Abstract
The pentatricopeptide repeat (PPR) protein family consists of organellar proteins predicted to bind to specific RNA sequences. Plants have hundreds of distinct PPR proteins, whereas other eukaryotes generally have many fewer. The genome of the parasitic protozoon Trypanosoma brucei is predicted to encode more than 30 different PPR proteins, which is an extraordinarily high number for a nonplant organism. Here we report the characterization T. brucei PPR9 (TbPPR9). Epitope tagging shows that the protein is exclusively mitochondrially localized. Interestingly, while in induced RNA interference cell lines TbPPR9 is efficiently downregulated, the level of its mRNA is not affected. Ablation of TbPPR9 selectively abolishes oxidative but not mitochondrial substrate-level phosphorylation. The molecular basis of this phenotype is the fact that TbPPR9 is required for the stability of the cytochrome oxidase subunit 1 (COX1) and COX2 mRNAs. This is supported by the observation that ablation of TbPPR9 destabilizes the COX complex but not the cytochrome bc1 or the ATP synthase complex. Moreover, it was shown by blue native gel electrophoresis that TbPPR9 is present in a large complex of unknown composition.
INTRODUCTION
Sequencing of the Arabidopsis thaliana genome led to the discovery of a very large protein family that is defined by a degenerate 35-amino-acid pentatricopeptide repeat (PPR) motif (38). PPR proteins contain 2 to 30 PPR motifs that are usually arranged in tandem. Essentially all PPR proteins are organellar proteins. In plants, approximately 75% are localized to mitochondria and 25% to plastids. PPR proteins function at all levels of organellar gene expression, from transcription to translation. To explain these diverse functions, it has been proposed that they are sequence-specific RNA-binding proteins that recruit effector enzymes to defined sites on organellar RNAs (32). An alternative model suggests that the function of PPR proteins can be explained as a passive consequence of the ability of the proteins to bind to single-stranded RNA in a sequence-specific way. This binding could mask interaction with other proteins and/or remodel the local RNA structure (29).
One of the most intriguing aspects of PPR proteins is their phylogenetic distribution. They are found in all eukaryotes analyzed to date. However, there is an extraordinary discrepancy in their numbers between plants, which have 450 to 600 PPR proteins, and nonplant eukaryotes, including Saccharomyces cerevisiae yeast and humans, which have a much smaller number (38).
One of the first PPR proteins that has been experimentally analyzed, before the PPR was recognized as a protein family-defining domain, was Pet309 of Saccharomyces cerevisiae. Pet309 activates the translation of cytochrome oxidase subunit 1 (COX1) encoded in the mitochondrion by binding to the 5′ untranslated region of its mRNA (7, 20). Moreover, it also stabilizes the COX1 mRNA. Interestingly, only the translation activation and not the mRNA stabilization function of Pet309 appears to require PPRs (41). The human orthologue of Pet309 is the RNA-binding protein Lrp130. It has a similar function as Pet309 and if mutated causes one type of the Leigh syndrome, a disease in which COX activity is greatly reduced (26, 45).
The parasitic protozoon Trypanosoma brucei and related trypanosomatids are exceptional since their genomes encode many more PPR proteins than essentially any other eukaryote outside the plant lineage. Trypanosomal PPR proteins were first identified in two studies by bioinformatic screening of the predicted T. brucei proteome. Interestingly, one study identified 23 (25) and the other one 28 (30) PPR protein candidates. There was a large but incomplete overlap between the two sets of proteins identified in the two studies. Moreover, a more recent study even identified 36 putative PPR proteins (2). The differences between these studies can be explained by the fact that PPR domains are not always easy to recognize and are sometimes difficult to distinguish from the related tetratricopeptide repeat (TPR) motif. The large majority of putative trypanosomal PPR proteins are predicted to be mitochondrially localized and have orthologues within but not outside the trypanosomatid lineage (30). The number of identified PPR motifs in trypanosomal proteins ranges from 1 to 15, but in virtually all cases, the sequence alignments and secondary structure predictions suggest that this is an underestimation.
Recently, 8 T. brucei PPR proteins (TbPPR1 to TbPPR8) were subjected to a comparative experimental analysis (30). Seven of them (TbPPR1 to TbPPR7) were mitochondrially localized and essential for oxidative phosphorylation (OXPHOS). Ablation of six out of these seven proteins (TbPPR2 to TbPPR7) led to degradation of the mitochondrial rRNAs but did not affect the levels of other mitochondrial RNAs (30). Later studies confirmed that three of the rRNA-stabilizing PPR proteins (TbPPR2, -3, and -5) could be copurified with the large or the small subunit of mitochondrial ribosomes (46). Four more PPR proteins that were not included in the comparative experimental analysis were also recovered in the ribosomal fraction of T. brucei (46), as well as that of Leishmania tarentolae (21, 22). However, stabilization of mitochondrial rRNAs is not the only function of trypanosomal PPR proteins. Ablation of one of the PPR proteins included in the experimental analyses discussed above, TbPPR1, caused a specific but moderate reduction of the level of COX1 mRNA (25, 30). Moreover, a more recent study showed that a heterodimer of two trypanosomal PPR proteins, one of which is identical to the TbPPR1 mentioned above, stimulates mRNA polyadenylation and uridylation catalyzed by poly(A) polymerase and the terminal uridyltransferase (2). This indicates that some PPR proteins might have a more general function that is not transcript specific.
RNA interference (RNAi)-mediated ablation of most trypanosomal PPR proteins causes degradation of few specific mitochondrial RNA species (25, 30). This is in line with the postulated role of PPR proteins as sequence-specific RNA-binding proteins and suggests that the global function of the PPR protein family—mediating organellar gene expression—is conserved in trypanosomes.
Here we report the experimental characterization of trypanosomal PPR protein, which is not associated with the mitochondrial ribosome and does not appear to be a component of the mitochondrial polyadenylation or genomic RNA (gRNA)-binding complex. The protein selectively stabilizes COX1 and COX2 mRNAs and therefore is essential for oxidative phosphorylation.
MATERIALS AND METHODS
Epitope tagging.
To localize TbPPR9, we produced two transgenic cell lines expressing a protein variant containing either the carboxy-terminal Ty1 tag or the hemagglutinin (HA) tag. Both tags are routinely used in T. brucei. The Ty1 tag is detected by the monoclonal BB2 antibody, and the HA tag is visualized by the monoclonal antibody HA11. The Ty1-tagged variant of the TbPPR9 gene was cloned into a derivative of plasmid pLew-100 that allows tetracycline-inducible overexpression of the tagged protein (44), whereas the variant containing the C-terminal HA epitope was produced by in situ tagging (36).
Production and analysis of TbPPR9 RNAi cell line.
RNAi was performed using stem-loop constructs containing the puromycin resistance gene as described previously (4). As an insert, we used a 505-bp fragment (nucleotides 273 to 777) of the open reading frame (ORF) Tb11.01.7930. Transfection of T. brucei, selection with antibiotics, cloning, and induction with tetracycline were done as described previously (24). All transgenic cell lines used in this study are based on T. brucei strain 29-13, which was grown in SDM-79 standard medium (5) supplemented with 15% fetal calf serum, 50 μg/ml hygromycin, and 15 μg/ml G-418. Growth of RNAi cells was also analyzed in SDM-80 medium, a modified SDM-79 medium that lacks glucose and which is supplemented with 10% dialyzed fetal calf serum as described previously (18) both in the presence and in the absence of 1 μg/ml of tetracycline.
Northern blots of mitochondrial mRNAs.
Total RNA (5 μg each) was separated on 1% (wt/vol) agarose gels containing 0.2 M formaldehyde and electrophoresed in 20 mM morpholinepropanesulfonic acid, pH 7.0, and 0.2 M formaldehyde. After transfer, GeneScreen Plus membranes were UV cross-linked. As probes, we used random hexamer [α-32P]dCTP-labeled double-stranded DNA fragments. Two DNA fragments, corresponding to nucleotides 273 to 777 (corresponding to the region targeted by RNAi) and 1038 to 1437 (corresponding to a region that was not targeted by RNAi) of the ORF Tb11.01.7930, were used detect the mRNA of TbPPR9.
The fragments used to detect mitochondrial mRNAs have been described before (30). The 9S and 12S rRNAs were detected by specific oligonucleotide hybridization (30).
Reverse transcriptase PCR (RT-PCR) was done using the ImProm-II reverse transcription system (Promega). The following primers were used to amplify the cDNA corresponding to the fully edited COX3 mRNA: 5′-GATTTGTTTTGTGATTTTTTACG-3′ (forward) and 5′-CATAACACATAAAACATCAAAATAAAC-3′ (reverse).
Blue native gel electrophoresis.
Mitochondria were isolated under hypotonic conditions and resuspended in 20 mM Tris-HCl, pH 8.0, 0.25 M sucrose, 2 mM EDTA (33). Fifty micrograms of mitochondria was solubilized with 1% digitonin in lysis buffer (20 mM Tris-HCl [pH 7.4], 50 mM NaCl, 0.1 mM EDTA, 10% [vol/vol] glycerol), loaded on a 4 to 13% native polyacrylamide gel, and subjected to blue native gel electrophoresis (39). HA-tagged PPR proteins were detected by immunoblotting. T. brucei MRPL21 (TbMRPL21; Tb927.7.4140), a mitochondrial ribosomal protein of the large subunit containing three copies of the c-myc tag at its C terminus, was used as a marker for the ribosome.
Miscellaneous.
Polyclonal rabbit antisera of trypanosomal voltage-dependent anion channel (VDAC), COX4, and cytochrome c1 (CYTC1) were prepared using the carboxy-terminal peptides of the proteins as antigens. The trypanosomal F1β subunit of the ATP synthase complex was detected using a polyclonal antiserum directed against the ATP synthase of Crithidia fasciculata (generous gift of D. Speijer, AMC, Amsterdam, Netherlands). α-Ketoglutarate dehydrogenase (KDH) of trypanosomes was detected using a polyclonal antiserum raised against yeast KDH. Monoclonal antibodies against elongation factor 1a (eEF-1a) were from Santa Cruz Biotechnology. The monoclonal BB2 antibody recognizing the Ty1 epitope was from Diagenode, Belgium. All antibodies were used at 1:1,000 dilutions in immunoblots. Immunofluorescence was done as described previously (34).
All RNA isolations were done by the acid guanidinium method (6). ATP production assays using digitonin-purified mitochondria from RNAi cell lines were done as described previously (4). Subfractionation of mitochondria and carbonate extraction of mitochondrial membranes were done as described previously (30).
RESULTS
A novel trypanosomal PPR protein.
In a previous study, we presented a list of 28 predicted trypanosomal PPR proteins (30). This list was obtained by screening the T. brucei proteome for PPR motifs using TPRpred, an algorithm that detects both TPR and PPR motifs (15). In this early study, a TPRpred probability of less than 25% was considered not significant and proteins below that threshold were removed from the list of putative PPR proteins. However, it cannot be excluded that some of these proteins might nevertheless be bona fide PPR proteins. The present study focuses on one such example, the protein encoded by ORF Tb11.01.7930, which has a TPRpred probability of 2.3% only. Interestingly, the same protein had been identified before in the list of trypanosomal PPR proteins provided by Mingler et al. (25) that is based on BLAST searches using HMMER package-derived consensus sequences. However, in agreement with the low TPR probability found in our study, the BLAST score for the protein was low and the E value was high. In the present study, we show that the protein encoded by Tb11.01.7930 has all the features expected for a PPR protein, which is why we termed it TbPPR9.
TbPPR9 is conserved within trypanosomatids, and a global transcriptome analysis suggests that in T. brucei mainly a gene-internal ATG (position 124 in the nucleotide sequence) is used as a start codon, in which case the protein would have a molecular mass of 63.5 kDa (27, 37). Outside the putative PPR repeats, the protein does not show any homology to any proteins outside the trypanosomatid lineage.
TbPPR9 is mitochondrially localized.
Bioinformatic analysis predicts that TbPPR9 has no mitochondrial targeting signal. However, in T. brucei such predictions are difficult since mitochondrial presequences are often very short (11). Thus, in order to determine the localization of the protein, we prepared transgenic cell lines allowing expression of variants of TbPPR9 that carry the Ty1 peptide or the hemagglutinin (HA) tag at their carboxy termini (3). The transgenic cell line expressing the Ty1-tagged TbPPR9 was then subjected to a biochemical fractionation. Digitonin extractions (40) showed that essentially all of the tagged TbPPR9 is recovered in the pellet together with the mitochondrial marker (Fig. 1A). This was confirmed by immunofluorescence analysis using the cell line expressing the HA-tagged TbPPR9. The results in Fig. 1B also show that in this experiment the HA-tagged TbPPR9 colocalizes with the mitochondrial marker. In summary, these results show that TbPPR9 can be detected only in the mitochondrion.
Fig 1.
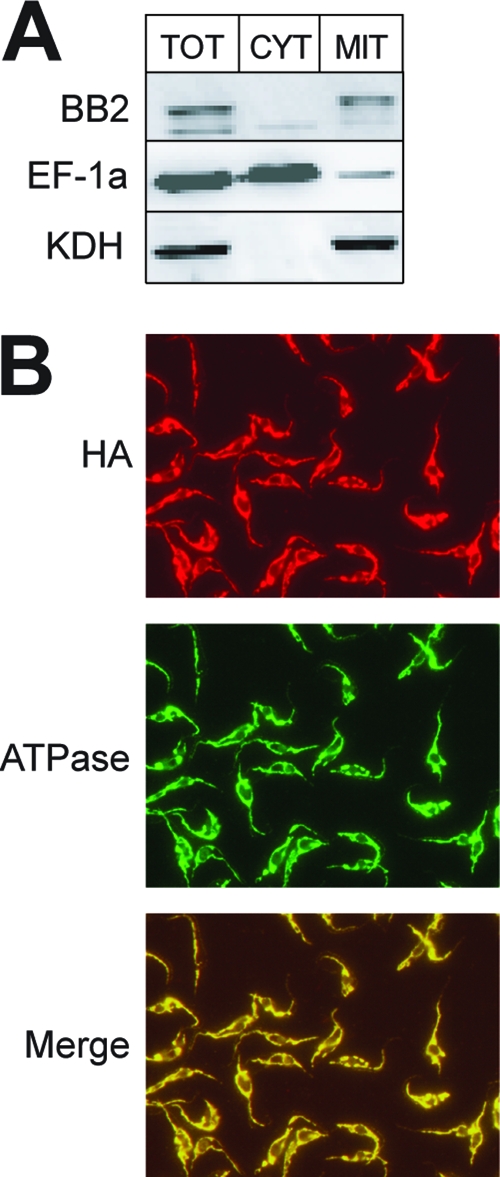
Localization of epitope-tagged TbPPR9. (A) Immunoblot analysis of 0.3 × 107 cell equivalents each of total cellular (TOT), crude cytosolic (CYT), and crude mitochondrial extracts (MIT) for the presence of the Ty1-tagged TbPPR9 protein (BB2). Only the relevant regions of the blots are shown. Comparison with molecular mass markers showed that the sizes of the tagged proteins were consistent with the prediction. eEF-1a served as a cytosolic marker (middle), and KDH was used as a mitochondrial marker (bottom). (B) Double immunofluorescence analysis of a T. brucei cell line expressing TbPPR9 carrying an HA tag. The cells were double stained with monoclonal anti-tag antibodies (HA) and a polyclonal antiserum directed against a subunit of the mitochondrial ATPase. A merged picture of the anti-tag antibody and the ATPase staining is also shown.
RNAi ablates tagged TbPPR9 but not the corresponding mRNA.
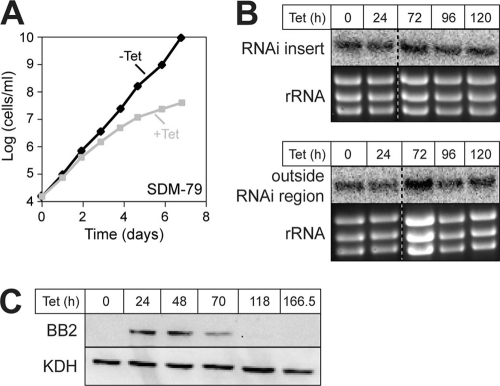
To study the function of TbPPR9, we established a stable transgenic cell line that allows inducible RNAi-mediated ablation of the protein. Figure 2A shows that ablation of TbPPR9 caused a slow-growth phenotype in SDM-79 standard medium (5) starting 3 to 4 days after induction. However, even after longer induction times, the cells kept growing, albeit at a much reduced rate. To assess the efficiency of RNAi, we determined the level of the TbPPR9 mRNA in both uninduced and induced cells. However, the Northern blots in Fig. 2B show that the level of the TbPPR9 mRNA is not affected even after prolonged induction times, a result unlike the one expected. The probe used in this analysis corresponded to the sequence that was targeted by the RNAi construct. In order to verify this unexpected result, we repeated the Northern analysis using a second probe that does not overlap with the first one and whose sequence is not targeted by the RNAi, and we obtained the same result. However, the fact that we reproducibly observe a slow-growth phenotype after addition of tetracycline suggests that TbPPR9 becomes downregulated in the RNAi cells, even though the levels of the TbPPR9 mRNA remain unchanged.
Fig 2.
Inducible RNAi-mediated ablation of TbPPR9. (A) Representative growth curve of tetracycline (Tet)-uninduced (−Tet) and -induced (+Tet) representative clonal T. brucei TbPPR9 RNAi cell line in standard culture medium SDM-79. (B) Northern analysis of total RNA isolated from the TbPPR9 RNAi cell line induced for the indicated times. The Northern blot on the top was probed with a labeled DNA fragment covering the region of the mRNA that is targeted by the RNAi (RNAi insert), whereas the blot on the bottom was probed with a labeled DNA fragment that recognizes a region in the TbPPR9 mRNA outside the region that is targeted by the RNAi (outside RNAi region). In both cases, ethidium bromide stains of the rRNA region are shown as loading controls. (C) Combination of tetracycline-inducible expression of the Ty1 tagged TbPPR9 and the tetracycline-inducible TbPPR9 RNAi in the same cell line. Total proteins were analyzed by immunoblotting at the indicated time points after tetracycline addition using the BB2 antiserum or KDH antiserum (KDH serves as a loading control).
To test this directly, we performed the RNAi in cells expressing the tetracycline-inducible tagged version of TbPPR9 that was used for the localization studies. In these cells, addition of tetracycline induces not only expression of the RNAi-mediating stem-loop RNA but also expression of the Ty1-tagged TbPPR9. Immunoblot analysis of protein extracts from these cells shows that the level of the tagged TbPPR9 declines after 3 days of induction and that after 5 days it is hardly detectable anymore (Fig. 2C). However, the mitochondrial marker α-ketoglutarate dehydrogenase, which serves as a loading control, is not affected. Northern blots confirmed that in this cell line, as in the original RNAi cell line, the levels of the endogenously expressed TbPPR9 mRNA do not decrease upon induction of RNAi. Interestingly, however, in the case of the ectopically expressed TbPPR9 mRNA, a slight reduction of 25% is observed (see Fig. S1 in the supplemental material).
These results show that while the levels of the TbPPR9 mRNA are not affected in induced TbPPR9 RNAi cell lines, the protein nevertheless gets downregulated, suggesting that expression of the stem-loop RNA somehow interferes with the translation of the TbPPR9 mRNA. A similar case has previously been described for an RNAi cell line directed against T. brucei CPSF30 (TbCPSF30), a trypanosomal zinc finger protein involved in nuclear RNA processing (13). However, in contrast to TbPPR9, where the same levels of mRNAs were found in uninduced and induced cells, induction of RNAi for TbCPSF30 resulted in an accumulation of a large amount of an RNA variant that was slightly longer than TbCPSF30 mRNA.
In summary, these results show that RNAi is a suitable method to study the function of TbPPR9. The mechanism that is responsible for the selective protein but not mRNA knockdown of TbPPR9 is of high interest but beyond the scope of the present study.
TbPPR9 is required for efficient oxidative phosphorylation.
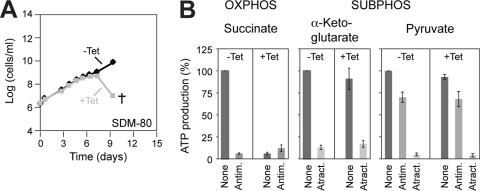
Ablation of TbPPR9 causes a slow-growth phenotype in SDM-79 medium, which contains proline and glucose as the major energy sources. When tested in SDM-80, a modified version of SDM-79 that lacks glucose (18), we obtained a growth phenotype stronger than the slow-growth phenotype observed in SDM-79, and TbPPR9-ablated cells abruptly stopped growing after 7 days and died thereafter (Fig. 3A). Thus, TbPPR9 is essential for survival in SDM-80. The fact that the onset of the growth phenotype occurs later in the glucose-free SDM-80 than in SDM-79 can be explained by the much longer generation time that is observed in the former. In procyclic T. brucei, glucose—after conversion into pyruvate—is used for mitochondrial substrate-level phosphorylation (SUBPHOS) in the trypanosome-specific acetate:succinate coenzyme A (CoA) transferase/succinyl-CoA synthetase (ASCTS) cycle (31). When grown in SDM-79, where glucose is available, the energy needs of T. brucei can largely be met by substrate-level phosphorylation alone (4, 18). In the glucose-free SDM-80, however, the sole energy source is proline, which can be utilized only by oxidative phosphorylation (OXPHOS) (18). Growth in SDM-80 therefore selects for cells capable of performing efficient OXPHOS. All mitochondrial gene products of T. brucei either function in OXPHOS or are components of the mitochondrial translation machinery whose function is to produce components of the OXPHOS complexes. The lack of growth of TbPPR9-ablated cells in SDM-80 lacking glucose is therefore in line with the proposed function of PPR proteins in mediating mitochondrial gene expression.
Fig 3.
Ablation of TbPPR9 abolishes OXPHOS. (A) Representative growth curve of tetracycline-uninduced (−Tet) and -induced (+Tet) representative clonal T. brucei TbPPR9 RNAi cell line in glucose-free SDM-80 culture medium. The cross indicates that further incubation led to the death of the whole population. (B) In organello mitochondrial ATP production induced by succinate, α-ketoglutarate, and pyruvate of a tetracycline-uninduced (−Tet) and -induced (+Tet) TbPPR9 cell line was determined using luciferase. The substrates tested and the additions of antimycin (Antim.) and atractyloside (Atract.) are indicated at the top and the bottom, respectively. ATP production in mitochondria isolated from uninduced cells tested without antimycin or atractyloside is set to 100%. The bars represent means expressed as percentages. Standard errors of three independent biological replicates are indicated.
We have recently established an assay that allows the quantification of the different modes of ATP production in isolated mitochondria of T. brucei (4). Using this assay, it was possible to test whether the lack of growth of induced TbPPR9 RNAi cell lines on glucose-free SDM-80 is indeed due to deficient OXPHOS. To measure OXPHOS, mitochondria are incubated with ADP and the substrate succinate. To measure SUBPHOS in the citric acid cycle, α-ketoglutarate is used as a substrate, whereas measuring SUBPHOS in the ASCTS cycle requires the addition of pyruvate as well as the cosubstrate succinate (4). Atractyloside prevents mitochondrial import of the added ADP and thus inhibits all forms of mitochondrial ATP production. OXPHOS, in contrast to either form of SUBPHOS, is antimycin sensitive. Figure 3B shows that ablation of TbPPR9 selectively abolishes OXPHOS (induced by succinate) but does not interfere with either of the two forms of SUBPHOS (induced by α-ketoglutarate and pyruvate, respectively). These results are consistent with the strong growth phenotype that is observed in the induced RNAi cell line in the absence of glucose and indicate that TbPPR9 is ultimately required for OXPHOS.
Ablation of TbPPR9 causes a selective decline of COX1 and COX2 mRNAs.
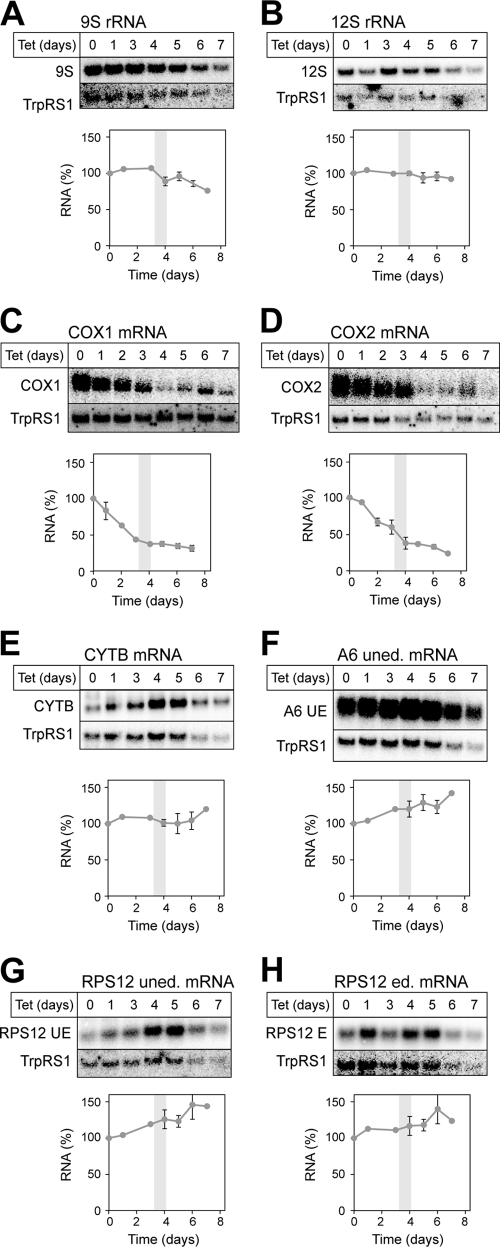
In order to investigate the molecular basis leading to the loss of OXPHOS in the absence of TbPPR9, we isolated total RNA from the uninduced and induced TbPPR9 RNAi cell line at various times after induction. Subsequently, Northern hybridization was used to determine the steady-state levels of RNAs encoded in the mitochondrion. Blots were probed for 9S and 12S rRNA and for the transcripts of the COX1, COX2, cytochrome b (CYTB), ribosomal protein 12 (RPS12), and ATPase subunit 6 (A6) genes. COX2 and CYTB transcripts are edited in a small domain only. The hybridization probes therefore detected both edited and unedited RNAs. RPS12 and A6 transcripts, on the other hand, are extensively edited. Thus, in the case of RPS12, two probes were used: one that detects unedited and minimally edited RNAs (RPS12 uned.) and another one that recognizes extensively and fully edited molecules (RPS12 ed.). For A6, only the unedited transcript (A6 uned.) was analyzed. The results are shown in Fig. 4. Only the relevant portions of the Northern blots are shown. No accumulation of precursor RNAs was seen for any of the tested RNA species, indicating that PPR9 is unlikely to be involved in RNA processing. However, ablation of TbPPR9 did selectively reduce the levels of COX1 and COX2 mRNAs. The levels of all other tested RNAs were either not affected (12S rRNA and CYTB), slightly decreased at late time points after induction (9S rRNA), or slightly increased (A6 uned., RPS12 ed., and RPS12 uned.). The decrease of COX1 and COX2 mRNAs is already evident 48 h after induction, well before the slow-growth phenotype of the cells becomes apparent, suggesting that it is a direct effect of the lack of TbPPR9.
Fig 4.
Lack of TbPPR9 selectively affects the level of the COX1 and COX2 mRNAs. Total RNA from the uninduced TbPPR9 cell line and total RNA from the TbPPR9 RNAi cell line induced for the indicated times were analyzed for the levels of 9S rRNA (A) and 12S rRNA (B), as well as for the levels of COX1 (C), COX2 (D), CYTB (E), preedited A6 (F), preedited RPS12 (PE) (G), and edited RPS12 (H) RNAs, using Northern hybridization (upper part of top panels). To normalize for loading differences, each filter was reprobed for the mRNA of the cytosolic tryptophanyl-tRNA synthetase (TrpRS1) (lower part of top panels). The graphs show a quantification of the signals shown on the corresponding Northern blots. The level of the RNAs in uninduced cells was set to 100%. All RNA levels were normalized by using cytosolic TrpRS. The experiments for COX1 and COX2 were done in triplicate, using data from three independently induced cell lines, and the standard errors are indicated. The same applies for the time points of 0, 4, 5, and 6 days for all the other RNAs. For each cell line, the growth phenotype was monitored in parallel. The gray bar indicates the time interval when the growth phenotype became apparent. E, edited; UE, unedited.
Many mitochondrial mRNAs exist in two populations having poly(A) tails of different lengths. Two PPR proteins are known to be directly involved in the polyadenylation of mitochondrial mRNAs (2). However, it has also been shown that ablation of many other trypanosomal PPR proteins caused a global effect on polyadenylation due to ATP depletion (see Fig. 5 in reference 30). The analysis of the TbPPR9 RNAi cell line shown in Fig. 4 does not have the resolution required to precisely monitor the fate of the short and the long poly(A) tails. Thus, while it appears that the decrease of the COX1 and COX2 mRNAs is preceded by a loss of the long poly(A) tail, it is much less clear whether the poly(A) tails of mRNAs whose levels remain stable in the induced TbPPR9 RNAi cell line are also affected.
Fig 5.
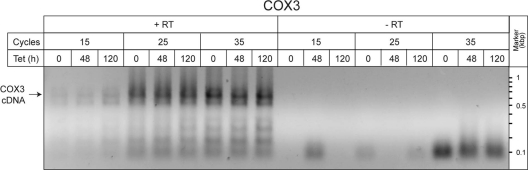
RT-PCR analysis of the edited COX3 mRNA. Primers designed to preferentially amplify the fully edited COX3 mRNA were used for RT-PCR of RNA samples that were isolated from uninduced cells and from TbPPR9 RNAi cells that were induced for 48 and 120 h. The Northern blots detecting the TrpRS1 mRNA in Fig. 4 were used to normalize the concentration of the RNA samples that were used for the RT-PCR analysis. After 15, 25, and 35 cycles, aliquots were analyzed on an agarose gel and stained with ethidium bromide. All reactions were performed in the presence (+RT) and absence (−RT) of reverse transcriptase. The expected length of the cDNA corresponding to the fully edited COX3 mRNA of 654 nucleotides is indicated by the arrow.
Besides COX1 and COX2, the mitochondrial DNA of T. brucei also encodes COX3. The final COX3 mRNA is extensively edited. In order to quantify the levels of the edited COX3 mRNAs, we therefore relied on RT-PCR using primers that are specific for the edited form. The results in Fig. 5 show that the same levels of COX3 cDNA are amplified in uninduced cell lines and in cell lines induced for 2 and 5 days, irrespective of whether the PCR was done for 15, 25, or 35 cycles.
In summary these results show that TbPPR9 is essential for the stabilization of COX1 and COX2 mRNAs, whereas the stability of all other tested RNAs, including the edited COX3 mRNA, does not depend on the presence of TbPPR9.
Ablation of TbPPR9 selectively destabilizes the COX complex.
Ablation of TbPPR9 selectively destabilizes the COX1 and COX2 mRNAs (Fig. 4). We would therefore expect that the observed loss of OXPHOS (Fig. 3) is due to a deficient COX complex. In order to investigate this question, we determined how the levels of marker proteins for the different respiratory complexes are affected during TbPPR9 RNAi. The immunoblot in Fig. 6 shows that the ablation TbPPR9 is paralleled by a reduction in the levels of COX4, a subunit of the COX complex encoded in the nucleus. These results are consistent with the idea that due to the decline of the COX1 and COX2 mRNAs and their corresponding gene products, the entire COX complex becomes destabilized. This may in turn prevent assembly of subunits such as COX4 encoded in the nucleus and may ultimately lead to their degradation. The levels of cytochrome c1, a component of the cytochrome bc1 complex, and the F1β subunit of the ATP synthase complex, in contrast, are not affected, suggesting that the other respiratory complexes remain intact in the absence of TbPPR9. In summary, these results support the proposed function of TbPPR9 in stabilizing COX1 and COX2 mRNAs.
Fig 6.
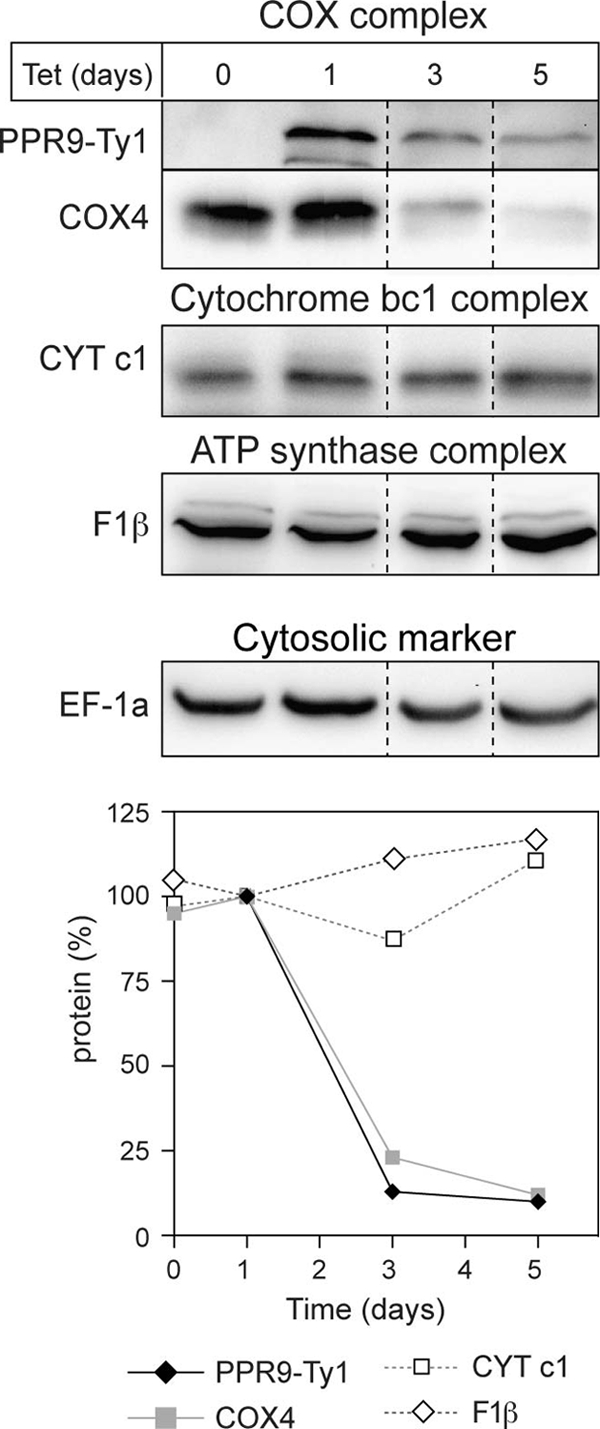
Lack of TbPPR9 affects the COX complex but not the cytochrome bc1 or the ATP synthase complex. Total protein from the uninduced and induced TbPPR9 RNAi cell line (days of induction are indicated at the top) was analyzed for the levels of Ty1-tagged TbPPR9 (PPR9-Ty1), COX4 of the COX complex, cytochrome c1 (CYT c1) of the cytochrome bc1 complex, and the F1β subunit of the ATP synthase complex (F1β) using immunoblots and specific antisera. Each signal was normalized to the levels of cytosolic translation elongation factor 1a (EF-1a) to correct for loading differences. The graph shows a quantification of the normalized signals. The level of the proteins in cells induced for 1 day was set to 100%.
TbPPR9 is part of a large complex.
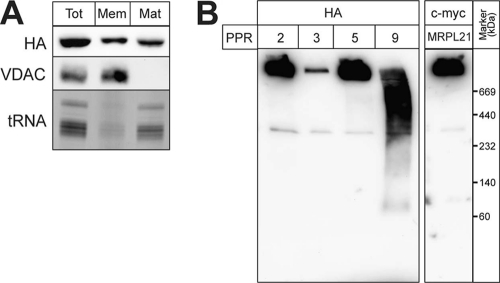
Essentially all trypanosomal PPR proteins are predicted to be soluble. However, in a previous analysis we showed that among 8 analyzed trypanosomal PPR proteins, only one behaved like a soluble matrix protein. All the other ones were peripherally associated with the mitochondrial inner membrane, probably via membrane-bound ribosomes (30). Freeze-thaw cycles and subsequent centrifugation allow subfractionation of mitochondria into a matrix and a membrane fraction. In such an analysis, TbPPR9 essentially behaves like a soluble protein. However, a small fraction of the protein appears to be membrane associated and thus might be associated with the mitochondrial ribosomes (Fig. 7A). In order to see whether it is associated with a protein complex, we subjected mitochondrial extracts containing tagged TbPPR9 to blue native gel electrophoresis. Figure 7B shows that the tagged TbPPR9 migrates as a smear indicative of a large complex with a molecular mass of between 440 and 800 kDa. The mitochondrial ribosomes can also be resolved by blue native gels, as evidenced by the high-molecular-mass band that contains a tagged large-subunit ribosomal protein. Moreover, as expected, the ribosomal complex also contains tagged TbPPR2, TbPPR3, and TbPPR5, whose ablation, as previously shown, destabilizes the mitochondrial rRNAs. Thus, TbPPR9 comigrates with a large protein complex that is distinct from the mitochondrial ribosomes.
Fig 7.
TbPPR9 is a component of a large soluble complex. (A) Mitochondria of a cell line expressing HA-tagged TbPPR9 were fractionated into membrane (Mem) and matrix (Mat) fractions. (Top) Immunoblot stained with the HA antiserum; (middle) immunoblot analysis for the outer membrane protein VDAC, which serves as a membrane marker; (bottom) an ethidium bromide-stained agarose gel containing RNA isolated from the three fractions; only the region corresponding to tRNAs that serve as a matrix marker is shown. (B) Mitochondrial digitonin extracts from cell lines expressing HA-tagged TbPPR2, -3, -5 (29), and -9 as well as from a cell line expressing c-myc-tagged mitochondrial large-subunit ribosomal protein 21 (MRPL21) (29) were resolved on the same blue native gel, blotted, and analyzed using anti-HA and anti-c-myc antisera, respectively.
Moreover, TbPPR9 was not found to be associated with the polyadenylation complex whose composition was recently determined (8) and which contains a number of other PPR proteins (2, 25, 30).
To test whether the COX1 and COX2 mRNAs cofractionate with the TbPPR9-containing complex, we performed immunoprecipitations of the tagged protein. Tagged TbPPR9 was efficiently recovered in the pellet fraction in such experiments, and Northern blots showed that the pellet also contained COX1 and COX2 mRNAs. However, a similar percentage of CYTB mRNA that is not affected by TbPPR9 RNAi was also recovered in the same fraction (data not shown). It is therefore presently not possible to decide whether the complex contains more mRNAs than the ones for COX1 and COX2 or whether the associated mRNAs simply reflect unspecific binding.
DISCUSSION
Here we present the characterization of TbPPR9, which was detected in two independent genomic screens for PPR proteins but obtained low scores in both analyses (25). We now show that the protein has an exclusive mitochondrial localization and that its ablation causes a decrease of COX1 and COX2 mRNAs encoded in the mitochondrion but not of any other tested RNA species. These results strongly suggest that TbPPR9 is a bona fide PPR protein. The fact that the protein alone could not be unambiguously identified by bioinformatic analysis illustrates the difficulty of recognition of members of the PPR protein family and indicates that the 28 predicted trypanosomal PPR proteins represent an underestimation (30). A recent characterization of PPR proteins required for polyadenylation reached the same conclusion (2). Moreover, using more sophisticated bioinformatic protocols, the set of known PPR proteins has also been expanded in other organisms, such as Saccharomyces cerevisiae (19) and Schizosaccharomyces pombe (17). Thus, it is likely that mammals also have a higher number of PPR proteins than is anticipated right now.
As in other organisms, the trypanosomal COX complex has a highly complicated structure. It is a mosaic of 3 subunits encoded in the mitochondrion (COX1 to COX3) and at least 10 subunits encoded in the nucleus (23). Moreover, in T. brucei the COX complex is developmentally regulated. It is active in the procyclic form of T. brucei, which produces much of its ATP by OXPHOS, but is absent from the bloodstream form, which produces ATP by glycolysis only (28, 42). Thus, T. brucei requires not only sophisticated pathways that coordinate the levels of the subunits produced in the nucleus and mitochondrion but also mechanisms that mediate the stage-specific regulation of the COX complex during the life cycle.
The TbPPR9 mRNA is present in comparable amounts in procyclic and bloodstream forms. However, consistent with its role in COX complex biogenesis, a transient approximately 2-fold increase of the TbPPR9 mRNA is observed during differentiation from the stumpy bloodstream form to the procyclic form (14). How much of the protein is present and whether it is essential in the bloodstream form are unknown.
In T. brucei, transcription of both nuclear and mitochondrial protein-coding genes is polycistronic. Thus, their expression is primarily controlled on a posttranscriptional level. The fact that ablation of TbPPR9 selectively reduces the level of COX1 and of both edited and unedited COX2 mRNAs indicates that it plays a role in the stabilization of these two mRNAs.
This is reminiscent of the yeast PPR protein Pet309, which also stabilizes COX mRNAs encoded in the mitochondrion. Pet309 in addition functions as a translational activator (7, 20). Whether this might also be the case for TbPPR9 is unknown at present, but the possibility cannot be excluded. However, despite these functional similarities, the two proteins are not orthologues and the sequence similarity between them is not higher than expected for two PPR proteins from different species.
PPR proteins are predicted to bind to specific RNA sequences. It is therefore likely that TbPPR9 exerts its function by direct binding to the corresponding RNAs. As it is the case for many PPR proteins, we were not able to obtain soluble recombinant TbPPR9, which prevented us from directly assessing its RNA-binding capability. However, the fact that the protein was recovered in a large complex that among other RNAs contained the COX1 and COX2 mRNAs is consistent with the idea that it binds to these two RNAs, although unspecific binding cannot be excluded. It has been shown in plants that a single PPR protein can bind to multiple targets by recognizing a degenerate consensus sequence defined by conserved pyrimidines and purines as well as a few invariant nucleotides (10). However, in the case of TbPPR9 the length of the putative target sequence is unknown, which makes the bioinformatic identification of putative TbPPR9 binding sites essentially impossible.
Other mitochondrial proteins of T. brucei whose ablation influences the levels of the COX mRNAs have been described. The best-studied ones are MRP1/2, which build a heterotetrameric complex that can promote annealing of complementary RNAs (1, 35), and RBP16, an RNA-binding protein of the Y-box family (12). Both proteins are essential in procyclic T. brucei. Ablation of either of the two proteins causes the same phenotype: it interferes with editing of the CYTB transcripts and causes the loss of the ND4 and COX1 mRNAs (9). Thus, MRP1/2 and RBP16 are ultimately required for the biogenesis of three respiratory complexes, whereas the function of TbPPR9, from all that we know, is limited to the stabilization of two of the three mRNAs that encode components of the COX complex.
To date, 10 different mitochondrially localized trypanosomal PPR proteins have been studied experimentally in some detail. The function of 6 of these 10 could be linked to the rRNAs.
Two, TbPPR1 (25, 30) and TbPPR9 (the focus of the present study), are required for the stabilization of transcripts encoding subunits of the COX complex. Two trypanosomal PPR proteins (one of which is identical to TbPPR1) were found in the mitochondrial polyadenylation complex (8) and shown to be required for the regulation of polyadenylation.
TbPPR9 RNAi cell lines show pronounced and very early reduction of COX1 and COX2 mRNAs, indicating that the protein is directly involved in the stabilization of the two mRNAs. The COX3 mRNA, however, was not affected. Its level must therefore be regulated by other factors. The COX3 transcript needs to be extensively edited. It is therefore possible that the abundance of its mRNA is regulated on the level of RNA editing.
In summary, it appears that one of the main functions of the trypanosomal PPR proteins that have been analyzed experimentally is the stabilization of small sets of functionally related RNAs. However, the function of more than two-thirds of all identified trypanosomal PPR proteins has not been elucidated yet. Interestingly, some of them were found to be associated with large protein complexes that are involved in mitochondrial RNA processing. Thus, 5 trypanosomatid PPR proteins, including the previously characterized TbPPR1, were recovered in a large complex involved in polyadenylation of mitochondrial mRNAs (8). A subset of the same PPR proteins was also found in a gRNA-binding complex that is required for RNA editing (43), as well as in a complex that is required for the stability of edited mRNA (43). Thus, many of the same PPR proteins appear to be associated with multiple RNA-processing complexes (16). Blue native gel electrophoresis shows that TbPPR9 is also associated with a large complex of unknown composition and function that appears to be distinct from the ones described above.
Ablation of the complexes mentioned above, in line with their proposed function, generally affects many different RNAs. Ablation of PPR proteins, on the other hand, as would be expected for proteins that bind specific RNA sequences, often affects specific transcripts only. The stable association of PPR proteins with these general RNA-processing complexes therefore appears to be counterintuitive. An attractive idea in line with the fact that some of the same PPR proteins are found in different complexes would be that they function as adaptors for functional integration of specific transcripts into the corresponding RNA-processing complexes. Thus, we would expect that many of the trypanosomal PPR proteins that have not yet been studied experimentally might also be found in large complexes devoted to general RNA-processing steps. However, despite their association with these complexes, we expect their function to be specific for single or small groups of mitochondrial transcripts. Further work will show to what extent these predictions can be confirmed.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elke K. Horn and Katharina Schmid-Lüdi for excellent technical assistance.
This work was supported by grant 3100A0_121937 (to A.S.) from the Swiss National Foundation.
Footnotes
Published ahead of print 4 November 2011
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. 2003. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA 9: 62–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R. 2011. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol. Cell 42: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bastin P, Bagherzadeh A, Matthews KR, Gull K. 1996. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77: 235–239 [DOI] [PubMed] [Google Scholar]
- 4. Bochud-Allemann N, Schneider A. 2002. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 277: 32849–32854 [DOI] [PubMed] [Google Scholar]
- 5. Brun R, Schönenberger M. 1979. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36: 289–292 [PubMed] [Google Scholar]
- 6. Chomczyinski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159 [DOI] [PubMed] [Google Scholar]
- 7. Coffin JW, Dhillon R, Ritzel RG, Nargang FE. 1997. The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr. Genet. 32: 273–280 [DOI] [PubMed] [Google Scholar]
- 8. Etheridge RD, Aphasizheva I, Gershon PD, Aphasizhev R. 2008. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 27: 1596–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fisk JC, Presnyak V, Ammerman ML, Read LK. 2009. Distinct and overlapping functions of MRP1/2 and RBP16 in mitochondrial RNA metabolism. Mol. Cell. Biol. 29: 5214–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hammani K, et al. 2009. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21: 3686–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Häusler T, Stierhof Y-D, Blattner J, Clayton C. 1997. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur. J. Cell Biol. 73: 240–251 [PubMed] [Google Scholar]
- 12. Hayman ML, Read LK. 1999. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem. 274: 12067–12074 [DOI] [PubMed] [Google Scholar]
- 13. Hendriks EF, Abdul-Razak A, Matthews KR. 2003. tbCPSF30 depletion by RNA interference disrupts polycistronic RNA processing in Trypanosoma brucei. J. Biol. Chem. 278: 26870–26878 [DOI] [PubMed] [Google Scholar]
- 14. Kabani S, et al. 2009. Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics 11: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karpenahalli MR, Lupas AN, Soding J. 2007. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinform. 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koslowsky DJ. 2009. Complex interactions in the regulation of trypanosome mitochondrial gene expression. Trends Parasitol. 25: 252–255 [DOI] [PubMed] [Google Scholar]
- 17. Kühl I, Dujeancourt L, Gaisne M, Herbert CJ, Bonnefoy N. 2011. A genome wide study in fission yeast reveals nine PPR proteins that regulate mitochondrial gene expression. Nucleic Acids Res. 39: 8029–8041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lamour N, et al. 2005. Proline metabolism in procyclic Trypanosoma brucei is down-regulated in the presence of glucose. J. Biol. Chem. 280: 11902–11910 [DOI] [PubMed] [Google Scholar]
- 19. Lipinski KA, Puchta O, Surandranath V, Kudla M, Golik P. 2011. Revisiting the yeast PPR proteins—application of an iterative hidden Markov model algorithm reveals new members of the rapidly evolving family. Mol. Biol. Evol. 28: 2935–2948 [DOI] [PubMed] [Google Scholar]
- 20. Manthey GM, McEwen JE. 1995. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14: 4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maslov DA, et al. 2006. Isolation and characterization of mitochondrial ribosomes and ribosomal subunits from Leishmania tarentolae. Mol. Biochem. Parasitol. 148: 69–78 [DOI] [PubMed] [Google Scholar]
- 22. Maslov DA, et al. 2007. Proteomics and electron microscopic characterization of the unusual mitochondrial ribosome-related 45S complex in Leishmania tarentolae. Mol. Biochem. Parasitol. 152: 203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mayho M, Fenn K, Craddy P, Crosthwaite S, Matthews K. 2006. Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei: evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res. 34: 5312–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McCulloch R, Vassella E, Burton P, Boshart M, Barry JD. 2004. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol. Biol. 262: 53–86 [DOI] [PubMed] [Google Scholar]
- 25. Mingler MK, et al. 2006. Identification of pentatricopeptide repeat proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 150: 37–45 [DOI] [PubMed] [Google Scholar]
- 26. Mootha VK, et al. 2003. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. U. S. A. 100: 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nilsson D, et al. 2010. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 6: e1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Priest JW, Hajduk SL. 1994. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J. Bioenerg. Biomembr. 26: 179–191 [DOI] [PubMed] [Google Scholar]
- 29. Prikryl J, Rojas M, Schuster G, Barkan A. 2011. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. U. S. A. 108: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pusnik M, Small I, Read LK, Fabbro T, Schneider A. 2007. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol. Cell. Biol. 27: 6876–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Riviere L, et al. 2004. Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism. J. Biol. Chem. 279: 45337–45346 [DOI] [PubMed] [Google Scholar]
- 32. Schmitz-Linneweber C, Small I. 2008. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13: 663–670 [DOI] [PubMed] [Google Scholar]
- 33. Schneider A, Charrière F, Pusnik M, Horn EK. 2007. Isolation of mitochondria from procyclic Trypanosoma brucei. Methods Mol. Biol. 372: 67–80 [DOI] [PubMed] [Google Scholar]
- 34. Schneider A, et al. 1987. Subpellicular and flagellar microtubules of Trypanosoma brucei brucei contain the same alpha tubulin isotypes. J. Cell Biol. 104: 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schumacher MA, Karamooz E, Zíkov́ A A, Trantírek L, Lukes J. 2006. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell 126: 701–711 [DOI] [PubMed] [Google Scholar]
- 36. Shen S, Arhin GK, Ullu E, Tschudi C. 2001. In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 113: 171–173 [DOI] [PubMed] [Google Scholar]
- 37. Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. 2010. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 38: 4946–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Small ID, Peeters N. 2000. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25: 46–47 [DOI] [PubMed] [Google Scholar]
- 39. Stojanovski D, Pfanner N, Wiedemann N. 2007. Import of proteins into mitochondria. Methods Cell Biol. 80: 783–806 [DOI] [PubMed] [Google Scholar]
- 40. Tan THP, Pach R, Crausaz A, Ivens A, Schneider A. 2002. tRNAs in Trypanosoma brucei: genomic organization, expression and mitochondrial import. Mol. Cell. Biol. 22: 3707–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tavares-Carreón F, et al. 2008. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 283: 1472–1479 [DOI] [PubMed] [Google Scholar]
- 42. Tielens AGM, VanHellemond JJ. 1998. Differences in energy metabolism between Trypanosomatidae. Parasitol. Today 14: 265–271 [DOI] [PubMed] [Google Scholar]
- 43. Weng J, et al. 2008. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol. Cell 32: 198–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99: 89–101 [DOI] [PubMed] [Google Scholar]
- 45. Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. 2004. The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 382: 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zíkov́ A A, et al. 2008. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol. Cell. Proteomics 7: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.