Scholar]
- 13. Day CL, et al. 2001. Relative dominance of epitope-specific cytotoxic T-lymphocyte responses in human immunodeficiency virus type 1-infected persons with shared HLA alleles. J. Virol. 75:6279–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goecks J, Nekrutenko A, Taylor J. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goulder PJ, Watkins DI. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4:630–640 [DOI] [PubMed] [Google Scholar]
- 16. Goulder PJ, Watkins DI. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8:619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greene JM, et al. 2010. Extralymphoid CD8+ T cells resident in tissue from simian immunodeficiency virus SIVmac239{Delta}nef-vaccinated macaques suppress SIVmac239 replication ex vivo. J. Virol. 84:3362–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lichterfeld M, Yu XG, Le Gall S, Altfeld M. 2005. Immunodominance of HIV-1-specific CD8(+) T-cell responses in acute HIV-1 infection: at the crossroads of viral and host genetics. Trends Immunol. 26:166–171 [DOI] [PubMed] [Google Scholar]
- 20. Loffredo JT, et al. 2008. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J. Virol. 82:1723–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Newberg MH, et al. 2006. Immunodomination in the evolution of dominant epitope-specific CD8+ T lymphocyte responses in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 176:319–328 [DOI] [PubMed] [Google Scholar]
- 22. O'Connor DH, et al. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493–499 [DOI] [PubMed] [Google Scholar]
- 23. O'Connor SL, et al. 2007. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics 59:449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Streeck H, Nixon DF. 2010. T cell immunity in acute HIV-1 infection. J. Infect. Dis. 202(Suppl. 2):S302–S308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valentine LE, et al. 2008. Recognition of escape variants in ELISPOT does not always predict CD8+ T-cell recognition of simian immunodeficiency virus-infected cells expressing the same variant sequences. J. Virol. 82:575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vanderford TH, et al. 2011. Viral CTL escape mutants are generated in lymph nodes and subsequently become fixed in plasma and rectal mucosa during acute SIV infection of macaques. PLoS Pathog. 7:e1002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weidt G, Utermohlen O, Heukeshoven J, Lehmann-Grube F, Deppert W. 1998. Relationship among immunodominance of single CD8+ T cell epitopes, virus load, and kinetics of primary antiviral CTL response. J. Immunol. 160:2923–2931 [PubMed] [Google Scholar]
- 28. Yang OO. 2003. Will we be able to ‘spot’ an effective HIV-1 vaccine? Trends Immunol. 24:67–72 [DOI] [PubMed] [Google Scholar]
- 29. Yang OO. 2004. CTL ontogeny and viral escape: implications for HIV-1 vaccine design. Trends Immunol. 25:138–142 [DOI] [PubMed] [Google Scholar]