Abstract
The genus Trichoderma is one of the most widely used biological control agents of plant-pathogenic fungi. The main mechanism for survival and dispersal of Trichoderma is through the production of asexual spores (conidia). The transition from filamentous growth to conidiation can be triggered by light, nutrient deprivation, and mechanical damage of the mycelium. We conducted proteomic profiling analyses of Trichoderma atroviride after a blue light pulse. The use of two-dimensional electrophoresis (2-DE) and mass spectrometry (MS) analysis allowed us to identify 72 proteins whose expression was affected by blue light. Functional category analysis showed that the various proteins are involved in metabolism, cell rescue, and protein synthesis. We determined the relationship between mRNA levels of selected genes 30 min after a light pulse and protein expression levels at different times after the pulse and found this correlation to be very weak. The correlation was highest when protein and mRNA levels were compared for the same time point. The transcription factors BLR-1 and BLR-2 are vital to the photoconidiation process; here we demonstrate that both BLR proteins are active in darkness and affect several elements at both the transcript and protein levels. Unexpectedly, in darkness, downregulation of proteins prevailed in the Δblr-1 mutant, while upregulation of proteins predominated in the Δblr-2 mutant. Our data demonstrate that the BLR proteins play roles individually and as a complex.
INTRODUCTION
Trichoderma spp., endophytic plant symbionts that are widely used to control plant diseases, have been used for decades to improve plant growth and yield (27, 38). These fungi have the capacity to attack a range of economically important aerial and soilborne plant pathogens by means of enzymatic lysis, antibiotic production, and competition for space and nutrients (17, 28). Another mechanism of plant protection observed upon application of Trichoderma is the induction of plant defense responses (55, 63). One of the most-studied species of the genus Trichoderma is Trichoderma atroviride, a mycoparasite widely used as a biological control agent. Its main mechanism of survival and dispersal is through the development of asexual spores (conidia). In this fungus, conidiation is stimulated by several factors, such as pH, light, nutrient exhaustion, and mechanical damage (13, 14).
Light affects fungi by influencing their germination, vegetative and reproductive growth, metabolism, morphology, and circadian rhythms (10, 56, 57). In the interest of providing enhanced biocontrol agents, intensive research is still being conducted to understand the molecular factors that control the morphogenetic switch from mycelia to conidia.
In total darkness, when nutrients are not limited, T. atroviride grows continuously as mycelia. Under these conditions, a short pulse of blue light given to a growing colony induces a synchronous mechanism that leads to conidiation. The complete conidiogenesis process in T. atroviride occurs within 24 h after light exposure. From traditional photobiology studies using T. atroviride as a model, a single photoreceptor was proposed to induce conidiation after a blue light pulse (52). However, recent biochemical and molecular data suggest the involvement of at least two perception systems regulating this response (13, 14, 47). Biochemical changes triggered by light in dark-grown colonies of Trichoderma include shifts in membrane potential and ATP levels, a transitory biphasic oscillation in intracellular cyclic AMP levels, activation of adenylyl cyclase, and phosphorylation of proteins (52).
In Neurospora crassa, a complex formed by the white collar (WC) proteins controls all known light responses. White collar 1 (WC-1) is both a GATA-type zinc finger transcription factor and a flavin adenine dinucleotide (FAD)-binding photoreceptor (2, 23). Using their PAS domains (16), WC-1 and WC-2, another GATA-type zinc finger transcription factor, form the white collar complex (WCC), which controls most light responses in this fungus (15). Besides its capacity to initiate light responses in Neurospora, the WCC functions in the dark as an essential circadian clock component, controlling the expression of the frequency (frq) gene (36). Proteins similar to the blue light photoreceptor WC-1 have been identified in most fungal models, highlighting its importance (32). Vivid (VVD), another photoreceptor of UV/blue light, acts as a common repressor of WCC activity for most light responses, leading to photoadaptation (54).
Blue light regulator 1 (BLR-1) and BLR-2 are the T. atroviride homologues of the white collar proteins of N. crassa. As in Neurospora, they most likely act as a complex (BLRC), as deletion of either of them results in loss of known light responses. Therefore, both proteins are essential for photoconidiation and gene expression modulated by blue light (13, 47). A critical difference of the BLRC compared with the WCC is its capacity to activate and repress gene expression (47). Additionally, the BLRC appears to have functions in the dark that are not related to circadian rhythms, since strains with mutations in the corresponding genes do not undergo conidiation in response to carbon starvation in darkness (14).
In T. atroviride, red light causes a reduction of vegetative growth and affects transcriptional regulation of some genes, suggesting the participation of additional photoreceptors (13). Several genes encoding putative photoreceptors have been identified in the T. atroviride genome, such as the genes for a CPD (cyclobutane pyrimidine dimer) photolyase, a cryptochrome-DASH, a 6-4 photolyase, a phytochrome, and an opsin (52).
The use of modern sequencing technologies makes the establishment of comprehensive and quantitative mRNA expression profiles feasible for species with sequenced genomes. Microarray analysis of transcript profiles indicated that approximately 2.8% of T. atroviride genes are responsive to light (47). Furthermore, a recent high-throughput pyrosequencing approach allowed the identification of 331 transcripts regulated by white light and 204 transcripts responsive to blue light, suggesting the contributions of several photoreceptors (U. E. Esquivel-Naranjo, M. A. Hernandez-Oñate, and A. Herrera-Estrella, submitted for publication). Although mRNA expression profiles are indispensable, by themselves they are only part of the quantitative description of biological systems. Posttranscriptional mechanisms controlling protein half-lives, translation rates, and subcellular localization (41, 42, 51, 58, 62) play essential regulatory functions. For example, several polypeptides vary in abundance before and after structural changes are visible in Trichoderma strains after treatment with blue light (4).
To gain further insight into the blue light response at the level of the proteome, we characterized the protein expression profile of T. atroviride following a blue light pulse. This proteomic analysis allowed us to identify and categorize a number of dynamic proteins involved in the blue light response in T. atroviride. Additionally, this study explored the correlation between transcript and protein expression levels in a particular metabolic state, allowing us to detect the weak correlation between these two features and indicating that both analyses are necessary and complementary. Furthermore, we analyzed the blue light response in BLR-1 and BLR-2 mutants, which showed different protein profiles even in darkness, suggesting individual functions of these proteins under the study conditions. Additionally, blue light provoked a proteomic response in the deletion mutants, reinforcing the notion of the existence of an additional light perception system. Finally, we analyzed the transcriptional regulation of several BLR-dark-regulated genes, finding that deletion of either blr-1 or blr-2 differentially modified the expression levels of several genes, supporting the view that BLR proteins are essential for the blue light response not only by forming the BLRC but also by playing important roles individually.
MATERIALS AND METHODS
Strains, culture media, and reagents.
Trichoderma atroviride wild-type (WT) strain IMI 206040 and the Δblr-1 and Δblr-2 mutant strains (13) were maintained on potato dextrose agar (PDA) plates (Difco, Sparks, MD). Urea, thiourea, 425- to 600-μm glass beads, trichloroacetic acid (TCA), polyvinyl-polypyrrolidone (PVPP), and diethyl pyrocarbonate (DEPC) were purchased from Sigma (St. Louis, MO). Thirteen-centimeter IPG dry strips (pH 3 to 10 and pH 4 to 7), IPG buffers (pH 3 to 10 and pH 4 to 7), dry strip cover fluid, electrode papers, iodoacetamide, and bromophenol blue were obtained from GE Healthcare (Uppsala, Sweden). Acrylamide-bisacrylamide (29:1) (30%) and protein assay dye reagent were obtained from Bio-Rad (Hercules, CA). CHAPS [3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate] was purchased from USB (Cleveland, OH). Ultrapure agarose, Tris, glycine, glycerol, dithiothreitol (DTT), TRIzol reagent, and phenol-chloroform-isoamyl alcohol (25:24:1) were obtained from Invitrogen (Carlsbad, CA). Sypro Ruby was obtained from Molecular Probes (Eugene, OR). SDS, hydrochloric acid, formamide, and β-mercaptoethanol were obtained from Merck (Darmstadt, Germany). Acetone, formaldehyde, glacial acetic acid, ethanol, and methanol were obtained from Karal (Guanajuato, Mexico). Ammonium persulfate and TEMED (N,N,N′,N′-tetramethylethylenediamine) were obtained from Gibco BRL (Gaithersburg, MD). MOPS (morpholinepropanesulfonic acid) and bovine serum albumin were purchased from Roche (Mannheim, Germany). Trypsin Gold was obtained from Promega (Madison, WI). All reagents and enzymes for reverse transcription-PCR (RT-PCR) and conventional PCR were obtained from Invitrogen (Carlsbad, CA).
Culture conditions and blue light induction assays.
T. atroviride wild-type and Δblr-1 and Δblr-2 mutant strains were grown on PDA plates at 28°C and exposed to white light or damaged with a scalpel to induce conidiation. A drop containing 1,000 conidia was inoculated at the center of a PDA plate as a preculture and then incubated in the dark for 36 h at 28°C. Fresh PDA plates covered with cellophane sheets were inoculated with an agar plug taken from the growing edge of the dark-developed colony, covered with aluminum foil, and incubated for 36 h under the same conditions as the inoculums. Thirty-six-hour-old colonies were subjected to a brief pulse of blue light (1,200 μmol m−2 s−1) in a light-emitting diode (LED) chamber (Percival) at 28°C and plates taken back to darkness for 30, 45, 90, or 150 min or 8 or 16 h. Only the perimeter of the colonies exposed to light was collected. Dark-growing colonies were also sampled and used as controls. Samples of mycelium were collected, frozen immediately in liquid nitrogen, and stored in a deep freezer for protein and RNA extraction.
Sample preparation and 2D.
Frozen mycelium was ground until a fine powder was obtained and was then transferred to a 50-ml centrifuge tube. Extraction solution (10% TCA, 2% β-mercaptoethanol, 1% PVPP) and 1 volume of sterile glass beads were added. Samples were mixed by 10 cycles of 1 min of vortexing and 1 min of incubation in an ice bath. Extracted proteins were recovered by centrifuging samples for 10 min at 5,500 rpm and 4°C. Clarified supernatants were transferred to a fresh tube and proteins allowed to precipitate for 4 h at −20°C. Proteins were recovered by centrifugation for 20 min at 18,000 rpm at 4°C and then washed with absolute acetone and absolute ethanol. After washes, proteins were allowed to air dry, resuspended in rehydrating solution (7 M urea, 2 M thiourea, 2% CHAPS, 65 mM DTT, 1% IPG buffer [pH 3 to 10 or 4 to 7]) by mixing in a vortex machine, and transferred to a microcentrifuge tube. Proteins were quantified using the method of Bradford (8), using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) and a working wavelength of 595 nm. A total of 150 μg of sample was loaded onto 13-cm IPG dry strips with a pH range of 3 to 10 or 4 to 7 in a reswelling tray (GE Healthcare), covered with 3 ml of dry strip cover fluid, and allowed to rehydrate passively for 16 h. Rehydrated strips were placed on an IPGphor electrofocusing unit (GE Healthcare). Electrofocusing for each pH range was carried out with the following programs: for the pH range of 3 to 10, step 1 was 500 V for 500 V-h, followed by gradient 1 (1,000 V for 800 V-h) and gradient 2 (8,000 V for 11,300 V-h), and step 2 was 8,000 V for 2,900 V-h; for the pH range of 4 to 7, step 1 and gradients 1 and 2 were the same as those described above, and step 2 was different (8,000 V for 5,400 V-h). The protein strips were equilibrated in two steps: 5 ml of denaturing solution (6 M urea, 50 mM Tris-HCl, pH 8.8, 30% glycerol, 2% SDS, 0.01% DTT) was added and the strips gently stirred for 15 min, and the solution was discarded and replaced by 5 ml alkylating solution (6 M urea, 50 mM Tris-HCl, pH 8.8, 30% glycerol, 2% SDS, 0.025% iodoacetamide), with the strips gently stirred for 15 min. Equilibrated strips were placed on top of 25- by 20-cm 12.5% denaturing polyacrylamide gels. SDS-PAGE was carried out on an Ettan Dalt Six unit (GE Healthcare) with a constant current of 40 mA per gel until the bromophenol blue tracking dye reached the bottom of the gels. The gels were then fixated and stained with Sypro Ruby for 30 min. Destained gels were imaged using a GelDoc photo documenter (Bio-Rad), and acquired images were converted into TIFF files. Triplicates of each condition were obtained.
Image acquisition and identification of differentially expressed proteins.
Image analyses were carried out with Melanie 7 software (GeneBio, GE Healthcare, Swiss-Prot). Spot detection parameters were adjusted for each triplicate. Total spots per gel, the correlation between replicates, and the spot percent volume (%vol) were obtained. The experimental design used in this study allowed us to guarantee that all protein variations could be attributed to light exposure. Spot %vol values were considered the most adequate quantitative parameter because they represent the fraction of a single spot with respect to the global volume of all spots present on each gel; hence, analysis was carried out with normalized values of spot intensities. Only those spot sets present/absent in all images of the same condition were considered for the analysis, as well as the behavior in terms of pixel Gauss distribution to avoid misdetection of artifacts. Pair comparisons were carried out between each time after blue light pulse and the darkness condition by determining the %vol mean for each spot set and calculating the corresponding ratio (time after blue light pulse/darkness [ABLP/D]). Data are listed as mean ABLP/D values ± standard deviations (SD). Because the main variable affecting mean values for the ABLP/D ratio is exposure to blue light, one-way analysis of variance (ANOVA) sufficiently satisfies the requirements of assessment of differential expression of paired comparisons, with P values of ≤0.05 indicating statistical significance. This direct statistical evaluation was thus performed, rendering a range of nonsignificant differences in protein abundance (0.66 ≤ x ≤ 1.5), to determine those %vol values that were statistically different.
In-gel digestion, MS analysis, and protein identification.
Protein spots were excised from the gels and transferred to a microcentrifuge tube containing 200 μl of sterile deionized water. Spots were destained and trypsin digested according to the manufacturer's recommendations. Identification of differentially expressed proteins by the peptide mass fingerprinting (PMF) method was carried out with a Voyager-DE Pro matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) instrument (Applied Biosystems) operated in reflector mode at 337 nm and 3Hz in the mass-to-charge (m/z) range of 600 to 4,000. Monoisotopic peaks of trypsin autodigestion (m/z 842.5100) and adrenocorticotropin (ACTH) 18-39 (m/z 2,465.1989) were used for internal calibration. The m/z ratio of each peak present in the mass spectra obtained was used to identify each spot, using the online software Mascot (Matrix Science), which carries out a comparison of experimental m/z data introduced by the user with theoretical m/z values for the in silico trypsin-generated peptide collection of the annotated protein database derived from the T. atroviride genome sequencing project (http://genome.jgi-psf.org/Triat2/Triat2.home.html), based on the following parameters: mass tolerance of 50 ppm, maximum of 1 missed cleavage, oxidation at methionine residues, and fixed modification of carbamidomethylation of cysteine residues.
Comparison of protein and mRNA expression.
For comparison of protein and mRNA expression, we made use of data sets from 454 sequencing analyses recently conducted in our laboratory (Esquivel-Naranjo et al., submitted). Nonparametric Spearman's rank correlation values (Rs) between the proteome and the transcriptome were calculated with R software (46).
RNA extraction and Northern blotting.
Frozen mycelium samples treated as described above were ground with a mortar and pestle in liquid nitrogen and RNA extracted using TRIzol reagent following the manufacturer's recommendations. RNA samples from different dark-grown samples were separated in denaturing agarose gels, transferred to nylon membranes (GE Healthcare), and subjected to hybridization following standard procedures (50), using fragments ranging from 140 to 200 bp, amplified by PCRs corresponding to blr-dark-regulated genes, as probes and using the 28S rRNA gene as a loading control.
RESULTS
Effect of blue light on protein profile of T. atroviride.
As mentioned above, blue light exposure of T. atroviride elicits a fast and synchronized response at the transcriptional level, resulting in the production of green conidia. Therefore, we were interested in the response of T. atroviride to light at the protein level. The expression pattern of light-regulated genes has been described in previous studies, indicating that transcripts begin accumulating immediately after the light pulse, reaching maximum induction by 30 min and decreasing 120 min after light exposure (6, 13, 19, 47). Consequently, we decided to study the proteome response around this time, selecting 30, 45, 90, and 150 min after the light pulse, and to expand our proteome screening we analyzed two samples corresponding to later stages (8 and 16 h after the blue light pulse), when conidiophores start appearing.
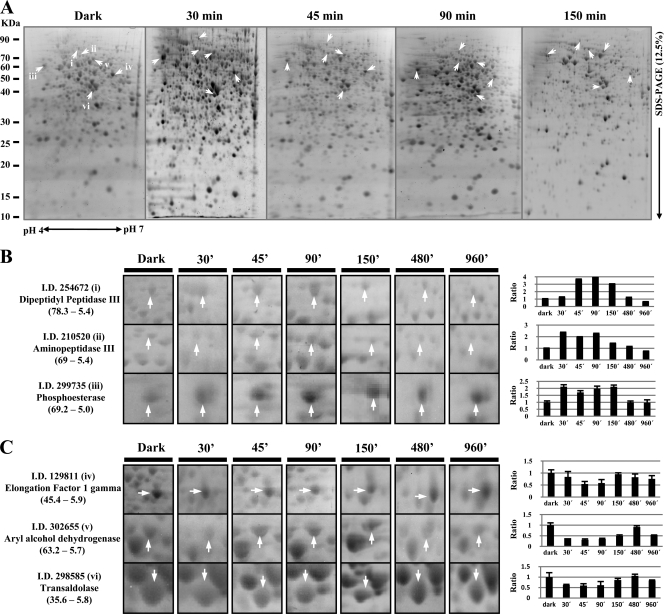
T. atroviride extracts obtained from mycelia containing soluble proteins were employed. Preliminary experiments using IPG strips (pH 3 to 10) showed that most of the proteins extracted under these conditions migrated in the pH range from 4 to 7 and had a molecular mass between 10 and 90 kDa (data no shown). Therefore, for the two-dimensional electrophoresis (2-DE) system, a 13-cm, pH 4 to 7 IPG strip combined with a vertical multigel electrophoresis system was used. For each sample, at least triplicate gels were used, and they were highly reproducible. Representative protein profiles corresponding to the early light responses are shown in Fig. 1A: an average of 800 protein spots were detected consistently in every gel, using Melanie, version 7.0, 2DE gel analysis software. Protein expression was considered altered after the blue light pulse if the mean spot density for replicate gels was significantly different (P < 0.05) from the mean spot density for replicate gels from dark growth sampling. For quantitative analysis of differently expressed proteins after the blue light pulse, protein spots were considered overexpressed if the ratios of their intensities were ≥1.5 and underexpressed if the ratios were ≤0.66.
Fig 1.
2-DE proteome analyses of T. atroviride strains subjected to a blue light pulse. (A) Total protein profiles. Proteins extracted from T. atroviride grown in the dark and 30, 45, 90, and 150 min after a light pulse were separated by 2-DE and visualized with Sypro Ruby stain. (B and C) Close-ups of several increased spots (B) and decreased spots (C) after the blue light pulse (from left to right, dark conditions and 30, 45, 90, 150, 480, and 960 min after the light pulse). The right panels show relative abundances in terms of the fold change of each spot in the gel (x axis, sampling time; y axis, fold change). Arrows indicate the proteins expressed differentially in response to blue light. Ratio, relationship between treatment and dark control protein levels. Numbers in parentheses indicate the molecular weight and isoelectric point, respectively.
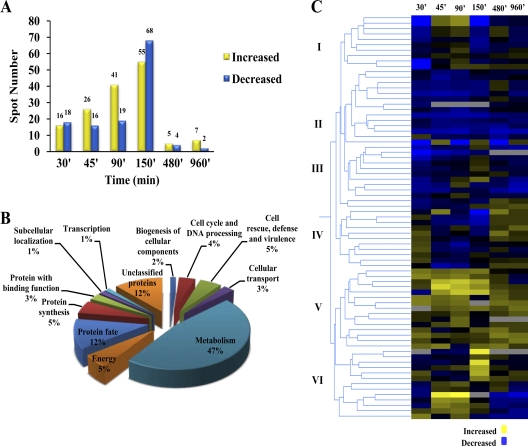
Two hundred proteins were selected for identification by mass spectrometry. Examples of differentially regulated protein spots are shown in Fig. 1B and C. Among the spots visualized in each gel, quantitative intensity analysis and statistics revealed 34 protein spots whose levels were significantly altered 30 min after the light pulse versus the dark growth control. By 45 min, 42 protein spots were responsive, 60 spots were altered by 90 min, and 123 protein spots were altered by 150 min. Only 9 spots were altered 8 and 16 h after the blue light pulse (Fig. 2A), at which times aerial hyphae and conidiophores with immature conidia are observed.
Fig 2.
Proteomic profiling of T. atroviride after a blue light pulse. (A) Dynamics of differentially expressed proteins after a blue light pulse in T. atroviride. (B) Functional classification of identified proteins that showed significantly altered protein levels after a blue light pulse in T. atroviride, categorized according to the MIPS Functional Catalogue (FunCat description). The listed percentage corresponds solely to the proportion of identified proteins. (C) Protein clustering showing expression profiles of 72 proteins after a blue light pulse.
Cellular functions affected by blue light.
Following spot scission and in-gel digestion, differentially expressed proteins were analyzed by electrospray ionization tandem MS (ESI MS/MS) and MALDI MS. Proteins were identified on the basis of the observed peptide sequences obtained by PMF and de novo sequencing matching those found in the T. atroviride protein database (http://genome.jgi-psf.org/Triat2/Triat2.home.html). A total of 72 proteins were successfully identified and classified in terms of their cellular function. According to the MIPS Functional Catalogue (49), the majority of the identified proteins were classified in a least one functional category. The most highly represented proteins were those falling in the metabolism category (47%), such as proteins involved in carbohydrate metabolism, amino acid metabolism, and fatty acid metabolism. There were also proteins (12%) related to protein fate. Other differentially expressed proteins (5%) were involved in protein synthesis. The same percentage (5%) was estimated to be proteins involved in cellular energy production. Four percent of proteins were identified in the category of cell cycle and DNA processing. There were also proteins (5%) implicated in cell rescue, defense, and virulence. The remaining functional categories included a smaller number of proteins related to transcription, biogenesis of cellular components, and cellular transport, proteins with binding functions, and proteins involved in subcellular localization. Finally, among the identified proteins, the remainder (12%) could not be classified using this database (Fig. 2B; see Table S1 in the supplemental material).
A dynamic differential proteomic response is activated by blue light in T. atroviride.
To dissect the proteomic responses after a pulse of blue light, a protein clustering analysis was performed on 66 protein spots that were identified and found to be differentially regulated. We performed a hierarchical clustering search based on the relative abundances of these proteins through the time course analyzed. We made the assumption that proteins strictly responsive to light should behave similarly across time and may participate together in a multimolecule-driven physiological response. Two major clusters and six subclusters were identified, comprising downregulated (subclusters I, II, III, and IV) and upregulated (subclusters V and VI) responses to light (Fig. 2C).
We defined these protein clusters as molecular phenotypes representing characteristic proteomic responses at specific times after a light pulse. For the 9 proteins in subcluster I, downregulation was seen early, followed by upregulation, with a second downregulation peak. For most of the proteins (24 of 26 proteins) in subclusters II and III, downregulation predominated throughout the entire time analyzed, with a couple of exceptions in subcluster III. Proteins in subcluster IV represent a rapid, transient positive response to light: they showed increased abundance 30 min after the light pulse, and their abundance dropped by 45 min and increased again at times when morphological changes become evident. In subcluster V, comprised of 15 proteins, upregulation prevailed at all times. Thirteen proteins belonging to subcluster VI showed a sharp decrease 30 min after the light pulse, increased during the early phase of the response to light (45 to 150 min), and decreased at the stages where morphological changes are visible. Protein identification and fold changes throughout the time course for each protein identified are shown in Table S2 in the supplemental material.
A low correlation between transcriptome and proteome data tendencies was detected after the light pulse was given.
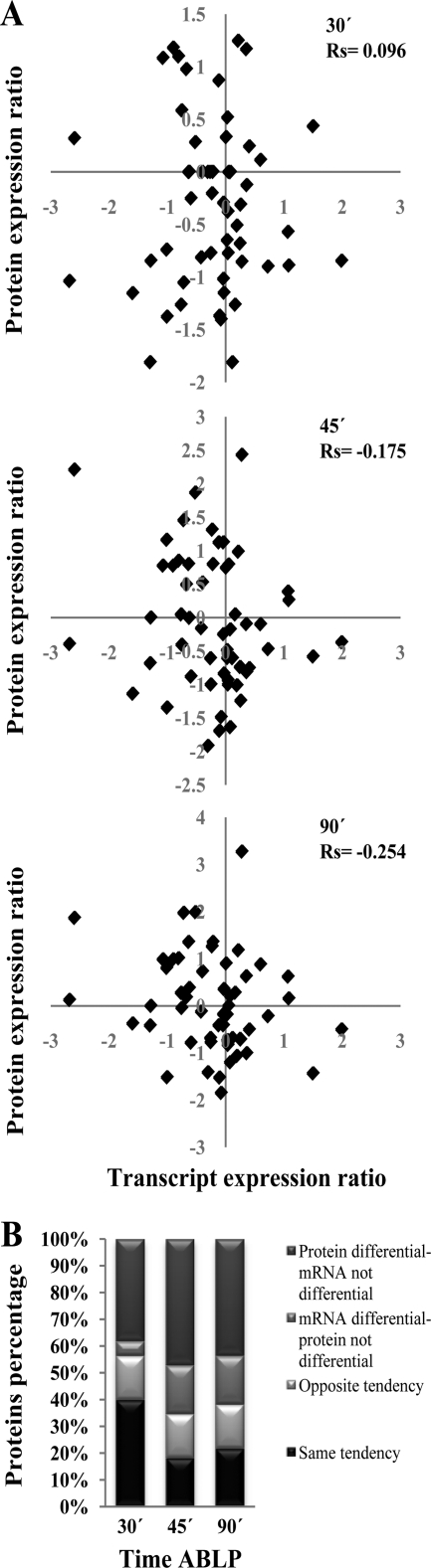
The proteome of a cell is the product of controlled biosynthesis and is regulated largely (not exclusively) by transcription; conversely, the transcriptome can be regarded as a sensitive readout of the proteome or the biochemical status of the cell. Hence, the proteome and the transcriptome are interconnected in an extremely complex way. In order to acquire a deeper understanding of the complexity between these two processes, we monitored expression at the RNA level 30 min after the blue light pulse and measured protein levels at various times after the pulse (see Table S3 in the supplemental material). We used this set of differential expression profiles as a tool to address one major question: does protein expression correlate with transcriptional regulation for the majority of differentially expressed proteins? To answer this question, we used previously generated RNA pyrosequencing data and assessed differential expression at the RNA level for those proteins that were identified as differentially expressed in T. atroviride 30, 45, and 90 min after a light pulse. The ratios of protein and RNA expression showed an overall weak correlation along time (Fig. 3A), with a low correlation 30 min after the blue light pulse (Rs = 0.096) (only 40% of total protein and mRNA tested followed the same trend) and a decreasing correlation at 45 min (Rs = −0.175) and 90 min (Rs = −0.254), with only 18 and 21% of compared ratios, respectively, changed in the same fashion (Fig. 3B; see Table S3). Classification of proteins by functional categorization revealed that several proteins involved in metabolism and protein fate had the best correlation between protein and transcript regulation. Sixteen percent of the protein changes occurred in the opposite direction of that observed for mRNA levels after 30, 45, and 90 min, but they did not correspond to the same proteins in all three cases. Metabolism was the functional category to which almost all proteins following this trend corresponded. It is important that many proteins identified as differentially regulated by light are not regulated by this stimulus at the transcriptional level, as the corresponding transcripts showed no changes after light exposure, suggesting that an important posttranscriptional regulation takes places in the photoconidiation process. Conversely, several genes transcriptionally regulated by light were not affected at the protein level by this environmental signal, reinforcing the notion of the existence of several checkpoints at different levels to tightly control light responses.
Fig 3.
Correlation between transcriptomic and proteomic data for selected proteins. (A) RNA expression ratios at 30 min versus protein expression ratios 30, 45, and 90 min after a blue light pulse (values are expressed as log2 values). (B) Distribution of differentially expressed proteins based on their behavior with respect to the corresponding transcript at the indicated time points after a blue light pulse.
The putative photoreceptor complex proteins BLR-1 and BLR-2 differentially regulate protein levels not only after a blue light pulse but also in the dark.
We also explored the responses of Δblr-1 and Δblr-2 deletion mutants to blue light in a proteomic framework to identify protein patterns that represent responses of the fungus independent of the BLRC. In T. atroviride, the response to blue light is mediated primarily via the blue light regulators BLR-1 and BLR-2. However, it has been proposed that T. atroviride has a second, as yet undiscovered light receptor. Furthermore, the existence of two types of light response has been demonstrated by microarray analysis of gene expression of blr deletion mutants, showing that there are blr-dependent and unpredicted blr-independent responses (47).
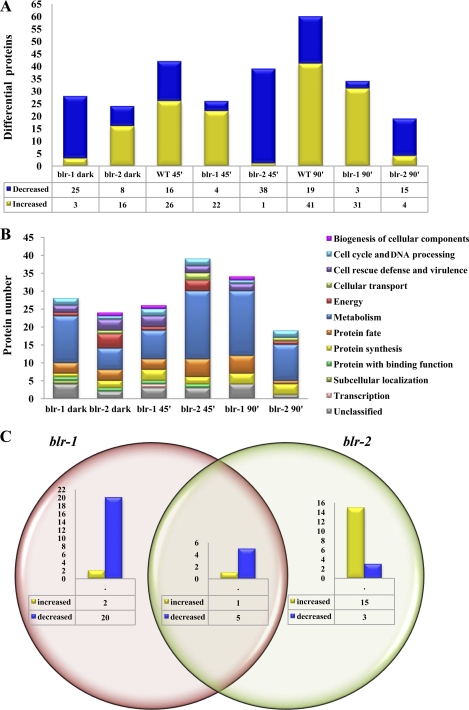
Therefore, to decipher general behavior at the protein level, protein profiles of the blr deletion mutants (13) grown in the dark and 45 min and 90 min after a light pulse were analyzed in 2-DE gels. The analysis of the corresponding sets of gels revealed an average of 800 protein spots stained with Sypro Ruby. The results from these comparisons are shown in Table S4 in the supplemental material. In the comparison between the WT and Δblr-1 strains in darkness, 29 differentially expressed proteins were found, most of them downregulated (25 proteins). Conversely, under the same conditions, 19 of 27 differentially expressed proteins detected were upregulated in the Δblr-2 strain. These results were unexpected, since the view is that these proteins act together in a complex to control light responses. Forty-five minutes after the blue light pulse, 26 proteins were differentially regulated in the Δblr-1 mutant: 22 were increased and 4 were decreased compared with the same strain grown in the dark. In the Δblr-2 mutant, 39 proteins were differentially expressed: 38 of them were downregulated 45 min after the light pulse. Similarly, 90 min after the light pulse, most of the differentially expressed proteins were upregulated in the Δblr-1 mutant, and most of them were downregulated in the Δblr-2 mutant (Fig. 4A). It is noteworthy that opposite patterns of protein accumulation were observed in the blr mutant strains. The identified differentially expressed proteins of the Δblr-1 and Δblr-2 mutants were categorized into several functional groups, including metabolism, cell rescue and defense, protein synthesis, energy production, and transcription, among others (Fig. 4B).
Fig 4.
Proteomic profiling of photoreceptor complex mutants of T. atroviride compared with the wild type in the dark and after a blue light pulse. (A) Differentially expressed proteins in the Δblr-1 and Δblr-2 mutants in the dark compared with the wild-type strain in the dark and differentially expressed proteins after a blue light pulse in the WT, Δblr-1, and Δblr-2 strains compared with themselves in the dark. (B) Functional classification of identified proteins that showed a significantly altered protein level (>1.5-fold) after a blue light pulse in the Δblr-1 and Δblr-2 mutants, categorized according to the MIPS Functional Catalogue (FunCat description). (C) Venn diagram of differentially expressed proteins observed in the blr deletion mutants in the dark.
Several proteins were differentially expressed in the dark and had the opposite behavior depending on the mutation. In order to further understand these data, the behavior of the differentially expressed proteins is depicted in more detail in Fig. 4C. In the Δblr-1 mutant, 28 proteins were differentially expressed, while in the Δblr-2 mutant, 24 proteins displayed differential behavior; 6 proteins were differentially regulated in both strains. In the Δblr-1 strain, 20 proteins were downregulated. Some of these are related to C compound and carbohydrate metabolism and other proteins related to protein processing. Other downregulated proteins in this mutant are involved in amino acid metabolism. Another group of proteins that decreased in the dark in the Δblr-1 mutant compared to the wild type are involved in fatty acid metabolism, the cytoskeleton, and defense, among other categories. In contrast, only two proteins were upregulated in the Δblr-1 mutant strain (one unclassified protein and another related to biosynthesis of histidine). Moreover, in the Δblr-2 strain, 18 proteins were differentially regulated, with 3 proteins related to protein synthesis, transcription, and the oxidative stress response being downregulated. In contrast with what was observed in the Δblr-1 mutant, the majority of the differentially expressed proteins in this mutant were upregulated. In total, 15 proteins showed increased levels. Several of them were linked to protein fate, and others were involved in sugar catabolism. Six proteins differentially regulated in both mutants were identified, one with increased levels involved in protein fate (aminopeptidase P) and five with decreased levels associated with energy, cell rescue and defense, the cell cycle, and DNA processing. These data indicate that differences in the proteome regulation level can be observed between the blr mutant strains and strongly indicate that BLR proteins have independent roles and also work at least partially as a complex in the dark.
Altered gene expression of the Δblr-1 and Δblr-2 mutants in the dark.
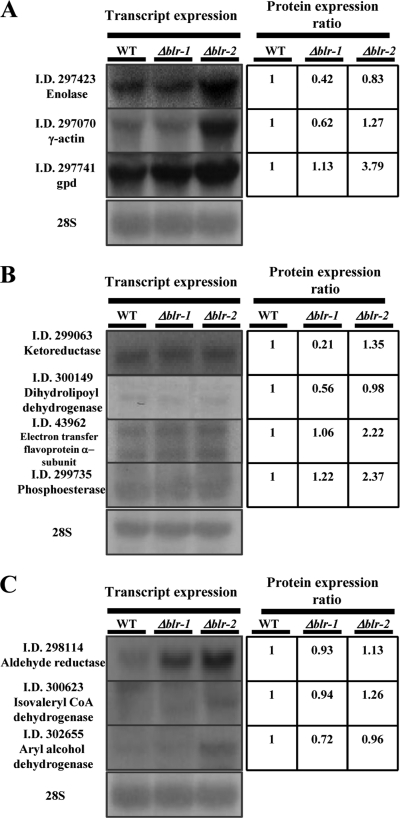
Based on the proteomic results, it is possible that BLR proteins might have independent roles yet function as a complex independently of light. The first issue we wanted to address was whether the observed changes in protein level in the dark were a reflection of changes occurring at the transcript level. Seven genes for which the corresponding proteins appeared to be either induced or repressed in the blr mutants relative to the wild type in the dark were analyzed by Northern blotting (Fig. 5A and B). In this analysis, we also included three genes that corresponded to proteins without significant variation relative to the WT in the dark (Fig. 5C). Figure 5A shows that whereas enolase and γ-actin were present at lower levels in the Δblr-1 mutant and no significant changes were observed in the Δblr-2 mutant, the corresponding transcripts were only slightly affected in the Δblr-1 mutant and were clearly elevated in the Δblr-2 mutant. In the case of glyceraldehyde-3-phosphate dehydrogenase, protein levels were found to be increased 3-fold in the Δblr-2 mutant; this change appeared to be a reflection of an increase in mRNA levels (Fig. 5A). When we analyzed ketoreductase and dihydrolipoyl dehydrogenase, two proteins present at clearly significantly lower levels in the Δblr-1 mutant, no differences were found at the mRNA level. Similarly, analysis of the electron transfer flavoprotein alpha subunit and phosphoesterase found both at higher levels in the Δblr-2 mutant, but no changes were detected at the mRNA level (Fig. 5B). Unexpectedly, in the case of aldehyde reductase, isovaleryl coenzyme A (CoA) dehydrogenase, and aryl alcohol dehydrogenase, with no changes observed at the protein level, all three showed higher transcript levels in the Δblr-2 mutant, whereas transcript levels were affected in the Δblr-1 mutant only for the gene encoding aldehyde reductase (Fig. 5C). These results provide further evidence of the previously introduced concept that whole transcription patterns do not always correlate directly with protein expression levels and of the independent roles of BLR-1 and BLR-2 even in the dark (26).
Fig 5.
Altered gene expression of Δblr-1 and Δblr-2 mutants in the dark. The left panels show Northern blot analyses of selected T. atroviride genes from strains grown in the dark. Twenty micrograms of total RNA from mycelia grown in the dark was separated by electrophoresis in a denaturing agarose gel, blotted on a nylon membrane, and hybridized with 32P-labeled gene-specific cDNA probes targeting the indicated genes. Hybridization with a 28S rRNA gene probe was used as a loading control. The right panels show the expression ratios of the proteins encoded by the genes shown in the left panels to the wild type in the dark. (A) Genes for which mRNA levels correspond with protein level changes in the Δblr-1 or Δblr-2 mutant. (B) Genes for which mRNA levels do not correlate with protein abundance changes. (C) Genes for which mRNA levels change in the dark but the corresponding proteins are not regulated in the dark.
DISCUSSION
In T. atroviride, blue light stimulates the synchronous production of conidia. Recently, it was shown that 254 genes are responsive specifically to blue light. Additionally, 331 genes undergo expression changes in response to white light, strongly suggesting the existence of additional, functional light receptors (Esquivel-Naranjo et al., submitted). The light signal, in consequence, is expected to affect protein expression levels in the mycelium, where the response is observed. Therefore, it was of major interest to study how and to what extent proteins are expressed differentially during the early stages of photoconidiation and throughout the whole phenomenon. This question was addressed by analyzing protein profiles at six different times after exposure of T. atroviride strains to a blue light pulse. Our time course analysis provides a view of protein expression changes during asexual reproduction induced by blue light in a filamentous fungus.
A rapid illumination change elicits a fast alteration in protein expression, and massive changes were already detectable after 30 min. The relative abundances of transcripts peaked between 30 and 60 min (19). Nevertheless, protein levels at the early stages of the response showed a dynamic pattern of increasing protein expression, beginning with 34 differentially expressed proteins, as many as 123 proteins after 150 min of the light pulse, and only 9 differentially expressed proteins at late stages of the photoconidiation process. These data indicate that the blue light response takes place mainly in the first hours after the signal. The number of upregulated spots exceeds that of downregulated ones, indicating that more active Trichoderma physiological processes might be required when Trichoderma faces the challenge of blue light stress, in agreement with the results of the transcriptome analysis. In contrast, relatively few proteins were differentially regulated in the late stages of conidiation. This could be due to the fact that at those times the metabolic conditions of the cell are comparable to when it grows vegetatively in the dark. An alternative explanation is that we analyzed soluble (cytoplasmic) proteins and that the most dramatic changes expected are at the structural level.
Reactive oxygen species (ROS) are generated during mitochondrial respiration and in response to various environmental stressors, including blue light irradiation (31, 37, 43). Oxygen radicals have been identified as key actors in the stress response, and their role as secondary messengers has been established (44). Furthermore, ROS are scavenged by antioxidant enzymes such as catalase, superoxide dismutase (SOD), and glutathione S-transferase (GST). Any failure in this process might result in a state of oxidative stress leading to the oxidation of lipids, proteins, and DNA. In our study, we found that catalase and superoxide dismutase were downregulated. In agreement with the low levels of catalase found in our study it has been reported for N. crassa that due to the lack of catalase, hyphal adhesion and aerial hypha and conidium production are induced (39). Similarly, downregulated levels of SOD were detected in T. atroviride after exposure to the light pulse. In this regard, a sod-1 mutant of N. crassa was affected in the light-induced occurrence of polarity of the perithecia (64).
We also found Hsp70 and Hsp80 to be expressed differentially in our gels. Heat shock proteins or chaperones participate in several stress responses and are part of a general stress response (34). Hsp70 proteins are able to bind damaged or abnormal protein complexes and aggregates to resolve and refold them (11). A simultaneous regulation of both proteins might imply their synergistic role in proper protein folding and maturation of polypeptides in cells after the light pulse. In addition to ROS scavengers and chaperones, exposure of T. atroviride to blue light induced enzymes involved in the degradation of proteins.
A 20S proteasome beta subunit was detected as upregulated. Ubiquitin-mediated proteolysis plays significant roles in the cell cycle, in cell differentiation and development, in the cellular response to extracellular effectors and stress, in the modulation of cell surface receptors and ion channels, in DNA repair, and in the biogenesis of diverse organelles. All of these processes are related to the photoconidiation response of T. atroviride, as evidenced by transcriptome analyses (Esquivel-Naranjo et al., submitted). In plants, the ubiquitin-proteasome pathway has been related broadly to light responses and plant development (40); additionally, the conserved COP9 signalosome (CSN) is required for light-controlled responses in filamentous fungi (9). In addition, many of the differentially regulated proteins were implicated in protein synthesis and protein fate, and most of these enzymes showed the highest levels of upregulation. We found cyclophilin to be regulated differentially by blue light. Cyclophilins participate in the protein folding process not only as peptidyl prolyl isomerases but also as chaperones (20). The increase in abundance of these proteins indicates the importance and extent of protein synthesis and protein turnover in dealing with a stress situation. Accordingly, upregulation of genes involved in protein turnover was also observed upon illumination of mycelia of Trichoderma reesei and T. atroviride (53).
Following the light pulse, radical changes occurred to adapt to the new developmental program, and adaptation of metabolism was the main response found in our investigation. In this sense, large amounts of energy are needed to ensure asexual development and to maintain vegetative growth at the same time (52). Accordingly, many differentially regulated proteins were involved in energy production and in carbohydrate and amino acid metabolism: a regulation in the abundance of proteins implicated in glycolysis might provide energy needed for ATP-dependent protein synthesis. After illumination, amino acid metabolism has significant alterations, as indicated by the seven differentially expressed proteins involved in this process identified here. The behavior of these proteins suggests that both quality and quantity are important for the light response.
The adaptation and response mechanisms in the presence of light also involve fatty acid metabolism. For Aspergillus nidulans, it has been reported that fatty acid composition is influenced by light (12), and in N. crassa, it was found that the concentration of lipids increased in mycelia grown in constant light compared to the organism cultivated in darkness (45). Changes in lipid content in response to light have been determined for Trichoderma viride (7). We identified two differentially expressed proteins related to lipid metabolism and one connected with fatty acid transport. This is in agreement with changes in fatty acid composition dependent on the developmental stage of sexual development reported for N. crassa (25). Hence, asexual development requires fatty acid synthesis, corroborating transcriptomic studies of this and other filamentous fungi.
The Spitzenkörper, a dynamic structure present at the tips of hyphal cells with a single polarized growth site, is linked directly with cell morphogenesis and polar growth (60). This cell apparatus works by incorporating cell wall components into vesicles and carrying these vesicles to the plasma membrane at the hyphal tip for exocytosis and assembly of fresh wall components (3). Microtubules and actin microfilaments have important roles in this vesicle trafficking (29, 59). As expected, in our study we found the levels of several proteins directly involved in polarized growth to be affected suitably for the morphological changes going on through the process of photoconidiation. Every essential pathway is adjusted to the requirements of protection from light and preparation for reproductive growth. It is important that T. atroviride adjusts to blue light during photoconidiation by adjusting the expression of oxidative stress-related proteins, chaperones, and metabolism.
A protein may not be regulated at exactly the same time point as its transcript, due to the fact that translation follows transcription. In our experimental approach, we analyzed the expression of differentially expressed proteins at the transcript level in an attempt to establish the lag phase between transcription of responsive genes and changes in the corresponding protein levels. However, the largest proportion of proteins whose levels correlated with changes in mRNA levels was found 30 min after light exposure, whereas this correlation decreased at later times. We hypothesize that the group of genes whose changes at the transcript level are reflected at the protein level represents the core of the light response, due to the fact that transcriptional changes directly affect the level of protein, thereby regulating a functional element at once. A large group of proteins showed the opposite behavior to that observed at the mRNA level, allowing us to hypothesize that whereas mRNA increases, many protein species are degraded, possibly due to oxidative damage as a consequence of the well-established generation of ROS provoked by blue light exposure (43, 44). Conversely, mRNA accumulation is negatively regulated by light, but some proteins are protected from degradation, with the increase in half-life resulting in their accumulation. Similarly, when proteome data were compared with corresponding transcriptome data during fermentation, a low level of correlation was found in Saccharomyces cerevisiae (48). Similar observations were made in protein and mRNA abundance studies of S. cerevisiae growing at mid-log phase (26) and after heat shock of A. fumigatus (1). In some eukaryotes, many stress conditions lead to a repression of translation initiation and an accumulation of translationally repressed mRNA in stress granules or P bodies. Under these conditions, the translational machinery selectively translates mRNAs specific to the stress response while retaining the bulk of the mRNA cytoplasmic pool for later reuse and recovery from stress (33), or the translational machinery might be saturated and unable to process the translation of mRNA at the same rate (61). Together, these results indicate that simple deduction of protein actions from mRNA transcript analysis is insufficient and that both analyses are complementary.
In N. crassa, the transcription factors WC-1 and WC-2 form a heterodimeric complex which is responsible for all blue light responses. WC-1 and WC-2, along with FRQ, are the key elements for maintaining circadian rhythm in Neurospora (2, 18). The WC proteins have been found in the dark as a heterodimer capable of binding light-responsive elements (LREs). They form a bulky WCC that transiently binds to LREs in the frq promoter and the promoters of other light-responsive genes (23, 24, 30). The DNA binding domain of WC-1 is required for the function of the WCC in the circadian clock in constant darkness, but it is not required for induction of genes by light (16). In T. atroviride, the orthologues of wc-1 and wc-2, blr-1 and blr-2, regulate a substantial group of light-responsive genes and photoconidiation (13, 47). Although BLR-1 and BLR-2 form a potential UV-A receptor, these proteins also have light-independent functions (21). Besides conidiation, vegetative growth of T. atroviride on different carbon sources is influenced by light, and blr mutants show different remaining responses to this signal (22). Another role in the dark for the BLR proteins was shown when the corresponding mutants were exposed to carbon deprivation and did not undergo conidiation (14). Surprisingly, we identified from the proteomic profiles that the blr-1 and blr-2 mutations elicit the down- and upregulation of different kinds of functional proteins even in the dark, and this was supported by the transcription analyses in this study. The fact that various proteins involved in a variety of cellular processes were found to be regulated inversely depending on the mutation clearly indicates that they have a function even in darkness. Although the targets of the BLR proteins include a number of transcription factors after a blue light pulse (Esquivel-Naranjo et al., submitted), it seems that the BLR proteins might regulate those or other transcription factors in the dark. Here we showed for T. atroviride that blr-1 deletion affects the levels of several proteins, with a main tendency of downregulation, and that blr-2 mutation generally exhibits the opposite effect, while there are several proteins regulated by both, suggesting that BLR-1 may act directly as an activator and BLR-2 may act as a repressor. According to our data, it seems that BLR-1 and BLR-2 do not always work as a complex, may have independent functions, and might play only a minor role as a complex in the dark. Nevertheless, the BLR proteins appear to be essential in the dark to maintain the metabolic balance of the cell.
As reported above, drastic changes in the proteome of the wild type occurred through the first hours of analysis after a blue light pulse. It is noteworthy that in addition to regulation in the dark, the deletion mutants showed substantial light responses. The Δblr-1 strain in some way reflected the behavior of upregulation of protein levels observed in the WT both 45 and 90 min after a light pulse. It seems that BLR-1 has opposite roles under dark and light conditions, acting as an activator in the dark and as a repressor after a light pulse. Our data suggest that the mechanism underlying the coordinated regulation of asexual development and metabolism in T. atroviride might be the interaction between BLR-1 and -2 and other elements that maintain cellular balance: when any of them is defective, the complex cannot be formed. For A. nidulans, it has been reported that Velvet acts in a comparable way: in the dark, it remains in the nucleus in complex with VelB and LaeA and induces sexual development, but in the light, it is exported to the cytoplasm and secondary metabolism is affected (5). However, the Δblr-2 mutant showed the opposite behavior in its proteomic profiles, as downregulation dominated following the light pulse. In this respect, overexpression of blr-1 caused a decrease in conidiation, and conversely, overexpression of blr-2 resulted in increased levels of transcripts regulated by blue light and in an enhanced conidiation pattern (19). The signal involved in the blue light response is complex, and it seems possible that it may be generated in part by one of the flavoproteins in the electron transport chain, since they absorb blue light (35). The response in the deletion mutants could therefore partially be the result of electron transport inhibition, although we cannot discard the participation of an additional photoreceptor system.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alicia Chagolla for assistance with mass spectrometry and Stewart Gillmore for critical reading of the manuscript.
A.S.-A. and A.S.P.-M. are indebted to CONACYT for a doctoral and a postdoctoral fellowship, respectively. This work was supported in part by a CONACYT grant (I0110/193/10 FONS.INST.-30-10) and SEP-CONACYT (83798).
Footnotes
Published ahead of print 4 November 2011
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Albrecht D, Gunthke R, Brakhage AA, Kniemeyer O. 2010. Integrative analysis of the heat shock response in Aspergillus fumigatus. BMC Genomics 11: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ballario P, et al. 1996. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15: 1650–1657 [PMC free article] [PubMed] [Google Scholar]
- 3. Bartnicki-Garcia S, Hegert E, Gierz G. 1990. A novel computer model for generating cell shape: application to fungal morphogenesis, p 43–60 In Kuhn PJ, Trinci APJ, Jung MJ, Goosey MW, Coping LG. (ed), Biochemistry of cell walls and membranes in fungi. Springer-Verlag, Berlin, Germany: [Google Scholar]
- 4. Baum D, Horwitz BA. 1991. Changes in synthesis and abundance of specific polypeptides at early and late stages of blue-light-induced sporulation of Trichoderma. J. Photochem. Photobiol. B 11: 117–127 [Google Scholar]
- 5. Bayram O, et al. 2008. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320: 1504–1506 [DOI] [PubMed] [Google Scholar]
- 6. Berrocal-Tito G, Rosales-Saavedra T, Herrera-Estrella A, Horwitz BA. 2000. Characterization of blue-light and developmental regulation of the photolyase gene phr1 in Trichoderma harzianum. Photochem. Photobiol. 71: 662–668 [DOI] [PubMed] [Google Scholar]
- 7. Betina V, Koman V. 1980. Changes in the lipid composition during the photo-induced conidiation of Trichoderma viride. Folia Microbiol. 25: 295–300 [DOI] [PubMed] [Google Scholar]
- 8. Bradford MM. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- 9. Brauss G, Irniger S, Bayram O. 2010. Fungal development and the COP9 signalosome. Curr. Opin. Microbiol. 13: 672–676 [DOI] [PubMed] [Google Scholar]
- 10. Buck JW, Dong W, Mueller DS. 2010. Effect of light exposure on in vitro germination and germ tube growth of eight species of rust fungi. Mycologia 102: 1134–1140 [DOI] [PubMed] [Google Scholar]
- 11. Burnie JP, Carter TL, Hodgetts SJ, Matthews RC. 2006. Fungal heat-shock proteins in human disease. FEMS Microbiol. 30: 53–88 [DOI] [PubMed] [Google Scholar]
- 12. Calvo AM, Gardner HW, Keller NP. 2001. Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans. J. Biol. Chem. 276: 25766–25774 [DOI] [PubMed] [Google Scholar]
- 13. Casas-Flores S, Rios-Momberg M, Bibbins M, Ponce-Noyola P, Herrera-Estrella A. 2004. BLR-1 and BLR-2 are key regulatory elements for photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology 150: 3561–3569 [DOI] [PubMed] [Google Scholar]
- 14. Casas-Flores S, et al. 2006. Cross talk between a fungal blue-light perception system and the cyclic AMP signaling pathway. Eukaryot. Cell 5: 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. 2009. Genome-wide analysis of light-inducible responses reveals hierarchical light signaling in Neurospora. EMBO J. 28: 1029–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng P, Yang Y, Gardner KH, Liu Y. 2002. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol. Cell. Biol. 22: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chet I. 1987. Trichoderma—application, mode of action and potential as a biocontrol agent of soilborne plant pathogenic fungi, p 137–160 In Chet I. (ed), Innovative approaches to plant disease control. J. Wiley and Sons, New York, NY: [Google Scholar]
- 18. Crosthwaite SK, Dunlap JC, Loros JJ. 1997. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science 276: 763–769 [DOI] [PubMed] [Google Scholar]
- 19. Esquivel-Naranjo EU, Herrera-Estrella A. 2007. Enhanced responsiveness and sensitivity to blue light by blr-2 overexpression in Trichoderma atroviride. Microbiology 153: 3909–3922 [DOI] [PubMed] [Google Scholar]
- 20. Freskgard PO, Bergenhen N, Jonhsson BH, Svensson M, Carlsson U. 1992. Isomerase and chaperone activity of prolyl isomerase in the folding of carbonic anhydrase. Science 258: 464–466 [DOI] [PubMed] [Google Scholar]
- 21. Friedl MA, Kubicek CP, Druzhinina IS. 2008. Carbon source dependence and photostimulation of conidiation in Hypocrea atroviridis. Appl. Environ. Microbiol. 74: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Friedl MA, Schmoll M, Kubicek CP, Druzhinina IS. 2008. Photostimulation of Hypocrea atroviridis growth occurs due to a crosstalk of carbon metabolism, blue light receptors and response to oxidative stress. Microbiology 154: 1229–1241 [DOI] [PubMed] [Google Scholar]
- 23. Froehlich AC, Liu Y, Loros JJ, Dunlap JC. 2002. White collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297: 815–819 [DOI] [PubMed] [Google Scholar]
- 24. Froehlich AC, Loros JJ, Dunlap JC. 2003. Rhythmic binding of a White collar-containing complex to the frequency promoter is inhibited by Frequency. Proc. Natl. Acad. Sci. U. S. A. 100: 5914–5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goodrich-Tanrikulu M, Howe K, Stafford A, Nelson MA. 1998. Changes in fatty acid composition of Neurospora crassa accompany sexual development and ascospore germination. Microbiology 144: 1713–1720 [DOI] [PubMed] [Google Scholar]
- 26. Gygi SP, Rochon Y, Franza BR, Aebersold R. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harman GE. 2000. Myths and dogmas of biocontrol. Changes in perceptions derived from research on Trichoderma harzianum T-22. Plant Dis. 84: 377–393 [DOI] [PubMed] [Google Scholar]
- 28. Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. 2004. Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2: 43–56 [DOI] [PubMed] [Google Scholar]
- 29. Harris SD, et al. 2005. Spitzenkorper meets polarisome: microscopy, genetics, and genomics coverage. Eukaryot. Cell 4: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. He Q, Liu Y. 2005. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 19: 2888–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He YY, Häder DP. 2002. UV-B-induced formation of reactive oxygen species and oxidative damage of the cyanobacterium Anabaena sp. protective effects of ascorbic acid and N-acetyl-l-cysteine. J. Photochem. Photobiol. 66: 115–124 [DOI] [PubMed] [Google Scholar]
- 32. Herrera-Estrella A, Horwitz BA. 2007. Looking through the eyes of fungi: molecular genetics of photoreception. Mol. Microbiol. 64: 5–15 [DOI] [PubMed] [Google Scholar]
- 33. Hilgers C, Teixeira D, Parker R. 2006. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA 12: 1835–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hohmann S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66: 300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laloraya MM, Kumar GP, Laloraya M. 1994. Photochemical reaction sequences of blue light activated flavins: sensory transduction through free radical messengers. Biochem. Mol. Biol. Int. 33: 543–551 [PubMed] [Google Scholar]
- 36. Liu Y, Bell-Pedersen D. 2006. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot. Cell 5: 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Y, Fiskum G, Schubert D. 2002. Generation of reactive oxygen species by the mitochondrial electron transport chain. J. Neurochem. 80: 780–787 [DOI] [PubMed] [Google Scholar]
- 38. Mastouri F, Bjorkman T, Harman GE. 2010. Seed treatment with Trichoderma harzianum alleviates biotic, abiotic, and physiological stresses in germinating seeds and seedlings. Phytopathology 100: 1213–1221 [DOI] [PubMed] [Google Scholar]
- 39. Michan S, Lledias F, Hansberg W. 2003. Asexual development is increased in Neurospora crassa cat-3-null mutant strains. Eukaryot. Cell 2: 798–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moon J, Parry G, Estelle M. 2004. The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moore MJ. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309: 1514–1518 [DOI] [PubMed] [Google Scholar]
- 42. Neduva V, Russell RB. 2006. Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 17: 465–471 [DOI] [PubMed] [Google Scholar]
- 43. Omata Y, et al. 2006. Intra- and extracellular reactive oxygen species generated by blue light. J. Biomed. Mater. Res. 77: 470–477 [DOI] [PubMed] [Google Scholar]
- 44. Parent C, Capelli N, Dat J. 2008. Reactive oxygen species, stress and cell death in plants. C. R. Biol. 331: 255–261 [DOI] [PubMed] [Google Scholar]
- 45. Ram S, Nair BG, Chhatpar HS. 1984. Photoregulation of some enzymes from Neurospora crassa. Experientia 40: 1382–1384. [DOI] [PubMed] [Google Scholar]
- 46. R Development Core Team 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project. [Google Scholar]
- 47. Rosales-Saavedra T, et al. 2006. Novel light-regulated genes in T. atroviride: a dissection by cDNA microarrays. Microbiology 152: 3305–3317 [DOI] [PubMed] [Google Scholar]
- 48. Rossignol T, Kobi D, Jacquet-Gutfreund L, Blondin B. 2009. The proteome of a wine yeast strain during fermentation, correlation with the transcriptome. J. Appl. Microbiol. 107: 47–55 [DOI] [PubMed] [Google Scholar]
- 49. Ruepp A, et al. 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32: 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed [Google Scholar]
- 51. Schafmeier T, et al. 2005. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation dependent inactivation of its transcription factor. Cell 122: 235–246 [DOI] [PubMed] [Google Scholar]
- 52. Schmoll M, Esquivel-Naranjo EU, Herrera-Estrella A. 2010. Trichoderma in the light of day—physiology and development. Fungal Genet. Biol. 47: 909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schuster A, Kubicek CP, Friedl MA, Druzhinina IS, Schmoll M. 2007. Impact of light on Hypocrea jecorina and the multiple cellular roles of Envoy in this process. BMC Genomics 8: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schwerdtfeger C, Linden H. 2003. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22: 4846–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shoresh M, Harman GE. 2008. The molecular basis of shoot responses of maize seedlings to Trichoderma harzianum T22 inoculation of the root: a proteomic approach. Plant Physiol. 147: 2147–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Steyaert JM, Weld RJ, Mendoza-Mendoza A, Stewart A. 2010. Reproduction without sex: conidiation in the filamentous fungus Trichoderma. Microbiology 156: 2887–2900 [DOI] [PubMed] [Google Scholar]
- 57. Tish D, Schmoll M. 2010. Light regulation of metabolic pathways in fungi. Appl. Microbiol. Biotechnol. 85: 1259–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Urlinger S, Kuchler K, Meyer TH, Uebel S, Tampe R. 1997. Intracellular location, complex formation, and function of the transporter associated with antigen processing in yeast. Eur. J. Biochem. 245: 266–272 [DOI] [PubMed] [Google Scholar]
- 59. Virag A, Griffiths AJ. 2004. A mutation in the Neurospora crassa actin gene results in multiple defects in tip growth and branching. Fungal Genet. Biol. 41: 213–225 [DOI] [PubMed] [Google Scholar]
- 60. Virag A, Harris SD. 2006. The spitzenkörper: a molecular perspective. Mycol. Res. 110: 4–13 [DOI] [PubMed] [Google Scholar]
- 61. Warringer J, Hult M, Regot S, Posas F, Sunnerhagen P. 2010. The HOG pathway dictates the short term translational response after hyperosmotic shock. Mol. Biol. Cell 21: 3080–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang Y, et al. 2004. Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes Dev. 18: 255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yedidia I, et al. 2003. Concomitant induction of systemic resistance to Pseudomonas syringae pv. lachrymans in cucumber by Trichoderma asperellum (T-203) and accumulation of phytoalexins. Appl. Environ. Microbiol. 69: 7343–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yoshida Y, Hasunuma K. 2004. Reactive oxygen species affect photomorphogenesis in Neurospora crassa. J. Biol. Chem. 279: 6986–6993 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.