Abstract
SCF (Skp1–cullin/Cdc53–F-box protein) ubiquitin ligases bind substrates via the variable F-box protein and, in conjunction with the RING domain protein Rbx1 and the ubiquitin-conjugating enzyme Ubc3/Cdc34, catalyze substrate ubiquitination. The cullin subunit can be modified covalently by conjugation of the ubiquitin-like protein Rub1/NEDD8 (neddylation) or bound noncovalently by the protein CAND1 (cullin-associated, neddylation-dissociated). Expression of the Candida albicans CAND1 gene homolog CaTIP120 in Saccharomyces cerevisiae is toxic only in the presence of CaCdc53, consistent with a specific interaction between CaTip120 and CaCdc53. To genetically analyze this system in C. albicans, we deleted the homologs of RUB1/NEDD8, TIP120/CAND1, and the deneddylase gene JAB1, and we also generated a temperature-sensitive allele of the essential CaCDC53 gene by knock-in site-directed mutagenesis. Deletion of CaRUB1 and CaTIP120 caused morphological, growth, and protein degradation phenotypes consistent with a reduction in SCF ubiquitin ligase activity. Furthermore, the double Carub1−/− Catip120−/− mutant was more defective in SCF activity than either individual deletion mutant. These results indicate that CAND1 stimulates SCF ubiquitin ligase activity and that it does so independently of neddylation. Our data do not support a role for CAND1 in the protection of either the F-box protein or cullin from degradation but are consistent with the suggested role of CAND1 in SCF complex remodeling.
INTRODUCTION
Cullin-RING ligases (CRLs), the largest family of ubiquitin ligases, mediate the specific degradation of a variety of cellular proteins. The largest and most versatile subset of CRLs contains the SCF (Skp1-cullin-F box) ubiquitin ligases, consisting of four subunits: Skp1, Cul1 (Cdc53 in Saccharomyces cerevisiae), the RING finger protein Rbx1 (also called Roc1 or Hrt1), and one of several F-box proteins (FBPs) (8, 54, 66). The crystal structure of the SCF complex reveals that Cul1 is an elongated protein that forms a scaffold on which the ubiquitin-conjugating enzyme (Ubc), substrate, and other subunits are positioned (49, 70). Rbx1 binds near the C terminus of Cul1 and recruits the Ubc. Skp1 binds near the N terminus of Cul1 and in turn recruits the different FBPs via the N-terminal F-box domain (49, 70). The C-terminal domain of the FBP binds to the substrate and tethers it near the catalytic center of the core complex (49, 61, 70). Accordingly, the substrate specificity of the complex is determined by the identity of the FBP (53). For S. cerevisiae, sequence analysis identified some 21 putative FBPs (66), the vast majority of which were found experimentally to interact with Skp1 and Cdc53 (e.g., see references 36 and 58). In the Candida albicans genomic sequence, genes for at least 13 FBPs can be detected by sequence homology (our unpublished observations), among which 2, Cdc4 and Grr1, were found experimentally to function in degradation of specific proteins (2, 40).
The ubiquitination activity of the SCF complex can be modulated by reversible conjugation of the ubiquitin-related protein NEDD8/Rub1 to the cullin subunit, at a single lysine residue located in the C-terminal domain, in a process called neddylation (51). Neddylation is mechanistically similar to ubiquitination, in that NEDD8/Rub1 is activated by E1 (Uba3/Ula1) and E2 (Ubc12) enzymes (37, 41) and requires Rbx1 (30) and an E3-like protein, Dcn1 (34, 35). Deneddylation of cullins is promoted by Csn5/Jab1, a subunit of another conserved multiprotein complex, the COP9 signalosome (CSN) (9, 15, 27). Neddylation is essential in mammalian cells (10, 26), Caenorhabditis elegans (29), Arabidopsis thaliana (4), and fission yeast (Schizosaccharomyces pombe) (50), whereas deletion of its pathway has minimal discernible phenotypes in budding yeast (S. cerevisiae) (37, 41). Biochemical data suggest that neddylation increases SCF activity by increasing its affinity for the Ubc (31, 55). Recently, neddylation was also shown to induce significant conformational flexibility in the SCF complex, possibly facilitating contact between the E2 enzyme and the acceptor lysines on the substrate and on the elongating ubiquitin chain (18, 55).
The role of neddylation has also been linked functionally to CAND1 (cullin-associated, Nedd8-dissociated), a protein that binds unneddylated cullin and competes with Skp1-FBP binding (43, 69). CAND1 has two binding sites, on either end of the cullin: one overlaps with the SKP1 binding site, and one overlaps with the neddylation site (24). Whereas CAND1 suppresses SCF ubiquitination activity in vitro (43, 47, 69), mutant analyses of both Arabidopsis (11, 14, 20) and, more recently, C. elegans (5) are consistent with CAND1 promoting the activity of the SCF and of additional CRLs in vivo. Interestingly, the deneddylation complex CSN also exhibits a similar inconsistency between its negative activity on CRL function in vitro and its promotion of CRL function in vivo. To explain these discrepancies, it was proposed that both CSN and CAND1 function to protect FBPs (12, 16, 28, 63) or cullin (32, 68) from autocatalytic ubiquitination and degradation in the absence of substrate. An alternative model suggests that CAND1, in combination with the neddylation/deneddylation cycle, promotes the redistribution of the FBPs bound to the complexes according to cellular needs (6, 54, 56).
The opportunistic pathogen Candida albicans is a dimorphic fungus, able to switch between two distinct growth forms: a yeast form and a hyphal (or mold) form (65). The switch to hyphal growth occurs in response to various extracellular stimuli and involves transcriptional (42) and posttranscriptional (45, 71) mechanisms. SCF-mediated protein degradation also plays a role in C. albicans morphogenesis, as evidenced by the hyphal phenotype of the Cacdc4−/− mutant (2, 59) and the pseudohyphal phenotype of the Cagrr1−/− mutant (7, 40) and of CaCdc53 depletion (62). Like most fungal genomes, the C. albicans genome also contains easily discernible homologs of the NEDD8, CAND1, and CSN5 genes. In order to analyze the functions of CAND1 and neddylation in SCF activity in C. albicans, we generated mutants of the core SCF component Cdc53 (CUL1) and of its modifiers Rub1 (NEDD8), Jab1 (CSN5), and Tip120 (CAND1). The phenotypes of these mutants, as well as genetic epistasis analysis, enabled us to probe the functional relationships between these regulators of the SCF complex in vivo.
MATERIALS AND METHODS
Plasmids and strains.
The C. albicans and S. cerevisiae strains used in this study are listed in Tables 1 and 2. Plasmid and strain construction is described in the supplemental material.
Table 1.
C. albicans strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| KC2 (CAF3) | ura3Δ::imm434/ura3Δ::imm434 | 21 |
| KC190 | KC2 rub1Δ::hisG-URA3-hisG/rub1Δ::hisG | This work |
| KC208 | KC2 rub1Δ::hisG/rub1Δ::hisG | This work |
| KC203 | KC2 jab1Δ::hisG-URA3-hisG/jab1Δ::hisG | This work |
| KC213 | KC2 jab1Δ::hisG/jab1Δ::hisG | This work |
| KC218 | KC2 CDC53/CDC53-3×HA::URA3 | This work |
| KC219 | KC213 CDC53/CDC53-3×HA::URA3 | This work |
| KC220 | KC208 CDC53/CDC53-3×HA::URA3 | This work |
| KC298 | KC2 tip120Δ::hisG-URA3-hisG/tip120Δ::hisG | This work |
| KC299 | KC2 tip120Δ::hisG/tip120Δ::hisG | This work |
| KC321 | KC208 tip120Δ::hisG/tip120Δ::hisG | This work |
| KC354 | KC299 CDC53/CDC53-3×HA::URA3 | This work |
| KC355 | KC321 CDC53/CDC53-3× HA::URA3 | This work |
| KC274 (SN148) | ura3Δ::imm434/ura3Δ::imm434 his1/his1 leu2/leu2 arg4/arg4 | 48 |
| KC325 | KC274 cdc53Δ::LEU2/CDC53 | This work |
| KC326 | KC274 cdc53Δ::LEU2/CDC53::URA3 | This work |
| KC327 | KC274 cdc53Δ::LEU2/cdc53RRR::URA3 | This work |
| KC329 | KC274 cdc53Δ::LEU2/CDC53-3× HA::URA3 | This work |
| KC362 | KC274 cdc53Δ::LEU2/cdc53-1::URA3 | This work |
| KC363 | KC274 cdc53Δ::LEU2/cdc53-1 | This work |
| KC395 | KC274 rub1Δ::hisG-URA3-hisG/rub1Δ::hisG | This work |
| KC424 | KC274 tip120Δ::hisG-URA3-hisG/tip120Δ::hisG | This work |
| KC427 | KC274 rub1Δ::hisG/rub1Δ::hisG tip120Δ::hisG-URA3-hisG/tip120Δ::hisG | This work |
| KC471 | KC274 URA3 | This work |
Table 2.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MATaura3-1 leu2,3-112 trp1-1 ade2-1 his3-11,15 | R. Rothstein |
| MTY740 | W303-1A cdc53-1 | M. Tyers |
| BY4742 | MATα ura3Δ0 lys2Δ0 leu2Δ0 his3Δ1 | EUROSCARF |
| KY1293 | BY4742 cdc53Δ::CaCDC53 HIS3 | This work |
| KY1294 | BY4742 cdc53Δ::CaCDC53RRRHIS3 | This work |
| KY1306 | ura3Δ0 lys2Δ0 leu2Δ0 his3Δ1 dcn1Δ::KANrcdc53Δ::CaCDC53 HIS3 | This work |
| KY1309 | ura3Δ0 met15Δ0 leu2Δ0 his3Δ1 jab1Δ::KANrcdc53Δ::CaCDC53 HIS3 | This work |
| KY1310 | ura3Δ0 lys2Δ0 leu2Δ0 his3Δ1 rub1Δ::KANrcdc53Δ::CaCDC53 HIS3 | This work |
| KY1352 | BY4742 cdc53Δ::FLAG-CaCDC53HIS3 | This work |
Protein analysis.
Protein levels were assayed by Western blotting using the following monoclonal antibodies: 9E10 to detect the Myc epitope, anti-FLAG (Sigma) to detect the FLAG epitope, and HA.11 (Covance) to detect the hemagglutinin epitope. Proteins were extracted by the quantitative NaOH–2-mercaptoethanol method, as described previously (64). To compare steady-state protein levels, equal protein amounts were loaded; to monitor protein disappearance after promoter shutoff, equal culture volume equivalents were loaded. Loading and transfer were monitored by Ponceau staining of the membrane and by actin quantitation using an anti-β-actin antibody (ab8224; Abcam). Enhanced chemiluminescence (ECL) signals were quantitated with a Bio-Rad Chemidoc apparatus. Pulse-chase analysis of proteins in C. albicans was performed as described in reference 33. For FLAG-CaCdc53 immunoprecipitation, exponentially growing cultures induced for 1 h with galactose and then shifted to glucose for 2 h were spun down, resuspended in 1 ml lysis buffer (0.5% NP-40, 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], and a 1:500 dilution of antiprotease cocktail containing chymostatin, pepstatin A, leupeptin, and antipain [10 mg/ml of each in dimethyl sulfoxide]), and broken with glass beads (0.5 mm) in a BeadBeater apparatus for 3 pulses of 1.5 min each. The extracts were cleared from nonspecific interactions by a 15-min incubation with protein A Sepharose beads prior to immunoprecipitation with M2 FLAG Sepharose beads (Sigma). Beads were washed 3 times in lysis buffer, and proteins were eluted in gel loading buffer.
Microscopy and flow cytometry.
Cells were fixed in 70% ethanol and visualized with a Zeiss AxioImager M1 microscope equipped with differential interference contrast (DIC) optics, using a 40× or 100× objective. Colonies were visualized with a Zeiss Stemi 2000C binocular microscope. For flow cytometry, cells were prepared as described previously (25), with some modifications. A total of 107 to 108 cells were fixed in 70% ethanol for 1 h to overnight, washed with 0.2 M Tris, pH 7.5, and then incubated overnight in a shaker at 37°C in 0.2 M Tris, pH 7.5, 10 mM EDTA, 1 mg/ml RNase A. The cells were then spun down, resuspended in 50 mM HCl with 5 mg/ml pepsin, incubated for another 2 h at 37°C, and washed with 0.2 M Tris, pH 7.5. Approximately 5 × 106 cells were then incubated for at least 15 min at 30°C while shaking in 0.3 ml 0.2 M Tris, pH 7.5, with 2 μM SYTOX (Molecular Probes, Inc.). The cells were diluted 1:10 in 0.2 M Tris, pH 7.5, just before injection into the flow cytometer (Beckton-Dickinson FACScalibur). A total of 20,000 ungated events were recorded for each run.
RESULTS
Generation of a C. albicans CDC53 ts allele.
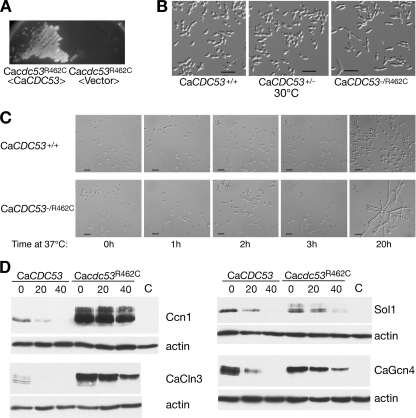
We identified, by homology searching, C. albicans orf19.1674 as the closest bidirectional homolog of S. cerevisiae CDC53. In order to experimentally validate this assignment, orf19.1674 was cloned into an S. cerevisiae vector and introduced into the S. cerevisiae cdc53-1 mutant, which is unable to grow at 37°C. The orf19.1674/CaCDC53 plasmid suppressed the temperature sensitivity of that strain (data not shown); furthermore, we subsequently found that substitution of the ScCDC53 coding sequence with the CaCDC53 coding sequence yielded a viable strain (see below and Fig. 3), thereby confirming that CaCDC53 is a functional homolog of ScCDC53. While in S. cerevisiae the core SCF subunit genes CDC53, SKP1, and RBX1 and the cell cycle-related F-box protein gene CDC4 are all essential, for C. albicans, a mutant with deletion of the CDC4 homolog is viable (2). Assuming that in C. albicans the core SCF complex may, like CaCDC4, be nonessential, we initially attempted to delete both alleles of CaCDC53. Although the first allele was readily deleted, we repeatedly failed to obtain a deletion of the second allele, suggesting that CaCDC53 may in fact be essential. In order to study its function, we proceeded to create a temperature-sensitive (ts) allele in C. albicans, modeled on the S. cerevisiae cdc53-1 allele. The cdc53-1 mutation causes an arginine-to-cysteine change at position 488 (52), in a region of the protein highly conserved between ScCdc53 and CaCdc53. The homologous arginine residue encoded by CaCDC53, R462, was mutated to cysteine. The remaining allele of the CaCDC53/Cacdc53Δ::LEU2 strain was then replaced with CaCDC53R462C (see Materials and Methods). The resulting strain was unable to grow at 37°C, but this growth defect was complemented by reintroducing a wild-type CaCDC53 copy into the strain, indicating that the CaCDC53 mutation was responsible for the temperature sensitivity of the strain (Fig. 1A). At 24°C and 30°C, the CaCDC53R462C strain grew as well as the wild-type strain; however, even at the growth-permissive temperature of 30°C, the mutant already showed a clear morphological phenotype, consisting of an elongated cell shape (Fig. 1B). When shifted from 24°C to 37°C, the mutant strain rapidly ceased to proliferate, and microscopic examination of the cells after the shift to 37°C indicated a rapid induction of germ tube-like extensions (Fig. 1C). Even at the semipermissive temperature of 30°C, Ccn1 and CaCln3 (both substrates of SCFCaGRR1) (40), Sol1 (a substrate of SCFCaCDC4) (2), and CaGcn4 (a rapidly degraded homolog of the S. cerevisiae SCFCDC4 substrate Gcn4) (23) were all significantly stabilized (Fig. 1D). In all cases, slower-migrating forms of the proteins accumulated in the mutant. These bands may correspond to phosphorylated forms of the proteins that are normally recognized by the SCF complexes and rapidly degraded in the wild-type strain.
Fig 3.
CaTIP120 expression in S. cerevisiae. (A) The CaTIP120-expressing plasmid KB1958 was transformed into strains KY1068 (wild type; ScCDC53), KY1293 (Sccdc53Δ::CaCDC53), KY1294 (Sccdc53Δ::CaCDC53RRR), KY1310 (Sccdc53Δ::CaCDC53 rub1Δ), KY1309 (Sccdc53Δ::CaCDC53 jab1Δ), and KY1306 (Sccdc53Δ::CaCDC53 dcn1Δ). Serial 5-fold dilutions of overnight cultures were spotted onto SC plates without uracil and with the indicated carbon source and were incubated at 30°C for 3 days (galactose) or 2 days (glucose). (B) FACS analysis of strains KY1068, KY1293, and KY1294 harboring plasmid KB1586 at the indicated times of galactose induction. (C) Coimmunoprecipitation of CaTip120 with CaCdc53. Plasmid KB1960, expressing Myc-CaTip120 under the control of the GAL1 promoter (+), or a vector plasmid (−) was transformed into strain KY1352 expressing FLAG-tagged CaCdc53 under the control of the ScCDC53 promoter (+) or expressing untagged CaCdc53 (−). (Top) Whole-cell extracts (WCE) were analyzed for Myc-CaTip120 expression by Western blotting (W.B.). FLAG-CaCdc53 was immunoprecipitated (i.p.) from the extracts, and the precipitate was analyzed by Western blotting for the presence of Myc-CaTip120 (middle) or FLAG-CaCdc53 (bottom). The asterisk indicates a band cross-reacting with the anti-FLAG antibody. Note that CaCdc53 runs as two bands, representing the neddylated and unneddylated species.
Fig 1.
Phenotype of Cacdc53-1 temperature-sensitive mutant. (A) Strain KC363 (Cacdc53Δ/Cacdc53R462C), harboring a vector plasmid or a reintroduced wild-type copy of CaCDC53 (KB1869), was grown on a YPD plate for 24 h at 37°C. (B) Morphologies of wild-type, CaCDC53+/− heterozygous (KC326), and CaCDC53R462C/− mutant strains grown to log phase at 30°C. Bar = 20 μm. (C) Strains KC471 (WT URA+) and KC363 (CaCDC53R462C/−) were grown in liquid YPD medium to log phase at 24°C, shifted to 37°C, and photographed with Nomarski optics at the indicated times. Bar = 20 μm. (D) Degradation of Ccn1, CaCln3, Sol1, and CaGcn4 in the CaCDC53+/− heterozygote (KC325) versus the Cacdc53-1 mutant (KC363). The indicated substrates were tagged with a 6× Myc epitope at the C terminus and expressed from the glucose-repressible MAL2 promoter in plasmids KB1698, KB1697, KB1578, and KB1192. Cells were grown overnight in maltose and then diluted and grown to mid-log phase. Glucose (2%) was added to repress the promoter. Aliquots were taken at the indicated times after glucose addition. Western blots were incubated sequentially with the 9E10 and anti-β-actin antibodies.
Identification of C. albicans RUB1 and JAB1/CSN5.
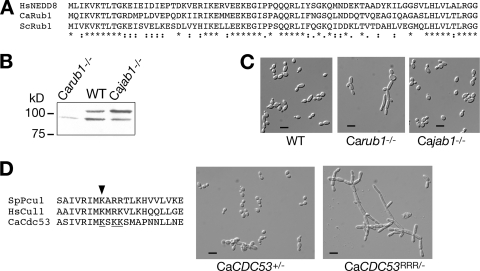
A homology search of the C. albicans genomic database identified orf19.330.1 as the closest bidirectional homolog of S. cerevisiae RUB1. Sequence comparison between CaRub1, ScRub1, and human NEDD8 (Fig. 2A) indicated that NEDD8 is equally related to either fungal protein (about 60% identity), and surprisingly, the two fungal Rub1 proteins are slightly less related to each other (55% identity, in contrast with 100% identity between S. cerevisiae and C. albicans ubiquitins). Since the identity between the NEDD8 and Rub1 proteins and ubiquitin is also around 55%, we needed to provide functional evidence that CaRub1 indeed functions as a NEDD8 homolog. The best-characterized function of Rub1/NEDD8 is the formation of a covalent adduct with cullins such as Cdc53 (41). This adduct can be detected in protein gels as an additional, slower-migrating species of Cdc53. In order to test whether CaRub1 similarly modifies CaCdc53, the latter protein was tagged at its C terminus with a three-hemagglutinin (3× HA) epitope tag and visualized in a wild-type strain versus a strain deleted for both alleles of CaRUB1. As shown in Fig. 2B, for the wild-type strain, CaCdc53-3xHA did in fact migrate as two bands; the upper band is undetectable in the Carub1−/− background. We also tested whether the extent of CaCdc53 modification is affected by the activity of the deneddylation complex CSN. Jab1/Csn5 carries the catalytic activity of the S. cerevisiae and mammalian CSN complexes: in the absence of Jab1 and other components of the CSN complex, the Rub1-modified form of Cdc53 accumulates (17, 46). orf19.3371 was identified as the closest homolog of S. cerevisiae JAB1. We therefore called this open reading frame (ORF) CaJAB1 and deleted both copies from the C. albicans genome. As shown in Fig. 2B, the upper band of CaCdc53-3xHA was relatively more prominent for the Cajab1−/− mutant. These data confirm that CaRub1 is a covalent modifier of CaCdc53 and that CaJab1 decreases the extent of CaRub1 modification of CaCdc53.
Fig 2.
Effect of CaCdc53 neddylation on C. albicans morphology. (A) Sequence alignment of C. albicans Rub1 with human NEDD8 and S. cerevisiae Rub1. (B) CaCdc53 is unneddylated in the Carub1−/− strain and hyperneddylated in the Cajab1−/− strain. One CaCDC53 allele in strains KC2, KC208, and KC213 was fused to the 3× HA epitope tag by using plasmid KB1570. Proteins were extracted from cells in log phase, electrophoresed, and incubated with the 12CA5 antibody. (C) Cell morphology of the wild-type (WT) (KC2), Carub1−/− (KC208), and Cajab1−/− (KC213) strains grown to log phase at 30°C. Magnification, ×40. Bar = 10 μm. (D) Phenotype of CaCDC53RRR mutant. (Left) Sequence alignment of the neddylation sites of hCul1, SpPcu1, and CaCdc53. The arrowhead indicates the site of neddylation in SpPcu1 and HsCul1. The lysine residues that were mutated to arginine in CaCdc53 are underlined. (Right) Micrographs of the CaCDC53+/− (KC326) and CaCDC53RRR/− (KC327) strains grown to log phase at 30°C. Magnification, ×40. Bar = 10 μm.
Phenotypes of neddylation and deneddylation mutants.
Cullin neddylation is necessary for optimal SCF activity in most organisms (66). We found that the Carub1−/− mutant grew markedly slower than the wild-type strain in liquid. On agar plates, the Carub1−/− strain formed smaller, rougher colonies. The Cajab1−/− mutant, in contrast, did not exhibit any growth or colony morphology phenotypes. Microscopic observation indicated that the Carub1−/− mutant—but not the Cajab1−/− mutant—exhibited an elongated cell morphology (Fig. 2C). This could be due to failure to neddylate CaCdc53. However, in S. cerevisiae, Rub1 is known to modify other proteins in addition to Cdc53 (38). In order to test by an alternative approach whether CaCdc53 neddylation per se causes the Carub1−/− morphogenetic phenotype, we constructed a CaCdc53 allele that is mutated in the predicted neddylation target site. In human Cul1 (hCul1), neddylation was found to occur on residue K720 (51), and in the S. pombe Cul1 homolog Pcu1, it was found to occur on K713 (50). Alignment of CaCdc53 with hCul1 and Pcu1 strongly suggests that lysine 699 is homologous to the neddylation targets in hCul1 and Pcu1 (Fig. 2D). Since two adjacent residues in CaCdc53, at positions 701 and 702, are also lysines, we were concerned that if K699 were mutated, these residues may serve as alternative Rub1 acceptors. Therefore, we mutated all three lysines—at positions 699, 701, and 702—to arginines, generating the CaCdc53RRR allele. This construct was used to replace the remaining CaCDC53 allele in a CaCDC53+/− heterozygote, generating a CaCDC53RRR/− mutant. The cell morphology phenotype of this strain resembled the morphology of the Carub1−/− mutant and was in fact more filamentous than that of the Carub1−/− strain (Fig. 2D), possibly due to the added haploinsufficiency of CaCDC53 in the CaCDC53RRR/− strain versus the Carub1−/− strain. These results are consistent with the lack of CaCdc53 neddylation being responsible for the morphogenetic phenotype of the Carub1−/− mutant.
Identification of the C. albicans CAND1 homolog CaTip120.
It is striking that prevention of CaCdc53 neddylation shows a clear morphogenetic phenotype in C. albicans, suggesting that neddylation is required for optimal SCF function in this organism, whereas deletion of the neddylation pathway in wild-type S. cerevisiae causes no morphological or substrate degradation defects (37, 41; our unpublished results). Comparison of the genomes of the two organisms suggested a possible reason for this difference: C. albicans contains an open reading frame with significant (P = 10−12) homology to human CAND1/TIP120, whereas in the S. cerevisiae genome, no obvious CAND1 ortholog could be detected (recently, Lag2 was identified as an S. cerevisiae protein able to bind Cdc53 [44, 60], but it is not clear yet whether Lag2 fulfills a similar function to that of CAND1 in other organisms). CAND1 (cullin-associated, Nedd8-dissociated) was shown to inhibit SCF activity in vitro by binding to cullin; this binding is counteracted by neddylation of cullin (43, 69). Thus, this raises the possibility that in C. albicans, CaCdc53 modification by CaRub1 is required to counteract the inhibitory activity of CAND1. Conversely, expressing CAND1 in S. cerevisiae may render cullin neddylation essential in this organism (in keeping with standard yeast gene nomenclature, the C. albicans CAND1/TIP120 gene homolog, orf19.6729, was named CaTIP120).
To test whether orf19.6729 in fact expresses a functional CAND1 homolog with respect to interaction with Cul1/Cdc53, we expressed it in S. cerevisiae under the control of the inducible GAL1 promoter. Protein gel electrophoresis suggested that the translation start site of orf19.6729 is misannotated and that the actual start site is 139 nucleotides (nt) downstream (see Fig. S1 in the supplemental material). We accordingly expressed the shortened version of orf19.6729 as the CaTIP120 homolog for further analyses in S. cerevisiae.
If CaTip120 inhibits SCF activity by binding to the cullin, and since SCF activity is essential, overexpression of CaTip120 should be toxic. However, expression of the GAL1-CaTIP120 construct had no effect on growth of wild-type S. cerevisiae (Fig. 3A, upper row). We surmised that this might be due to an inability of C. albicans Tip120 to interact with the S. cerevisiae cullin, Cdc53. Therefore, an S. cerevisiae strain was built in which the resident CDC53 gene was replaced with CaCDC53, either wild type or CaCDC53RRR, under the control of the ScCDC53 promoter. The viability of those strains confirmed that CaCdc53 was able to stand in functionally for ScCdc53 and to interact with the basic components of the essential SCF complex. Notably, however, the S. cerevisiae cdc53Δ::CaCDC53 strain was unable to grow at 37°C, whereas a cdc53Δ strain carrying the CaCDC53 gene with its native promoter on a plasmid grew well at that temperature (not shown). This suggests either that CaCDC53 is not expressed efficiently from the ScCDC53 promoter or that S. cerevisiae SCF function is not optimal with CaCdc53.
Induction of GAL1-CaTIP120 in the CaCDC53 strain was toxic, as expected if CaTip120 was a negative regulator of CaCdc53 (Fig. 3A). In addition, as expected if neddylation was able to partially counteract CaTip120 binding to CaCdc53, the CaCDC53RRR strain, carrying the presumably unneddylatable allele of CaCDC53, was even more sensitive to CaTip120 expression (Fig. 3A). Similarly, S. cerevisiae CaCDC53 strains deleted for RUB1 or for the Rub1 ligase gene DCN1 were also hypersensitive to expression of CaTip120 (Fig. 3A). In contrast, the JAB1 deletion strain was not more sensitive and may be marginally more resistant to CaTip120 expression. Taken together, these data are consistent with the toxicity of CaTip120 on CaCdc53 being counteracted by neddylation of CaCdc53. Since SCF function is required for progression through the cell cycle, specifically at the G1-to-S transition (19), we used fluorescence-activated cell sorter (FACS) analysis to assay the effect of CaTip120 expression on cell cycle distribution. The CaTip120-inhibited cultures accumulated in the G1 phase of the cell cycle (Fig. 3B), consistent with defects in SCF complex activity.
Since the S. cerevisiae CaCDC53 background may exhibit reduced SCF activity, based on its temperature sensitivity at 37°C, the toxicity of CaTip120 expression in this background could alternatively have been explained as an indirect synthetic effect of the reduction in SCF activity, not an effect of direct interaction with CaCdc53. We therefore checked that the same GAL1p-CaTIP120 construct, expressed in the S. cerevisiae cdc53-1 mutant at the semipermissive temperature of 30°C, had no effect on cell growth (see Fig. S2 in the supplemental material). Thus, the phenotype resulting from the expression of CaTip120 in S. cerevisiae is consistent with it being a negative regulator of the SCF complex by direct binding with its cognate cullin, CaCdc53.
Finally, to obtain more direct evidence for a physical interaction between CaCdc53 and CaTip120, FLAG-tagged CaCdc53 was immunoprecipitated from cells expressing Myc-tagged CaTip120. As shown in Fig. 3C, Myc-CaTip120 immunoprecipitated specifically with FLAG-CaCdc53, confirming that these proteins physically interact.
Phenotype of C. albicans TIP120 mutant.
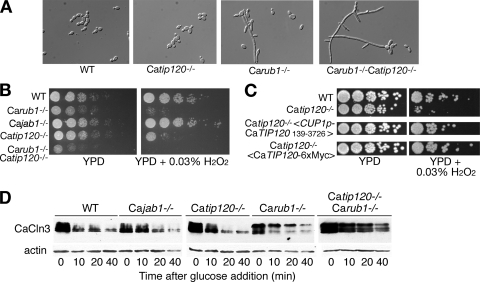
If an important role of neddylation is to counteract the inhibitory activity of CAND1, then the clear phenotypic effect of deletion of the NEDD8 homolog RUB1 in C. albicans, as opposed to the scant phenotypic effect of the homologous deletion in S. cerevisiae, may be linked to the absence of a CAND1 homolog in the latter organism. A straightforward prediction of this model is that deleting CaTIP120 should suppress the phenotype of the CaRUB1 deletion. To test this prediction, we deleted CaTIP120 in both the wild-type background and the Carub1−/− background. The Catip120−/− mutant by itself displayed no apparent morphological phenotypes (Fig. 4A; see Fig. S3 in the supplemental material). However, contrary to our prediction, the Carub1−/− Catip120−/− double mutant, far from suppressing the cell elongation phenotype of Carub1−/−, exacerbated it (Fig. 4A; see Fig. S3).
Fig 4.
Phenotype of C. albicans CaTIP120 deletion mutant alone and in combination with the CaRUB1 deletion. (A) Morphology of the wild-type (KC2), Carub1−/− (KC208), Catip120−/− (KC299), and Carub1−/− Catip120−/− (KC321) strains grown in liquid YPD to log phase at 30°C. (B) Growth of the same strains and the Cajab1−/− (KC213) strain on H2O2. The control plate was incubated for 2 days at 30°C, and the H2O2 plate was incubated for 3 days. (C) Complementation of the H2O2 sensitivity phenotype with the N-terminally truncated CaTIP120 open reading frame and with a C-terminally epitope-tagged CaTIP120 version. Strains used were KC2 (wild type), KC299 (Catip120−/−), and KC299 transformed with KB1962 (CUP1p-Myc-CaTIP120139-3726) or KB2029 (CaTIP120-6xMyc). (D) Degradation of SCF substrate CaCln3. The strains used for panel B were transformed with plasmid KB2152 (MAL2p-CaCLN3-3xHA). Degradation of CaCln3-3× HA upon promoter shutoff with glucose was followed by Western blotting. Equal loading was verified by incubating the same blot with an anti-actin monoclonal antibody.
Since the Catip120−/− mutant displayed no apparent morphology or growth phenotype under normal growth conditions (yeast extract-peptone-dextrose [YPD] medium, 30°C or 37°C), we also assayed for growth effects under stress conditions. The Carub1−/− and Catip120−/− single and double mutants, as well as the Cajab1−/− mutant, were grown in the presence of 0.03% H2O2. The Carub1−/− mutant displayed markedly reduced growth under these conditions (Fig. 4B), in excess of the relative growth reduction of the Carub1−/− versus wild-type strains observed under standard growth conditions. The Catip120−/− mutant also showed markedly reduced growth in 0.03% H2O2. The double mutant showed further growth reduction compared to each mutant separately. The Cajab1−/− mutant did not show any growth defect under these conditions. We also tested reversion of the H2O2 sensitivity phenotype by expressing the shortened CaTIP120 ORF (starting at nt +139) (see Fig. S1 in the supplemental material) or an epitope-tagged version of CaTip120 (Fig. 4C). This confirmed both that the H2O2 sensitivity phenotype was due to the loss of Tip120 and that the shorter ORF is functional in C. albicans.
We next tested the effects of CaRub1 and CaTip120 on SCF activity more directly, by assaying degradation of a known SCF substrate, CaCln3, which depends on the SCFCaGRR1 complex for its degradation (40). The epitope-tagged substrate was expressed from the repressible MAL2 promoter. Decay of the steady-state protein bands was followed after promoter repression with glucose. In the wild-type strain, CaCln3 exhibited initial fast decay followed by persistence of a small residual protein population (Fig. 4D). The decay in the Catip120−/− mutant was similar, except that the initial decay was somewhat retarded. A similar kinetics was detected in the Cajab1−/− mutant. In the Carub1−/− mutant, a significant stabilization was detected, accompanied by accumulation of a slower-migrating species of CaCln3. Finally, in the double Carub1−/− Catip120−/− mutant, the protein was further stabilized, but the migration pattern, a mixture of slower- and faster-migrating bands, was different from that of the Carub1−/− mutant and suggested the persistence of several phosphospecies of CaCln3 (Fig. 4D). Taken together, these data indicate that in vivo, CaRub1 and CaTip120 are not antagonistic but rather act synergistically on SCF function.
Effects of neddylation and CaTip120 on stability of the F-box protein CaGrr1.
One possible role of the neddylation/deneddylation cycle is that it is required to protect SCF complex components from ubiquitination and degradation in the absence of substrate. Several F-box proteins have been shown to be unstable (22, 72). Since we found that CaCln3 turnover was affected in the Cajab1−/− mutant and in the Carub1−/− and Catip120−/− single and double mutants (Fig. 4D), and since the CaCln3-specific F-box protein is CaGrr1 (40), we tested how CaGrr1 levels were affected in the various mutants. Steady-state levels of epitope-tagged CaGrr1, expressed from its native promoter, were quantitated by Western blotting. Strikingly, the protein levels were increased between 2- and 8-fold, rather than decreased, in all the mutants tested (Fig. 5A). To test whether this increase was due to metabolic stabilization of the protein rather than to increased expression, the stability of CaGrr1 was tested in various mutants by pulse-chase analysis. The results of this analysis indicated that whereas CaGrr1 was rapidly degraded in the wild-type background, to the point that it became undetectable after 60 min of chase, it was stabilized to various extents in the mutants tested (Fig. 5B). We thus concluded that the reduced CaCln3 degradation in the mutants tested cannot be attributed to a destabilization of the F-box protein.
Fig 5.
Stability of the F-box protein CaGrr1. A construct carrying 6× Myc-tagged CaGRR1 under the control of its own promoter (plasmid KB1799) was integrated into the same strains as those used for Fig. 4 and also into KC363 (Cacdc53-1). (A) Protein extracts from log-phase cells were Western blotted and sequentially incubated with anti-Myc (antibody 9E10; upper panel) and anti-β-actin (lower panel). Two independent transformants of the wild-type strain were tested. The numbers below the blots give the ratio of the Myc signal to the actin signal, normalized to the ratio for the wild-type strain (average for the two wild-type cultures; the variance is indicated). (B) [35S]methionine pulse-chase analysis of the same strains. C, no-tag control.
Stability and neddylation kinetics of the cullin CaCdc53.
Data obtained from Neurospora and Drosophila suggest that neddylation might affect the turnover of cullins (28, 68). We therefore tested whether neddylation and CaTip120 might affect SCF function via CaCdc53 stability. Initially, we had used a C-terminal 3× HA tag integrated into one of the two CaCDC53 alleles to demonstrate CaCdc53 neddylation by CaRub1 (Fig. 2B). However, the CaCDC53-3xHA allele is only partially active, based on the phenotype of a C. albicans strain expressing CaCDC53-3xHA only (see Fig. S5 in the supplemental material) and on the inability of this allele to complement the S. cerevisiae CDC53 deletion (see Fig. S4). Therefore, we constructed a CaCdc53 allele tagged N-terminally with the Myc epitope tag. This allele expressed in S. cerevisiae was able to complement the CDC53 deletion (see Fig. S4).
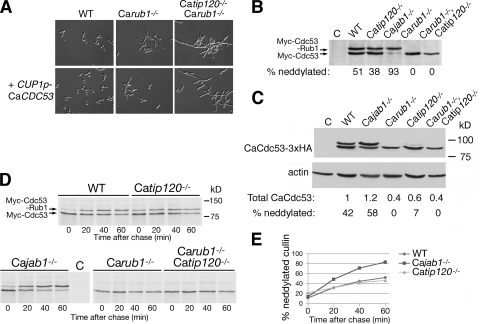
The expression of CaCdc53 under the control of the heterologous CUP1 promoter enabled us first to test the simple hypothesis that the Carub1−/− and Catip120−/− mutants were defective in SCF activity due to reduced CaCdc53 levels, because if this were the case, then the Carub1−/− and Carub1−/− Catip120−/− morphological phenotypes might be suppressed by CaCDC53 overexpression from the extra CaCDC53 copy under control of the CUP1 promoter. As shown in Fig. 6A, no such suppression of the morphological phenotype was detected in the presence of the added CUP1p-Myc-CaCDC53 allele. On the other hand, Western blot analysis of Myc-CaCdc53 did reveal some differences in steady-state levels of the protein in the mutants (Fig. 6B). Since the variations in Myc-CaCdc53 levels could be attributed to either variations in cullin stability or indirect effects on CUP1 promoter activity, we performed two additional experiments. In the first, we analyzed the steady-state levels of the CaCdc53-3xHA construct, which is under regulation of the native CaCDC53 promoter and is thus expected to more faithfully represent the levels of native CaCdc53. Normalized to β-actin, the differences between the various strains did indicate a 2.5-fold reduction in CaCdc53 level in the Carub1−/− mutants (Fig. 6C). However, when we directly assayed the stability of Myc-CaCdc53 by pulse-chase analysis with radioactive methionine, the protein appeared largely stable in all strains tested (Fig. 6D), suggesting that reduced cullin stability cannot account for the phenotypes of the Carub1−/− and Catip120−/− mutants.
Fig 6.
Neddylation and stability of CaCdc53. (A) Introduction of an additional allele of CaCDC53 under the control of the CUP1 promoter does not ameliorate the morphology phenotype of the Carub1−/− and Carub1−/− Catip120−/− mutants. Cells with the indicated genotype, without (top) or with (bottom) addition of the CaCUP1-CaCDC53 plasmid (KB1897), were grown to logarithmic phase and photographed at a magnification of ×40 with DIC optics. (B) Cells of the indicated genotype, transformed with KB1897, were grown to logarithmic phase and analyzed by Western blotting with an anti-Myc (9E10) monoclonal antibody. (C) Cells of the indicated genotype, transformed with the 3× HA-URA3 construct integrated after CaCDC53, were grown to logarithmic phase and analyzed by Western blotting with an anti-HA (HA.11) monoclonal antibody. Anti-β-actin was used as a loading control (lower panel). For quantitation, the intensities of the bands of neddylated and unneddylated CaCdc53 were summed, divided by the actin band intensity, and normalized to the quantitation in the wild-type background. (D) [35S]methionine pulse-chase analysis of the same strains as in panel B. Cells were grown to logarithmic phase, induced for 15 min with 0.1 mM copper, labeled for 10 min with [35S]methionine, and chased with cold methionine. (E) Graph showing the fraction of neddylated CaCdc53 versus time, as derived from phosphorimager quantitation of the experiment shown in panel D.
Western blot analysis of CUP1p-Myc-CaCDC53 further revealed an increased fraction of neddylated Myc-CaCdc53 in the Cajab1−/− deneddylase mutant, as expected, but also, unexpectedly, a slight decrease in the fraction of neddylated Myc-CaCdc53 in the Catip120−/− mutant (38% versus 51% in the wild type) (Fig. 6B). In the strain set expressing the CaCdc53-3xHA allele, overall levels of neddylation were lower, and in the Catip120−/− strain in particular, neddylation was barely detectable (Fig. 6C).
Whereas Western blotting revealed the steady-state distribution of neddylated versus unneddylated CaCdc53, the pulse-chase experiment shown in Fig. 6D enabled us to follow the kinetics of neddylation of newly synthesized Myc-CaCdc53. The fraction of neddylated cullin took some 60 min to approach a steady-state level of 50% in the wild-type background (Fig. 6E). In the Cajab1−/− mutant, as expected, neddylation occurred faster and reached a higher level before reaching a plateau at over 80%. The kinetics of neddylation in the Catip120−/− mutant, however, was more complex. The initial fraction of neddylated Myc-CaCdc53 observable right after labeling was slightly but reproducibly (in 2 independent experiments) higher in the Catip120−/− strain. Subsequently, neddylation in the Catip120−/− strain proceeded more slowly and eventually reached a plateau at a level just below that of the wild-type strain (Fig. 6E), in accordance with the lower steady-state fraction of neddylated CaCdc53 detected by Western blotting in the Catip120−/− strain (Fig. 6B).
DISCUSSION
C. albicans as a model system for studying the SCF ubiquitin ligase.
SCF complexes represent the most versatile class of ubiquitin ligases. In spite of the availability of crystal structures which provide high-resolution pictures of the cullin in association with the other SCF components (49, 70) and with CAND1 (24), as well as the effects of cullin neddylation (18), questions remain regarding the mechanics of SCF ubiquitin ligase activity and, notably, regarding the interrelated roles of the SCF modifiers Nedd8 and CAND1. It is likely that the roles of neddylation, deneddylation, and CAND1 are related to the dynamics of SCF function in the cellular context, and these roles may thus be studied usefully in a genetic model system. Here we identified and analyzed the functions of the SCF complex and of NEDD8/Rub1 and CAND1/Tip120 in the pathogenic yeast Candida albicans. C. albicans might be particularly suited to the study of SCF modifiers because unlike S. cerevisiae, it does appear to possess a clear CAND1 homolog, based on sequence homology and functional analysis, and because the C. albicans RUB1 deletion mutant, while viable, still exhibits a detectable morphological and biochemical phenotype.
In order to establish C. albicans as a model for the study of SCF function, we complemented previous studies of individual F-box proteins with an analysis of the C. albicans Cul1 homolog CaCDC53. Since CaCDC53, like its S. cerevisiae counterpart, was found to be essential, we created a conditional mutant by knocking in the point mutation of the Sccdc53-1 allele into the homologous position of CaCDC53. This approach may prove generally useful for the study of essential genes in C. albicans. Trunk et al. (62) recently applied an alternative approach, using a conditional promoter, to analyze the effect of CaCdc53 depletion. They found that the CaCdc53-depleted cells arrested with a pseudohypha-like morphology, rather than the hyphal morphology shown for the Cacdc53-1 mutant (Fig. 1B). This difference could be due to the different incubation temperatures at which the two experiments were performed (30°C versus 37°C) or to different levels of residual CaCdc53 activity remaining in both cases. Interestingly, they predicted, based on global transcription analysis of the CaCdc53-depleted cells, that CaGcn4 is a likely target of CaCdc53-mediated degradation (62), a prediction that was borne out by our experiment (Fig. 1C).
Dissociation of CAND1 from cullin in the absence of neddylation.
Expression of the C. albicans CDC53 sequence ortholog in S. cerevisiae enabled us to confirm its functional homology. Furthermore, by revealing the genetic interaction between CaCdc53 and CaTip120 in S. cerevisiae, this ectopic reconstitution indicated that these proteins interact specifically in vivo (Fig. 3). The relation of CaTip120 toxicity to neddylation in S. cerevisiae, i.e., the hypersensitivity of the strain to CaTip120 in the absence of neddylation, further correlated with the antagonism between cullin neddylation and CAND1 binding that was characterized in vitro with the homologous human proteins (24, 43, 69). However, genetic analysis in C. albicans indicated that CaTip120 and neddylation (CaRub1) are both synergistic with SCF function in vivo, rather than antagonistic, similar to observations in other genetic systems (5, 14). It is possible that in S. cerevisiae, the high levels of CaTIP120 expression under the control of the GAL1 promoter turn CaTip120 into a repressor rather than an activator of SCF activity; however, we noted that ectopic overexpression of CaTip120 in C. albicans is not deleterious (our unpublished observations). Furthermore, expression of CaTIP120 from promoters that are considerably weaker than that of GAL1 did not relieve its toxicity in the S. cerevisiae CaCDC53 rub1Δ strain, let alone transform it into an SCF activator (our unpublished data). Thus, an alternative possibility is that an additional C. albicans factor(s) that is missing from S. cerevisiae turns CaTip120 into an activator, e.g., by allowing its dissociation from CaCdc53.
Indeed, recent reports show that in mammalian cell culture, CAND1 binds weakly to Cul1 in spite of low levels of neddylation (13), and inhibition of neddylation does not significantly increase the fraction of CAND1-bound cullin (3, 39), suggesting that neddylation cannot constitute the only factor limiting CAND1-cullin association. Similarly, our observation that CAND1 promotes SCF activity in C. albicans even in the Carub1−/− mutant background implies that removal of neddylation does not immediately lead to sequestration of the cullin in an inactive cullin-CAND1 complex. Thus, any model of SCF function should accommodate a mechanism for CAND1 association with and dissociation from the cullin in the absence of neddylation.
CAND1 promotes SCF activity in the absence of neddylation.
The Catip120−/− mutant phenotype matches the phenotypes of mutants in Arabidopsis thaliana CAND1/ETA2/HVE, in the sense that the phenotypes of the plant mutants are consistent with a decrease in SCF activity and thus that, in plants as well, CAND1 appears to promote SCF activity in vivo (1, 11, 14, 20). Among their phenotypes, mutants in AtCAND1 show reduced auxin signaling and stabilization of the SCFTIR1 substrate Axr2. Regarding the interaction between neddylation and CAND1, Chuang et al. (14) notably showed by double mutant analysis that the axr1-12 mutant, a hypomorphic mutant of the Rub1 activation enzyme, was essentially epistatic to eta2-1. The conclusion of this study was that the role of neddylation cannot be solely to counteract sequestration of the cullin subunit by CAND1. Whether CAND1 has a role in the absence of neddylation could not be addressed in these studies, because neddylation is essential in plants. We could address this question rigorously in C. albicans by deleting CaTIP120 in the Carub1−/− background. The increased severity of the double mutant phenotype, at the levels of both morphology and H2O2 sensitivity, suggests that Tip120/CAND1 in fact stimulates SCF activity independently of neddylation.
The complexity of the interaction between neddylation and CAND1 is underscored by the observation that CaCdc53 neddylation levels are reduced, rather than induced, in the absence of CaTip120 (Fig. 6). This effect is exacerbated with the CaCdc53-3xHA allele. In this construct, a 33-amino-acid extension is added to the C terminus of Cul1. We noticed that the C terminus of Cul1, which is highly conserved among all orthologs, is adjacent (<1 nm) to the neddylation site, according to the Cul1 crystal structure (24). Thus, the addition of the epitope tag sequence to the C terminus may partially hinder neddylation, although we cannot explain why CaCdc53-3xHA neddylation would be specifically sensitive to the loss of CaTip120.
Role of CAND1 in promoting SCF activity.
Neddylation was found to promote ubiquitination activity of the SCF complex (51). Deneddylation would thus be expected to reduce SCF activity; paradoxically, however, similar to the CAND1 mutants, mutants of the CSN complex showed reduced SCF activity in vivo (15, 57). Two alternative models were suggested to resolve the so-called “CSN/CAND1 paradox.” One is that deneddylation and cullin sequestration by CAND1 protect SCF components such as the cullin and F-box proteins from adventitious ubiquitination and degradation in the absence of substrate (16, 63, 67, 68). The second is that the neddylation/deneddylation cycle, together with CAND1, promotes the exchange of FBPs on the cullin platform (6, 15, 56).
We did not see evidence that either the cullin or the FBP was destabilized in any of our C. albicans mutants. To the contrary, the stability of CaGrr1, the specific receptor for CaCln3, was increased in all mutants tested. Thus, protection of the F-box protein by CAND1 cannot be invoked to explain the Catip120−/− phenotype, at least in the case of CaGrr1. However, our data are consistent with a role for neddylation and CAND1 in coordinating disassembly of SCF complexes and reassembly of alternative complexes, enabling exchange of the F-box protein component on the cullin. In the wild-type background, SCF activity is responsible for the basal level of CaGrr1 turnover, as evidenced by its stabilization in the Cacdc53-1 mutant, i.e., some level of CaGrr1 attrition may be a natural consequence of its incorporation into active ubiquitination complexes. Stabilization of CaGrr1 in the Carub1−/− and Catip120−/− mutants may thus reflect a defect in incorporation into the SCF complex, which would also explain stabilization of the substrate CaCln3.
The kinetics of CaCdc53 neddylation could similarly be explained by a role for CAND1 in SCF complex assembly. In the wild-type strain, the slow kinetics of Rub1-CaCdc53 conjugate formation suggests that cullin neddylation may depend—at least in part—on a rate-limiting event, such as, e.g., incorporation of the cullin into an active SCF complex. The more complex kinetics of CaCdc53 neddylation in the Catip120−/− mutant, with a higher initial neddylation rate followed by a slower second step of neddylation, may reflect two different modes of cullin neddylation: (i) initial neddylation of newly synthesized, uncomplexed cullin, which may normally be held in check by CaTip120/CAND1 binding and, in the Catip120−/− mutant, may directly reflect the balance of neddylation and deneddylation activity on the free cullin; and (ii) neddylation of the cullin as a consequence of incorporation into active SCF complexes, which in the Catip120−/− strain may be partially inhibited due to slower FBP exchange and SCF complex formation. The proposed role of CaTip120/CAND1 in SCF complex remodeling may also explain the H2O2 sensitivity of the Catip120−/− strain: to the extent that FBP exchange may acquire added importance under changing external conditions, the sensitivity of the Catip120−/− strain to H2O2 stress might be due to a failure to assemble specific SCF complexes required under these circumstances.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mike Tyers for the S. cerevisiae cdc53-1 strain, Ariel Stanhill for anti-FLAG antiserum, and Sara Selig for a critical reading of the manuscript.
This work was supported by grants from the U.S.-Israel Binational Science Foundation and the Israel Science Foundation to D.K.
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Alonso-Peral MM, et al. 2006. The HVE/CAND1 gene is required for the early patterning of leaf venation in Arabidopsis. Development 133: 3755–3766 [DOI] [PubMed] [Google Scholar]
- 2. Atir-Lande A, Gildor T, Kornitzer D. 2005. Role for the SCF(CDC4) ubiquitin ligase in Candida albicans morphogenesis. Mol. Biol. Cell 16: 2772–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bennett EJ, Rush J, Gygi SP, Harper JW. 2010. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143: 951–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bostick M, Lochhead SR, Honda A, Palmer S, Callis J. 2004. Related to ubiquitin 1 and 2 are redundant and essential and regulate vegetative growth, auxin signaling, and ethylene production in Arabidopsis. Plant Cell 16: 2418–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bosu DR, et al. 2010. C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues. Dev. Biol. 346: 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bosu DR, Kipreos ET. 2008. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butler DK, et al. 2006. The GRR1 gene of Candida albicans is involved in the negative control of pseudohyphal morphogenesis. Fungal Genet. Biol. 43: 573–582 [DOI] [PubMed] [Google Scholar]
- 8. Cardozo T, Pagano M. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell. Biol. 5: 739–751 [DOI] [PubMed] [Google Scholar]
- 9. Chamovitz DA, et al. 1996. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86: 115–121 [DOI] [PubMed] [Google Scholar]
- 10. Chen Y, McPhie DL, Hirschberg J, Neve RL. 2000. The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S-M checkpoint and causes apoptosis in neurons. J. Biol. Chem. 275: 8929–8935 [DOI] [PubMed] [Google Scholar]
- 11. Cheng Y, Dai X, Zhao Y. 2004. AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol. 135: 1020–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chew EH, Poobalasingam T, Hawkey CJ, Hagen T. 2007. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells—evidence for cullin dimerization. Cell. Signal. 19: 1071–1080 [DOI] [PubMed] [Google Scholar]
- 13. Chua YS, Boh BK, Ponyeam W, Hagen T. 2011. Regulation of cullin RING E3 ubiquitin ligases by CAND1 in vivo. PLoS One 6: e16071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chuang HW, Zhang W, Gray WM. 2004. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell 16: 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cope GA, Deshaies RJ. 2003. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114: 663–671 [DOI] [PubMed] [Google Scholar]
- 16. Cope GA, Deshaies RJ. 2006. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cope GA, et al. 2002. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298: 608–611 [DOI] [PubMed] [Google Scholar]
- 18. Duda DM, et al. 2008. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91: 221–230 [DOI] [PubMed] [Google Scholar]
- 20. Feng S, et al. 2004. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16: 1870–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134: 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galan JM, Peter M. 1999. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. U. S. A. 96: 9124–9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gildor T, Shemer R, Atir-Lande A, Kornitzer D. 2005. Coevolution of cyclin pcl5 and its substrate gcn4. Eukaryot. Cell 4: 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldenberg SJ, et al. 2004. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119: 517–528 [DOI] [PubMed] [Google Scholar]
- 25. Haase SB, Reed SI. 2002. Improved flow cytometric analysis of the budding yeast cell cycle. Cell Cycle 1: 132–136 [PubMed] [Google Scholar]
- 26. Handeli S, Weintraub H. 1992. The ts41 mutation in Chinese hamster cells leads to successive S phases in the absence of intervening G2, M, and G1. Cell 71: 599–611 [DOI] [PubMed] [Google Scholar]
- 27. Harari-Steinberg O, Chamovitz DA. 2004. The COP9 signalosome: mediating between kinase signaling and protein degradation. Curr. Protein Pept. Sci. 5: 185–189 [DOI] [PubMed] [Google Scholar]
- 28. He Q, Cheng P, He Q, Liu Y. 2005. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 19: 1518–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones D, Crowe E, Stevens TA, Candido EP. 2002. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol. 3: RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. 1999. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13: 2928–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawakami T, et al. 2001. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20: 4003–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim SH, Kim HJ, Kim S, Yim J. 2010. Drosophila Cand1 regulates Cullin3-dependent E3 ligases by affecting the neddylation of Cullin3 and by controlling the stability of Cullin3 and adaptor protein. Dev. Biol. 346: 247–257 [DOI] [PubMed] [Google Scholar]
- 33. Kornitzer D. 2002. Monitoring protein degradation. Methods Enzymol. 351: 639–647 [DOI] [PubMed] [Google Scholar]
- 34. Kurz T, et al. 2008. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell 29: 23–35 [DOI] [PubMed] [Google Scholar]
- 35. Kurz T, et al. 2005. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435: 1257–1261 [DOI] [PubMed] [Google Scholar]
- 36. Kus BM, Caldon CE, Andorn-Broza R, Edwards AM. 2004. Functional interaction of 13 yeast SCF complexes with a set of yeast E2 enzymes in vitro. Proteins 54: 455–467 [DOI] [PubMed] [Google Scholar]
- 37. Lammer D, et al. 1998. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12: 914–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laplaza JM, Bostick M, Scholes DT, Curcio MJ, Callis J. 2004. Saccharomyces cerevisiae ubiquitin-like protein Rub1 conjugates to cullin proteins Rtt101 and Cul3 in vivo. Biochem. J. 377: 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee JE, et al. 2011. The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol. Cell. Proteomics 10: M110.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li WJ, et al. 2006. The F-box protein Grr1 regulates the stability of Ccn1, Cln3 and Hof1 and cell morphogenesis in Candida albicans. Mol. Microbiol. 62: 212–226 [DOI] [PubMed] [Google Scholar]
- 41. Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. 1998. A novel protein modification pathway related to the ubiquitin system. EMBO J. 17: 2208–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4: 728–735 [DOI] [PubMed] [Google Scholar]
- 43. Liu J, Furukawa M, Matsumoto T, Xiong Y. 2002. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell 10: 1511–1518 [DOI] [PubMed] [Google Scholar]
- 44. Liu Y, Mimura S, Kishi T, Kamura T. 2009. A longevity protein, Lag2, interacts with SCF complex and regulates SCF function. EMBO J. 28: 3366–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Loeb JD, Sepulveda-Becerra M, Hazan I, Liu H. 1999. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol. Cell. Biol. 19: 4019–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maytal-Kivity V, Piran R, Pick E, Hofmann K, Glickman MH. 2002. COP9 signalosome components play a role in the mating pheromone response of S. cerevisiae. EMBO Rep. 3: 1215–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Min KW, et al. 2003. TIP120A associates with cullins and modulates ubiquitin ligase activity. J. Biol. Chem. 278: 15905–15910 [DOI] [PubMed] [Google Scholar]
- 48. Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4: 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112: 243–256 [DOI] [PubMed] [Google Scholar]
- 50. Osaka F, et al. 2000. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19: 3475–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. 2004. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23: 1985–1997 [DOI] [PubMed] [Google Scholar]
- 52. Patton E, et al. 1998. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 12: 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Patton EE, Willems AR, Tyers M. 1998. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14: 236–243 [DOI] [PubMed] [Google Scholar]
- 54. Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 6: 9–20 [DOI] [PubMed] [Google Scholar]
- 55. Saha A, Deshaies RJ. 2008. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. 2009. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol. Cell 35: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schwechheimer C. 2004. The COP9 signalosome (CSN): an evolutionary conserved proteolysis regulator in eukaryotic development. Biochim. Biophys. Acta 1695: 45–54 [DOI] [PubMed] [Google Scholar]
- 58. Seol JH, Shevchenko A, Deshaies RJ. 2001. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3: 384–391 [DOI] [PubMed] [Google Scholar]
- 59. Shieh JC, White A, Cheng YC, Rosamond J. 2005. Identification and functional characterization of Candida albicans CDC4. J. Biomed. Sci. 12: 913–924 [DOI] [PubMed] [Google Scholar]
- 60. Siergiejuk E, et al. 2009. Cullin neddylation and substrate-adaptors counteract SCF inhibition by the CAND1-like protein Lag2 in Saccharomyces cerevisiae. EMBO J. 28: 3845–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91: 209–219 [DOI] [PubMed] [Google Scholar]
- 62. Trunk K, et al. 2009. Depletion of the cullin Cdc53p induces morphogenetic changes in Candida albicans. Eukaryot. Cell 8: 756–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wee S, Geyer RK, Toda T, Wolf DA. 2005. CSN facilitates cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 7: 387–391 [DOI] [PubMed] [Google Scholar]
- 64. Weissman Z, Kornitzer D. 2004. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol. Microbiol. 53: 1209–1220 [DOI] [PubMed] [Google Scholar]
- 65. Whiteway M, Bachewich C. 2007. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61: 529–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Willems AR, Schwab M, Tyers M. 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695: 133–170 [DOI] [PubMed] [Google Scholar]
- 67. Wu JT, Chan YR, Chien CT. 2006. Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol. 16: 362–369 [DOI] [PubMed] [Google Scholar]
- 68. Wu JT, Lin HC, Hu YC, Chien CT. 2005. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat. Cell Biol. 7: 1014–1020 [DOI] [PubMed] [Google Scholar]
- 69. Zheng J, et al. 2002. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell 10: 1519–1526 [DOI] [PubMed] [Google Scholar]
- 70. Zheng N, et al. 2002. Structure of the Cul1-Rbx1-Skp1-F box-Skp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]
- 71. Zheng X, Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23: 1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhou P, Howley PM. 1998. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2: 571–580 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.